Abstract
Objectives
SARS-CoV-2 infections with Omicron variants have a high capability of human-to-human transmission. Nevertheless, the duration of isolation for mild cases was shortened to 5 to 7 days. We aimed to detect the duration of viral shedding among healthcare workers (HCWs) with Omicron by using viral culture.
Methods
We prospectively included newly diagnosed nonsevere, symptomatic SARS-CoV-2 positive HCWs. Nasopharyngeal swab samples were obtained consecutively on days 5, 7,10, and 14 of onset of symptoms. The samples were examined by nucleic acid amplification test and viral culture.
Results
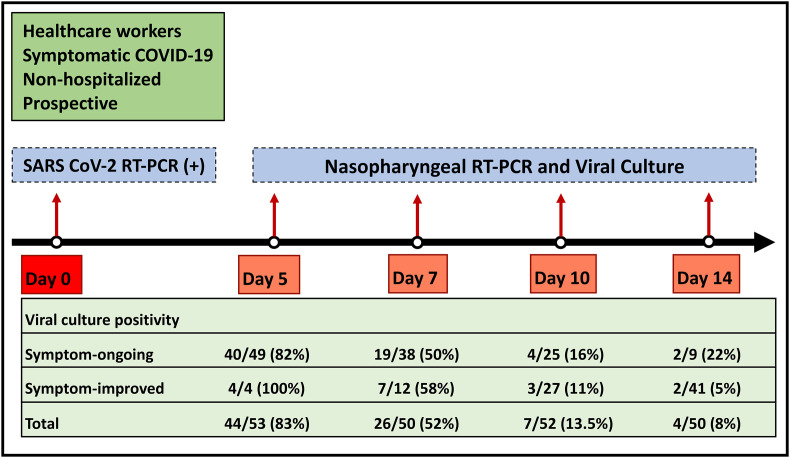
In total, 55 non-severe patients with SARS-CoV-2 Omicron variant were included. The mean age of the population was 34 years (range, 23 to 54) and 78% (43/55) were female. The PCR positivity rate on days 5, 7, 10, and 14 was 96.4% (53/55), 87.3% (48/55), 74.545% (41/55), and 41.8% (23/55) consecutively, whereas the viral culture positivity rates were 83% (44/53), 52% (26/50), 13.5% (7/52), and 8% (4/50). Among the patients who became symptom-free, the viral culture positivity rates were 100% (4/4), 58% (7/12), 11% (3/27), and 5% (2/41).
Discussion
We showed that among the SARS-CoV-2 Omicron variant infected patients, viral shedding continues for ≥10 days in 13.5% of all cases and 11% in symptom-free cases. The decision for cessation of isolation according to the presence of symptoms could be reconsidered until further studies disapprove of our results. Meanwhile, the infected HCWs who give care to high-risk patients for severe COVID-19 might extend their isolations ≤10 days after the onset of symptoms, regardless of their symptoms.
Keywords: COVID-19, PCR, SARS CoV-2, Shedding, Viral culture
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; COVID-19, CoronavirusDisease 2019; PCR, Polymerase Chain Reaction
Introduction
The details of transmission dynamics of SARS-CoV-2 are critical for containment of the virus, including the isolation rules and their implementation. The variants of SARS-CoV-2 were reported to have slight differences in transmission rate, clinical features, complications, and vaccine coverage [1]. However, soon after the Omicron variant of SARS-CoV-2 was reported on 17 December 2021, significant changes in isolation rules have been implemented by the CDC [2].
The duration of isolation for mild cases was shortened to 5 to 7 days, depending on the presence of symptoms and vaccination status in most countries [2]. However, this decision was based on the first studies on the viral shedding of the Omicron variant, with the major limitation of a small sample size [3]. Although high vaccination rates among the population and decreased mortality were reported [4], poor outcomes could still be seen among high-risk groups, such as haematological malignancies [5], and healthcare workers (HCWs) are critical since they give care to vulnerable patients. Therefore, the characteristics of the Omicron variant need to be detailed. In this study, we aimed to describe the duration of viral shedding among nonhospitalized HCWs infected with the Omicron variant of SARS-CoV-2.
Methods
Study population and design
This prospective study included SARS-CoV-2 PCR positive HCWs from 8 January 2022, to 17 February 2022. All of the participants were symptomatic and nonhospitalized. Because the HCWs were detected as SARS-CoV-2 positive in the occupational health and safety unit, they were asked to participate in the study. The first day of the onset of symptoms was defined as “Day 0″, and nasopharyngeal samples were collected on “Day 5,” “Day 7,” “Day 10,” and “Day 14.” The same team collected the samples in the viral transport medium (DMEM high glucose, 5% heat-inactivated fetal bovine serum, and 1% penicillin/streptomycin/amphotericin B) and transferred them to the microbiology laboratory within two hours. The information about the patient's demographic features, chronic illness history, symptoms, and vaccination status were recorded. All of the samples were studied for SARS-CoV-2 by RT-PCR. Only Omicron-positive patients were included in the final analysis. The viral culture was performed on PCR-positive samples.
Microbiology analysis
Viral nucleic acid extraction was performed using EZ1 Virus Mini Kit v2 (Qiagen, Hilden, Germany) on the EasyOne DNA isolation system (Qiagen). Bio-Speedy SARS-CoV-2 + Omicron RT-qPCR (Bioeksen R&D Technologies, Istanbul, Turkey) was employed for the detection of SARS-CoV-2 ORF1ab + N genes and NSP6 LSG105-107del mutation; PCR amplification and analysis were performed on a Bio-Rad CFX96 Real-time System (BioRad, Hercules, CA, USA). The samples with the cycle threshold (Ct) value < 33 for SARS-CoV-2 ORF1ab + N genes were considered positive for SARS-CoV-2 RNA. For the SARS-CoV RNA positive samples, if the difference between Ct values of NSP6 105-107/106-108del and Ct ofORF1ab + N genes is < 6, the sample is considered as an Omicron variant according to manufacturer instructions.
Viral culture
Vero E6 Cells (ATCC, Manassas, VA, USA; No. CRL-1586) and/or HEK293 expressed ACE2 and TMPRSS2 (National Institutes of Health, Bethesda, MA, USA) cell lines were plated on 96 well plates in DMEM high glucose (Sigma, St. Louis, MO, USA) with a mixture of 10% fetal bovine serum (Gibco; ThermoFisher Scientific, Waltham, MA, USA), 1% penicillin-Streptomycin (Hyclone; Cytiva Life Sciences, Marlborough, MA, USA), and amphotericin B (Hyclone). Nasopharyngeal samples were added to cells with 90% confluency and incubated for 1 hour at 37°C and 5% CO2. The culture supernatants were discarded and a fresh culture medium (DMEM high glucose supplemented with 5% fetal bovine serum, 1% penicillin-streptomycin, and amphotericin B) was added to the cell line. Cytopathic effect was monitored on day 6 and culture supernatant was collected on day 7. Viral growth was confirmed with quantitative RT-PCR from culture supernatants that induced visible cytopathic effect under the inverted tissue microscope. Briefly, viral RNA was extracted using EZNA Viral RNA Kit (Omega, Norcross, Georgia, US) according to the manufacturer's recommendation. Quantitative RT-PCR was conducted with primers and TaqMan probes targeting nucleocapsid N1 and RNA-dependent RNA polymerase genes using QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems, ThermoFisher Scientific).
Descriptive statistics were performed by using STATA version 16 (College Station, TX, USA). The categorical variables were compared by using the χ2 test. Koç University Institutional Review Board approved the study (reference number 2020.145.IRB1.034). All participants signed the informed consent form.
Results
During the study period (8 January through 17 February 2022), the HCWs who fulfilled the inclusion criteria were asked to be recruited to the study. In total, 95 HCWs were diagnosed with COVID-19 were asked to be included, 65 (68%) consented and 60 HCWs completed the study process. Five of them were infected with non-Omicron SARS-CoV-2, so 55 cases with Omicron variants were included in the analysis. The mean age was 34 years (range, 23 to 54 years), and 78% (43/55) were female. Forty-two percent (23/55) of the population were nurses, and 16% (9/55) were physicians. Only one patient was unvaccinated, and 54 patients were vaccinated; 27 (49%) had two CoronaVac and two BioNTech, seven (13%) had two CoronaVac and three BioNTech, five (9%) had two BioNTech, three (5.5%) had three BioNTech, three (5.5%) had two CoronaVac and one BioNTech, and three (5.5%) had two CoronaVac.
None of the patients were hospitalized. During the study period, favipiravir was suggested by the Ministry of Health of Turkey, but the HCWs in our hospital did not receive it. During the study period, molnupiravir was not available. Two HCWs had hypertension but under control, one had obesity, and one had breast cancer in remission.
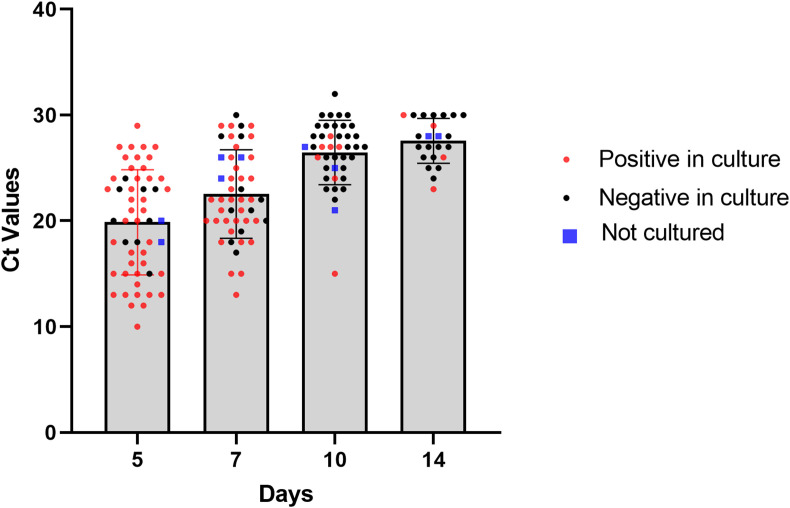
The PCR positivity rate on day 5, 7, 10, and 14 were 96.4% (53/55), 87.3% (48/55), 74.545% (41/55), and 41.8% (23/55), consecutively. The duration of shedding, which was defined by viral culture positivity rates, were 83% (44/53), 52% (26/50), 13.5% (7/52), and 8% (4/50) on defined days (Fig. 1, Fig. 2 ). Among four cases whose SARS CoV-2 viral culture tests were positive on day 14, one was unvaccinated. Three were previously healthy and vaccinated (one with two CoronaVac and two BioNTech; one with three BioNTech and one with two CoronaVac).
Fig. 1.
Viral culture positivity, negativity, and cycle threshold values of SARS-CoV-2 RT-PCR positive samples on 5, 7, 10, and 14 days after the onset of symptoms. Each dot shows one subject (viral culture positive, negative or not cultured, and cycle threshold values on y axis). Ct, cycle threshold.
Fig. 2.
SARS-CoV-2 culture positivity and its relationship with the presence of the symptom.
In 19% (10/53) of the cases, the duration of shedding was longer than the duration of the symptoms. On day 5, four patients declared no more symptoms, although viruses were isolated by the culture. The rate of viral culture positivity was 82% (40/49) among the patients who declared that they have the symptom. The rate of viral shedding among symptom-free cases were 58% (7/12) on day 7, whereas they were 11% (3/27) on day 10 and 5% (2/41) on day 14 (Fig. 2).
Discussion
In this prospective study, we aimed to show the viral shedding among nonhospitalized HCWs infected with the SARS-CoV-2 Omicron variant. Isolation of the virus in cell culture remains the most reliable method to assess the infectivity of SARS-CoV positive individuals [6], although PCR is used in routine daily practice. Prospectively collected nasopharyngeal samples from HCWs on days 5, 7, 10, and 14 were studied for RT-PCR, variant, and viral culture. We showed that among the SARS-CoV-2 Omicron variant infected patients, 52% shed infectious viral particles on day 7, 13.5% on day 10, and 8.5% on day 14.
The duration of viral shedding was calculated as 16.8 days in a recent meta-analysis; however, they used RT-PCR which is not a proper parameter to define viral shedding [7]. In a recent report, the viral kinetics of symptomatic nonsevere patients with Delta and Omicron variants were compared, and no difference was detected in terms of viral shedding regardless of the vaccination status. They found that >50% had a positive viral culture on day 5 and 25% on day 8. However, the number of cases with Omicron was 19, and the number of Delta cases was 37 in that study [8]. In another study including 18 Omicron cases, the virus was detected until 9 days, and no virus was detected in culture 10 days after the onset of symptoms [3]. In our study, live viral shedding was detected in 13.5% of all the cases on day 10.
After the emergence of the Omicron variant, the CDC recommendations for isolation in COVID-19 were updated regarding a symptom-based approach. The CDC suggested ending isolation if a patient does not have fever and if the patient's symptoms improved on day 5 [9]. Although the CDC suggests wearing a well-fitting mask until day 10, after the cessation of isolation on day 5, it is not possible to follow up on the cases whether they maintain isolation precautions while working. Among HCWs who were cleared from symptoms on day five, viral shedding was detected in all. The rate of viral shedding was 58% on day 7, 11% on day 10, and 5% on day 14 among the patients who stated that their symptoms had resolved. Our study showed that resolving of the symptoms does not mean that viral shedding discontinues. Because the symptom-based approach for cessation of isolation may not be reliable, it might be better to change isolation rules regardless of the continuation of symptoms.
The strengths of our study were being prospective and using viral culture for defining the presence of infectious viral particles. We had some limitations. Our study included nonsevere HCWs with Omicron and might not be generalized to patients with high-risk factors for severe COVID-19. We focused on HCWs because of their critical role in infection control, and also easier accessibility in longitudinal follow-up. Our study included vaccinated HCWs, however the shedding of the live virus might be even longer among unvaccinated HCWs. Thus, the shedding duration might be longer in the community especially among unvaccinated people and also the patients with older age and immunocompromised conditions, such as malignancy using chemotherapy.
In conclusion, we showed that viral shedding continues for ≥10 days among mild cases infected with Omicron, and a symptom-based approach might not be a good approach for the cessation of isolation. The HCWs who ended their isolation and turned back to work should be alerted of their contagiousness until 10 to 14 days. The infected HCWs who give care to the high-risk patients for severe COVID-19 might extend their isolations ≤10 days after the onset of symptoms, regardless of their symptoms. If they need to start working earlier than 10 days, their adherence to infection control measures should be followed up closely.
Author's contributions
ŞK, ÖE, and FC conceptualized this study. ZEK, GGE, CV, YB, ZN, TB, ÖŞ, MAK, and EP gathered the resources. ŞK, ÖE, GGE, FC, and YB are responsible for the writing of the study.
Transparency declaration
None of the authors declared conflict of interest. This study was supported by KUISCID (Koç University-Isbank Center for Infectious Diseases).
Acknowledgements
The authors would like to thank Sibel Tanır, “Koç Healthcare Unit,” and the hospital administration for their significant support in this study.
Editor: L. Leibovici
References
- 1.Fiolet T., Kherabi Y., MacDonald C.J., Ghosn J., Peiffer-Smadja N. Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: a narrative review. Clin Microbiol Infect. 2022;28:202–221. doi: 10.1016/j.cmi.2021.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention . 2021. CDC updates and shortens recommended isolation and quarantine period for general population.https://www.cdc.gov/media/releases/2021/s1227-isolation-quarantine-guidance.html [cited 2021 Dec 27]. Available from: [Google Scholar]
- 3.Takahashi K., Ishikane M., Ujiie M., Iwamoto N., Okumura N., Sato T., et al. Duration of infectious virus shedding by SARS-CoV-2 Omicron variant-infected vaccinees. Emerg Infect Dis. 2022;28:998–1001. doi: 10.3201/eid2805.220197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization WHO coronavirus (COVID-19) dashboard [updated 2022 Jul 25; cited 2022 May 3] https://covid19.who.int Available from:
- 5.Attaway A.H., Scheraga R.G., Bhimraj A., Biehl M., Hatipoğlu U. Severe COVID-19 pneumonia: pathogenesis and clinical management. BMJ. 2021;372:n436. doi: 10.1136/bmj.n436. [DOI] [PubMed] [Google Scholar]
- 6.Bal A., Brengel-Pesce K., Gaymard A., Quéromès G., Guibert N., Frobert E., et al. Clinical and laboratory characteristics of symptomatic healthcare workers with suspected COVID-19: a prospective cohort study. Sci Rep. 2021;11:14977. doi: 10.1038/s41598-021-93828-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan D., Zhang X., Chen C., Jiang D., Liu X., Zhou Y., et al. Characteristics of viral shedding time in SARS-CoV-2 infections: a systematic review and meta-analysis. Front Public Health. 2021;9:652842. doi: 10.3389/fpubh.2021.652842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boucau J., Marino C., Regan J., Uddin R., Choudhary M.C., Flynn J.P., et al. Duration of viable virus shedding in SARS-CoV-2 Omicron variant infection. medRxiv. 2022 [Google Scholar]
- 9.Centers for Disease Control and Prevention Ending isolation and precautions for people with COVID-19: interim guidance [updated and cited 2022 Jan 14] https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html Available from: