Abstract
The impact of three different soil DNA extraction methods on bacterial diversity was evaluated using PCR-based 16S ribosomal DNA analysis. DNA extracted directly from three soils showing contrasting physicochemical properties was subjected to amplified ribosomal DNA restriction analysis and ribosomal intergenic spacer analysis (RISA). The obtained RISA patterns revealed clearly that both the phylotype abundance and the composition of the indigenous bacterial community are dependent on the DNA recovery method used. In addition, this effect was also shown in the context of an experimental study aiming to estimate the impact on soil biodiversity of the application of farmyard manure or sewage sludge onto a monoculture of maize for 15 years.
Up to now, most of the microbial diversity studies conducted in complex ecosystems, such as soil, have been biased essentially by the unculturability of many microorganisms and the lack of sensitivity of traditional microbiological methods (4). In the past decade, applications of new molecular biology methods based primarily on amplification of soil-extracted nucleic acids have provided a pertinent alternative to classical culture-based microbiological methods, providing unique insight into the composition, richness, and structure of microbial communities (3, 6, 9, 11). However, the results of molecular analysis of microbial communities rely not only on the extraction of DNAs representative of the indigenous bacterial community composition but also on factors related to PCR, such as the choice of primers, the concentration of amplified DNA, errors in the PCR, or even the method chosen for analysis. Recently, numerous studies have investigated new methods to improve extraction, purification, amplification, and quantification of DNA from soils (8, 13, 14). Comparative studies have been performed to analyze the efficiency of methods for extraction and purification of soil DNA recovered, revealing that these methods suffer from low efficiency, mainly due to incomplete cell lysis and DNA sorption to soil particles (1, 5). However, the impact of the extraction method on the outcome of indigenous microbial community analysis has not been clearly established (5).
The goal of this study was to evaluate the effect of DNA extraction methods on the bacterial diversity detected within DNA extracted from three soils exhibiting contrasting physicochemical characteristics and, in the context of an experimental study, from unamended soil and soils amended for 15 years with farmyard manure or sewage sludge. Two commercial DNA purification kits and a laboratory-devised method based on mechanical lysis were used to extract DNA directly from soils. Amplified ribosomal DNA restriction analysis (ARDRA) and ribosomal intergenic spacer analysis (RISA) were performed to estimate the effect of the DNA extraction procedure used on the bacterial diversity revealed.
DNA extraction from soils.
The physicochemical properties of the three soils used in this study are presented in Table 1. The field experiment was conducted in the Institut National de la Recherche Agronomique domain of Bordeaux, France (45°N, 1°W) on an acid, coarse, sandy soil (7). This experiment was limited to continuously growing corn (Zea maize, variety INRA 160), with four treatments, as follows: none (i) (i.e., N-P-K fertilizers only, with soil considered unamended), (ii) farmyard manure (10 tons of dry matter per ha each year), (iii) sewage sludge (10 tons of dry matter per ha each year), and (iv) sewage sludge (100 tons of dry matter per ha every 2 years), with blocks as for treatment in a randomized manner. Fresh soils samples were sieved (2-mm mesh) and stored at 4°C. Nucleic acids were extracted from three 250-mg aliquots of soils using two commercial kits, the UltraClean Soil DNA kit (MoBio Laboratories, Inc., Solana Beach, Calif.) and the Fast DNA spin sample kit (for soil; Bio 101, Lajolla, Calif.), according to the manufacturers' recommendations and using a procedure developed in our laboratory. Briefly, 1 ml of a solution containing 100 mM Tris (pH 8.0), 100 mM EDTA, 100 mM NaCl, 1% (wt/vol) polyvinylpyrrolidone, and 2% (wt/vol) sodium dodecyl sulfate was added to 250 mg of soil in a 2-ml mini-bead-beater tube containing 0.5 and 0.1 g of 106-μm- and 2-mm-diameter glass beads, respectively. Samples were then homogenized for 30 s at 1,600 rpm in a mini-bead-beater cell disruptor (Mikro-Dismembrator S; B. Braun Biotech International), after which the samples were centrifuged at 14,000 × g for 1 min at 4°C. The collected supernatants were incubated for 10 min on ice with 1/10 volume of 5 M sodium acetate and centrifuged at 14,000 × g for 5 min. After precipitation with 1 volume of ice-cold isopropanol, the nucleic acids were washed with 70% ethanol and purified using a Sepharose 4B spin column. The quality and the size of the soil DNAs were checked by electrophoresis on 1% agarose gels. DNA was quantified using a BioPhotometer (Eppendorf, Hamburg, Germany). Three replicates were used for DNA evaluation. A double-factor analysis of variance was performed to determine significant differences in the yield of soil DNA. The Newman-Keuls procedure (P < 0.05) was used to evaluate the impact of the different soils and extraction methods used on the DNA extraction yield.
TABLE 1.
Properties of soils used in DNA extraction methods
| Soil sampling location | Soil type | Amt (%) of:
|
% Organic C | pH | ||
|---|---|---|---|---|---|---|
| Clay | Sand | Silt | ||||
| Dijon | Eutric calcaric cambisoil | 22 | 38 | 40 | 15.8 | 7.8 |
| Couhins | Podzol | 4.2 | 83.3 | 12.5 | 1.3 | 5.3 |
| Epoisses | Eutric calcaric cambisoil | 48 | 7.2 | 44.6 | 20 | 7.5 |
ARDRA and RISA.
To test the possible selectivity of the DNA extraction method, ARDRA and RISA were performed on DNA extracted directly from soils. Three independent replicates were included in each analysis, and the results were examined separately. Small subunit (SSU) ribosomal DNA (rDNA) genes were amplified from each soil replicate in thermocycler (PTC 200 gradient cycler; MJ Research, Waltham, Mass.). PCR amplification from 50 ng of extracted soil DNA was conducted with a total volume of 50 μl by using 1 μM concentrations of the universal primers 27f (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 1492r (5′-TAC GGH TAC CTT GTT ACG ACT T-3′) (2) and 2.5 U of Taq DNA polymerase (Appligene) under the following conditions: 5 min at 94°C, 35 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C, plus an additional 15-min cycle at 72°C. The PCR products were then separated by electrophoresis on a 1% agarose gel. Bands were extracted and purified from the gel using the Qiaex II kit (Qiagen, Hilden, Germany) as recommended by the manufacturer. Ten microliters of purified 16S rDNA from each sample was digested with 15 U of RsaI, HhaI, or MspI in 15-μl reaction mixtures.
The intergenic spacer region between the small and large subunits of the ribosomal genes was amplified from 50 ng of DNA extracted from soil with a total volume of 50 μl by using the universal primers 38r (5′-CCG GGT TTC CCC ATT CGG-3′) and 72f (5′-TGC GGC TGG ATC TCC TT-3′) (2) under the conditions described above. For both ARDRA and RISA, 15-μl aliquots were separated by electrophoresis on a native 6% acrylamide gel run for 12 or 17 h at 8 mA, respectively. Gels were stained with SYBR green II (Molecular Probes, Leiden, The Netherlands) by following the recommendations of the manufacturer.
PCR efficiency analysis.
Three microliters of 16S rDNA PCR product was separated by electrophoresis on a 1% agarose gel run for 1 h at 100 V. Gels were stained with ethidium bromide and analyzed with the ImageQuant program. 16S-23S intergenic rDNA PCR products were purified with columns (Microcon PCR/Amicon Bioseparations; Millipore, Bedford, Mass.) according to the manufacturers' recommendations. A 2-μl volume of each purified PCR product was then diluted into 48 μl of sterile water, and the nucleic acid content was determined using a BioPhotometer (Eppendorf). For both 16S and 16S-23S intergenic rDNA PCR products, three repeats were carried out. Data were analyzed statistically as described above.
Effect of extraction methods on bacterial diversity analysis.
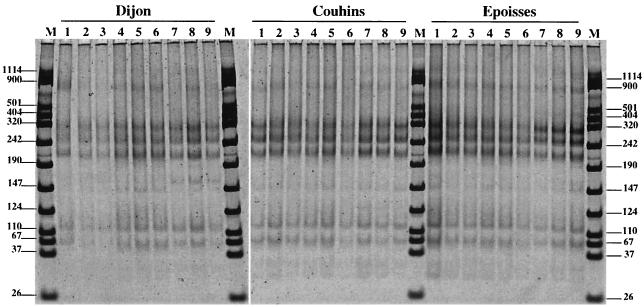
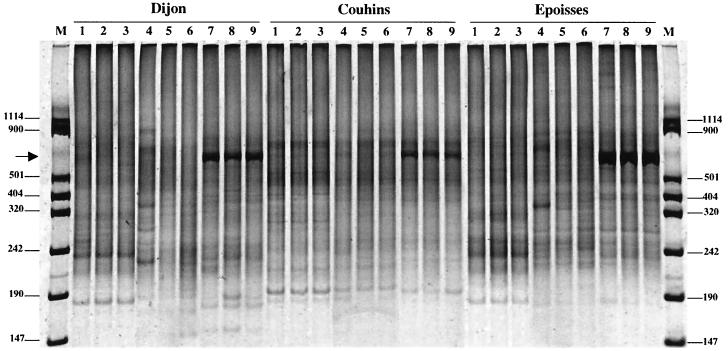
The DNA yields obtained ranged between 0.2 and 2.5 μg g of soil−1 from sample to sample and differed with the extraction method (Table 2). The MS laboratory method yielded an amount of DNA that was significantly higher than those obtained with the kits from MoBio Laboratories and Bio 101 for Dijon and Epoisses soils. However, the quantity of DNA extracted from the sandy soil of Couhins was not significantly affected by the extraction method used. These results therefore suggest that the yield of DNA per gram of soil depends on the method used and on the properties of the soil considered. Patterns of restriction fragment length polymorphism of amplified 16S rDNA genes digested with RsaI restriction enzyme indicate that amplified 16S rDNA genes were very similar whatever extraction method used or soil tested (Fig. 1). Other enzymes (HhaI and MspI) were tested and also provided identical patterns for the three soils (data not shown). It is commonly admitted that ARDRA resolves microbial diversity up to the genus level. Our results therefore confirm that some genera are widely distributed and dominant in the three agricultural soils tested here. In addition, they suggest that at the genus level the particular DNA extraction method used does not influence bacterial diversity analysis. Evaluation of the impact of extraction methods on bacterial diversity was further conducted by means of RISA. Demonstration of the relevance and the sensitivity of the RISA approach for microbial diversity analysis has been previously reported (10). Similar RISA patterns were visualized for the three soil replicates, illustrating the reproducibility of the results for both DNA extraction and amplification (Fig. 2). However, it has to be noted that some differences between soils were observed: for DNAs extracted from the Dijon soil, additional bands were observable at 900, 300, and 220 bp (Fig. 2, lane 4 for Dijon soil). In addition, for each soil many differences were observed between the RISA patterns obtained for DNA extracted using different methods. For instance, a strong band between 242 and 320 bp and another one just above 501 bp were consistently observed in the RISA patterns generated for DNA extracted from the soil of Couhins using the MoBio Laboratories kit (Fig. 2, lanes 4 to 6). However, these two bands were not shown in the RISA patterns obtained with DNA extracted using the MS laboratory method (Fig. 2, lanes 1 to 3) and the Bio 101 kit (Fig. 2, lanes 7 to 9). Therefore, differences in the location of the dominant bands were evident for both Epoisses and Dijon soils when the RISA patterns obtained with the three different DNA extraction methods were compared. RISA patterns obtained with the soil of Couhins appeared to be more similar regardless of the extraction method used. This probably reflects the homogeneity of this soil due to its high percentage of sand (Table 1). Surprisingly, a strong-dominant band at 800 bp was observed for the three soils studied only in the RISA patterns obtained with DNA recovered with the Bio 101 kit, while it was not possible to distinguish this band with the two other extraction methods. Assuming that the brightness of this 800-bp band was expected to be identical for the same soil whatever the extraction method used, the discrepancies (variations in the positions and brightness of the bands) in the results obtained for the same soil using different DNA extraction methods strongly suggest a preferential DNA recovery and/or preferential DNA amplification, depending on the method used. It must be emphasized that such discrepancies were observed in comparing DNA extracted from soils using only methods based on mechanical lysis by bead beating, while other methods, based on sonication or lysozyme lysis, have been reported (5, 8). Diversity analyses based on cultivation methods are biased by the fact that cultivatable bacteria represent only a small proportion (<0.1%) of the soil microbial community (12). Similarly, our data show that DNA extraction from soil also suffers from biases that can distort the revealed community composition, richness, and structure and that the outcome of microbial community analysis is dependent on the DNA recovery method used. Because important variations in the brightness of the bands appeared in the RISA patterns for the different soil DNA extraction methods, it seems that the relative abundance of phylotypes in soil cannot be accurately estimated with these direct molecular approaches.
TABLE 2.
Yield of DNA extracted from Dijon, Couhins, and Epoisses soils using the MoBio Laboratories kit, the Bio 101 kit, or the MS laboratory method
| Soil sampling location | Yield of DNA (μg/g of soil) witha:
|
||
|---|---|---|---|
| MoBio Laboratories kit | Bio 101 kit | MS laboratory method | |
| Dijon | 0.19 ± 0.11 a | 0.92 ± 0.46 ab | 2.01 ± 1.07 b |
| Couhins | 0.70 ± 0.23 ab | 0.79 ± 0.36 ab | 0.97 ± 0.10 b |
| Epoisses | 0.47 ± 0.13 ab | 1.01 ± 0.86 ab | 2.52 ± 1.09 b |
Values are means ± standard errors. (SE). Values followed by the same letter do not differ significantly (P < 0.05).
FIG. 1.
ARDRA. Shown is a SYBR green II-stained gel (6% acrylamide) of RsaI-digested PCR products amplified with 16S rDNA universal primers (27f and 1492r) from DNA extracted from Dijon, Couhins, and Epoisses soils following three different extraction methods: the MS laboratory method (lanes 1 to 3), the MoBio kit method (lanes 4 to 6), and the Bio 101 kit method (lanes 7 to 9). Lanes M, VIII Boehringer Mannheim molecular size markers (sizes indicated in base pairs at left and right).
FIG. 2.
RISA shown is a SYBR green II-stained gel (6% acryamide) of PCR products amplified with 16S rDNA intergenic spacer universal primers (38r and 72f) from DNA extracted from Dijon, Couhins, and Epoisses soils following three different extraction methods: the MS laboratory method (lanes 1 to 3), the MoBio kit method (lanes 4 to 6), and the Bio 101 kit method (lanes 7 to 9). Lanes M, VIII Boehringer Mannheim molecular size markers (sizes indicated in base pairs at left and right). Arrow, 800-bp band.
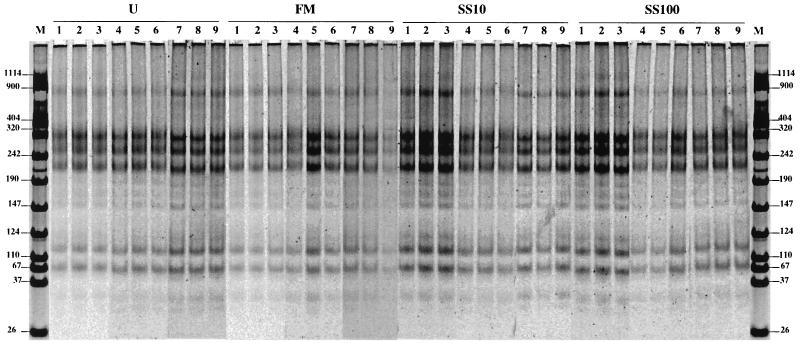
In order to evaluate more precisely the biases introduced by the soil DNA extraction methods, we have conducted ARDRA and RISA in the context of experimental studies on DNA extracted from the soil of Couhins treated with farmyard manure and sewage sludge or not treated for 15 years. ARDRA produced patterns identical to those shown in Fig. 1 (Fig. 3). These results indicate that the dominant microbial genera were apparently not affected by the application of either farmyard manure or sewage sludge. However, despite the fact that almost identical RISA patterns were previously obtained with the soil of Couhins whatever the DNA extraction method used (Fig. 2), RISA conducted on DNA extracted directly from unamended soil or farmyard manure- or sewage sludge-treated soil of Couhins confirmed that the obtained fingerprints also depend on the extraction method used (Fig. 4). Notably, the strong-dominant band at 800 bp previously observed only with DNA recovered with the Bio 101 kit was detected again, with the same intensity, in analyses of unamended and farmyard manure- and sewage sludge-treated soils.
FIG. 3.
ARDRA. Shown is a SYBR green II-stained gel (6% acrylamide) of RsaI-digested PCR products amplified with 16S rDNA universal primers (27f and 1492r) from DNA extracted from unamended soil (U) and farmyard manure (FM)- and sewage sludge (SS10 and SS100)-treated plots of Couhins following three different extraction methods: the MS laboratory method (lanes 1 to 3), the MoBio kit method (lanes 4 to 6), and the Bio 101 kit method (lanes 7 to 9). Lanes M, VIII Boehringer Mannheim molecular size markers (sizes indicated in base pairs at left and right). SS10, 10 tons of dry matter/ha/years. SS100, 100 tons of dry matter/ha/2 years.
FIG. 4.
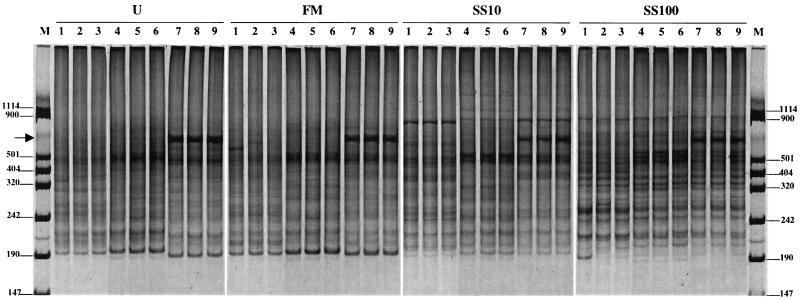
RISA. Shown is SYBR green II-stained gel (6% acryamide) of PCR products amplified with 16S ribosomal universal primers (38r and 72f) from DNA extracted from unamended soil (U) and farmyard manure (FM)- or sewage sludge (SS10 and SS100)-treated plots of Couhins soil following three different extraction methods: the MS laboratory method (MS) (lanes 1 to 3), the MoBio kit method (lanes 4 to 6), and the Bio 101 kit method (lanes 7 to 9). Lanes M, VIII Boehringer Mannheim molecular size markers (sizes indicated in base pairs at left and right). Arrow, 800-bp band.
Further, no matter what the extraction method used, the impact on soil biodiversity of the application of either farmyard manure or sewage sludge could be shown clearly. Regardless of the extraction method used, the RISA patterns for unamended soil and farmyard manure (FM)-treated soil were very similar, indicating that the application of farmyard manure did not affect the soil biodiversity (Fig. 4). On the contrary, a comparison of RISA patterns for unamended soil and SS10 or SS100 sewage sludge-treated soil showed clear differences, indicating that the application of sewage sludge dramatically modified the soil biodiversity. More precisely, when comparing the RISA fingerprint for unamended soil with that for sewage sludge-treated soil, two major effects can be shown, as follows: (i) a strong diminution of the number of phylotypes (i.e., bands at 190 bp and between 242 and 340 bp) and (ii) an important modification of RISA fingerprints due to both the reinforcement of existing phylotypes (i.e., bands below 242 bp and at 320 bp) and the appearance of new phylotypes (i.e., bands at 900 bp, above 501 bp, and at 320 bp) (Fig. 4). In addition, it can be noted that the higher dose of sewage sludge (i.e., 100 tons/ha every 2 years) produced a stronger alteration of the microbial biodiversity of the soil of Couhins. Nevertheless, again the extraction method used affected the biodiversity as revealed by RISA. Despite the bias introduced by the DNA extraction used, the impact of agricultural practices on soil biodiversity can be measured thanks to this molecular approach, though the relative abundance of phylotypes in soils cannot be accurately estimated.
In addition, the yield of the 16S rDNA and 16S-23S intergenic rDNA PCR reactions was measured in order to estimate the impact of the extraction method and/or the soil matrix on PCR efficiency. The 16S rDNA amplification efficiency was estimated by image analysis. In fact, no matter what the soil matrix, the samples extracted with the MS laboratory method yielded lower amounts of 16S rDNA PCR product than those extracted with the Bio 101 kit (Table 3). The two-way analysis of variance revealed that the 16S rDNA amplification was significantly affected by the extraction method but not by the soil matrix. In addition, no significant interaction between the extraction method and the soil matrix was shown by the Newman-Keuls procedure. The effect of the extraction method on the efficiency of the 16S rDNA is probably due to differential coextraction of impurities which may affect the activity of the Taq polymerase. However, despite the alteration of the amplification efficiency, no clear differences have been observed in ARDRA (Fig. 3). This contradictory result might be due to the fact that ARDRA patterns reveal only dominant microbial genera, hiding much of the existing diversity and probably masking the effect of the alteration of the amplification efficiency.
TABLE 3.
Yield of 16S rDNA PCR product amplified from DNA extracted from unamended soil and farmyard manure- and sewage sludge-treated soils following three different extraction methods
| Soila | Yield of 16S rDNA PCR product (arbitrary units) withb:
|
||
|---|---|---|---|
| MS laboratory method | MoBio Laboratories kit | Bio 101 kit | |
| Unamended | 172.5 ± 20.5 a | 194.0 ± 22.6 ab | 223.0 ± 7.7 b |
| Treated with: | |||
| Farmyard manure | 163.4 ± 21.8 a | 163.4 ± 6.9 a | 227.3 ± 9.4 b |
| Sewage sludge (SS10) | 174.5 ± 18.1 a | 192.4 ± 7.5 ab | 218.5 ± 4.5 b |
| Sewage sludge (SS100) | 171.8 ± 21.7 a | 197.8 ± 15.4 ab | 207.0 ± 16.2 ab |
SS10, 10 tons of dry matter/ha/yr. SS100, 100 tons of dry matter/ha/2 yr.
Values are means ± SE. Values followed by the same letter do not differ significantly (P < 0.05).
The yield of the 16S-23S intergenic rDNA PCR was also determined. In fact, no matter what extraction method was used, the PCR yield resulting from the amplification of DNA extracted from FM-treated soil was significantly higher than that resulting from the amplification of DNA extracted from SS100 sewage sludge-treated soil (Table 4). Therefore, the soil matrix significantly affected the yield of the 16S-23S rDNA amplification and the extraction method did not affect it significantly. Again, PCR efficiency alterations due to soil matrices would be in contradiction with the RISA results, which showed that the microbial diversity of unamended soil was very similar to that of the FM-treated soil but very different from that of either the SS10 or the SS100 sewage sludge-treated soil. Therefore, despite the observed soil matrix effect on the efficiency of 16S-23S rDNA amplification, the biodiversity revealed by RISA was not affected in the same way, suggesting that the relation between PCR efficiency and the results of RISA is not obvious.
TABLE 4.
Yield of the RISA PCR product amplified from DNA extracted from unamended soil or farmyard manure- or sewage sludge-treated soil following three different extraction methods
| Soila | Yield of RISA PCR product (ng/μl) withb:
|
||
|---|---|---|---|
| MS laboratory method | MoBio Laboratories kit | Bio 101 kit | |
| Unamended | 61.4 ± 12.4 b | 41.0 ± 10.4 ab | 33.9 ± 7.4 a |
| Treated with: | |||
| Farmyard manure | 52.3 ± 8.6 ab | 67.3 ± 14.7 b | 61.4 ± 16.0 b |
| Sewage sludge (SS10) | 47.1 ± 4.5 ab | 63.1 ± 21.3 ab | 58.6 ± 11.8 ab |
| Sewage sludge (SS100) | 31.4 ± 8.6 a | 39.2 ± 15.6 a | 54.1 ± 4.4 ab |
SS10, 10 tons of dry matter/ha/yr. SS100, 100 tons of dry matter/ha/2 yr.
Values are means ± SE. Values followed by the same letter do not differ significantly at (P < 0.05).
In conclusion, the results presented here clearly demonstrate that soil DNA extraction methods can affect both phylotype abundance and composition of the indigenous bacterial community. PCR biases also occur. Notably, the PCR efficiency of 16S or 16S-23S rDNA was affected by the extraction method or the soil matrix, respectively. Overall, assuming that the biases operated uniformly for all samples examined using the same DNA extraction method, our RISA data also indicated that these direct molecular methods allowed (i) the differentiation of soils according to their bacterial communities and (ii) the monitoring of differences in the bacterial communities in a soil in response to a stress. However, all the problems described above need to be considered before drawing conclusions concerning relative abundance of microbial phylotypes in soils. Additionally, our work suggests that the use of standard soil DNA extraction and PCR methods by soil microbiologists could provide a more complete understanding of the composition and diversity of soil microbial communities.
Acknowledgments
We thank Jean-Claude Fournier and Bernard Lagacherie for helpful discussions. This study was supported by the MATE and the Burgundy Région (contract number B03039).
REFERENCES
- 1.Frostegard A, Courtois S, Ramisse V, Clerc S, Bernillon D, LeGall F, Jeanin P, Nesme X, Simonet P. Quantification of bias related to the extraction of DNA directly from soils. Appl Environ Microbiol. 1999;65:5409–5420. doi: 10.1128/aem.65.12.5409-5420.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gurtler V, Stanisich V A. New approaches to typing and identification of bacteria using the 16S–23S rDNA spacer region. Microbiology. 1996;142:3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- 3.Hill G T, Mitkowski N A, Aldrich-Wolfe L, Emele L R, Jurkonie D D, Ficke A, Maldona-Ramirez S, Lynch S T, Nelson E B. Methods for assessing the composition and diversity of soil microbial communities. Appl Soil Ecol. 2000;15:25–36. [Google Scholar]
- 4.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kresk M, Wellington E M H. Comparison of different methods for the isolation and purification of total community DNA from soil. J Microbiol Methods. 1999;39:1–16. doi: 10.1016/s0167-7012(99)00093-7. [DOI] [PubMed] [Google Scholar]
- 6.Kuske C R, Barns S M, Bush J D. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl Environ Microbiol. 1997;63:3614–3621. doi: 10.1128/aem.63.9.3614-3621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lineres I, Chaussod R, Juste C, Solda P. Microbial biomass and biological activities in an acid sandy soil treated with sewage sludge or farmyard manure in a long term field experiment. In: Dirkzwager A H, L'Hermite P, editors. Sewage sludge treatment and use: new developments, technological aspects and environmental effects. London, United Kingdom: Elsevier Applied Science; 1989. pp. 517–520. [Google Scholar]
- 8.Miller D N, Bryant J E, Madsen E L, Ghiorse W C. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl Environ Microbiol. 1999;65:4715–4724. doi: 10.1128/aem.65.11.4715-4724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nusslein K, Tiedje J M. Characterization of the dominant and rare members of a young Hawaiian soil bacterial community with small-subunit ribosmal DNA from DNA fractionated on the basis of its guanine and cytosine composition. Appl Environ Microbiol. 1998;64:1283–1289. doi: 10.1128/aem.64.4.1283-1289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ranjard L, Poly F, Combrisson J, Richaume A, Gourbiere F, Thioulouse J, Nazaret S. Heterogeneous cell density and genetic structure of bacterial pools associated with various soil microenvironments as determined by enumeration and DNA fingerprinting approach (RISA) Microb Ecol. 2000;39:263–272. [PubMed] [Google Scholar]
- 11.Stackebrandt E, Liesack W, Goebel B M. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J. 1993;7:232–236. doi: 10.1096/fasebj.7.1.8422969. [DOI] [PubMed] [Google Scholar]
- 12.Torsvik V, Golsoyr J, Daae F. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai Y-L, Olson B H. Rapid method for direct extraction of DNA from soil and sediment. Appl Environ Microbiol. 1991;57:1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou J, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]