Summary
In this article, human semen samples from the Visem dataset are automatically assessed with machine learning methods for their quality with respect to sperm motility. Several regression models are trained to automatically predict the percentage (0–100) of progressive, non-progressive, and immotile spermatozoa. The videos are adopted for unsupervised tracking and two different feature extraction methods—in particular custom movement statistics and displacement features. We train multiple neural networks and support vector regression models on the extracted features. Best results are achieved using a linear Support Vector Regressor with an aggregated and quantized representation of individual displacement features of each sperm cell. Compared to the best submission of the Medico Multimedia for Medicine challenge, which used the same dataset and splits, the mean absolute error (MAE) could be reduced from 8.83 to 7.31. We provide the source code for our experiments on GitHub (Code available at: https://github.com/EIHW/motilitAI).
Subject areas: Health sciences, Medicine, Reproductive medicine, Bioinformatics, Biocomputational method, Artificial intelligence
Graphical abstract
Highlights
-
•
Improvements to state of the art in automatic human sperm motility prediction
-
•
Unsupervised feature quantization used with off-the-shelf tracking algorithms
-
•
Framework publicly available on GitHub: https://github.com/EIHW/motilitAI
Health sciences; Medicine; Reproductive medicine; Bioinformatics; Biocomputational method; Artificial intelligence
Introduction
eHealth or “the use of information and communications technology in support of health and health-related fields” (World Health Organization, 2017) has been a prioritized item on the agenda of the World Health Organization (WHO) since 2005 (World Health Organization, 2005a). From then until 2016, the percentage of WHO member states that have a national eHealth policy in place has risen to (World Health Organization, 2016). eHealth is further considered an important factor for improving both the quality and availability of affordable health care, moving countries closer toward achieving universal health coverage (World Health Organization, 2005b).
One such issue can, for example, be found with fertility-related problems (Yee et al., 2013). Across the globe, approximately 8%–12% of couples are affected by infertility (Stephen and Chandra, 1998; Kumar and Singh, 2015) which is defined as the inability to achieve a clinical pregnancy after 12 or more months of regular unprotected sexual intercourse (Zegers-Hochschild et al., 2009; Practice Committee of the American Society for Reproductive Medicine, 2008). The issue can be a result of both male and female factor infertility (Kumar and Singh, 2015). In males, infertility is often related to deficiencies in sperm quality measured by characteristics and reference values defined by the WHO (Cooper et al., 2010). The attributes most strongly associated with fertility can be found in the concentration, motility, and morphology of sperm (Kumar and Singh, 2015). The analysis of these characteristics can serve as a valuable baseline for diagnosis and treatment of patients but requires either specialized, expensive medical equipment or manual annotation by trained medical staff (David et al., 1981; Mortimer et al., 2015). In this respect, machine learning approaches that use video recordings of semen (the seminal fluid which contains the sperm) samples to detect morphology and motility of the included spermatozoa could assist physicians in their work. To work toward this goal, the Visem dataset (Haugen et al., 2019) collected and released by the Simula Research Laboratory contains microscopic recordings of semen samples which are additionally annotated with regards to the mentioned characteristics of spermatozoa quality. In our work, we make use of the Visem dataset. It is a freely available dataset on which a range of state-of-the-art machine learning methods have been applied facilitating a better comparison of our framework’s efficacy with a wide variety of methods.
For this paper, a novel combination of unsupervised tracking, feature extraction and quantization methods, and machine learning models is investigated to perform automatic analysis of the motility of recorded spermatozoa cells. Motility means observing the speed and way of movement of sperm, i.e., if they travel on a straight path or in a circle. Furthermore, before extracting features, the data from the Visem dataset are preprocessed to minimize the negative impact that might come from blurred camera settings and numerous cuts within each video. The effectiveness of the applied feature extraction methods and machine learning models are compared to the approaches provided by the data organizers and state-of-the-art deep learning-based methodologies from various research groups. These contributions have been submitted to the Medico: Multimedia for Medicine sub-challenge (Hicks et al., 2019b) that was part of the 2019 edition of the Medieval challenge.
The remainder of this paper is structured as follows. The proceeding section reviews the related work on computer-aided sperm analysis (CASA), and more specifically on automated prediction of sperm motility. In “sec:dataset”, we describe the dataset. “sec:expset” follows with the illustration of our approach, including preprocessing, particle tracking algorithms, feature extraction, and machine learning models. All accomplished results are listed in “sec:results” and discussed in “sec:discussion”. Finally, we give a conclusion and suggestions for future work in “sec:conclusion”.
Related work
Sperm motility characteristics have been defined in the official WHO lab manual (World Health Organization, 1999). The motility of sperm cells can further be analyzed by classifying them according to multiple categories (Björndahl et al., 2010). Spermatozoa can either be immotile or motile, where for motile sperm, further categorization can be applied. A particular cell is motile if its tail is beating. The beating of the tail alone, however, does not translate to effective movement. Therefore, motile sperm cells are additionally grouped according to the progressivity of their movement. Non-progressive spermatozoa beat their tails without any net gain in space, whereas progressive cells do gain space in the process (Björndahl et al., 2010).
Traditionally, these characteristics had to be assessed manually by trained clinical staff (Mortimer et al., 2015; David et al., 1981), but advancements in computational hardware led to the introductions of CASA (Mortimer, 1990). CASA works well for many non-human species (Van der Horst et al., 2009; Lueders et al., 2012), but has traditionally struggled with the accurate assessment of male fertility characteristics from microscopic video recordings of human sperm cells (Björndahl et al., 2010). This discrepancy is caused both by biological as well as technical limitations (Mortimer, 1994; Mortimer and Mortimer, 1998; Mortimer et al., 1995). First of all, from a biological perspective, human sperm has many characteristics that are detrimental to automatic analysis, such as high amounts of debris particles, generally lower sperm motility and concentration, and many dead spermatozoa which are often also clumped together (Mortimer et al., 2015). As a consequence, while progressive movement can be detected quite accurately, non-progressive motile sperm cells are very hard to automatically differentiate from drifting debris or dead spermatozoa (Mortimer et al., 1995). Furthermore, the clumping of alive cells with debris or dead spermatozoa can negatively affect the automatic tracking, leading to missing and interrupted tracks (Mortimer et al., 2015). Morphology is especially hard to assess by commercial CASA systems, as accurate analysis is only possible for sperm heads (Mortimer and Mortimer, 1998). Especially for motility, the CASA systems base their analysis on computing various kinematic statistics about each sperm track and then using those for determining progressive and non-progressive motility based on agreed-upon rules and thresholds (Mortimer et al., 2015). Therefore, advancements made for these systems are mainly aimed at mitigating the problems and limitations that arise from the general quality of human sperm, such as eliminating drift, recovering sperm tracks through collision, or detecting cells that are clumped together (Mortimer et al., 2015). Urbano et al. (2016), for example, implemented a robust multi-target sperm tracking algorithm that is able to effectively deal with collisions based on the joint probabilistic data association filter (Bar-Shalom et al., 2009). Hidayatullah et al. (2021) have proposed a machine learning framework for the prediction of bull sperm motility using a Support Vector Machine (SVM) classifier combined with three CASA parameters: curvilinear velocity, straight-line velocity, and linearity. The authors have demonstrated the efficacy of their approach and indicated that their method could be utilized for examining human sperm (Hidayatullah et al., 2021). Apart from the video-based CASA systems, signal processing-based machines, such as the SQA-V Gold Semen Analyzer (SQA-Vision – The Ultimate Automated Semen Analysis Solution for Hospitals, Reproductive Centers, Free Standing Labs, and Cryobanks, available at http://mes-global.com/analyzers), exist that provide more accurate results but are expensive, prohibiting their use in developing countries. Many CASA systems used in research and medical applications are closed-source, proprietary software, or integrated hardware-software solutions. However, recently, developments toward the introduction of open-source alternatives into the field have been made, e. g., with openCASA (Alquézar-Baeta et al., 2019). Furthermore, applications that solve individual parts of the automatic sperm analysis task can be found with particle tracking software, such as Trackpy (Allan et al., 2019), or motility analysis toolkits for inference of cell state (Kimmel et al., 2018).
The advancements in the field of machine learning, especially deep learning (DL) for image analysis, also made an impact on the field, leading to new possibilities for micro cinematographic approaches. Recently, Valiuškaitė et al. (2020) have applied region-based convolutional neural networks (RCNNs) to evaluate sperm head motility in human semen videos. In particular, the authors first applied a Faster R-CNN—with ImageNet (Huang et al., 2017; Deng et al., 2009) pre-trained convolutional neural networks (CNNs)—for sperm head segmentation and then used a heuristic algorithm for sperm motility calculation. Compared to the above methods, the approach taken in our work does not focus on achieving the best possible tracking accuracy. Rather, we show that through the use of unsupervised feature learning and quantization, noisy or inaccurate sperm tracks can still perform well in downstream motility prediction tasks. The tracking methods used in our experiments work off-the-shelf, i.e., they are not adapted to the particularities of a specific dataset, as would be the case when using deep neural networks (DNNs) that are trained on the database at hand. In 2019, the Medico Multimedia for Medicine challenge (Hicks et al., 2019b) presented researchers with the opportunity to develop automatic analysis systems for the assessment of human semen quality. The challenge dataset, Visem, contains 85 video recordings of semen samples which are annotated with regard to morphology and motility of the recorded sperm cells on a per-video basis. While there are only a handful of challenge submissions (Hicks et al., 2019c; Thambawita et al., 2019a, b), they all used current deep learning approaches and showed that video-based analysis can provide insight into important characteristics of spermatozoa health. For the task of motility prediction, their CNN-based models could improve significantly over both a ZeroR baseline as well as models based on traditional image features and regression algorithms (Hicks et al., 2019c, a).
More related to the methodology applied in this paper, a feature representation of textual documents from the field of Natural Language Processing, namely the Bag-of-Words model, has recently been applied to other domains. One such example can be found in the study by (Amiriparian et al., 2018; Amiriparian, 2019), where deep feature vectors are aggregated and quantized in an unsupervised fashion to form noise-robust feature representations for a number of audio analysis tasks. Similarly, Amiriparian et al. (2017) applied Bags-of-Deep-Features for the task of video sentiment analysis. In this work, a similar model is employed to generate feature representations for entire sperm samples from individual per-track movement statistics.
Results
Dataset
The data used for the experiments come from the so-called Visem dataset (Haugen et al., 2019) collected by the Simula Research Laboratory (Visem dataset available at: https://datasets.simula.no/visem/). This dataset consists of 85 videos of live spermatozoa from men aged 18 years or older. Each video has a resolution of pixels, captured with 400x magnification using an Olympus C×31 microscope and runs at 50 frames-per-second. The name of each video file is composed of the patient ID, the date of recording, a short optional description, and the code of the assessing person (e. g. 1_09.09.02_SSW). Each sample is annotated with both motility and morphology characteristics. For motility, the percentages (0–100) of progressive, non-progressive, and immotile particles are given. These values form the ground truth for the experiments conducted in this paper.
Further, the dataset includes the results of a standard semen analysis and a set of sperm characteristics, i.e., the level of sex hormones measured in the blood of the participants, levels of fatty acids in spermatozoa or of phospholipids (measured in the blood). Besides, general, anonymized study participant-related data such as age, abstinence time, and body mass index (BMI) are given by the sixth csv-file. Additionally, WHO analysis data, e.g., the ground truth for sperm quality assessment could be accessed.
The Medico Multimedia for Medicine challenge’s provided subject independent 3-fold cross-validation setup (Hicks et al., 2019b) was adopted for our experiments.
Approach and experimental settings
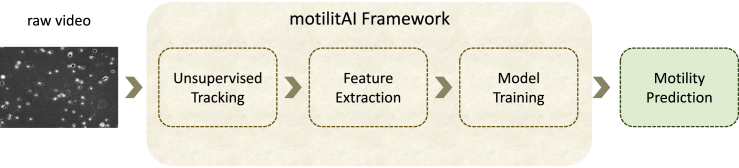
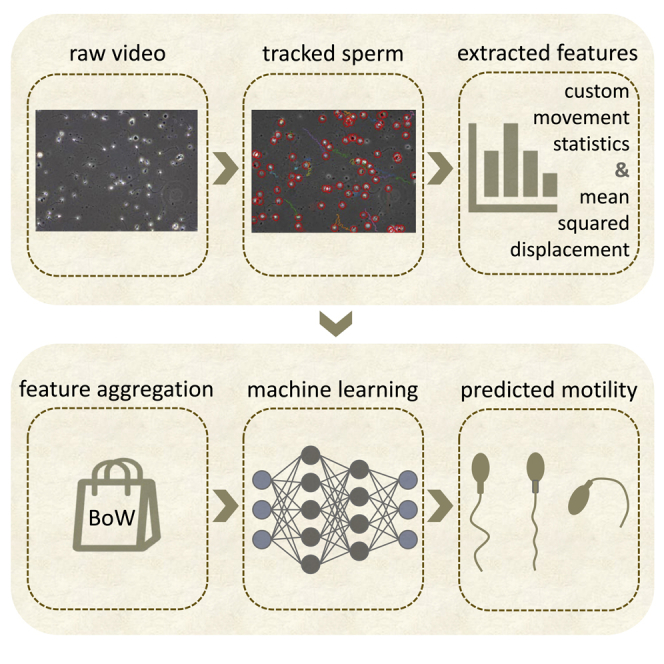
The different aspects of the overall approach of this paper are depicted in Figure 1. The videos from the dataset are preprocessed and subsequently tracking is applied to them to extract features. These features, i.e., Mean Squared Displacement (MSD) and movement statistics, are aggregated using Bag-of-Words (BoW) and, in the case of MSD, their mean values. We make use of all of the video material, collecting detected tracks and displacement features along with the whole duration of every clip, before aggregating them to video-level feature representations. Afterward, different models, i.e., a linear Support Vector Regressor (SVR), Multilayer Perceptron (MLP) regressor, CNN, and long short-term memory (LSTM) network, are trained on those features to predict motility. We systematically evaluate all combinations of feature extraction and machine learning models, as far as applicable, i.e., the BoW features that form an aggregated, sparse representation of entire video samples are not combined with recurrent neural networks (RNNs) or CNNs.
Figure 1.
Our proposed framework for motility predictions consists of the following steps
First, preprocessing is applied to the videos after which the spermatozoa are tracked. From these tracks, features are extracted in the form of custom movement statistics (CMS) and mean squared displacement (MSD). Finally, we aggregate those features and use them to train different networks for motility prediction.
Tracking
To achieve spermatozoa tracking, two different approaches are pursued. On the one hand, sparse optical flow with the Lucas-Kanade algorithm is applied for this purpose, see “sparse optical flow with Lucas-Kanade algorithm”. On the other hand, the Crocker-Grier algorithm that is used in the so-called Trackpy tool is a second method to track sperm, as can be seen, in “Crocker-Grier algorithm”. It should be noted that both of these algorithms are quite old and not tuned to the particularities of tracking spermatozoa. However, our work on the problem focuses on harnessing unsupervised representation learning to extract useful and performant features even from noise or imperfect sperm tracks.
Sparse optical flow with Lucas-Kanade algorithm
The Lucas-Kanade method falls into the latter category as a differential approach for estimating sparse optical flow (Lucas and Kanade, 1981). A basic assumption made for computing optical flow is that the brightness of the image is constant across all recorded frames, i.e., pixel intensities are merely translated according to their respective velocities between consecutive video images (Fleet and Weiss, 2006). Although this assumption rarely holds for real-world video sequences, it nevertheless works well in practice to estimate optical flow (Fleet and Weiss, 2006). The Lucas-Kanade method introduces the additional constraint that the optical flow is constant for any small subspace of the image. Together with Tomasi (Tomasi and Kanade, 1991), Kanade improved this tracking algorithm by detecting good image patches from the eigenvalues of the gradient matrix based on certain thresholds. Shi and Tomasi finally also introduced a method of filtering out bad features, by comparing affine compensated tracked image patches between non-consecutive frames, the assumption being that translation should be enough to account for dissimilarities in image patches along a detected track (Shi and Tomasi, 1994).
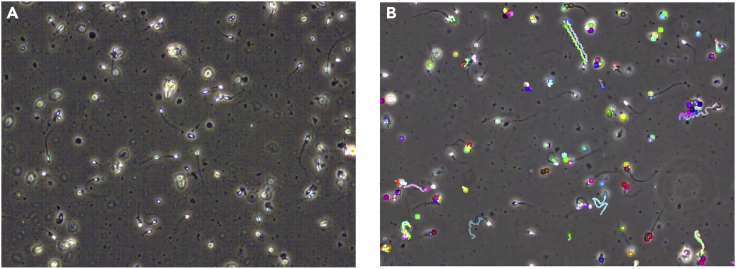
Implementations of all the components used in this tracking algorithm are available in the open-source computer vision library OpenCV (Lucas-Kanade Tracker: https://github.com/opencv/opencv/blob/master/samples/python/lk_track.py) (Bradski, 2000). To achieve better results in detecting sperm particles and their positions over time, different values for the feature detection hyperparameters maxCorners, minDistance, and blockSize are optimized to smaller values of 100, 10, and 10, respectively. An example of the sperm tracks detected by this method is visualized in “Microscopic recording of a sperm sample contained in the Visem dataset (Figure 2A). Figure 2B depicts spermatozoa tracks as detected by the Lukas- Kanade method—one of two unsupervised tracking algorithms utilized in this work.”
Figure 2.
An example frame from the Visem dataset and the result of automated sperm cell tracking
(A) Microscopic recording of a sperm sample contained in the Visem dataset.
(B) depicts spermatozoa tracks as detected by the Lukas-Kanade method—one of two unsupervised tracking algorithms utilized in this work.
However, besides identifying suitable parameter values for tracking the sperm cells, it is necessary to extract information about the sperm’s position over time to be able to compute different statistics, such as a certain sperm’s speed over a specific time interval. For this purpose, different information on every tracked sperm particle had to be computed and stored. These data include the number of the first and the last frame of a sperm’s track, the position of the sperm in every frame of the track, and the distance the sperm has moved in total. With the information stored about all sperm particles in a certain video, features describing a particular spermatozoon’s movement can be extracted. The particularities of these features will be discussed in “custom movement statistics”.
Crocker-Grier algorithm
The second tracking method employed for tracking sperm particles in the videos of the Visem dataset comes in the form of the Crocker-Grier algorithm for microscopic particle tracking and analysis in colloidal studies (Crocker and Grier, 1996). Therefore, the target application of this approach is more closely related to the task of spermatozoa tracking from video recordings. The algorithm can track colloidal spheres—Gaussian-like blobs of a certain total brightness—across frames of microscopic video recordings of particles, and consists of a number of distinct consecutive steps. First of all, geometric distortion, non-uniform contrast, and noise are alleviated by spatial warping, background removal, and filtering, respectively (Crocker and Grier, 1996; Jain, 1989; Pratt, 2013). After these preprocessing steps, candidate particle centroids can be detected by finding local brightness maxima (Crocker and Grier, 1996), e.g., computed by grayscale dilation (Jain, 1989). These maxima are further filtered by considering only those in the upper 30th percentile of brightness (Crocker and Grier, 1996), and refined according to the brightness-weighted centroids in their immediate vicinity. Afterward, particle positions can be linked probabilistically considering the dynamics of Brownian particle diffusion (Crocker and Grier, 1996), . At this stage, tracks can also be interrupted or terminated if, for example, particles leave the video frame. However, past locations are kept in memory so that relinking is possible, should the particle reappear.
The open-source python library Trackpy (Trackpy tool: https://github.com/soft-matter/trackpy) (Allan et al., 2019) provides an implementation of this algorithm and additional tools to process and extract features from particle tracks. Parameters regarding the location and linking of particles into trajectories had to be adjusted in order to improve the tracking accuracy. To reduce the hyperparameter space of our experimental pipeline, we chose to find these parameters manually by qualitative analysis of a few samples of the Visem dataset. First, an estimate of 11 pixels for the size and a minimum mass of 900 spermatozoa heads is found to lead to accurate detection of sperm cells. For linking the locations, a maximum travel distance of five pixels per frame resulted in consistent, uninterrupted tracking. Furthermore, a maximum of three frame-positions are kept in memory for cells that disappeared. Some of the detected trajectories ( frames) are too short for analysis and are therefore filtered out. As some videos contain camera drift, Trackpy’s built-in drift subtraction is applied. The drift-subtracted and filtered tracks for each video scene then served as basis for feature extraction, as will be explained in “feature extraction”.
Feature extraction
For the task of predicting motility statistics for input semen samples, features are extracted based on the spermatozoa tracks obtained with the methods described in “Tracking”. Three feature descriptors are considered in the experiments. Custom movement statistics are computed from the tracks generated with the basic Lucas-Kanade tracker, mean squared displacement vectors are extracted directly with Trackpy, and finally, a range of more involved and computationally heavier particle motility statistics is created.
Custom movement statistics
The first feature representation chosen for performing motility prediction on the dataset is constructed by computing a set of statistics from the tracks detected with the adapted Lucas-K. tracker. Based on the nature of the task at hand for which it is important to differentiate between progressive and non-progressive movement of sperm cells, both the total amount of movement by spermatozoa in particular time frames as well as the actual distances covered by them are of interest. The first aspect can be calculated for a specific window by accumulating the number of pixels a particular cell moved between each consecutive video frame while the second metric looks at the Euclidean distance between the positions of the cell at the start and end of the time window. These calculations can then be carried out for sliding windows of different sizes, and statistical functionals can be applied to their results. This leads to feature vectors of fixed length for each sperm track found by the Lucas-Kanade tracking algorithm. Specifically, these window sizes are used (measured in number of frames): 5, 10, 20, 50, 80, 100, 150, 200, 250, 300, 400, 500, 750, and 1000. After computing both metrics as described above for the whole sample by sliding each of the windows over a particular track with a hop size of one frame, mean, maximum, and minimum functionals are applied to the resulting series of motility calculations. Two additional features are computed as the total distance covered by a single sperm cell during the whole video sample and its average speed in pixels moved per frame. In total, the approach leads to numerical feature vectors of size 14 × 2 × 3 + 2 = 86 for each detected sperm track. Before being applicable to the task of motility analysis on a per-sample basis, these vectors can be further processed and aggregated per video clip. Here, two possibilities are explored. First, feature vectors of a single video sample are reduced by their feature-wise mean. Secondly, a BoW approach is applied to the vectors that both quantizes and summarizes them in an unsupervised manner.
Displacement features
A common statistical measure that is employed to characterize the random movement of particles can be found with the MSD (Frenkel and Smit, 2001). It can be used to describe the explorative behavior of particles in a system, i.e., if movement is restricted to diffusion or affected by some sort of force. The displacement of a single particle j is defined as the distance it traveled in a particular time frame of duration l (lag-time) ti to ti + l measured as the square of the Euclidean distance between its positions at the start (xj(ti)) and end (xj(ti + l)) of the frame. For a set of N particles, the ensemble mean displacement for a specific time interval can then be computed as:
| (Equation 1) |
When observing a longer period of time (T0 to T1), an average of MSD can further be computed from sliding windows of particular lag times over the whole segment. This can be done for each individual particle (mean squared displacement of each particle (imsd)) or again as an average for all of the particles (ensemble mean squared displacement of all particles (emsd)). Finally, computing these displacement values for a range of different lag times can capture more detailed information about particle movement. For the application of automated sperm motility analysis, mean squared displacement of spermatozoa in a given sample for different sized time windows could give insight into the amount of progressive and non-progressive motility. Given enough time, a progressive sperm cell would travel across a larger distance, whereas a sperm that is merely moving in place would display the same amount of displacement for both short and long time frames. Trackpy provides interfaces to compute both imsd and emsd for a range of increasing time frames. Specifically, it considers lag-times up to a user definable maximum that are increased in framewise step sizes, i.e., in the case of the Visem dataset that is recorded at 50 fps, the consecutive window sizes grow by , each. When considering a maximum lag-time of 10 s for example, 500 mean squared displacement values are computed from the sperm tracks. As emsd is computed as an aggregated measurement for all sperm cells of a given sample in a particular time frame, it can be directly used as input for the machine learning algorithms described in “regression models”. Also, imsd feature vectors, which are extracted on a per-track basis, can be further quantized and aggregated using the Bag-of-Words framework described in “Bag-of-Words” to form a clip level representation. In this article, three different combinations of window and hop sizes are considered for the extraction of emsd feature vectors: a window size of 2 s with a 1 s hop and 10 s windows with either 1 s or 5 s hops. Based on the motility prediction performance achieved using the different emsd feature configurations, hop and window sizes for imsd prediction are chosen.
Bag-of-Words
The use of unsupervised tracking algorithms allows the extraction of useful features on a more granular, per-spermatozoon basis. As the sperm cell count varies heavily between the different samples in the Visem dataset and annotations are further only available on a per-sample level, a type of feature aggregation mechanism has to be implemented to leverage per-cell information. In “Displacement Features”, regarding the mean displacement of all spermatozoa during a given time frame of a specific recording has been introduced as a first, baseline method for this problem. However, simply averaging the displacement of all cells might lead to the loss of more granular information. For this reason, a histogram representation based on the famous BoW model extended to be used with arbitrary numerical input features will be employed. For the experiments, the input feature vectors belonging to individual sperm cell tracks are first standardized to zero mean and unit variance before a random subset is chosen to form a codebook. Afterward, a fixed number of the top nearest vectors from the codebook is computed for each input feature vector. Aggregated over all sperm tracks belonging to a given sample recording, the counts of these assigned vectors form a histogram representation which is further processed by term frequencyinverse document frequency (tf-idf). Furthermore, the number of codebook vectors N and assigned vectors a is optimized by evaluating all combinations of and on the given data using different machine learning models.
Regression models
The features that are described in “feature extraction” are used as input for various machine learning approaches. The extraction methods described above lead to variable numbers of feature vectors for each original video sample, e. g., displacement vectors are extracted for overlapping windows. To enable comparisons between all implemented approaches and the methods applied by the participants of the Medico challenge, the predictions of each model are mean aggregated on a per-sample basis, i. e., each model produces a single prediction for each of the 85 patients contained in the Visem dataset. The models are outlined in the following. As metrics, we utilize both the MAE as well as the root-mean-square error (RMSE) which is more sensitive to outliers. The metrics are computed as follows:
| (Equation 2) |
| (Equation 3) |
We compute these metrics for each of the three challenge folds and present the mean of their values.
Linear support vector regressor
The first method to predict the motility of spermatozoa is a linear SVR. Here, different scaling options, i. e., StandardScaler, MinMaxScaler, and no scaler, are tested. Five distinct complexity values c equally distributed between and are evaluated. The best value for this is found by the MAE obtained in an internal 5-fold cross-validation on each fold’s training data.
Multilayer perceptron
The architecture of the MLP model contains multiple fully connected layers with batch normalization applied before the activation function. The model is trained with the Adam optimizer in order to minimize the mean squared error (MSE) and an additional L2 weight regularization term. Exponential linear unit (ELU) and rectified linear unit (ReLU) are evaluated as choices for the activation functions of the layers. A random search is performed over different parameters, including learning rate, number of layers and units per layer, batch size, and dropout that are listed in Table 1. The best parameters are determined by the MAE achieved on the random 20% validation splits of each fold’s training data.
Table 1.
All hyperparameters and their values that are optimized for the different machine learning models
| hyper-parameter | MLP | RNN | CNN |
|---|---|---|---|
| batch size | 16, 32, 64 | 16, 32, 64 | 16, 32, 64 |
| Dropout | 0.2, 0.4 | 0.2, 0.4 | 0.2, 0.4 |
| kernel regularizer | 10−4, 10−3, 10−2 | 10−4, 10−3, 10−2 | 10−4, 10−3, 10−2 |
| activation dense | ELU, ReLU | ELU, ReLU | ELU, ReLU |
| number of layers | 2, 4, 8 | 2, 4, 8 | 2, 4, 8 |
| learning rate | 10−4, 10−3, 10−2 | 10−4, 10−3, 10−2 | 10−4, 10−3, 10−2 |
| no. of units/filters | 256, 512, 1024 | 32, 64 | 64, 128, 256 |
| cell type | – | – | GRU, LSTM |
| recurrent dropout | – | – | 0, 0.2, 0.4 |
| bidirectional | – | – | true, false |
Convolutional neural network
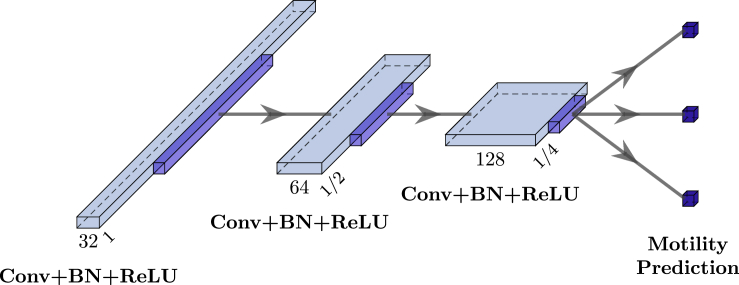
Another method used for the prediction of motility of spermatozoa is a 1-dimensional CNN. Its model architecture is constructed by multiple convolutional blocks which are stacked on top of each other. Each convolutional block consists of the following parts. First, a 1-dimensional convolutional layer with a kernel size of three and stride of one extracts features from the input. Batch normalization is then applied before the non-linear activation function. Afterward, the output is max-pooled and neurons are randomly dropped out to prevent overfitting. Furthermore, the number of filters in the convolutional layer is doubled for each consecutive block. After the last block, a fully connected layer with linear activation predicts the three target values for the regression problems. Figure 3 depicts an example of such a CNN with 32 filters in the first layer and three convolutional blocks. The model is trained with the Adam optimizer to minimize the MSE and an additional L2 weight regularization term. In order to optimize both model architecture and training settings, a random parameter search is performed. Here, the learning rate of the Adam optimizer, different functions for the activation, the number of layers, filters and batch size, dropout, and kernel regularizer are adjusted, as can be seen in “All hyperparameters and their values that are optimized for the different machine learning models.”. 50 different combinations of those parameters are tested and the best one is chosen according to validation MAE. The network is trained for an indefinite number of epochs, stopping early if the validation MAE has not increased for 100 epochs.
Figure 3.
This figure shows the architecture of the model used for the CNN
Three similar blocks of layers with an increasing number of output filters are stacked consecutively. Finally, the output of the last block is fed into a fully connected layer with output neurons for each of the predicted motility characteristics percentage of immotile sperm, percentage of progressive motility, and percentage of non-progressive motility.
Recurrent neural network
The model architecture of the considered recurrent neural network (RNN) consists of multiple recurrent layers. Each of those layers contains either a gated recurrent unit (GRU) or LSTM cell. Bidirectional variants where the input is processed both in the forward and backward direction are also tested. Dropout is applied both within (between time steps) and after each recurrent layer. Both GRUs and LSTMs use hyperbolic tangent activation (hyperbolic tangent (tanh)) for their recurrent layers and sigmoid as their gate activation functions. In order to minimize the MSE and an additional L2 weight regularization term, the model is trained with the RMSProp optimizer. Parameters such as learning rate, number of layers and recurrent units, and batch size are optimized, as listed in Table 1.
Evaluation
Motility of spermatozoa is predicted by a linear SVR (“Linear Support Vector Regressor”), an MLP (“Multilayer Perceptron”), a CNN (“Convolutional Neural Network”), and an LSTM (“Long Short-term Memory network”), where all models are trained on emsd features extracted with Trackpy. Moreover, motility prediction is achieved by BoW with a linear SVR (“Bag-of-Words with Support Vector Regressor”) and with an MLP regressor (“Bag-of-Words with Multilayer Perceptron”), both trained on imsd features extracted with the help of Trackpy, and on features created by computations on a set of statistics from tracks detected with the customized Lucas-Kanade tracker. We did not train any CNN or RNN models on the BoWs, as they are sparse quantizations of entire video samples, thus containing neither structural nor temporal information that could be exploited by those types of neural networks. Since BoWs are sparse quantizations of entire video samples, they contain neither structural nor temporal information. For this reason, we refrained to train CNN or RNN models on BoW features.
MAE and RMSE results for both validation and evaluation are outlined in Tables 2 and 3. However, for purposes of readability, in the text, we mainly remark on MAE results achieved on evaluation using the same parameters found for the best results on validation. As described in “sec:dataset”, we used the 3-fold cross-validation setup of the Medico Multimedia for Medicine challenge. Therefore, the reported MAEs and RMSEs are mean values computed over 3-folds.
Table 2.
Mean absolute error (MAE) and root-mean-square error (RMSE) results of proposed experiments using four machine learning models on emsd features
| metric | hop | window | SVR |
MLP |
CNN |
RNN |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| val | eval | val | eval | val | eval | val | eval | |||
| MAE | 1s | 2s | 11.13 | 10.91 | 6.49 | 11.56 | 6.29 | 10.48 | 6.55 | 12.97 |
| 1s | 10s | 10.16 | 8.36 | 5.19 | 8.83 | 5.03 | 8.44 | 6.59 | 8.49 | |
| 5s | 10s | 10.12 | 8.60 | 5.26 | 8.13 | 7.21 | 8.74 | 6.45 | 8.13 | |
| RMSE | 1s | 2s | 14.02 | 14.30 | 8.04 | 15.13 | 8.21 | 14.17 | 10.27 | 16.17 |
| 1s | 10s | 12.87 | 11.06 | 6.84 | 11.48 | 6.55 | 10.82 | 9.18 | 11.41 | |
| 5s | 10s | 12.85 | 11.56 | 6.95 | 10.56 | 7.21 | 11.31 | 9.49 | 10.79 | |
Three different hop and window size combinations are evaluated for the extraction of the emsd feature vectors. hop size: refers to the difference between two adjacent window centers. For example, for a hop size of 1 s and a window size of 10 s two adjacent widows are for 90% overlapped. val: results on the (val)idation set of the data. eval: results on the unseen (eval)uation set of the data. Best results for each metric are highlighted in gray shading.
Table 3.
Mean absolute error (MAE) results of proposed experiments using a BoW with SVR and MLP on custom movement statistics (CMS) and mean squared displacement (MSD) features
| codebook size | assigned vectors | SVR + CMS |
SVR + msd |
MLP + CMS |
MLP + msd |
||||
|---|---|---|---|---|---|---|---|---|---|
| val | eval | val | eval | val | eval | val | eval | ||
| 2 500 | 1 | 8.34 | 8.00 | 7.71 | 7.55 | 7.63 | 8.11 | 6.70 | 8.01 |
| 10 | 8.31 | 7.91 | 7.85 | 7.73 | 7.37 | 8.37 | 6.19 | 8.74 | |
| 50 | 8.33 | 8.03 | 8.10 | 8.01 | 7.52 | 8.50 | 6.29 | 7.95 | |
| 100 | 8.28 | 8.00 | 8.18 | 8.06 | 7.18 | 8.64 | 6.47 | 8.35 | |
| 200 | 8.23 | 8.05 | 8.26 | 8.09 | 7.15 | 8.39 | 6.41 | 8.16 | |
| 500 | 8.26 | 8.31 | 8.27 | 8.03 | 6.75 | 8.11 | 5.91 | 7.85 | |
| 5 000 | 1 | 8.42 | 8.26 | 7.87 | 7.81 | 7.99 | 8.29 | 6.70 | 8.04 |
| 10 | 8.22 | 7.93 | 7.69 | 7.43 | 7.12 | 8.73 | 6.65 | 8.54 | |
| 50 | 8.38 | 8.07 | 8.05 | 7.99 | 7.50 | 8.42 | 6.30 | 8.18 | |
| 100 | 8.34 | 8.03 | 8.11 | 8.03 | 7.39 | 8.40 | 6.14 | 8.54 | |
| 200 | 8.29 | 8.00 | 8.19 | 8.07 | 7.31 | 8.40 | 6.35 | 8.68 | |
| 500 | 8.22 | 8.08 | 8.28 | 8.10 | 7.05 | 7.54 | 6.23 | 8.40 | |
| 10 000 | 1 | 8.56 | 8.73 | 8.08 | 8.18 | 7.85 | 7.92 | 7.03 | 8.11 |
| 10 | 8.19 | 7.86 | 7.56 | 7.31 | 7.41 | 8.17 | 6.47 | 8.27 | |
| 50 | 8.40 | 7.98 | 7.92 | 7.86 | 7.41 | 8.17 | 6.27 | 8.07 | |
| 100 | 8.38 | 8.07 | 8.05 | 7.99 | 7.42 | 7.95 | 6.12 | 8.04 | |
| 200 | 8.34 | 8.03 | 8.11 | 8.03 | 7.26 | 8.26 | 6.28 | 8.19 | |
| 500 | 8.27 | 8.03 | 8.23 | 8.09 | 7.13 | 7.67 | 6.34 | 7.91 | |
18 different codebook sizes and number of assigned vectors combinations, all with a window size of 5 s are evaluated. Best results for each codebook size are highlighted in gray shading.
Linear support vector regressor
The first set of results comes from training a linear SVR on the emsd feature vectors extracted with Trackpy. As described in “displacement features”, three different combinations of window and hop size are evaluated for feature extraction. Training and optimization of the regressor is further done as outlined in “Linear Support Vector Regressor”. Table 2 shows both MAEs and RMSEs achieved during validation and evaluation using 3-fold cross-validation. It is apparent from both the validation and evaluation results that choosing only a small window size of 2 s for computing the displacement statistics leads to feature representations that lack useful information for predicting motility characteristics of the sperm cells for each sample. For the configurations, using a larger window size of 10 s and a larger hop size of 5 s leads to slightly better results during validation but decreased evaluation performance. Considering the best validation result, a minimum MAE of 8.60 is obtained on evaluation with the SVR trained on emsd features. Measured against the state of the art, this result shows a relative improvement of 2.6% (Thambawita et al., 2019b).
Multilayer perceptron
Secondly, an MLP is trained on the emsd feature vectors that have been extracted with Trackpy. Again, following the procedure described in “Displacement features”, features are extracted by three combinations of window and hop size. In “Multilayer Perceptron”, it is shown how the network is trained and optimized. Best results are achieved with a learning rate of , a batch size of 16, and a dropout of 0.2. For a window size of 2 s and 1 s hop, best results are obtained with the ReLU activation function, eight layers, a factor of for the L2 weight regularization, and units on each layer. The model trained on features extracted with a window size of 10 s and 1 and 5 s hop is performing best for choosing ELU as activation function, four layers, as factor for the L2 weight regularization, and 512 units on each layer. In Table 2, an overview of MAE and RMSE results coming from validation and evaluation with 3-fold cross-validation is given. Same as with the SVR (see “linear support vector regressor”), choosing a window size of 2 s for the computation of displacement statistics performs the worst. For choosing a larger window size of 10 s, validation performance is somewhat better for applying a hop size of 1 s than 5 s. However, the larger hop size of 5 s is performing slightly better on evaluation. The minimum MAE value of 8.83 is achieved by the MLP trained on displacement features, which is as good as the findings of state of the art (SOTA) (Thambawita et al., 2019b).
Convolutional neural network
Features extracted with the help of the Trackpy tool are used for a third set of experiments, this time applying a CNN. Here, features are extracted by three combinations of window and hop size, as can be read in “Displacement Features”. How the network is trained and optimized is explained in “Convolutional Neural Network”. Best results are achieved with a learning rate of , ELU as activation function, and a factor of for the L2 weight regularization. Training the network on features extracted with a window size of 2 s and 1 s hop performed best with four layers, starting with 32 filters for the first layer, a batch size of 64, and a dropout of . Best results with a window size of 10 s and hop size of 1 and 5 s are for a number of eight layers, starting with 32 filters for the first layer, a batch size of 32, and a dropout of 0.4. As can be observed in Table 2, using the emsd features obtained with 1 s overlapping 2 s segments of one video is not quite keeping pace with the best results of previous papers on this kind of experiment. emsd features with overlapping 10 s parts of the videos with a hop size of 1 and 5 s are more promising. Experiments with 1 s hop are achieving best results for MAE and RMSE values on validation and evaluation. Going by the best validation result, the minimum MAE is at 8.44 for training a CNN on emsd features. These results show a relative improvement of 4.4% against state-of-the-art results by Thambawita et al. (2019b). Moreover, the CNN performs slightly better than the previous models—SVR achieving 8.60 MAE, see “linear support vector regressor”, and MLP resulting in 8.83 MAE, see “Multilayer perceptron”. As the improvements are only marginal at best, it is questionable if structural dependencies which could be exploited by the CNN can be found in the emsd feature vectors.
Recurrent neural network
A fourth set of experiments is done with training an RNN on the features extracted with the Trackpy tool. As described in “displacement features”, three different combinations of window and hop size are assessed for feature extraction. Now, contrary to the other experiments, the feature sequences are formed from the vectors extracted from consecutive overlapping windows and the RNN uses all of the information in the sequence to make a prediction. The network is trained and optimized according to “Recurrent Neural Network”. Best results are obtained with bidirectional LSTM cells with a number of 256 recurrent units on each layer. Further, applying dropout to the activations in the recurrent layers decreases performance in all cases. For training this network on features of 2 and 10 s windows and 1 s hop, the best hyperparameters are a learning rate of , a number of two layers, a batch size of 16, a dropout of 0.4, and a factor of for the L2 weight regularization. With a window size of 10 s and 5 s hop, best performance is achieved with a learning rate of , a number of four layers, a batch size of 64, a dropout of 0.2, and a factor of for the L2 weight regularization. Validation and evaluation with 3-fold cross-validation scored the MAE and RMSE values displayed in Table 2. Choosing a window size of 10 s and a hop size of 5 s achieves evaluation results that are slightly better than state-of-the-art results. The minimum MAE is at 8.13 MAE for evaluation, a relative improvement of 7.8% against state-of-the-art results by Thambawita et al. (2019b), as well as 3.6% against previous experiments, for example those using a CNN on the same emsd features, cf. “Convolutional Neural Network”. Considering the temporal dependencies within sequences of emsd vectors therefore seems to improve on regression performance if ever so slightly. Furthermore, the RNN experiments enforce the notion that emsd features computed over longer time intervals contain more information regarding the motility of sperm cells, as even when taking a sequence of shorter frames into account as a whole, results are better with the greater window size.
Bag-of-Words with support vector regressor
A possible drawback of the initial experiments with emsd features might be that they aggregate information about the movement across all spermatozoa in a given sample in a very primitive fashion with mean values. Therefore, the experiments with unsupervised feature quantization and aggregation via BoW of single-spermatozoon-based features investigate a more sophisticated approach of analyzing a variable number of sperm cells.
Here, the prediction of the motility of spermatozoa is accomplished by generating BoWs from the features described in “feature extraction” that serve as input for training an SVR.
Custom movement statistics features
In the first set of experiments for predicting motility using a BoW, the BoW is generated from movement statistics coming from the adapted Lucas-Kanade tracker, as discussed in “Custom Movement Statistics”. As shown in “Bag-of-Words”, for training and optimization of the model, codebook sizes of 2500, 5000, and 10000, assigning 1, 10, 50, 100, 200, and 500 vectors, and complexity values between and are considered. Best results are detected with a complexity of 10. Validation and evaluation achieved MAE results are shown in Table 3. Choosing any of the investigated combinations of codebook size and the number of assigned vectors leads to evaluation results that are slightly better than state-of-the-art results by Thambawita et al. (2019b). The best result on the validation, achieved with a codebook size of and 10 assigned vectors is 7.86 MAE on evaluation, a relative improvement of 10.9% against state-of-the-art results, and 6.8% against the best result of previous experiments with a CNN cf. “Convolutional Neural Network”. These results suggest the superiority of the BoW approach to simple mean aggregation. Tuning the BoW hyperparameters shows that choosing a smaller number of codebook vectors to assign to each input sample leads to improved results. Furthermore, a marginal performance gain can be achieved with larger codebooks.
Displacement features
The same model as in the preceding part, the BoW with a linear SVR, is now trained on imsd features extracted with Trackpy, described in “displacement features”. As outlined in “Bag-of-Words”, codebook sizes of 2500, 5000, and 10000, assigning 1, 10, 50, 100, 200, and 500 vectors and complexity values between and are tested to extract features in order to train and optimize the model. A complexity of showed the best results. In Table 3, the validation and evaluation for MAE values are reported. Evaluation results for any tested combination of codebook size and number of assigned vectors are better than or at least equally good as all previous experiments in this article and state-of-the-art results. A codebook size of 10000 and 10 assigned vectors achieves the overall minimum evaluation MAE of 7.31. This is a relative improvement of over 17.2% compared to the best submission (Thambawita et al., 2019b) and is outperforming the results provided in “Custom movement statistics features”.
The imsd features extracted with Trackpy therefore serve as a more powerful basis for feature creation than the custom statistics generated from movement tracks. The observations about codebook sizes and number of assigned vectors also hold for this set of experiments, with larger codebooks and fewer vector assignments leading to the best results.
Bag-of-Words with multilayer perceptron
Motility prediction of spermatozoa is additionally achieved by training a BoW with an MLP on features created with both Trackpy and calculations coming from the adapted Lucas-Kanade tracker.
Custom movement statistics features
Experiments for this model are started by training the BoW with an MLP regressor on custom movement statistics features created with the help of the tailored Lucas-Kanade tracker that has been explained in “Custom Movement Statistics”. Codebook sizes of 2500, 5000, and 10000 and 1, 10, 50, 100, 200, and 500 assigned vectors are tested for feature extraction, so that the model can be optimized, see “Bag-of-Words”. For further optimization, various values for different hyperparameters are assessed as shown in “multilayer perceptron”. Best results are accomplished with a learning rate of , a batch size of 64, a dropout of 0.4, two layers, and units per layer. ReLU is the best performing activation function and a factor of proved best for the L2 weight regularization. MAE results for validation and evaluation are listed in Table 3. The best MAE results for evaluation of 8.11 MAE are achieved for a codebook size of 2500 and 500 assigned vectors.
Displacement features
The same model of a BoW with an MLP regressor as in the preceding part in this section is additionally trained on displacement features extracted with Trackpy, as shown in “displacement features”. As described in “Bag-of-Words”, codebook sizes of 2500, 5000, and 10000 and 1, 10, 50, 100, 200, and 500 assigned vectors are evaluated for feature extraction. This model is further trained and optimized according to “Multilayer perceptron”, obtaining best results with a learning rate of , a batch size of 16, a dropout of 0.2, four layers, and 256 units in each of those layers. Here, ELU activation function and a factor of for the L2 weight regularization performed best. Table 3 illustrates MAE results for the validation and evaluation. A codebook size of 2500 and assigning 500 vectors achieves 7.85 MAE on evaluation.
Discussion
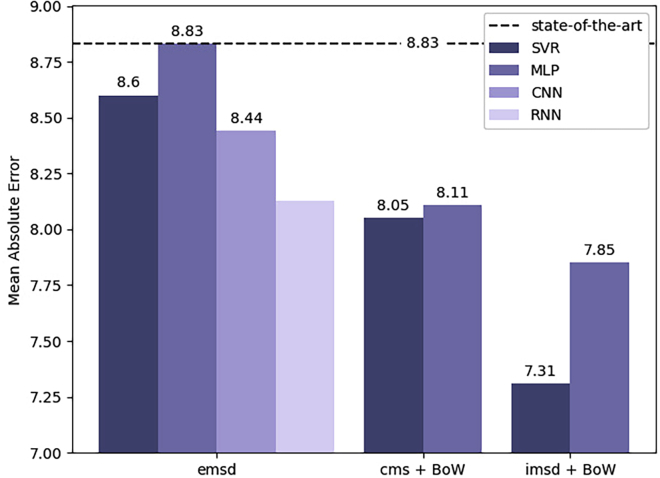
The large number of experiments conducted and evaluated in this article (summarised in Table 4) additionally requires a high-level overview which discusses individual strengths and weaknesses. For predicting motility, almost every experiment in this article improved upon the state of the art (Thambawita et al., 2019b) (The MAE provided by Thambawita et al. (2019b) was at the time of writing this manuscript the best available MAE result. However, it should be noted that we did not find a peer-reviewed publication of this paper. The best (peer-reviewed) published MAE value for the task of sperm motility prediction based on the Visem dataset is provided by Hicks et al. (2019d).) which is already better than the ZeroR baseline. The best results of every investigated combination of feature representation and machine learning algorithm are displayed in Figure 4. Using emsd feature vectors extracted from overlapping windows of the input videos already leads to better results with almost every machine learning model. Comparing the different algorithms for this feature type shows that more involved neural network architectures, i. e., CNNs and RNNs, are able to extract additional information from the features, by either considering structural dependencies within a single vector (CNN) or exploiting long(er) term temporal dependencies between consecutive emsd measurements (RNN)—with the latter leading to the best results with these kinds of features. However, an MLP trained on emsd vectors is inferior to the more robust SVR. The remaining three types of features all aggregate sperm-level information into subject-level sparse feature representations via BoW. Therefore, they are no longer suitable candidates for the CNN and RNN models. Overall, the BoW methodology still leads to stronger results considering the simpler machine learning strategies applied in those experiments. Furthermore, the strongest feature representations can be extracted by constructing BoWs from imsd vectors with the overall best MAE of 7.31. This result is further significantly better than state-of-the-art results at measured by a one-tailed T-Test than by Hicks et al. (2019c) at . For these experiments, the SVR outperformed the MLP and therefore, the latter is not applied in the very last experiment. This observation can be explained by the small size of the training dataset in the BoW experiments where features are aggregated per patient. As deep learning models generally require larger amounts of data to perform well, this circumstance might have prevented the MLP from achieving better results.
Table 4.
Mean absolute error (MAE) and root-mean-square error (RMSE) results of proposed experiments using eight different machine learning models with SVR, MLP, CNN, and RNN on emsd, imsd, and custom movement statistics (CMS) features
| emsd |
CMS |
imsd |
SOTA | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SVR | MLP | CNN | RNN | SVR | MLP | SVR | MLP | |||
| MAE | val | 10.12 | 5.19 | 5.03 | 6.45 | 8.19 | 6.75 | 7.56 | 5.91 | – |
| eval | 8.60 | 8.83 | 8.44 | 8.13 | 7.86 | 8.11 | 7.31 | 7.85 | 8.83 | |
| RMSE | val | 12.85 | 6.84 | 6.55 | 9.18 | 10.16 | 9.04 | 9.39 | 8.74 | – |
| eval | 11.56 | 11.48 | 10.82 | 11.41 | 10.38 | 10.81 | 9.56 | 10.49 | 12.05 | |
The best overall result is highlighted in bold.
Figure 4.
Best results for motility prediction and state of the art (Thambawita et al., 2019b)
The lower the mean absolute error (MAE) the better, showing that SVR models achieve better results than the other models for every type of features. BoWs outperform models trained on emsd features.
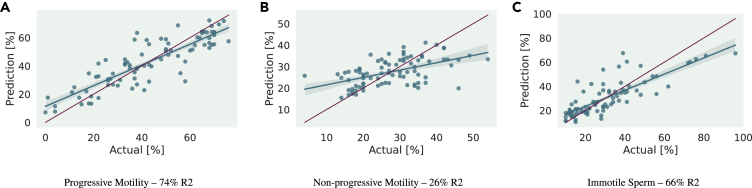
For our best model, an SVR trained on quantized, BoW representations learn from per-sperm displacement measurements, we further perform an analysis on the individual dimensions of motility—progressive and non-progressive motility as well as percentage of immotile sperm cells. Figure 5 plots the model’s predictions for each motility metric against its true value. It is clear that progressive motility of sperm cells is most easily detected by our framework, achieving a coefficient of determination of . For the other two dimensions, our model did not perform as well. Detection of non-progressive motility is closer to a mean prediction baseline than the actual distribution, achieving an R2 score of merely . However, this is the dimension exhibiting the lowest amount of variance in the dataset, making it harder for the models to learn important discriminating features about the underlying data distribution. Lastly, for most patients, the amount of immotile sperm cells in their samples was below , leading to a condensed data distribution. Nonetheless, immotile sperm cells were still quite reliably detected by our framework, yielding an R2 score of .
Figure 5.
Visualization of our best model’s predictions for the three dimensions of sperm-motility—percentage of progressively motile (A), non-progressively motile (B), and immotile spermatozoa (C)
The green line represents a linear regression fitted to relate the model’s predictions to the manually annotated ground-truth motility labels. The shaded margin around the regression line visualizes a 95% confidence interval obtained by bootstrapping. A red identity line is further added to the plot for easier assessment of the overall performance. Finally, coefficients of determination (R2 scores) are given for each result. For a detailed account about the error analysis please refer to “Discussion”.
Limitations of the study
When looking at the computational effort required to run our motility prediction pipeline, real-time analysis (at least 30 fps) is not yet possible. The main bottleneck can be found with the extraction of the sperm tracks which was performed for the entirety of each video. Here, especially the Crocker-Grier algorithm is quite slow, requiring a runtime longer than each video’s duration for tracking the spermatozoa. For example, extracting tracks and displacement features for a 20 s video clip with a sliding window of 10 s with 5 s hop, takes around 22 s on a desktop Intel i9 processor (8 cores, 16 threads). The rest of the pipeline can afterward be run in under 2 s. An investigation into more efficient tracking algorithms is required for improving the pipeline’s performance.
Conclusion and future work
In this article, the task of automatic sperm quality assessment from microscopic video recordings is addressed by applying a framework of unsupervised tracking, feature quantization, and machine learning. The publicly available Visem dataset served as the basis for predicting the motility of spermatozoa. Two different tracking algorithms are utilized in order to enable extraction of features on a per-sperm cell basis. The features are then quantized and aggregated with a BoW approach and used as input for machine learning models. All methods herein achieved improvements for motility prediction over the submissions to the Medico Multimedia for Medicine challenge. The overall best results are achieved by unsupervised tracking of sperm cells with the Crocker-Grier (Crocker and Grier, 1996) algorithm, extracting imsd features for each detected track and aggregating those features into a histogram representation using BoW. With this feature representation, a linear SVR improved the mean (3-fold) MAE from 8.83 to 7.31, a decrease of over . The results further show that the unsupervised feature quantization helps to achieve more consistent and robust results, regardless of which feature representation is chosen as input. For future work, the presented framework can be extended and improved upon by pursuing a number of additional research directions. First of all, other methods of feature extraction from sperm tracks can be explored. During the experiments in this article, a more involved and computationally heavy set of features in the form of sperm motility parameters, such as curve linear velocities and coefficients obtained from regression analysis, are evaluated. Combined with the BoW feature quantization, however, these are less successful than the simpler imsd vectors. More interesting could be to integrate unsupervised representation learning into the process. A direct approach, for example, could train an autoencoder directly on the video content. Considering the noisy nature of the sperm sample recordings which contain lots of debris and background contrast variation, and furthermore exhibit very sparse motion characteristics, this seems hardly feasible with state-of-the-art deep learning methods. Instead, convolutional and recurrent autoencoders could be applied to suitable transformations of the detected tracks, as has already been done for single tracks of myogenic cells (Kimmel et al., 2019). Here, all tracks could be considered together or individually in an unsupervised training procedure. Using motilitAI, our low-resource AI-based method for automatic sperm motility recognition, we hope for its integration in digital microscopes and making our solution reachable for everyone at low cost.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| VISEM | https://zenodo.org/record/2640506 | https://doi.org/10.5281/zenodo.2640506 |
| Software and algorithms | ||
| motiliAI | https://github.com/EIHW/motilitAI | |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Shahin Amiriparian.
Materials availability
This study did not generate new unique reagents.
Method details
The whole machine learning pipeline reported in this article can be found at https://github.com/EIHW/motilitAI. A snapshot of the code is further included as supplemental material Data S1.
Quantification and statistical analysis
-
•
Samples of 85 participants have been used for machine learning methods
-
•
Statistical significance over baseline results was determined via three-fold cross-validation and Student’s t test.
Acknowledgment
This research was partially supported by Zentrales Innovationsprogramm Mittelstand (ZIM) under grant agreement No. 16KN069455 (KIRun), Deutsche Forschungsgemeinschaft (DFG) under grant agreement No. 421613952 (ParaStiChaD), and SCHU2508/12-1 (Leader Humor).
Author contributions
Conceptualization, S.O. and S.A.; Methodology, S.O. and S.A.; Software, S.O. and M.G.; Investigation, S.O.; Writing – Original Draft, S.O., S.A., and M.G.; Writing – Review & Editing, B.S.; Supervision, S.A. and B.S.
Declaration of interests
The authors declare no competing interests.
Published: August 19, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104644.
Supplemental information
Data and code availability
-
•
Data: This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
-
•
Code: All original code is available in this paper’s supplemental information.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Allan D., van der Wel C., Keim N., Caswell T.A., Wieker D., Verweij R., Reid C., Thierry Grueter L., Ramos K., Apiszcz Z., et al. Soft-Matter/Trackpy: Trackpy v0.4.2. 2019. [DOI]
- Alquézar-Baeta C., Gimeno-Martos S., Miguel-Jiménez S., Santolaria P., Yániz J., Palacín I., Casao A., Cebrián-Pérez J.Á., Muiño-Blanco T., Pérez-Pé R. Opencasa: a new open-source and scalable tool for sperm quality analysis. PLoS Comput. Biol. 2019:1–18. doi: 10.1371/journal.pcbi.1006691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiriparian S. Deep Representation Learning Techniques for Audio Signal Processing. Technische Universität München. 2019 Ph.D. Thesis. [Google Scholar]
- Amiriparian S., Cummins N., Ottl S., Gerczuk M., Schuller B. Proceedings of the 7th International Conference on Affective Computing and Intelligent Interaction Workshops and Demos (ACIIW) IEEE; 2017. Sentiment analysis using image-based deep spectrum features; pp. 26–29. [Google Scholar]
- Amiriparian S., Gerczuk M., Ottl S., Cummins N., Pugachevskiy S., Schuller B. Proceedings of the International Joint Conference on Neural Networks (IJCNN) IEEE; 2018. Bag-of-deep-features: noise-robust deep feature representations for audio analysis; pp. 1–7. [Google Scholar]
- Bar-Shalom Y., Daum F., Huang J. Control Systems Magazine. IEEE; 2009. The probabilistic data association filter; pp. 82–100. [Google Scholar]
- Björndahl L., Mortimer D., Barratt C.L., Castilla J.A., Menkveld R., Kvist U., Alvarez J.G., Haugen T.B. Cambridge University Press; 2010. A Practical Guide to Basic Laboratory Andrology. [Google Scholar]
- Bradski G. Dr. Dobb’s Journal of Software Tools; 2000. The OpenCV Library. [Google Scholar]
- Cooper T.G., Noonan E., Von Eckardstein S., Auger J., Baker H., Behre H.M., Haugen T.B., Kruger T., Wang C., Mbizvo M.T., Vogelsong K.M. World health organization reference values for human semen characteristics. Hum. Reprod. Update. 2010:231–245. doi: 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]
- Crocker J.C., Grier D.G. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 1996:298–310. [Google Scholar]
- David G., Serres C., Jouannet P. Kinematics of human spermatozoa. Gamete Res. 1981:83–95. [Google Scholar]
- Deng J., Dong W., Socher R., Li L.J., Li K., Fei-Fei L. Proceedings of the Conference on Computer Vision and Pattern Recognition (CVPR) IEEE; 2009. Imagenet: a large-scale hierarchical image database; pp. 248–255. [Google Scholar]
- Fleet D., Weiss Y. Handbook of Mathematical Models in Computer Vision. Springer; 2006. Optical flow estimation; pp. 237–257. [Google Scholar]
- Frenkel D., Smit B. volume 1. Elsevier; 2001. (Understanding Molecular Simulation: From Algorithms to Applications). [Google Scholar]
- Haugen T.B., Hicks S.A., Andersen J.M., Witczak O., Hammer H.L., Borgli R., Halvorsen P., Riegler M.A. Proceedings of the 10th ACM on Multimedia Systems Conference, ACM. 2019. Visem: a multimodal video dataset of human spermatozoa. [DOI] [Google Scholar]
- Hicks S., Halvorsen P., Haugen T.B., Andersen J.M., Witczak O., Pogorelov K., Hammer H.L., D-T D.N., Lux M., Riegler M. Proceedings of the CEUR Workshop on Multimedia Benchmark Workshop. MediaEval; 2019. Predicting sperm motility and morphology using deep learning and handcrafted features. [Google Scholar]
- Hicks S., Halvorsen P., Haugen T.B., Andersen J.M., Witczak O., Pogorelov K., Hammer H.L., Dang-Nguyen D.T., Lux M., Riegler M. Proceedings of the CEUR Workshop on Multimedia Benchmark Workshop. MediaEval; 2019. Medico multimedia task at mediaeval 2019. [Google Scholar]
- Hicks S., Haugen T.B., Halvorsen P., Riegler M. Proceedings of the CEUR Workshop on Multimedia Benchmark Workshop. MediaEval; 2019. Using deep learning to predict motility and morphology of human sperm. [Google Scholar]
- Hicks S.A., Andersen J.M., Witczak O., Thambawita V., Halvorsen P., Hammer H.L., Haugen T.B., Riegler M.A. Machine learning-based analysis of sperm videos and participant data for male fertility prediction. Sci. Rep. 2019:1–10. doi: 10.1038/s41598-019-53217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidayatullah P., Mengko T.L.E.R., Munir R., Barlian A. Bull sperm tracking and machine learning-based motility classification. IEEE Access. 2021;9:61159–61170. doi: 10.1109/access.2021.3074127. [DOI] [Google Scholar]
- Van der Horst G., Kitchin R., Van der Horst M., Atherton R. The effect of the breeding season, cryopreservation and physiological extender on selected sperm and semen parameters of four ferret species: implications for captive breeding in the endangered black-footed ferret. Reprod. Fertil. Dev. 2009:351–363. doi: 10.1071/rd08075. [DOI] [PubMed] [Google Scholar]
- Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Proceedings of the Conference on Computer Vision and Pattern Recognition (CVPR) IEEE; 2017. Densely connected convolutional networks; pp. 4700–4708. [Google Scholar]
- Jain A.K. Prentice Hall; 1989. Fundamentals of Digital Image Processing. [Google Scholar]
- Kimmel J.C., Brack A.S., Marshall W.F. Deep convolutional and recurrent neural networks for cell motility discrimination and prediction. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019;18:562–574. doi: 10.1109/TCBB.2019.2919307. [DOI] [PubMed] [Google Scholar]
- Kimmel J.C., Chang A.Y., Brack A.S., Marshall W.F. Inferring cell state by quantitative motility analysis reveals a dynamic state system and broken detailed balance. PLoS Comput. Biol. 2018:1–29. doi: 10.1371/journal.pcbi.1005927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N., Singh A.K. Trends of male factor infertility, an important cause of infertility: a review of literature. J. Hum. Reprod. Sci. 2015:191. doi: 10.4103/0974-1208.170370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas B.D., Kanade T. 1981. An Iterative Image Registration Technique with an Application to Stereo Vision. [Google Scholar]
- Lueders I., Luther I., Scheepers G., Van der Horst G. Improved semen collection method for wild felids: urethral catheterization yields high sperm quality in african lions (panthera leo) Theriogenology. 2012:696–701. doi: 10.1016/j.theriogenology.2012.02.026. [DOI] [PubMed] [Google Scholar]
- Mortimer D. Handbook of the Laboratory Diagnosis and Treatment of Infertility. CRC Press, Inc.; 1990. Objective analysis of sperm motility and kinematics; pp. 97–133. [Google Scholar]
- Mortimer D. Oxford University Press on Demand; 1994. Practical Laboratory Andrology. [Google Scholar]
- Mortimer D., Aitken R., Mortimer S., Pacey A. CSIRO; 1995. Workshop Report: Clinical Casa – the Quest for Consensus; pp. 951–959. [DOI] [PubMed] [Google Scholar]
- Mortimer D., Mortimer S. Studies in Profertility Series. The Parthenon Publishing Group Limited; 1998. Value and reliability of casa systems; pp. 73–90. [Google Scholar]
- Mortimer S.T., van der Horst G., Mortimer D. The future of computer-aided sperm analysis. Asian J. Androl. 2015:545. doi: 10.4103/1008-682X.154312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Practice Committee of the American Society for Reproductive Medicine Definitions of infertility and recurrent pregnancy loss. Fertil. Steril. 2008:1603. doi: 10.1016/j.fertnstert.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Pratt W.K. CRC press; 2013. Introduction to Digital Image Processing. [Google Scholar]
- Shi J., Tomasi . Proceedings of the Conference on Computer Vision and Pattern Recognition (CVPR) IEEE; 1994. Good features to track; pp. 593–600. [Google Scholar]
- Stephen E.H., Chandra A. Updated projections of infertility in the United States: 1995–2025. Fertil. Steril. 1998:30–34. doi: 10.1016/s0015-0282(98)00103-4. [DOI] [PubMed] [Google Scholar]
- Thambawita V., Halvorsen P., Hammer H., Riegler M., Haugen T.B. CEUR Workshop Proceedings. 2019. Extracting temporal features into a spatial domain using autoencoders for sperm video analysis. [Google Scholar]
- Thambawita V., Halvorsen P., Hammer H., Riegler M., Haugen T.B. CEUR Workshop Proceedings. 2019. Stacked dense optical flows and dropout layers to predict sperm motility and morphology. [Google Scholar]
- Tomasi C., Kanade T. Carnegie Mellon University; 1991. Detection and Tracking of Point Features, School of Computer Science. [Google Scholar]
- Urbano L.F., Masson P., VerMilyea M., Kam M. IEEE Transactions on Medical Imaging. IEEE; 2016. Automatic tracking and motility analysis of human sperm in time-lapse images; pp. 792–801. [DOI] [PubMed] [Google Scholar]
- Valiuškaitė V., Raudonis V., Maskeliūnas R., Damaševičius R., Krilavičius T. Deep learning based evaluation of spermatozoid motility for artificial insemination. Sensors. 2020;21:72. doi: 10.3390/s21010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . Cambridge University Press; 1999. WHO Laboratory Manual for the Examination of Human Semen and Sperm-Cervical Mucus Interaction. [Google Scholar]
- World Health Organization ehealth. Resolution. 2005:16–25. [Google Scholar]
- World Health Organization Sustainable health financing, universal coverage and social health insurance. Resolution. 2005;58:33. [Google Scholar]
- World Health Organization . volume 3. World Health Organization; 2016. (Atlas of EHealth Country Profiles: The Use of EHealth in Support of Universal Health Coverage: Based on the Findings of the Third Global Survery on EHealth 2015). [Google Scholar]
- World Health Organization . World Health Organization; 2017. Global Diffusion of eHealth: Making Universal Health Coverage Achievable: Report of the Third Global Survey on eHealth. [Google Scholar]
- Yee W.K., Sutton K.L., Dowling D.K. In vivo male fertility is affected by naturally occurring mitochondrial haplotypes. Curr. Biol. 2013;23:R55–R56. doi: 10.1016/j.cub.2012.12.002. [DOI] [PubMed] [Google Scholar]
- Zegers-Hochschild F., Adamson G.D., de Mouzon J., Ishihara O., Mansour R., Nygren K., Sullivan E., van der Poel S., ICMART. WHO International committee for monitoring assisted reproductive technology (ICMART) and the world health organization (WHO) revised glossary of art terminology. Hum. Reprod. 2009:2683–2687. doi: 10.1093/humrep/dep343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data: This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
-
•
Code: All original code is available in this paper’s supplemental information.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.