Abstract
Due to the possibility that bacteria could be involved in the clearance of paralytic shellfish toxins (PST) from bivalve molluscs, investigations into which, if any, bacteria were able to grow at the expense of PST focused on several common shellfish species. These species were blue mussels, oysters, razor fish, cockles, and queen and king scallops. Bacteria associated with these shellfish were isolated on marine agar 2216 and characterized by their carbon utilization profiles (BIOLOG). Selected isolates from groups demonstrating 90% similarity were screened for their ability to metabolize a range of PST (gonyautoxins 1 and 4 [GTX 1/4], GTX 2/3, GTX 5, saxitoxin, and neosaxitoxin) using a novel screening method and confirming its results by high-performance liquid chromatography. Results suggest that molluscan bacteria have different capacities to utilize and transform PST analogues. For example, isolates M12 and R65 were able to reductively transform GTX 1/4 with concomitant production of GTX 2/3, while isolate Q5 apparently degraded GTX 1/4 without the appearance of other GTXs. Other observed possible mechanisms of PST transformations include decarbamoylation by isolate M12 and sulfation of GTXs by isolates Q5, R65, M12, and C3. These findings raise questions as to the possible role of bacteria resident in the shellfish food transport system. Some researchers have suggested that the microflora play a role in supplying nutritional requirements of the host. This study demonstrates that bacteria may also be involved in PST transformation and elimination in molluscan species.
Paralytic shellfish toxins (PST) are potent neurotoxins produced by some strains of dinoflagellates, such as Alexandrium spp., Pyrodinium bahamense var. compressum, and Gymnodinium catenatum (7). When passed through the marine food web, these toxins can lead to human disease through consumption of contaminated shellfish. For example, filter-feeding bivalves, such as mussels, cockles, oysters, and scallops, feed on dinoflagellates, transferring them from the gills to digestive organs where the toxins become concentrated (6, 21).
The incidence of human PST poisoning has increased markedly since the early 1970s, with approximately 2,000 cases reported annually worldwide (13). This represents a serious health risk, as mortality rates in humans are reported to be between 10 and 20% (14), and although molluscan shellfish are themselves relatively unaffected by PST, outbreaks of PST poisoning in humans result in detrimental economic impacts on both the fish and shellfish industries.
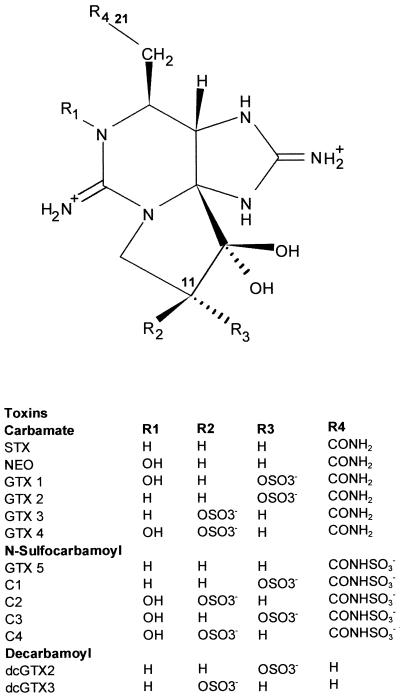
The structures of PST are based on a tetrahydropurine skeleton with two permanent guanidinium functions. Substitutions at four distinct positions around the basic PST structure categorize different PST analogues (Fig. 1). Currently, there are 22 known PST derivatives split into three categories: carbamate (with saxitoxin [STX] generally being considered the most potent), N-sulfocarbamoyl, and decarbamoyl (10, 23). Although PST analogues have different toxicities, they have the same action of blocking sodium channels in mammalian nerve cells, preventing conductance of signals along the neuron, with the net result being paralysis of mammals and, possibly, death occurring from respiratory failure (2).
FIG. 1.
Structures of PST analogues used in this study (adapted from references 10 and 11).
An understanding of the interactions between shellfish and PST is therefore essential, but despite the known toxicological properties of PST, our knowledge of the physiology of their degradation is limited. Enzymatic activities of shellfish undoubtedly play a role in interconversion of PST following accumulation (6, 8, 31), but reports by Kotaki et al. (17–19) also suggest that marine bacteria are capable of transforming these marine neurotoxins. Precursory studies by Kotaki and colleagues suggested that Vibrio and Pseudomonas spp. isolated from the viscera of marine crabs, snails, and a marine red alga, a Jania sp., were capable of transforming hydroxysulfate carbamate derivatives to STX through reductive eliminations (17–19).
Using a rapid screening BIOLOG method developed to test bacterial metabolic activity in the presence of PST and high-performance liquid chromatography (HPLC) for confirmation of PST transformation, the aim of this study was to further examine the capacity of the bacterial flora associated with several common bivalve species to utilize a range of PST.
MATERIALS AND METHODS
Isolation and enumeration of bacteria from shellfish.
Flesh from shellfish harvested from areas in West Scotland was extracted, under aseptic conditions, from mussels (Mytilus edulis), cockles (Cerastoderma edule), oysters (Ostrea edulis), razor fish (Ensis arcuata), queen scallops (Chlamys opercularis), and king scallops (Pecten maximus). Bacteria were enumerated using plate count methods as follows. Shellfish homogenates, 50% (wt/vol) in peptone water (CM9) (Oxoid Ltd., Basingstoke, United Kingdom), were serially diluted in sterile seawater, and suitable dilutions were plated onto marine agar 2216 (Becton-Dickinson, Oxford, United Kingdom) and incubated aerobically for up to 7 days at 20°C. All bacteria from plates containing approximately 30 colonies and bacteria showing distinct morphotypes were purified by repeated passage on marine agar. Bacteria were stored in marine broth 2216 (Becton Dickinson) plus 20% glycerol (wt/vol) at −70°C prior to further investigation.
Cluster analysis of shellfish isolates using BIOLOG
Pure bacteria isolated from the dilution plates were categorized according to their metabolic profiles. The profiles were generated using 96-well BIOLOG-GN MicroPlates (BIOLOG Inc., Hayward, Calif.). The BIOLOG-GN method simultaneously determines the bacterial oxidation of 95 different carbon sources (3). Bacteria were grown on marine agar for 24 h at 20°C prior to suspension in inoculation medium of the following composition: NaCl, 25 g liter−1; MgCl2, 8 g liter−1; KCl, 0.5 g liter−1; and carrageenan type II, 1.5 g liter−1 . Turbidity was adjusted to an A590 of 0.35, and plates were inoculated with 125 μl of the cell suspension in each well. Plates were incubated in a moist container for 48 h at 20°C, and carbon oxidation profiles were determined using a BIOLOG MicroPlate reader at 590 nm. Results were converted to a binary scale, according to the metabolic capacity of each isolate. Isolates unable to oxidize the available carbon compound were given a score of 0, whereas a positive result, demonstrated by the appearance of purple in an active well, was given a score of 1. A matrix measuring the similarity between samples was formed using simple matching distance. Cluster analysis was done using average-linkage clustering. All analysis was carried out using the Genstat 5, version 4.1, statistical package. Bacterial numbers in each cluster were enumerated by determining the number of isolates from the serial dilutions of each shellfish species which clustered at the 90% similarity level. A randomly selected isolate from each cluster formed at the 90% similarity level was subsequently screened for the ability to utilize PST.
Screening for bacterial utilization of PST using BIOLOG-MT microplates
Bacteria were added to various concentrations of commercially available carbamate and carbamoyl PST in BIOLOG-MT microplates. Bacteria were cultured for 24 h on marine agar plates at 20°C prior to suspension in sterile BIOLOG inoculation medium (20 ml) as previously described. Bacterial suspensions were subsequently transferred to BIOLOG-MT microplates, to which 1 or 2 μl of different toxin standards (gonyautoxins 1 and 4 [GTX 1/4], GTX 2/3, GTX 5, saxitoxin [STX], and neosaxitoxin [NEO] in 0.1 M acetic acid) were added, to give a final volume of 100 μl. The final concentrations (in nanograms per microliter) of the individual toxins follow: GTX 1, 0.74; GTX 4, 0.32; GTX 2, 1.20; GTX 3, 0.29; GTX 5, 1.11; STX, 1.40; and NEO, 1.40 at the lowest concentration examined and double this amount for the highest levels. All toxins were obtained from the Canadian National Research Council (CNRC) (Halifax, Nova Scotia, Canada). Control wells contained inoculation media and toxin standards or bacterial suspensions plus 0.1 M acetic acid. BIOLOG-MT microplates were incubated at 20°C for 3 days in a moist container and read at 590 nm. All isolates showing a 50% or greater increase in absorbancy above that for control wells were further investigated using HPLC analysis. Selected isolates showing no increase and increases below 50% were also analyzed.
HPLC analysis of PST transformations.
A mixture of 24-h-old bacterial cultures (196 μl) grown in marine broth and individual toxin standards (4 μl) were incubated at 20°C in an orbital shaker at 100 to 120 rpm for 0, 6, 12, 24, 48, 72, and 144 h. The final concentrations (in nanograms per liter) of the individual toxins were as follows: GTX 1, 1.48; GTX 4, 0.64; GTX 2, 2.40; GTX 3, 0.58; GTX 5, 2.22; STX, 2.80; and NEO, 2.80. An aliquot (2 μl) of each culture was spread onto marine agar after 144 h to ensure that cultures were still viable. Controls sampled over the same time course consisted of bacterial cultures in marine broth with no toxin added and toxin standards in marine broth (no bacteria). Spent media from the bacterial cultures were subsequently transferred to Ultrafree centrifugal filter units (10,000 nominal molecular weight limit) (Millipore Co., Bedford, Mass.) and centrifuged at 13,000 × g for 15 min to remove particulate matter. Supernatants were retained and frozen at −20°C for subsequent analysis of residual substrates and their products by HPLC.
HPLC analysis of toxins.
Concentrations of PST in culture supernatants were determined by HPLC, applying the isocratic methods of Franco and Fernández-Vila (10), using ion pair chromatography with postcolumn derivatization and fluorimetric detection. Ion pair chromatography was performed with a Spectra Physics autosampler and Spectra System P4000 pumps (Thermo Separation Products, Fremont, Calif.) and a FP1520 Intelligent fluorescence detector (Jasco, Ltd., Essex, United Kingdom) with computer integration (PC1000 version 3.5; Thermo Separation Products). A silica-based reverse-phase Merck Purospher C18 column (250 mm by 4 mm [inner diameter]; Merck, Darmstadt, Germany) was used for toxin separations. Authentic external PST standards (CNRC) were run prior to sample analysis and after every fifth sample. All samples were quantitated by comparison of sample peak areas with those of the standards.
RESULTS
Enumeration and cluster analysis of shellfish bacteria.
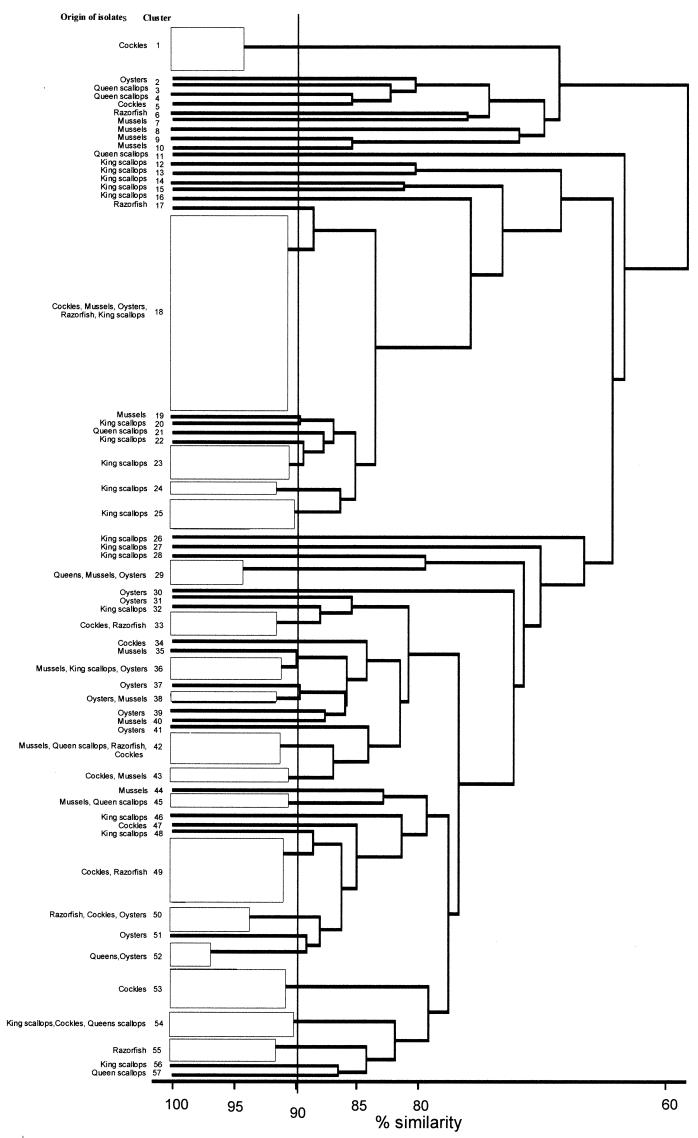
Bacteria intrinsic to shellfish were isolated on marine agar; log10 bacterial numbers ranged from 5.6 (g of shellfish flesh)−1 in queen scallops to 4.3 (g of shellfish flesh)−1 in razor fish (Table 1). Microscopic observation showed all isolates to be gram-negative, rod-shaped bacteria. On repeated passage on marine agar, four isolates present in oysters, razor fish, and cockles did not grow. A total of 147 isolates from shellfish continued to thrive, giving satisfactory metabolic profiles on BIOLOG-GN plates (3). Cluster analysis at the 90% similarity level separated these bacteria into 57 clusters (Fig. 2 and Table 1) and demonstrated that culturable bacterial populations associated with different shellfish exhibited considerable variation. For example, bacterial isolates forming clusters 7 to 10 were detected only in mussels; clusters 2, 30, 31, 37, 39, 41, and 51 consisted of bacterial isolates obtained from oysters; clusters 6, 17, and 55 comprised isolates from razor fish; clusters 1, 5, 47, and 53 were from cockles, and clusters 3, 4, 11, 21, and 57 were unique to the queen scallop microflora. A more diverse microbial population was isolated from king scallops, with their unique bacteria falling into clusters 12 to 16, 20, 22 to 28, 32, 46, 48, and 56 (Fig. 2 and Table 1). Bacteria forming cluster 18 were common to all molluscs sampled except queen scallops, but these bacteria were present at vastly different densities, ranging from 0.56% in cockles to ca. 37% of the culturable population in mussels.
TABLE 1.
Enumeration of molluscan bacteria based on carbon utilization profiles generated by BIOLOG
| Molluscan sample | Cluster associated with molluscan samples (log10 no. of isolates/g of shellfish flesh) | Total no. of bacteria (log10)/g of shellfish flesh |
|---|---|---|
| Mussels | 7 (3.6); 8 (4.6); 9 (2.5); 10 (2.5); 18 (4.6); 19 (4.0); 29 (2.4); 35 (2.1); 36 (2.1); 38 (2.1); 40 (3.6); 42 (3.7); 43 (3.4); 44 (2.3); 45 (2.3) | 5.1 |
| Oysters | 2 (2.1); 18 (3.0); 29 (4.6); 30 (4.2); 31 (4.2); 36 (2.3); 37 (2.3); 38 (2.6); 39 (3.6); 41 (2.1); 50 (2.5); 51 (2.5); 52 (1.9) | 4.9 |
| Razor fish | 6 (1.9); 17 (1.9); 18 (2.4); 33 (1.9); 42 (2.7); 49 (2.7); 50 (1.7); 55 (4.3) | 4.3 |
| Cockles | 1 (3.4); 5 (4.6); 18 (2.7); 33 (3.6); 42 (3.6); 43 (1.9); 47 (3.6); 49 (3.0); 50 (2.6); 53 (3.5); 54 (2.9) | 4.7 |
| Queen scallops | 3 (4.5); 4 (4.0); 11 (5.11); 21 (4.2); 29 (4.0); 42 (4.0); 45 (3.9); 52 (5.3); 54 (3.9); 57 (4.0) | 5.6 |
| King scallops | 12 (3.8); 13 (4.0); 14 (3.4); 15 (4.0); 16 (4.2); 18 (4.3); 20 (4.0); 22 (3.9); 23 (4.9); 24 (4.3); 25 (4.6); 26 (4.0); 27 (4.0); 28 (4.0); 32 (4.0); 36 (4.0); 46 (4.5); 48 (4.5); 56 (4.0) | 5.5 |
FIG. 2.
Cluster analysis using carbon utilization profiles (BIOLOG-GN) of the microflora associated with bivalve shellfish. The dendrogram was generated using average-linkage clustering.
Toxin utilization by pure cultures isolated from shellfish.
Selected bacterial isolates from clusters demonstrating 90% similarity were screened for their ability to utilize PST using 96-well BIOLOG-MT microplates. These plates contain a minimal salts solution plus an indicator dye with no additional carbon and energy source, as supplied by BIOLOG. Hence, a carbon and energy source must be added to cause a metabolic reaction and subsequent color change (12). In these experiments, a range of carbamate and carbamoyl PST were used. The results shown in Fig. 3 are expressed as percentage increases in optical density in wells containing bacterial suspensions plus PST compared to the optical density in control wells containing bacterial suspensions without the addition of toxins. The addition of toxin standards alone to BIOLOG-MT plates did not cause any color reaction.
FIG. 3.
Utilization of PST by shellfish bacteria determined using BIOLOG-MT microplates. The final concentrations (in nanograms per microliter) of the toxins in 100 μl were as follows: GTX 1, 0.74; GTX 4, 0.32; GTX 2, 1.20; GTX 3, 0.29; STX, 1.40; NEO, 1.40; and GTX 5, 1.11 when 1 μl of toxin was added to the wells (indicated by 1* and 1 under the bars) and double this amount with the addition of 2 μl (indicated by 2). The sources of the bacterial isolates are indicated by the prefix in the designation as follows: C for cockles, R for razor fish, M for mussels, Q for queen scallops, and K for king scallops. Results are mean values from two experiments.
Not all isolates tested grew at the expense of PST in BIOLOG-MT plates. Bacterial isolates exhibiting the greatest growth enhancement were C3, R69, M12, Q5, R78, K9, M7, M11, and R65, which were obtained from clusters 1, 6, 10, 11, 18, 28, 29, 36, and 49, respectively, ostensibly using a wide range of different toxins (Fig. 3).
In some cases, enhanced reactions were noted where lower concentrations of toxins had been added, while higher concentrations led to inhibition in several wells. For example, M11, which utilizes GTX 1/4, showed nearly an 100% increase in optical density at lower concentrations and only a 45% increase when higher concentrations of toxins were added. In contrast, addition of higher concentrations of GTXs to cultures of Q5, C3, and R65 demonstrated heightened color reactions.
Analysis of bacterial transformation of PST by HPLC.
To determine possible reactions occurring in BIOLOG-MT plates, bacteria showing the greatest increases in absorbancy (i.e., above 50%) (Fig. 3) with individual toxin standards in BIOLOG-MT plates were incubated either without any added toxin or with PST standards in marine broth and toxin profiles were monitored over time by HPLC.
HPLC analysis of bacterial supernatants which were incubated in the absence of toxins did not show the presence of quantifiable amounts of toxins. However, a peak at ca. 1.5 min (U1) was consistently observed in all cultures, regardless of whether PST were added to the cultures. In some cases, this peak increased when bacteria were incubated with PST (see Fig. 4 to 6), indicating the possible presence of C toxins (10).
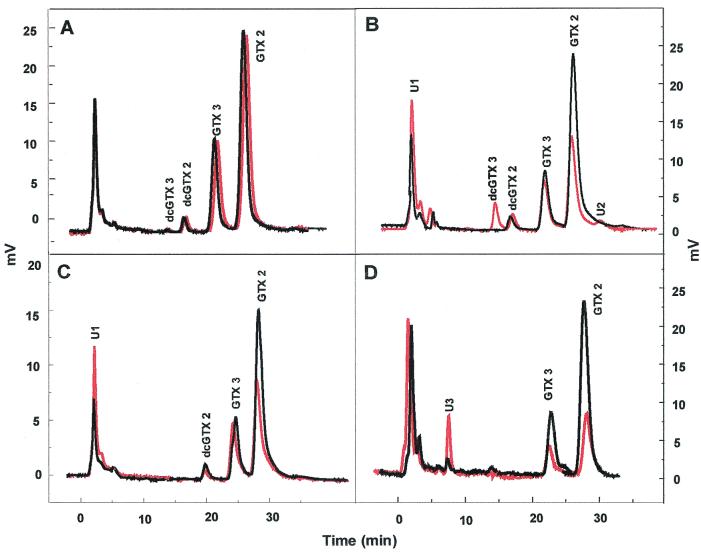
FIG. 4.
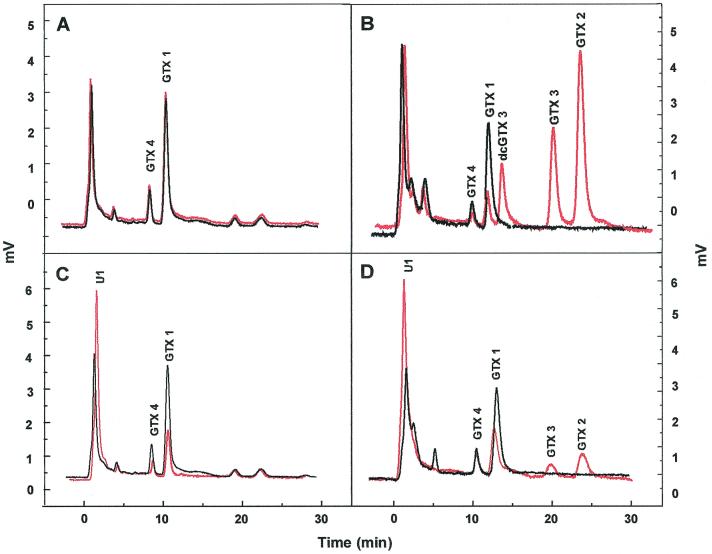
Chromatograms showing utilization of GTX 1/4 by bacterial isolates. (A) Control (marine broth plus toxin), (B) M12 (cluster 10), (C) Q5 (cluster 11), and (D) R65 (cluster 40). Samples shown were taken at 0 h (black lines) and 144 h (red lines).
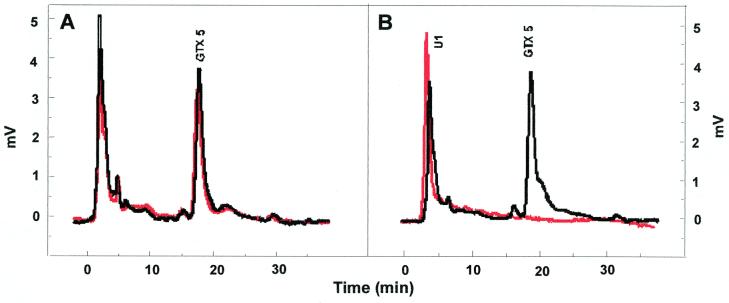
FIG. 6.
Chromatograms showing utilization of GTX 5 by bacteria. (A) Control and (B) C3 (cluster 1). Samples shown were taken at 0 h (black lines) and 144 h (red lines).
To ascertain the efficacy of the BIOLOG-MT plates, randomly selected isolates showing less than 50% increases in absorbancy were also examined, these bacterial isolates did not show any significant differences in toxin profiles by HPLC (data not shown), suggesting that these bacteria were not metabolically active in the BIOLOG-MT plates. Of the isolates showing increases above 50% (Fig. 3), changes in toxin profiles were generally observed. Exceptions to this were isolates K9 incubated with GTX 2/3 and 5, R69 with GTX 5, M11 with GTX 1/4 and all bacterial isolates grown with addition of STX and NEO which did not show any reduction in levels of these toxins below the control samples (Table 2).
TABLE 2.
Utilization rates of PST by molluscan bacteria
| Representative isolate | Utilization rate (pg h−1 107 cells−1)a
|
||||||
|---|---|---|---|---|---|---|---|
| GTX 1 | GTX 4 | GTX 2 | GTX 3 | GTX 5 | STX | NEO | |
| C3 (cluster 1) | − | − | − | − | 92.5 ± 3.35 | + | − |
| R69 (cluster 6) | − | − | − | − | + | − | + |
| M12 (cluster 10) | 10.42 ± 1.44 | 0.83 ± 0.06 | 19.21 ± 1.01 | 0.42 ± 0.02 | − | − | − |
| Q5 (cluster 11) | 8.75 ± 1.22 | 2.40 ± 0.45 | 12.36 ± 0.83 | 0.56 ± 0.12 | − | − | + |
| R78 (cluster 18) | − | − | − | − | − | − | + |
| K9 (cluster 28) | − | − | + | + | + | − | + |
| M7 (cluster 29) | − | − | 60.00 ± 3.33 | 14.60 ± 1.57 | − | − | + |
| M11 (cluster 36) | + | + | − | − | − | − | − |
| R65 (cluster 49) | 0.63 ± 0.09 | + | − | − | − | − | − |
Rates are determined after 48 h of incubation for all isolates except C3 and M7, which were determined after 24 h of incubation. Results are the means ± standard errors of the means for three experiments. Symbols: −, no utilization as determined by BIOLOG-MT microplates; +, isolates demonstrated a 50% or greater increase in absorbance above control wells in BIOLOG-MT microplates.
Biotransformations of GTX 1/4.
Quantitative assessment of control samples containing marine broth and GTX 1/4 showed that high percentages (ca. 98%) of these toxins were recovered after 144 h (Fig. 4A).
The main products of GTX 1/4 breakdown by M12 were GTX 3 and 2 with accumulation of a product corresponding to decarbamoyl GTX 3 (dcGTX 3) after 24 h. These products could be seen clearly as separate peaks in time sequence HPLC chromatograms after 144 h (Fig. 4B). Reduction of GTX 1 in the cultures was fast (65% decrease overall), while utilization of GTX 4 was much slower (Table 2). Overall, ca. 45% of the net accumulated GTX 1/4 was transformed to GTX 2/3.
Isolate Q5 again demonstrated faster utilization of GTX 1 (42% decrease) than GTX 4 (Table 2). However, with this isolate there was no concomitant increase in other GTX analogues in any of the time interval samples. The initial peak (U1) observed at 0 h, however, increased by ca. 59% after 144 h (Fig. 4C).
Isolate R65 assimilated GTX 1 at a much lower rate than M12 and Q5, with slight increases in GTX 2 and 3 observed. However, a 67% increase in U1 was detected (Table 2). Levels of GTX 4 were not reduced over the time period examined (Fig. 4D).
Biotransformations of GTX 2/3.
In control samples containing marine broth and GTX 2/3, toxin profiles did not alter after 144 h, with ca. 97% of these toxins being recovered (Fig. 5A).
FIG. 5.
Chromatograms showing utilization of GTX 2/3 by bacterial isolates. (A) Control, (B) M12 (cluster 10), (C) Q5 (cluster 11), and (D) M7 (cluster 29). Samples shown were taken at 0 h (black line) and 144 h (red lines).
Incubations of bacterial isolate M12 spiked with GTX 2/3 showed rapid utilization of GTX 2 (38% decrease overall), with only negligible (3%) decreases in the amounts of GTX 3 observed (Table 2 and Fig. 5B). After 24 h of incubation, a peak corresponding to dcGTX 3 appeared, with slight increases in dcGTX 2 and the initial peak (U1). After 48 h of incubation, an additional small peak (U2) with a retention time of 31.5 min was detected. With continued incubation, no other change in toxin profile was seen (Fig. 5B).
Isolate Q5 also demonstrated rapid utilization of GTX 2 (37% decrease overall) with slower degradation of GTX 3 (Table 2). However, there was no concomitant increase in other GTX analogues in any of the samples. Higher yields of U1 were again observed after 72 h and remained stable to 144 h (Fig. 5C).
Approximately 60% of available GTX 2 and 3 was utilized after only 24 h by isolate M7 at accelerated rates (Table 2). Accumulation of an unknown compound (U3) with a retention time of 9.5 min was apparent after 6 h of incubation (Fig. 5D).
Degradation of GTX 5.
Control samples did not show any changes in toxin profiles, with ca. 96% recovery of GTX 5 after 144 h (Fig. 6A). GTX 5 degradation by isolate C3 was very rapid (ca. 93 pg h−1 107 cells−1) (Table 2), and after only 24 h, no residual toxin was detected. After 48 h, an increase in the initial peak U1 was observed which remained stable over the time period examined (144h) (Fig. 6B). No other bacterial isolates under these experimental conditions were capable of GTX 5 degradation.
DISCUSSION
There have been many reports that toxin profiles of contaminated shellfish vary considerably from those of the causative dinoflagellate (1, 4, 5, 9, 20, 25, 26), indicating that toxin transformation occurs during accumulation and depuration processes in shellfish. Changes in toxin profiles of shellfish tissues may arise from selective retention or elimination of individual toxins, by epimerization, or by a variety of enzymatic conversions performed by the molluscs (6, 8, 24). Bacteria resident in shellfish digestive systems are also prime choices to mediate biotransformations due to their diverse metabolic capacities (16, 29). Hence, in this study, the microflora resident in different molluscan shellfish were investigated to determine the pertinence of bacterial PST metabolism and to try to deduce reactions which may be involved.
From our data, it would appear that the shellfish examined possessed varied bacterial floras. The bacteria isolated are representative of the proportion of bacteria which can be cultivated on marine agar and cannot be described as the native microflora. However, it would appear that each shellfish examined had a unique flora based on its metabolic capacity to utilize the carbon sources supplied in BIOLOG-GN plates (Table 1).
When these shellfish bacteria were exposed to PST in BIOLOG-MT plates, endogenous metabolism of many of the bacteria tested was inhibited, as shown by the absence of a change in the color. A positive color reaction observed in BIOLOG-MT plates suggests that the toxins had been used as a carbon or energy source by the bacteria, with possible transformation of the toxins into more or less noxious compounds. In some cases, enhanced color reactions were noted where lower concentration of toxins were added, while higher concentrations of the toxins led to inhibition in several wells (Fig. 3).
We have not yet tested the possibility of cometabolism of PST in the presence of other specific substrates; other substrates may serve as inducers, thereby enhancing degradation of PST. It is also feasible that enzymes responsible for PST transformations are susceptible to catabolite repression and that degradative activity will not develop until other carbon sources have been exhausted, as observed in the degradation of the cyanobacterial hepatotoxin, microcystin, by aquatic bacteria (15). This may in part explain why some of our isolates appeared to be metabolically active in BIOLOG-MT plates but did not demonstrate the ability to transform PST by HPLC analysis when grown in nutrient-rich marine broth, as other carbon sources may not have been exhausted (Table 2).
It is feasible that shellfish contaminated with PST provide natural enrichment cultures for bacteria possessing the ability to utilize PST. Stewart et al. (30), studying microbial utilization of the neurotoxin domoic acid, noted that the metabolic capacity for this toxin was higher in bacteria isolated from blue mussels than those found in sea scallops, which are notorious for retaining toxins, suggesting that stored domoic acid is generally made unavailable in its digestive gland and hence is not available to bacteria following entry into the digestive system. He concluded that the relationship between the presence and absence of domoic acid-utilizing bacteria and the capacities for the clearance of domoic acid by different shellfish must be inextricably linked. All six shellfish species we examined are suspension-type filter feeders and are known to have different capacities for accumulating and retaining PST (F. G. Howard, personal communication). It is therefore conceivable that these shellfish will select and enrich for growth of bacterial species with various capacities for PST metabolism.
Using HPLC to monitor utilization of PST in this study, it has been possible to demonstrate potential biotransformation mechanisms. These mechanisms include reductive elimination, demonstrated by de novo appearance of GTX 2 and 3 following incubation with GTX 1/4 by bacterial isolates M12 and R65 (Fig. 4B and D). Previously, Kotaki et al. (17–19) demonstrated transformation of GTX 1, 2, and 3 and NEO to STX via reactions of reductive elimination of C-11 hydroxysulfate (OHSO3) and N-1 hydroxyl (OH) moieties by bacteria isolated from mussels, Vibrio and Pseudomonas spp. It was suggested that in these reactions, reduction of N-1 OH proceeded faster than that of C-11 OHSO3, as NEO was not detected in the reaction mixtures. It was also suggested that reductive eliminations proceeded much faster under anaerobic conditions than those under aerobic conditions, paralleling the reductive nature of the reaction. In our study, we did not detect NEO or STX in samples incubated aerobically with GTX 1/4 analogues. Furthermore, when isolate M12 was incubated with GTX 2/3, reduction of C-11 OHSO3 did not give rise to STX, which supports these earlier findings. However, recently the activity of bacterial extracts to transform GTXs to STXs was found to be due to the presence of the natural reductant glutathione (28). HPLC analysis of isolates showing increased reactions in BIOLOG-MT plates with NEO (R69, Q5, R78, K9, and M7) did not detect STX, which would arise by the reductive elimination of N-1 OH. Exposure of isolate C3 to STX alone did not result in the formation of any other PST analogues. Kotaki and colleagues (18, 19) judged NEO to be more resistant than GTX 1 to bacterial transformation despite the possession of N-1 OH.
Other possible mechanisms observed in this study include the decarbamoylation of GTX 3 at N-21, as shown by an increase in a peak corresponding to dcGTX 3 from incubations of bacterial isolate M12 with GTX 2/3 and GTX 1/4 (Fig. 4B and 5B). dcGTX 3 and 2 are minor contaminants in GTX 2/3 standards supplied by the CNRC, and in our chromatograms, increased peaks are observed at the same retention time as that of the dcGTX 3 contaminants (Fig. 4B and 5B). As such, we can only speculate on the presence of dcGTX3 in these cultures and cannot at this stage demonstrate stoichiometric responses.
Other bacterial activity observed may involve sulfation of GTX 2/3 and GTX 1/4 at N-21, giving rise to C 1/C 2 and C 3/C 4, respectively. However, at this point we can only speculate about the formation of C toxins from GTXs in our samples, because by using the isocratic HPLC methodology of Franco and Fernández-Vila (10), the more polar C toxins are known to elute in the region where we observed increased levels of U1 (Fig. 4C and D, 5B and C, and 6B). Toxins having an N-sulfocarbamoyl moiety are converted to carbamate counterparts when heated at low pH (10), and verification of production of C toxins by our bacterial isolates by converting them into their GTX derivatives is currently in progress. In animals and microorganisms, sulfotransferases play an important role in the detoxification of bioactive compounds (22, 27), and it is possible that bacterial sulfotransferases have the capacity to sulfate STX analogues, transforming them into less toxic sulfocarbamoyl derivatives. Sulfotransferase activity has been implicated with respect to the production of sulfocarbamoyl toxins in toxic strains of the dinoflagellate G. catenatum and Alexandrium catenella (32, 33) with the formation of GTX 5, C1, and C2 from STX, GTX 2, and GTX 3, respectively, following incubation with 3′-phosphate-5′-phosphosulfate. Interestingly, N-sulfotransferase activity has been detected in both toxic and nontoxic G. catenatum (24). None of our bacterial isolates showing PST transformation demonstrated the ability to autonomously produce PST, indicating that the enzymes related to toxin transformation in dinoflagellates and bacteria may not link directly to synthesis of the STX skeleton.
Incubation of dialyzed extracts of dinoflagellates with different toxins has shown that A. tamarense can transform GTX 2 and 3 to GTX 1 and 4, indicating the presence of an oxidase (24). This route of PST transformation was not, however, demonstrated by any bacterial isolates in this study.
In these experiments, reaction rates as well as reaction products varied significantly between bacterial isolates (Table 2 and Fig. 4 to 6). As bacterial species and their population densities may differ among shellfish, bacterial toxin conversions in the digestive systems of shellfish will undoubtedly be more variable than shown by the in vitro experiments in this study and will be dependent on many other factors, including the rate of PST accumulation and retention and detoxifying enzymes possessed by individual shellfish species. However, our experiments demonstrate that the microflora of a variety of shellfish possess adequate enzymatic capacity to carry out extensive side chain modifications of PST, yet the biological role of these transformations remains unclear. Bacteria may play a significant part in PST elimination in molluscan species; as such, possible practical applications are feasible in terms of manipulating the bacterial flora of shellfish to depurate shellfish of these toxins.
ACKNOWLEDGMENTS
We thank Robert Fryer for assistance with the cluster analysis program used in this study and Matt Gubbins for useful discussions.
This work was supported in part by funds from Fisheries Research Services (United Kingdom) and by a grant from the European Union (FAIR CT96–1558).
REFERENCES
- 1.Asakawa M, Miyazawa K, Takayama H, Noguchi T. Dinoflagellate Alexandrium tamarense as the source of paralytic shellfish poison (PSP) contained in bivalves from Hiroshima Bay, Hiroshima Prefecture, Japan. Toxicon. 1995;33:691–697. doi: 10.1016/0041-0101(94)00177-a. [DOI] [PubMed] [Google Scholar]
- 2.Baden G D, Trainer V L. Mode of action of toxins of seafood poisoning. In: Falconer I R, editor. Algal toxins in seafood and drinking water. London, United Kingdom: Academic Press, Ltd.; 1993. p. 4974. [Google Scholar]
- 3.Bochner B. Breathprints at the microbial level. ASM News. 1989;55:536–539. [Google Scholar]
- 4.Bricelj V M, Lee J H, Cembella A D. Influence of dinoflagellate cell toxicity on uptake and loss of paralytic shellfish toxins in the northern quahog Mercenaria mercenaria. Mar Ecol Prog Ser. 1991;74:33–46. [Google Scholar]
- 5.Bricelj V M, Lee J H, Cembella A D, Anderson D M. Uptake kinetics of paralytic shellfish toxins from the dinoflagellate Alexandrium fundyense in the mussel Mytilus edulis. Mar Ecol Prog Ser. 1990;63:177–188. [Google Scholar]
- 6.Bricelj V M, Shumway S E. Paralytic shellfish toxins in bivalve molluscs: occurrence, transfer kinetics, and biotransformations. Rev Fish Sci. 1998;6:315–383. [Google Scholar]
- 7.Cembella A D. Ecophysiology and metabolism of paralytic shellfish toxins in marine microalgae. NATO ASI Ser Ecol Sci. 1998;41:381–403. [Google Scholar]
- 8.Cembella A D, Shumway S E, Larocque R. Sequestering and putative biotransformation of paralytic shellfish toxins by the sea scallop Placopecten magellanicus: seasonal and spatial scales in natural populations. J Exp Mar Biol Ecol. 1994;180:1–22. [Google Scholar]
- 9.Cembella A D, Shumway S E, Lewis N I. Anatomical distribution and spatio-temporal variation in paralytic shellfish toxin composition in two bivalve species from the Gulf of Maine. J Shellfish Res. 1993;12:389–403. [Google Scholar]
- 10.Franco J M, Fernández-Vila P. Separation of paralytic shellfish toxins by reversed phase high performance liquid chromatography, with postcolumn reaction and fluorimetric detection. Chromatographia. 1993;35:613–620. [Google Scholar]
- 11.Gallacher S, Flynn K J, Franco J M, Brueggemann E E, Hines H B. Evidence for production of paralytic shellfish toxins by bacteria associated with Alexandrium spp. (Dinophyta) in culture. Appl Environ Microbiol. 1997;63:239–245. doi: 10.1128/aem.63.1.239-245.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon R W, Hazen T C, Fliermans C B. Rapid screening for bacteria capable of degrading toxic organic compounds. J Microbiol Methods. 1993;18:339–347. [Google Scholar]
- 13.Hallegraeff G M. Harmful algal blooms: a global overview. In: Hallegraeff G M, Anderson D M, Cembella A D, editors. Manual on harmful marine microalgae. IOC manual and guide no. 33. Paris, France: United Nations Educational, Scientific, and Cultural Organization; 1995. pp. 1–22. [Google Scholar]
- 14.Jay J M. Modern food microbiology. New York, N.Y: Van Nostran Reinhold Ltd.; 1970. pp. 267–268. [Google Scholar]
- 15.Jones G J, Bourne D G, Blakeley R L, Doelle H. Degradation of the cyanobacterial hepatotoxin microcystin by aquatic bacteria. Nat Toxins. 1994;2:228–235. doi: 10.1002/nt.2620020412. [DOI] [PubMed] [Google Scholar]
- 16.Koch A L. The macroeconomics of bacterial growth. In: Fletcher M, Floodgate G D, editors. Bacteria in their natural environments. London, United Kingdom: Academic Press, Ltd.; 1985. pp. 1–42. [Google Scholar]
- 17.Kotaki Y. Screening of bacteria which convert gonyautoxin 2,3 to saxitoxin. Nippon Suisan Gakkaishi. 1989;55:1239. [Google Scholar]
- 18.Kotaki Y, Oshima Y, Yasumoto T. Bacterial transformation of paralytic shellfish toxins. In: Anderson D M, White A W, Baden D G, editors. Toxic dinoflagellates. New York, N.Y: Elsevier Science Publishers; 1985. pp. 287–292. [Google Scholar]
- 19.Kotaki Y, Oshima Y, Yasumoto T. Bacterial transformation of paralytic shellfish toxins in coral reef crabs and a marine snail. Bull Jpn Soc Sci Fish. 1985;51:1009–1013. [Google Scholar]
- 20.Lassus P, Bardouill M, Massselin P, Naviner P, Truquet P. Comparative efficiencies of different non-toxic microalgal diets in detoxification of PSP-contaminated oysters (Crassoatrea gigas Thunberg) J Nat Toxins. 2000;9:1–12. [PubMed] [Google Scholar]
- 21.Lassus P, Ledoux M, Bardouill M, Bohec M. Influence of initial toxicity and extraction procedure on paralytic toxin changes in the mussel. Toxicon. 1992;31:237–242. doi: 10.1016/0041-0101(93)90142-6. [DOI] [PubMed] [Google Scholar]
- 22.Lee N-S, Kim B-T, Kim D-H, Kobashi K. Purification and reaction mechanism of arylsulfate sulfotransferase from Haemophilus K-12, a mouse intestinal bacterium. J Biochem. 1995;118:796–801. doi: 10.1093/oxfordjournals.jbchem.a124982. [DOI] [PubMed] [Google Scholar]
- 23.Oshima Y. Postcolumn derivatization liquid chromatographic method for paralytic shellfish toxins. J AOAC Int. 1995;78:528–532. [Google Scholar]
- 24.Oshima Y. Chemical and enzymatic transformation of paralytic shellfish toxins in marine organisms. In: Lassus P, Arzul G, Gentien P, Marcaillou C, editors. Harmful marine algal blooms. Paris, France: Lavoisier Publishers; 1995. pp. 475–480. [Google Scholar]
- 25.Oshima Y, Fallon W E, Shimizu Y, Noguchi T, Hashimoto Y. Toxins of the Gonyaulax sp. and infested bivalves in Owase Bay. Bull Jpn Soc Sci Fish. 1976;42:851–856. [Google Scholar]
- 26.Oshima Y, Sugino K, Itakura H, Hirota M, Yasumoto T. Comparative studies on paralytic shellfish toxin profile of dinoflagellates and bivalves. In: Granéli E, Sundström B, Edler L, Anderson D M, editors. Toxic marine phytoplankton. New York, N.Y: Elsevier Science Publishers; 1990. pp. 391–396. [Google Scholar]
- 27.Roy A B. Sulfotransferases. In: Mulder G J, editor. Sulfation of drugs and related compounds. Boca Raton, Fla: CRC Press; 1981. pp. 131–185. [Google Scholar]
- 28.Sakomoto S, Sato S, Ogata T, Kodama M. Formation of intermediate conjugates in the reductive transformation of gonyautoxins to saxitoxins by thiol compounds. Fish Sci. 2000;66:136–141. [Google Scholar]
- 29.Stanier R Y, Doudoroff M, Adelberg E A. The microbial world. 2nd ed. Englewood Cliffs, N.J: Prentice-Hall Inc.; 1963. [Google Scholar]
- 30.Stewart J E, Marks L J, Gilgan M W, Pfeiffer E, Zwicker B M. Microbial utilization of the neurotoxin domoic acid: blue mussels (Mytilus edulis) and soft shell clams (Mya arenaria) as sources of the microorganisms. Can J Microbiol. 1998;44:456–464. [PubMed] [Google Scholar]
- 31.Sullivan J J, Iwaoka W T, Liston J. Enzymatic transformation of PSP toxins in the littleneck clam (Protothaca staminea) Biochem Biophys Res Commun. 1983;114:465–472. doi: 10.1016/0006-291x(83)90803-3. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida T, Sako Y, Kakutani T, Fujii A, Uchida A, Ishida Y, Arakawa O, Noguchi T. Comparative study of two sulfotransferases for sulfation to N-21 of Gymnodinium catenatum and Alexandrium catenella toxins. In: Reguera B, Blanco J, Fernández M L, Wyatt T, editors. Harmful algae. Grafisant, Spain: Xunta de Galicia and IOC, United Nations Educational, Scientific, and Cultural Organization; 1998. pp. 366–369. [Google Scholar]
- 33.Yoshida T, Sako Y, Uchida A, Ishida Y, Arakawa O, Noguchi T. Purification and properties of paralytic shellfish poisoning toxins sulfotransferase from toxic dinoflagellates Gymnodinium catenatum. In: Yasumoto T, Oshima Y, Fukuyo Y, editors. Harmful and toxic algal blooms. 1996. pp. 499–502. IOC, United Nations Educational, Scientific, and Cultural Organization, Grafisant, Spain. [Google Scholar]