Summary
Background
Middle ear diseases such as otitis media and middle ear effusion, for which diagnoses are often delayed or misdiagnosed, are among the most common issues faced by clinicians providing primary care for children and adolescents. Artificial intelligence (AI) has the potential to assist clinicians in the detection and diagnosis of middle ear diseases through imaging.
Methods
Otoendoscopic images obtained by otolaryngologists from Taipei Veterans General Hospital in Taiwan between Jany 1, 2011 to Dec 31, 2019 were collected retrospectively and de-identified. The images were entered into convolutional neural network (CNN) training models after data pre-processing, augmentation and splitting. To differentiate sophisticated middle ear diseases, nine CNN-based models were constructed to recognize middle ear diseases. The best-performing models were chosen and ensembled in a small CNN for mobile device use. The pretrained model was converted into the smartphone-based program, and the utility was evaluated in terms of detecting and classifying ten middle ear diseases based on otoendoscopic images. A class activation map (CAM) was also used to identify key features for CNN classification. The performance of each classifier was determined by its accuracy, precision, recall, and F1-score.
Findings
A total of 2820 clinical eardrum images were collected for model training. The programme achieved a high detection accuracy for binary outcomes (pass/refer) of otoendoscopic images and ten different disease categories, with an accuracy reaching 98.0% after model optimisation. Furthermore, the application presented a smooth recognition process and a user-friendly interface and demonstrated excellent performance, with an accuracy of 97.6%. A fifty-question questionnaire related to middle ear diseases was designed for practitioners with different levels of clinical experience. The AI-empowered mobile algorithm's detection accuracy was generally superior to that of general physicians, resident doctors, and otolaryngology specialists (36.0%, 80.0% and 90.0%, respectively). Our results show that the proposed method provides sufficient treatment recommendations that are comparable to those of specialists.
Interpretation
We developed a deep learning model that can detect and classify middle ear diseases. The use of smartphone-based point-of-care diagnostic devices with AI-empowered automated classification can provide real-world smart medical solutions for the diagnosis of middle ear diseases and telemedicine.
Funding
This study was supported by grants from the Ministry of Science and Technology (MOST110-2622-8-075-001, MOST110-2320-B-075-004-MY3, MOST-110-2634-F-A49 -005, MOST110-2745-B-075A-001A and MOST110-2221-E-075-005), Veterans General Hospitals and University System of Taiwan Joint Research Program (VGHUST111-G6-11-2, VGHUST111c-140), and Taipei Veterans General Hospital (V111E-002-3).
Keywords: Middle ear diseases, Deep learning, Convolutional neural network, Transfer learning, Mobile applications, Edge computing
Research in context.
Evidence before this study
Deep learning methods have been used to train learning models to predict middle ear diseases, which are among the most common issues faced by clinicians around the world. We searched PubMed without language constraints for peer-reviewed research articles up to Dec 31, 2021, using the search terms (“tympanic membrane”[MeSH Terms]) AND (“artificial intelligence” [MeSH Terms]). Neither of the studies mentioned a small-sized model applicable to mobile devices had been developed.
Added value of this study
This is the first study to develop a smartphone-based edge AI application to assist clinicians in detecting and categorising eardrum or middle ear diseases. This study applied the transfer learning model to develop a mobile device application to detect middle ear diseases. Our model successfully distinguished normal eardrums and diseased eardrums and achieved 98.9% detection accuracy; when the data were divided into ten common middle ear diseases, the accuracy reached 97.6%.
Implications of all the available evidence
This AI application, which is based on edge computing, is low-cost, portable, and functional in real time and thus innovative and practical. The application could be executed without additional cloud resource requirements. The proposed model can be easily integrated into mobile devices and used in telemedicine for detecting middle ear disorders.
Alt-text: Unlabelled box
Introduction
Middle ear diseases such as otitis media and middle ear effusion are amongst the most common issues faced by clinicians providing primary care to children and adolescents. The worldwide prevalence of otitis media and otitis media with effusion are 2.85% and 0.6%, respectively, in the general population and 25.3% and 17.2%, respectively, in childhood.1, 2, 3 Making correct diagnoses through the inspection of ear drums is still a challenge for most general practitioners due to lack of specialty training, and ancillary hearing tests are often needed to achieve better accuracy.4 Severe complications such as acute mastoiditis, labyrinthitis or meningitis may develop if middle ear diseases are not properly diagnosed and treated in time, and these complications may cause sequalae such as hearing impairment or neurological deficits.5 Furthermore, some of these middle ear diseases are treated with unnecessary medications, such as broad-spectrum antibiotics, due to the difficulty of making prompt diagnoses.6 It is not surprising that even experienced otolaryngologists can only reach a diagnostic accuracy of 75% for some middle ear diseases, such as otitis media with effusion.7
To overcome this diagnostic dilemma and improve diagnostic accuracy, several methods, such as decision trees, support vector machines (SVMs), neural networks and Bayesian decision approaches, have been used to train different learning models and predict whether an image corresponds to a normal ear or an otitis media case.8,9 Myburgh et al. utilized a video endoscope to acquire images and a cloud server for pre-processing and entered the images into a smartphone-based neural network program for diagnosis. This system can distinguish five different middle ear diseases with a diagnosis rate of 86%.10 Seok et al. created a deep learning model for detection and identified the fine structures of the tympanic membrane; the interpretation accuracy of the sides of the tympanic membrane can reach 88.6%, and the diagnosis rate of tympanic membrane perforation is 91.4%.11
Deep learning algorithms with convolutional neural networks (CNNs) have recently been successfully utilized in many medical specialties.12 Lightweight CNNs such as MobileNet are models derived from CNNs that are widely used in image analysis. The main advantage of using the lightweight CNNs is that they require fewer computing resources than the traditional CNN models, making them suitable for running on mobile devices such as smartphones.13
Transfer learning and ensemble learning are the new trends for increasing performance and have been proven to be efficient in handling relatively small datasets.14 While transfer learning algorithms are built for classification, the ensemble learning technique can be further implemented to obtain a more accurate classification result with a weighted voting approach. Transfer learning is a type of learning framework that transfers the parameters of a pretrained model to a new model and can therefore perform a large amount of time-consuming data labelling work and improve the learning performance.15 Such a lightweight network contains fewer parameters and smaller scales of specific problems and learns the patterns contained in the input data through pretrained models; therefore, it is suitable for mobile devices with constrained computing resources.16
In this study, we developed a deep learning model adapted to smartphones for the detection and diagnosis of middle ear diseases. The pretrained model with the best performance was converted into a smartphone-based program by transfer learning. The accuracy of the proposed algorithm was compared against a cohort of professional otolaryngologists and other practitioners.
Methods
Data acquisition
Our data were acquired from patients with middle ear diseases who sought medical help in the Department of Otolaryngology at Taipei Veterans General Hospital (TVGH) in Taiwan from January 1st, 2011 to December 31st, 2019. The study protocol was approved by the Institutional Review Board of Taipei Veterans General Hospital (no. 2020-03-001CCF), and the study adhered to the tenets of the Declaration of Helsinki. Informed consent was not applicable due to de-identified data.
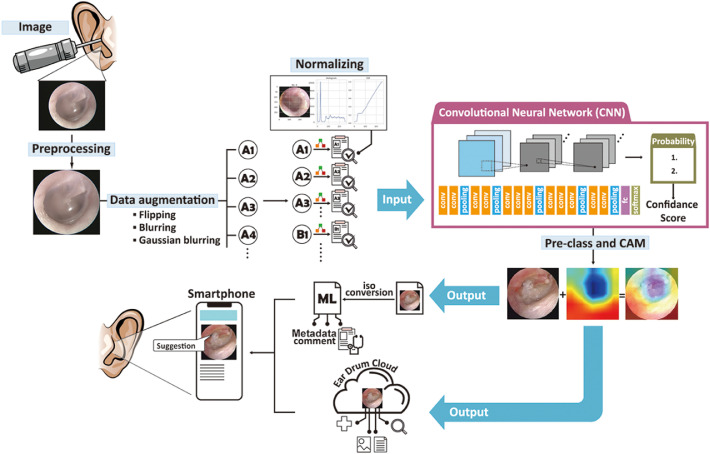
The human- and computer-aided diagnosis system implemented in this study is summarized in Figure 1.
Figure 1.
Developing a smartphone-based computing program for diagnosing middle ear disease and providing medical suggestions.
The main architecture of our AI model is a CNN with transfer learning. Each layer extracts different tympanic membrane/intermediate image features, and all the extracted features are integrated to determine the type of middle ear disease and the corresponding treatment. Subsequently, a Core ML model was developed for this new smartphone-based eardrum application, where users can upload eardrum images to the cloud. The eardrum app analyses the input image and indicates the type of middle ear disease and the treatment to be performed based on the results.
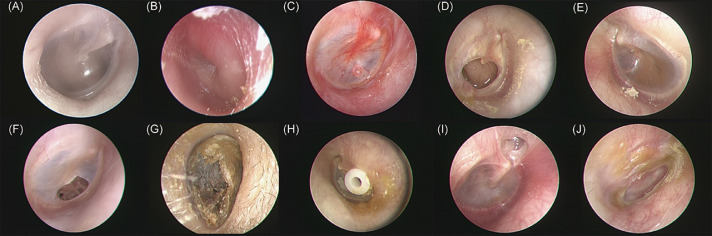
Data attributes and types include eardrum images and categories (Figure 2). The ground-truth eardrum classification was annotated by three otolaryngology specialists. First, the otolaryngology specialists determined if the acquired otoendoscopic images were normal or diseased. Next, we checked if there were space-occupying abnormalities or foreign bodies (such as cerumen impactions, otomycosis or ventilation tubes) and then inspected if there were any perforations in the eardrum (chronic suppurative otitis media or tympanic perforation). We then scrutinized whether the middle ear was currently experiencing acute inflammation (acute otitis media or acute myringitis) or chronic inflammation (middle ear effusion or tympanic retraction). Ten classes of common middle ear conditions/diseases were selected and annotated, including (A) normal tympanic membrane, (B) acute otitis media, (C) acute myringitis, (D) chronic suppurative otitis media, (E) otitis media with effusion, (F) tympanic membrane perforation, (G) cerumen impaction, (H) ventilation tube, (I) tympanic membrane retraction, and (J) otomycosis. Because the initial image data were obtained in different sizes, we conducted an initial quality control step, filtering out low-resolution or incorrectly formatted images. The inclusion standard was 224 × 224 pixels, which is a 32-bit original image format.
Figure 2.
Representative images of ten classes of common middle ear conditions/diseases.
(A) normal tympanic membrane, (B) acute otitis media, (C) acute myringitis, (D) chronic suppurative otitis media, (E) otitis media with effusion, (F) tympanic membrane perforation, (G) cerumen impaction, (H) ventilation tube, (I) tympanic membrane retraction, and (J) otomycosis.
Data preparation
Pre-processing is the process of preparing raw data into a format that is suitable for machine learning. The processes, including image classification, the removal of images that cannot be judged by the human eye and the removal of black edges from images, were performed using the OpenCV libraries and Canny edge detection function.
Data augmentation expands the amount of data by generating modified data from an original dataset. As the normal eardrum category accounts for more than half of the original classification data, we deployed several transformation techniques using the OpenCV library, including flips (vertical, horizontal and both) and colour transformations (histogram normalisation, gamma correction, and Gaussian blurring). Images were randomly assigned to either the training set and validation dataset with a ratio of 8:2 ratio using the Scikit-Learn library.
Development of the deep learning algorithms
Several standard CNN architectures were used to train the middle ear disease detection and classification model, including VGG16, VGG19, Xception, InceptionV3, NASNetLarge, and ResNet50. The training was performed with the Keras library, with TensorFlow as the backend. The final softmax layer was added with two units to fit the number of different target categories in the middle ear disease classification task. Because the weight initialisation technique with an ImageNet pretrained network has been shown to improve the convergence speed, we applied this procedure in the initial training of the middle ear disease training model. All the layers of the proposed CNNs were fine-tuned by performing training for 50 epochs using a stochastic gradient descent (SGD) optimiser (a batch size of 32 images, a learning rate of 0.0002, and a moment of 0.8). Our model uses an independent test dataset to evaluate the performance, and both the accuracy and the loss converged during the model training and model validation stages. The same experimental learning rate policy was applied to train all of the tested CNN models. All the AI model training and validation were performed on an IBM AC-922 Linux-based server with a 20-core Power CPU, an NVIDIA V100 SMX2 32-GB GPU card, and 1,024 GB of available RAM.
After the CNN training was completed, the best-performing model was selected as the pretrained model, and the weights were entered into the lightweight mobile CNNs, including NASNetMobile, MobileNetV2, and DenseNet201. The three mobile CNNs with transfer learning were then built for further assessment.
Model assessment
We evaluated the performance of the models in terms of their ability to detect and classify middle ear diseases using the following metrics: ROC (receiver operating characteristic) curves (true versus false-positive rate), AUC (area under the receiver operating characteristic curve), accuracy, precision and recall. AccuracyTPTNTPTNFP; precisionTP(TPFP; recall or true-positive rate TP(TPFN; false-positive rate = FPFPTN (TP: true-positive; TN: true-negative; FP: false-positive; FN: false-negative).
The F1-score is the harmonic mean of recall and precision, which are the sum of true positives divided by the sum of all positives and the sum of true positives divided by the sum of all relevant samples. The F1-score considers the number of each label when calculating the recall and precision and is therefore suitable for evaluating the performance on multi-label classification tasks with imbalanced datasets. The model selection was based on the F1-score.
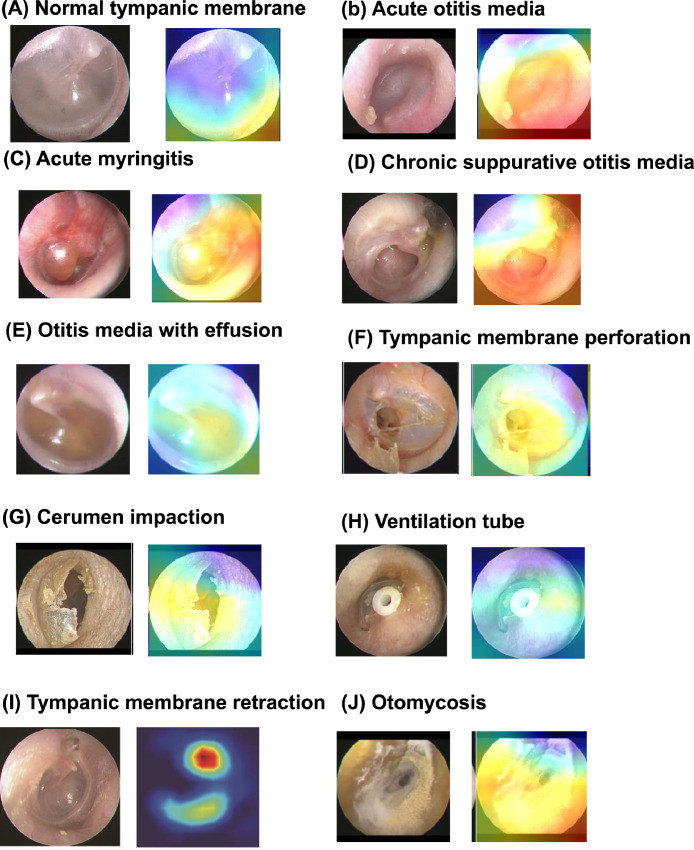
Class activation map
To interpret the prediction decision, a class activation map (CAM) was deployed to recognize the CNN results. The CAM is presented with a rainbow colormap scale, where the red colour stands for high relevance, yellow stands for medium relevance, and blue stands for low relevance.17 This process can effectively visualize feature variations in illuminated conditions, normal structures, and lesions, which are commonly observed in middle ear images. CAMs can provide supplementary analysis for the CNN model so that the location of the disease pre-set by the program can be clearly understood instead of being recognized as an indistinct judgement.17
Model conversion and integration into a smartphone application
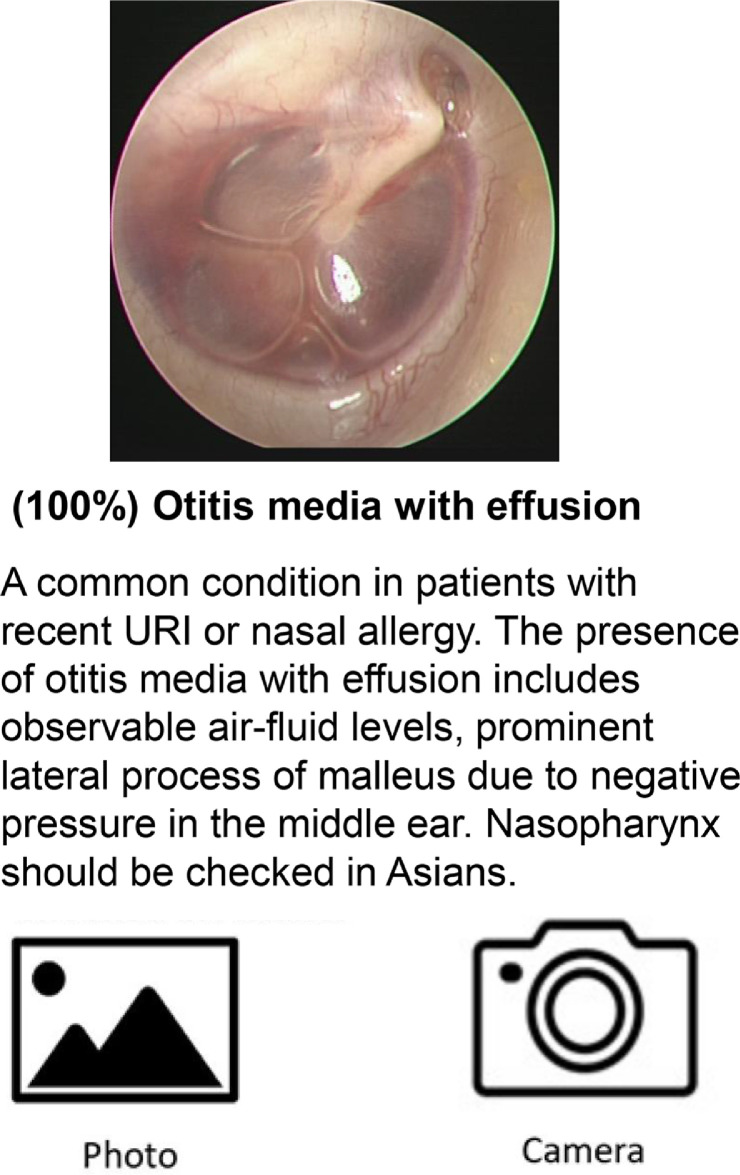
The pretrained model was integrated into the smartphone application by using the Python library Core ML Tools. Figure 3 demonstrates the application's interface (designed for the Apple iOS system). A user provides a middle ear disease image as input. This image is then transformed into a probability distribution over the considered categories of middle ear lesions. The most probable classification (e.g., normal eardrum, acute otitis media, chronic otitis media, tympanic perforation, otitis media with effusion, cerumen impaction, acute myringitis, ventilation tube, eardrum atelectasis, or otomycosis) is then displayed on the screen. The inference process is executed in near-real-time.
Figure 3.
Representative demonstration of middle ear disease detection in the mobile application.
Evaluating the performance of clinicians
To compare the performance of our AI program with that of clinicians, 6 practitioners with different levels of expertise, including 2 board-certified otolaryngology specialists (over 5 years of expertise), 2 otolaryngology residents (1-4 years of expertise or training in otolaryngology) and 2 general practitioners (GPs) (less than 1 year of expertise or training in otolaryngology), were recruited. A set of questionnaires was designed with a total of fifty questions, each containing a picture of the endoscopic view of an eardrum from our preliminary data bank of images, and the responders were asked to choose the most suitable option from ten middle ear disease categories.
Statistical analysis
The performance of our model was evaluated by the indices of accuracy, precision, recall, and F1-score. The ROC curve and AUC were plotted using the Python Scikit-Learn library. An ROC curve plots true positive rate versus the false positive rate at different classification thresholds. The AUC scales with overall performance across all possible classification thresholds. All statistical tests were two-sided permutation tests. A p-value <0.05 was considered to be statistically significant. The obtained data were entered into Excel and analysed using SPSS (version 24 for Windows; IBM Company, Chicago, USA).
Role of the funding source
The funder of the study has no role in study design, the collection, analysis and interpretation of data, the writing of the report, or the decision to submit the paper for publication. All authors had full access to all data within the study. The corresponding authors have final responsibility for the decision to submit for publication.
Results
Establishing and pre-processing an eardrum image dataset for CNN model training
A total of 4,954 pictures were initially included in the study. Next, 2,134 pictures were excluded because the photos were not suitable for analysis due to issues such as blurring, duplications, inappropriate positions, and missing medical records. The remaining 2,820 clinical eardrum images were collected and divided into 10 categories. After pre-processing, 2,161 images were selected for further CNN training. To improve the performance of deep learning, the dataset was increased by data augmentation, and the total number of images was expanded to 10,465. The image number of each category is detailed in Supplementary Table 1.
Diagnostic performance of the CNN models and mobile CNN models
Six different CNN architectures (Xception, ResNet50, NASNetLarge, VGG16, VGG19, and InceptionV3) were tested on the training dataset, and the performance of the different models based on these ten types of normal and middle ear disease images was verified using the validation dataset. The training process was run for 50 epochs, and the best models with the minimal loss values (corresponding to the 34th, 50th, and 42nd epochs for VGG16, InceptionV3 and ResNet50, respectively) were selected and used for verification (Supplementary Figure 2). After parameter adjustment, our results show that among the six tested CNN architectures, InceptionV3 exhibited the highest F1-score during validation (98%) and was selected for further transfer learning (Table 1). Our results showed that almost all CNNs achieved a high detection accuracy in binary outcome cases (pass/refer) of middle ear disease images and successfully subclassified the majority of the images. By making use of the confusion matrix and classification report to examine the training results, the average preliminary accuracy achieved on the test dataset reached 91.0%, and the average training time was 6,575 seconds.
Table 1.
Image characteristics and diagnostic performance.
| Evaluation Indicators |
Acc (%) | Recall (%) | Precision (%) | Sen (%) | Sp (%) | F1-Score (%) | AUC Score (95%CI) |
Training/Test Time (sec) | Best Number of Epochs |
|---|---|---|---|---|---|---|---|---|---|
| CNN models | |||||||||
| VGG16 | 97.3 | 97.1 | 97.2 | 97.1 | 99.7 | 97.1 | 0.984(0.979-0.989) | 1200 | 34 |
| VGG19 | 96.7 | 96.4 | 96.5 | 96.4 | 99.6 | 96.5 | 0.998(0.996-0.999) | 1650 | 41 |
| Xception | 76.6 | 75.8 | 81.7 | 75.8 | 97.4 | 77.0 | 0.866(0.853–0.879) | 2550 | 49 |
| InceptionV3* | 98.0 | 97.9 | 98.1 | 97.9 | 99.8 | 98.0 | 0.989(0.985–0.992) | 9000 | 50 |
| NASNetLarge | 80.8 | 80.7 | 81.4 | 80.7 | 97.9 | 80.0 | 0.893(0.888–0.898) | 23,200 | 47 |
| ResNet50 | 96.8 | 96.5 | 96.7 | 96.5 | 96.8 | 96.5 | 0.981(0.977–0.984) | 1850 | 42 |
| Lightweight CNN models | |||||||||
| NASNetMobile(TL) | 97.2 | 97.1 | 97.2 | 97.1 | 99.7 | 97.1 | 0.984(0.980–0.988) | 1550 | 2 |
| MobileNetV2(TL)⁎⁎ | 97.6 | 97.4 | 97.4 | 97.4 | 99.7 | 97.4 | 0.985(0.980–0.990) | 4500 | 9 |
| DenseNet201(TL) | 97.3 | 97.1 | 97.2 | 97.1 | 99.7 | 97.1 | 0.984(0.979–0.989) | 1600 | 2 |
Acc = accuracy; Sen = sensitivity; Sp = specificity; AUC = area under the receiver operating characteristic curve; TL= Transfer learning with InceptionV3.
Champion model for transfer learning.
Champion model integrated into smartphone.
To enable the use of the CNN model on a mobile phone without affecting the accuracy of the model, we further shifted the information contained in different but relatively large source domains by transfer training with three lightweight CNNs, including NASNetMobile, MobileNetV2 and DenseNet201. MobileNetV2 exhibited the highest F1-score during validation (97.6%). The average accuracies and AUCs of the lightweight CNNs transferred with InceptionV3 were 97.3% and 0.98, respectively. The accuracies, recall rates, and F1-scores reached 0.99 for most disease classifications in MobileNetV2 (Supplementary Figure 2). A CAM, which is a representation of the data output by global average pooling, was used to analyse and identify the key features for deep learning image classifications (Figure 4).
Figure 4.
Representative class activation maps (CAMs) of 10 common ear drum/middle ear diseases. A CAM is a heatmap-like representation of the data output by global average pooling. The hot spots (red) generated by the CAM represent more important parts of the object, rather than all of it, and it does not produce a segmentation result with fine boundaries.
Achieving specialist levels of middle ear disease detection performance: interpretation accuracy comparison among general physicians, specialists and AI models
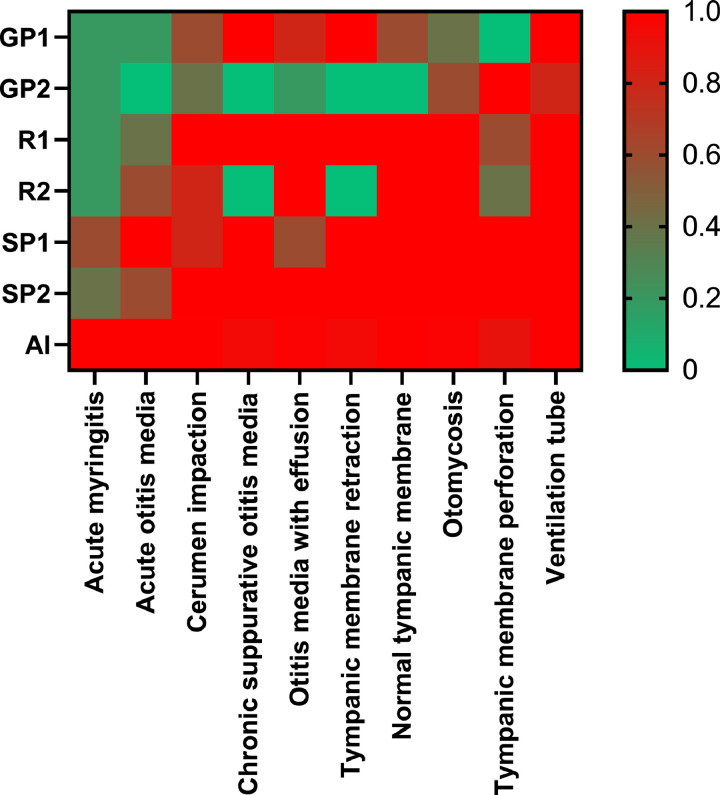
A fifty-question questionnaire was designed for practitioners with different levels of clinical experience to assess middle ear diseases in cases selected from the database. The average accuracy rates were 36.0%, 80.0% and 90.0% for the general practitioners, otolaryngologic residents and specialists, respectively. The difference between the specialist and general practitioner groups reached statistical significance (p<0.005); however, the difference between the specialist and resident groups was not significant (p=0.136). The AI model and specialists had similarly high prediction accuracy scores. On the other hand, less experienced reviewers demonstrated markedly worse prediction accuracy scores, which were separate from the cluster of AI and experienced reviewer scores. For acute myringitis and acute otitis media, AI obviously outperformed the reviewers with all levels of expertise (Figure 5).
Figure 5.
Heatmap comparing the results produced by the AI and the human practitioners. GP=general practitioner, R=resident doctor, SP=otolaryngology specialist, AI= artificial intelligence.
Discussion
In this study, we developed the first smartphone-based edge AI system to assist clinicians in detecting and categorising ear drum or middle ear diseases, to our knowledge. Our model successfully distinguished normal eardrums and diseased eardrums and achieved a high detection accuracy. When the data were divided into ten common middle ear diseases, the accuracy reached 97.6%. The model showed a high degree of accuracy in disease prediction and provided the most probable diagnoses ranked by the probability of various diseases on a smartphone interface. In addition, this model is a “device-side AI” (also called “edge AI”); because the computations can be performed locally on the images acquired on mobile devices, this method has the advantage of offline connection with a fast-computing power, which reduces latency delays and protects patient privacy. This study is also the first to investigate several mainstream CNN methods with lightweight architectures for ear drum/middle ear disease detection and diagnosis on mobile devices.
In the past, CNNs were experimentally used for otitis media diagnosis tasks, although most current databases and codes were not available for public use, and only limited disease categories were available to support empirical clinical diagnosis.18 We divided all collected otoendoscopic eardrum pictures into 10 categories (normal ears and common ear diseases) based on past research and clinical experience. To improve the accuracy of system interpretation, we deleted pictures with a poor image quality, blurry images and less representative images, aggregated the results into 2161 pictures with a better quality, and then analysed them via the artificial intelligence system. Machine learning techniques such as SVMs, k-nearest neighbours (K-NN) and decision trees are supervised machine learning models with simple processes and high levels of execution efficiency. These models are suitable for the prediction and classification and regression data; however, their model training processes may be time-consuming and GPU-consuming, as deep learning approaches must be trained on a very large dataset. To overcome this drawback, a deep CNN was pretrained via transfer learning, which applies the knowledge learned by certain models to another task.
Due to the advancement of image analysis, we found that MobileNet had fewer parameters than other CNN models and was more feasible in mobile devices with less processing power, such as smartphones. Recent studies have shown that although MobileNet is a lightweight network model, its classification accuracy was only 1% less than that of the conventional CNN after conducting the transfer learning-based training technique, and this result was similar to that obtained in our study.19 Eventually, we successfully developed the CNN system with transfer learning and placed it in a mobile application to make it more convenient and accessible.
Some relevant studies utilizing mobile devices for the detection and diagnosis of middle ear diseases have been performed, as summarized in Table 2. Wu et al. used a digital handheld otoscope connected to a smartphone and developed a CNN model for the automated classification of paediatric acute otitis media and otitis media with effusion, achieving an accuracy of 90.7%.20 By using smartphone-enabled otoscopes, Moshtaghi et al. reported a 100% sensitivity rate for the detection of abnormal middle ear conditions and developed a model to achieve a 98.7% overall accuracy on intact tympanic membranes and a 91% overall accuracy in terms of perforation.21 Myburgh et al. reported on a smartphone application for loading ear drum images and obtained an accuracy of 86.8% with a neural network.10 Viscaino et al. developed a computer-aided support system based on digital otoscope images and machine learning algorithms such as SVM, KNN and decision tree.22 With otoendoscopic images, Khan et al. also developed a CNN model that reached a 95% accuracy in differentiating otitis media with effusion (OME) from normal eardrum conditions.23 To our knowledge, we are the first to utilize transfer learning for smartphone applications to develop device-side AI for detecting and diagnosing middle ear diseases. Edge computing, which is also called “edge AI” or “device AI”, is meant to deal with the constraints faced by conventional deep learning-based architectures deployed in a centralized cloud environment, such as the considerable latency of the mobile network, energy consumption and the financial costs affecting the overall performance of the system.24 Our results show that the developed algorithm achieved a high detection accuracy in binary outcome cases (pass/refer) of middle ear disease images and successfully subclassified the majority.
Table 2.
Comparison of the proposed model with previous studies.
| N | Classification | Accuracy | Year | Algorithm | Device | |
|---|---|---|---|---|---|---|
| Senaras et al.33 | 247 | 2 | 84.6% | 2017 | CNN | PC |
| Myburgh et al.10 | 389 | 5 | 81.6%∼ 86.8% | 2018 | DT+ CNN | PC |
| Cha et al.32 | 10,544 | 6 | 93.7% | 2019 | CNN+ETL | PC |
| Livingstone et al.26 | 1366 | 14 | 88.7% | 2020 | AutoML | PC |
| Khan et al.23 | 2484 | 3 | 95.0% | 2020 | CNN | PC |
| Wu et al.20 | 12,230 | 2 | 90.7% | 2021 | CNN+TL | Smartphone-otoscope |
| Zafer et al.8 | 857 | 4 | 99.5% | 2020 | DCNN, SVM | PC (public dataset) |
| Viscaino et al.22 | 1060 | 4 | 93.9% | 2020 | SVM, K-NN, DT | PC |
| Zeng et al.34 | 20,542 | 6 | 95.6% | 2021 | CNN+ETL | Real-time detection device |
| Current study | 2171 | 10 | 97.6% | 2022 | CNN+TL | Smartphone application |
N= number of images in dataset; DT= decision tree; AutoML = automated machine learning; CNN=convolutional neural network; DCNN= deep convolutional neural network; K-NN= K-nearest neighbour; PC = personal computer; ETL=ensemble transfer learning.
In our study, the diagnostic accuracy of the proposed AI algorithm was comparable to that of otolaryngology specialists and superior to that of non-experts, including general practitioners and otolaryngologic residents. Khan et al. developed an expert judgment system to differentiate 2 different middle ear diseases (normal, chronic otitis media with tympanic perforation, and otitis media with effusion).23 The average accuracy score for medical professionals with mixed expertise was 74%, which is inferior to that of the proposed CNN model (95%). Byun et al. reported an average accuracy score of 82.9% in diagnosing three common middle ear diseases (otitis media with effusion, chronic otitis media, and cholesteatoma) for junior otolaryngology resident doctors, which is inferior to the results of the proposed AI algorithm (97.2%).25 A similar trend was observed in the study by Livingston et al., who found a lower accuracy of 58.9% for non-experts compared to the proposed AI algorithm (88.7%).26 While AI algorithms have achieved performances that are comparable to those of medical experts in diagnosing common middle ear disorders, the difference between a diagnosis made by AI and that made by a clinician with different types of expertise still needs to be further explored.
The utilisation of artificial intelligence coupled with the CNN strategies of deep learning for automated disease detection is made possible by the fact that each ear drum/middle ear disease has a unique visual feature. Ear drum/middle ear diseases are usually detected and diagnosed through visual examination by clinicians based on otoscopy. However, it is challenging for general practitioners or non-otolaryngologists to make proper diagnoses due to limited training and experience in otoscopy. In addition, due to economic or geographical barriers, access to otolaryngology specialists for an adequate diagnosis and a proper subsequent treatment may be limited.27 The burden of hearing loss has attracted increasing attention worldwide, and hearing damage caused by middle ear disease can be reversed if diagnosed promptly.28 The recent COVID-19 pandemic has also posed challenges in terms of providing adequate health services, rendering telemedicine a useful tool in detecting and diagnosing diseases that previously required close contact and on-site inspection.29 Telemedicine for middle ear diseases is made possible for clinicians by using otoscopic images obtained from smartphone-enabled otoscopy.30 However, conducting telemedicine for otoscopic inspection may be difficult in areas or countries with poor mobile internet infrastructure or limited mobile internet access. Our system can be simply embedded in smartphones, and it enables the detection and recognition of ear drum/middle ear diseases in a timely manner without experts or online consultations.
The training results of the CNN models showed that the detection and diagnosis of middle ear disease could be achieved and simplified by transfer learning. Therefore, a lightweight model such as MobileNetV2 can also have a comparable accuracy with that of other large models. Using MobileNetV2 and TensorFlow Serving or Core ML technology, the model can be put on a mobile device to assist doctors in the diagnosis of middle ear diseases. The program also revealed a user-friendly interface for assisting diagnoses. For example, the intricate interrogation showed that 9 of the 180 images of chronic suppurative otitis media were diagnosed as tympanic perforation, while 7 of the 155 images of tympanic retraction were diagnosed as otitis media with effusion. In real-world practice, both diseases sometimes coexist and are diagnosed simultaneously. Our solution provides clinicians with the most likely diagnosis by ranking percentages and including the coexisting disease in the first two orders.
In this study, the transfer learning model was used to develop a mobile device application to detect middle ear diseases, so that model inference could be performed without additional cloud resource requirements. Other studies were based on cloud computing methods to perform model inference, which requires internet access and long-latency connections, and personal privacy could be violated. However, our system does not require internet access. In places where medical resources are scarce or mobile internet access is limited, clinicians can make good use of this device to assist in proper the diagnosis of middle ear diseases.
Current AI systems have achieved diagnostic performances that are comparable to those of medical experts in many medical specialties, especially in image recognition-related fields.31 Smartphones with features such as high-resolution cameras and powerful computation chips have been extensively used worldwide. The diagnostic accuracy of our smartphone-based AI application was comparable to that of otolaryngology experts. The diagnostic suggestions generated by medical AI may be beneficial for medical professionals with different levels and types of expertise. For non-otolaryngology medical professionals, the proposed algorithm may assist in expert-level diagnosis, leading to seamless diagnoses, increasing the speed of treatment or ensuring a proper referral. For otolaryngology experts, the proposed algorithm may provide an opportunity for shared decision making by providing easily accessible smartphone tools, thereby improving the doctor-patient relationship.
Our study has some limitations that should be noted. First, the model cannot cover all eardrum/middle ear diseases, such as complicated cholesteatoma and rare middle ear tumours. However, our model is sufficient for the interpretation of the most common eardrum/middle ear diseases and the decision regarding further referral. Second, despite the high accuracy, the final diagnoses still require further inspection by medical personnel to prevent misjudgements. Procrastination with regard to treating some middle ear diseases, such as acute otitis media, may lead to fatal complications in rare cases. Third, all of our image data were retrieved from a single medical centre with high-resolution otoendoscopy, and further expansion of the image database or labelling of the disease targets may be required to extend its applicability and publicity. Finally, although the smartphone application can either take an existing middle ear image or a snapshot provided by the mobile device as input, the quality of the images obtained via the latter approach may not be satisfactory for further processing. Further studies are required to examine the coupled otoscopic application to better acquire eardrum images.
In this study, we developed a system for detecting and categorizing common eardrum or middle ear diseases by artificial intelligence and integrated it into a smartphone application to detect ten common ear conditions by transfer learning. The accuracy of the proposed model is comparable to that of otolaryngology specialists in the detection or diagnosis of common middle ear diseases. Due to its compactness, the developed model can easily be integrated into a mobile device and used in telemedicine for detecting middle ear disorders.
Contributors
YC Chen, YF Cheng and YC Chu participated in writing the final draft. YC Chen, YF Cheng, WH Liao, CY Huang and YC Chu designed the study, interpreted the results, and wrote the draft. Albert Yang and YF Cheng conceptualized and designed the study, interpreted the data, and critically revised the manuscript. YC Chen, YT Lee and WY Lee reviewed and verified the raw data. CY Hsu, YT Lee and YC Chen helped review all of the images in our study. All authors had full access to all the data in the study and accept responsibility to submit for publication.
Data sharing statement
The analysed data in this study can be obtained from Yen-Fu Cheng at yfcheng2@vghtpe.gov.tw.
Editor note
The Lancet Group takes a neutral position with respect to territorial claims in published maps and institutional affiliations.
Declaration of interests
Yen-Chi Chen, Yuan-Chia Chu, Chii-Yuan Huang, Albert C Yang, Yen-Ting Lee, Wen-Ya Lee, Chien-Yeh Hsu, Wen-Huei Liao, Yen-Fu Cheng have no conflict of interest to declare.
Acknowledgements
We would like to thank the Big Data Centre, Taipei Veterans General Hospital for data arrangement and Ms. Jue-Ni Huang, Shang-Liang Wu, PhD for the statistical analysis. This study was supported by grants from the Ministry of Science and Technology (MOST110-2622-8-075-001, MOST110-2320-B-075-004-MY3, MOST-110-2634-F-A49 -005, MOST110-2745-B-075A-001A and MOST110-2221-E-075-005), Veterans General Hospitals and University System of Taiwan Joint Research Program (VGHUST111-G6-11-2, VGHUST111c-140), and Taipei Veterans General Hospital (V111E-002-3).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.eclinm.2022.101543.
Contributor Information
Albert C. Yang, Email: ccyang10@vghtpe.gov.tw.
Wen-Huei Liao, Email: whliao@vghtpe.gov.tw.
Yen-Fu Cheng, Email: yfcheng2@vghtpe.gov.tw.
Appendix. Supplementary materials
References
- 1.Kim CS, Jung HW, Yoo KY. Prevalence of otitis media and allied diseases in Korea–results of a nation-wide survey, 1991. J Korean Med Sci. 1993;8(1):34–40. doi: 10.3346/jkms.1993.8.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paradise JL, Rockette HE, Colborn DK, et al. Otitis media in 2253 Pittsburgh-area infants: prevalence and risk factors during the first two years of life. Pediatrics. 1997;99(3):318–333. doi: 10.1542/peds.99.3.318. [DOI] [PubMed] [Google Scholar]
- 3.Libwea JN, Kobela M, Ndombo PK, et al. The prevalence of otitis media in 2–3 year old Cameroonian children estimated by tympanometry. Int J Pediatr Otorhinolaryngol. 2018;115:181–187. doi: 10.1016/j.ijporl.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Legros J-M, Hitoto H, Garnier F, Dagorne C, Parot-Schinkel E, Fanello S. Clinical qualitative evaluation of the diagnosis of acute otitis media in general practice. Int J Pediatr Otorhinolaryngol. 2008;72(1):23–30. doi: 10.1016/j.ijporl.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 5.Leskinen K, Jero J. Acute complications of otitis media in adults. Clin Otolaryngol. 2005;30(6):511–516. doi: 10.1111/j.1749-4486.2005.01085.x. [DOI] [PubMed] [Google Scholar]
- 6.Principi N, Marchisio P, Esposito S. Otitis media with effusion: benefits and harms of strategies in use for treatment and prevention. Expert Rev Anti-infective Therapy. 2016;14(4):415–423. doi: 10.1586/14787210.2016.1150781. [DOI] [PubMed] [Google Scholar]
- 7.Pichichero ME, Poole MD. Comparison of performance by otolaryngologists, pediatricians, and general practioners on an otoendoscopic diagnostic video examination. Int J Pediatr Otorhinolaryngol. 2005;69(3):361–366. doi: 10.1016/j.ijporl.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 8.Zafer C. Fusing fine-tuned deep features for recognizing different tympanic membranes. Biocybernet Biomed Eng. 2020;40(1):40–51. [Google Scholar]
- 9.Viscaino M, Maass JC, Delano PH, Cheein FA. Computer-aided ear diagnosis system based on CNN-LSTM hybrid learning framework for video otoscopy examination. IEEE Access. 2021;9:161292–161304. doi: 10.1109/ACCESS.2021.3132133. [DOI] [Google Scholar]
- 10.Myburgh HC, Jose S, Swanepoel DW, Laurent C. Towards low cost automated smartphone-and cloud-based otitis media diagnosis. Biomed Signal Process Control. 2018;39:34–52. [Google Scholar]
- 11.Seok J, Song J-J, Koo J-W, Kim HC, Choi BY. The semantic segmentation approach for normal and pathologic tympanic membrane using deep learning. BioRxiv. 2019 doi: 10.1101/515007. In preparation. [DOI] [Google Scholar]
- 12.Anwar SM, Majid M, Qayyum A, Awais M, Alnowami M, Khan MK. Medical image analysis using convolutional neural networks: a review. 2018;42(11):1–13. [DOI] [PubMed]
- 13.Srinivasu PN, SivaSai JG, Ijaz MF, Bhoi AK, Kim W, Kang JJ. Classification of Skin Disease Using Deep Learning Neural Networks with MobileNet V2 and LSTM. Sensors. 2021;21(8):2852. doi: 10.3390/s21082852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Liu Z, Wang G, Cai Z, Zhang H. Ensemble transfer learning algorithm. IEEE Access. 2017;6:2389–2396. [Google Scholar]
- 15.Howard D, Maslej MM, Lee J, Ritchie J, Woollard G, French L. Transfer learning for risk classification of social media posts: Model evaluation study. J Med Internet Res. 2020;2(5):e15371. doi: 10.2196/15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xue D, Zhou X, Li C, et al. An application of transfer learning and ensemble learning techniques for cervical histopathology image classification. IEEE Access. 2020;8:104603–104618. [Google Scholar]
- 17.Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A. Proceedings of the IEEE conference on computer vision and pattern recognition. 2016. Learning deep features for discriminative localization; pp. 2921–2929. [Google Scholar]
- 18.Livingstone D, Talai AS, Chau J, Forkert ND. Building an Otoscopic screening prototype tool using deep learning. J Otolaryngol-Head Neck Surg. 2019;48(1):1–5. doi: 10.1186/s40463-019-0389-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W, Li Y, Zou T, Wang X, You J, Luo Y. “A Novel Image Classification Approach via Dense-MobileNet Models”. Mob Inf Syst. 2020;2020(1) doi: 10.1155/2020/7602384. [DOI] [Google Scholar]
- 20.Wu Z, Lin Z, Li L, et al. Deep learning for classification of pediatric otitis media. The Laryngoscope. 2021;131(7):E2344–E2351. doi: 10.1002/lary.29302. [DOI] [PubMed] [Google Scholar]
- 21.Moshtaghi O, Sahyouni R, Haidar YM, et al. Smartphone-enabled otoscopy in neurotology/otology. Otolaryngology–Head and Neck Surgery. 2017;156(3):554–558. doi: 10.1177/0194599816687740. [DOI] [PubMed] [Google Scholar]
- 22.Viscaino M, Maass JC, Delano PH, Torrente M, Stott C, Auat Cheein F. Computer-aided diagnosis of external and middle ear conditions: A machine learning approach. PLoS One. 2020;15(3) doi: 10.1371/journal.pone.0229226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan MA, Kwon S, Choo J, et al. Automatic detection of tympanic membrane and middle ear infection from oto-endoscopic images via convolutional neural networks. Neural Networks. 2020;126:384–394. doi: 10.1016/j.neunet.2020.03.023. [DOI] [PubMed] [Google Scholar]
- 24.Mazzia V, Khaliq A, Salvetti F, Chiaberge M. An edge AI application. IEEE Access; 2020. (Real-time apple detection system using embedded systems with hardware accelerators). 8:9102–9114. [Google Scholar]
- 25.Byun H, Yu S, Oh J, et al. An assistive role of a machine learning network in diagnosis of middle ear diseases. J Clin Med Res. 2021;10(15):3198. doi: 10.3390/jcm10153198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livingstone D, Chau J. Otoscopic diagnosis using computer vision: An automated machine learning approach. Laryngoscope. 2020;130(6):1408–1413. doi: 10.1002/lary.28292. [DOI] [PubMed] [Google Scholar]
- 27.Crowson MG, Ranisau J, Eskander A, et al. A contemporary review of machine learning in otolaryngology–head and neck surgery. Laryngoscope. 2020;130(1):45–51. doi: 10.1002/lary.27850. [DOI] [PubMed] [Google Scholar]
- 28.Haile LM, Kamenov K, Briant PS, et al. Hearing loss prevalence and years lived with disability, 1990–2019: findings from the Global Burden of Disease Study 2019. Lancet. 2021;397(10278):996–1009. doi: 10.1016/S0140-6736(21)00516-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith AC, Thomas E, Snoswell CL, et al. Telehealth for global emergencies: Implications for coronavirus disease 2019 (COVID-19) J Telemed Telecare. 2020;26(5):309–313. doi: 10.1177/1357633X20916567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meng X, Dai Z, Hang C, Wang Y. Smartphone-enabled wireless otoscope-assisted online telemedicine during the COVID-19 outbreak. Am J Otolaryngol. 2020;41(3):102476. doi: 10.1016/j.amjoto.2020.102476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X, Faes L, Kale AU, et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digital Health. 2019;1(6):e271–e297. doi: 10.1016/S2589-7500(19)30123-2. [DOI] [PubMed] [Google Scholar]
- 32.Cha D, Pae C, Seong SB, Choi JY, Park HJ. Automated diagnosis of ear disease using ensemble deep learning with a big otoendoscopy image database. EBioMedicine. 2019;45:606–614. doi: 10.1016/j.ebiom.2019.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Senaras C, Moberly AC, Teknos T, et al. Medical Imaging 2017: Computer-Aided Diagnosis; 2017. SPIE; 2017. Autoscope: automated otoscopy image analysis to diagnose ear pathology and use of clinically motivated eardrum features; pp. 500–507. [Google Scholar]
- 34.Zeng X, Jiang Z, Luo W, Li H, Li H, Li G, Li Z, et al. Efficient and accurate identification of ear diseases using an ensemble deep learning model. Sci Rep. 2021;11(1):1–10. doi: 10.1038/s41598-021-90345-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.