Abstract
The viability of lactic acid bacteria is crucial for their applications as dairy starters and as probiotics. We investigated the usefulness of flow cytometry (FCM) for viability assessment of lactic acid bacteria. The esterase substrate carboxyfluorescein diacetate (cFDA) and the dye exclusion DNA binding probes propidium iodide (PI) and TOTO-1 were tested for live/dead discrimination using a Lactococcus, a Streptococcus, three Lactobacillus, two Leuconostoc, an Enterococcus, and a Pediococcus species. Plate count experiments were performed to validate the results of the FCM assays. The results showed that cFDA was an accurate stain for live cells; in exponential-phase cultures almost all cells were labeled, while 70°C heat-killed cultures were left unstained. PI did not give clear live/dead discrimination for some of the species. TOTO-1, on the other hand, gave clear discrimination between live and dead cells. The combination of cFDA and TOTO-1 gave the best results. Well-separated subpopulations of live and dead cells could be detected with FCM. Cell sorting of the subpopulations and subsequent plating on agar medium provided direct evidence that cFDA labels the culturable subpopulation and that TOTO-1 labels the nonculturable subpopulation. Applied to cultures exposed to deconjugated bile salts or to acid, cFDA and TOTO-1 proved to be accurate indicators of culturability. Our experiments with lactic acid bacteria demonstrated that the combination of cFDA and TOTO-1 makes an excellent live/dead assay with versatile applications.
Lactic acid bacteria (LAB) are applied in food production for their useful metabolic properties. They are used as starters and as probiotics. However, these applications imply that the LAB are exposed to various stress conditions that may affect the physiological status of the microbes. LAB are employed as starter cultures in the production of fermented foods, such as cheese, yogurts, wines, and fermented meats. The starter cultures are often stored in freeze-dried form, which decreases the number of CFU significantly (6). Cell proliferation and metabolic activity are crucial for success of fermentation processes, such as in cheese production. The starter bacteria multiply after being added to the curd, convert lactose to lactic acid, and degrade casein to peptides and amino acids. These are essential functions for the development of texture and flavor (9). At the same time the conditions of the fermentation process, in particular the decline of the pH, the temperature, and the high salt concentration, affect the physiological status of the bacteria.
Besides being used in dairy fermentations, several LAB species are employed as probiotics. Probiotics are living microorganisms which upon ingestion in certain numbers should exert health effects beyond inherent basic nutrition (17). The species are selected mainly on the basis of their potential health-associated properties, but it is well recognized that further criteria should also be fulfilled (14, 17, 20, 39). One of the requirements is resistance to technological processes, such as survival in fermented milk to provide a suitable shelf life period for the product. Another requirement is resistance to gastric acid and bile. This is necessary for persistence in the gastrointestinal tract to perform health-promoting actions (7, 13, 14). In studies on survival and stress response, the quantitative assessment of viability is important.
In concept, bacterial viability is the reproductive capacity, and survival is the maintenance of the viability (1). In operation, viability has to be demonstrated by replication in a validated laboratory system (1, 22). The conventional method for quantitative survival studies is the plate count technique, in which replication on an appropriate agar medium is tested. Although this is the only direct proof of culturability (1, 22), the plate count method has major drawbacks (3). For many species there is not (yet) a good growth medium. Furthermore, the plate count technique requires long incubation times (2 days to a few weeks). Alternative techniques for viability assessment are desired for fundamental as well as routine microbiology research, although they have to be rapid and reliable. Flow cytometry (FCM) is an appealing technique for fast viability assessment.
FCM is a rapid technique for cell-by-cell multiparameter analysis that is often used in combination with fluorescent labeling (37). Cells are analyzed at rates of 100 to 1,000 per s as they are carried within a fast-flowing fluid stream that passes a focused light beam. The forward-angle light scatter (FSC), the side-angle light scatter (SSC), and the fluorescence at selected wavelengths are measured. The analyses are done on large populations of cells, typically 5,000 to 10,000. Subpopulations can be identified and distinguished when they differ in light scatter or fluorescence characteristics. Also, subpopulations can be physically selected (sorted) for further study. FCM in combination with fluorescent labeling is increasingly applied in microbiology. It is used in counting the total number of bacteria and in detecting specific strains by 16S rRNA sequence or by antigen expression. It is also used for characterizing and quantifying cellular physiological parameters such as DNA content, enzyme activity, respiration, membrane potential, intracellular pH, and membrane integrity (11, 16, 27, 34, 35).
Various fluorescent probes are used for viability assessment (3, 18, 30, 35). Redox probes are used, such as tetrazolium salts that are reduced by the electron transfer chain. Also, membrane potential probes are used, such as anionic oxonol dyes and the cationic dye rhodamine 123. Furthermore, esterase substrates are used, such as fluorescein diacetate and calcein AM. These are nonfluorescent precursors that are taken up by the cell. The fluorescent products are positively charged, so they are retained in the cell provided that the membrane is intact. Thus, labeling indicates enzymatic activity and membrane integrity (4, 12). Finally, dye exclusion probes are used extensively, especially DNA binding compounds. The exclusion of such impermeant probes by cells with intact membranes is taken as an indicator of viability.
Two groups of dye exclusion probes are of importance: phenanthridium nucleic acid dyes and cyanine nucleic acid dyes. The phenanthridium nucleic acid dyes include ethidium bromide, propidium iodide (PI), and ethidium homodimer-1. These probes have been used almost exclusively to evaluate cell membrane integrity of bacterial as well as eucaryotic cells (5, 18, 28). Cyanine nucleic acid dyes are compounds that also have the chemical characteristics necessary for a viability assay based on dye exclusion. The group comprises the compounds of the monomeric TO-PRO series, the dimeric TOTO series, and the SYTOX series (Molecular Probes Inc., Eugene, Oreg.). These probes bind to DNA with little specificity, have very high fluorescence enhancement factors, and have a range of spectra covering the entire visible spectrum (18, 19). SYTOX-Green was described as a probe for bacterial viability and antibiotic susceptibility testing and has been applied and further investigated in several studies (25, 32, 36). In contrast, the TO-PRO and TOTO series compounds have been described mainly as probes for DNA gel electrophoresis and DNA analysis by FCM and laser confocal microscopy (18), whereas reports on their use as viability indicators are scarce. YOYO-1 and YOYO-3 have been applied for eucaryotic cells (2, 23). TO-PRO-3 was used for investigating starving and resuscitating cultures of Micrococcus luteus (40). TO-PRO-1 was compared to PI and SYTOX-Green to study injured Escherichia coli (32).
The subject of this study was the rapid FCM analysis of LAB, in particular, the assessment of survival when LAB are exposed to bile salts or to acid. We aimed for an FCM assay that accurately indicates culturability, with proven validity for the given stress conditions. Plate counts were performed to ensure that the populations indicated as live by the FCM viability assay were indeed culturable while populations indicated as dead were not culturable. A selection of nine LAB species was tested, including species from the different genera of dairy LAB as well as species used in various dairy products and probiotics (8, 26, 33). We evaluated carboxyfluorescein diacetate (cFDA) as a live stain using the labeling protocol for Lactococcus lactis analysis by fluorescence microscopy developed in an earlier study (5). Furthermore, the impermeant nucleic acid stains PI and TOTO-1 were evaluated for their capacity to stain dead LAB cells using FCM. PI was included because it is the probe most used for detection of dead cells and its spectroscopic properties make it suitable for FCM (18). TOTO-1 was chosen because the excitation and emission spectra are suitable for FCM, it has a high fluorescence enhancement, and its molecular mass is approximately twice as high as that of PI (18, 19). Possible applications of the developed FCM live/dead assay are discussed.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The species used are listed in Table 1. Leuconostoc lactis L60, Lactobacillus helveticus T97, Lactobacillus casei R, and Lactobacillus delbrueckii subsp. bulgaricus 2 were supplied by NIZO Food Research, Ede, The Netherlands. The other strains are from the strain collection of our laboratory. The strains were maintained as freezer stocks at −80°C in 40% glycerol. After inoculation from the freezer stocks, the cultures were grown to stationary phase (16 to 30 h). Lactococcus lactis, Enterococcus faecium, and Pediococcus acidilacti were grown at 30°C in M17 broth (Unipath Oxoid, Basingstoke, United Kingdom) supplemented with 0.5% (wt/vol) lactose (LM17). Streptococcus salivarius subsp. thermophilus was grown at 42°C in LM17. Leuconostoc mesenteroides and Leuconostoc lactis were grown at 30°C in MRS broth (Merck, Darmstadt, Germany). Lactobacillus casei was grown at 37°C in MRS broth. Lactobacillus delbrueckii subsp. bulgaricus and Lactobacillus helveticus were grown at 42°C in MRS broth. The cultures were then diluted (1:9) in fresh medium and grown to mid-exponential phase. The cultures were harvested at an optical density at 620 nm (OD620) of approximately 0.7 by centrifugation at 4,000 × g for 10 min at 10°C. Unless mentioned otherwise, 50 mM potassium phosphate (KPi) buffer adjusted to pH 7.0 was used for suspending, washing, and incubating cells. Harvested cells were washed twice, concentrated to an OD620 of 20, and kept on ice until use.
TABLE 1.
Evaluation of FCM in combination with cFDA, PI, and TOTO-1 for identifying live and dead cellsa
| Organism | Resultb with probe:
|
||
|---|---|---|---|
| cFDA | PI | TOTO-1 | |
| Lactococcus lactis NCDO 712 | + | − | + |
| Streptococcus salivarius subsp. thermophilus | + | + | + |
| Lactobacillus delbrueckii subsp. bulgaricus 2 | + | + | +/− |
| Lactobacillus casei R | + | + | + |
| Lactobacillus helveticus T97 | + | − | + |
| Leuconostoc mesenteroides DSM 20343 | + | + | + |
| Leuconostoc lactis L60 | + | − | +/− |
| Enterococcus faecium 93-828-3 | + | − | + |
| Pediococcus acidilacti | + | +/− | + |
Nine LAB species were tested using exponential-phase cells that were not treated and cells that were heat killed at 70°C.
The live and dead populations were well separated (+), overlapped each other somewhat (+/−), or overlapped each other considerably (−).
Treatments.
Portions of 200 μl of concentrated cell suspension (OD620 of 20) were exposed to heat, acid, or bile salts. Cells were heat killed by exposure to 70°C for 10 min. Treatments with acid were done by incubating cells at 30°C for 60 min in 10 mM KPi adjusted with hydrochloric acid to pH 2.0, 3.0, or 4.0, or in 50 mM KPi adjusted to pH 5.0 or 6.0. Treatments with bile salts were done by incubation of cells at 30°C for 60 min with a final concentration of 0.05, 0.10, 0.25, 0.50, or 1.00% (wt/wt) deconjugated bile salts (50% sodium cholate, 50% sodium deoxycholate [Sigma-Aldrich, Steinheim, Germany]). As a control, cells were incubated at pH 7.0 and 30°C for 60 min. After the incubations, the cells were spun down, resuspended in buffer, and put on ice until use.
Measurement of culturability.
Tenfold serial dilutions of control and treated samples were made in buffer, and triplicate aliquots of 100 μl of the appropriate dilution were spread out on agar plates. Lactococcus lactis, E. faecium, and P. acidilacti were plated on LM17 medium. The other species were plated on MRS medium. After 3 days of aerobic incubation at 30°C, the colonies were counted.
Fluorescence labeling.
cFDA, PI, and TOTO-1 were purchased from Molecular Probes, Inc. TOTO-1 is 1,1′(4,4,7,7-tetramethyl-4,7-diazaundecamethylene)-bis-4-[3-methyl-2,3dihydro(benzo-1,3-oxazole)-2-methylidene]-1-(3′-tri-methylammoniumpropyl)-pyridinium tetraiodide. cFDA is an esterase substrate yielding the fluorescent carboxyfluorescein (cF) upon hydrolysis. cF has an excitation maximum (λex) of 492 nm and an emission maximum (λem) of 517 nm. PI and TOTO-1 bind to DNA. PI has a molecular mass of 668 g per mol and a fluorescence enhancement of 20- to 30-fold upon binding. The PI-DNA complex has a λex of 535 nm and a λem of 617 nm. TOTO-1 has a molecular mass of 1,303 g per mol and a very high fluorescence enhancement of 1,400-fold. The TOTO-1–DNA complex has a λex of 514 nm and a λem of 533 nm. Stock solutions of 100 μM cFDA in 50 mM KPi buffer (pH 7.0), 1.5 mM PI in distilled water, and 100 μM TOTO-1 in dimethyl sulfoxide were prepared. For single-probe labeling, concentrated cell suspensions (OD620 of 10) were incubated with 50 μM cFDA, 30 μM PI, or 1 μM TOTO-1 at 30°C for 10 min. After incubation with cFDA, the cells were washed once. For double labeling, concentrated cell suspensions (OD620 of 10) were incubated with 50 μM cFDA and 1 μM TOTO-1 simultaneously at 30°C for 10 min, after which the cells were washed once. All labeled cell suspensions were kept on ice until use.
Fluorescence microscopy.
Labeled cell suspensions were diluted to approximately 109 cells per ml and microscopically analyzed with an Axioskop epifluorescence microscope equipped with a 12-V, 50-W halogen lamp for transmitted-light illumination, a 50-W mercury arc lamp for epifluorescence illumination, a fluorescein isothiocyanate filter set (excitation wavelength, 450 to 490 nm; emission wavelength, >520 nm), a 100× 1.3-numerical-aperture Plan-Neofluar objective lens, and an MC80 camera (Carl Zeiss, Oberkochen, Germany). Photomicrographs were made with simultaneous light and epifluorescence microscopy, a low transmitted-light intensity, and an exposure time of 15 s on Kodak 400 ASA color films. In these photomicrographs both the labeled cells and the nonlabeled cells were visible.
Spectrofluorimetry.
To measure the cF labeling capacity, i.e., the amount of cF in the cells per milligram of protein, labeled cells were lysed by incubation at 70°C for 15 min and the debris was removed by centrifugation. The fluorescence of the supernatant was measured fluorimetrically (excitation at 490 ± 5 nm and emission at 515 ± 5 nm) with a Perkin-Elmer LS 50B luminescence spectrometer equipped with a plate reader by using computer-controlled data acquisition. The cF concentration was calculated from a calibration curve with a cF concentration range from 0 to 1.5 μM in 50 mM KPi buffer (pH 7.0). The cell protein concentrations were analyzed by the Lowry method.
FCM.
FCM analyses were performed on a FACSCalibur flow cytometer (Becton Dickinson Immunocytometry Systems, San Jose, Calif.) equipped with a 15-mW, 488-nm, air-cooled argon ion laser and a cell-sorting catcher tube. Cell samples were diluted to approximately 106 cells per ml and delivered at the low flow rate, corresponding to 150 to 500 cells per s. FSC, SSC, and three fluorescence signals were measured. A band pass filter of 530 nm (515 to 545 nm) was used to collect the green fluorescence (FL1), a band pass filter of 585 nm (564 to 606 nm) was used to collect the yellow-orange fluorescence (FL2), and a long-pass filter of 670 nm was used to collect the red fluorescence (FL3). FSC was collected with a diode detector. SSC and the three fluorescence signals were collected with photomultiplier tubes. All signals were collected by using logarithmic amplifications. A combination of FSC and SSC was used to discriminate bacteria from background. Data were analyzed with the CELLQuest program (version 3.1f; Becton Dickinson) and the WinMDI program (version 2.8; Joseph Trotter, John Curtin School of Medical Research, Canberra, Australia [http://jcsmr.anu.edu.au]).
Sorting.
Sorting experiments were performed with Lactococcus lactis. Exponential-phase cell suspensions that were left unstained or incubated with cFDA, as well as 1:1 mixtures of exponential-phase cells and 70°C heat-killed cells labeled with cFDA or TOTO-1, were used. Furthermore, cell suspensions exposed to 0.10% bile salts labeled with cFDA or TOTO-1 were used. All sample handling was done aseptically. In the dot plot of FL1 and FL2, regions of nonlabeled, cF-labeled, and TOTO-1-labeled cells were defined to use as sort gates. One region was sorted at a time. Culturability was tested by plating 100-μl samples of the sorted subpopulations directly out of the sort collection tubes.
RESULTS
Staining with cFDA, PI, or TOTO-1.
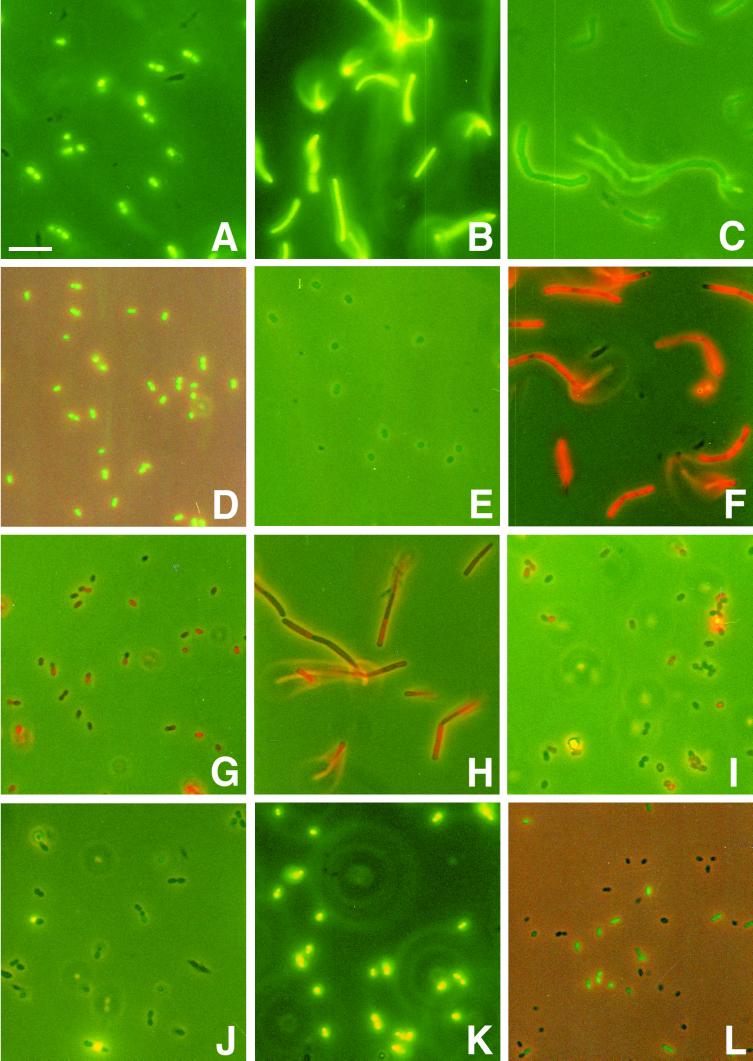
Differential staining of live and dead cells by cFDA, PI, and TOTO-1 was investigated by testing the probes on exponential-phase cells that were not treated and cells that were heated at 70°C for 10 min. The 70°C treatment killed all cells, as was confirmed by plating 100 μl of cell suspensions that contained 1010 CFU per ml before heat treatment. Samples were analyzed by FCM. For visual reference, fluorescence microscopic photographs were made (Fig. 1A to K).
FIG. 1.
Labeling of LAB with fluorescent probes. (A to E) Evaluation of cFDA as a stain for live cells. S. salivarius subsp. thermophilus (A), Lactobacillus delbrueckii subsp. bulgaricus (B), Lactobacillus casei (C), Leuconostoc mesenteroides (D), and P. acidilacti (E) exponential-phase cell suspensions were incubated with 50 μM cFDA and washed once. (F to I) Evaluation of PI as a stain for dead cells. Lactobacillus delbrueckii (F) and E. faecium (G) 70°C heat-killed cell suspensions and Lactobacillus helveticus (H) and Leuconostoc lactis (I) exponential-phase cell suspensions were incubated with 30 μM PI. (J and K) Evaluation of TOTO-1 as a stain for dead cells. Exponential-phase cells (J) and 70°C heat-killed cells (K) of S. salivarius subsp. thermophilus were incubated with 1 μM TOTO-1. (L) Application of labeling for viability assessment after bile stress. Lactococcus lactis was exposed to 0.10% deconjugated bile salts at 30°C for 60 min. After being washed, the cell suspension was incubated with 50 μM cFDA. All labeling incubations were at 30°C for 10 min. All photographs were made with simultaneous phase-contrast illumination and epifluorescence excitation and an exposure time of 15 s to visualize both labeled and nonlabeled cells. The composite was made with Adobe Photoshop 5.5. Bar, 10 μm.
cFDA gave good results (Table 1). Incubation of exponential-phase cells of the nine selected LAB species with cFDA resulted in a high fraction of cF-labeled cells, in most cases above 90% (Fig. 1A to E), whereas no cells of 70°C heat-killed suspensions were labeled. In the FCM analysis of all species the labeled population gave a peak in the green fluorescence histogram, which was resolved from the signal of nonlabeled cells.
Notably, the intensity of cF fluorescence differed among the LAB species. The FCM histograms of cF-labeled control samples of exponential-phase cells (Fig. 2), as well as the photographs (Fig. 1A to E), show the differences in cF labeling intensity when the same protocol is applied for all species. This was confirmed by spectrofluorimetry, which revealed a greater-than-10-fold difference between Lactobacillus delbrueckii subsp. bulgaricus and P. acidilacti, the species with the highest and the lowest cF labeling capacities, respectively (data not shown). However, the results showed that the same standard protocol for labeling enables discrimination of live and dead cells of all tested species by FCM analysis.
FIG. 2.
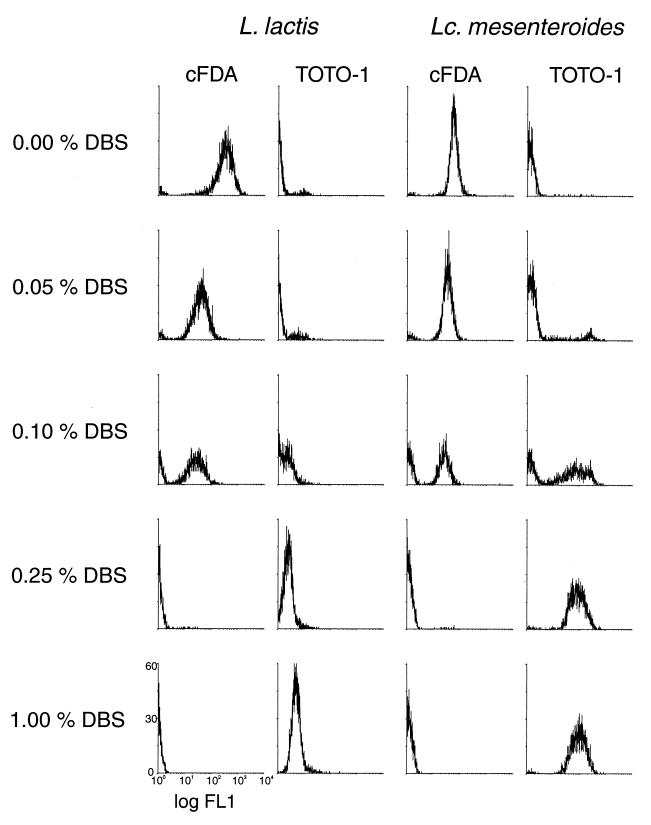
Flow cytometry histograms of FL1 of Lactococcus lactis and Leuconostoc mesenteroides cell suspensions stained with cFDA or with TOTO-1 after exposure to deconjugated bile salts (DBS).
PI gave satisfactory results for only some of the tested species (Table 1). Good results were obtained for the species S. salivarius subsp. thermophilus, Lactobacillus delbrueckii subsp. bulgaricus (Fig. 1F), Lactobacillus casei, and Lactobacillus mesenteroides. All cells of the 70°C heat-treated suspension were brightly labeled as observed with microscopy, and the labeled population gave a peak in the FCM histogram distinct from that of nonlabeled cells. Of the exponential-phase cells, only a small fraction was labeled. The FCM results agreed with microscopic observations. However, for the other species PI labeling did not give clear FCM results. In addition, the estimates of the living and dead fractions with microscopy did not agree with FCM results. For Lactococcus lactis and E. faecium (Fig. 1G) the labeling intensity of the heat-killed cells was too low, which caused overlap of the peak of labeled cells with the peak of nonlabeled cells in the FCM fluorescence histogram. On the other hand, untreated Lactobacillus helveticus (Fig. 1H), Leuconostoc lactis (Fig. 1I), and P. acidilacti cell suspensions showed PI labeling, although with low intensity. This low-intensity labeling resulted in peaks in the FCM fluorescence histogram distinct from that of nonlabeled cells and partly overlapping with the peak of 70°C heat-treated labeled cell suspensions.
TOTO-1 labeling enabled easy identification of the live and dead cells by FCM for most of the LAB species (Table 1). Dead cells were labeled with high-fluorescence intensity, while live cells were left unstained (Figs. 1J and K). The high fluorescence intensity of the dead cells resulted in peaks in the fluorescence histogram that were clearly resolved from the signal of nonlabeled cells. For Lactobacillus delbrueckii subsp. bulgaricus and Leuconostoc lactis there was some overlap between the FCM fluorescence histogram peaks of labeled and nonlabeled populations. In general, the TOTO-1 labeling gave clearer results than the PI labeling, and peaks of live and dead cells in the FCM fluorescence histograms were better resolved.
Because cFDA and TOTO-1 gave the best results and appeared to be useful for the different LAB species, these probes were chosen for further testing as stains for live and dead cells. The probes were tested separately and in combination using stressed cultures.
Viability assessment after treatment with bile salts or acid.
Three LAB species, Lactococcus lactis, Lactobacillus helveticus, and Leuconostoc mesenteroides, were selected for more detailed analysis. FCM was applied for viability assessment using cFDA and TOTO-1 after exposure to deconjugated bile salts or acid. The results of the FCM were compared with the survival tested by plate counts. For visual impressions of labeling after stress, the cell suspensions were also analyzed by fluorescence microscopy (Fig. 1L). The microscopic observations agreed with the FCM results.
In the FCM analyses the bacteria were identified by their light scatter. In the dot plot of the FSC and the SSC, a region that comprised the cell population was created. Interfering particles that also had an SSC above the threshold value but were not in the created region were thus disregarded. Both cF (λem, 517 nm) and TOTO-1 (λem, 533 nm) were best detected by the FL1 detector. Figures 2 and 3 display diagrams of the distribution of the cell population among 1,024 channels of fluorescence on a logarithmic scale. The height indicates the number of cells in a particular fluorescence channel.
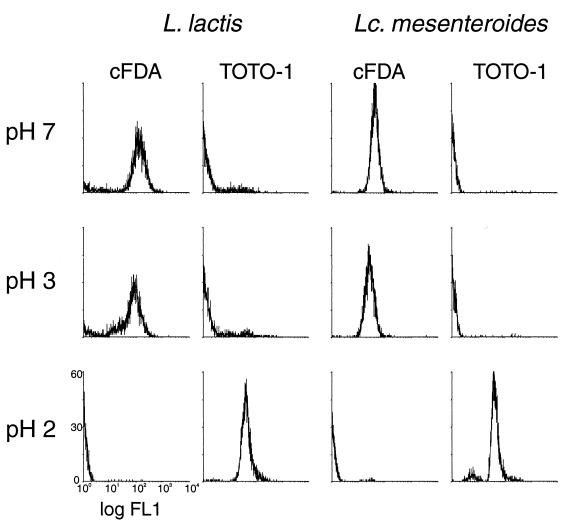
FIG. 3.
Flow cytometry histograms of FL1 of Lactococcus lactis and Leuconostoc mesenteroides cell suspensions stained with cFDA or with TOTO-1 after exposure to acid.
In cell suspensions incubated without bile salts, between 90 and 98% of the cells were labeled by cFDA. This varied between the experiments and between the species. The results of TOTO-1 labeling were complementary to the results of cF labeling: 2 to 10% of the cells were labeled. Exposure to 0.05% bile salts hardly changed the number of labeled cells. Also, the number of CFU was similar to that of nontreated cell suspensions. However, the average fluorescence of the cF-labeled population, especially that of Lactococcus lactis, was lower (Fig. 2). This suggests lower accumulation of cF caused by minor damage to the membrane. Exposure to higher bile salt concentrations resulted in lower fractions of cF-labeled cells and higher fractions of TOTO-1-labeled cells. Exposure to 0.10% bile salts resulted in heterogeneity in the populations. In Lactococcus lactis cell suspensions approximately 50% were labeled by cF. TOTO-1 staining resulted in overlapping peaks of labeled and nonlabeled cells (Fig. 2). The plate counts were at 42% compared to cell suspensions incubated without bile salts. In Leuconostoc mesenteroides cell suspensions both cFDA and TOTO-1 divided the population equally into labeled and nonlabeled subpopulations (Fig. 2). The plate counts were at 36%. In Lactobacillus helveticus cell suspensions cFDA labeled approximately 90% of the cells and TOTO-1 labeled 10% of the cells (not shown). The plate counts were at 89%. After exposure to 0.25% bile salts or more, almost no cells were labeled by cFDA. In agreement with this, almost all cells were labeled by TOTO-1. Correspondingly, the survival was low, 0.1% at maximum.
For cell suspensions exposed to acid the results of cF labeling and TOTO-1 labeling agreed with plate counts. The results for Lactococcus lactis, Lactobacillus helveticus, and Leuconostoc mesenteroides were similar. Labeling with cF or TOTO-1 after exposure to pH 6.0, 5.0, 4.0, or 3.0 did not result in a change of the distribution of the subpopulations compared to that at pH 7.0 (Fig. 3). However, after exposure to pH 2.0, almost no cells were labeled with cF whereas almost all cells were stained with TOTO-1 (Fig. 3). Accordingly, the culturability was not affected until pH 3.0, but exposure to pH 2.0 resulted in at least a 3-log-unit reduction of the plate counts.
Fluorescent labeling in combination with FCM revealed stress-induced heterogeneity in the cultures. Most importantly, live and dead subpopulations could be distinguished.
Double staining with cFDA and TOTO-1.
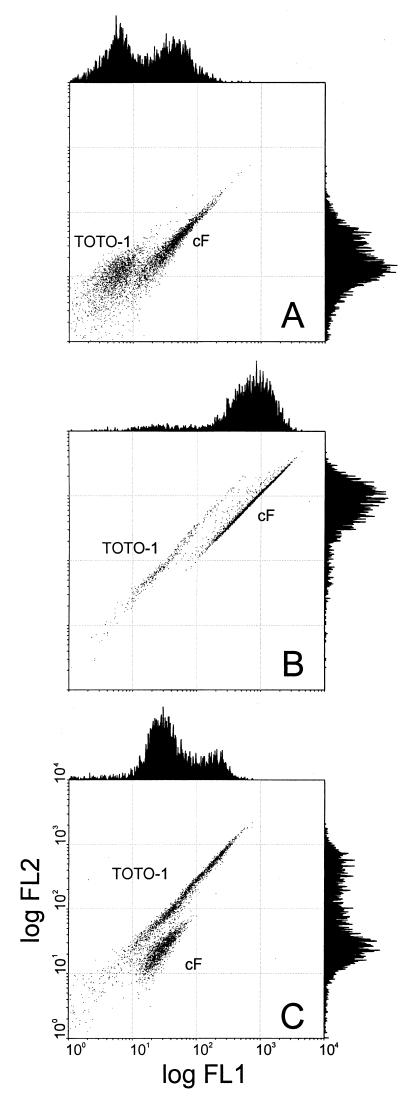
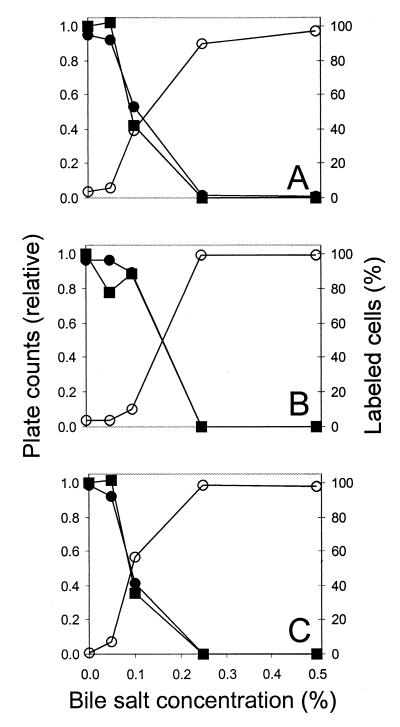
Live/dead assays with two differentially staining probes are attractive because detection is easier when all cells are labeled. Therefore, we tried double staining with cFDA and TOTO-1. Using FCM, the cF- and the TOTO-1-labeled populations could be spatially resolved in dot plots of FL1 and FL2, as illustrated by double-stained cultures that were stressed by exposure to 0.10% bile salts (Fig. 4). In the histograms of FL1 and FL2, there is considerable overlap between the peaks of cF-labeled and TOTO-1-labeled cells (Fig. 4). However, since the emission spectra of cF and TOTO-1 are different, the FL1/FL2 ratios are different. Therefore, the subpopulations are resolved in the dot plot of FL1 and FL2. For Lactococcus lactis exposed to 0.10% bile salts, the double labeling gave clear separation into two subpopulations (Fig. 4A), while single labeling with TOTO-1 did not give such clear results (Fig. 2). This illustrates the advantage of double staining. The cF-labeled cells and the TOTO-1-labeled cells were counted after double labeling by performing region analysis on FL1-FL2 dot plots. Figure 5 shows the results of these fluorescence counts in comparison with plate counts for cultures of Lactococcus lactis, Lactobacillus helveticus, and Leuconostoc mesenteroides that were exposed to bile salts. The results of the labeling are in agreement with the plate counts. For cultures exposed to low pH, the FCM counts also agreed with plate counts (data not shown).
FIG. 4.
Flow cytometry dot plots of FL1 and FL2 of Lactococcus lactis (A), Lactobacillus helveticus (B), and Leuconostoc mesenteroides (C) that were exposed to bile salts and stained with cFDA and TOTO-1. The cell suspensions were exposed to 0.10% deconjugated bile salts at 30°C for 60 min. After being washed, the cell suspensions were incubated with 50 μM cFDA and 1 μM TOTO-1. The cF-labeled and TOTO-1-labeled subpopulations are spatially resolved in the dot plots.
FIG. 5.
Comparison of cFDA and TOTO-1 labeling with plate counts for bile-stressed LAB. Lactococcus lactis (A), Lactobacillus helveticus (B), and Leuconostoc mesenteroides (C) were exposed to 0.05, 0.10, 0.25, or 0.50% bile salts at 30°C for 60 min or incubated without bile salts as a control. After washing, the culturability was tested by plate counts (■) and by FCM using cFDA staining (●) and TOTO-1 staining (○).
Cell sorting.
To establish a direct relationship between labeling and culturability, the labeled and nonlabeled populations were sorted and plated. Lactococcus lactis exponential-phase nonlabeled cell suspensions gave a number of colonies corresponding to approximately 80% of the number of cells actually sorted. Similarly, cFDA-stained nontreated cell suspensions sorted on cF labeling gave a number of colonies corresponding to approximately 80% of the number of cells recovered in the sorting tube. The somewhat lower plate counts may have been caused by stress imposed during cell sorting.
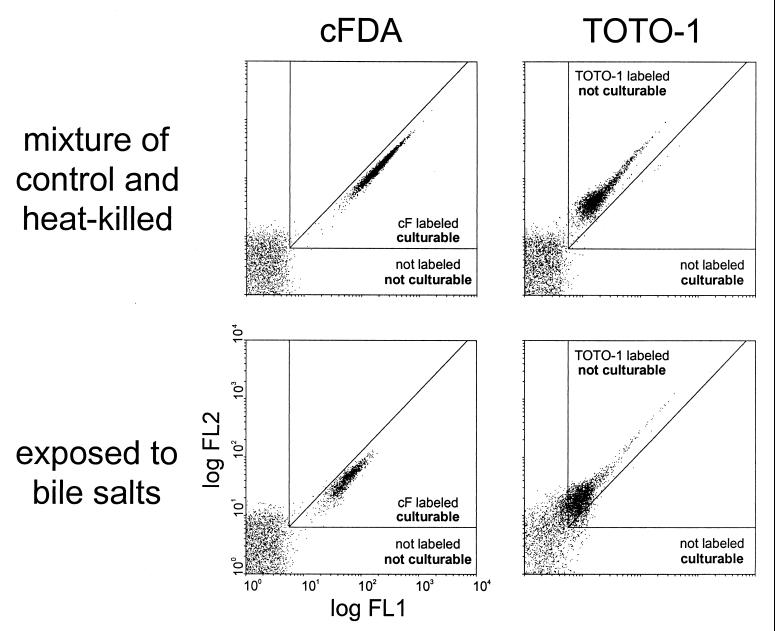
Standard regions for fluorescence-based sorting were then defined using mixtures of exponential-phase cells and 70°C heat-killed cells that were incubated either with cFDA or with TOTO-1 (Fig. 6). The labeled and nonlabeled subpopulations of the mixtures and of cell suspensions treated with 0.10% bile salts were then sorted and plated. When incubated with cFDA, the sorted cF-labeled subpopulation had high fractions of culturability, similar to that mentioned above. Accordingly, the nonlabeled subpopulation gave no colonies (Fig. 6). When incubated with TOTO-1, the labeled subpopulation was not culturable, whereas the nonlabeled subpopulation had a high culturability. The sorting experiments provided direct evidence that the FCM viability assay with cF and TOTO-1 indicates live and dead, i.e., culturable and nonculturable, subpopulations in stressed cultures.
FIG. 6.
Combined sorting and plating experiments with cFDA- and TOTO-1-stained Lactococcus lactis. Regions of nonlabeled cells, cF-labeled cells, and TOTO-1-labeled cells were defined using 1:1 mixtures of exponential-phase cells and 70°C heat-killed cells that were incubated with cFDA or TOTO-1. These mixtures and cell populations stressed with 0.10% bile salts were sorted using the defined regions as sort gates. The culturability of the sorted subpopulations was tested by plate counting.
DISCUSSION
We examined the usefulness of FCM for viability assessment of LAB. To be useful, the method has to be reliable and rapid. Furthermore, it has to be useful for LAB of different genera and food applications. This was taken into account in selecting species for this study (8, 26, 33). Three fluorescent probes were tested for their usefulness for live/dead discrimination: cFDA as a stain for live cells and PI and TOTO-1 as stains for dead cells. The probes were tested using exponential-phase cells as a positive control and 70°C heat-killed cells as a negative control. Flow cytometry results were compared with plate counts. Labeled cell suspensions were also visually inspected by fluorescence microscopy. The results showed that cFDA was successful in labeling the live cells and leaving the dead cells unstained. TOTO-1 appeared to be better than PI as a stain for dead cells. For double staining, cF and TOTO-1 were shown to be an excellent combination for a flow cytometric live/dead assay, as was supported by sorting experiments with Lactococcus lactis. In further experiments the assay was successfully applied to deconjugated bile salt-stressed cultures and to acid-stressed cultures of Lactococcus lactis, Lactobacillus helveticus, and Leuconostoc mesenteroides.
Exposure to hydrochloric acid is often used as an in vitro condition to investigate the resistance of bacteria to a passage through the stomach (10). Generally, the viability is not affected when LAB are incubated with hydrochloric acid at pH 3.5 or higher, while at lower pH the survival decreases to less than 1%, at pH values that are dependent on species and strain (7, 24, 29). We found no decrease of culturability when cells were exposed for 60 min at 30°C to hydrochloric acid solutions with pHs as low as 3.0. At pH 2.0 there were hardly any surviving cells. The results of the labeling indicate that at pH 3.0 the membrane stays intact, while after exposure to pH 2.0 the membrane is damaged. Further studies using acid solutions with pHs between 3.0 and 2.0 could elucidate at what concentration of acid the membrane becomes compromised and how that relates to culturability.
In addition to resistance to acid, resistance to bile is recognized as an important feature for LAB used as probiotics (14, 17, 33). In the human intestinal tract the concentration is variable, with a maximum of 2% (10). The conjugated bile salts that are excreted are deconjugated by intestinal microorganisms, which makes them less effective as a detergent, but the deconjugated bile salts do kill bacteria at concentrations of below 0.5% (10, 15). The results of our experiments on survival in buffer show that the concentration of 0.10% deconjugated bile salts falls within the critical range, but different survival fractions were found for the three species. At 0.25% almost no surviving cells were detected. Different strains of one species can also have different levels of tolerance to bile, as reported in a study of six Lactobacillus acidophilus strains (24). By selective bile pressure, variants of Lactobacillus acidophilus that have a higher resistance to bile salts and that may be considered candidates for probiotic strains could be obtained (7, 38). Our labeling experiments indicated that membrane integrity is crucial for bile resistance. The detection of damage to membranes is indicative of the culturability, as was shown by the agreement between labeling results and plate counts.
cFDA was tested as a live-cell stain for LAB. cFDA is an esterase substrate that needs enzyme activity to yield the fluorescent compound and membrane integrity to keep the compound in the cell. Labeled and nonlabeled cells were distinguished successfully by FCM. One standard protocol was used for all species, which resulted in different fluorescence intensities of the different species. This diversity in labeling capacity might be explained by differences in permeability affecting the diffusion of cFDA, differences in esterase activity, or differences in esterase specificity. Adjusting the protocol can in principle optimize cF labeling intensities. P. acidilacti had the lowest labeling intensity, which made examination by microscopy difficult with our standard labeling protocol. However, even this relatively low labeling intensity was sufficient for accurate FCM analysis. Besides differences in labeling intensity, the possibility of cF efflux is a point to keep in mind (5, 31). To prevent cF efflux, the experiments were performed with washed cells and without fermentable sugars in the buffer. Under these conditions, no significant loss of cF from the cells occurred, as was checked by spectrofluorimetry. The retention of cF under nonenergizing conditions enables accurate FCM assays for all species. The labeling with cF gave clear discrimination between live and dead cells, as was confirmed by the sorting experiments.
PI and TOTO-1 were tested as counterstains for cFDA in the FCM viability assay. PI is a red fluorescent phenanthridinium intercalating dye used extensively for detecting dead cells (5, 18, 28, 29, 32). TOTO-1 is a yellow fluorescent dimeric cyanine dye. These dyes have the necessary properties for a dye exclusion probe but are hardly used as such (2, 18, 32, 40). In our experiments TOTO-1 proved to be superior to PI in discriminating intact and damaged cells. This may be because TOTO-1 is larger than PI; the molecular masses are 1,303 and 668 g per mol, respectively. Also, a lower partitioning into the membrane could be a factor in favor of TOTO-1. Furthermore, the very high fluorescence enhancement of TOTO-1 enables good distinction of nonlabeled and labeled cells in the FCM. The labeling with TOTO-1 gave clear discrimination between live and dead cells, as was confirmed by sorting experiments.
Live/dead assays based on two probes have advantages over assays with one probe that labels either live or dead cells. In FCM, the identification of cells is facilitated when all cells are fluorescently labeled, especially when the background is high. In addition, total enumeration can be done together with viability assessment in the same assay when all of the cells are detectable. There are several probe kits from Molecular Probes developed for such assays (18). The live/dead viability/ cytotoxicity kit for animal cells is based on two probes discriminating between live and dead cells: calcein AM and ethidium homodimer-1. Unfortunately, this probe combination appeared not to be generally suitable for use with bacterial cells (18, 21). The live/dead BacLight viability kit for bacteria is also based on a combination of two probes, but only one of the probes, PI, discriminates between intact and damaged cells. SYTO 9 is a green fluorescent permeant nucleic acid stain that is included in the assay to have all of the cells labeled. PI is supposed to enter cells with compromised membranes only. PI displaces SYTO from the DNA because PI has a higher affinity for DNA. Both probes are excited by the blue laser used in many flow cytometers. The ViaGram Red+ bacterial Gram stain and viability kit combines Gram staining using Texas Red-X wheat germ agglutinin with a viability assay using the permeant DNA stain DAPI (4′,6′-diamidino-2-phenylindole) and the dead-cell stain SYTOX-Green. The combination of DAPI and SYTOX-Green acts on the same principles as the BacLight probe combination. UV light is needed for excitation, which makes it unsuitable for a flow cytometer equipped with only a blue laser. The assay developed in this study combines two probes that individually discriminate between live and dead cells. cFDA acts as a stain for live cells because it needs hydrolysis by intracellular esterases and retention by an intact membrane. TOTO-1 acts as stain for dead cells because it is a nucleic acid binding probe excluded by cells with intact membranes. Thus, this assay acts on the same principle as the successful live/dead viability kit for animal cells. Instead of using a counterstain only to have all of the cells labeled, both probes provide information on the cell status. Furthermore, this assay employs TOTO-1, which proved to be better than PI in our experiments. The green fluorescent cF-labeled cells and the yellow fluorescent TOTO-1-labeled cells are difficult to distinguish with microscopy; however, singly labeled cell samples can be used when a visual impression is required. In conclusion, the combination of cFDA and TOTO-1 makes a reliable live/dead assay for FCM assessment of bacterial viability.
This live/dead assay has many possible applications. In this study the application of viability assessment after exposure to bile or acid was validated. Likewise, the assay can be used for screening LAB that are possibly probiotic for tolerance against bile and acid under various conditions. FCM analyses are fast, and one standard labeling protocol appeared to be workable for all species in our selection, which makes the assay attractive for such studies. Furthermore, the viability of starters can be examined. Survival of starters after freeze-drying and after storage is of interest in dairy production (6). For the evaluation of different conditions FCM can be of use. The developed live/dead assay can also be applied as a fast screening method for assessment of susceptibility of bacteria to a wide range of antimicrobial compounds, including antibiotics. In summary, the probes cFDA and TOTO-1 make an excellent combination for bacterial live/dead assays by FCM with versatile applications.
ACKNOWLEDGMENTS
This work was financially supported by The Netherlands Technology Foundation (STW).
We thank Jeroen Hugenholtz from the NIZO Food Research Institute for useful discussion and provision of bacterial strains. We thank Boudewijn van Veen for technical assistance.
REFERENCES
- 1.Barer M R, Harwood C R. Bacterial viability and culturability. Adv Microb Physiol. 1999;41:93–137. doi: 10.1016/s0065-2911(08)60166-6. [DOI] [PubMed] [Google Scholar]
- 2.Becker B, Clapper J, Harkins K R, Olson J A. In situ screening assay for cell viability using a dimeric cyanine nucleic acid stain. Anal Biochem. 1994;221:78–84. doi: 10.1006/abio.1994.1382. [DOI] [PubMed] [Google Scholar]
- 3.Breeuwer P, Abee T. Assessment of viability of microorganisms employing fluorescence techniques. Int J Food Microbiol. 2000;55:193–200. doi: 10.1016/s0168-1605(00)00163-x. [DOI] [PubMed] [Google Scholar]
- 4.Bunthof C J, van den Braak S, Breeuwer P, Rombouts F M, Abee T. Fluorescence assessment of Lactococcus lactis viability. Int J Food Microbiol. 2000;55:291–294. doi: 10.1016/s0168-1605(00)00170-7. [DOI] [PubMed] [Google Scholar]
- 5.Bunthof C J, van den Braak S, Breeuwer P, Rombouts F M, Abee T. Rapid fluorescence assessment of the viability of stressed Lactococcus lactis. Appl Environ Microbiol. 1999;65:3681–3689. doi: 10.1128/aem.65.8.3681-3689.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Champagne C P, Mondou F, Raymond Y, Roy D. Effect of polymers and storage temperature on the stability of freeze-dried lactic acid bacteria. Food Res Int. 1996;29:555–562. [Google Scholar]
- 7.Chou L S, Weimer B. Isolation and characterization of acid- and bile-tolerant isolates from strains of Lactobacillus acidophilus. J Dairy Sci. 1999;82:23–31. doi: 10.3168/jds.S0022-0302(99)75204-5. [DOI] [PubMed] [Google Scholar]
- 8.Cogan T M. History and taxonomy of starter cultures. In: Cogan T M, Accolas J-P, editors. Dairy starter cultures. New York, N.Y: VCH Publishers, Inc.; 1996. pp. 1–24. [Google Scholar]
- 9.Crow V L, Coolbear T, Holland R, Pritchard G G, Martley F G. Starters as finishers: starter properties relevant to cheese ripening. Int Dairy J. 1993;3:423–460. [Google Scholar]
- 10.Davenport H W. Physiology of the digestive tract: an introductory text. 3rd ed. Chicago, Ill: Year Book Medical Publishers Inc.; 1971. [Google Scholar]
- 11.Davey H M, Kell D B. Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analyses. Microbiol Rev. 1996;60:641–696. doi: 10.1128/mr.60.4.641-696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaper J P, Edwards C. The use of fluorogenic esters to detect viable bacteria by flow cytometry. J Appl Bacteriol. 1994;77:221–228. [Google Scholar]
- 13.Drouault S, Corthier G, Ehrlich S D, Renault P. Survival, physiology, and lysis of Lactococcus lactis in the digestive tract. Appl Environ Microbiol. 1999;65:4881–4886. doi: 10.1128/aem.65.11.4881-4886.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunne C, Murphy L, Flynn S, O'Mahony L, O'Halloran S, Feeney M, Morrissey D, Thornton G, Fitzgerald G, Daly C, Kiely B, Quigley E M, O'Sullivan G C, Shanahan F, Collins J K. Probiotics: from myth to reality. Demonstration of functionality in animal models of disease and in human clinical trials. Antonie Leeuwenhoek. 1999;76:279–292. [PubMed] [Google Scholar]
- 15.Floch M H, Binder H J, Filburn B, Gershengoren W. The effect of bile acids on intestinal microflora. Am J Clin Nutr. 1972;25:1418–1426. doi: 10.1093/ajcn/25.12.1418. [DOI] [PubMed] [Google Scholar]
- 16.Fouchet P, Jayat C, Hechard Y, Ratinaud M H, Frelat G. Recent advances of flow cytometry in fundamental and applied microbiology. Biol Cell. 1993;78:95–109. doi: 10.1016/0248-4900(93)90120-4. [DOI] [PubMed] [Google Scholar]
- 17.Guarner F, Schaafsma G J. Probiotics. Int J Food Microbiol. 1998;39:237–238. doi: 10.1016/s0168-1605(97)00136-0. [DOI] [PubMed] [Google Scholar]
- 18.Haugland P. Molecular probes—handbook of fluorescent probes and research chemicals. 7th ed. 1999. www.probes.com [Online.] Molecular Probes, Inc., Eugene, Oreg. CD-ROM and www.probes.com. . [Google Scholar]
- 19.Hirons G T, Fawcett J J, Crissman H A. TOTO and YOYO: new very bright fluorochromes for DNA content analyses by flow cytometry. Cytometry. 1994;15:129–140. doi: 10.1002/cyto.990150206. [DOI] [PubMed] [Google Scholar]
- 20.Huis in 't Veld J, Shortt C. Selection criteria for probiotic micro-organisms. In: Leeds A R, Rowland I R, editors. Gut flora and health—past, present and future. London, United Kingdom: The Royal Society of Medicine Press; 1996. pp. 27–36. [Google Scholar]
- 21.Kaneshiro E S, Wyder M A, Wu Y P, Cushion M T. Reliability of calcein acetoxy methyl ester and ethidium homodimer or propidium iodide for viability assessment of microbes. J Microbiol Methods. 1993;17:1–16. [Google Scholar]
- 22.Kell D B, Kaprelyants A S, Weichart D H, Harwood C R, Barer M R. Viability and activity in readily culturable bacteria: a review and discussion of the practical issues. Antonie Leeuwenhoek. 1998;73:169–187. doi: 10.1023/a:1000664013047. [DOI] [PubMed] [Google Scholar]
- 23.Krylov S N, Zhang Z R, Chan N W C, Arriaga E, Palcic M M, Dovichi N J. Correlating cell cycle with metabolism in single cells: combination of image and metabolic cytometry. Cytometry. 1999;37:14–20. doi: 10.1002/(sici)1097-0320(19990901)37:1<14::aid-cyto2>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 24.Lankaputhra W E V, Shah N P. Survival of Lactobacillus acidophilus and Bifidobacterium spp. in the presence of acid and bile salts. Cultured Dairy Products J. 1995;30:2–7. [Google Scholar]
- 25.Lebaron P, Catala P, Parthuisot N. Effectiveness of SYTOX green stain for bacterial viability assessment. Appl Environ Microbiol. 1998;64:2697–2700. doi: 10.1128/aem.64.7.2697-2700.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Limsowtin G K Y, Powell I B. Types of starters. In: Cogan T M, Accolas J-P, editors. Dairy starter cultures. New York, N.Y: VCH Publishers, Inc.; 1996. pp. 101–130. [Google Scholar]
- 27.Lloyd D, editor. Flow cytometry in microbiology. London, United Kingdom: Springer-Verlag; 1993. [Google Scholar]
- 28.López-Amorós R, Comas J, Vives-Rego J. Flow cytometric assessment of Escherichia coli and Salmonella typhimurium starvation-survival in seawater using rhodamine 123, propidium iodide, and oxonol. Appl Environ Microbiol. 1995;61:2521–2526. doi: 10.1128/aem.61.7.2521-2526.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marteau P, Minekus M, Havenaar R, Huis in 't Veld J H J. Survival of lactic acid bacteria in a dynamic model of the stomach and small intestine: validation and the effects of bile. J Dairy Sci. 1997;80:1031–1037. doi: 10.3168/jds.S0022-0302(97)76027-2. [DOI] [PubMed] [Google Scholar]
- 30.McFeters G A, Yu F P, Pyle B H, Stewart P S. Physiological assessment of bacteria using fluorochromes. J Microbiol Methods. 1995;21:1–13. doi: 10.1016/0167-7012(94)00027-5. [DOI] [PubMed] [Google Scholar]
- 31.Molenaar D, Bolhuis H, Abee T, Poolman B, Konings W N. The efflux of a fluorescent probe is catalyzed by an ATP-driven extrusion system in Lactococcus lactis. J Bacteriol. 1992;174:3118–3124. doi: 10.1128/jb.174.10.3118-3124.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mortimer F C, Mason D J, Gant V A. Flow cytometric monitoring of antibiotic-induced injury in Escherichia coli using cell-impermeant fluorescent probes. Antimicrob Agents Chemother. 2000;44:676–681. doi: 10.1128/aac.44.3.676-681.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Sullivan M G, Thornton G, O'Sullivan G C, Collins J K. Probiotic bacteria: myth or reality? Trends Food Sci Technol. 1992;3:309–314. [Google Scholar]
- 34.Porter J, Deere D, Hardman M, Edwards C, Pickup R. Go with the flow—use of flow cytometry in environmental microbiology. FEMS Microbiol Ecol. 1997;24:93–101. [Google Scholar]
- 35.Porter J, Deere D, Pickup R, Edwards C. Fluorescent probes and flow cytometry: new insights into environmental bacteriology. Cytometry. 1996;23:91–96. doi: 10.1002/(SICI)1097-0320(19960201)23:2<91::AID-CYTO1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 36.Roth B L, Poot M, Yue S T, Millard P J. Bacterial viability and antibiotic susceptibility testing with SYTOX Green nucleic acid stain. Appl Environ Microbiol. 1997;63:2421–2431. doi: 10.1128/aem.63.6.2421-2431.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shapiro H M. Practical flow cytometry. 3rd ed. New York, N.Y: Wiley-Liss, Inc.; 1995. [Google Scholar]
- 38.Suskovic J, Brkic B, Matosic S, Maric V. Lactobacillus acidophilus M92 as potential probiotic strain. Milchwissenschaft. 1997;52:430–435. [Google Scholar]
- 39.Svenssson U. Industrial perspectives. In: Tannock G W, editor. Probiotics. A critical review. Wymondham, United Kingdom: Horizon Scientific Press; 1999. pp. 57–64. [Google Scholar]
- 40.Votyakova T V, Kaprelyants A S, Kell D B. Influence of viable cells on the resuscitation of dormant cells in Micrococcus luteus cultures held in an extended stationary phase: the population effect. Appl Environ Microbiol. 1994;60:3284–3291. doi: 10.1128/aem.60.9.3284-3291.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]