Abstract
Dysfunction of mRNA or RNA-binding proteins (RBPs) causes cellular aging and age-related degenerative diseases; however, information regarding the mechanism through which RBP-mediated posttranscriptional regulation affects cellular aging and related disease processes is limited. In this study, PUM1 was found to be associated with the self-renewal capacity and aging process of human mesenchymal stem cells (MSC). PUM1 interacted with the 3’-untranslated region of Toll-like receptor 4 (TLR4) to suppress TLR4 mRNA translation and regulate the activity of nuclear factor-κB (NF-κB), a master regulator of the aging process in MSCs. PUM1 overexpression protected MSCs against H2O2-induced cellular senescence by suppressing TLR4-mediated NF-κB activity. TLR4-mediated NF-κB activation is a key regulator in osteoarthritis (OA) pathogenesis. PUM1 overexpression enhanced the chondrogenic potential of MSCs even under the influence of inflammation-inducing factors, such as lipopolysaccharide (LPS) or interleukin-1β (IL-1β), whereas the chondrogenic potential was reduced following the PUM1 knockdown-mediated TLR4 activation. PUM1 levels decreased under inflammatory conditions in vitro and during OA progression in human and mouse disease models. PUM1 knockdown in human chondrocytes promoted chondrogenic phenotype loss, whereas PUM1 overexpression protected the cells from inflammation-mediated disruption of the chondrogenic phenotype. Gene therapy using a lentiviral vector encoding mouse PUM1 showed promise in preserving articular cartilage integrity in OA mouse models. In conclusion, PUM1 is a novel suppressor of MSC aging, and the PUM1-TLR4 regulatory axis represents a potential therapeutic target for OA.
Subject terms: Cell biology, Immunological disorders
Introduction
Mesenchymal stem cells (MSCs) have potential as a novel therapeutic option for regenerating damaged tissue. Due to their self-renewal and multipotent properties, MSCs may be used to treat several pathologies, especially skeletal diseases such as osteoarthritis (OA) and osteoporosis [1–3]. Promising outcomes with MSC use in musculoskeletal diseases, including those of bone and cartilage, have been demonstrated [4–6]. However, subculture for in vitro MSC expansion is associated with cellular senescence and self-renewal, and the long-term cultivation of MSCs in vitro decreases their multi-differentiation potential [7, 8], causing a loss of therapeutic potential. MSCs intrinsically show an age-associated decline in number and function [9, 10], leading to the loss of tissue homeostasis and the development of degenerative diseases [11, 12]. Cellular aging impairs the regenerative potential of MSCs [13], and the associated biological mechanisms are related to cartilage degeneration [14, 15]. Thus, at the molecular level, aging-associated changes are closely linked with the development of OA [16]. It has been shown that chondrocyte density in cartilage tissues decreases with age, and cellular defense mechanisms are impaired in aged chondrocytes [17]. During the aging process, the ability of chondrocytes to produce extracellular matrix proteins is weakened [18]. Therefore, understanding the aging mechanisms of MSCs and chondrocytes is pivotal to identifying novel therapeutic targets for age-related diseases such as OA.
Dysregulation of mRNA or RNA-binding proteins (RBPs) is related to cellular aging and age-related degenerative diseases [19, 20]; however, little is known about how RBP-mediated posttranscriptional regulation affects these processes. PUMILIO, an RBP, and translational repressor, directly binds to its target mRNAs in a sequence-specific manner. Its homology domain (HD) has eight imperfect tandem repeats in the C-terminus that create a repeating curved structure, which recognizes one RNA base within the conserved 8-nucleotide (UGUA_AUA) Pumilio/FBF (fem-3 mRNA binding factor) response element (PRE) in the 3’-untranslated region (UTR) of the target mRNAs [21, 22]. PUMILIO proteins are highly conserved evolutionarily [23], but little is known about their function in mammals. The human genome encodes two PUMILIO proteins, PUMILIO1 (PUM1) and PUMILIO2 (PUM2), with similar RNA-binding specificity. In mice, embryonic stem cells (ESCs) with Pum2 mutation do not have impaired self-renewal, and differentiation capacities [24], and Pum2 mutant mice are viable and fertile, although they have smaller testes [25]. PUM1 has been implicated in neurodegeneration [26] and genomic stability regulation [27]. In one study, null mutations in Pum1 or Pum2 were associated with dose-dependent decreases in body and organ size [28], and deleting both genes caused embryonic lethality [29], implying that mammalian PUM1 and PUM2 may relate to developmental processes. However, the function, targets, and regulatory mechanisms of PUM1 and PUM2 in human MSCs and chondrocytes remain unclear.
Here, we aimed to identify the role of the PUM1 protein in cellular senescence and OA progression by investigating its interaction with the 3ʹ-UTR of Toll-like receptor 4 (TLR4) and effects on the activity of nuclear factor-κB (NF-κB) in MSCs. Our findings can help us assess the self-renewal capacity and aging process of human MSCs and allow us to identify potential therapeutic targets for OA and prevent cellular aging.
Results
PUM1 is associated with cellular senescence in MSCs
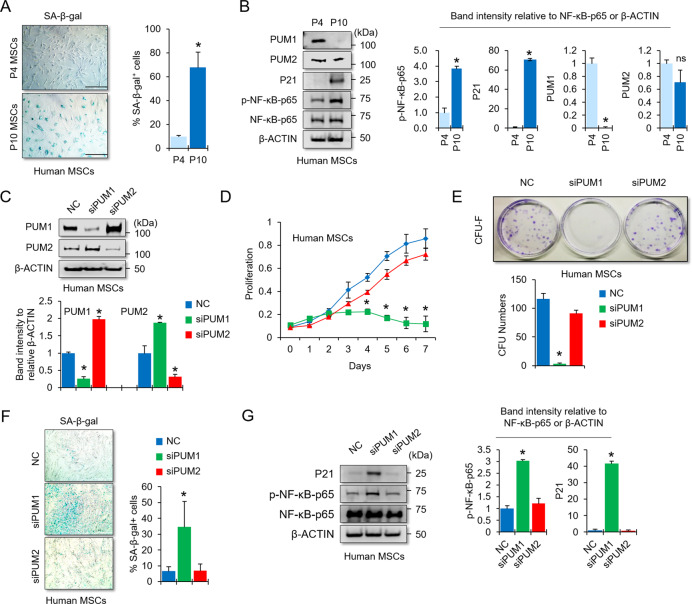
Senescence-associated (SA)-β-galactosidase activity was used to evaluate cellular senescence in early versus late passage MSCs. We found an increase in SA-β-galactosidase activity in late passage human MSCs (SA-β-gal; Fig. 1A) and subsequently performed a western blot analysis to observe whether protein levels of PUM1 and PUM2 changed during cellular senescence in human MSCs. Phosphorylated NF-κB, the master regulator of the senescence-associated secretory phenotype (SASP) transcriptional program, was used as an aging marker [30]. The NF-κB target P21 is a widely used aging marker in MSC biology [31, 32]. Our results showed that PUM1 was undetectable in late passage MSCs, whereas PUM2 was slightly decreased (Fig. 1B). siRNA-mediated knockdown (KD) experiments were performed to investigate whether the reduction of PUM1 and PUM2 in late passage MSCs was involved in the process of cellular senescence. Western blot results showed that the respective proteins in the PUM1 or PUM2 siRNA-transfected groups were decreased compared to those in the negative control siRNA-transfected group (Fig. 1C). The protein level of PUM1 was increased in the PUM2-KD group while the protein level of PUM2 was increased in the PUM1-KD group, indicating that PUM1 and PUM2 repressed each other. It is well known that PUM1 and PUM2 affect the expression of each other [33, 34] because of the presence of PREs in the 3’-UTR of PUM1 and PUM2 mRNAs [35]. PUM1-KD or PUM2-KD did not affect the CD105 and CD90 cell surface markers for MSCs (Fig. S1); however, PUM1 single KD significantly decreased MSC proliferation and self-renewal capacity (Fig. 1D and E). Given that MSCs transfected with PUM1 siRNA did not proliferate until day 7, the time point to assess the extent of cellular senescence, the cells were initially plated at 100% confluence on the culture dishes to compare SA-β-gal activity at the same density between groups tested. Additionally, cell plating densities with 100% confluence can cause cellular senescence and low proliferation rates in MSCs [7, 8, 36]. PUM1-KD MSCs showed increased SA-β-gal activity (Fig. 1F) and p-NF-κB and P21 protein levels (Fig. 1G). Next, we performed an additional experiment using the H2O2-induced premature senescence model (Fig. S2A) because it has been reported that H2O2 induces premature MSC senescence within one week [37, 38]. H2O2 treatment increased SA-β-gal activity of MSCs within four days and slightly exacerbated the cellular senescence of PUM1-KD MSCs although the cells were already senescent without H2O2 (Fig. S2B); Therefore, these data suggest that the reduction of PUM1 expression may be involved in the progression of MSC senescence.
Fig. 1. KD of PUM1 induces cellular senescence in mesenchymal stem cells.
A Early (passage 4) or late (passage 10) passage MSCs were stained with SA-β-gal (blue), and the intensity of SA-β-gal-positive staining in the cell population showed an increase in late passage MSCs. Scale bar = 350 μm. B Protein levels of PUM1, PUM2, and cellular senescence-related markers were analyzed in early and late passage MSCs using western blotting. C Protein levels of PUM1 and PUM2 were analyzed using western blotting in MSCs transfected with siRNA for negative control (NC), PUM1, or PUM2. D The cell proliferation assay was used to determine the proliferative capacities of negative control, PUM1, and PUM2 siRNA-transfected MSCs using the EZ-Cytox Kit. Each experiment was performed in triplicate (n = 3). *P < 0.05 in a comparison with NC siRNA-transfected cells. E The number of colony-forming cells was counted, and the average size of the colonies was measured in triplicate by three independent observers (n = 3). *P < 0.05 in a comparison with NC siRNA-transfected MSCs. F SA-β-gal staining was used to determine the extent of cellular senescence in the NC, PUM1, and PUM2 siRNA-treated MSCs. The SA-β-gal assay was performed 1 week after the MSCs were seeded onto a six-well plate. The SA-β-gal positive cells were quantified using ImageJ (n = 3, in triplicate). *P < 0.05 in a comparison with NC siRNA-transfected MSCs. Scale bar = 350 μm. G Protein levels of the senescence-related markers were analyzed in MSCs using western blotting in cells transfected with NC, PUM1, or PUM2 siRNAs.
PUM1 targets TLR4 in MSCs
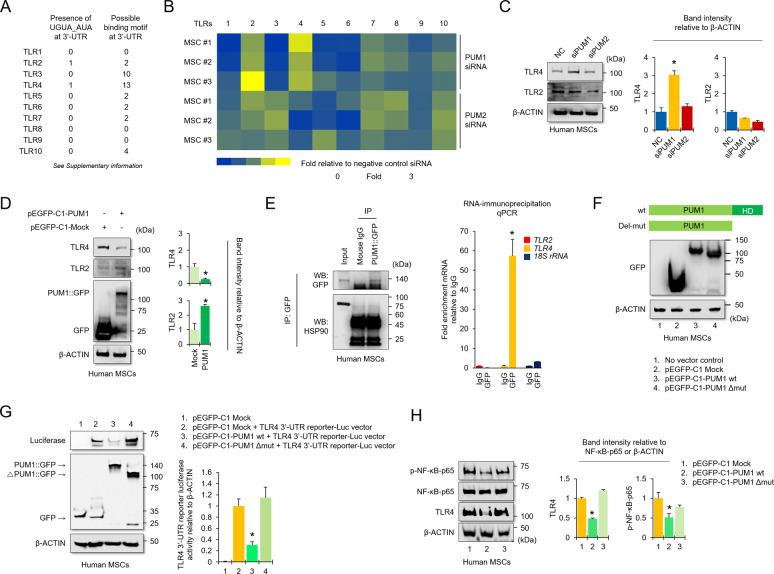
NF-κB induces the SASP transcriptional program in aged cells [30, 39]. The NF-κB pathway is evolutionarily conserved and can be triggered by diverse signals related to aging, cellular senescence, and immune activation [39]. NF-κB signaling activation links to several biological receptor pathways, such as Toll-like receptors (TLRs) and inflammatory signals [40–43]. Bohn et al. identified that human PUM1 and PUM2 target RNAs in HEK293 cells [44]. Using their RNA-seq analysis, we found that the TLR2, TLR3, TLR4, TLR7, and TLR9 mRNAs were partially stabilized by PUM1 and PUM2 KD [44], and the 3’-UTRs of TLR2 and TLR4 mRNAs have PUM binding elements (PBEs) (Fig. 2A and Table S1). To re-confirm changes to mRNA expression of TLRs by PUM1 or PUM2-KD in human MSCs, we performed quantitative PCR analysis using human MSCs from three donors. TLR2 and TLR4 mRNAs were increased in MSCs with PUM1-KD (Fig. 2B). Additionally, PUM1-KD upregulated the protein levels of TLR4 in MSCs, but TLR2 was not changed (Fig. 2C). PUM1 overexpression downregulated the protein levels of TLR4 whereas TLR2 was slightly upregulated (Fig. 2D). Therefore, we hypothesized that PUM1 might target TLR4 mRNA because the amount of TLR4 protein was reversely changed by modulating PUM1 expression whereas the change in the TLR2 protein level did not show a relevant difference. We next examined PUM1 binding to the TLR4 or TLR2 mRNAs via RNA-immunoprecipitation (RIP). TLR4 transcripts were significantly enriched in PUM1 RIP from human MSCs, indicating direct physical association whereas TLR2 transcripts were not enriched in the PUM1 RIP (Fig. 2E). We identified one PBE (UGUA_AUA) with an exact matching sequence in the human TLR4 3’-UTR and several PBEs with putative binding motifs (Fig. 2A and Table S1). To investigate whether a cause-and-effect relationship existed between the increase in PUM1 expression and decreased TLR4 mRNA, we prepared MSCs overexpressing pEGFP-C1-tagged PUM1 or a pEGFP-C1-tagged PUM1 mutant lacking the RNA-binding domain (ΔPUM1-HD) (Fig. 2F). The vectors were transfected to lentivirus-infected HEK 293 T cells containing the TLR4 3’-UTR downstream of a CMV-driven reporter (Luciferase). PUM1 binding to the TLR4 3’-UTR should repress luciferase expression, and we identified that the overexpression of PUM1, and not ΔPUM1-HD, significantly repressed luciferase activity (Fig. 2G). TLR4 and the TLR4 downstream target, p-NF-κB levels were not reduced by ΔPUM1-HD (Fig. 2H).
Fig. 2. PUM1 interacts with the TLR4 mRNA and downregulates NF-κB activity in MSCs.
A List of 3ʹ-UTRs for human Toll-like receptors (TLRs). Left panel indicates PBEs with the exact sequences (UGUA_AUA), whereas right panel means PBEs with possible binding motifs. (Source: https://genome.ucsc.edu/). B A heatmap was generated for real-time quantitative PCR data using RNA extracted from MSCs transfected with a single siRNA targeting PUM1 or PUM2. The results were compared to the group transfected with negative control (NC) siRNAs (n = 3 experimental replicates). C Protein levels for TLR2 and TLR4 were analyzed in MSCs using western blotting for cells transfected with NC, PUM1, or PUM2 siRNA. D Protein levels for PUM1::GFP, TLR2, and TLR4 were analyzed in MSCs using western blotting for cells transfected with the pEGFP-C1 vector control having no insert or the pEGFP-C1-PUM1 vector. E Blot data showed that PUM1::GFP-Immunoprecipitation was successful, and HSP90 was used as a loading control for the input group. The bar graph displays the RIP-qPCR fold enrichment of the indicated mRNAs relative to the IgG control. RIP was performed in MSCs transfected with the pEGFP-C1-PUM1 vector using electroporation with the Neon transfection system. *P < 0.05 in a comparison with the IgG control (n = 3 experimental replicates). F The protein level for GFP was analyzed in MSCs using western blotting in cells that were transfected with the pEGFP-C1 vector control with no insert, the pEGFP-C1-PUM1 vector, or pEGFP-C1-PUM1 mutant vector lacking the RNA-binding domain (ΔPUM1-HD). G Protein levels of GFP and luciferase were analyzed in HEK293T cells transduced with the TLR4 3’-UTR reporter-Luc vector. The cells were selected using puromycin (10 µg/mL) and transfected with the pEGFP-C1 vector control with no insert, the pEGFP-C1-PUM1 vector, or the pEGFP-C1-ΔPUM1-HD vector, and the cells were analyzed by western blot analysis to compare the luciferase activities between the groups. *P < 0.05 compared with the pEGFP-C1 vector control with no insert (n = 3 experimental replicates). H Protein levels for TLR4, P65, and p-P65 were analyzed in MSCs using western blotting in cells transfected with pEGFP-C1 vector control with no insert, the pEGFP-C1-PUM1 vector, or pEGFP-C1-ΔPUM1-HD.
PUM1 overexpression protects MSCs from H2O2-induced cellular senescence
An H2O2-induced premature cellular senescence model was employed. This treatment induced senescence in MSCs by increasing the SA-β-gal-positive cell population (%) and its activity (Fig. 3A–C). TLR4, NF-κB, and P21 were increased in H2O2-treated MSCs relative to the controls, whereas PUM1 was reduced (Fig. 3D). Additionally, 5-ethynyl-2´-deoxyuridine (EdU) staining was performed to evaluate the proliferation of H2O2-treated MSCs. We confirmed that H2O2 treatment considerably decreased the EdU-positive cell population (Fig. 3E). PUM1 overexpression or siRNA-mediated TLR4 KD in MSCs significantly reduced the SA-β-gal-positive cell population (%) and its activity against H2O2 treatment (Fig. 3F–H). H2O2 treatment did not significantly upregulate protein levels of pNF-κB and P21 in MSCs transfected with the pcDNA3.1-PUM1 vector or TLR4 siRNA (Fig. 3I). Correspondingly, the H2O2-induced reduction in the proliferation and colony-forming ability of MSCs was rescued by PUM1 overexpression or TLR4 KD (Fig. 3J and K). To clarify whether MSC senescence induced by PUM1-mediated TLR4 regulation depended on NF-κB, phorbol myristate acetate (PMA, potential NF-κB activator), an activator of protein kinase C (PKC) [45], was used to induce NF-κB activity in PUM1-overexpressing MSCs. PMA treatment abolished the decrease in SA-β-gal-positive cell populations and activity in H2O2-treated MSCs overexpressing PUM1 (Fig. S3A and B). Protein levels of pNF-κB and P21 were recovered by PMA treatment in H2O2-treated MSCs overexpressing PUM1 (Fig. S3C). Correspondingly, PMA abolished the PUM1-mediated rescue of the colony-forming capacity in H2O2-treated MSCs, despite the presence of PUM1 and TLR4 (Fig. S3D).
Fig. 3. PUM1 overexpression alleviates H2O2-induced cellular senescence.
A Representative SA-β-gal (green fluorescence) staining images of MSCs indicate senescent cells. B The bar graph indicates the percentage of SA-β-gal positive cells. The images were photographed and counted by the Countess II FL Automated Cell Counter. *P < 0.05 compared to the no H2O2 group (n = 3 experimental replicates). C The bar graph indicates the fluorescence-based activity of SA-β-gal. The fluorescence was measured at 360 nm/465 nm (excitation/emission). *P < 0.05 compared to the no H2O2 group (n = 3 experimental replicates). D Protein levels of PUM1, TLR4, and the senescence-related markers were analyzed in MSCs treated or untreated with H2O2 (200 μM) using western blotting. E Representative EdU (green) staining images of MSCs indicate a decrease in cell proliferation by H2O2 treatment. DAPI (blue) staining was performed to visualize all nuclei of the cells tested. The bar graph shows quantification (%) of the EdU-positive cell population. F MSCs were transfected with negative control siRNA, pcDNA control with no insert, pcDNA-PUM1, or siRNA targeting TLR4. The cells were subsequently cultured for 4 days with or without H2O2 (200 μM). Representative SA-β-gal (green fluorescence) staining images of MSCs indicate senescent cells. G The bar graph indicates the percentage of SA-β-gal positive cells. *P < 0.05 compared with the no H2O2 group (n = 3 experimental replicates). H The bar graph indicates the fluorescence-based activity of SA-β-gal. *P < 0.05 compared with the no H2O2 group (n = 3 experimental replicates). (I) Protein levels of PUM1, TLR4, and the senescence-related markers were analyzed in MSCs treated or untreated with H2O2 (200 μM) using western blotting. J Representative EdU (green) staining images of MSCs indicate a change in cell proliferation in the H2O2-treated MSCs. DAPI (blue) staining was performed to visualize all nuclei of the cells tested. The bar graph shows quantification (%) of the EdU-positive cell population. K The vector-transfected MSCs (1 × 103 cells per 100 mm dish) were maintained for 12 days with or without H2O2 in a 20% FBS-containing medium. The colony-forming abilities of the cells were analyzed via crystal violet (CV) staining. The bar graph indicates the colony numbers of the MSCs tested.
PUM1 preserves MSC chondrogenic potential under inflammatory microenvironment
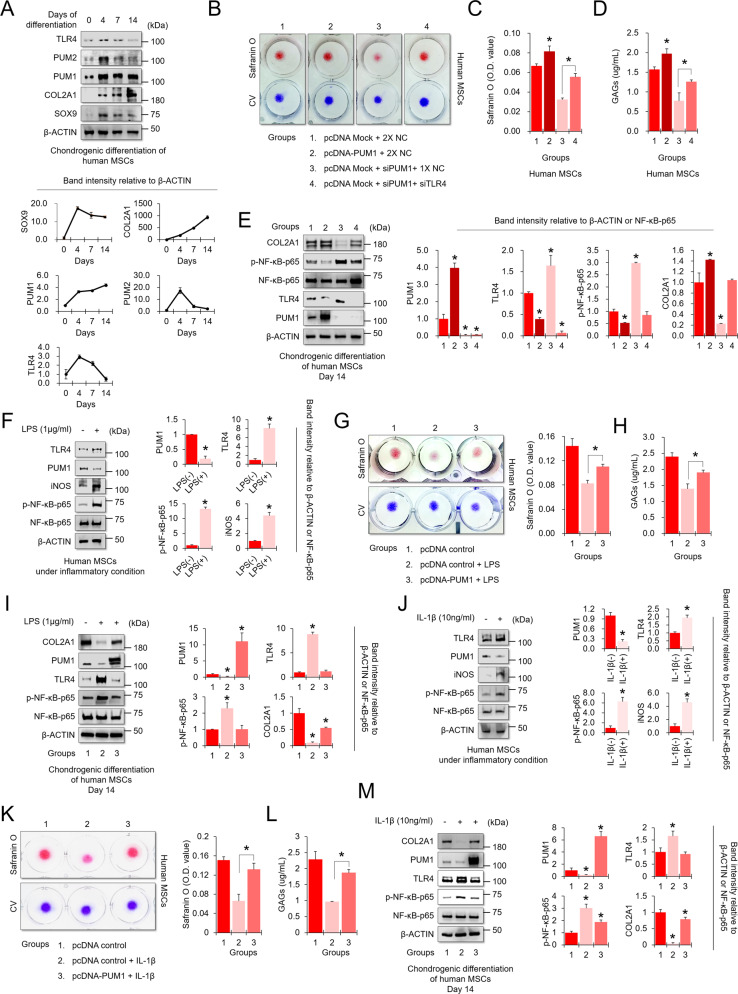
Studies have shown that TLR4 signaling is heavily involved in OA pathogenesis [46]. NF-κB activation, a signaling downstream of TLR4, influences OA-associated processes, including chondrocyte catabolism, survival, and synovial inflammation [47–49]. To investigate whether PUM1-mediated TLR4/NF-κB signaling regulation was associated with chondrogenic differentiation and homeostasis, we performed chondrogenic differentiation-related assays in MSCs. Protein levels of PUM1, SOX9, and COL2A1 were upregulated in the chondrogenic induction, whereas TLR4 and PUM2 levels were increased on day 4 of differentiation and subsequently decreased gradually during MSC chondrogenesis (Fig. 4A). PUM1 overexpression and KD enhanced and significantly reduced the chondrogenic potential of MSCs, respectively (Fig. 4B–E). Interestingly, the double KD of PUM1 and TLR4 rescued the MSC chondrogenic potential (Fig. 4B–E). We additionally tested the effects of PUM1 KD on the osteogenic and adipogenic differentiation of MSCs because the reduction of PUM1 was involved in the aging process of MSCs (Fig. 1) while cellular aging also affected osteogenic and adipogenic differentiation [50]. PUM1 KD significantly reduced the osteogenic potential of MSCs but did not alter the adipogenic differentiation capacity (Fig. S4). According to previous reports, the differentiation potential of MSCs during passaging into an adipogenic lineage was variable, i.e., unchanged, reduced or improved [31]. Therefore, the correlation between MSC senescence and adipogenic differentiation is considered still ambiguous. In our study, the adipogenic potential of PUM1-deficient MSCs remained unchanged, but the colony-forming ability and osteogenic and chondrogenic potential were clearly reduced by KD of PUM1.
Fig. 4. PUM1 overexpression preserves the high chondrogenic potential of MSCs in an inflammatory environment.
A MSCs were maintained in a chondrogenic medium in high-density culture conditions (dot culture in 1 × 105 cells per dot) for 4, 7, or 14 days. Undifferentiated MSCs were used as day 0 control. The protein levels of PUM1, PUM2, TLR4, and chondrogenic markers were analyzed in MSCs during MSC chondrogenesis. B MSCs were transfected with pcDNA control vector with no insert, pcDNA-PUM1 expression vector, negative control siRNA, or siRNAs targeting PUM1 and TLR4. The cells were maintained in a chondrogenic medium in high-density culture conditions for 14 days. Safranin O staining was performed to detect glycosaminoglycans (GAGs). C Stained cells were destained with 10% cetylpyridinium for quantitative analysis. The absorbance was measured at 490 nm. *P < 0.05 compared to groups 1 and 3 (n = 3 experimental replicates). D GAG content was measured using the Blyscan sulfate GAG assay. The sulfated GAGs were quantified in the supernatant and standardized using a chondroitin 4-sulfate standard solution. *P < 0.05 compared to groups 1 and 3 (n = 3 experimental replicates). E The expression levels of PUM1, TLR4, P65, p-P65, and COL2A1 were analyzed in chondrogenic MSCs by western blotting analysis of the cells transfected with pcDNA control vector with no insert, pcDNA-PUM1 expression vector, negative control siRNA, or siRNAs targeting PUM1 and TLR4. F MSCs were maintained in growth medium in high-density culture conditions (dot culture in 1 × 105 cells per dot), with or without LPS (1 μg/mL), for two days. The expression levels of PUM1, TLR4, P65, p-P65, and iNOS were analyzed in MSCs under LPS-induced inflammatory conditions. G MSCs were transfected with a pcDNA control vector with no insert or pcDNA-PUM1 expression vector. The cells were maintained in a chondrogenic medium under high-density culture conditions with or without LPS (1 μg/mL) for 14 days. Safranin O staining was performed to detect glycosaminoglycans (GAGs). Stained cells were destained with 10% cetylpyridinium for quantitative analysis. Absorbance was measured at 490 nm. *P < 0.05 compared to groups 1 and 3 (n = 3 experimental replicates). H The GAG contents were measured using the Blyscan sulfate GAG assay. The sulfated GAGs were quantified in the supernatant and standardized using a chondroitin 4-sulfate standard solution. *P < 0.05 compared to group 2 (n = 3 experimental replicates). I The expression levels of PUM1, TLR4, P65, p-P65, and COL2A1 were analyzed with western blotting analysis of the MSCs treated with or without LPS (1 μg/mL). J MSCs were maintained in growth medium under high-density culture conditions (dot culture in 1 × 105 cells per dot), with or without IL-1β (10 ng/mL), for 2 days. The expression levels of PUM1, TLR4, P65, p-P65, and iNOS were analyzed in MSCs under IL-1β-induced inflammatory conditions. K MSCs were transfected with a pcDNA control vector with no insert or pcDNA-PUM1 expression vector. The cells were maintained in a chondrogenic medium under high-density culture conditions, with or without IL-1β (10 ng/mL), for 14 days. Safranin O staining was performed to detect glycosaminoglycans (GAGs). Stained cells were destained with 10% cetylpyridinium for quantitative analysis. Absorbance was measured at 490 nm. *P < 0.05 compared to groups 1 and 3 (n = 3 experimental replicates). L The GAG contents were measured using the Blyscan sulfate GAG assay. The sulfated GAGs were quantified in the supernatant and standardized using a chondroitin 4-sulfate standard solution. *P < 0.05 compared to group 2 (n = 3 experimental replicates). M The expression levels of PUM1, TLR4, P65, p-P65, and COL2A1 were analyzed with western blotting analysis of the MSCs treated with or without IL-1β (10 ng/mL).
Chondrogenic induction of MSCs has commonly been employed for cartilage tissue engineering. Robust chondrogenic potential and cartilage matrix synthesis by MSCs are needed for a successful cartilage repair, even when a damaged or osteoarthritic joint is challenged by an inflammatory microenvironment [51]. We used two inflammatory factors, lipopolysaccharide (LPS) and interleukin-1β (IL-1β), to mimic the OA environment in vitro. LPS is well known as a strong TLR4 activator. PUM1 was decreased in LPS-induced inflammatory conditions, and TLR4 and pNF-κB were increased by LPS treatment (Fig. 4F). PUM1 was overexpressed during chondrogenic differentiation in LPS-induced inflammatory conditions to analyze whether chondrogenic potential loss was caused by LPS-mediated PUM1 decrease. LPS treatment reduced cartilage matrix formation and GAG synthesis in MSCs differentiated during chondrogenic differentiation, and PUM1 overexpression protected MSCs from the LPS challenge (Fig. 4G and H). Protein levels of TLR4 and p-NF-κB-p65 were upregulated by LPS during MSC chondrogenesis, whereas PUM1 and COL2A1 levels were downregulated by LPS (Fig. 4I). In the LPS-treated environment, PUM1 overexpression suppressed TLR4 and p-NF-κB-p65 levels and partially rescued COL2A1 (Fig. 4I). These results indicate that a decrease of PUM1 is associated with the LPS-induced reduction in MSC chondrogenic potential. It is well established that IL-1β plays an important role in OA pathogenesis [52]. We next determined the protein levels to clarify whether PUM1-mediated TLR4 regulation was involved in IL-1β-induced inflammatory responses. IL-1β treatment reduced the PUM1 protein levels while upregulating TLR4 expression (Fig. 4J). PUM1 overexpression protected MSC chondrogenic potential against IL-1β-mediated inflammatory responses (Fig. 4K and L), which was due to PUM1-mediated inhibition of TLR4 expression (Fig. 4M).
Reduction of PUM1 expression may be associated with OA pathogenesis
Patient-derived chondrocytes are a valuable model for studying OA pathogenesis; however, primary chondrocytes lose their chondrogenic potential and become senescent during in vitro cultivation which is an obstacle to studying OA pathogenesis [53]. Primary human chondrocytes became senescent, and COL2A1 was reduced during passaging (Fig. 5A). PUM1 and TLR4 levels were decreased and increased, respectively, during in vitro passaging (Fig. 5B). The normal chondrocyte cell line TC28a2 is widely used for studying normal or pathological cartilage disease mechanisms in vitro [54, 55]. To confirm whether PUM1 and TLR4 expression was affected under in vitro OA conditions, TC28a2 cells were treated with either LPS or IL-1β for 5 days. PUM1 protein levels were decreased, while TLR4 levels were dramatically increased by both LPS and IL-1β treatment (Fig. 5C, D). The in vitro observation of the changes in PUM1 and TLR4 expression levels was further verified in human cartilage tissues. Using human samples from OA patients, we performed immunohistochemistry, western blot, and reverse transcription-quantitative PCR (qPCR) analyses to observe PUM1 and TLR4 expression levels. Cartilage tissues were divided into intact or damaged area. Safranin O staining showed a decrease in proteoglycan in the damaged cartilage tissue area, where PUM1 was sparsely detected but TLR4 was highly detected (Fig. 5E, F). The western blotting and qPCR results of chondrocytes isolated from intact or damaged cartilage tissues showed both mRNA and protein levels of PUM1 were downregulated in the damaged cartilage tissues, whereas TLR4 was upregulated (Fig. 5G, H).
Fig. 5. PUM1 and TLR4 are inversely detected in the inflammatory environment in vitro and human OA cartilage tissues.
A The SA-β-gal staining (Scale bar = 350 μm) and (B) western blot analysis during passaging of primary chondrocytes. The expression levels of PUM1, PUM2, TLR4, COL2A1, and COL1A1 (one of the dedifferentiation markers in primary chondrocytes) were analyzed in primary chondrocytes by western blot analysis. C The expression levels of PUM1, TLR4, P65, p-P65, and COL2A1 were analyzed in cells from the human chondrocyte cell line TC28a2 treated with LPS (5 μg/mL), using western blot analysis. LPS treatment was maintained for five days. D The expression levels of PUM1, TLR4, P65, p-P65, and COL2A1 were analyzed in cells from the human chondrocyte cell line TC28a2 treated with IL-1β (10 ng/mL), using western blot analysis. IL-1β treatment was maintained for five days. E Representative images of safranin O/fast green staining and PUM1 and TLR4 immunostaining in intact and damaged cartilage tissues from patients with OA. Scale bar = 200 μm or 100 μm. F Quantification (%) of PUM1- or TLR4-positive cells from total cell population per field in immunohistochemical sections. G The expression levels of PUM1, TLR4, and COL2A1 were analyzed in primary chondrocytes isolated from cartilage tissues of human patients with OA, using western blot analysis. The obtained cartilage tissues were divided into intact or damaged parts, and then the tissues were minced, homogenized, and lysed. H The mRNA levels of PUM1 and TLR4 were analyzed in human primary chondrocytes isolated in cartilage tissues from OA patients, using reverse transcription-qPCR. The human cartilage tissues were divided into intact or damaged, and the tissues were then incubated with type 2 collagenase to isolate chondrocytes.
PUM1 protects chondrocytes from inflammatory responses, and non-coding RNA activated by DNA damage (NORAD) appears to act as an upstream regulator of PUM1 in osteoarthritis
To evaluate whether PUM1 affects chondrocyte homeostasis under inflammatory conditions, TC28a2 cells were transfected with a pEGFP-C1 PUM1 expression vector (Fig. S5). PUM1 siRNA, and TLR4 siRNA, then LPS or IL-1β was treated to evaluate the effect of the PUM1-mediated TLR4 regulation under in vitro inflammatory conditions. TC28a2 cells showed weak staining with safranin O after LPS treatment, whereas PUM1-overexpressing LPS-treated cells showed safranin O staining intensity like that of the control group (Fig. 6A), indicating that PUM1 overexpression rescued the chondrocyte role in matrix production from inflammation. However, PUM1 KD increased sensitivity to inflammation; conversely, double KD of PUM1 and TLR4 decreased this sensitivity (Fig. 6A). Western blot analysis showed that LPS treatment decreased PUM1 and increased TLR4 and pNF-κB levels. PUM1 overexpression suppressed the LPS-mediated increase in TLR4 and pNF-κB levels, whereas COL2A1 level was slightly rescued (Fig. 6B). Conversely, TLR4 and pNF-κB levels were increased by PUM1 KD in LPS-treated TC28a2 cells, and double KD of PUM1 and TLR4 rescued the effect of PUM1 single KD (Fig. 6B). Likewise, IL-1β treatment showed similar results in safranin O staining and western blot analysis (Fig. 6C and D), indicating that the PUM1-mediated TLR4 regulation may be intertwined with OA pathogenesis.
Fig. 6. PUM1 protects chondrocytes from inflammation, and NORAD may be an upstream regulator of the PUM1-TLR4 axis.
A TC28a2 cells were transfected with pEGFP-C1 control vector with no insert, the pEGFP-C1-PUM1 expression vector, negative control (NC) siRNA, or siRNAs targeting PUM1 and TLR4. The cells were maintained under the LPS (5 μg/mL)-induced inflammatory condition for five days. Safranin O staining was performed to detect glycosaminoglycans (GAGs). The stained cells were destained with 10% cetylpyridinium for quantitative analysis. The absorbance was measured at 490 nm. *P < 0.05 compared to groups 1 and 4 (n = 3 experimental replicates). B The expression levels of PUM1, TLR4, P65, p-P65, and COL2A1 were analyzed in TC28a2 cells in LPS-induced inflammatory conditions using western blotting. C TC28a2 cells were transfected with pEGFP-C1 control vector with no insert, the pEGFP-C1-PUM1 expression vector, NC siRNA, or siRNAs targeting PUM1 and TLR4. The cells were maintained under the IL-1β (10 ng/mL)-induced inflammatory condition for five days. Safranin O staining was performed to detect glycosaminoglycans (GAGs). The stained cells were destained with 10% cetylpyridinium for quantitative analysis. The absorbance was measured at 490 nm. *P < 0.05 compared to the groups 1 or 4 (n = 3 experimental replicates). D The expression levels of PUM1, TLR4, P65, p-P65, and COL2A1 were analyzed in TC28a2 cells under IL-1β-induced inflammatory conditions using western blotting. NORAD mRNA levels were examined in the (E) LPS- or (F) IL-1β-treated TC28a2 cells using reverse qPCR. G NORAD mRNA levels were also measured using RNA samples extracted from primary chondrocytes of OA patients (n = 3 experimental replicates). H RIP was performed in TC28a2 cells transfected with the pEGFP-C1-PUM1 vector. PUM1-IP was confirmed with western blot analysis, and HSP90 was used to confirm the presence of the input sample. The bar graph displays the RIP-qPCR fold enrichment of the indicated mRNAs relative to the IgG control. *P < 0.05 in comparison with the IgG control (n = 3 experimental replicates). (I) Reverse transcription-qPCR was performed to confirm the transfection efficiency of NORAD siRNA (100 nM). The expression levels of PUM1, TLR4, P65, p-P65, and COL2A1 were analyzed in NC siRNA- or NORAD siRNA-transfected TC28a2 cells under (J) LPS- or (K) IL-1β-induced inflammatory conditions, using western blotting. *P < 0.05 in comparison with group 2 (n = 3 experimental replicates).
NORAD is reported to bind to the PUMILIO proteins and repress their activity [27, 56]. A recent study reported that NORAD has been one of the most upregulated long non-coding RNAs in hip OA [57]. Thus, upregulated NORAD in the OA environment may affect the expression level of PUMILIO. However, there is no report related to this hypothesis. Therefore, we first observed how NORAD expression changed in the inflammatory environment in vitro after LPS and IL-1β treatment. LPS or IL-1β treatment increased NORAD expression in TC28a2 cells (Fig. 6E, F). To extend these results to the human OA model, we first confirmed their presence in the primary chondrocytes obtained from OA patients. NORAD levels were upregulated in the chondrocytes isolated from damaged cartilage tissues (Fig. 6G). We also confirmed that NORAD bound to PUM1 in TC28a2 cells using RNA-IP (Fig. 6H). In addition, NORAD KD rescued PUM1 protein levels under LPS- or IL-1β-induced inflammatory conditions (Fig. 6I, J). TLR4 and pNF-κB protein levels were also downregulated by NORAD KD (Fig. 6I, J).
Gene therapy with PUM1 preserves articular cartilage integrity in mouse OA model
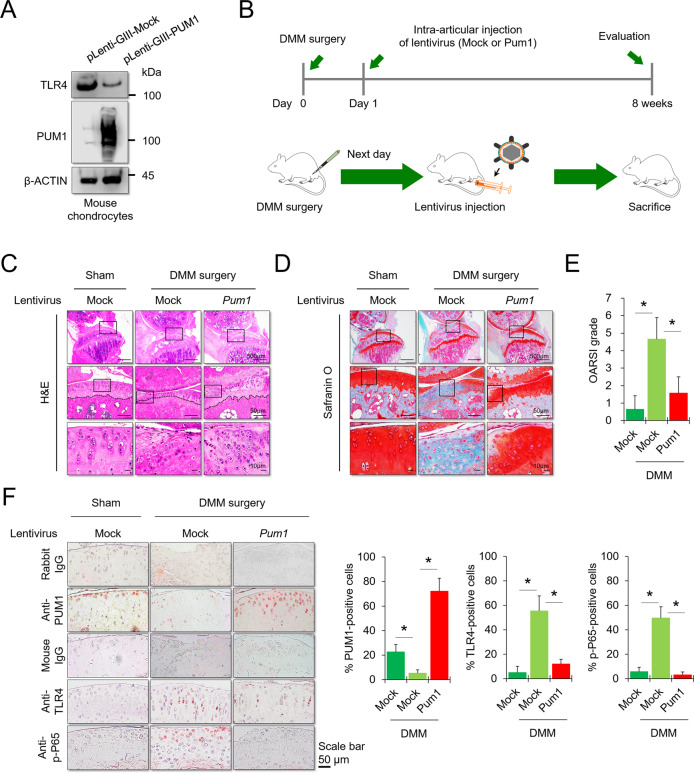
The mechanisms of action identified in the current study could be applied for OA treatment. To evaluate whether PUM1 can serve as a target for OA therapy, a lentiviral vector encoding mouse Pum1 and the surgical destabilization of the medial meniscus (DMM) model of osteoarthritis were employed for animal studies in vivo. The transduction efficiency of the lentivirus encoding Pum1 was confirmed in mouse chondrocyte ATDC5 cell line by western blot analysis (Fig. 7A). Next, we assessed the effects of PUM1 overexpression on cartilage matrix homeostasis in the DMM-induced OA mouse model in vivo. The lentivirus encoding Pum1 was injected one day after DMM surgery (Fig. 7B). Cartilage destruction was assessed 8 weeks after DMM surgery in the weight-bearing region of the medial tibial plateau, by hematoxylin and eosin (H&E) staining, safranin O staining, and OARSI grade [58]. Eight weeks after DMM surgery, H&E staining showed the loss of cartilage matrix at the superficial region of mouse knee cartilage in the DMM group, whereas Pum1-lentivirus injection prevented DMM-induced cartilage matrix destruction (Fig. 7C). Particularly, DMM surgery caused a severe loss of chondrocytes in the superficial region at the contact area between the medial femoral condyle and medial tibial plateau of the mouse knee. In contrast, injection of the lentivirus encoding Pum1 prevented the loss of chondrocytes caused by DMM surgery (Fig. 7C, middle and lower). DMM surgery significantly destroyed the mouse knee cartilage matrix, whereas injection of Pum1-encoding lentivirus resulted in reduced cartilage destruction relative to that in mock lentivirus-injected mice (Fig. 7D). In the Pum1 lentivirus-injected DMM mice, the OARSI score was significantly reduced compared to that in the mock vector-injected DMM group (Fig. 7E). We further evaluated whether intra-articular injection of Pum1 lentivirus affected TLR4 expression and NF-κB-P65 phosphorylation levels in cartilage tissues after DMM surgery. PUM1 was almost undetectable in the DMM-induced OA knee but was highly detected in mice injected with the lentivirus encoding mouse Pum1. Pum1-encoding lentivirus injection further suppressed the DMM-mediated TLR4 upregulation and prevented NF-κB-P65 phosphorylation (Fig. 7F). These findings indicate that the intra-articular PUM1 delivery could be a potential target for OA treatment, as it preserves cartilage integrity in the OA environment in vivo.
Fig. 7. Gene therapy with a lentiviral vector encoding PUM1 in a mouse model for OA.
A Protein levels of mouse PUM1 and TLR4 were analyzed in the mouse chondrocyte ATDC5 cell line infected with a lentiviral vector encoding pLenti-GiII-Mock or -mouse PUM1 gene. B Schematic of the time course used for the DMM-induced in vivo osteoarthritis experiments. The following figures indicate histological analyses of the changes in joints after DMM surgery. C H&E staining was performed to observe the cell morphology and tissue integrity in the articular cartilage tissues of the mouse knee undergoing DMM surgery. The figures located at the top show the extent of damage to the entire cartilage tissue in the weight-bearing area of the medial tibial plateau. The middle and bottom figures show the extent of damage and the morphology of chondrocytes in the contact area of the weight-bearing region between the medial tibial plateau and the medial femoral condyle. Scale bar = 500, 100, or 10 μm. D Likewise, Safranin O and fast green staining of articular cartilage tissues from mice that underwent sham or DMM surgery is indicated. Scale bar = 500, 100, or 10 μm. Safranin O and fast green staining demonstrated OA progression through the 8-week time course in the medial tibial plateau. E OARSI scores of the medial tibial plateau of DMM mice (Sham-Mock, n = 6; DMM-Mock, n = 6; DMM-PUM1, n = 6). Data are expressed as means, and the error bars indicate the standard deviation. F Representative images for PUM1 (red), TLR4 (red), and p-P65 (red) immunostaining in cartilage tissues obtained from sham or DMM mouse knees (Sham-Mock, n = 6; DMM-Mock, n = 6; DMM-PUM1, n = 6). Scale bar = 50 μm. The bar graphs show quantification (%) of the PUM1-, TLR4-, or p-P65-positive cells from total cell population per field, in immunohistochemical sections.
Discussion
MSCs have therapeutic potential for treating various skeletal diseases but there are obstacles to their implementation. For example, the ex vivo expansion of MSCs causes oxidative stress and cellular senescence, leading to deteriorated in vivo engraftment and functions [59]. In particular, the premature aging of MSCs remains a major challenge for clinical applications, as aged MSC-derived chondrocytes have low chondrogenic potential and hypertrophic phenotype, limiting the regenerative capacity for forming a hyaline cartilage tissue [60]. Therefore, the prevention of cellular senescence in cultured MSCs is required to harness their regenerative potential for cartilage therapies, and understanding the mechanisms related to cellular aging is essential. The NF-κB signaling pathway is critically involved in the cellular senescence of MSCs as well as inflammatory activation in many types of cells [61]. Discovering novel signaling pathways involved with NF-κB activation and clarifying the related molecular mechanisms would be essential in preventing or treating various diseases initiated by this signaling pathway. Moreover, the genetic mechanisms involved in the NF-κB signaling can provide important clues for the development of specific technologies to control cellular aging. In this study, we showed that PUM1 is essential for healthy MSCs during cultivation in vitro. PUM1 overexpression in MSCs during in vitro expansion protected MSCs against the H2O2-induced cellular senescence and improved their chondrogenic potential. Thus, the overall therapeutic efficacy is considered to increase when the PUM1 expression is preserved during the in vitro MSC expansion because the MSC-based transplantation has been common approach for cartilage repair [62]. The present study demonstrates that the genetically modified MSCs with constant PUM1 expression may be effective for the regeneration of damaged cartilage using in vitro cell culture system, suggesting that their possible applications in highly inflamed OA models which yet needs further studies and in this case the MSCs overexpressing PUM1 might be applied to augment the roles of PUM1.
Mechanistically, PUM1 binds to TLR4 mRNA to repress translation. TLR4 is an upstream regulator of NF-κB and increases its activity [63]. Interestingly, molecular genetic changes associated with aging share various common pathways in the OA progression [16], meaning that the TLR4-mediated NF-κB activation may be an essential target for OA treatment. Indeed, TLR4 inhibition by a small molecule inhibitor, resatorvid, successfully suppressed the effects of tissue damage-related molecules in chondrocytes. Moreover, TLR4 expression levels were higher in the chondrocytes from OA patients than in those from healthy individuals [64]. In this study, we also confirmed that TLR4 was higher expressed in damaged OA cartilage tissues than in intact cartilage tissues. Thus, TLR4 upregulation may act as a molecular initiator of aging and OA progression. Comprehensively, TLR4 expression modulation during aging or OA can be an important target for treating related diseases. Finding relevant upstream regulators would help understand the cause of aging and OA and lead to future treatments, such as gene therapy. In this respect, PUM1 may be an important target for the treatment and etiology of TLR4 activation-mediated aging and OA.
The decrease in PUM1 expression during aging and OA progression may be the leading cause of TLR4 activation, thereby aggravating related diseases. This suggests that TLR4 regulation through PUM1 expression may be the key to treat and delay aging-related diseases. However, the reason for PUM1 expression decrease during MSC senescence and OA pathogenesis was not studied here. Mechanistically, one example of ROS-mediated cellular senescence occurs via DNA damage [65]. Studies have shown that ROS generated by Ox-LDL upregulates non-coding RNA NORAD activated by DNA damage [66]. NORAD plays a negative regulator of PUMILIO activity or expression. Global transcriptome analysis by Lee et al. indicated that PUMILIO and NORAD have opposing effects on an overlapping set of genes [27]. Overall, DNA damage-activated NORAD may induce TLR4-mediated activation of NF-κB by inhibiting PUM1 activity during MSC aging. PUM1 expression was significantly downregulated in human and mouse OA tissues. A recent study showed that NORAD expression was highly upregulated in patients with hip OA [57], which supports our results for the downregulation of PUM1 expression in LPS or IL-1β-treated chondrocytes and DMM surgery-induced mouse OA models. In this study, we first reported the relationship between NORAD and PUM1 in OA, indicating that NOARD may serve as a novel biomarker for OA pathogenesis. Although it is recognized that NORAD targets the PUM proteins, it is still unclear how NORAD regulates PUM1 or PUM2 [67]. Therefore, further studies are warranted to clarify the roles of NORAD in aging and OA pathogenesis.
NF-κB may serve as a biomarker for aging, indicating potential targets for developing novel therapeutic strategies to improve the quality of life for the elderly [68]. In one study, TLR4-mediated NF-κB-p53/p21 signaling activation contributed to the senescence of dental pulp stem cells, a type of MSC [69]. Further, H2O2-mediated TLR4 activation increased pNF-κB and P21 levels, causing the senescence of MSCs. This signaling pathway is involved in OA development [46]. Thus, TLR4 targeting may help develop new drugs for aging and age-related diseases, and studies regarding this are currently ongoing. However, many TLR4-targeting drugs have failed in phase 3 clinical trials [70]. TLR4 plays an important role in innate immunity; therefore, side effects may occur upon reduction of TLR4 expression in non-target tissues. PUM1 targeting may be an alternative for the development of drugs to modulate TLR4. We performed gene therapy to prevent OA using PUM1 gene-encoding lentivirus and observed a reduction in TLR4 expression in OA cartilage tissues, preventing OA progression. Therefore, unwanted side effects might be reduced by injecting PUM1-encoding lentivirus into a local site (knee joint with OA) to modulate abnormally elevated TLR4 expression in OA tissues.
Overall, PUM1 expression was downregulated, and TLR4 and NF-κB expression was upregulated during MSC aging and OA development. We demonstrated the PUM1-mediated posttranscriptional inhibition of TLR4-NF-κB signaling regulated TLR4 expression and PUM1 interaction with the TLR4 3’-UTR, causing the downregulation of NF-κB activity. Thus, inducing PUM1 expression to reduce the inflammatory effects of TLR4-NF-κB in OA cartilage may provide a novel approach for treating OA and age-associated diseases resulting from TLR4-NF-κB signaling. In conclusion, the PUM1-mediated regulation of TLR4 expression has the potential as a clinically suitable target for OA treatment. Therefore, intra-articular delivery of the PUM1 gene may be a promising therapeutic approach to prevent OA progression.
Materials and methods
Study design
This study was conducted to evaluate (i) whether modulation of PUM1 expression could prevent cellular senescence and (ii) whether the associated mechanism could be a therapeutic target protecting articular cartilage from OA degeneration. Human bone marrow-derived MSCs were used for in vitro experiments on cellular senescence. Bone marrow aspirates were obtained from the posterior iliac crests of 12 adult donors, with the approval of the Institutional Review Board (IRB; 2017-0308-001) of the Yonsei University College of Medicine. MSCs isolated from the bone marrow were selected based on their ability to adhere to plastic cell culture dishes. Almost all the cultured cells (98%) were positive for CD90 and CD105 and negative for CD34 and CD45. The cell culture protocol has been described previously [71]. Cryopreserved vials of additional human MSCs were purchased from ATCC (PCS500012, Manassas, VA, USA). For all in vitro assays, at least three independent experiments were carried out, and in the case of primary cells, such as MSCs and patient-derived chondrocytes, at least three donors were tested per experiment. To minimize donor variation in experimental results obtained from patient-derived chondrocytes, the TC28a2 cell line was used for mechanism studies, and at least three independent experiments were carried out. Sample sizes and the characteristics of the MSCs and primary chondrocytes for each experiment are included in the figure legends and Supplementary Table S2. For immunostaining, western blot, and qPCR analyses in human cartilage tissues, the cartilage samples were obtained from five patients with OA undergoing total knee arthroplasty, with approval from the Institutional Review Board (IRB) (2019-1374-002) of the Yonsei University College of Medicine. Exclusion criteria for total knee arthroplasty were history of rheumatoid arthritis, inflammatory arthritis, autoimmune disease, ankylosing spondylitis, cancer, and long-term corticosteroid therapy (Please see Supplementary Table S2 for more detail). The histological results were analyzed by the investigators and orthopedic surgeons in a blinded manner.
Cell culture
MSCs were maintained in low-glucose Dulbecco’s Modified Eagle’s Medium (DMEM-LG; Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island NY, USA) and 1% antibiotic–antimycotic solution (Invitrogen) at 37 °C and 5% CO2. The MSCs were replated at a density of 5000 cells/cm2 and were subcultured at 80% confluency. To isolate chondrocytes, articular cartilages obtained from the knee joints of patients were first divided into intact and damaged parts. The cartilage tissues were shaved from the articular surface under sterile conditions, completely minced, and washed three times in phosphate-buffered saline (PBS). The minced tissues were then incubated in DMEM high-glucose medium (DMEM-HG; Invitrogen) supplemented with 2% collagenase (Sigma-Aldrich, St. Louis, MO, USA) and 0.5% dispase (Sigma-Aldrich) at 37 °C and 5% CO2 for 4 h. After that, the cell suspensions were passed through a 40 µm cell strainer to remove debris, and then the cells were cultured in DMEM-HG containing 10% FBS and 1% antibiotic–antimycotic solution at 37 °C and 5% CO2. The TC28a2 normal human and ATDC5 mouse chondrogenic cell lines were purchased from Sigma-Aldrich (Sigma-Aldrich). Primary chondrocytes and chondrocyte cell lines were maintained in DMEM-HGsupplemented with 10% FBS and 1% antibiotic–antimycotic solution at 37 °C and 5% CO2.
Animal experiments
All animal use procedures were conducted as per the approval of the Institutional Animal Care and Use Committee (IACUC) protocol of the Yonsei University College of Medicine (Approval number: IACUC-2016-0099). DMM surgery was conducted on the right knee joint of 8-week-old male C57BL/6 mice to induce OA by dissecting the medial meniscus-ligament to destabilize the medial meniscus. The sham group was surgically treated as described above, but without dissecting the medial meniscus-ligament. The next day, the lentivirus (1 × 1011 virus particles /10 μL) encoding pLenti-GIII-CMV-PUM1 or pLenti-GIII-CMV-no insert was injected intra-articularly to test whether PUM1 overexpression in mouse cartilage tissue preserved cartilage integrity under the DMM-induced OA model. At week 8, the mice were euthanized, and the joints were collected for histological assessment (n = 6 per group).
Senescence-associated β-galactosidase (SA-β-gal) assays
The SA-β-gal assay, using a cellular senescence assay kit (Millipore, Temecula, CA, USA), has been described previously. Positive cells were counted using a phase-contrast microscope or an image analyzer (MetaMorph; Universal Imaging Corp., Downingtown, PA, USA). The fluorescence-based SA-β-gal assays were performed using the Cellular Senescence Live Cell Analysis Assay Kit (ENZ-KIT130; Enzo Life Science, Lausen, Switzerland) and Cellular Senescence Activity Assay Kit (ENZ-KIT129; Enzo Life Science), following the manufacturer’s protocols. For SA-β-gal live cell analysis, the pretreatment solution provided in the kit was added to the senescent cells, and these were incubated at 37 °C for 2 h. SA-β-gal substrate solution was directly added to the cells, subsequently incubated at 37 °C for 4 h. After washing thrice with PBS, the cells were analyzed using the Countess II FL Automated Cell Counter (Thermo Fisher Scientific, Rockford, IL, USA). For the SA-β-gal activity assay, cell lysis buffer provided from the kit was used to treat senescent cells, which were incubated at 4 °C for 5 min. The whole lysate was transferred to a microcentrifuge tube and centrifuged to collect the supernatant. After determining the total protein concentration of each lysate sample, the lysate was mixed with the assay buffer provided with the kit, and the samples were incubated at 37 °C for 1 h. After adding stop solution, fluorescence was measured with a fluorescence plate reader at 360 nm/465 nm (excitation/emission).
RNA interference (RNAi)
Negative control, PUM1, PUM2, TLR4, and NORAD siRNAs were purchased from Bioneer (Daejeon, South Korea). Information on the siRNAs used in this study is provided in Table S3. Briefly, cells were plated to obtain 70–80% confluent growth in 6-well plates and transfected with 100 nM of each siRNA using Lipofectamine 2000 (Invitrogen). After 6 h of transfection, the medium was replaced with fresh medium.
Quantitative real-time polymerase chain reaction (qPCR)
PCR analysis was performed as described previously. Briefly, total RNA was isolated using an RNeasy kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. One microgram of total RNA was reverse-transcribed using the Omniscript kit (Qiagen). Primer sets were validated and purchased from Bioneer. The validated or designed primer sets used are listed in Table S4. Mean cycle threshold values from triplicate (n = 3) measurements were used to calculate gene expression, with normalization to ACTB (β-ACTIN) as the internal control.
Cell proliferation
Cell proliferation assays were performed using an EZ-Cytox Kit (Daeil Lab Service, Seoul, Korea). Cells transfected with siRNAs or plasmid expression vectors were seeded in 12-well culture plates at a density of 1 × 104 cells per well. The cells were maintained in a growth medium for 7–10 days, and the cell culture media were replaced once a day during cell viability assays. Briefly, after washing cells in PBS, 20 μL of EZ-Cytox (tetrazolium salts) solution was added to each well and incubated at 37 °C for 4 h. After incubation, the conditioned medium was transferred to 96-well plates. The absorbance was measured at 450 nm. All samples were tested in triplicate.
Western blotting
MSCs or chondrocytes isolated from cartilage tissues were lysed in the PRO-PREPTM Protein Extraction Solution (iNtRON Biotechnology, Seongnam, South Korea). Protein concentrations were determined using the Bio-Rad Protein Assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Approximately 30 μg of protein were analyzed via 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE; Sigma-Aldrich). The resolved proteins were transferred to membranes blocked with 5% skim milk (BD, Sparks, MD, USA) for 1 h at room temperature. The membranes were incubated for ~12 h with antibodies against PUM1 (Abcam, Cambridge, UK), PUM2 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), P65 (Cell Signaling Technology, Danvers, MA, USA), phosphorylated P65 (Cell Signaling Technology), P21 (Santa Cruz Biotechnology), TLR2 (Cell Signaling), TLR4 (Santa Cruz Biotechnology), GFP (Santa Cruz Biotechnology), Luciferase (Santa Cruz Biotechnology), SOX9 (Santa Cruz Biotechnology), COL2A1 (Santa Cruz Biotechnology), and iNOS (Abcam). Membranes were further probed with the antibodies against β-ACTIN (Santa Cruz Biotechnology), which served as a loading control.
Crystal violet (CV) staining for the colony-forming unit fibroblast (CFU-F) assay
Cells were seeded at a density of 1 × 103 cells in 100 mm culture dishes according to their conditions and maintained in DMEM-LG supplemented with 20% FBS for 12 days. The medium was replaced every two days during colony formation. Subsequently, the cells were fixed in a 1:1 acetone/methanol fixative, stained with a 20% CV solution (Merck, Darmstadt, Germany) for 30 min, and washed in distilled water.
EdU assay
The EdU proliferation assay was performed using the EdU Proliferation Kit (ab219801, Abcam) according to the manufacturer’s instructions. Before the EdU assay, MSCs (2 × 104 cells per well in 12-well culture plates) were incubated with or without 200 μM H2O2 for four days. The cells were incubated with EdU for 2 h under normal cell growth conditions and fixed with 4% formaldehyde. After washing, the cells were incubated with permeabilization buffer for 20 min. Followed by another step of washing, the cells were incubated with the EdU reaction buffer and then stained with DAPI. EdU-stained cells were observed under a fluorescence microscope and counted for quantification.
Vector preparation and transfection of plasmid DNA using electroporation
The PUM1 gene was synthesized using the cloning services of AbClon (Seoul, South Korea) and was cloned into pcDNA between NheI and XhoI (Takara Bio, Inc., Shiga, Japan) to generate pcDNA-PUM1 that expresses the PUM1-HA protein, or pEGFP-C1 vector between the NheI and AgeI sites (Takara Bio, Inc.) to generate pEGFPC1-PUM1 that expressed a green fluorescent protein (GFP)-PUM1 fusion protein. A mock vector with no insert was used as a control. PUM1 mutant lacking the RNA-binding domain (ΔPUM1-HD) was cloned into the pEGFP-C1 vector between the NheI and AgeI sites (Takara Bio, Inc.) to generate pEGFPC1-ΔPUM1-HD that expressed a green fluorescent protein (GFP)-PUM1 fusion protein (see Fig. S6). Each vector was transfected with MSCs or TC28a2 cells using electroporation with the neon transfection system according to the manufacturer’s instructions (Cat no, MPK5000; Invitrogen). After transfection of plasmids into MSCs or TC28a2 cells, they were washed with PBS and detached using the trypsin/EDTA solution. Approximately 1 × 106 cells were suspended in 90 µL of Transfection Resuspension Buffer R and were mixed with 5 g of plasmid DNA in 10 µL volume. Next, a 100 µL aliquot was placed in the electroporator Neon Gold Pipette Tips provided in the kit and pulsed (Pulse voltage: 990 V, pulse width: 40 ms). The contents of the transfection pipette were placed onto a 100 mm cell culture dish. Transfection efficiency was confirmed by fluorescence microscopy and western blot analysis 24 h after electroporation.
RNA-immunoprecipitation (RIP)
RIP was performed to know to determine potential interactions between PUM1 protein and TLR4 mRNA or lncRNA NORAD. Approximately 1 × 107 MSCs or TC28a2 cells transfected with pEGFP-C1-PUM1 vector were used per experiment. PUM1-bound RNA-IP was performed according to the RiboCluster Profiler RIP assay kit protocol (MBL International Corporation, Woburn, MA, USA). The cells were lysed by dithiothreitol (DTT; Sigma)-added lysis buffer provided by the manufacturer, and then the lysates were precleared with protein G plus agarose beads (Thermo Fisher Scientific) in DTT-added wash buffer provided by the manufacturer. The precleared lysates were transferred to the prepared tubes containing GFP antibody or IgG-immobilized beads. After approximately 12 h, GFP antibody or IgG-immobilized protein G agarose beads-RNA/protein complexes were separated to extract proteins and PUM1-bound RNAs. Eluted RNA was analyzed by qPCR. The GFP antibody-bound RNA was isolated and purified with provided kit according to the manufacturer’s instructions.
TLR4 3ʹ-UTR luciferase analysis
TLR4 3ʹ-UTR Lenti-reporter-Luc vector was purchased commercially from Applied Biological Materials Inc. (abm, Richmond, BC, Canada). To obtain lentiviral particles with TLR4 3ʹUTR-reporter-luciferase, HEK293T cells were seeded in 100 mm culture dishes at a density of 3 × 106 cells per dish. The next day, cells were transfected with TLR4 3ʹ-UTR Lenti-reporter-Luc vector with psPAX2 (Addgene #12260) and pMD2.G (Addgene #12259) using Lipofectamine 2000 (Invitrogen). After 6 h of transfection, the medium was replaced. The lentiviral vector-transfected HEK293T cells were maintained for two days, and subsequently, the supernatants were collected and stored at −70 °C. To transduce the vector expressing TLR4 3ʹUTR-reporter-luciferase, HEK293T cells were seeded in 6-well plates at a density of 5 × 104 cells per well. After 48 h of transduction, the medium was replaced afresh with 10 μg/mL puromycin dihydrochloride (Sigma), and the cells were maintained for three days. The selected cells were used in the TLR4 3ʹ-UTR luciferase reporter assay. The pEGFP-C1-mock, pEGFP-C1-PUM1, or pEGFP-C1-ΔPUM1-HD vectors were transfected with the selected cells by puromycin dihydrochloride using Lipofectamine 2000, and the cells were then maintained with a growth medium for 2 days after which the cells were lysed using PRO-PREP Protein Extraction Solution, and the protein was analyzed by western blotting to compare the luciferase activities of TLR4 3’-UTR. The band intensity on the western blot was quantified using ImageJ (n = 3, in triplicate).
Chondrogenic differentiation
The differentiation medium has been described previously [8]. MSCs were dotted on the center of individual wells of 24-well plates at 1 × 105 cells per well in a 10 μL volume. The cells were allowed to adhere for 2 h in the cell culture incubator. Next, a chondrogenic medium consisting of DMEM-HG supplemented with 1% Insulin Transferrin Selenium-A (ITS; Invitrogen), 50 mg/mL ascorbic acid (Invitrogen), and 10 ng/mL TGF-β3 (R&D Systems), was added gently. The chondrogenic media containing 10 ng/mL of TGF-β3 was changed every two days during in vitro differentiation periods. To detect proteoglycan synthesis, 0.1% safranin O solution (Sigma) was added, and the cells were incubated for 1 h after fixing with 4% paraformaldehyde. For quantitative analysis, absorbance was detected at 490 nm, followed by destaining with 100% ethanol for 20 min. The safranin O values were normalized to absorbance from the CV staining.
Glycosaminoglycans (GAG) assay
The amounts of sulfated glycosaminoglycans (GAGs) in the medium obtained from chondrogenic cultures were determined using the Blyscan kit (Biocolor) according to the manufacturer’s instructions. Briefly, 500 μL of the culture medium was mixed with 1 mL of Blyscan dye reagent by shaking for 30 min to complete the GAG–dye binding. After centrifugation, the dye bound to the GAGs was dissolved in a dissociation reagent. The recovered dye concentration was determined at 656 nm, and a chondroitin 4-sulfate standard solution (Biocolor) was used to generate the standard curves. All samples and standards were tested in triplicate.
Mouse PUM1 lentivirus
Mouse PUM1 lentiviral vector was purchased from abm (Cat No. 382050640495). pLenti-GIII-CMV vector with no insert was used as a control. For packaging of the control or PUM1 lentivirus, HEK293T cells were co-transfected with the lentiviral packaging vectors psPAX2, and pMD2.G. Medium containing the viral particles was collected by ultracentrifugation at 19400 × g for 2.5 h. Transduction efficiency of the lentivirus encoding mouse PUM1 was confirmed in the ATDC5 mouse chondrocyte cell line by western blot analysis.
Histology and immunohistochemistry
Cartilage tissue sections were fixed for seven days in 10% formalin at room temperature. After fixation, the formalin-fixed specimens were embedded in paraffin. The paraffin-embedded sections were deparaffinized, rehydrated, and washed with PBS, and the tissue sections were used to evaluate the disruption of cartilage tissue in the human cartilage or DMM-induced OA. The prepared tissue samples were sliced with a thickness of 4 mm and stained using H&E and safranin O solution to observe the OA progression or incubated with anti-TLR4 (Santa Cruz Biotechnology), anti-phosphorylated NF-κB-P65 and anti-PUM1 (Cell Signaling Technology) antibodies to detect PUM1, phosphorylated NF-κB-P65, and TLR4 levels. The stained tissues were observed using a VS120 virtual microscope (Olympus, Tokyo, Japan), and images of the sections were analyzed using the OlyVIA 2.5 program (Olympus). Safranin O-stained histological sections were used to assess cartilage degeneration (OARSI score).
Statistical analysis
All experiments were performed in triplicate using samples from at least three donors. The Student’s t-test was used to assess the significance of the statistical difference for two-group comparisons. The significance of the statistical differences among three or more groups was calculated using a one-way analysis of variance and post-hoc Bonferroni correction. Data are shown as the mean ± standard deviation. For all tests, P < 0.05 was considered statistically significant.
Supplementary information
Author contributions
DSY. designed the study, performed the experiments, analyzed the data, and wrote the paper. K-ML, YC, EAK, N-HL, SC, KHP, and J-HL performed in vitro and in vivo experiments. H-WK and JWL designed the study, interpreted the data, and revised the paper.
Funding
This study was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (NRF-2019R1I1A1A01060928, NRF-2018K1A4A3A01064257, NRF-2018R1A2B3003446, and NRF-2021R1A5A2022318).
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request.
Competing interests
The authors declare no competing interests.
Ethical approval
Animal handling and procedures were approved and conducted as per the approval of the Institutional Animal Care and Use Committee (IACUC) protocol of the Yonsei University College of Medicine (Approval number: IACUC-2016-0099).
Footnotes
The original online version of this article was revised: Due to a missing grant number.
Edited by M. Sibilia
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
2/21/2022
A Correction to this paper has been published: 10.1038/s41418-022-00945-w
Contributor Information
Hae-Won Kim, Email: kimhw@dku.edu.
Jin Woo Lee, Email: ljwos@yuhs.ac.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-021-00925-6.
References
- 1.Gupta PK, Das AK, Chullikana A, Majumdar AS. Mesenchymal stem cells for cartilage repair in osteoarthritis. Stem Cell Res Ther. 2012;3:25. doi: 10.1186/scrt116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu L, Yin C, Zhao F, Ali A, Ma J, Qian A. Mesenchymal stem cells: cell fate decision to osteoblast or adipocyte and application in osteoporosis treatment. Int J Mol Sci. 2018;19:360. doi: 10.3390/ijms19020360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veronesi F, Torricelli P, Borsari V, Tschon M, Rimondini L, Fini M. Mesenchymal stem cells in the aging and osteoporotic population. Crit Rev Eukaryot Gene Expr. 2011;21:363–77. doi: 10.1615/CritRevEukarGeneExpr.v21.i4.60. [DOI] [PubMed] [Google Scholar]
- 4.Lee WY, Wang B. Cartilage repair by mesenchymal stem cells: clinical trial update and perspectives. J Orthop Transl. 2017;9:76–88. doi: 10.1016/j.jot.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kabat M, Bobkov I, Kumar S, Grumet M. Trends in mesenchymal stem cell clinical trials 2004-2018: Is efficacy optimal in a narrow dose range? Stem Cells Transl Med. 2020;9:17–27. doi: 10.1002/sctm.19-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin YZ, Lee JH. Mesenchymal stem cell therapy for bone regeneration. Clin Orthop Surg. 2018;10:271–8. doi: 10.4055/cios.2018.10.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoon DS, Kim YH, Lee S, Lee KM, Park KH, Jang Y, et al. Interleukin-6 induces the lineage commitment of bone marrow-derived mesenchymal multipotent cells through down-regulation of Sox2 by osteogenic transcription factors. FASEB J. 2014;28:3273–86. doi: 10.1096/fj.13-248567. [DOI] [PubMed] [Google Scholar]
- 8.Yoon DS, Kim YH, Jung HS, Paik S, Lee JW. Importance of Sox2 in maintenance of cell proliferation and multipotency of mesenchymal stem cells in low-density culture. Cell Prolif. 2011;44:428–40. doi: 10.1111/j.1365-2184.2011.00770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou S, Greenberger JS, Epperly MW, Goff JP, Adler C, Leboff MS, et al. Age-related intrinsic changes in human bone-marrow-derived mesenchymal stem cells and their differentiation to osteoblasts. Aging Cell. 2008;7:335–43. doi: 10.1111/j.1474-9726.2008.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stolzing A, Jones E, McGonagle D, Scutt A. Age-related changes in human bone marrow-derived mesenchymal stem cells: consequences for cell therapies. Mech Ageing Dev. 2008;129:163–73. doi: 10.1016/j.mad.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Yang YK. Aging of mesenchymal stem cells: Implication in regenerative medicine. Regen Ther. 2018;9:120–2. doi: 10.1016/j.reth.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodell MA, Rando TA. Stem cells and healthy aging. Science. 2015;350:1199–204. doi: 10.1126/science.aab3388. [DOI] [PubMed] [Google Scholar]
- 13.Ksiazek K. A comprehensive review on mesenchymal stem cell growth and senescence. Rejuvenation Res. 2009;12:105–16. doi: 10.1089/rej.2009.0830. [DOI] [PubMed] [Google Scholar]
- 14.Martin JA, Buckwalter JA. Roles of articular cartilage aging and chondrocyte senescence in the pathogenesis of osteoarthritis. Iowa Orthop J. 2001;21:1–7. [PMC free article] [PubMed] [Google Scholar]
- 15.Martin JA, Buckwalter JA. Aging, articular cartilage chondrocyte senescence and osteoarthritis. Biogerontology. 2002;3:257–64. doi: 10.1023/A:1020185404126. [DOI] [PubMed] [Google Scholar]
- 16.Loeser RF, Collins JA, Diekman BO. Ageing and the pathogenesis of osteoarthritis. Nat Rev Rheumatol. 2016;12:412–20. doi: 10.1038/nrrheum.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lotz M, Loeser RF. Effects of aging on articular cartilage homeostasis. Bone. 2012;51:241–8. doi: 10.1016/j.bone.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loeser RF. Aging processes and the development of osteoarthritis. Curr Opin Rheumatol. 2013;25:108–13. doi: 10.1097/BOR.0b013e32835a9428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deschenes M, Chabot B. The emerging role of alternative splicing in senescence and aging. Aging Cell. 2017;16:918–33. doi: 10.1111/acel.12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong Q, Wei L, Zhang MQ, Wang X. Regulatory RNA binding proteins contribute to the transcriptome-wide splicing alterations in human cellular senescence. Aging (Albany NY) 2018;10:1489–505. doi: 10.18632/aging.101485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards TA, Pyle SE, Wharton RP, Aggarwal AK. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105:281–9. doi: 10.1016/S0092-8674(01)00318-X. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Zamore PD, Hall TM. Crystal structure of a Pumilio homology domain. Mol Cell. 2001;7:855–65. doi: 10.1016/S1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- 23.Moore FL, Jaruzelska J, Fox MS, Urano J, Firpo MT, Turek PJ, et al. Human Pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with DAZ (Deleted in AZoospermia) and DAZ-like proteins. Proc Natl Acad Sci USA. 2003;100:538–43. doi: 10.1073/pnas.0234478100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siemen H, Colas D, Heller HC, Brustle O, Pera RA. Pumilio-2 function in the mouse nervous system. PLoS One. 2011;6:e25932. doi: 10.1371/journal.pone.0025932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu EY, Chang R, Salmon NA, Reijo, Pera RA. A gene trap mutation of a murine homolog of the Drosophila stem cell factor Pumilio results in smaller testes but does not affect litter size or fertility. Mol Reprod Dev. 2007;74:912–21. doi: 10.1002/mrd.20687. [DOI] [PubMed] [Google Scholar]
- 26.Gennarino VA, Singh RK, White JJ, De Maio A, Han K, Kim JY, et al. Pumilio1 haploinsufficiency leads to SCA1-like neurodegeneration by increasing wild-type Ataxin1 levels. Cell. 2015;160:1087–98. doi: 10.1016/j.cell.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S, et al. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell. 2016;164:69–80. doi: 10.1016/j.cell.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin K, Qiang W, Zhu M, Ding Y, Shi Q, Chen X, et al. Mammalian Pum1 and Pum2 Control Body Size via Translational Regulation of the Cell Cycle Inhibitor Cdkn1b. Cell Rep. 2019;26:2434–50 e2436. doi: 10.1016/j.celrep.2019.01.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin K, Zhang S, Shi Q, Zhu M, Gao L, Xia W, et al. Essential requirement of mammalian Pumilio family in embryonic development. Mol Biol Cell. 2018;29:2922–32. doi: 10.1091/mbc.E18-06-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chien Y, Scuoppo C, Wang X, Fang X, Balgley B, Bolden JE, et al. Control of the senescence-associated secretory phenotype by NF-kappaB promotes senescence and enhances chemosensitivity. Genes Dev. 2011;25:2125–36. doi: 10.1101/gad.17276711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Wu Q, Wang Y, Li L, Bu H, Bao J. Senescence of mesenchymal stem cells (Review) Int J Mol Med. 2017;39:775–82. doi: 10.3892/ijmm.2017.2912. [DOI] [PubMed] [Google Scholar]
- 32.Nicolae CM, O’Connor MJ, Constantin D, Moldovan GL. NFkappaB regulates p21 expression and controls DNA damage-induced leukemic differentiation. Oncogene. 2018;37:3647–56. doi: 10.1038/s41388-018-0219-y. [DOI] [PubMed] [Google Scholar]
- 33.Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3’ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010;12:1014–20. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- 34.Uyhazi KE, Yang Y, Liu N, Qi H, Huang XA, Mak W, et al. Pumilio proteins utilize distinct regulatory mechanisms to achieve complementary functions required for pluripotency and embryogenesis. Proc Natl Acad Sci USA. 2020;117:7851–62. doi: 10.1073/pnas.1916471117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS One. 2008;3:e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mareschi K, Rustichelli D, Calabrese R, Gunetti M, Sanavio F, Castiglia S, et al. Multipotent mesenchymal stromal stem cell expansion by plating whole bone marrow at a low cellular density: a more advantageous method for clinical use. Stem Cells Int. 2012;2012:920581. doi: 10.1155/2012/920581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu Y, Li T, Ding Y, Sun L, Tu T, Zhu W, et al. Changes in mesenchymal stem cells following long-term culture in vitro. Mol Med Rep. 2016;13:5207–15. doi: 10.3892/mmr.2016.5169. [DOI] [PubMed] [Google Scholar]
- 38.Zhou L, Chen X, Liu T, Gong Y, Chen S, Pan G, et al. Melatonin reverses H2 O2 -induced premature senescence in mesenchymal stem cells via the SIRT1-dependent pathway. J Pineal Res. 2015;59:190–205. doi: 10.1111/jpi.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salminen A, Kauppinen A, Kaarniranta K. Emerging role of NF-kappaB signaling in the induction of senescence-associated secretory phenotype (SASP) Cell Signal. 2012;24:835–45. doi: 10.1016/j.cellsig.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 40.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 41.Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-kappaB signaling pathways. Nat Immunol. 2011;12:695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- 42.Shih VF, Tsui R, Caldwell A, Hoffmann A. A single NFkappaB system for both canonical and non-canonical signaling. Cell Res. 2011;21:86–102. doi: 10.1038/cr.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawai T, Akira S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med. 2007;13:460–9. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 44.Bohn JA, Van Etten JL, Schagat TL, Bowman BM, McEachin RC, Freddolino PL, et al. Identification of diverse target RNAs that are functionally regulated by human Pumilio proteins. Nucleic Acids Res. 2018;46:362–86. doi: 10.1093/nar/gkx1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hellweg CE, Arenz A, Bogner S, Schmitz C, Baumstark-Khan C. Activation of nuclear factor kappa B by different agents: influence of culture conditions in a cell-based assay. Ann N. Y Acad Sci. 2006;1091:191–204. doi: 10.1196/annals.1378.066. [DOI] [PubMed] [Google Scholar]
- 46.Gomez R, Villalvilla A, Largo R, Gualillo O, Herrero-Beaumont G. TLR4 signalling in osteoarthritis-finding targets for candidate DMOADs. Nat Rev Rheumatol. 2015;11:159–70. doi: 10.1038/nrrheum.2014.209. [DOI] [PubMed] [Google Scholar]
- 47.Roman-Blas JA, Jimenez SA. NF-kappaB as a potential therapeutic target in osteoarthritis and rheumatoid arthritis. Osteoarthr Cartil. 2006;14:839–48. doi: 10.1016/j.joca.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 48.Jimi E, Fei H, Nakatomi C. NF-kappaB signaling regulates physiological and pathological chondrogenesis. Int J Mol Sci. 2019;20:6275. doi: 10.3390/ijms20246275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sitcheran R, Cogswell PC, Baldwin AS., Jr NF-kappaB mediates inhibition of mesenchymal cell differentiation through a posttranscriptional gene silencing mechanism. Genes Dev. 2003;17:2368–73. doi: 10.1101/gad.1114503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang YK, Ogando CR, Wang See C, Chang TY, Barabino GA. Changes in phenotype and differentiation potential of human mesenchymal stem cells aging in vitro. Stem Cell Res Ther. 2018;9:131. doi: 10.1186/s13287-018-0876-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cooke ME, Allon AA, Cheng T, Kuo AC, Kim HT, Vail TP, et al. Structured three-dimensional co-culture of mesenchymal stem cells with chondrocytes promotes chondrogenic differentiation without hypertrophy. Osteoarthr Cartil. 2011;19:1210–8. doi: 10.1016/j.joca.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson CI, Argyle DJ, Clements DN. In vitro models for the study of osteoarthritis. Vet J. 2016;209:40–49. doi: 10.1016/j.tvjl.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 53.Loeser RF. Aging and osteoarthritis: the role of chondrocyte senescence and aging changes in the cartilage matrix. Osteoarthr Cartil. 2009;17:971–9. doi: 10.1016/j.joca.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Claassen H, Schicht M, Brandt J, Reuse K, Schadlich R, Goldring MB, et al. C-28/I2 and T/C-28a2 chondrocytes as well as human primary articular chondrocytes express sex hormone and insulin receptors-Useful cells in study of cartilage metabolism. Ann Anat. 2011;193:23–29. doi: 10.1016/j.aanat.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goldring MB. Culture of immortalized chondrocytes and their use as models of chondrocyte function. Methods Mol Med. 2004;100:37–52. doi: 10.1385/1-59259-810-2:037. [DOI] [PubMed] [Google Scholar]
- 56.Tichon A, Gil N, Lubelsky Y, Havkin Solomon T, Lemze D, Itzkovitz S, et al. A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat Commun. 2016;7:12209. doi: 10.1038/ncomms12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ajekigbe B, Cheung K, Xu Y, Skelton AJ, Panagiotopoulos A, Soul J, et al. Identification of long non-coding RNAs expressed in knee and hip osteoarthritic cartilage. Osteoarthr Cartil. 2019;27:694–702. doi: 10.1016/j.joca.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pritzker KP, Gay S, Jimenez SA, Ostergaard K, Pelletier JP, Revell PA, et al. Osteoarthritis cartilage histopathology: grading and staging. Osteoarthr Cartil. 2006;14:13–29. doi: 10.1016/j.joca.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 59.Denu RA, Hematti P. Effects of oxidative stress on mesenchymal stem cell biology. Oxid Med Cell Longev. 2016;2016:2989076. doi: 10.1155/2016/2989076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Szychlinska MA, Stoddart MJ, D’Amora U, Ambrosio L, Alini M, Musumeci G. Mesenchymal Stem Cell-Based Cartilage Regeneration Approach and Cell Senescence: Can We Manipulate Cell Aging and Function? Tissue Eng Part B Rev. 2017;23:529–39. doi: 10.1089/ten.teb.2017.0083. [DOI] [PubMed] [Google Scholar]
- 61.Mato-Basalo R, Morente-Lopez M, Arntz OJ, van de Loo FAJ, Fafian-Labora J, Arufe MC. Therapeutic potential for regulation of the nuclear factor kappa-B transcription factor p65 to prevent cellular senescence and activation of pro-inflammatory in mesenchymal stem cells. Int J Mol Sci. 2021;22:3367. doi: 10.3390/ijms22073367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lam ATL, Reuveny S, Oh SK. Human mesenchymal stem cell therapy for cartilage repair: Review on isolation, expansion, and constructs. Stem Cell Res. 2020;44:101738. doi: 10.1016/j.scr.2020.101738. [DOI] [PubMed] [Google Scholar]
- 63.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–51. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 64.Schelbergen RF, Blom AB, van den Bosch MH, Sloetjes A, Abdollahi-Roodsaz S, Schreurs BW, et al. Alarmins S100A8 and S100A9 elicit a catabolic effect in human osteoarthritic chondrocytes that is dependent on Toll-like receptor 4. Arthritis Rheum. 2012;64:1477–87. doi: 10.1002/art.33495. [DOI] [PubMed] [Google Scholar]
- 65.Passos JF, Nelson G, Wang C, Richter T, Simillion C, Proctor CJ, et al. Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol Syst Biol. 2010;6:347. doi: 10.1038/msb.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bian W, Jing X, Yang Z, Shi Z, Chen R, Xu A, et al. Downregulation of LncRNA NORAD promotes Ox-LDL-induced vascular endothelial cell injury and atherosclerosis. Aging (Albany NY) 2020;12:6385–6400. doi: 10.18632/aging.103034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marques FC, Ulitsky I. Aging well with Norad. Elife. 2019;8:e45974. doi: 10.7554/eLife.45974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Balistreri CR, Candore G, Accardi G, Colonna-Romano G, Lio D. NF-kappaB pathway activators as potential ageing biomarkers: targets for new therapeutic strategies. Immun Ageing. 2013;10:24. doi: 10.1186/1742-4933-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feng G, Zheng K, Cao T, Zhang J, Lian M, Huang D, et al. Repeated stimulation by LPS promotes the senescence of DPSCs via TLR4/MyD88-NF-kappaB-p53/p21 signaling. Cytotechnology. 2018;70:1023–35. doi: 10.1007/s10616-017-0180-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ain QU, Batool M, Choi S. TLR4-targeting therapeutics: structural basis and computer-aided drug discovery approaches. Molecules. 2020;25:627. doi: 10.3390/molecules25030627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee S, Yoon DS, Paik S, Lee KM, Jang Y, Lee JW. microRNA-495 inhibits chondrogenic differentiation in human mesenchymal stem cells by targeting Sox9. Stem cells Dev. 2014;23:1798–808. doi: 10.1089/scd.2013.0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request.