Abstract
Rationale
Nox4 is a constitutively active NADPH oxidase that constantly produces low levels of H2O2. Thereby, Nox4 contributes to cell homeostasis and long-term processes, such as differentiation. The high expression of Nox4 seen in endothelial cells contrasts with the low abundance of Nox4 in stem cells, which are accordingly characterized by low levels of H2O2. We hypothesize that Nox4 is a major contributor to endothelial differentiation, is induced during the process of differentiation, and facilitates homeostasis of the resulting endothelial cells.
Objective
To determine the role of No×4 in differentiation of murine inducible pluripotent stem cells (miPSC) into endothelial cells (ECs).
Methods and results
miPSC, generated from mouse embryonic wildtype (WT) and Nox4−/− fibroblasts, were differentiated into endothelial cells (miPSC-EC) by stimulation with BMP4 and VEGF. During this process, Nox4 expression increased and knockout of Nox4 prolonged the abundance of pluripotency markers, while expression of endothelial markers was delayed in differentiating Nox4-depleted iPSCs. Eventually, angiogenic capacity of iPSC-ECs is reduced in Nox4 deficient cells, indicating that an absence of Nox4 diminishes stability of the reached phenotype. As an underlying mechanism, we identified JmjD3 as a redox target of Nox4. iPSC-ECs lacking Nox4 display a lower nuclear abundance of the histone demethylase JmjD3, resulting in an increased triple methylation of histone 3 (H3K27me3), which serves as a repressive mark for several genes involved in differentiation.
Conclusions
Nox4 promotes differentiation of miPSCs into ECs by oxidation of JmjD3 and subsequent demethylation of H3K27me3, which forced endothelial differentiation and stability.
Keywords: Nox4, iPSC, Differentiation, Redox-switch, JmjD3, H3K27me3
Nonstandard abbreviations and acronyms
- Nox4
NADPH oxidase Nox4 knockout
- iPSC
Induced pluripotent stem cell derived
- iPSC-EC
Induced pluripotent stem cell derived endothelial cells
- WT
Wild type
- EC
Endothelial cell
1. Introduction
The vascular system that supplies the organism with oxygen and nutrients is lined with endothelial cells. In the process of vasculogenesis, endothelial cells differentiate from progenitor cells [1].
Several essential stimuli and factors required for differentiation have been identified, and in vitro-protocols to differentiate stem cells into endothelial cells have been developed [2]. Reactive oxygen species (ROS) belong to the stimuli identified [3]. It is interesting to note, that stem cells reside in a low ROS environment [4,5] and are characterized by high antioxidant enzyme expression [4] and low ROS formation. In the course of stem cell differentiation, ROS formation increases, which has been largely attributed to mitochondrial expansion [6]. Besides mitochondria NADPH oxidases are well-characterized ROS producing enzymes, allowing a controlled timely and localized ROS formation. They can be categorized into three groups: those dependent on cytosolic subunits (Nox1, Nox2, Nox3), those that are calcium-dependent (Nox5 and DUOX), and those which are constitutively active, such as Nox4.
Different from most other NADPH oxidases, Nox4 produces H2O2, which has protective vascular functions, like activation of protein kinase G or induction of eNOS expression [7]. H2O2 elicits its effects i.e. by oxidation of cysteine residues in proteins. As a consequence of H2O2 mediated oxidation, disulfide bonds, sulfenic, sulfinic or sulfonic acid are formed, which depending on their localizations, can change enzyme activity, protein-protein interaction and protein folding. Oxidation in the active center of enzymes usually results in inactivation. Importantly, formation of disulfide bonds and sulfenic acid are reversible and therefore such modifications may have strong regulatory implications [8]. Among those proteins modified by H2O2 are epigenetic enzymes, like histone deacetylases or demethylases [9]. These enzymes control gene expression, and their differential activation is required for cellular differentiation [10].
Nox4 has been linked to differentiation of cardiac myocytes [11], neuronal stem cells [[12], [13], [14]], astrocytes [15], and embryonic bodies [15]. Down-regulation of Nox4 has been suggested to attenuate renewal of mouse iPSCs [16]. Differentiation of embryonic stem cells into smooth muscle cells requires Nox4 [17], and Nox4 maintains smooth muscle cell differentiation [18]. Although these studies provide a solid basis to infer a role of Nox4 in differentiation, the highest Nox4 expression besides the kidney, is present in the endothelium [7]. Based on these observations, we hypothesize that Nox4 is required for the differentiation of endothelial cells. To address this aspect, murine iPSCs from WT and Nox4−/− mice were generated and subsequently differentiated into the endothelial lineage.
2. Methods
2.1. Mice
C57Bl/6 WT and global Nox4 knockout (Nox4−/−) mice, as generated and reported previously by our group [7] were kept under standard conditions. Pregnant female mice were sacrificed by cervical dislocation at day 12.5 post conception to harvest the embryos and mouse embryonic fibroblasts from five different litters – WT and Nox4−/− – were isolated and genotyped.
2.2. Viral transduction of fibroblasts, reprogramming into miPSC & maintenance
Isolated WT & Nox4 −/− cells were reprogrammed into induced pluripotent stem cells using the STEMCCA system [19]. Briefly, 20.000 mouse embryonic fibroblast (MEF) were seeded into culture dishes. After 24 h cells were infected with STEMCCA virus for 24. Media was changed for five consecutive days. Cells were then reseeded on mitomycin (Serva, 10 μg/mL, 3 h) -inactivated feeder cells until murine induced pluripotent colonies emerged. Individual colonies were subsequently picked and expanded. Abundance of pluripotency markers such as alkaline phosphatase activity and expression of Oct4, Nanog, Sox2 and Ssea1 indicate the success of the procedure. Cells were passaged (Stem cell media: DMEM GlutaMAX, # 10566016, Gibco; 20 % Knockout Serum Replacement, # 10828028, 25 μg mLIF, # 250–02, Peprotech; 0.1% β-mercaptoethanol, 200 μM Glutamine, #11539876, Gibco) by 70% before colonies touched and reseeded between 2.5 × 103 and 5 × 103 cells per cm2.
2.3. Differentiation of miPSCs to endothelial cells
Murine induced pluripotent stem cells (miPSC) were differentiated for up to 14 days into endothelial cells [20]. Briefly, miPSC were detached from cell culture dishes by accutase (#A6964, Sigma-Aldrich), centrifuged and resuspended in differentiation media (alpha MEM #22571–020, Gibco, 50 ng/mL Vegf/Bmp4, #100–20, #120–05, Peprotech, 0.1% β-mercaptoethanol, 200 μM Glutamine, #11539876, Gibco). Hanging drops were formed for four days, with 500 cells per drop and 144 drops per dish. On day four formed embryonic bodies (EBs) were transferred to a low adherent dish to further differentiate in suspension. On day 7 EBs were transferred to 0,2 % gelatine coated cell culture dishes for the duration of 7 days. Media was exchanged then every other day. A time scale is represented in Supp. Fig. 4A.
2.4. General cell culture techniques
Human embryonic kidney (Hek) 293 cells were purchased from ATCC (Manassas, USA) and used as an artificial model system. Hek293 cells overexpressing Nox4 in a tetracycline-inducible manner (Nox4-tet-on) were kindly provided by K.H. Krause, University of Geneva, Switzerland [21]. All Hek cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM), high glucose, GlutaMAX (Gibco, Carlsbad, USA), supplemented with 10% fetal calf serum (FCS, #S0115, Biochrom, Berlin, Germany), 1% MEM Non-essential Amino Acid Solution (100x), 1 mM Na-Pyruvate (both from Merck, Darmstadt, M7145, S8636) and gentamicin (50 μg/mL, Gibco). Human umbilical vein endothelial cells (HUVECs) were obtained from Lonza (#CC-2519, Lot No. 371074, 369146, 314457, 192485, 186864, 171772, Walkersville, USA) and PeloBiotech (#PB-CH-190-813, Lot No. QC-18P13F11, Planegg, Germany). HUVECs were cultured in dishes coated with gelatin in endothelial growth medium (EGM). EGM was composed of endothelial basal medium (EBM) supplemented with human recombinant epidermal growth factor (EGF), EndoCGS-Heparin (PeloBiotech), 8% FCS, penicillin (50 U/mL) and streptomycin (50 μg/mL) (#15140–122, Gibco).
2.5. Lipofection of Hek293 cells
Hek293 cells were transiently transfected with Lipofectamin 2000 reagent (#11668019, Thermo Fisher Scientific, Carlsbad, USA). Per 6 cm dish 50 μL transfection medium (Hek293 medium without antibiotics) including 1.5 μg DNA were mixed with 35 μL transfection medium including 3.5 μL Lipofectamin 2000. The transfection mixture was then added dropwise and after 4 h the medium was changed to Hek293 growth medium. pCMV-HA-JMJD3 was a gift from Kristian Helin (Addgene plasmid # 24167; http://n2t.net/addgene:24167; RRID:Addgene_24167).
2.5.1. Quantitative real-time polymerase chain reaction (qRT-PCR)
Isolation of mRNA was performed with an RNA-Mini-kit (Bio&Sell, Feucht, Germany) according to the manufacturer's protocol. Genomic DNA was digested with DNaseI (#4536282001, Roche, Basel, Switzerland). For cDNA synthesis, Random hexamer primers (Promega, Madison, USA) together with Superscript III Reverse Transcriptase (Invitrogen, Darmstadt, Germany) were used. qRT-PCR was performed with the aid of an AriaMx qPCR cycler (Agilent Technologie, Santa Clara, USA). Primers are listed in Table 1. DNA was detected using iQ™ SYBR® Green in a Supermix (BioRad, Hercules, USA). DNA levels were calculated using the ΔΔct method and gene expression was normalised to elongation factor (EF).
Table 1.
qRT-PCR primers.
| Gene | Forward primer (5’→3′) | Reverse primer (5’→3′) |
|---|---|---|
| EF | GACATCACCAAGGGTGTGCAG | GCGGTCAGCACACTGGCATA |
| Cybb | GTGCACCATGATGAGGAGAA | TTGCAATGGTCTTGAACTCG |
| Nox4 | TGTTGGGCCTAGGATTGTGTT | TGTTGGGCCTAGGATTGTGTT |
| Nox1 | CCTCCTGACTGTGCCAAAGG | ATTTGAACAACAGCACTCACCAA |
| Noxo1 | TGGAGGAGGTAGCAACGTGC | AGAGCGACTGCCCTCGTAGG |
| Duox1 | ATTCCTCGACCAAAGTGCGT | TTCTCCTCTGGGGAGCCTTT |
| Duox2 | TCTTCACCATGATGCGGTCC | GGAGTCCGGTTGATGAACGA |
| Cyba | TGGACTCCCATTGAGCCTAAACC | GTTGGTAGGTGGTTGCTTGATG |
| Kdr | TGCGCAGCAGGATGGCAAAGAC | TCTGGCTGTCATCTGGGATCACTT |
| NOS3 | CTCACCATAGCTGTGCTGGCTTAC | GATGCAGGGCAAGTTAGGATCAGG |
| VE-Cad. | GGCAGGTAGCATGTTGGGG | ATGGCGTTTGACAGCACTTG |
| CD31 | GGACAGACCCTTCCACCAAG | CTGTTTGGCCTTGGCTTTCC |
| Oct4 | CTGAAGCAGAAGAGGATCAC | TTGTTGTCGGCTTCCTCCAC |
| Sox2 | TGCTGCCTCTTTAAGACTAGGG | TCTGGCGGAGAATAGTTGGG |
| Klf4 | AACTCGCTAACCCACCAGGAATCG | CATTGATGTCCGCCAGGTTGAAGG |
| Nanog | AATTCTGGGAACGCCTCATC | CTTGTCAGCCTCAGGACTTG |
| Esg1 | GAAGTCTGGTTCCTTGGCAGGATG | ACTCGATACACTGGCCTAGC |
| Eras | ACTGCCCCTCATCAGACTGCTACT | CACTGCCTTGTACTCGGGTAGCTG |
| Gdf3 | GTTCCAACCTGTGCCTCGCGTCTT | AGCGAGGCATGGAGAGAGCGGAGCAG |
| Fgf4 | CGTGGTGAGCATCTTCGGAGTGG | CCTTCTTGGTCCGCCCGTTCTTA |
| Tdgf1 | ATGGACGCAACTGTGAACATGATGTTCGCA | CTTTGAGGTCCTGGTCCATCACGTGACCAT |
| Dax1 | TGCTGCGGTCCAGGCCATCAAGAG | GGGCACTGTTCAGTTCAGCGGATC |
| Zfp296 | CCATTAGGGGCCATCATCGCTTTC | CACTGCTCACTGGAGGGGGCTTGC |
| Actb | AGATCAAGATCATTGCTCCTCCT | ACGCAGCTCAGTAACAGTCC |
2.6. RNA sequencing
RNA was isolated from miPSC/miPSC-ECs using the miRNeasy micro Kit (Qiagen) combined with on-column DNase digestion (DNase-Free DNase Set, Qiagen) to avoid contamination by genomic DNA. RNA and library preparation integrity were verified with LabChip Gx Touch 24 (PerkinElmer). 600 ng of total RNA was used as input for SMARTer Stranded Total RNA Sample Prep Kit - HI Mammalian (Clontech). Sequencing was performed on the NextSeq500 instrument (Illumina) using v2 chemistry, resulting in average of 26 M reads per library with 1x75bp single end setup. The resulting raw reads were assessed for quality, adapter content and duplication rates with FastQC [22]Trimmomatic version 0.38 was employed to trim reads after a quality drop below a mean of Q20 in a window of 10 nucleotides [23]. Only reads between 30 and 150 nucleotides were cleared for further analyses. Trimmed and filtered reads were aligned against the Ensembl mouse genome version mm10 (release 99) using STAR 2.7.3a with the parameter “--outFilterMismatchNoverLmax 0.1” to increase the maximum ratio of mismatches to mapped length to 10% [24]. The number of reads aligning to genes was counted with featureCounts 1.6.5 tool from the Subread package [25]. Only reads mapping at least partially inside exons were admitted and aggregated per gene. Reads overlapping multiple genes or aligning to multiple regions were excluded.
2.7. Single cell RNA sequencing
Days 0, 4, 7 and 14 of differentiation were sequenced of WT and Nox4−/− cells. In brief, cells were dissociated by accutase filtered through strainer (70 μM) and resuspended in 0.1 % EDTA/PBS. Cells were stained with trypan blue counted in Neubauer chamber and diluted to 1000 cells/μL. Cellular suspensions were loaded on a 10X Chromium Controller (10X Genomics) according to manufacturer's protocol based on the 10X Genomics proprietary technology. All scRNA-seq libraries were prepared using Chromium Single-Cell 3′ v3.1 Reagent Kit (10X Genomics) according to manufacturer's protocol. Briefly, the initial step consisted of performing an emulsion where individual cells were isolated into droplets together with gel beads coated with unique primers bearing 10X cell barcodes, unique molecular identifiers, and poly(dT) sequences. Reverse transcription reactions were engaged to generate barcoded full-length cDNA followed by the disruption of emulsions using the recovery agent and cDNA clean up with DynaBeads MyOne Silane Beads (Thermo Fisher Scientific). Bulk cDNA was amplified using a Biometra Thermocycler TProfessional Basic Gradient with 96-Well Sample Block (98 °C for 3 min; cycled 12 × : 98 °C for 15 s, 63 °C for 20 s, and 72 °C for 1 min; 72 °C for 1 min; held at 4 °C). Amplified cDNA product was cleaned with the SPRIselect Reagent Kit (Beckman Coulter). Indexed sequencing libraries were constructed using the reagents from the Chromium Single-Cell 3′ v3.1 Reagent Kit as follows: fragmentation, end repair, and A-tailing; size selection with SPRIselect; adaptor ligation; postligation clean up with SPRIselect; sample index polymerase chain reaction; and clean up with SPRI select beads. Library quantification and quality assessment were performed using Bioanalyzer Agilent 2100 using a High-Sensitivity DNA chip (Agilent Genomics). Indexed libraries were equimolarly pooled and sequenced on 2 Illumina NovaSeq 6000 using paired-end 26 × 98 bp as sequencing mode by GenomeScan (Leiden, the Netherlands).
Outputs from STARsolo [26] were used to create Seurat (version 4.0.4) [27] objects per sample using the functions Read10X and CreateSeuratObject with the parameters min.cells = 3 and min.features = 200 in R (version 4.1.1, [28]) The separate Seurat objects were then merged to a single dataset. Mitochondrial read percentages were calculated with the function PercentageFeatureSet and the pattern ^mt-. The percentages of mitochondrial reads were subsequently used in the transformation of the object with SCTransform. The Seurat object was then subjected to principle component analysis, uniform manifold approximation, neighbour identification and cluster identification using the functions RunPCA, RunUMAP, FindNeighbors and FindClusters, respectively. Cluster markers were then identified using FindAllMarkers with default parameters. A cluster of feeder cells unconnected to the differentiation process was removed from the analysis. Slingshot (2.0.0) [29] was used to learn a pseudotime trajectory through the remaining clusters, using day 0 as a starting point with otherwise default parameters. Canonical cell marker genes were taken from literature [30] and PanglaoDB [31], a curated database of marker genes. Expression of different marker genes was then computed along the length of the Slingshot trajectory, either as individual genes or average expression of all genes relating to particular cell types. Proportions of cells expressing marker genes were computed using DotPlot in the Seurat package. Resulting data were then included in further downstream analysis and plots, where expression values and uniform manifold application embeddings were taken from the Seurat object and used to generate plots with ggplot2 (3.3.5).
2.8. ChIP sequencing
Cells were isolated using the ChIP-IT Kit from active motiv Sequencing on Nextseq500 with paired end mode using V2.5 chemistry. ChIP was perfomed on Day 7 of endothelial differentiation protocol from the whole cell population H3K27me3 antibody (#pAb-069-050, Diogenode). Sequencing reads were aligned to the mm10 genome build using Bowtie2 (2.4.4) with default parameters. Bigwig files were generated and manually analyzed in the IGV Browser (2.8.12; [32].) and comparing these with the RNA Bulk sequencing data.
2.8.1. Nucleocounter
NucleoCounter® NC-3000™ "Advanced Image Cytometer" (Chemometec) was used according to manufacture manual. Briefly, cells were digested with accutase and resuspended until single cell suspension was achieved. Cells were then stained with Hoechst (#33342, Thermoscientific) and with fluorophore coupled antibody CD31 (#102509, Biolegend), VEGFR2 (#555308, BD) or isotype control (#400512, Biolegend, #553930, BD).
2.9. Western blot
Cells were lysed in Triton X buffer containing Tris-HCl (100 mM), NaCl (750 mM), NaPPi (50 mM), NaF (100 mM) Triton X-100 (10%), phenylmethylsulfonyl fluoride (PMSF, 1 mM), orthovanadate (OV, 2 mM), okadaic acid (OA, 10 nM) and protease inhibitor mix (PIM). The lysate was centrifuged (17,000 g, 10 min, 4 °C) and the supernatant was collected. Protein concentration was determined using Bradford assay. Samples were boiled in LämmLi buffer under reducing conditions, separated by SDS-PAGE and transferred to a nitrocellulose membrane by Western blotting. Fluorescence-based detection of secondary antibodies was performed using the Odyssey CLx imaging system (Licor, Bad Homburg, Germany). Primary antibodies: eNOS (#334600, Invitrogen), H3K27ac (#pAb-174-050, Diagenode), H3K27me2 (#C15410046-50, Diagenode) H3K27m3, H1 (#05–457, Millipore), H3 (#C15200011, Diagenode), mJmjD3 (#AP1022b, Abgent), hJmjD3 (#AP1022a, Abgent), UTX (#33510, Cell Signaling), EZH2 (#3147, Cell Signaling), topoisomerase I (#sc-5342, Santa Cruz) and β-actin (#A1978, Sigma-Aldrich). Fluorescence-labelled secondary antibodies: IRDye® RD680 Donkey anti-Mouse, RD680 Donkey anti-Rabbit, CW800 Donkey anti-Mouse, CW800 Donkey anti-Rabbit, RD680 Donkey anti-Goat (#926–68072, #926–68073, #926–32212, #926–32213, #926–68074 Licor).
2.10. Nuclear extraction
Cells were scratched from the plate in Hank's buffer and pelleted by centrifugation (17,000 g, 1 min, 4 °C). The pellet was resuspended in nuclear extraction buffer containing HEPES (10 mM), KCl (10 mM), EDTA (0.1 mM), EGTA (0.1 mM), PMSF (1 mM) and PIM. After incubation on ice for 15 min, Nonidet P40 (1.5%) was added, samples were centrifuged (17,000 g, 1 min, 4 °C) and the cytosolic protein lysate (supernatant) was transferred into a new tube. The lysate and the pellet, containing nuclear proteins, were boiled in LämmLi buffer and used for Western blot.
2.11. BIAM switch assay
Alkylation of thiol groups was performed in living cells before harvest. N-ethylmaleimide (NEM, 50 mM, #4259, Sigma-Aldrich) was added to the medium and left for 5 min at room temperature. Cells were then washed with PBS containing 100 mM NEM and 500 μL trichloroacetic acid (TCA, 20%) was added. Cells were scratched, transferred into a tube and incubated on ice for 15 min. Samples were centrifuged (13000 g, 30 min, 4 °C) and washed with 10% and then 5% TCA. The pellet was resuspended in denaturing buffer (DB, pH 8.5, 100 mM Tris-HCl, 8 M Urea, 5 mM EDTA, 0.5% SDS) with NEM (25 mM) and incubated for 1 h at 37 °C. After addition of 800 μL acetone, proteins were precipitated over night at −20 °C. Samples were centrifuged (13000 g, 30 min, 4 °C) and washed twice with acetone. Then pellets were resuspended in DB with 5 mM DTT for 5 min at 37 °C to reduce oxidized thiols, which were subsequently labelled by adding DB with EZ-Link Iodoacetyl-LC-Biotin (BIAM, 2 mg/mL, #21333, Thermo Fisher Scientific) for 1 h at 37 °C and 850 rpm. Proteins were precipitated by adding 800 μL acetone and stored over night at −20 °C. Samples were centrifuged (13000 g, 30 min, 4 °C) and washed twice with acetone. The pellet was resuspended with lysis buffer composed of Tris-HCl (50 mM), EDTA (5 mM), Triton X-100 (1%), SDS (1%), PMSF (1 mM), OV (2 mM), OA (10 nM) and PIM. After incubation for 1 h at 37 °C the protein amount was determined by Lowry assay. 500 μg protein were adjusted to 1 μg/μL with lysis buffer. Labelled proteins were pulled down with 100 μL Pierce™ Streptavidin Agarose beads (#20353, Thermo Fisher Scientific) slurry overnight at 4 °C. Beads were washed five times with lysis buffer and boiled with 1.5x LämmLi buffer. Samples were analyzed on Western blot.
2.12. Immunofluorescence staining
Cells were fixed with 4% Paraformaldehyde in cell culture media, permeabelised with 0.05% Triton-X 100. Cells were blocked 30 min in 3% BSA and subsequently incubated over night at 4 °C with primary antibodies. The next day, cells were washed and incubated for 30 min with fluorophore coupled-secondary antibody in the dark and either immediately imaged or stored at 4 °C in the dark. Fluorescence images were imaged with LSM 800 from Zeiss and quantified with ImageJ software. Matrigel plugs were imaged with the light sheet microscopy and quantified with Imaris software.
2.12.1. Proximity ligation assay
Cells were treated in the same fashion as with the immunofluorescence protocol according to the instruction of the kit unless indicated otherwise (#DUO92007-100RXN, Sigma-Aldrich). In Brief after permeabilisation step, cells were blocked with blocking buffer as well as the antibodies. Afterwards cells were incubated 24 h with primary antibodies. Cells were then ligated and polymerase was used to amplify the signal. Signal was positive if proteins of interest were in proximity of one another.
2.12.2. Tube formation assay
Tube formation assays were performed in μ-Slide Angiogenesis coverslips (Ibidi, Planegg, Germany). Matrigel was thawed on ice. 10 μL of Matrigel (#356231, Corning, Corning, USA) were added per well and allowed to polymerized for 30 min at 37 °C. 5.000 cells in 50 μL Diff. media with 1% FCS were seeded onto the Matrigel and after 4 h incubation in a cell culture-incubator at 37 °C, 30 μL medium were replaced by 30 μL PBS with 4% PFA in order to fix the cells. Tube formation was analyzed by counting branching points and measuring total tube length using ImageJ [33].
2.13. Spheroids outgrowth assay
Outgrowth assay was prepared in a two-day protocol. Cells were differentiated until day seven of differentiation and dissociated to single cells. 400 cells were mixed with 25 μL differentiation media (20 % Methocell; #M0512, Sigma) with the goal of 100 drops per square plate (#688102, Greiner). Plates were set upside down in incubator for 24 h. The next day spheroids were rinsed with PBS (#10010056, Gibco) and centrifuged for 4 min at 1200 rpm without brake. Supernatant was discarded and spheroids resuspended in equal portions Methocell & collagen (20 % FCS). Further on distributed into 48-well plates, where they were then either imaged or stimulated with Vegf165 and then imaged.
2.14. Matrigel plug preparation
All experimental procedures were approved by the local governmental authorities and were performed in accordance with the local animal protection guidelines. WT & Nox4 knockout endothelial cells were isolated at day 14 of differentiation with biotinylated isolectin B4 beads (#CatNoB-1205, Linaris). 150 cells were formed to embryonic bodies with the hanging drop technic. After 24h, hanging drops spheroids were stained with Vybrant Dil [34]. Spheroids were mixed with EBM methocel and fibrinogen 1:1 with Matrigel and subcutaneously injected into SCID-mice. After 3 weeks the mice were sacrificed and perfused with an isolectin B4 Alexa 647 staining solution to visualize the newly formed vessels in the plugs. Transplanted plugs were further processed by clearing method. The clearing method was conducted according to 3Disco metheod [35]. In brief, Matrigel plugs were dehydrated in 50% THF (Tetrahydrofuran # 186562 Sigma) for 24 h at 4 °C, followed by 2 h dehydration phases 70%–100% THF at room temperature (RT). Another 100 % THF step 24 h at 4 °C, followed by 100% DCM (Dichlormethan #270997 Sigma) for 20-30 min at RT until the plugs completely submerge and sink to the bottom of the flask. Change to 100% DBE (Benzyl Ether #108014 Sigma) over night at RT until plugs are transparent. Keep in 100% DBE in light protected flasks until imaging. Images were taken by the Ultramicroscope II (UM-II, LaVision Biotec, Bielefeld) at 20× magnification (10 Zoom body + 2× Objective). Pictures were taken with Neo 5.5 (3-tap) sCOMs Camera (Andor, Mod.No.: DC-152q-C00-FI). The ImSpectorPro Version 5.0.110 was used. 3D Images and quantification were performed with Imaris (Bitplane Version 7.6). Auto fluorescence signals were deleted manually with the surface function. Background subtraction was done with the background subtraction algorithm (filter width = 2000 μm). Cells were detected and counted with the SpotsAlgorithm (Estimated Diameter = 10.0 μm; Background Subtraction = true; Intensity Center Ch = 3″ above 720; Region Growing Type = Local Contrast). Lower threshold was chosen depending to the background signal.
Statistics
Statistical analysis was performed with GraphPad Prism 9.1.2. Means with standard deviation (SD) are shown. Number of biological replicates are considered as “n”, if not stated differently. Bonferroni correction was applied in case of multiple testing, for multiple group comparisons ANOVA followed by post hoc testing was performed. For testing individual statistics of dependent samples paired t-test was used, in case of Unpaired t-test with Welch's correction. Samples with non-normal distributions were probed by Mann-Whitney test. A p value of <0.05 was considered significant.
Results
Nox4 increases during endothelial cell differentiation
According to the GTex database (GTexportal, Supp. Fig. 1A&B), Nox4 is highly expressed in blood vessel and kidney. Lymphatic endothelial cells, vascular endothelial cells, and fibroblasts exhibit the highest single-cell expression of Nox4. In order to verify the hypothesis that Nox4 is upregulated in the process of endothelial differentiation, a dataset of seven different protocols for differentiation of iPSCs into endothelial cells, published by Belt et al. [36] was re-analyzed (Supp. Fig. 1C). With decreased expression of stem cell gene (Sox2, Nanog, Pou5f1 (Oct4)) during differentiation, endothelial markers (Kdr (VEGFR2), Nos3, CD31 (Pecam1), CD34 and Cdh5 (VE-Cadherin)) increased. Within the Nox related genes analyzed, only Nox4 was consistently increased throughout all differentiation protocols.
Nox4 has no significant effect on reprogramming murine induced pluripotent stem cells
Reprogramming efficiency of murine WT- and Nox4−/−-fibroblasts was similar (Supp. Fig. 2A). Stem cells genes Sox2, Oct4, Klf4 & Nanog decreased in a similar fashion during differentiation (Supp. Fig. 2B). Comparison of miPSCs to murine embryonic stem cells reviled homogeneous expression of stem genes throughout cell types (Supp. Fig. 2C). Stem cell marker protein expression (Ssea-1, Sox2, Klf4 and Oct3/4) was similar in WT- and Nox4−/−-miPSCs as shown by immunofluorescence staining (Supp. Fig. 3A). Finally, when comparing proliferation capacity of WT- and Nox4−/−-miPSCs, by cell count for 33 passages, no significant difference was observed (Supp. Fig. 3B).
Nox4 promotes endothelial cell differentiation
In contrast to similar efficiencies in reprogramming and maintenance of the stem cell phenotype, forced in vitro endothelial differentiation was more efficient in WT-, when compared to Nox4−/−-miPSCs.
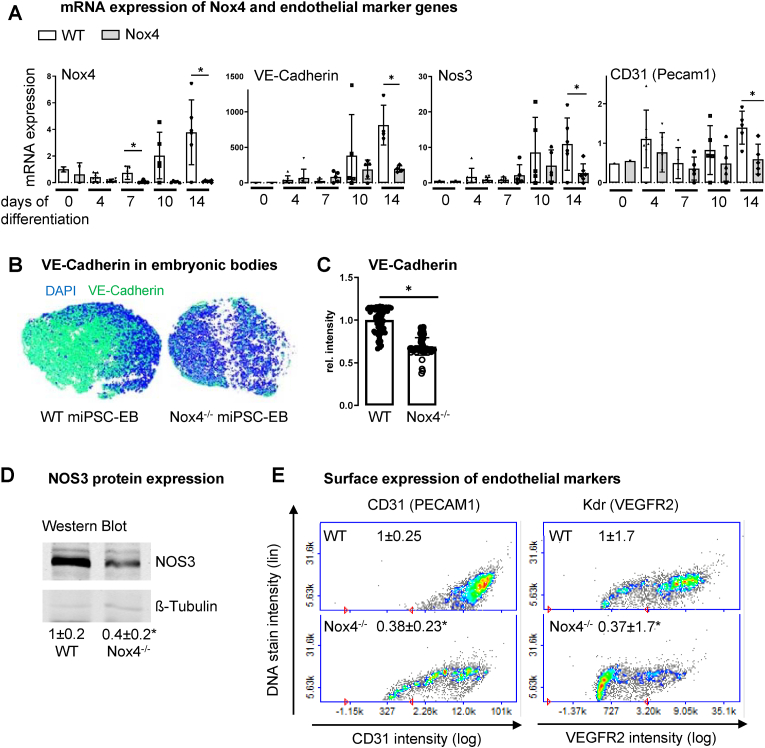
In WT-miPSCs Nox4 expression increased in the course of differentiation (approx. 4 fold from Day 7 to day 14). As Nox4 expression did not increase in Nox4−/−-cells, we analyzed for a compensatory increase of other Nox genes, which was not observed (Supp. Fig. 4B&C). Endothelial genes VE-Cadherin, Nos3, and CD31 (Pecam1) remained unchanged within the first 7 days of differentiation and subsequently increased until day 14 in WT-cells. This was most impressive for Cdh5, which increased 1000 fold (Fig. 1A). In contrast, differentiation into the endothelial lineage was attenuated in Nox4-deficient cells: mRNA expression of VE-Cadherin, Nos3, and CD31 (Pecam1) was significantly lower in Nox4−/− when compared to WT cells on day 14 of differentiation (Fig. 1A). These results were verified on protein level. VE-cadherin protein abundance was lower in embryonic bodies formed by Nox4−/−-miPSCs when compared to WT (Fig. 1C&D). Both, CD31 surface expression as well as Nos3 expression were attenuated in differentiating Nox4−/−-miPSCs (Fig. 1B&C). Interestingly, although Kdr (VEGFR2) mRNA expression was similar in WT- and Nox4−/−-miPSCs (Supp. Fig. 4A), surface abundance of Kdr (VEGFR2) protein was significantly lower in Nox4−/− when compared to WT cells (Fig. 1B). Surface expression may serve as an indicator of Kdr (VEGFR2) protein expression per cell, while mRNA is measured throughout the whole population in a dish. Accordingly, it is possible that mRNA expression indicates a general success of differentiation induction in both genotypes, which however in Nox4−/−-miPSCs is not stable and stops shortly after induction, while in WT-miPSCs pro-differentiating stimuli result in enhanced Kdr (VEGFR2) protein and eventually surface expression. The non-significant (p = 0.17 on day 14) trend of more Kdr (VEGFR2) mRNA expression in WT-miPSCs could support this guess. Further studies are needed to clarify this issue.
Fig. 1.
Loss of Nox4 expression suppresses differentiation of miPSCs into miPSC-ECs (A) Real-time qPCR for Nox4 and endothelial markers in the course of differentiation (n = 5–7; *p<0.05 WT vs. Nox4−/−). (B&C) Staining and statistics for VE-Cadherin expression in embryonic bodies of differentiating miPSC-EC in the course of differentiation on day7 (miPSC-EB). (B) 3D-reconstruction of z-Stack images from immunofluorescence stainings for VE-Cadherin (green) and DAPI (blue). (C) Z-stack analysis of maximum intensity projection in rel. intensity (*p<0.05). (D) Western blot for NOS3 on day 14 of differentiation with numbers indicating the statistics. (E) Advanced Image Cytometer measurement for Kdr (VEGFR2) and CD31 (PECAM1) at Day 7 of differentiation with numbers indicating the statistics of relative intensity (n = 4; *p<0.05 WT vs. Nox4−/−). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
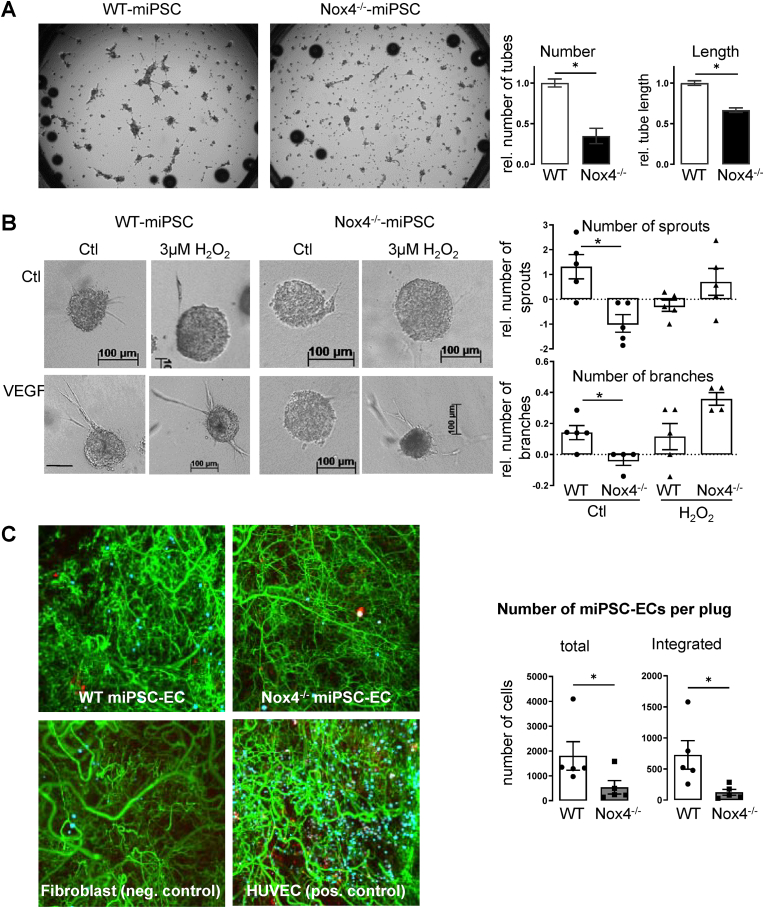
To study the possibility of impaired Kdr (VEGFR2) signaling in Nox4−/−-miPSCs, VEGF-induced angiogenic capacity was studied on day 7 of differentiation. Differentiating miPSCs from WT and Nox4−/− embryonic bodies were analyzed for tube formation as well as for spheroid outgrowth. As embryonic bodies contain a mixture of cells at several states of differentiation, basal tube formation and sprouting can be performed by any of them even without external stimuli. In order to focus on the effect of VEGF treatment only, statistics was executed for relative differences between unstimulated and VEGF treated cells and spheroids. Both assays reviled less VEGF response in Nox4−/−-miPSCs when compared to WT-miPSCs (Fig. 2A&B). In the spheroid assay, treatment with H2O2, the product of Nox4, had little effect in WT cells, while it rescued angiogenic function of Nox4−/−-miPSCs back to WT level.
Fig. 2.
Nox4 promotes angiogenic potential in the course of differentiation. (A&B) VEGF-induced effects in cells, derived from embryonic bodies at day 7 of differentiation in (A) tube formation assay with statistics for number of tubes and tube length and (B) Spheroid outgrowth assay with statistics for sprouting & branching with and without 3 μM H2O2 (n = 3–8; *p<0.05 WT vs. Nox4−/−). (C) Integration of purified WT- or Nox4−/−-miPSC-ECs into newly formed endothelial networks in Matrigel plugs 3 weeks after implantation into SCID-mice. (green: newly formed vessel staine with Isolectin B4; red: cells still present in the plug stained with Vybrant Dil; blue: integrated cells detected by algorithm); fibroblasts used as negative control and HUVECs as positive control (n = 5; *p<0.05 WT vs. Nox4−/−). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
As pointed out above we hypothesized that absence of Nox4 may alter the stability of differentiation states reached by the cells. Accordingly, we analyzed for angiogenic potential of cells isolated from the differentiating population by isolectin B4 on day 14. Purified miPSC-ECs where stained with Vybrant dil, used to form spheroids, which then were suspended in Matrigel. Eventually, the prepared Matrigel was implanted into SCID-mice. After 3 weeks, WT-miPSC-ECs and human umbilical vein endothelial cells integrated well into the newly formed network. Integration was defined as near contact of the stained miPSC-EC and the newly formed vessel. In contrast, Nox4−/−-miPSC-ECs and fibroblasts did not integrate (Fig. 2C). These data suggest that immature endothelial cells formed from Nox4−/− miPSCs exhibit an attenuated angiogenic capacity.
Lack of Nox4 delays maturation of miPSC into endothelial cells
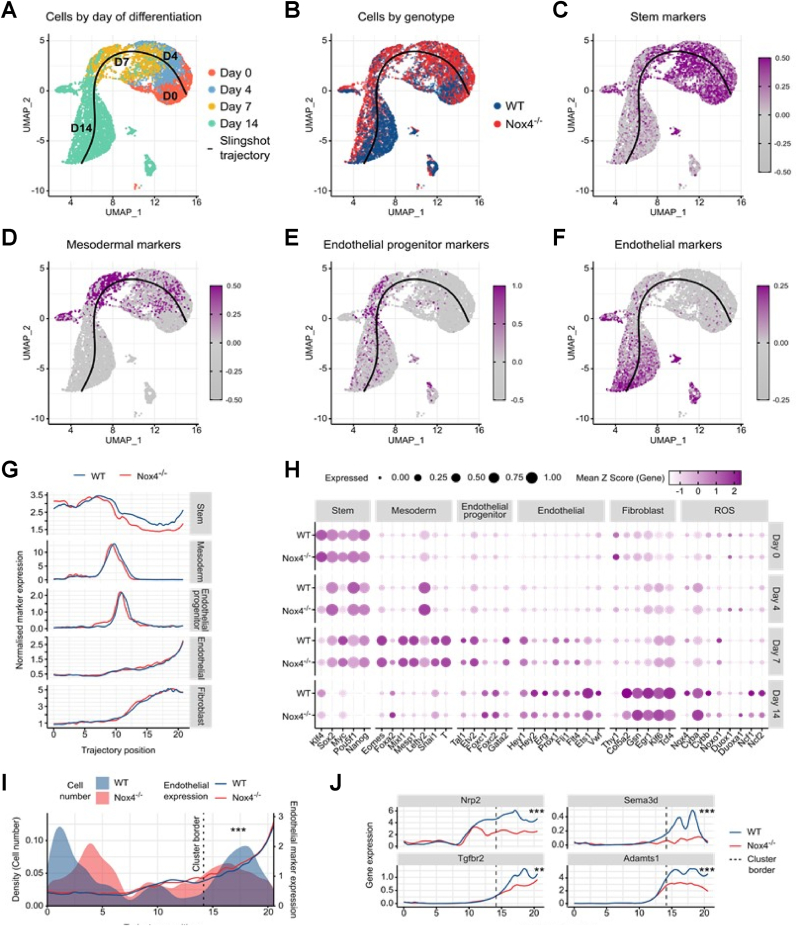
A limitation of bulk RNA-sequencing studies is that they represent a snapshot of mRNA expression in a population of cells, which is a problem when the cells being examined are highly diverse, as seen in differentiating cells. Single cell RNA-sequencing (scRNAseq) is a powerful tool to determine cell specific gene expression and identify differences in lineage development, if applied on a time-course. To take advantage of these possibilities, WT- and Nox4−/−-miPSCs differentiating towards the endothelial linage were subjected to scRNAseq on days 0, 4, 7, and 14 of differentiation. The whole dataset, encompassing both WT and Nox4−/− cells (Supp. Fig. 5A) was subset to the endothelial lineage (Fig. 3A&B) and further analyzed by Uniform Manifold Approximation and Projection (UMAP). Slingshot [29] was used to infer a pseudotime trajectory spanning from day 0 to day 14 of differentiation (Fig. 3A). The progressive differentiation across the course of the trajectory could be observed in the sequential gene expression of stem cell, mesodermal, endothelial progenitor and finally endothelial markers (Fig. 3C–F). By examining marker gene expression, it initially appeared that both WT and Nox4−/− cells were capable of progressing through the course differentiation, with no obvious differences in expression patterns across the pseudotime trajectory (Fig. 3G&H). An increase in Nox4 expression during differentiation in WT-miPSCs could be recapitulated in this data (Fig. 3H), but only subtle differences in expression could be observed in other redox-related genes (Supp. Fig. 5B). In the absence of wholescale changes in marker gene expression between WT and Nox4−/− cells, analysis instead showed that a significantly greater proportion of WT cells advance further along the endothelial trajectory as compared to Nox4−/− cells (Fig. 3I). This stark difference suggests that although Nox4−/− cells were capable of differentiating to mature endothelial cells – as evidenced by their gene expression patterns - the differentiation of Nox4−/− cells seems to stall more often than that of WT cells. In order to identify genes whose expression was affected by this, differential gene expression analysis was carried out on the cluster of cells at the endothelial extreme of the trajectory. Arising from this were a number of endothelial-related genes displaying significant changes in expression between WT and Nox4−/− cells. Amongst these were neuropilin 2 (Nrp2), semaphoring 3d (Sema3d), transforming growth factor beta receptor 2 (Tgfbr2) and a disintegrin and metalloproteinase with thrombospondin motifs 1 (Adamts1), each of which were lower-expressed in Nox4−/−, when compared to WT cells (Fig. 3J). Similar trends were detected for the genes kinase insert domain receptor (Kdr/VEGFR2), von Willebrand factor (Vwf), cadherin 5 (Cdh5/VE-cadherin) and ETS-related gene (Erg) (Supp. Fig. 5C), although after Bonferroni adjustment these changes were not statistically significant. Accordingly, despite a similar course of differentiation of WT and Nox4−/− cells, in terms of marker gene expression, more WT cells appeared to move further along the differentiation trajectory when compared to Nox4−/− cells (Fig. 2F,I,J). Collectively, these data suggest that the differentiation protocol used here yields endothelial cells. The lack of Nox4 however, delays endothelial differentiation and results in less mature endothelial cells.
Fig. 3.
Lack of Nox4 delays maturation of miPSC into endothelial cells as determined by scRNAseq. (A–F) Uniform manifold approximation and projection (UMAP) plots of cells at days 0, 4, 7 and 14 of differentiation from miPSC to EC. Day of differentiation, cell genotype, stem cell marker expression, mesodermal marker expression, endothelial progenitor marker expression, and endothelial cell marker expression superimposed on cellular embedding, respectively. (G) Normalised expression of marker genes for stem cells, mesodermal cells, endothelial progenitor cells, endothelial cells and fibroblasts in WT and Nox4−/− cells, along the fitted pseudotime trajectory. (H) Percentages of wildtype and Nox4−/− cells from each day of differentiation expressing subsets of marker genes, in combination with Z-score normalised expression of each gene across all cells. (I) Density of WT and Nox4−/− cell positions along the fitted pseudotime trajectory, in combination with mean EC marker expression. ***p< 0.001, Fishers Exact Test. (J) Normalised gene expression of Nrp2, Sema3d, Tgfbr2 and Adamts1 along the trajectory fitted to the differentiation process, in WT and Nox4−/− cells. *** Padj <0.001, ** Padj <0.01, Wilcoxon Rank Sum test, Bonferroni corrected.
Nox4 is required for the epigenetic gene activation during endothelial linage differentiation.
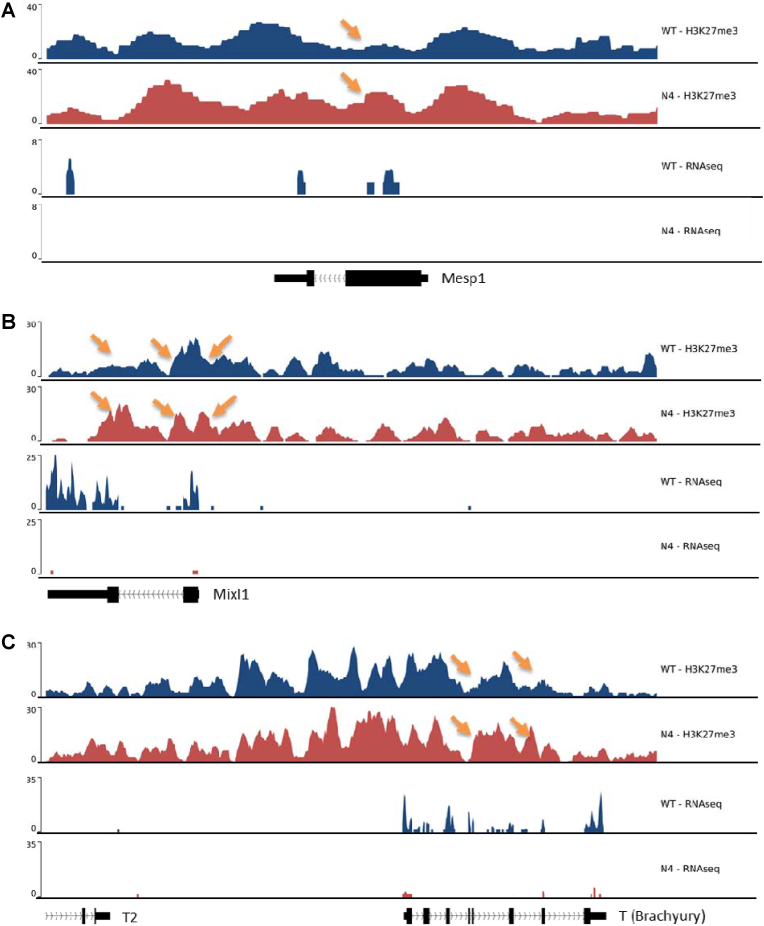
Endothelial linage commitment requires activation of endothelial gene programs. Earlier studies have shown that methylation status of lysine 27 of histone 3 (H3K27) regulates embryonic development and mesodermal linage commitment [10,37]. Chromatin immune-precipitation for H3K27 was performed looking at tri-methylation (H3K27me3), as a mark for silenced gene expression. Genome traces of ChIPseq and NGS-RNAseq data at gene loci of transcription factors were analyzed at day 7 of differentiation (Fig. 4A-C). One of which is Mesp1, which is essential for early mesodermal differentiation [38]. Indeed, Mesp1 was not only higher expressed in WT cells, the gene was also more strongly decorated with H3K27me3 in Nox4−/− cells (orange arrow as indicators). Similar findings were noted at the Mixl1 locus, required for patterning of the murine embryo [39,40] and T (Brachyury) one of the most important markers for mesodermal differentiation and early organogenesis [41].
Fig. 4.
H3K27me3 regulated chromatin accessibility on mesodermal transcription factor loci differs between WT- and Nox4−/−-miPSCs. (A–C) IGV original traces showing H3K27me3 ChIP- and RNA-Seq in WTand Nox4−/−-miPSCs on day 7 of differentiation. Loci for Mesp1 (A, chr7:79,786,926–79,798,903), Mixl1 (B, chr1:180,691,043–180,711,055) and T (C, chr17:8,421,042–8,445,355) are shown. Arrows indicate higher H3K27me3 abundance at exon proximity of genes in Nox4−/−. Numbers in square brackets indicate data range values.
Nox4 deficiency increases H3K27me3 due to reduced nuclear JmjD3 abundance
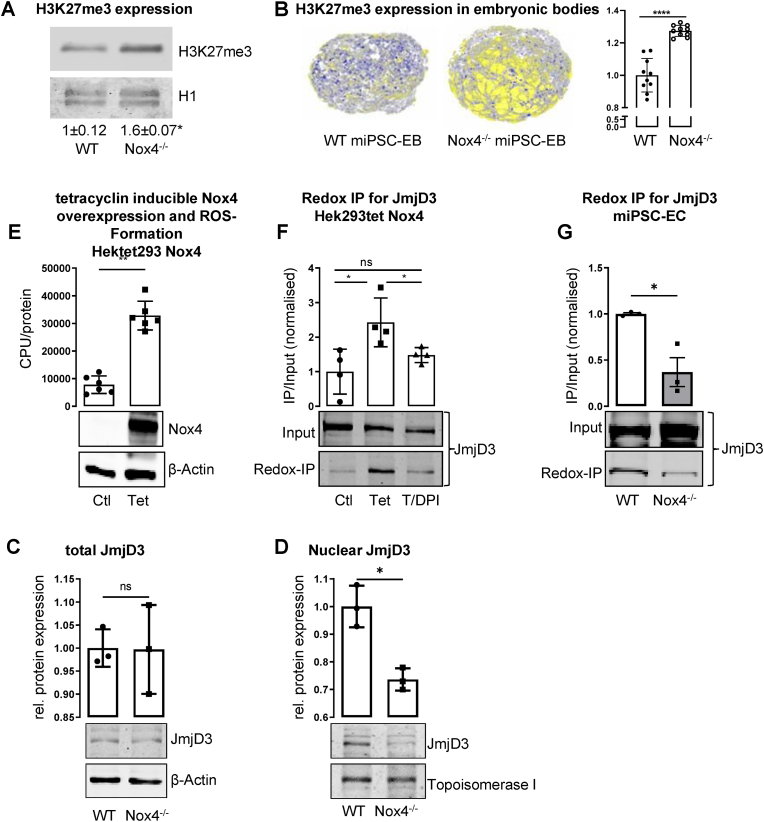
Given that Nox4 constitutively produces low level of ROS, it is likely that abundance of epigenetic marks is affected in general. Accordingly, we hypothesized that Nox4 affects not only H3K27me3 decoration of individual genes, but also whole cell level of this epigenetic mark. Indeed, whereas global levels of H3K27ac and H3K27me2 were similar between WT and Nox4−/− on day 7 of differentiation (Supp. Fig. 6A&B), H3K27me3 was significantly more abundant in cells lacking Nox4 (Fig 5A&B).
Fig. 5.
Nox4 maintains nuclear JmjD3 and reduces H3K7me3 (A) Western Blot for H3K27me3 in WT vs. Nox4−/− (n = 3; *p<0.05). (B) Immunofluorescence staining of differentiating miPSC-EC embryonic bodies for H3K27me3 (yellow) and DAPI (blue); 3D reconstruction and statistics (*p<0.05). (C&D) Western Blot and statistics for JmjD3 in whole cell lysates (C) and nuclear extracts (D, n = 3; *p<0.05). (D) Reactive Oxygen Species (ROS) measurement of Hek293 tetracyclin inducible Nox4 overexpression cells. (E) BIAM switch redox assay of JmjD3 in Nox4 overexpressing human embryonic kidney cell shows high abundance of oxidized JmjD3, Tetracyclin 1 μg/ml, 24h, DPI 3 μM, 3h (n = 4; *p<0.05). (F) BIAM switch redox assay of JmjD3 shows less oxidized JmjD3 in Nox4−/− vs. WT miPSCECs (n = 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
H3K27 triple methylation is dependent on histone demethylases, in particular JmjD3 and histone methyl transferases, such as Ezh2 [42]. We therefore hypothesized that Nox4-derived H2O2 may oxidize one of these proteins. As a model to test this hypothesis we utilized HEK/Tet (described earlier [43]). Those cells overexpress Nox4 and produce ROS via a tetracycline-inducible mechanism (Fig. 5E). In this system, activation of Nox4 dependent ROS formation resulted in an increased oxidation of JmjD3 but not of Ezh2. The pan-Nox inhibitor DPI (Diphenylene iodonium) prevented Nox4 mediated oxidation of JmjD3, but had no effect on oxidation status of Ezh2 (Fig. 5F, Supp. Fig. 6C&D). We conclude that differential oxidation of JmjD3 in this system indeed is a consequence of increased formation of H2O2 by Nox4. The impact of Nox4 mediated H2O2 formation on the redox-state of JmjD3 was determined in miPSC on day 7 of differentiation into endothelial cells. Oxidation of JmjD3 was lower in Nox4−/− as compared to WT cells, clearly demonstrating that Nox4-derived H2O2 acts as the physiological oxidation mechanism of JmjD3 in miPSCs as well (Fig. 5F). The findings of increased H2K37me3 in Nox4 deficient cells together with less oxidation of the demethylase suggests that the oxidation of JmjD3 enables activity of the enzyme. A possible mechanism underlying such an effect would be an increased nuclear abundance and subsequent physical vicinity of H3 and JmjD3. Nuclear import has been shown to control activity of other epigenetic enzymes, like histone deacetylases (HDACs) [9,44]. Despite similar total cellular levels of JmjD3, fractionation experiments showed reduced nuclear abundance of JmjD3 in Nox4−/− when compared to WT cells. (Fig. 6C&D). Collectively, these data suggest that Nox4 facilitates oxidation and nuclear translocation of JmjD3, which subsequently demethylates H3K27me3.
Discussion
Several studies support a link between Nox4 and differentiation [11,13,14,17,45]. Nox4 has a protective role in the cardiovascular system, which at least in part is mediated by an healthy endothelium [[46], [47], [48]]. Nox4 further supports angiogenic signaling and angiogenesis itself [[49], [50], [51], [52]]. Despite the well-established role of Nox4 in endothelial function, its impact on endothelial differentiation remains less clear. The present study, underlines the importance of Nox4 for differentiation, maturation and stability of endothelial cells. In fact, lack of Nox4 results in attenuated, incomplete and unstable differentiation of miPSCs into endothelial cells, which eventually end up with attenuated angiogenic capacity of Nox4 deficient miPSC-ECs. A possible mechanism is an altered chromatin remodeling which correlates with a reduced nuclear level of JmjD3.
In the course of differentiation, Nox4 expression increased significantly, while expression of other NADPH oxidases remained unchanged during the differentiation process. Induction of Nox4 in the differentiating process is accompanied by expression for endothelial markers such as CD31, Nos3 and VE-Cadherin. ScRNAseq is a powerful tool [54] to analyze the differentiation process of miPSCs to endothelial cells in more detail and to remove population biases. Data processing and visualization of pseudotime trajectory reviled subtle differences in differentiation of WT- and Nox4−/−-miPSCs rather than fundamental variances. Nevertheless those analyses gave rise to the notion endothelial maturation was attenuated after deletion of Nox4, reflected by an attenuated angiogenic capacity of Nox4−/−-miPSC-ECs.
Subtle but consistent alterations in gene expression may be a consequence of epigenetics and chromatin remodeling as a dynamic process, essential for developmental and differentiation. The chromatin repressive mark H3K27me3 has been described to be important in mesodermal [37] and cardiovascular [10] differentiation as well as endothelial lineage committing procedures [58]. A recent study suggests H3K27me3 is not essential for early embryonic stem cell differentiation, but is required for maintenance of differentiated cells [59]. Small changes in chromatin status can have an impact on the developmental process of cells. Indeed, small increases in H3K27me3 at gene loci of mesodermal transcriptions factors such as Mesp1, Mixl1 and T (Brachyury), can influence expression of these genes and therefore influence differentiation [[39], [40], [41]].
H2O2 decreases abundance H3K27me3 in murine embryonic fibroblasts [60]. Identification of redox-targets of Nox4 is a long-standing interest of our group [9,43]. Previous work discovered proteins involved in epigenetic regulations such as Gab1 and HDAC4 as differentially oxidized by Nox4 [9,43]. Herein we identified Jmjd3 as a target of Nox4 derived H2O2. JmjD3 is a demethylase that removes methylations from H3K27me3 and others and whose intracellular localization is dynamic [63]. JmjD3 has two N-terminal nuclear localization signal sites (NLS) essential for JmjD3 to translocate into the nucleus [63]. One of the two NLS has a cysteine, which may represent a target for Nox4 mediated redox modification. Consequently, it is possible that Nox4-dependent oxidation of this cysteine is required for effective nuclear import of JmjD3. This hypothesis fits the assumption of Nox4 being localized in the endoplasmic reticulum (ER) and perinuclear spaces [61,62]. JmjD3 may need to pass microdomains with high H2O2 concentrations [64] and subsequent oxidation of JmjD3 supports its nuclear translocation. Alternatively, intra-nuclear oxidation of JmjD3 by Nox4D, a truncated version of Nox4 found in the nucleus [65], may force its nuclear export. Further experimental work is required to prove this.
In summary, present study identified that Nox4 promotes the differentiation process of miPSCs into endothelial cells. Mechanistically, Nox4 oxidized Jmjd3, leading to its nuclear retention and limiting effect on H3K27 methylation at gene loci of transcription factors important to endothelial differentiation.
Author contributions
FM, FH and KS designed the experiments. FM, FH, GKB, TS,JE performed the experiments. FM, FH, GKB, TW analyzed the data. FH, SG, WA TW and RG performed bioinformatics and/or sequenced samples. FH, RPB and KS wrote the manuscript. All authors interpreted the data and approved the manuscript.
Declaration of competing interest
The authors have declared that no conflict of interest exists.
Acknowledgments
We thank Manuela Spaeth, Katalin Pálfi, Dr. Bianca Schumacher, Christina Reschke and Sabine Harenkamp for excellent technical assistance. We thank James Alexander Oo, Dr. Flávia Rezende and Dr. Matthias Leisegang for excellent scientific discussions and input for the project. This work was supported by the Goethe University Frankfurt am Main, the German Centre for Cardiovascular Research (DZHK), the DFG excellence cluster Cardiopulmonary Institute (CPI), the DFG SFB815, SFB834 Dr. Rolf M. Schwiete Stiftung.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102381.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Marcelo K.L., Goldie L.C., Hirschi K.K. Regulation of endothelial cell differentiation and specification. Circ. Res. 2013;112:1272–1287. doi: 10.1161/CIRCRESAHA.113.300506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Wang K., Zhang T., Dong Q., Nice E.C., Huang C., Wei Y. Redox homeostasis: the linchpin in stem cell self-renewal and differentiation. Cell Death Dis. 2013;4:e537. doi: 10.1038/cddis.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyublinskaya O.G., Ivanova J.S., Pugovkina N.A., Kozhukharova I.V., Kovaleva Z.V., Shatrova A.N., Aksenov N.D., Zenin V.V., Kaulin Y.A., Gamaley I.A., Nikolsky N.N. Redox environment in stem and differentiated cells: a quantitative approach. Redox Biol. 2017;12:758–769. doi: 10.1016/j.redox.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cieślar-Pobuda A., Yue J., Lee H.-C., Skonieczna M., Wei Y.-H. ROS and oxidative stress in stem cells. Oxid. Med. Cell. Longev. 2017 doi: 10.1155/2017/5047168. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bigarella C.L., Liang R., Ghaffari S. Stem cells and the impact of ROS signaling. Development. 2014;141:4206–4218. doi: 10.1242/dev.107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schröder K., Zhang M., Benkhoff S., Mieth A., Pliquett R., Kosowski J., Kruse C., Luedike P., Michaelis U.R., Weissmann N., Dimmeler S., Shah A.M., Brandes R.P. Nox4 is a protective reactive oxygen species generating vascular NADPH oxidase. Circ. Res. 2012;110:1217–1225. doi: 10.1161/CIRCRESAHA.112.267054. [DOI] [PubMed] [Google Scholar]
- 8.Lim J.C., Choi H.-I., Park Y.S., Nam H.W., Woo H.A., Kwon K.-S., Kim Y.S., Rhee S.G., Kim K., Chae H.Z. Irreversible oxidation of the active-site cysteine of peroxiredoxin to cysteine sulfonic acid for enhanced molecular chaperone activity. J. Biol. Chem. 2008;283:28873–28880. doi: 10.1074/jbc.M804087200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schader T., Löwe O., Reschke C., Malacarne P., Hahner F., Müller N., Gajos-Draus A., Backs J., Schröder K. Oxidation of HDAC4 by Nox4-derived H2O2 maintains tube formation by endothelial cells. Redox Biol. 2020;36 doi: 10.1016/j.redox.2020.101669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohtani K., Zhao C., Dobreva G., Manavski Y., Kluge B., Braun T., Rieger M.A., Zeiher A.M., Dimmeler S. Jmjd3 controls mesodermal and cardiovascular differentiation of embryonic stem cells. Circ. Res. 2013;113:856–862. doi: 10.1161/CIRCRESAHA.113.302035. [DOI] [PubMed] [Google Scholar]
- 11.Li J., Stouffs M., Serrander L., Banfi B., Bettiol E., Charnay Y., Steger K., Krause K.-H., Jaconi M.E. The NADPH oxidase NOX4 drives cardiac differentiation: role in regulating cardiac transcription factors and MAP kinase activation. Mol. Biol. Cell. 2006;17:3978–3988. doi: 10.1091/mbc.e05-06-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshikawa Y., Ago T., Kuroda J., Wakisaka Y., Tachibana M., Komori M., Shibahara T., Nakashima H., Nakashima K., Kitazono T. Nox4 promotes neural stem/precursor cell proliferation and neurogenesis in the Hippocampus and restores memory function following trimethyltin-induced injury. Neuroscience. 2019;398:193–205. doi: 10.1016/j.neuroscience.2018.11.046. [DOI] [PubMed] [Google Scholar]
- 13.Lee J.-E., Cho K.E., Lee K.E., Kim J., Bae Y.S. Nox4-mediated cell signaling regulates differentiation and survival of neural crest stem cells. Mol. Cell. 2014;37:907–911. doi: 10.14348/molcells.2014.0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nadworny A.S., Guruju M.R., Poor D., Doran R.M., Sharma R.V., Kotlikoff M.I., Davisson R.L. Nox2 and Nox4 influence neonatal c-kit(+) cardiac precursor cell status and differentiation. Am. J. Physiol. Heart Circ. Physiol. 2013;305:H829–H842. doi: 10.1152/ajpheart.00761.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J., Kim J., Lim H.J., Lee S., Bae Y.S., Kim J. Nox4-IGF2 Axis promotes differentiation of embryoid body cells into derivatives of the three embryonic germ layers. Stem Cell Rev. Rep. 2021 doi: 10.1007/s12015-021-10303-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang X., Wei X., Jiang L., Niu C., Zhang J., Chen S., Meng D. Nox2 and Nox4 regulate self-renewal of murine induced-pluripotent stem cells. IUBMB Life. 2016;68:963–970. doi: 10.1002/iub.1574. [DOI] [PubMed] [Google Scholar]
- 17.Xiao Q., Luo Z., Pepe A.E., Margariti A., Zeng L., Xu Q. Embryonic stem cell differentiation into smooth muscle cells is mediated by Nox4-produced H2O2. Am. J. Physiol. Cell Physiol. 2009;296:C711–C723. doi: 10.1152/ajpcell.00442.2008. [DOI] [PubMed] [Google Scholar]
- 18.Clempus R.E., Sorescu D., Dikalova A.E., Pounkova L., Jo P., Sorescu G.P., Schmidt H.H.H., Lassègue B., Griendling K.K. Nox4 is required for maintenance of the differentiated vascular smooth muscle cell phenotype. Arterioscler. Thromb. Vasc. Biol. 2007;27:42–48. doi: 10.1161/01.ATV.0000251500.94478.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sommer C.A., Stadtfeld M., Murphy G.J., Hochedlinger K., Kotton D.N., Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cell. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blancas A.A., Lauer N.E., McCloskey K.E. Endothelial differentiation of embryonic stem cells. Curr. Protoc. Stem Cell Biol. 2008 doi: 10.1002/9780470151808.sc01f05s6. (Chapter 1) Unit 1F.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serrander L., Cartier L., Bedard K., Banfi B., Lardy B., Plastre O., Sienkiewicz A., Fórró L., Schlegel W., Krause K.-H. NOX4 activity is determined by mRNA levels and reveals a unique pattern of ROS generation. Biochem. J. 2007;406:105–114. doi: 10.1042/BJ20061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Sena Brandine G., Smith A.D. Falco: high-speed FastQC emulation for quality control of sequencing data. F1000Res. 2019;8:1874. doi: 10.12688/f1000research.21142.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 26.Kaminow B., Yunusov D., Dobin A. 2021. STARsolo: Accurate, Fast and Versatile Mapping/quantification of Single-Cell and Single-Nucleus RNA-Seq Data. [Google Scholar]
- 27.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R: R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, R Core Team; 2021. p. 2021. [Google Scholar]
- 29.Street K., Risso D., Fletcher R., Das D., Ngai J., Yosef N., Purdom E., Dudoit S. slingshot: Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics., Bioconductor. BMC Genom. 2018;477 doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mittnenzweig M., Mayshar Y., Cheng S., Ben-Yair R., Hadas R., Rais Y., Chomsky E., Reines N., Uzonyi A., Lumerman L., Lifshitz A., Mukamel Z., Orenbuch A.-H., Tanay A., Stelzer Y. A single-embryo, single-cell time-resolved model for mouse gastrulation. Cell. 2021;184:2825–2842. doi: 10.1016/j.cell.2021.04.004. e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oscar Franzén, Li-Ming Gan, Johan L M Björkegren, PanglaoDB: PanglaoDB: a Web Server for Exploration of Mouse and Human Single-Cell RNA Sequencing Data.. [DOI] [PMC free article] [PubMed]
- 32.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carpentier G., Martinelli M., Courty J., Cascone I., editors. 24. – 26. Mondorf-les-Bains; Luxembourg]: 2012. (Proceedings of the ImageJ User and Developer Conference). Oktober 2012. CRP, Luxembourg, 2012. [Google Scholar]
- 34.Laib A.M., Bartol A., Alajati A., Korff T., Weber H., Augustin H.G. Spheroid-based human endothelial cell microvessel formation in vivo. Nat. Protoc. 2009;4:1202–1215. doi: 10.1038/nprot.2009.96. [DOI] [PubMed] [Google Scholar]
- 35.Ertürk A., Becker K., Jährling N., Mauch C.P., Hojer C.D., Egen J.G., Hellal F., Bradke F., Sheng M., Dodt H.-U. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protoc. 2012;7:1983–1995. doi: 10.1038/nprot.2012.119. [DOI] [PubMed] [Google Scholar]
- 36.Belt H., Koponen J.K., Kekarainen T., Puttonen K.A., Mäkinen P.I., Niskanen H., Oja J., Wirth G., Koistinaho J., Kaikkonen M.U., Ylä-Herttuala S. Temporal dynamics of gene expression during endothelial cell differentiation from human iPS cells: a comparison study of signalling factors and small molecules. Front. Cardiovasc. Med. 2018;5:16. doi: 10.3389/fcvm.2018.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C., Lee J.-E., Cho Y.-W., Xiao Y., Jin Q., Liu C., Ge K. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc. Natl. Acad. Sci. U.S.A. 2012;109:15324–15329. doi: 10.1073/pnas.1204166109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saga Y., Hata N., Kobayashi S., Magnuson T., Seldin M.F., Taketo M.M. MesP1: a novel basic helix-loop-helix protein expressed in the nascent mesodermal cells during mouse gastrulation. Development. 1996;122:2769–2778. doi: 10.1242/dev.122.9.2769. [DOI] [PubMed] [Google Scholar]
- 39.Hart A.H., Hartley L., Sourris K., Stadler E.S., Li R., Stanley E.G., Tam P.P.L., Elefanty A.G., Robb L. Mixl1 is required for axial mesendoderm morphogenesis and patterning in the murine embryo. Development. 2002;129:3597–3608. doi: 10.1242/dev.129.15.3597. [DOI] [PubMed] [Google Scholar]
- 40.Ng E.S., Azzola L., Sourris K., Robb L., Stanley E.G., Elefanty A.G. The primitive streak gene Mixl1 is required for efficient haematopoiesis and BMP4-induced ventral mesoderm patterning in differentiating ES cells. Development. 2005;132:873–884. doi: 10.1242/dev.01657. [DOI] [PubMed] [Google Scholar]
- 41.Beddington R.S., Rashbass P., Wilson V. Brachyury--a gene affecting mouse gastrulation and early organogenesis. Dev. Suppl. 1992:157–165. [PubMed] [Google Scholar]
- 42.Lavarone E., Barbieri C.M., Pasini D. Dissecting the role of H3K27 acetylation and methylation in PRC2 mediated control of cellular identity. Nat. Commun. 2019;10:1679. doi: 10.1038/s41467-019-09624-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Löwe O., Rezende F., Heidler J., Wittig I., Helfinger V., Brandes R.P., Schröder K. BIAM switch assay coupled to mass spectrometry identifies novel redox targets of NADPH oxidase 4. Redox Biol. 2019;21 doi: 10.1016/j.redox.2019.101125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKinsey T.A., Zhang C.L., Lu J., Olson E.N. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schröder K., Wandzioch K., Helmcke I., Brandes R.P. Nox4 acts as a switch between differentiation and proliferation in preadipocytes. Arterioscler. Thromb. Vasc. Biol. 2009;29:239–245. doi: 10.1161/ATVBAHA.108.174219. [DOI] [PubMed] [Google Scholar]
- 46.Zhang M., Mongue-Din H., Martin D., Catibog N., Smyrnias I., Zhang X., Yu B., Wang M., Brandes R.P., Schröder K., Shah A.M. Both cardiomyocyte and endothelial cell Nox4 mediate protection against hemodynamic overload-induced remodelling. Cardiovasc. Res. 2018;114:401–408. doi: 10.1093/cvr/cvx204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang M., Brewer A.C., Schröder K., Santos C.X.C., Grieve D.J., Wang M., Anilkumar N., Yu B., Dong X., Walker S.J., Brandes R.P., Shah A.M. NADPH oxidase-4 mediates protection against chronic load-induced stress in mouse hearts by enhancing angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18121–18126. doi: 10.1073/pnas.1009700107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schürmann C., Rezende F., Kruse C., Yasar Y., Löwe O., Fork C., van de Sluis B., Bremer R., Weissmann N., Shah A.M., Jo H., Brandes R.P., Schröder K. The NADPH oxidase Nox4 has anti-atherosclerotic functions. Eur. Heart J. 2015;36:3447–3456. doi: 10.1093/eurheartj/ehv460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harel S., Mayaki D., Sanchez V., Hussain S.N.A. NOX2, NOX4, and mitochondrial-derived reactive oxygen species contribute to angiopoietin-1 signaling and angiogenic responses in endothelial cells. Vascul. Pharmacol. 2017;92:22–32. doi: 10.1016/j.vph.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 50.Vogel J., Kruse C., Zhang M., Schröder K. Nox4 supports proper capillary growth in exercise and retina neo-vascularization. J. Physiol. 2015;593:2145–2154. doi: 10.1113/jphysiol.2014.284901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Craige S.M., Chen K., Pei Y., Li C., Huang X., Chen C., Shibata R., Sato K., Walsh K., Keaney J.F. NADPH oxidase 4 promotes endothelial angiogenesis through endothelial nitric oxide synthase activation. Circulation. 2011;124:731–740. doi: 10.1161/CIRCULATIONAHA.111.030775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim Y.-M., Kim S.-J., Tatsunami R., Yamamura H., Fukai T., Ushio-Fukai M. ROS-induced ROS release orchestrated by Nox4, Nox2, and mitochondria in VEGF signaling and angiogenesis. Am. J. Physiol. Cell Physiol. 2017;312:C749–C764. doi: 10.1152/ajpcell.00346.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ziegenhain C., Vieth B., Parekh S., Reinius B., Guillaumet-Adkins A., Smets M., Leonhardt H., Heyn H., Hellmann I., Enard W. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell. 2017;65:631–643. doi: 10.1016/j.molcel.2017.01.023. e4. [DOI] [PubMed] [Google Scholar]
- 58.Ohtani K., Vlachojannis G.J., Koyanagi M., Boeckel J.-N., Urbich C., Farcas R., Bonig H., Marquez V.E., Zeiher A.M., Dimmeler S. Epigenetic regulation of endothelial lineage committed genes in pro-angiogenic hematopoietic and endothelial progenitor cells. Circ. Res. 2011;109:1219–1229. doi: 10.1161/CIRCRESAHA.111.247304. [DOI] [PubMed] [Google Scholar]
- 59.Miller S.A., Damle M., Kim J., Kingston R.E. Full methylation of H3K27 by PRC2 is dispensable for initial embryoid body formation but required to maintain differentiated cell identity. Development. 2021;148 doi: 10.1242/dev.196329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Negishi M., Saraya A., Mochizuki S., Helin K., Koseki H., Iwama A. A novel zinc finger protein Zfp277 mediates transcriptional repression of the Ink4a/arf locus through polycomb repressive complex 1. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petry A., Djordjevic T., Weitnauer M., Kietzmann T., Hess J., Görlach A. NOX2 and NOX4 mediate proliferative response in endothelial cells, Antioxid. Redox Signal. 2006;8:1473–1484. doi: 10.1089/ars.2006.8.1473. [DOI] [PubMed] [Google Scholar]
- 62.Helmcke I., Heumüller S., Tikkanen R., Schröder K., Brandes R.P. Identification of structural elements in Nox1 and Nox4 controlling localization and activity. Antioxidants Redox Signal. 2009;11:1279–1287. doi: 10.1089/ars.2008.2383. [DOI] [PubMed] [Google Scholar]
- 63.Kamikawa Y.F., Donohoe M.E. The localization of histone H3K27me3 demethylase Jmjd3 is dynamically regulated. Epigenetics. 2014;9:834–841. doi: 10.4161/epi.28524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schröder K. NADPH oxidase-derived reactive oxygen species: dosis facit venenum. Exp. Physiol. 2019;104:447–452. doi: 10.1113/EP087125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anilkumar N., San Jose G., Sawyer I., Santos C.X.C., Sand C., Brewer A.C., Warren D., Shah A.M. A 28-kDa splice variant of NADPH oxidase-4 is nuclear-localized and involved in redox signaling in vascular cells. Arterioscler. Thromb. Vasc. Biol. 2013;33:e104–e112. doi: 10.1161/ATVBAHA.112.300956. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.