Abstract
The redox homeostasis system regulates many biological processes, intracellular antioxidant production and redox signaling. However, long noncoding RNAs (lncRNAs) involved in redox regulation have rarely been reported. Herein, we reported that downregulation of MAGI2-AS3 decreased the superoxide level in Human fibroblasts (Fbs), a replicative aging model, as detected by the fluorescent probes dihydroethidium (DHE) and MitoSOX™ Red. RNA pulldown combined with mass spectrometry showed that HSPA8 is a novel interacting protein of MAGI2-AS3, which was further confirmed by photoactivatable ribonucleoside–enhanced crosslinking and immunoprecipitation (PAR-CLIP). Downregulation of MAGI2-AS3 decreased the hydrogen peroxide (H2O2) content by stabilizing the HSPA8 protein level via inhibiting the protesome degradation of HSPA8. Further evidence showed that MAGI2-AS3 interacted with the C-terminal domain (CTD) of HSPA8. Downregulation of MAGI2-AS3 delayed cell senescence, while this antiaging effect was abolished by HSPA8 knockdown. The underlying molecular mechanism by which MAGI2-AS3 knockdown inhibited cell senescence was mediated via suppression of the ROS/MAP2K6/p38 signaling pathway. Taken together, these findings revealed that downregulation of lncRNA MAGI2-AS3 decreased the H2O2 content and delayed cell senescence by stabilizing the HSPA8 protein level, identifying a potential antiaging application.
Keywords: Long noncoding RNA, MAGI2-AS3, HSPA8, Cell senescence
Graphical abstract

Highlights
-
•
MAGI2-AS3 is a new regulator of H2O2 in Human fibroblasts.
-
•
MAGI2-AS3 regulates H2O2 through interaction with the HSPA8 protein in its CTD.
-
•
MAGI2-AS3 contributes to human fibroblast senescence in a manner dependent on its regulation of HSPA8.
1. Introduction
Redox-regulated events are fundamental for cellular processes, such as cell growth and death, cell differentiation and senescence and so on. Many diseases and pathological processes, including diabetes, arthritis, osteoporosis, cataracts and tumorigenesis, are caused by a redox imbalance [[1], [2], [3], [4], [5], [6]]; therefore, research on redox regulation is very important. With global aging, health management based on the redox theory of aging has begun to provide potential strategies to prevent disease progression [7]. Cellular redox levels are widely reported to be regulated by proteins and genes [8], but only ∼2% of the human genome encodes proteins and a large amount of our DNA produces thousands of uncharacterized RNAs [9]. In recent years, noncoding RNAs (ncRNAs) have received wide appreciation; among these transcripts, long noncoding RNAs (lncRNAs) are noncoding transcripts that are transcribed by RNA polymerase II; frequently 5’ capped, spliced and polyadenylated and longer than 200 nucleotides [10].
LncRNAs participate in biological processes via various mechanisms, including affecting gene expression and epigenetics. However, there are only a few reports about redox regulation by lncRNAs. Downregulation of the lncRNA LINC00963 was found to suppress oxidative stress in chronic renal failure (CRF) by elevating the enzymatic activity of glutathione peroxidase (GSH-Px) and superoxidase dismutase (SOD) [11]. Overexpression of the lncRNA SLC-AS6 decreased the glutathione level and concomitantly increased reactive oxygen species (ROS) levels and enhanced lipid peroxidation [12]. lncRNA-HCP5 was reported to promote the stemness and chemoresistance of gastric cancer cells by driving fatty acid oxidation [13]. Silencing of the lncRNA ANRIL was found to reduce myocardial injury in diabetes by inhibiting myocardial oxidative stress [14]. The lncRNA LINC00116 encodes mitoregulin (Mtln), which increases mitochondrial respiration rates and decreases mitochondrial ROS [15]. Although a few lncRNAs have been reported to regulate the redox process, the underlying mechanism remains incompletely understood.
MAGI2-AS3 is a single-copy RNA being located in chromosome 7, as antisense transcript of coding-gene MAGI2. It is reported to participate in the suppression of multiple tumors, such as bladder cancer, glioma, non-small-cell lung carcinoma, ovarian cancer, breast cancer and lung squamous cell carcinoma [16]. MAGI2-AS3 has also been reported to be associated with and affect the occurrence and development of some other diseases. Reduction of MAGI2-AS3 expression increased neuronal viability and reduced inflammation in Alzheimer disease (AD) model cells [17] and the lncRNA MAGI2-AS3 expression level was lower in the plasma of intervertebral disc degeneration (IDD) patients [18]. However, the effects of MAGI2-AS3 on redox and aging have not been studied. We first report that downregulation of MAGI2-AS3 decreases superoxide levels in Fbs, a replicative aging model, that MAGI2-AS3 expression is increased during cell senescence and that downregulation of MAGI2-AS3 delays cell senescence in a manner dependent on the HSPA8 protein.
2. Materials and methods
2.1. Cell culture
Human fibroblasts (Fbs) were obtained from the laboratory of Guanghui Liu (Institute of Zoology, Chinese Academy of Sciences) and 293T human embryonic kidney cells (293T cells) were obtained from ATCC. Fbs and HEK293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Gibco) and 100 μg/mL penicillin/streptomycin (HyClone) at 37 °C in 5% CO2.
2.2. Lentivirus production
The MAGI2-AS3 short hairpin RNA (shRNA), HSPA8-shRNA and negative control (NC) shRNA sequences were cloned into the pLVTHM plasmid. The MAP2K6 gene sequence was inserted into the vector pLE4 to obtain the overexpression plasmids (a gift from the Guanghui Liu laboratory). 293T cells were transfected with these plasmids together with the envelope plasmid pMD2. G and packaging plasmid psPAX2 (Addgene). Lentiviral particles were collected and concentrated after 48 h and were used for transduction in the presence of 4 μg/mL polybrene. shRNA sequences are listed in Table 1.
Table 1.
Sequences of short hairpin RNAs (shRNAs).
| Targeted gene | Targeted sequence |
|---|---|
| HSPA8 | GCTGGTCTCAATGTACTTA |
| MAGI2-AS3 | GTTTATAGCAGCTAAGTTTG |
| MAGI2-AS3 | GCAATCTTATGCAGTCATACT |
| CAT | CCGGATCTCACTTGGCGGC |
| Negative control | GATGAAATGGGTAAGTACA |
2.3. Real-time qPCR (RT-qPCR)
Total RNA was extracted with TRIzol reagent (Invitrogen) and reverse transcribed using a cDNA synthesis kit with dsDNase (Invitrogen). Then, real-time PCR was performed in a 7500 Real-Time PCR System (Applied Biosystems) with qPCR Mix (Genestar) and GAPDH or β-ACTIN was used as the reference gene. The primers are listed in Table 2.
Table 2.
Primers used for qPCR.
| Primer | Sequence (5′-3′) |
|---|---|
| 36B4 (F) | CAGCAAGTGGGAAGGTGTAATCC |
| 36B4 (R) | CCCATTCTATCATCAACGGGTACAA |
| GAPDH (F) | AGTCCACTGGCGTCTTCA |
| GAPDH (R) | GAGTCCTTCCACGATACCAA |
| HSPA8 (F) | TGCTGCTCTTGGATGTCACT |
| HSPA8 (R) | AAGGTCTGTGTCTGCTTGGT |
| MAGI2-AS3 (F) | ACTGGAAATACAAGCCCAAGT |
| MAGI2-AS3 (R) | TTTCCACTCTGCTGGTTATGG |
| P16 (F) | GCCTTTTCACTGTGTTGGAG |
| P16 (R) | TGCCATTTGCTAGCAGTGTG |
| P21 (F) | GCGACTGTGATGCGCTAATG |
| P21 (R) | GAAGGTAGAGCTTGGGCAGG |
| TELE (F) | CGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT |
| TELE (R) | GCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCCT |
| U1 (F) | GGGAGATACCATGATCACGAAGGT |
| U1 (R) | CCACAAATTATGCAGTCGAGTTTCCC |
| β-ACTIN (F) | GTCCACCGCAAATGCTTCTA |
| β-ACTIN (R) | TGCTGTCACCTTCACCGTTC |
| MAP2K6 (F) | TAGCCAGGAACAGAAACGGC |
| MAP2k6 (R) | ACACATCACCCTCCCGAAAC |
| CAT (F) | TAAGACTGACCAGGGCATC |
| CAT (R) | CAACCTTGGTGAGATCGAA |
| DLL3(F) | CAACTGTGAGAAGAGGGTGGA |
| DLL3 (R) | CCAGGTCCAGGCAGAGTC |
| IL1B(F) | TTCGAGGCACAAGGCACAAC |
| IL1B (R) | GTGGTGGTCGGAGATTCGTA |
| PECAM1(F) | ATGCCAGTGGAAATGTCC |
| PECAM1 (R) | TCAGAAGTGGTACTGGTG |
| ELN(F) | CTAAATACGGTGCTGCTGGC |
| ELN (R) | CATGGGATGGGGTTACAAAG |
| VWA5A(F) | TGCTTCTGCCCCATTGAAGA |
| VWA5A (R) | ACTGTGCTGGTCCTTGTGAC |
| MMP12(F) | CGCCTCTCTGCTGATGACATAC |
| MMP12 (R) | GGTAGTGACAGCATCAAAACTCAAA |
| FLT1(F) | GAAAACGCATAATCTGGGACAGT |
| FLT1 (R) | GCGTGGTGTGCTTATTTGGA |
| p66Shc (F) | GCCGAGTATGTCGCCTATGT |
| p66Shc (R) | GGGTGGGTTCCTGAGGTATT |
2.4. 5′ and 3’ Rapid Amplification of cDNA ends (RACE)
The 5′ and 3′ RACE assay were performed using the RLM-RACE Kit (Ambion) according to the manufacturer's instructions. The RACE PCR products were cloned into pGEM-T vector (Promega) and sequenced.
2.5. RNA sequencing (RNA-seq) and analysis
Fbs from the NC and MAGI2-AS3 knockdown groups were harvested and RNA was extracted by the procedure described above. RNA quality control (determination of the concentration, RNA integrity number [RIN], 28S/18S ratio and fragment size) was conducted using an Agilent 2100 Bioanalyzer (Agilent RNA 6000 Nano Kit). Total RNA was incubated with oligo(dT) magnetic beads for enrichment of poly(A)-containing mRNA and simultaneous hybridization with rRNA. DNA/RNA hybrids were digested by using RNase H and the DNA probe was digested with DNase I. The RNA was cleaved into short fragments and was then reverse transcribed to cDNA using N6 random primers. The ends of the DNA duplexes ends were repaired to form blunt ends and sticky ends with redundant “A” nucleotides at the 3′ end were generated by 5′-end phosphorylation for ligation to a bubbling joint with redundant “T” nucleotides at the 3′ end. The ligation product was amplified using specific primers. Then, the PCR product was degenerated to a single strand and was then cyclized to construct a circular DNA library using bridge-type primers. Data were analyzed after library sequencing on the BGISEQ-500 (BGI) platform.
2.6. Western blot analysis
Cells were lysed in RIPA buffer (Genestar) supplemented with protease inhibitor cocktail (MCE) and proteins were extracted after centrifugation of the supernatant at 12,000×g and 4 °C. Proteins were loaded into an electrophoresis chamber for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were then transferred to nitrocellulose membranes. Membranes were blocked in TBST containing 5% skim milk powder for 1 h and were then incubated with the following primary antibodies overnight at 4 °C, anti-HSPA8 (Abcam, Cat. # ab51052); anti-P16 (ABclonal, Cat. # A0262), anti-P21 (ABclonal, Cat. # A19094), anti-P38 (Cell Signaling Technology, Cat. # 9212S), anti-pp38 (Cell Signaling Technology, Cat. # 9211S), LaminB1 (Sino Biological, Cat. # 101237-T32), anti-actin (Sungene Biotech, Cat. # KM9001) and anti-tubulin (Sungene Biotech, Cat. # KM9003T). After washing with TBST, membranes were incubated for 1 h at room temperature with the corresponding secondary antibody. After washing with TBST three times, membranes were incubated with ECL solution (Thermo) and chemical signals were acquired using a ChemiDoc XRS + molecular imager (BioRad). The band signals were analyzed with Image Lab software.
2.7. β-Galactosidase staining
Cells were seeded in 6-well plates. Cells were stained with a β-galactosidase staining kit following the protocol (Beyotime) and were then placed in an incubator at 37 °C for 16–18 h. Images were acquired by microscopy (Zeiss LSM710) and β-galactosidase-positive cell rates were determined with ImageJ software.
2.8. Measurement of superoxide anion levels with MitoSOX™ Red and dihydroethidium (DHE)
Fbs were washed with PBS and digested with trypsin. Then, the cells were washed with PBS and stained with 5 μM MitoSOX ™ Red and DHE probes (diluted in HBSS supplemented with Ca2+ and Mg2+) for 10 min at 37 °C and 5% CO2. Then, the cells were washed with PBS twice. For the MitoSOX ™ Red probe, fluorescence was detected at excitation/emission wavelengths of 510 nm/590 nm in a microplate reader; for the DHE probe, fluorescence was detected at excitation/emission wavelengths of 300 nm/610 nm in a microplate reader. Protein concentrations were measured with a BCA protein assay kit. The mean fluorescence intensity of cells was calculated (fluorescence value divided by protein concentration).
Fbs were grown in glass-bottom dishes and stained with the MitoSOX ™ Red probe. Images were acquired with a laser scanning confocal microscopy system (LSM 710, Carl Zeiss) at excitation/emission wavelengths of 510 nm/590 nm and mean fluorescence intensities were calculated with ImageJ software.
2.9. Detection of relative H2O2 level with HyPerRed probe
Fbs were transduced with lentiviruses expressing HyPerRed probes which is a genetically encoded red fluorescent sensor for H2O2 detection. The relative H2O2 level were detected using a microplate reader (Thermo Scientific Varioskan LUX) and HyperRed was excited at 575 nm and emission at 620 nm.
2.10. Determination of lncRNA MAGI2-AS3 subcellular location
Fbs were seeded in 10-cm cell culture plates and harvested at a confluence of approximately 80%. The cytoplasmic and nuclear fractions were separated using a cytoplasmic and nuclear preparation kit (Applygen) according to the instructions. The components were divided into two equal parts for protein and RNA extraction. The proteins Lamin B1 and β-tubulin were used as the nuclear and cytoplasmic markers respectively. The cytoplasmic and nuclear proteins were loaded onto gels for Western blotting using anti-β-tubulin and anti-Lamin B1 primary antibodies. RNA was reverse transcribed into cDNA for q-PCR. GAPDH and β-ACTIN were used as the two internal references for cytoplasmic transcripts and U1 was used as the internal reference for nuclear transcripts.
2.11. Telomere detection by qPCR
Genomic DNA was extracted from cells with a TIANamp genomic DNA kit (Tiangen). A standard curve was generated before every reaction and the mean Ct value of the telomere gene TELE and internal gene 36B4 were measured by qPCR using a 7500 Real-Time PCR System (Applied Biosystems). The following equations were used for analysis: ΔCt = Ct (TELE)-Ct(36B4), T/S = [ 2 Ct(telomere)/2Ct (BTF)]−1 = 2 −ΔCt, relative T/S = 2-(ΔCt1−ΔCt2) = 2–ΔΔCt. The telomere restriction fragment (TRF) length was proportional to the relative T/S ratio.
2.12. Photoactivatable ribonucleoside–enhanced crosslinking and immunoprecipitation (PAR-CLIP)
After Fbs were 80% confluent, 100 μM 4-thiouridine (4-SU) was added to the culture medium and incubated for 16 h and cellular DNA and protein were then crosslinked with UV radiation at 365 nm. Cells were lysed using lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 0.05% Igepal, 0.5% NP-40 and 1% cocktail) on ice for 30 min. The lysate was centrifuged at 12,000×g and the supernatant containing proteins was obtained. Input samples were set aside for RNA and protein analysis and the remaining supernatant was divided into two equal parts. Protein G beads (Invitrogen) were prepared according to the instructions. The two parts were incubated with rabbit IgG (3 μg) or an anti-HSPA8 antibody (3 μg) overnight at 4 °C. Then, the samples were incubated with beads for 2h and set on a magnetic separator, the supernatant was discarded and the beads were washed twice with buffer A (1 × PBS, 0.1% SDS, 0.3% deoxycholate, 0.3% NP-40), twice with buffer B (1 × PBS, 0.1% SDS, 0.5% deoxycholate, 0.5% NP-40) and twice with buffer C (50 mM Tris-HCl [pH 7.4], 10 mM MgCl2, 0.5% NP-40). One-quarter of the beads were mixed with 5 × SDS loading buffer for protein analysis and the remaining beads were put into 142 μL buffer containing 107 μL NT-2 buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40), 15 μL 10% SDS, 2 μL RNase inhibitor and 18 μL protease K at 55 °C. RNA was extracted with Trizol (Invitrogen) and subsequently analyzed to detect MAGI2-AS3 expression.
2.13. RNA pulldown
Cell lysates were freshly prepared in cell lysis buffer supplemented with proteinase inhibitor cocktail (MCE). Biotin-labeled MAGI2-AS3 RNA transcripts or truncated MAGI2-AS3 fragments and control RNA transcript were synthesized by in vitro transcription using T7 RNA polymerase (Roche) and biotin RNA labeling mix (Roche). The RNA transcripts were purified with RNA Clean & Concentrator™-5 (Zymo Research). RNA (3 μg) was incubated in structure buffer at 95 °C for 2 min and was then recovered to room temperature slowly. The cell lysate supernatant was incubated with RNA with slow rotation overnight at 4 °C. Streptavidin magnetic beads (Invitrogen) were precleared with lysis buffer. The beads were incubated with the RNA-protein mixture for 2 h with rotation and were then washed twice with NT2 buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40), twice with NT2 high-salt buffer containing 500 mM NaCl and once with NT2 high-salt buffer containing 1 M NaCl. The RNA-protein complexes were eluted and digested for mass spectrometry (MS) analysis.
2.14. In-gel digestion of proteins
The gel bands containing the protein sample were manually excised. Each of the protein bands was then digested individually as below. The protein bands were cut into small plugs, washed twice in 200 μL of distilled water for 10 min each time. The gel bands were dehydrated in 100% acetonitrile for 10 min and dried in a speedvac for approximately 15 min. Reduction (10 mM DTT in 25 mM NH4HCO3 [pH 8.0] for 45 min at 56 °C) and alkylation (40 mM iodoacetamide in 25 mM NH4HCO3 for 45 min at room temperature in the dark) were performed, followed by washing of the gel plugs with 50% acetonitrile in 25 mM ammonium bicarbonate twice. The gel plugs were then dried using a speedvac and digested with sequence-grade modified trypsin (40 ng for each band) in 25 mM NH4HCO3 overnight at 37 °C. The enzymatic reaction was stopped by adding formic acid to a 1% final concentration. The solution was then transferred to a sample vial for LC-MS/MS analysis.
2.15. LC-MS/MS analysis
All nanoLC-MS/MS experiments were performed on a Q Exactive (Thermo Scientific) equipped with an Easy n-LC 1000 HPLC system (Thermo Scientific). The peptides were loaded onto a 100 μm id × 2 cm fused silica trap column packed in-house with reversed phase silica (Reprosil-Pur C18 AQ, 5 μm, Dr. Maisch GmbH) and then separated on an a 75 μm id × 20 cm C18 column packed with reversed phase silica (Reprosil-Pur C18 AQ, 3 μm, Dr. Maisch GmbH). The peptides bounded on the column were eluted with a 78-min linear gradient. The solvent A consisted of 0.1% formic acid in water solution and the solvent B consisted of 0.1% formic acid in acetonitrile solution. The segmented gradient was 4–8% B, 8 min; 8–22% B, 50 min; 22–32% B, 12 min; 32–90% B, 1 min; 90% B, 7 min at a flow rate of 300 nl/min.
The MS analysis was performed with Q Exactive mass spectrometer (Thermo Scientific). With the data-dependent acquisition mode, the MS data were acquired at a high resolution 70,000 (m/z 200) across the mass range of 300–1600 m/z. The target value was 3.00E+06 with a maximum injection time of 60 ms. The top 20 precursor ions were selected from each MS full scan with isolation width of 2 m/z for fragmentation in the HCD collision cell with normalized collision energy of 30%. Subsequently, MS/MS spectra were acquired at resolution 17,500 at m/z 200. The target value was 5.00E+04 with a maximum injection time of 80 ms. The dynamic exclusion time was 40 s. For nano electrospray ion source setting, the spray voltage was 2.0 kV; no sheath gas flow; the heated capillary temperature was 320 °C.
2.16. Protein identification and quantification analysis
The raw data from Q Exactive were analyzed with Proteome Discovery version 2.2.0.388 using Sequest HT search engine for protein identification and Percolator for FDR (false discovery rate) analysis. The Uniprot human protein database was used for searching the data from Fbs cell sample. Some important searching parameters were set as following: trypsin was selected as enzyme and two missed cleavages were allowed for searching; the mass tolerance of precursor was set as 10 ppm and the product ions tolerance was 0.02 Da; the cysteine carbamidomethylation were specified as fixed modifications; The methionine oxidation was chosen as variable modifications. FDR analysis was performed with Percolator and FDR <1% was set for protein identification. The peptides confidence was set as high for peptides filter. Proteins label free quantification was also performed on Proteome Discovery using the areas of identified peptides. Only unique and razor peptides of proteins were selected for protein relative quantification. The normalization mode was selected as total peptide amount to corrected experimental bias.
2.17. Statistical analysis
The results were repeated in three experiments. All data were analyzed by unpaired two-tail t-test or Kruskal-Wallis test. All differences were considered statistically significant when the p value was less than 0.05.
3. Results
3.1. Downregulation of MAGI2-AS3 decreased the superoxide level in Fbs cell
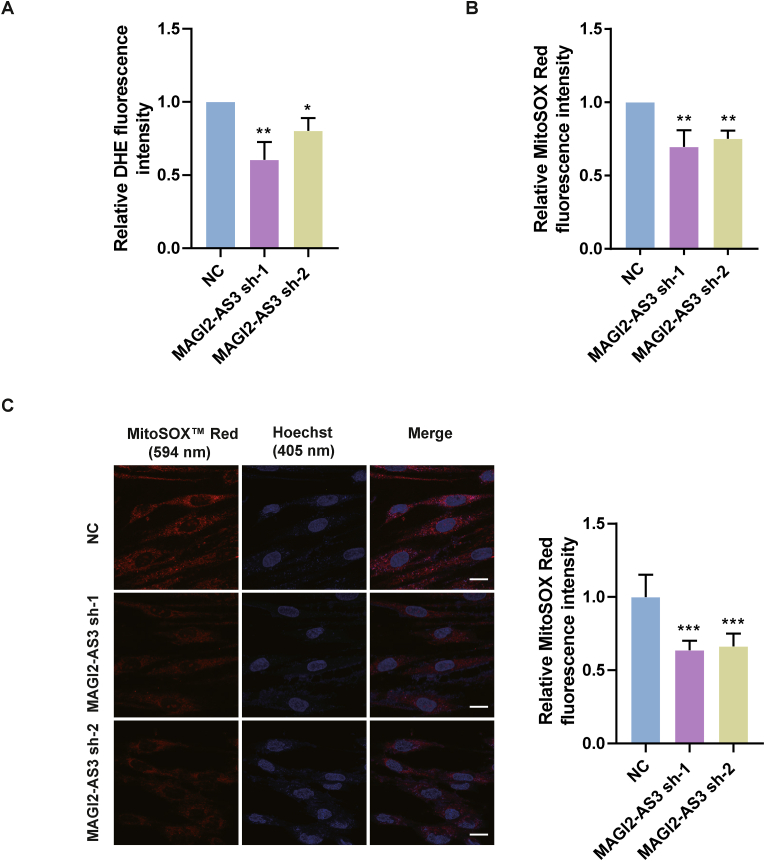
To delineate MAGI2-AS3 function, we characterized the sequence of MAGI-AS3 by performing 5′ and 3’ Rapid Amplification of cDNA Ends (RACE) in HEK293T. The result showed that MAGI2-AS3 consisted of four exons, containing 3975 nucleotides (Fig. S1A). We then examined the coding probability of MAGI2-AS3 by in silico prediction with two powerful computational tools for distinguishing coding and non-coding transcripts, Coding Potential Calculator (CPC) and Coding-Potential Assessment Tool (CPAT) [19,20]. The results obtained with these tools showed that MAGI2-AS3 has no ability to code a protein (Table S1). To investigate the effect of MAGI2-AS3 on redox conditions in Fbs, we knocked down MAGI2-AS3 using MAGI2-AS3 shRNA-1 and MAGI2-AS3 shRNA-2 which were abbreviated as MAGI2-AS3 sh-1 and MAGI2-AS3 sh-2 in this study in Fbs (Fig. S1B) and then used the fluorescent probes DHE and MitoSOX™ Red to measure the superoxide content. DHE mainly reacts with O2˙− to form 2-hydroxyethidium, which intercalates into DNA and produces red fluorescence (excitation at 300 nm and emission at 610 nm) in cells [21]. MitoSOX™ Red is a cationic derivative (triphenylphosphonium) of DHE that is directly targeted to mitochondria to specifically detect O2˙−generated in mitochondria in live cells (excitation: 510 nm and emission: 590 nm) [22]. We used the CAT gene knockdown cells as the positive control, which lead to the increase of O2˙− level in mitochondria (Figs. S1C and S1D). The microplate reader measurements showed that the DHE and MitoSOX™ Red fluorescence values were decreased significantly in Fbs overexpressing MAGI2-AS3 sh-1 and MAGI2-AS3 sh-2 compared with Fbs overexpressing MAGI2-AS3 NC-shRNA (Fig. 1A and B).
Fig. 1.
Downregulation of the lncRNA MAGI2-AS3 decreased the superoxide level in Fbs. (A) The total O2˙− content in NC, MAGI2-AS3 sh-1 and MAGI2-AS3 sh-2 cells was measured using the O2˙− sensitive fluorescent dye DHE with a microplate reader. The data are shown as the mean ± SEM (n = 3). *: p < 0.05, **: p < 0.01 by unpaired t-test. (B) Mitochondrial O2˙− in NC, MAGI2-AS3 sh-1, MAGI2-AS3 sh-2 cells was measured using the O2˙− sensitive fluorescent dye MitoSOX ™ Red with a microplate reader. The data are shown as the mean ± SEM (n = 3). *: p < 0.05, **: p < 0.01 by unpaired t-test. (C) Left: Production of the mitochondrial superoxide anion (mitochondrial O2˙−) in the NC, MAGI2-AS3 sh-1 and MAGI2-AS3 sh-2 groups was measured using MitoSOX ™ Red with confocal microscopy. Red: MitoSOX ™ Red, blue: Hoechst (nucleus). The data are shown as the mean ± SEM (n = 3) (10 cells/group, 100 cells), bar = 20 μM ***: p < 0.001 by unpaired t-test. Right: Quantitation of the mitochondrial O2˙− level based on the fluorescence intensity at 594 nm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We further confirmed the results with MitoSOX™ Red in cells by confocal imaging. Consistent with the microplate reader measurements, the O2˙− level in mitochondria was indeed lower in the MAGI2-AS3 knockdown group than in the NC-shRNA group (Fig. 1C). These results demonstrated that downregulation of MAGI2-AS3 decreased the superoxide level in Fbs.
3.2. MAGI2-AS3 interacted with the HSPA8 protein via its CTD
To evaluate the potential mechanisms by which MAGI2-AS3 regulates the superoxide anion level in Fbs, we first determined the subcellular localization of MAGI2-AS3, because knowing the subcellular location of lncRNAs greatly facilitates the interpretation and understanding of lncRNA functions. A one-step method was used to isolate cytoplasmic RNA and nuclear RNA simultaneously (Fig. 2A). We determined the transcriptional level of the lncRNA MAGI2-AS3 by qPCR after reverse transcription and found that lncRNA MAGI2-AS3 was localized mainly in the cytoplasm in Fbs (Fig. 2A), consistent with previous reports of its localization in tumor cells [23]. We further performed an RNA pulldown assay to identify the interacting proteins of MAGI2-AS3. Sense and antisense strands of MAGI2-AS3 were synthesized by in vitro transcription and were then biotinylated. The sense and antisense strands were incubated with fibroblast lysates and RNA-protein complexes were separated on SDS-PAGE gels (Fig. 2B). Protein bands from the different fractions in the sense and antisense lanes were subjected to mass spectrometry analysis. Differentially expressed proteins between the two groups were analyzed according to the functional qualification of “redox” and “RNA binding protein” and HSPA8 was selected from the list of the top qualified proteins (Table 3). The interaction between MAGI2-AS3 and HSPA8 was also confirmed by RNA-pulldown followed by Western blot analysis (Fig. 2C) and by PAR-CLIP followed by RT-qPCR (Fig. 2D). Moreover, in vitro RNA-protein binding assay with GST-tagged HSPA8 and MAGI2-AS3 transcribed in vitro revealed that HSPA8 directly interacted with MAGI2-AS3 (Fig. 2E). Collectively, these data showed that HSPA8 indeed interacted with MAGI2-AS3. To understand the molecular mechanism of MAGI2-AS3, we further explored the region involved in the binding of the HSPA8 protein to MAGI2-AS3. The HSPA8 protein contains 646 amino acids. Amino acids 1–386 constitute the N-terminal binding region of the protein (NBD), amino acids 394–508 constitute the C-terminal substrate binding region of the protein (CBD) and amino acids 604–646 constitute the C-terminal binding region of the protein (C-terminal domain, CTD) [24]. To confirm the MAGI2-AS3 binding site in HSPA8, we permuted and combined these three regions into five truncated protein regions (Fig. 2F). The five truncated fragments of HSPA8 that interacted with MAGI2-AS3 were analyzed by RNA pulldown combined with Western blotting. Truncated fragments of 387–646 and 509–646 containing the CTD, interacted with MAGI2-AS3, while the others without the CTD, did not, indicating that MAGI2-AS3 interacted with HSPA8 via the CTD of HSPA8. (Fig. 2F). Additionally, we truncated MAGI2-AS3 into 4 fragments denoted as F1–F4 and performed RNA-pull down, showing that F1 and F2 were the main binding fragments with HSPA8 and F3 also had a weak binding with HSPA8 (Fig. 2G). Collectively, these data showed that HSPA8 mainly interacted with 5’ region of MAGI2-AS3 through its CTD and may mediate the regulation of superoxide anion and H2O2 level in Fbs cells.
Fig. 2.
MAGI2-AS3 interacted with the HSPA8 protein via its CTD. (A) Left: Marker proteins for nuclear (LAMINB1) and cytosolic (β-tubulin) proteins were evaluated using Western blot analysis after separation of the nuclear and cytoplasmic fractions of Fbs. Right: Subcellular localization of MAGI2-AS3. RT-qPCR analysis of MAGI2-AS3, U1, GAPDH and β-ACTIN in the nuclear and cytoplasmic fractions (U1 is located in the nucleus; GAPDH and β-ACTIN are located in the cytosol). The data are shown as the mean ± SEM (n = 3). (B) Proteins that interacted with MAGI2-AS3 were analyzed by RNA pulldown combined with mass spectrometry. Gel image showing proteins pulled down by MAGI2-AS3 sense and antisense RNAs from human fibroblast lysates. The bands in the regions enclosed in the red frames were subjected to mass spectrometry analysis. (C) Proteins were pulled down with MAGI2-AS3 sense and antisense RNAs from human fibroblast lysates, followed by Western blot analysis with an anti-HSPA8 antibody. The densitometry analysis was shown below the band. (D) The HSPA8 protein was immunoprecipitated from UV-crosslinked human fibroblast lysates and MAGI2-AS3 mRNA expression was detected by RT-qPCR. Left: Western blot analysis of the HSPA8 protein level in the IgG control and anti-HSPA8 antibody incubation groups. Right: RT-qPCR analysis of the MAGI2-AS3 level in the IgG control and anti-HSPA8 antibody incubation groups. The data are shown as the mean ± SEM (n = 3). **: p < 0.01 by unpaired t-test. (E) GST pull-down assay of the binding of MAGI2-AS3 to GST-tagged HSPA8. The data are shown as the mean ± SEM (n = 3). ***: p < 0.001 by unpaired t-test. (F) Construction of plasmids containing HSPA8 truncation fragments (pLE4-flag-truncated fragment-GFP). The HSPA8 protein consists of 646 amino acids. Amino acids 1–386 constitute the N-terminal binding region of the protein (NBD), amino acids 394–508 constitute the C-terminal substrate binding region of the protein (SBD) and amino acids 604–646 constitute the CTD. Based on this structure, we constructed five kinds of plasmids containing truncated fragments of the HSPA8 protein (HSPA8 A1:387–646, A2:509–646, A3:1–508, A4:1–386, A5:387-508). Left: Analysis of input for WT HSPA8 and the five truncated fragments of HSPA8 by Western blotting. Right: The truncated fragments of HSPA8 that interacted with MAGI2-AS3 were analyzed by RNA pulldown combined with Western blotting. (G) RNA pull-down assay of the binding of truncated MAGI2-AS3 to HSPA8 (MAGI2-AS3 F1:1–1491, F2:1485–2313, F3:2314–3130, F4:3021-3975, FL:1–3975) followed by Western blot with HSPA8 antibody, LacZ as a negative control transcript. The densitometry analysis was shown below the band. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 3.
Mass spectrometry results.
3.3. Downregulation of MAGI2-AS3 decreased the H2O2 content by stabilizing the HSPA8 protein level
LncRNA and protein interactions may alter the stability and translation of proteins [25]. We found that the HSPA8 mRNA levels were not significantly different among Fbs overexpressing MAGI2-AS3 sh-1, MAGI2-AS3 sh-2 and NC shRNA (Fig. 3A), but the HSPA8 protein levels were significantly higher in the MAGI2-AS3 knockdown groups than that in the control group (Fig. 3B). So we speculate that MAGI2-AS3 knock down decreased the HSPA8 degradation, contributing to stabilization of HSPA8 protein level. In order to study the pathway by which MAGI2-AS3 may interfere HSPA8 stability, we first explored the half-life of HSPA8. We treated Fbs cells with cycloheximide (CHX) to inhibit protein synthesis and measured the level of HSPA8 at 0,2,4,6,8 h. From the results we could see that the half-life of HSPA8 is about 4 h (Fig. S1E). The following experiments were carried out based on this result. We next study whether MAGI2-AS3 promotes degradation of HSPA8 through the proteasome pathway. Fbs cells overexpressing NC shRNA and MAGI2-AS3 sh-1 under CHX treatment were treated with or without MG132 for 4 h and then the level of HSPA8 was detected. MG132 is a potent proteasome inhibitor and it effectively blocks the proteolytic activity of the 26S proteasome complex. The result showed that MG132 treatment caused HSPA8 accumulation in cells expressing NC-shRNA. However, there was no difference of the HSPA8 level between Fbs cells overexpressing MAGI2-AS3 sh-1 with MG132 treatment and without MG132 treatment (Fig. 3C), indicating that MAGI2-AS3 promotes degradation of HSPA8 possibly through the proteasome pathway. In consideration of MG132 is not just a potent proteasome inhibitor but also a calpain inhibitor, we treated Fbs cells with CHX accompanied with or without MG101 for 4 h and found that MG101 treatment did not cause HSPA8 accumulation in Fbs cells (Fig. S1F), suggesting that MG132 inhibited HSPA8 protein degradation mainly through its proteasome inhibitor function. We further sought to determine whether the regulation of the superoxide level by MAGI2-AS3 knockdown is dependent on the regulation of HSPA8 by MAGI2-AS3 in Fbs. In cells with MAGI2-AS3 knockdown, we knocked down HSPA8 and found that the effect of superoxide deregulation in cells caused by MAGI2-AS3 knockdown was abolished (Fig. 3D and E), indicating that downregulation of MAGI2-AS3 decreased the superoxide anion level by stabilizing the HSPA8 protein level. We further measured the H2O2 level with the HyPerRed probe and found that MAGI2-AS3 knockdown decreased the H2O2 level in Fbs (Fig. 3F). It has reported that p66Shc can increase the level of ROS and produce oxidative stress [26]. p66shc mRNA expression was also decreased significantly in the MAGI2-AS3 knockdown group compared with the control group (Fig. 3G). These results demonstrated that downregulation of MAGI2-AS3 decreased the H2O2 level by stabilizing the HSPA8 protein level.
Fig. 3.
Downregulation of MAGI2-AS3 decreased the H2O2content by stabilizing the HSPA8 protein level. (A) RT-qPCR analysis of HSPA8 mRNA expression in control and Fbs transduced with lentiviral vectors expressing MAGI2-AS3 NC shRNA, MAGI2-AS3 sh-1 or MAGI2-AS3 sh-2. The data are shown as the mean ± SEM (n = 3). NS: not significant by unpaired t-test. (B) Western blot analysis of the HSPA8 protein level in control and Fbs transduced with lentiviral vectors expressing NC-shRNA, MAGI2-AS3 sh-1 or MAGI2-AS3 sh-2. The data are shown as the mean ± SEM (n = 3). *: p < 0.05, ***: p < 0.001 by unpaired t-test. (C) Western blot analysis of the HSPA8 protein level in Fbs transduced with lentiviral vectors expressing NC-shRNA and MAGI2-AS3 sh-1 under 10 μg/mL CHX treatment accompanied with or without 10 μM MG132 treatment for 4 h. The data are shown as the mean ± SEM (n = 3). *: p < 0.05 by unpaired t-test, NS: not significant. (D–E) Relative mitochondrial O2˙− levels in Fbs transduced with lentiviral vectors expressing NC-shRNA, NC and HSPA8 shRNA, MAGI2-AS3 sh-1 (or MAGI2-AS3 sh-2), MAGI2-AS3 sh-1 (or MAGI2-AS3 sh-2) and HSPA8 shRNA. The data are shown as the mean ± SEM (n = 3). #: p < 0.05, **: p < 0.01, ***: p < 0.001 by unpaired t-test. (F) Excitation (575 nm) and emission (620 nm) of HyPerRed in Fbs expressing NC-shRNA and MAGI2-AS3 sh-1 were detected with a microplate reader. The data are shown as the mean ± SEM (n = 3). *: p < 0.05, **: p < 0.01 by unpaired t-test. (G) RT-qPCR analysis of p66shc mRNA expression in Fbs transduced with lentiviral vectors expressing NC-shRNA and MAGI2-AS3 sh-1. The data are shown as the mean ± SEM (n = 3). ***: p < 0.001 by unpaired t-test.
3.4. Downregulation of MAGI2-AS3 delayed cell senescence
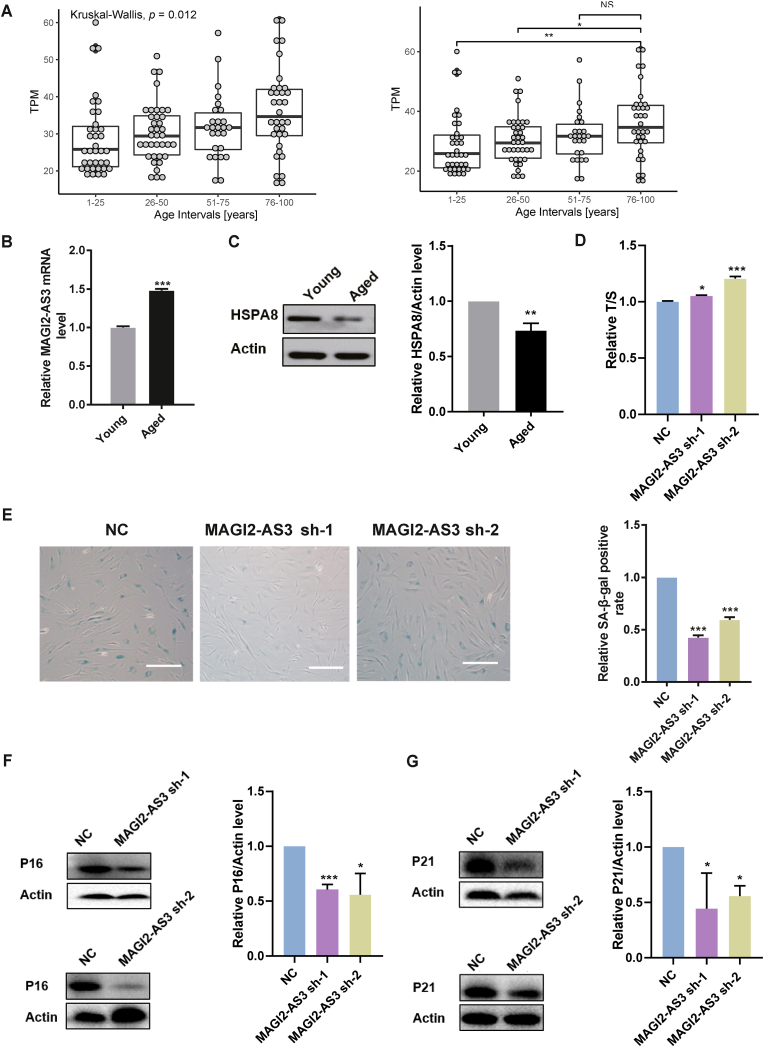
To further study whether MAGI2-AS3 is a senescence-related lncRNA (or to reveal the potential role of MAGI2-AS3 in cellular senescence), we analyzed its expression level in fibroblasts from 133 donors of all ages based on RNA-seq dataset GSE113957 [27] from the GEO database and found that its expression level was increased with age (Fig. 4A). We then used the replicative senescence model to study the function of MAGI2-AS3 in aging. P32–P34 Fbs were defined as young cells, while P40–P42 Fbs were defined as aged cells. Senescence-related phenotypes were characterized by the increasing prevalence of senescence-associated β-galactosidase (SA-β-Gal)-positive cells (Fig. S2A), telomere shortening in aged Fbs (Fig. S2B) and comparative increases in the mRNA and protein levels of P16 and P21 in aged Fbs (Figure S2C, S2D and S2E). We first measured the MAGI2-AS3 level during the cell senescence process and found that it was markedly increased in aged cells compared to young cells (Fig. 4B), while HSPA8 protein level is decreased markedly in aged cells (Fig. 4C). To confirm whether MAGI2-AS3 is a driver of or just an aging-related phenotype in the cell senescence process, we then studied the effect of MAGI2-AS3 downregulation on cell senescence in this model. Compared to NC-shRNA cells, Fbs overexpressing MAGI2-AS3 sh-1 and MAGI2-AS3 sh-2 presented markedly decreased senescence characteristics, as evidenced by the slower telomere shortening (Fig. 4D), lower proportion of SA-β-Gal-positive cells (Fig. 4E), lower P16 and P21 mRNA (Fig. S1G) and protein (Fig. 4F and G) levels. These results showed that MAGI2-AS3 was upregulated with age in human and senescent cells and downregulation of MAGI2-AS3 delayed cell senescence suggesting that MAGI2-AS3 is a driver of cell senescence.
Fig. 4.
Downregulation of MAGI2-AS3 delayed cell senescence. (A) Left: The expression level of MAGI2-AS3 is increased in human fibroblasts derived from aged donors. Differential expression across age groups is determined via a Kruskal-Wallis test, p = 0.012. Right: Horizontal lines denote the median and quantiles of transcripts per million (TPM) values in each group and outlier samples are labeled as dots. 1–25 years old (n = 36), 26–50 years old (n = 37), 51–75 years old (n = 26), 76–100 years old (n = 34). *: p < 0.05, **: p < 0.01, NS: not significant by Wilcoxon rank-sum test. (B) RT-qPCR analysis of MAGI2-AS3 mRNA expression in young and aged cells. The data are shown as the mean ± SEM (n = 3). ***: p < 0.001 by unpaired t-test. (C) Protein level of HSPA8 in young and aged cells. The data are shown as the mean ± SEM (n = 3). **: p < 0.01 by unpaired t-test. (D) The relative telomere length (T/S: ratio of the relative telomere length to single gene copy) in Fbs expressing NC-shRNA, MAGI2-AS3 sh-1 and MAGI2-AS3 sh-2 was measured by RT-qPCR. The data are shown as the mean ± SEM (n = 3). *: p < 0.05, ***: p < 0.001 by unpaired t-test. (E) Left: SA-β-Gal staining in Fbs expressing NC-shRNA, MAGI2-AS3 sh-1 and MAGI2-AS3 sh-2. Scale bar, 100 μm. Right: Statistical analysis of the percentages of SA-β-Gal-positive cells. The data are shown as mean ± SEM (n = 3). ***: p < 0.001 by unpaired t-test. (F–G) Western blot analysis of P16 and P21 protein expression in Fbs expressing NC-shRNA, MAGI2-AS3 sh-1. The data are shown as the mean ± SEM (n = 3). *: p < 0.05, ***: p < 0.001 by unpaired t-test.
3.5. HSPA8 knockdown blocked the antisenescence effect of MAGI2-AS3 downregulation
Downregulation of MAGI2-AS3 decreased the H2O2 level by stabilizing the HSPA8 protein level (Fig. 3). To confirm that downregulation of MAGI2-AS3 delayed cell senescence by stabilizing the protein level of HSPA8, we knocked down HSPA8 using HSPA8 shRNA (Fig. S1H) and knocked down MAGI2-AS3 by overexpressing MAGI2-AS3 sh-1 and MAGI2-AS3 sh-2 and found that the antisenescence effect caused by downregulation of MAGI2-AS3 was abolished. The proportion of SA-β-Gal-positive cells (Fig. 5A and B) and P21 mRNA level (Fig. 5C and D) did not differ between Fbs overexpressing NC-shRNA and those overexpressing MAGI2-AS3 sh-1 or MAGI2-AS3 sh-2 after HSPA8 knockdown, while the antisenescence effect was observed only in the MAGI2-AS3 knockdown groups (MAGI2-AS3 sh-1 and MAGI2-AS3 sh-2 groups). Downregulation of MAGI2-AS3 stabilized the HSPA8 protein level (Fig. 3C) and HSPA8 could weaken p66Shc signaling [28]. We further detected the level of p66Shc level in MAGI2-AS3 knockdown with or without HSPA8 interference condition. We could see that MAGI2-AS3 knock down decreased the p66Shc and O2˙− in mitochondria, while these effects disappeared when HSPA8 was knock down (Fig. 5E and F). These results indicated that the regulation of MAGI2-AS3 on p66Shc and O2˙− in mitochondria depended on the HSPA8 level and stabilizing the HSPA8 protein level plays a key role in the antisenescence effect of MAGI2-AS3 downregulation.
Fig. 5.
HSPA8 knockdown blocked the antisenescence effect of MAGI2-AS3 downregulation. (A) SA-β-Gal staining in Fbs transduced with lentiviral vectors expressing NC-shRNA, MAGI2-AS3 sh-1 (or MAGI2-AS3 sh-2), MAGI2-AS3 sh-1 (or MAGI2-AS3 sh-2) and HSPA8 shRNA. (B) Statistical analysis of the percentages of SA-β-Gal-positive cells. The data are shown as the mean ± SEM (n = 3). *: p < 0.05, #: p < 0.05, **: p < 0.01, ##: p < 0.01 by unpaired t-test. (C–D) RT-qPCR analysis of P21 mRNA expression in Fbs expressing NC-shRNA, MAGI2-AS3 sh-1 or MAGI2-AS3 sh-2 and HSPA8 shRNA. The data are shown as the mean ± SEM (n = 3). *: p < 0.05, ##: p < 0.01, ***: p < 0.001 by unpaired t-test. (E–F) RT-qPCR analysis of p66shc mRNA expression in Fbs transduced with lentiviral vectors expressing NC-shRNA, MAGI2-AS3 sh-1 or MAGI2-AS3 sh-2 and HSPA8 shRNA. The data are shown as the mean ± SEM (n = 3). #: p < 0.05, **: p < 0.01 by unpaired t-test.
3.6. Downregulation of MAGI2-AS3 delayed cell senescence mediated by the ROS/MAP2K6/p38 signaling pathway
To understand the molecular mechanism by which downregulation of MAGI2-AS3 delayed cell senescence, we performed genome-wide RNA-seq analysis of Fbs overexpressing NC-shRNA and MAGI2-AS3 sh-1. A total of 246 genes were upregulated and 193 genes were downregulated in the MAGI2-AS3 sh-1 group compared to the NC-shRNA group (Fig. 6A). Gene function analysis showed that 8 genes (MAP2K6, DLL3, IL1B, PECAM1, ELN, VWA5A, MMP12, FLT1) were related to aging and we further verified these genes by q-PCR and results showed that MAP2K6 which involved in the p38 MAP kinase pathway was markedly downregulated in MAGI2-AS3 knockdown Fbs (Fig. 6B and C). ROS/JNK/p38 signaling plays an important mediating role in senescent cells [29] and we also verified that the phosphorylation level of the p38 protein (pp38) was significantly increased in aged cells compared with young cells (Fig. 6D). Downregulation of MAGI2-AS3 decreased the H2O2 content by stabilizing the HSPA8 protein level (Fig. 3), thus, we measured ROS/MAP2K6/p38 signaling and found that the pp38 level was markedly decreased in Fbs overexpressing MAGI2-AS3 sh-1 compared to overexpressing NC-shRNA (Fig. 6E). We further studied the relationship of HSPA8 and pp38/p38 level which is the downstream of MAP2K6. p38 MAPK pathway activated by ROS plays an important role in accelerating aging [30]. We could see that MAGI2-AS3 knock down could decrease pp38/p38 level, while this effect disappeared when HSPA8 was knocked down (Fig. 6F), showing that MAGI2-AS3 knockdown decreased the pp38/p38 level depending on the HSPA8 level. To confirm that downregulation of MAGI2-AS3 delayed cell senescence mediated by the ROS/MAP2K6/p38 signaling pathway, we overexpressed MAP2K6 in cells with knockdown of MAGI2-AS3 (Fig. S1I) and found that the antisenescence effect caused by downregulation of MAGI2-AS3 was abolished as shown in telomere length detection and SA-β-Gal-positive cells (Fig. 6G and H). These results demonstrated that downregulation of MAGI2-AS3 delayed cell senescence mediated by the ROS/MAP2K6/p38 signaling pathway.
Fig. 6.
MAGI2-AS3 delayed cell senescence mediated by the ROS/MAP2K6/p38 MAPK signaling pathway. (A) RNA-seq results of differential gene expression in Fbs expressing NC-shRNA or MAGI2-AS3 sh-1. (B) GO analysis of differentially expressed genes (*p:< 0.05) in Fbs expressing NC-shRNA or MAGI2-AS3 sh-1. RT-qPCR analysis of 8 aging related genes (MAP2K6, DLL3, IL1B, PECAM1, ELN, VWA5A, MMP12, FLT1) expression in Fbs expressing NC-shRNA or MAGI2-AS3 sh-1. The data are shown as the mean ± SEM (n = 3). *: p < 0.05, **: p < 0.01, ***: p < 0.001 by unpaired t-test. (C) RT-qPCR analysis of MAP2K6 mRNA expression in Fbs expressing NC-shRNA, MAGI2-AS3 sh-1 or MAGI2-AS3 sh-2. The data are shown as the mean ± SEM (n = 3). *: p < 0.05, **: p < 0.01 by unpaired t-test. (D) Western blot analysis of the level of phosphorylated p38 protein in young and aged cells. The data are shown as the mean ± SEM (n = 3). *: p < 0.05 by unpaired t-test. (E) Western blot analysis of the p38 protein level and level of phosphorylated p38 protein in Fbs control and transduced with lentiviral vectors expressing NC-shRNA, MAGI2-AS3 sh-1. The data are shown as the mean ± SEM (n = 3). *: p < 0.05 by unpaired t-test. (F) Western blot analysis of the p38, phosphorylated p38 and HSPA8 proteins level in Fbs transduced with lentiviral vectors expressing NC-shRNA, MAGI2-AS3 sh-1, MAGI2-AS3 sh-1 and HSPA8-shRNA. The data are shown as the mean ± SEM (n = 3). *: p < 0.05, #: p < 0.05, **: p < 0.01, ##: p < 0.01 by unpaired t-test. (G) The relative telomere length (T/S: ratio of the relative telomere length to single gene copy) in Fbs expressing NC-shRNA, MAGI2-AS3 sh-1 with or without MAP2K6 overexpression was determined by RT-qPCR. The data are shown as the mean ± SEM (n = 3). *: p < 0.05, ##: p < 0.05 by unpaired t-test. (H) SA-β-Gal staining in Fbs transduced with lentiviral vectors expressing NC-shRNA, MAGI2-AS3 sh-1 with or without MAP2K6 overexpression. The data are shown as the mean ± SEM (n = 3). The data are shown as the mean ± SEM (n = 3). **: p < 0.01, ##: p < 0.01 by unpaired t-test. (I) Schematic diagram of downregulation of long non-coding RNA MAGI2-AS3 attenuates H2O2 level and delays cell senescence by reducing degradation of HSPA8.
4. Discussion
LncRNAs play important roles in diverse biological and pathological processes. However, there are few reports about the regulation of redox reactions by lncRNAs, let alone about the in-depth function and regulatory mechanism. In this study, we discovered that downregulation of MAGI2-AS3 decreased the superoxide level in Fbs, an effect that was mediated by reducing its interaction with the CTD of HSPA8. Further studies showed that MAGI2-AS3 was increased in senescent cells and that downregulation of MAGI2-AS3 delayed cell senescence by suppressing the ROS/MAP2K6/p38 signaling pathway (Fig. 6I). Our results showed that downregulation of the lncRNA MAGI2-AS3 reduced the superoxide anion level and delayed cell senescence, providing a potential target for suppressing cell senescence. This work augmented the relationship among lncRNAs, redox and aging.
Many factors regulate cell redox homeostasis, such as coding genes, antioxidant enzymes and exogenous small molecules. Redox regulation by lncRNAs has gradually been revealed recently. Our data showed that MAGI2-AS3 is involved in the regulation of the cellular superoxide anion level via the following mechanism. HSPA8 is decreased in Fbs cell senescence (Fig. 4C) and studies also showed that HSPA8 level decreased in human cerebrospinal fluid (CSF) and primate retina during aging [31,32]. MAGI2-AS3 knockdown stabilized the HSPA8 level by decreasing the interaction of its CTD with MAGI2-AS3 and HSPA8 played an important role in maintaining redox homeostasis. ROS can activate the phosphorylation of p66shc and further lead to an increase in H2O2. The chaperone HSPA8 can block p66shc signaling and prevent the production of ROS [28]. This regulatory mechanism is also supported by the latest reports. The lncRNA FENDRR binds to Nrf2, promotes its degradation and then promotes the apoptosis of Leydig cells in late-onset hyperopia [33]. Our study reports a new lncRNA, MAGI2-AS3, which regulates redox homeostasis and reveals a new regulatory mechanism. Although there have been some reports about the regulation of redox by lncRNAs, few studies demonstrate the mechanism and characteristics, for example, the direct interacting target, the specific redox species. Our work illustrates that lncRNAs may be an important part of the redox regulation network.
To date, most studies on the function of MAGI2-AS3 have focused on cancer [23,[34], [35], [36]]. However, this work revealed that MAGI2-AS3 is involved in the regulation of aging. We found that its expression was increased during cell senescence and that its downregulation delayed cell senescence. In addition, we provided a new target for antiaging. A similar study showed that the lncRNA AK156230 was downregulated during cell senescence in mouse embryonic fibroblast (MEFs) and that knockdown of AK156230 accelerated cell senescence [37]. This study supported the hypothesis that lncRNAs can participate in the aging process. The antiaging mechanisms of lncRNAs currently reported include modulation of telomere length [38,39], epigenetic alterations [[40], [41], [42], [43], [44]], proteostasis [[45], [46], [47], [48]] and moderation of stem cell properties [40]. Our report identified a new mechanism by which downregulation of MAGI2-AS3 could regulate the superoxide anion level to suppress the ROS/MAP2K6/p38 signaling pathway in order to exert an antiaging effect, providing a new target and mechanism by which lncRNAs regulate aging.
Since MAGI2-AS3 is expressed specifically in human and other primates such as rhesus while there is no homolog in mouse (Fig. S1A), the effect of MAGI2-AS3 knockdown on superoxide downregulation and antiaging need to be studied in primates in future research and it is important to validate these conclusions derived from replicative senescent cells to humans. Moreover, strategies to deregulate MAGI2-AS3 during aging or approaches to interfere with HSPA8 expression are urgently need study for application. Novel therapeutic strategies with antisense oligonucleotides (ASOs) have been used for disease therapy. ASOs can be delivered to the targeted organ with better delivery efficiency. This genetic therapeutic strategy has been utilized in diseases such as myotonic dystrophy [49], Hutchinson–Gilford progeria syndrome [50] and esophageal adenocarcinoma [51]. In these examples, ASOs of lncRNAs have also been used. With this method of lncRNA Malat1 knockdown by precise binding with the RNA of the myotonic dystrophy gene, myotonic muscular dystrophy could be fundamentally cured. If these strategies can be implemented, then our present study provides a new target and a rationale for antiaging application.
Declaration of competing interest
The authors declare no potential conflicts of interest.
Acknowledgments
We thank Dr. Tomoaki Hishida for kindly providing the pLE4 lentiviral vector. We are grateful Dr. Jifeng Wang (Laboratory of Proteomics, Institute of Biophysics, Chinese Academy of Sciences) for helping with MS analysis. This work was financially supported by the National Key R&D Program (2017YFA0504000), the National Natural Science Foundation of China (91849203, 31900893), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB39000000).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102383.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Barber S.R., et al. External national validation of the Leicester self-assessment score for type 2 diabetes using data from the English Longitudinal study of ageing. Diabet. Med. 2017;34(11):1575–1583. doi: 10.1111/dme.13433. [DOI] [PubMed] [Google Scholar]
- 2.Byun M., Kim J., Kim M. Physical and psychological factors affecting falls in older patients with arthritis. Int. J. Environ. Res. Publ. Health. 2020;17(3):1098. doi: 10.3390/ijerph17031098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engelbrecht C., Sardinha L.R., Rizzo L.V. Cytokine and chemokine concentration in the tear of patients with age-related cataract. Curr. Eye Res. 2020:1–6. doi: 10.1080/02713683.2020.1715445. [DOI] [PubMed] [Google Scholar]
- 4.Gupta S.K., Piccoli M.T., Thum T. Non-coding RNAs in cardiovascular ageing. Ageing Res. Rev. 2014;17:79–85. doi: 10.1016/j.arr.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Idda M.L., et al. Noncoding RNAs in Alzheimer’s disease. Wiley Interdiscip Rev RNA. 2018;9(2):1–10. doi: 10.1002/wrna.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siddique N., et al. Statistical analysis of fat and muscle mass in osteoporosis in elderly population using total body DXA scans. Ir. J. Med. Sci. 2020;189(3):1105–1113. doi: 10.1007/s11845-020-02177-8. [DOI] [PubMed] [Google Scholar]
- 7.Go Y.M., Jones D.P. Redox theory of aging: implications for health and disease. Clin. Sci. (Lond.) 2017;131(14):1669–1688. doi: 10.1042/CS20160897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen J.M., Jones D.P., Harris C. The redox theory of development. Antioxidants Redox Signal. 2020;32(10):715–740. doi: 10.1089/ars.2019.7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guttman M., et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W., et al. Effects of long non-coding RNA LINC00963 on renal interstitial fibrosis and oxidative stress of rats with chronic renal failure via the foxo signaling pathway. Cell. Physiol. Biochem. 2018;46(2):815–828. doi: 10.1159/000488739. [DOI] [PubMed] [Google Scholar]
- 12.Sanei-Ataabadi N., Mowla S.J., Nasr-Esfahani M.H. Transcript isoforms of SLC7A11-AS1 are associated with varicocele-related male infertility. Front. Genet. 2020;11:1015. doi: 10.3389/fgene.2020.01015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu H., et al. MSC-induced lncRNA HCP5 drove fatty acid oxidation through miR-3619-5p/AMPK/PGC1α/CEBPB axis to promote stemness and chemo-resistance of gastric cancer. Cell Death Dis. 2020;11(4):233. doi: 10.1038/s41419-020-2426-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai W., Lee D. Interfering with long chain noncoding RNA ANRIL expression reduces heart failure in rats with diabetes by inhibiting myocardial oxidative stress. J. Cell. Biochem. 2019;120(10):18446–18456. doi: 10.1002/jcb.29162. [DOI] [PubMed] [Google Scholar]
- 15.Stein C.S., et al. Mitoregulin: a lncRNA-encoded microprotein that supports mitochondrial supercomplexes and respiratory efficiency. Cell Rep. 2018;23(13):3710–3720. doi: 10.1016/j.celrep.2018.06.002. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kai-Xin L., et al. Roles of lncRNA MAGI2-AS3 in human cancers. Biomed. Pharmacother. 2021;141 doi: 10.1016/j.biopha.2021.111812. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J., Wang R. Deregulated lncRNA MAGI2-AS3 in Alzheimer's disease attenuates amyloid-β induced neurotoxicity and neuroinflammation by sponging miR-374b-5p. Exp. Gerontol. 2021;144 doi: 10.1016/j.exger.2020.111180. [DOI] [PubMed] [Google Scholar]
- 18.Poursheikhani A., et al. Integration analysis of long non-coding RNA (lncRNA) role in tumorigenesis of colon adenocarcinoma. BMC Med. Genom. 2020;13(1):108. doi: 10.1186/s12920-020-00757-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L., et al. CPAT: coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41(6):e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang Y.J., et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45(W1):W12–w16. doi: 10.1093/nar/gkx428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardiner, B., et al., Measurement of oxidative stress markers in vitro using commercially available kits, in Measuring Oxidants and Oxidative Stress in Biological Systems, L.J. Berliner and N.L. Parinandi, Editors. 2020, SpringerCopyright 2020, Springer Nature Switzerland AG.: Cham (CH). p. 39-60. [PubMed]
- 22.Kalyanaraman B. In: Measuring Oxidants and Oxidative Stress in Biological Systems. Berliner L.J., Parinandi N.L., editors. Springer Nature Switzerland AG; 2020. Pitfalls of reactive oxygen species (ROS) measurements by fluorescent probes and mitochondrial superoxide determination using MitoSOX; pp. 7–9. Cham (CH) Springer Copyright 2020. [PubMed] [Google Scholar]
- 23.Li D., et al. LncRNA MAGI2-AS3 is regulated by BRD4 and promotes gastric cancer progression via maintaining ZEB1 overexpression by sponging miR-141/200a. Mol. Ther. Nucleic Acids. 2020;19:109–123. doi: 10.1016/j.omtn.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rauch J.N., Zuiderweg E.R., Gestwicki J.E. Non-canonical interactions between heat shock cognate protein 70 (Hsc70) and Bcl2-associated anthanogene (BAG) Co-chaperones are important for client release. J. Biol. Chem. 2016;291(38):19848–19857. doi: 10.1074/jbc.M116.742502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Statello L., et al. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021;22(2):96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Migliaccio E., Giorgio M., Pelicci P.G. Apoptosis and aging: role of p66Shc redox protein. Antioxidants Redox Signal. 2006;8(3–4):600–608. doi: 10.1089/ars.2006.8.600. [DOI] [PubMed] [Google Scholar]
- 27.Fleischer J.G., et al. Predicting age from the transcriptome of human dermal fibroblasts. Genome Biol. 2018;19(1):221. doi: 10.1186/s13059-018-1599-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown J.E., et al. Essential role of the redox-sensitive kinase p66shc in determining energetic and oxidative status and cell fate in neuronal preconditioning. J. Neurosci. 2010;30(15):5242–5252. doi: 10.1523/JNEUROSCI.6366-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang D., et al. Autophagy inhibits the mesenchymal stem cell aging induced by D-galactose through ROS/JNK/p38 signalling. Clin. Exp. Pharmacol. Physiol. 2020;47(3):466–477. doi: 10.1111/1440-1681.13207. [DOI] [PubMed] [Google Scholar]
- 30.Hsieh C.C., et al. The ASK1-Signalosome regulates p38 MAPK activity in response to levels of endogenous oxidative stress in the Klotho mouse models of aging. Aging (Albany NY) 2010;2(9):597–611. doi: 10.18632/aging.100194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loeffler D.A., et al. Age-related decrease in heat shock 70-kDa protein 8 in cerebrospinal fluid is associated with increased oxidative stress. Front. Aging Neurosci. 2016;8:178. doi: 10.3389/fnagi.2016.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernstein S.L., et al. Heat shock cognate-70 gene expression declines during normal aging of the primate retina. Invest. Ophthalmol. Vis. Sci. 2000;41(10):2857–2862. [PubMed] [Google Scholar]
- 33.Liu Y., et al. LncRNA FENDRR promotes apoptosis of Leydig cells in late-onset hypogonadism by facilitating the degradation of Nrf2. Cell Tissue Res. 2021;386(2):379–389. doi: 10.1007/s00441-021-03497-6. [DOI] [PubMed] [Google Scholar]
- 34.Gokulnath P., et al. Long non-coding RNA MAGI2-AS3 is a new player with a tumor suppressive role in high grade serous ovarian carcinoma. Cancers. 2019;11(12):2008. doi: 10.3390/cancers11122008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu X., et al. MAGI2-AS3 inhibits breast cancer by downregulating DNA methylation of MAGI2. J. Cell. Physiol. 2021;236(2):1116–1130. doi: 10.1002/jcp.29922. [DOI] [PubMed] [Google Scholar]
- 36.Xue C., et al. Novel insights for lncRNA MAGI2-AS3 in solid tumors. Biomed. Pharmacother. 2021;137 doi: 10.1016/j.biopha.2021.111429. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y.N., et al. Identification of the lncRNA, AK156230, as a novel regulator of cellular senescence in mouse embryonic fibroblasts. Oncotarget. 2016;7(33):52673–52684. doi: 10.18632/oncotarget.10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Redon S., Reichenbach P., Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38(17):5797–5806. doi: 10.1093/nar/gkq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samper E., Flores J.M., Blasco M.A. Restoration of telomerase activity rescues chromosomal instability and premature aging in Terc-/- mice with short telomeres. EMBO Rep. 2001;2(9):800–807. doi: 10.1093/embo-reports/kve174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loewer S., et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 2010;42(12):1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Latos P.A., et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338(6113):1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 42.Di Ruscio A., et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503(7476):371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandey R.R., et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32(2):232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 44.Deng Z., et al. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell. 2009;35(4):403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoon J.H., et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47(4):648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abdelmohsen K., et al. 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Res. 2014;42(15):10099–10111. doi: 10.1093/nar/gku686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mourtada-Maarabouni M., et al. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28(2):195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 48.Carrieri C., et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491(7424):454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 49.Wheeler T.M., et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488(7409):111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erdos M.R., et al. A targeted antisense therapeutic approach for Hutchinson-Gilford progeria syndrome. Nat Med. 2021;27(3):536–545. doi: 10.1038/s41591-021-01274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu Y., et al. LncRNA PVT1 up-regulation is a poor prognosticator and serves as a therapeutic target in esophageal adenocarcinoma. Mol. Cancer. 2019;18(1):141. doi: 10.1186/s12943-019-1064-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.