Abstract
Positive single-strand RNA (+ RNA) viruses can remodel host cell membranes to induce a replication organelle (RO) isolating the replication of their genome from innate immunity mechanisms. Some of these viruses, including severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2), induce double-membrane vesicles (DMVs) for this purpose. Viral non-structural proteins are essential for DMV biogenesis, but they cannot form without an original membrane from a host cell organelle and a significant supply of lipids. The endoplasmic reticulum (ER) and the initial mechanisms of autophagic processes have been shown to be essential for the biogenesis of SARS-CoV-2 DMVs. However, by analogy with other DMV-inducing viruses, it seems likely that the Golgi apparatus, mitochondria and lipid droplets are also involved. As for hepatitis C virus (HCV), pores crossing both membranes of SARS-CoV-2-induced DMVs have been identified. These pores presumably allow the supply of metabolites essential for viral replication within the DMV, together with the export of the newly synthesized viral RNA to form the genome of future virions. It remains unknown whether, as for HCV, DMVs with open pores can coexist with the fully sealed DMVs required for the storage of large amounts of viral RNA. Interestingly, recent studies have revealed many similarities in the mechanisms of DMV biogenesis and morphology between these two phylogenetically distant viruses. An understanding of the mechanisms of DMV formation and their role in the infectious cycle of SARS-CoV-2 may be essential for the development of new antiviral approaches against this pathogen or other coronaviruses that may emerge in the future.
Keywords: COVID-19, SARS-CoV-2, Positive-sense single-stranded RNA virus, Virus/cell interactions, Double-membrane vesicle, Replication organelle
Introduction
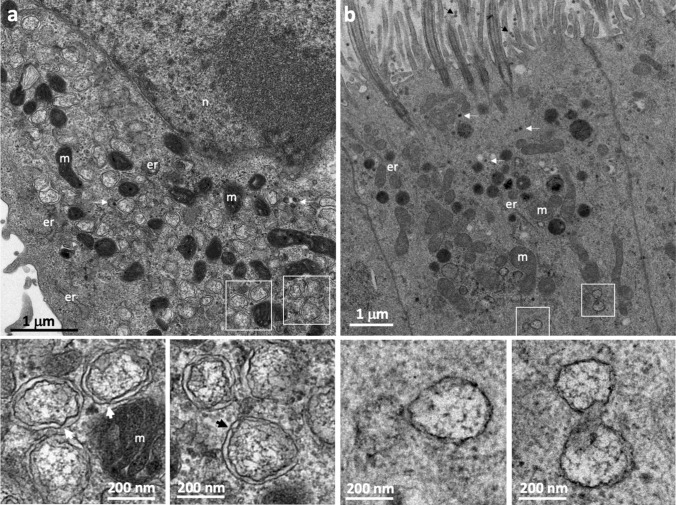
Positive single-strand RNA (+ RNA) viruses use their genomes directly as messenger RNA, producing a polyprotein from which the various viral proteins required for replication are derived. One of these proteins is the RNA-dependent RNA polymerase, which copies the + RNA genome to form a double-stranded (dsRNA) replicative template that is used to generate more + RNA for the translation of viral proteins and to constitute the genome of newly synthesized virions. The replication of + RNA viruses takes place in the cytoplasm, potentially exposing the viral RNA, and the replication intermediate constituted by a dsRNA in particular, to mechanisms of innate immunity engaged by the host cell [1, 2]. However, + RNA viruses have developed clever strategies for protecting themselves from such host cell defense mechanisms. The assembly of viral replication complexes involves associations with the cytoplasmic membranes of the host cell, with the virus manipulating host cell factors to induce the extensive remodeling of these membranes, thereby isolating viral replication from cell sensors. Virus-induced cellular organelles called replication organelles (ROs) are produced to concentrate the viral proteins and certain host cell factors essential for viral replication together in the same specific compartment, while isolating them from the rest of the cytoplasm. Some + RNA viruses, such as coronaviruses [3, 4], picornaviruses, including poliovirus, which is the most studied member of this family [5, 6], arteriviruses [7, 8], and noroviruses [9], induce a typical organelle called a double-membrane vesicle (DMV). This is also the case for the hepatitis C virus (HCV), which is, curiously, the only member of the Flaviviridae family known to induce DMVs, all other members of this family instead inducing single-membrane spherules generated by membrane invagination [10, 11]. Severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2), the causal agent responsible for the COVID-19 pandemic, was recently shown to induce such DMVs, like other members of the coronavirus family [12, 13]. DMVs are vesicles delimited by two closely associated concentric membranes. They are easily identifiable by electron microscopy (EM) in SARS-CoV-2-infected cells and are frequently observed in very large numbers, occupying a large portion of the cytoplasm (Fig. 1).
Fig. 1.
Morphology of the DMVs induced by SARS-CoV-2 (Wuhan strain) in Vero E6 cells (a) and cultured primary respiratory epithelium (b). The areas outlined by the white squares in a and b are shown at higher magnification in the lower panels. In both cases, the double membrane delineating the interior of the vesicle is visible. However, these two concentric membranes are separated by a wider space in Vero cells than in primary cultures of respiratory epithelium. The wider spacing of the two membranes in Vero cells makes it possible to identify the points of contact between the two membranes (large arrows on the high magnification images shown in the lower panels of a), probably corresponding to pores crossing the two membranes and allowing exchanges with the cytoplasm. These ultrastructural differences cannot be attributed to technical artifacts, as these two cell preparations were fixed and prepared for EM with the same protocol (described in detail in [13]). The small white arrows in a and b indicate the presence of virus particles, formed in the ERGIC compartment (intermediate between the ER and the Golgi apparatus). Some viruses secreted at the apical pole of the primary respiratory epithelium cells are also identified by small black arrows in b. m mitochondria; er endoplasmic reticulum; n nucleus
Morphogenesis and spatial organization of DMVs
All the DMVs induced by these different viruses are relatively similar in size (100–300 nm in diameter), but they display significant variations of spatial organization with the cytoplasm. The DMVs induced by arteriviruses appear to be closed compartments with outer membranes that are frequently connected to other virus-induced membrane structures or to the endoplasmic reticulum (ER), leading to the establishment of large reticulovesicular membrane networks [7, 8]. By contrast, picornavirus-induced DMVs are disconnected structures often observed in an open vase-like configuration [5, 6]. The DMVs induced by HCV [11] and noroviruses [9] seem to be organized in an intermediate manner. Furthermore, these viruses often induce other membrane structures that may appear at different stages of infection and probably correspond to precursor forms of DMVs. Thus, the morphology of picornavirus membrane rearrangements gradually changes during the course of viral infection, suggesting that this morphology is dynamic and undergoes a form of maturation. For poliovirus and coxsackie virus, another member of the picornavirus family, membrane remodeling begins with the formation of single-membrane structures, which are progressively transformed into DMVs [5, 6]. It has been suggested that these single-membrane structures initially form flattened cisternae that then curve to form DMVs. The presence of single-membrane vesicle clusters early in infection has also been observed for HCV, suggesting the possibility of a similar mechanism for the morphogenesis of the DMVs induced by this virus [14]. It is difficult to determine in which of these elements viral RNA replication actually occurs. The subcellular distribution of major viral components does not always provide a clear answer to this question, as only a small fraction of the structures in place may be engaged in active replication at a given time. Thus, experiments involving the metabolic labeling of newly synthesized viral RNA have shown that DMVs are, indeed, active sites of viral RNA synthesis for HCV [15], but there are probably differences between DMV-inducing viruses. For picornaviruses, the single-membrane structures seem to constitute the site of active replication, as these structures predominate at the peak of viral replication [6, 16, 17].
For SARS-CoV-2, discreet single-membrane vesicle clusters present at the very start of infection may serve as the precursors of DMVs [13]. However, for both coronaviruses and HCV, DMVs are clearly the site of active viral RNA synthesis, as suggested by experiments based on the metabolic labeling of newly synthesized viral RNA within the DMVs [12], and the correlation between the number of intracellular DMVs and viral replication rates [13]. Nevertheless, these experiments involving metabolic labelling have not demonstrated unequivocally whether replication takes place inside the DMVs, as newly synthesized RNA was observed both inside and outside the DMVs. The first DMVs are formed within 4 h of infection in SARS-CoV-2-infected cells [13, 18]. DMVs then rapidly become observable over large areas of the cytoplasm and are sometimes connected to the ER membrane, suggesting that the ER is probably the main source of membranes for DMVs, at least at the beginning of the infection [13, 18]. The primary targets of SARS-CoV-2 infection are nasal and respiratory epithelial cells, which form polarized monolayers with distinct apical and basolateral domains. Cultured primary respiratory epithelial cells are the closest model to the target tissues of SARS-CoV-2, but the Vero cell line, a monkey kidney epithelial cell line that is very easy to grow in the laboratory, has proved extremely valuable for improving our understanding of the cell biology of coronaviruses [3, 4, 12, 13]. The influence of cell type on the biogenesis of membrane rearrangements has not been studied, but minor morphological differences have been observed between these rearrangements in the two cell types (Fig. 1).
Role of viral proteins in DMV formation
The molecular composition of SARS-CoV-2 DMVs has yet to be determined, but studies on other coronaviruses, such as SARS-CoV and the Middle East Respiratory Syndrome (MERS) coronavirus, have shown that viral non-structural proteins play a key role in DMV formation. Coronaviruses have one of the largest known genomes among RNA viruses, ranging from 27 to 32 kb in length, and encode for 22–29 proteins, including at least 16 non-structural proteins (nsp). For the SARS-CoV, two of these nsp (nsp3 and nsp4) are particularly involved in DMV formation, as the interaction between the luminal loops of nsp3 and nsp4 has been shown to be important for initiating host cell membrane rearrangements [19]. Moreover, the expression of nsp3 and nsp4 is sufficient to induce DMV formation [20, 21]. Nsp3 is a multidomain protein with a papain-like activity and nsp4 is thought to anchor into a host-cell membrane the viral replication complex (or replisome) made of at least 9 other non-structural proteins [22]. Similarly, their functional equivalents in arteriviruses, the non-structural proteins nsp2 and nsp3, have also been shown to be necessary and sufficient for DMV formation [23]. This is also the case for two non-structural proteins of the poliovirus, 2BC and 3A [24]. For some viruses, the expression of a single non-structural protein seems to be sufficient for DMV formation. For HCV, whose genome encodes for a more limited number of non-structural proteins, the HCV RNA-dependent RNA polymerase (NS5A) is the sole viral protein required for this phenomenon [11]. Nevertheless, the DMVs observed following the expression of the minimal set of proteins required for vesicle formation are not entirely the same as those observed during a viral infection, indicating that additional factors may have a strong influence on DMV biogenesis. For example, other non-structural proteins, such as NS4B [25] and NS5B [11], participate in the formation of HCV-induced DMVs. This is also the case for the nsp6 protein of SARS-CoV [20], the NS1-2 and NS3 proteins of noroviruses [9] and the nsp5 protein of arteriviruses [26]. Moreover, other models of + RNA viruses have shown that replicating viral RNA is also a major player in the biogenesis of virus-induced membrane rearrangements [27].
Origin of the host cell membranes used for DMV formation
Studies on SARS-CoV-infected Vero cells have revealed that the DMVs are sometimes seen to be continuous with the ER, suggesting that DMVs are derived from the host ER membrane [14], as reported for HCV [11] and norovirus [9]. Recent studies with various cell types, including Vero cells and lung epithelial cells, have reported similar observations for SARS-CoV-2 [18]. However, markers of other organelles, including the Golgi apparatus, have also been identified in poliovirus DMVs [28]. Cocksackie viruses, which, like poliovirus, are picornaviruses, induce a remodeling of both ER and Golgi apparatus membranes [29]. Interestingly, one study based on computational analysis suggested that SARS-CoV-2 viral RNA may also localize to mitochondria [30], raising the possibility that SARS-CoV-2 may also remodel mitochondrial membranes to generate mitochondrion-derived DMVs. This possibility was further analyzed in a functional analysis of host cell proteins binding to SARS-CoV-2 RNA, which confirmed a physical and functional connection between SARS-CoV-2 and mitochondria [31]. EM images, such as that shown in Fig. 1a, in which large numbers of mitochondria are observed in close proximity to DMVs, support this hypothesis, which nevertheless remains to be confirmed, as a potential role for mitochondria in DMV biogenesis is to date based on indirect experiments.
Like mitochondria, autophagosomes are surrounded by a double membrane and a role for these organelles in the biogenesis of coronavirus membrane remodeling has been suggested [32]. Autophagy is part of the endogenous cell substrates recycling machinery and innate immunity strategies by targeting intracellular pathogens to lysosomes for degradation. Although many viruses have developed strategies to counteract the antiviral functions of the autophagy pathway, some of them on the contrary have acquired mechanisms to exploit specific functions of the host cell factors involved in this process, to promote viral replication [33]. Thus, the biogenesis of virus-induced DMVs is probably dependent on cellular factors essential for autophagosome formation. In the murine hepatitis virus (MHV), another coronavirus, and SARS-CoV-infected Vero cells, the viral replicase protein, nsp8, colocalizes with the autophagosome marker LC3 [34, 35]. LC3 also colocalizes with the viral replicases of many other DMV-inducing viruses, including arteriviruses [36] and HCV [37]. The isolation of HCV DMVs by cell fractionation and biochemical analysis showed that LC3 was present in the membranes of these vesicles [10]. In MHV-infected cells, two viral proteins that localize to the replication complex (p22 and N) were also found to be colocalized with LC3, and with ATG12, which, together with ATG5, promotes LC3 lipidation, an important step in autophagosome formation. However, while this study on MHV showed that LC3 was essential for viral replication, it also demonstrated that LC3 lipidation was not strictly necessary [35]. This finding suggests that DMV biogenesis is facilitated by the mechanisms underlying autophagosome formation, but not to the extent of lysosomal targeting and fusion. A recent study confirmed that an incomplete autophagy response is a feature of SARS-CoV-2 infection [38]. The expression of SARS-CoV-2 ORF3a was sufficient to trigger incomplete autophagy, in which autophagosome formation was induced but autophagosome maturation was impaired. It has also been shown that the lipid phosphatidic acid (PA) produced by acylglycerol phosphate acyltransferase (AGPAT) 1 and 2 activity in the ER is essential not only for autophagic vesicle formation, but also for the biogenesis of HCV- and SARS-CoV-2-induced DMVs [39]. Both these viruses also exploit other common factors involved in autophagosome formation, including class III phosphatidylinositol 3-kinase (PI3K), but without activating conventional autophagy pathways [40]. All these results suggest that SARS-CoV-2 and HCV can exploit autophagosome formation mechanisms to support DMV biogenesis, while blocking lysosome fusion to escape complete autophagy-mediated degradation. These observations reinforce the link between autophagy and DMV formation, revealing unexpected similarities in the mode of operation of two phylogenetically very different viruses [39, 40].
Lipid synthesis and redistribution for DMV biogenesis
The induction of host lipid synthesis by DMV-inducing viruses is required to provide the lipids necessary for these major membrane rearrangements [41]. Sharp differences in lipid composition have been found between the membranes of DMV and the ER membrane from which they are derived. For example, HCV-induced DMVs contain nine times more cholesterol than host cell ER membranes [15], and they are also enriched in phosphatidylinositol-4-phosphate, PI4P [42]. An enrichment in phosphatidyl-choline [43] and sphingomyelin [44] has also been observed in the membranes of HCV-induced DMVs. A lipidomic study on cells infected with a benign human coronavirus, coronavirus 229E, revealed global alterations to total intracellular lipid composition, with an increase in the levels of fatty acids and glycerophospholipid [45]. A recent functional analysis of a SARS-CoV-2/host protein interactome demonstrated a dependence of SARS-CoV-2 on host factors involved in cholesterol biosynthesis [46].
In addition to regulating lipogenesis, viral infection can also redistribute host cell lipids from other organelles. Connections between DMVs and host organelles have been observed at membrane contact sites (MCSs) [41]. MCSs are formed when the membranes of different organelles are in close proximity, but not close enough to achieve membrane fusion. Specific protein complexes act on these closely apposed membranes to regulate these MCSs. These complexes include lipid transport proteins (LTPs), which facilitate non-vesicular lipid transport between the two membranes [41]. For example, the Golgi-localized oxysterol-binding protein (OSBP) transports newly synthesized cholesterol from the ER to the Golgi apparatus at MCSs in a counter-exchange mechanism in which the phospholipid PI4P is transferred from the Golgi apparatus to the ER. This PI4P-cholesterol counterflow mechanism can be hijacked for the benefit of viral replication. Poliovirus [47] and HCV [48] hijack OSBP by recruiting PI4-kinase (PI4K) within their DMVs. The viral recruitment of PI4K thus leads to an enrichment of DMVs in PI4P, increasing OSBP recruitment, with PI4P-cholesterol counterexchange at ER contacts then providing cholesterol for the new membranes. There are four PI4K isoforms in human cells. Picornaviruses, such as coxsackieviruses, recruit PI4KIIIβ [49] to generate PI4P for the recruitment of OSBP. This exploitation of the MCS machinery to provide OSBP/PI4P-dependent cholesterol transport from the ER to DMVs is essential for viral replication [50]. Other picornaviruses and HCV recruit OSBP via PI4KIIIα [51]. The HCV NS5A protein binds to and activates PI4KIIIα, increasing PI4P levels on DMVs [42]. The structure of HCV DMVs is strongly altered by OSBP/PI4K depletion, suggesting that the action of OSBP is important for the structural characteristics of DMVs. The SARS-CoV-2 protein nsp4 was recently shown to interact with OSBP and PI4KIIIβ [52], suggesting that SARS-CoV-2 may also use OSBP-mediated lipid exchange to enrich its DMVs in cholesterol.
The exploitation of cellular lipid stores is another strategy of viruses that hijack host lipid transport mechanisms. Cholesterol is esterified in the ER transport to lipid droplets (LDs), dynamic organelles in which neutral lipids are stored, synthesized and mobilized according to cellular needs. Picornaviruses inhibit cholesterol esterification, preventing its transport to LDs, thereby increasing the availability of cellular cholesterol for use in DMVs [53]. They also induce the formation of MCSs between DMVs and existing LDs, providing platforms for fatty acid transport to DMVs. EM studies have shown that poliovirus and coxsackievirus DMVs form MCSs with LDs [29]. The inhibition of LD lipolysis disrupts both DMV formation and picornavirus replication, suggesting an important role for LDs in providing essential lipids for DMV biogenesis [54, 55]. HCV infection causes a subcellular redistribution of LDs [54], and DMVs are frequently observed in close proximity to LDs [56, 57]. However, for HCV, LDs have a role beyond contributing to lipid flow for DMV formation, as they also constitute an assembly platform for virus particle morphogenesis [58, 59]. Recent studies have suggested a possible role for LDs in the infectious cycle of SARS-CoV-2, as the viral capsid protein drives diacylglycerol acyltransferase (DGAT) gene expression to facilitate LD formation and associates with the adipocyte differentiation-related protein (ADRP) on the LD surface [60]. Another study has suggested that, like HCV, SARS-CoV-2 may also use LDs as assembly platforms [61]. However, our EM observations, and those of other groups, did not confirm a preferential juxtaposition of DMVs with LDs or the presence of SARS-CoV-2 virus particles close to LDs (Fig. 1). Further studies will be required to clarify the role of LDs in the various steps of the SARS-CoV-2 infectious cycle.
DMV functions
DMVs have a protective function, by concealing viral replication intermediates from cellular sensors. This was demonstrated with the HCV model, in which immune sensors for dsRNA, such as RIG-I (retinoic acid-inducible gene I) and MDA5 (melanoma differentiation-associated protein 5), were found to have a limited effect when dsRNA was present in DMVs [62]. In the poliovirus model, virus-induced membrane rearrangements have also been shown to have a vital function in protecting viral RNA from immune sensors [63]. However, it is unclear whether the presence of a double membrane is truly necessary for this protection, as several + RNA viruses induce membrane remodeling to generate single-membrane spherules through invagination, with similar effects [64]. DMVs also have large outer membrane surfaces to which replication complexes could potentially be anchored, but it seems more likely that replication occurs in association with the inner membrane facing the cytosolic interior of the DMV. This would clearly provide an isolated environment, which would be ideal for viral replication. A recent 3D tomographic cryo-EM study of SARS-CoV-2 infected cells showed that the interior of DMVs contained branched filamentous structures resembling dsRNAs [65]. However, such a compartment would need to remain capable of exchanging material with the cytosol, for the import of metabolites and the export of viral RNA for translation and encapsidation into newly synthesized virions. Closed DMVs with no apparent opening to the surrounding cytosol have regularly been observed in infected cells. In the case of picornaviruses, completely sealed DMVs coexist with open DMVs, which are probably their precursors. Completely sealed DMVs may represent the final stage, with which large amounts of viral RNA isolated in these structures to prevent contact with cellular sensors.
In the HCV model, in which only DMVs are encountered at peak viral replication, sealed DMVs seem to coexist with pore-containing DMVs (about 10% of the total) capable of exchanges with the cytoplasm [11]. It is, therefore, conceivable that productive viral RNA synthesis occurs within DMVs containing pores that eventually become sealed to isolate and store the excess viral RNA accumulated during infection. Cryo-EM experiments have recently shown that MHV- and SARS-CoV-2-induced DMVs, like those of HCV, have pores that span the double membrane, allowing the transport of newly synthesized viral RNA out of the DMV for translation and packaging into viral particles [66]. The nsp3 protein has been shown to be a major constituent of these pores, but the possibility of host cell factors also contributing to the formation of these pores cannot be excluded [66]. Despite having a lower resolution than cryo-EM, conventional EM studies have suggested that pores are present in SARS-CoV-2-induced DMVs (lower panels of Fig. 1a). Interestingly, components of the host cell nuclear transport machinery associated with DMVs have been shown to play a key role in protecting HCV-infected cells from innate immune sensors [62]. No such mechanism has yet been demonstrated for SARS-CoV-2, but the presence of annulate lamellae—storage forms of nuclear pore complexes (NPC) anchored in host cell membranes—has been shown to be an ultrastructural feature of infected cells for both these viruses [13, 67, 68]. This finding suggests a potential role for annulate lamellae, via the NPC components they contain, in the biogenesis of HCV- and SARS-CoV-2-induced DMVs or in the establishment of the pores crossing the DMV membranes. Interestingly, the potential involvement of NPC components in the DMV pores could suggest that these pores may also have a dynamic opening and closing function, as for NPCs.
Conclusion
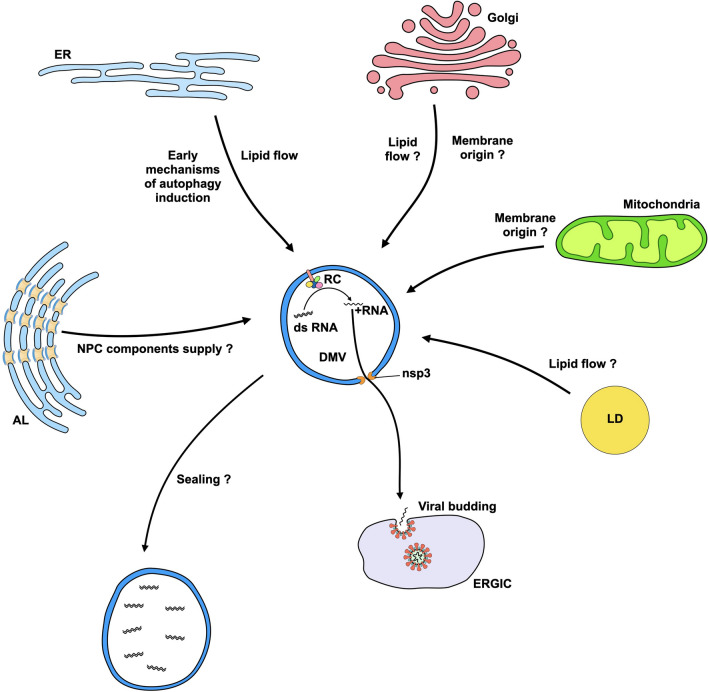
SARS-CoV-2, like other + RNA viruses, hijacks the organization and biogenesis of host cell membranes to form DMVs, in which its replication complexes are anchored. Figure 2 summarizes the mechanisms demonstrated or suspected to contribute to the biogenesis of SARS-CoV-2 DMVs and their potential role. Despite SARS-CoV-2 and HCV are phylogenetically unrelated, the DMVs of these two viruses are very similar. It will be interesting to determine in future studies whether the original observations made with the HCV model, such as the co-existence of pore-containing and sealed DMVs or the role of host cell NPC components in the biogenesis and/or function of DMVs, also apply to SARS-CoV-2. The anchoring of viral replication complexes to the inner membrane of DMVs and their direction toward the lumen of these DMVs are strongly suspected but have never been strictly demonstrated. Further studies of these aspects are therefore required. It will also be important to study the initial formation of these DMVs, at the beginning of the infection, and the transport of the newly synthesized viral RNA through the pores of the DMVs for constitution of the genome of future viral particles. These investigations could help to drive the design of new antiviral strategies against SARS-CoV-2 and other coronaviruses that might emerge in the future based on the inhibition of this replication organelle. In addition, molecular dynamics simulations by computational approaches that have been recently used to model the formation of a double membrane [69] could also help to reach this objective.
Fig. 2.
Model for SARS-CoV-2 DMV biogenesis and role of this virus-induced organelle in the viral life cycle. AL annulate lamella; ER endoplasmic reticulum; ERGIC ER-Golgi intermediate compartment; LD lipid droplet; NPC nuclear pore complex; nsp3 non-structural protein 3; RC replication complex
Author contributions
PR designed the manuscript. PR, SE, JBG and EB were involved in electron microscopy observations. PR wrote the first draft. SE, JBG, CH, RP and EB reviewed and edited the paper. CH designed the model. All authors read and approved the final manuscript.
Funding
Our research is supported by the University of Tours and INSERM, France.
Availability of data and material
Not applicable.
Declarations
Conflict of interest
The authors have no relevant financial and non-financial interests to disclose.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Blanchard E, Roingeard P. Virus-induced double-membrane vesicles. Cell Microbiol. 2015;17:45–50. doi: 10.1111/cmi.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolff G, Melia CE, Snijder EJ, Bárcena M. Double-membrane vesicles as platforms for viral replication. Trends Microbiol. 2020;28:1022–1033. doi: 10.1016/j.tim.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Wilde AH, Raj VS, Oudshoorn D, Bestebroer TM, van Nieuwkoop S, Limpens RWAL, Posthuma CC, van der Meer Y, Bárcena M, Haagmans BL, Snijder EJ, van den Hoogen BG. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-alpha treatment. J Gen Virol. 2013;94:1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Limpens RW, van der Schaar HM, Kumar D, Koster AJ, Snijder EJ, van Kuppeveld FJ, Bárcena M. The transformation of enterovirus replication structures: a three-dimensional study of single- and double-membrane compartments. MBio. 2011;2:e00166-11. doi: 10.1128/mBio.00166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belov GA, Nair V, Hansen BT, Hoyt FH, Fischer ER, Ehrenfeld E. Complex dynamic development of poliovirus membranous replication complexes. J Virol. 2012;86:302–312. doi: 10.1128/JVI.05937-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knoops K, Bárcena M, Limpens RW, Koster AJ, Mommaas AM, Snijder EJ. Ultrastructural characterization of arterivirus replication structures: reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J Virol. 2012;86:2474–2487. doi: 10.1128/JVI.06677-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang W, Chen K, Zhang X, Guo C, Chen Y, Liu X. An integrated analysis of membrane remodeling during porcine reproductive and respiratory syndrome virus replication and assembly. PLoS ONE. 2018;13:e0200919. doi: 10.1371/journal.pone.0200919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doerflinger SY, Cortese M, Romero-Brey I, Menne Z, Tubiana T, Schenk C, White PA, Bartenschlager R, Bressanelli S, Hansman GS, Lohmann V. Membrane alterations induced by nonstructural proteins of human norovirus. PLoS Pathog. 2017;13:e1006705. doi: 10.1371/journal.ppat.1006705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferraris P, Blanchard E, Roingeard P. Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J Gen Virol. 2010;91:2230–2237. doi: 10.1099/vir.0.022186-0. [DOI] [PubMed] [Google Scholar]
- 11.Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, Walther P, Antony C, Krijnse-Locker J, Bartenschlager R. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012;8:e1003056. doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snijder EJ, Limpens RWAL, de Wilde AH, de Jong AWM, Zevenhoven-Dobbe JC, Maier HJ, Faas FFGA, Koster AJ, Bárcena M. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol. 2020;18:e3000715. doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eymieux S, Rouillé Y, Terrier O, Seron K, Blanchard E, Rosa-Calatrava M, Dubuisson J, Belouzard S, Roingeard P. Ultrastructural modifications induced by SARS-CoV-2 in vero cells: a kinetic analysis of viral factory formation, viral particle morphogenesis and virion release. Cell Mol Life Sci. 2021;78:3565–3576. doi: 10.1007/s00018-020-03745-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferraris P, Beaumont E, Uzbekov R, Brand D, Gaillard J, Blanchard E, Roingeard P. Sequential biogenesis of host cell membrane rearrangements induced by hepatitis C virus infection. Cell Mol Life Sci. 2013;70:1297–1306. doi: 10.1007/s00018-012-1213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paul D, Hoppe S, Saher G, Krijnse-Locker J, Bartenschlager R. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J Virol. 2013;87:10612–10627. doi: 10.1128/JVI.01370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melia CE, van der Schaar HM, de Jong AWM, Lyoo HR, Snijder EJ, Koster AJ, van Kuppeveld FJM, Bárcena M. The origin, dynamic morphology, and PI4P-independent formation of encephalomyocarditis virus replication organelles. MBio. 2018;9:e00420-18. doi: 10.1128/mBio.00420-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melia CE, van der Schaar HM, Lyoo H, Limpens RWAL, Feng Q, Wahedi M, Overheul GJ, van Rij RP, Snijder EJ, Koster AJ, Bárcena M, van Kuppeveld FJM. Escaping host factor PI4KB inhibition: enterovirus genomic RNA replication in the absence of replication organelles. Cell Rep. 2017;21:587–599. doi: 10.1016/j.celrep.2017.09.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cortese M, Lee JY, Cerikan B, Neufeldt CJ, Oorschot VMJ, Köhrer S, Hennies J, Schieber NL, Ronchi P, Mizzon G, Romero-Brey I, Santarella-Mellwig R, Schorb M, Boermel M, Mocaer K, Beckwith MS, Templin RM, Gross V, Pape C, Tischer C, Frankish J, Horvat NK, Laketa V, Stanifer M, Boulant S, Ruggieri A, Chatel-Chaix L, Schwab Y, Bartenschlager R. Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies. Cell Host Microbe. 2020;28:853–866.e5. doi: 10.1016/j.chom.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagemeijer MC, Monastyrska I, Griffith J, van der Sluijs P, Voortman J, van Bergen en Henegouwen PM, Vonk AM, Rottier PJ, Reggiori F, de Haan CA. Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology. 2014;458–459:125–135. doi: 10.1016/j.virol.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier MJ. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. MBio. 2013;4:e00524-13. doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oudshoorn D, Rijs K, Limpens RWAL, Groen K, Koster AJ, Snijder EJ, Kikkert M, Bárcena M. Expression and cleavage of Middle East respiratory syndrome coronavirus nsp3–4 polyprotein induce the formation of double-membrane vesicles that mimic those associated with coronaviral RNA replication. MBio. 2017;8:e01658-17. doi: 10.1128/mBio.01658-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartenian E, Nandakumar D, Lari A, Ly M, Tucker JM, Glaunsinger BA. The molecular virology of coronaviruses. J Biol Chem. 2020;295:12910–12934. doi: 10.1074/jbc.REV120.013930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snijder EJ, van Tol H, Roos N, Pedersen KW. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J Gen Virol. 2001;82:985–994. doi: 10.1099/0022-1317-82-5-985. [DOI] [PubMed] [Google Scholar]
- 24.Suhy DA, Giddings TH, Jr, Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J Virol. 2000;74:8953–8965. doi: 10.1128/jvi.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paul D, Madan V, Ramirez O, Bencun M, Stoeck IK, Jirasko V, Bartenschlager R. Glycine zipper motifs in hepatitis C virus nonstructural protein 4B are required for the establishment of viral replication organelles. J Virol. 2018;92:e01890–e1917. doi: 10.1128/JVI.01890-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Hoeven B, Oudshoorn D, Koster AJ, Snijder EJ, Kikkert M, Bárcena M. Biogenesis and architecture of arterivirus replication organelles. Virus Res. 2016;220:70–90. doi: 10.1016/j.virusres.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopek BG, Settles EW, Friesen PD, Ahlquist P. Nodavirus-induced membrane rearrangement in replication complex assembly requires replicase protein a, RNA templates, and polymerase activity. J Virol. 2010;84:12492–12503. doi: 10.1128/JVI.01495-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlegel A, Giddings TH, Jr, Ladinsky MS, Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol. 1996;70:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melia CE, Peddie CJ, de Jong AWM, Snijder EJ, Collinson LM, Koster AJ, van der Schaar HM, van Kuppeveld FJM, Bárcena M. Origins of enterovirus replication organelles established by whole-cell electron microscopy. MBio. 2019;10:e00951-19. doi: 10.1128/mBio.00951-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu KE, Fazal FM, Parker KR, Zou J, Chang HY. RNA-GPS predicts SARS-CoV-2 RNA residency to host mitochondria and nucleolus. Cell Syst. 2020;11:102–108.e3. doi: 10.1016/j.cels.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flynn RA, Belk JA, Qi Y, Yasumoto Y, Wei J, Alfajaro MM, Shi Q, Mumbach MR, Limaye A, DeWeirdt PC, Schmitz CO, Parker KR, Woo E, Chang HY, Horvath TL, Carette JE, Bertozzi CR, Wilen CB, Satpathy AT. Discovery and functional interrogation of SARS-CoV-2 RNA-host protein interactions. Cell. 2021;184:2394–2411.e16. doi: 10.1016/j.cell.2021.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Z, Lu K, Mao B, Liu S, Trilling M, Huang A, Lu M, Lin Y. The interplay between emerging human coronavirus infections and autophagy. Emerg Microbes Infect. 2021;10:196–205. doi: 10.1080/22221751.2021.1872353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dinesh Kumar N, Smit JM, Reggiori F. Strategies employed by viruses to manipulate autophagy. Prog Mol Biol Transl Sci. 2020;172:203–237. doi: 10.1016/bs.pmbts.2020.01.004. [DOI] [PubMed] [Google Scholar]
- 34.Prentice E, Jerome WG, Yoshimori T, Mizushima N, Denison MR. Coronavirus replication complex formation utilizes components of cellular autophagy. J Biol Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reggiori F, Monastyrska I, Verheije MH, Calì T, Ulasli M, Bianchi S, Bernasconi R, de Haan CA, Molinari M. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monastyrska I, Ulasli M, Rottier PJ, Guan JL, Reggiori F, de Haan CA. An autophagy-independent role for LC3 in equine arteritis virus replication. Autophagy. 2013;9:164–174. doi: 10.4161/auto.22743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Kim JY, Liu HM, Lai MMC, Ou JJ. HCV-induced autophagosomes are generated via homotypic fusion of phagophores that mediate HCV RNA replication. PLoS Pathog. 2017;13:e1006609. doi: 10.1371/journal.ppat.1006609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qu Y, Wang X, Zhu Y, Wang W, Wang Y, Hu G, Liu C, Li J, Ren S, Xiao MZX, Liu Z, Wang C, Fu J, Zhang Y, Li P, Zhang R, Liang Q. ORF3a-mediated incomplete autophagy facilitates severe acute respiratory syndrome coronavirus-2 replication. Front Cell Dev Biol. 2021;9:716208. doi: 10.3389/fcell.2021.716208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tabata K, Prasad V, Paul D, Lee JY, Pham MT, Twu WI, Neufeldt CJ, Cortese M, Cerikan B, Stahl Y, Joecks S, Tran CS, Lüchtenborg C, Vovski P, Hörmann K, Müller AC, Zitzmann C, Haselmann U, Beneke J, Kaderali L, Erfle H, Thiel V, Lohmann V, Superti-Furga G, Brügger B, Bartenschlager R. Convergent use of phosphatidic acid for hepatitis C virus and SARS-CoV-2 replication organelle formation. Nat Commun. 2021;12:276. doi: 10.1038/s41467-021-27511-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Twu WI, Lee JY, Kim H, Prasad V, Cerikan B, Haselmann U, Tabata K, Bartenschlager R. Contribution of autophagy machinery factors to HCV and SARS-CoV-2 replication organelle formation. Cell Rep. 2021;37:110049. doi: 10.1016/j.celrep.2021.110049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong LH, Edgar JR, Martello A, Ferguson BJ, Eden ER. Exploiting connections for viral replication. Front Cell Dev Biol. 2021;9:640456. doi: 10.3389/fcell.2021.640456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS, Longerich T, Diehl S, Ramirez F, Balla T, Rohr K, Kaul A, Bühler S, Pepperkok R, Lengauer T, Albrecht M, Eils R, Schirmacher P, Lohmann V, Bartenschlager R. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J, Zhang Z, Chukkapalli V, Nchoutmboube JA, Li J, Randall G, Belov GA, Wang X. Positive-strand RNA viruses stimulate host phosphatidylcholine synthesis at viral replication sites. Proc Natl Acad Sci USA. 2016;113:e1064–e1073. doi: 10.1073/pnas.1519730113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gewaid H, Aoyagi H, Arita M, Watashi K, Suzuki R, Sakai S, Kumagai K, Yamaji T, Fukasawa M, Kato F, Hishiki T, Mimata A, Sakamaki Y, Ichinose S, Hanada K, Muramatsu M, Wakita T, Aizaki H. Sphingomyelin is essential for the structure and function of the double-membrane vesicles in hepatitis C virus RNA replication factories. J Virol. 2020;94:e01080–e1120. doi: 10.1128/JVI.01080-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan B, Chu H, Yang D, Sze KH, Lai PM, Yuan S, Shuai H, Wang Y, Kao RY, Chan JF, Yuen KY. Characterization of the lipidomic profile of human coronavirus-infected cells: implications for lipid metabolism remodeling upon coronavirus replication. Viruses. 2019;11:73. doi: 10.3390/v11010073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffmann HH, Sánchez-Rivera FJ, Schneider WM, Luna JM, Soto-Feliciano YM, Ashbrook AW, Le Pen J, Leal AA, Ricardo-Lax I, Michailidis E, Hao Y, Stenzel AF, Peace A, Zuber J, Allis CD, Lowe SW, MacDonald MR, Poirier JT, Rice CM. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host Microbe. 2021;29:267–280.e5. doi: 10.1016/j.chom.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arita M. Phosphatidylinositol-4 kinase III beta and oxysterol-binding protein accumulate unesterified cholesterol on poliovirus-induced membrane structure. Microbiol Immunol. 2014;58:239–256. doi: 10.1111/1348-0421.12144. [DOI] [PubMed] [Google Scholar]
- 48.Wang H, Perry JW, Lauring AS, Neddermann P, De Francesco R, Tai AW. Oxysterol-binding protein is a phosphatidylinositol 4-kinase effector required for HCV replication membrane integrity and cholesterol trafficking. Gastroenterology. 2014;146:1373–1385. doi: 10.1053/j.gastro.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJ, Altan-Bonnet N. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altan-Bonnet N. Lipid tales of viral replication and transmission. Trends Cell Biol. 2017;27:201–213. doi: 10.1016/j.tcb.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dorobantu CM, Albulescu L, Harak C, Feng Q, van Kampen M, Strating JR, Gorbalenya AE, Lohmann V, van der Schaar HM, van Kuppeveld FJ. Modulation of the host lipid landscape to promote RNA virus replication: the picornavirus encephalomyocarditis virus converges on the pathway used by hepatitis C virus. PLoS Pathog. 2015;11:e1005185. doi: 10.1371/journal.ppat.1005185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.St-Germain JR, Astori A, Samavarchi-Tehrani P, Abdouni H, Macwan V, Kim D-K, Knapp JJ, Roth FP, Gingras A-C, Raught B. A SARS-CoV-2 BioID-based virus-host membrane protein interactome and virus peptide compendium: new proteomics resources for COVID-19 research. BioRxiv. 2020 doi: 10.1101/2020.08.28.269175. [DOI] [Google Scholar]
- 53.Ilnytska O, Santiana M, Hsu NY, Du WL, Chen YH, Viktorova EG, Belov G, Brinker A, Storch J, Moore C, Dixon JL, Altan-Bonnet N. Enteroviruses harness the cellular endocytic machinery to remodel the host cell cholesterol landscape for effective viral replication. Cell Host Microbe. 2013;14:281–293. doi: 10.1016/j.chom.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Viktorova EG, Nchoutmboube JA, Ford-Siltz LA, Iverson E, Belov GA. Phospholipid synthesis fueled by lipid droplets drives the structural development of poliovirus replication organelles. PLoS Pathog. 2018;14:e1007280. doi: 10.1371/journal.ppat.1007280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laufman O, Perrino J, Andino R. Viral generated inter-organelle contacts redirect lipid flux for genome replication. Cell. 2019;178:275–289.e16. doi: 10.1016/j.cell.2019.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Depla M, Uzbekov R, Hourioux C, Blanchard E, Le Gouge A, Gillet L, Roingeard P. Ultrastructural and quantitative analysis of the lipid droplet clustering induced by hepatitis C virus core protein. Cell Mol Life Sci. 2010;67:3151–3161. doi: 10.1007/s00018-010-0373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee JY, Cortese M, Haselmann U, Tabata K, Romero-Brey I, Funaya C, Schieber NL, Qiang Y, Bartenschlager M, Kallis S, Ritter C, Rohr K, Schwab Y, Ruggieri A, Bartenschlager R. Spatiotemporal coupling of the hepatitis C virus replication cycle by creating a lipid droplet-proximal membranous replication compartment. Cell Rep. 2019;27:3602–3617.e5. doi: 10.1016/j.celrep.2019.05.063. [DOI] [PubMed] [Google Scholar]
- 58.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 59.Roingeard P, Melo RC. Lipid droplet hijacking by intracellular pathogens. Cell Microbiol. 2017;19:e12688. doi: 10.1111/cmi.12688. [DOI] [PubMed] [Google Scholar]
- 60.Yuan S, Yan B, Cao J, Ye ZW, Liang R, Tang K, Luo C, Cai J, Chu H, Chung TW, To KK, Hung IF, Jin DY, Chan JF, Yuen KY. SARS-CoV-2 exploits host DGAT and ADRP for efficient replication. Cell Discov. 2021;7:100. doi: 10.1038/s41421-021-00338-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dias SSG, Soares VC, Ferreira AC, Sacramento CQ, Fintelman-Rodrigues N, Temerozo JR, Teixeira L, Nunes da Silva MA, Barreto E, Mattos M, de Freitas CS, Azevedo-Quintanilha IG, Manso PPA, Miranda MD, Siqueira MM, Hottz ED, Pão CRR, Bou-Habib DC, Barreto-Vieira DF, Bozza FA, Souza TML, Bozza PT. Lipid droplets fuel SARS-CoV-2 replication and production of inflammatory mediators. PLoS Pathog. 2020;16:e1009127. doi: 10.1371/journal.ppat.1009127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neufeldt CJ, Joyce MA, Van Buuren N, Levin A, Kirkegaard K, Gale M, Jr, Tyrrell DL, Wozniak RW. The hepatitis C virus-induced membranous web and associated nuclear transport machinery limit access of pattern recognition receptors to viral replication sites. PLoS Pathog. 2016;12:e1005428. doi: 10.1371/journal.ppat.1005428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Du X, Zhang Y, Zou J, Yuan Z, Yi Z. Replicase-mediated shielding of the poliovirus replicative double-stranded RNA to avoid recognition by MDA5. J Gen Virol. 2018;99:1199–1209. doi: 10.1099/jgv.0.001111. [DOI] [PubMed] [Google Scholar]
- 64.Romero-Brey I, Bartenschlager R. Endoplasmic reticulum: the favorite intracellular niche for viral replication and assembly. Viruses. 2016;8:160. doi: 10.3390/v8060160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klein S, Cortese M, Winter SL, Wachsmuth-Melm M, Neufeldt CJ, Cerikan B, Stanifer ML, Boulant S, Bartenschlager R, Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat Commun. 2020;11:5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wolff G, Limpens RWAL, Zevenhoven-Dobbe JC, Laugks U, Zheng S, de Jong AWM, Koning RI, Agard DA, Grünewald K, Koster AJ, Snijder EJ, Bárcena M. A molecular pore spans the double membrane of the coronavirus replication organelle. Science. 2020;369:1395–1398. doi: 10.1126/science.abd3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eymieux S, Blanchard E, Uzbekov R, Hourioux C, Roingeard P. Annulate lamellae and intracellular pathogens. Cell Microbiol. 2021;23:e13328. doi: 10.1111/cmi.13328. [DOI] [PubMed] [Google Scholar]
- 68.Boson B, Mialon C, Schichl K, Denolly S, Cosset FL. Nup98 is subverted from annulate lamellae by hepatitis C virus core protein to foster viral assembly. MBio. 2022;8:92321. doi: 10.1128/mbio.02923-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bolmatov D, Carrillo JY, Sumpter BG, Katsaras J, Lavrentovich MO. Double membrane formation in heterogeneous vesicles. Soft Matter. 2020;16:8806–8817. doi: 10.1039/d0sm01167c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.