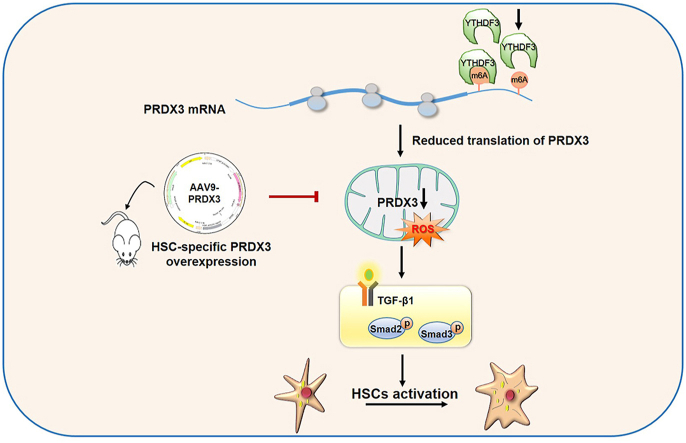
Abstract
Peroxiredoxin 3 (PRDX3) acts as a master regulator of mitochondrial oxidative stress and exerts hepatoprotective effects, but the role of PRDX3 in liver fibrosis is not well understood. N6-methyladenosine (m6A) is considered the most prevalent posttranscriptional modification of mRNA. This study aimed to elucidate the effect of PRDX3 on liver fibrosis and the potential mechanism through which the m6A modification regulates PRDX3. PRDX3 expression was found to be negatively correlated with liver fibrosis in both animal models and clinical specimens from patients. We performed adeno-associated virus 9 (AAV9)-PRDX3 knockdown and AAV9-PRDX3 HSC-specific overexpression in mice to clarify the role of PRDX3 in liver fibrosis. PRDX3 silencing exacerbated hepatic fibrogenesis and hepatic stellate cell (HSC) activation, whereas HSC-specific PRDX3 overexpression attenuated liver fibrosis. Mechanistically, PRDX3 suppressed HSC activation at least partially via the mitochondrial reactive oxygen species (ROS)/TGF-β1/Smad2/3 pathway. Furthermore, PRDX3 mRNA was modified by m6A and interacted with the m6A readers YTH domain family proteins 1–3 (YTHDF1-3), as evidenced by RNA pull-down/mass spectrometry. More importantly, PRDX3 expression was suppressed when YTHDF3, but not YTHDF1/2, was knocked down. Moreover, PRDX3 translation was directly regulated by YTHDF3 in an m6A-dependent manner and thereby affected its function in liver fibrosis. Collectively, the results indicate that PRDX3 is a crucial regulator of liver fibrosis and that targeting the YTHDF3/PRDX3 axis in HSCs may be a promising therapeutic approach for liver fibrosis.
Graphical abstract
1. Introduction
Liver fibrosis is a common pathophysiological process resulting from chronic liver injury and is characterized by excessive deposition of extracellular matrix (ECM) [1]. Hepatic stellate cells (HSCs) play an indispensable role in liver fibrosis because activated HSCs continuously deposit collagen and induce ECM accumulation in fibrotic livers [2]. Accumulated reactive oxidative species (ROS), such as superoxide radical anion (O2·−) and hydrogen peroxide (H2O2), mediate HSC activation and transdifferentiation into myofibroblasts, which subsequently induces ECM production [3,4]. Thus, an exploration of the precise antioxidant molecules that specifically target HSCs may provide a beneficial strategy for the inhibition of liver fibrosis.
Peroxiredoxins (PRDXs) are the most abundant superfamily of peroxidases that catalyze the reduction of H2O2 and play a major role in suppressing oxidative stress [5]. Among the six isoforms (PRDX1–PRDX6), PRDX3 is unique because it is abundantly and exclusively localized in mitochondria, which are the main source of intracellular ROS production [6]. Recent studies have shown that PRDX3 protects cells from excess mitochondrial ROS accumulation by eliminating approximately 90% of mitochondrial H2O27. For instance, the overexpression of PRDX3 reportedly prevents mitochondrial oxidative stress-induced damage in a variety of diseases, such as chronic kidney injury and diabetes [8,9]. Moreover, PRDX3 exerts hepatoprotective effects on nonalcoholic fatty liver disease, alcoholic liver injury and acetaminophen-induced liver injury [[10], [11], [12]]. However, whether PRDX3 is a promising therapeutic target in liver fibrosis and the regulatory mechanism through which PRDX3 is modulated remain unclear.
N6-methyladenosine (m6A) is considered the most prevalent chemical modification of mRNA and is mainly present within the consensus sequence of RRACH (where R = G or A and H = A, U or C) [13]. The m6A modification is reversible, dynamically installed by m6A methyltransferases (“writers”) and removed by demethylases (“erasers”) [14]. Additionally, m6A exerts multiple biological effects through “reader” proteins that directly bind and recognize m6A in mRNAs and affect various aspects of these m6A-modified mRNAs, including their stability, translation, and export [15]. Notably, recent studies indicate that m6A readers play a critical role in the development of liver diseases. YTH domain family proteins, including YTHDF1-3, accelerate the progression of hepatocellular carcinoma (HCC) by regulating the m6A methylation of target mRNAs [[16], [17], [18]]. In addition, YTHDC2 recognizes and binds to lipogenic genes, which subsequently prevents hepatic steatosis [19]. Although the differential m6A modification of related genes occurs in liver fibrosis and the m6A-modified genes are enriched mainly in biological processes such as oxidative stress [20], the regulatory mechanism through which m6A readers affect PRDX3 and its effects on liver fibrosis are far from being understood.
Our present study verified that PRDX3 expression was negatively correlated with liver fibrosis in both animal models and patient specimens. Mechanistically, PRDX3 knockdown induced mitochondrial ROS accumulation and thereby triggered HSC activation and liver fibrosis. Notably, therapies targeting PRDX3 in HSCs alleviated liver fibrosis in mice. Moreover, YTHDF3 recognized and bound to m6A in PRDX3 mRNA, which resulted in the regulation of PRDX3 expression and effects on liver fibrosis. This study provides the first demonstration that the regulation of PRDX3 depends on YTHDF3-mediated m6A modification and indicates that targeting the YTHDF3/PRDX3 axis in HSCs might represent a novel therapeutic approach for liver fibrosis.
2. Results
-
1.
PRDX3 expression is decreased in fibrotic liver tissues from humans and mice.
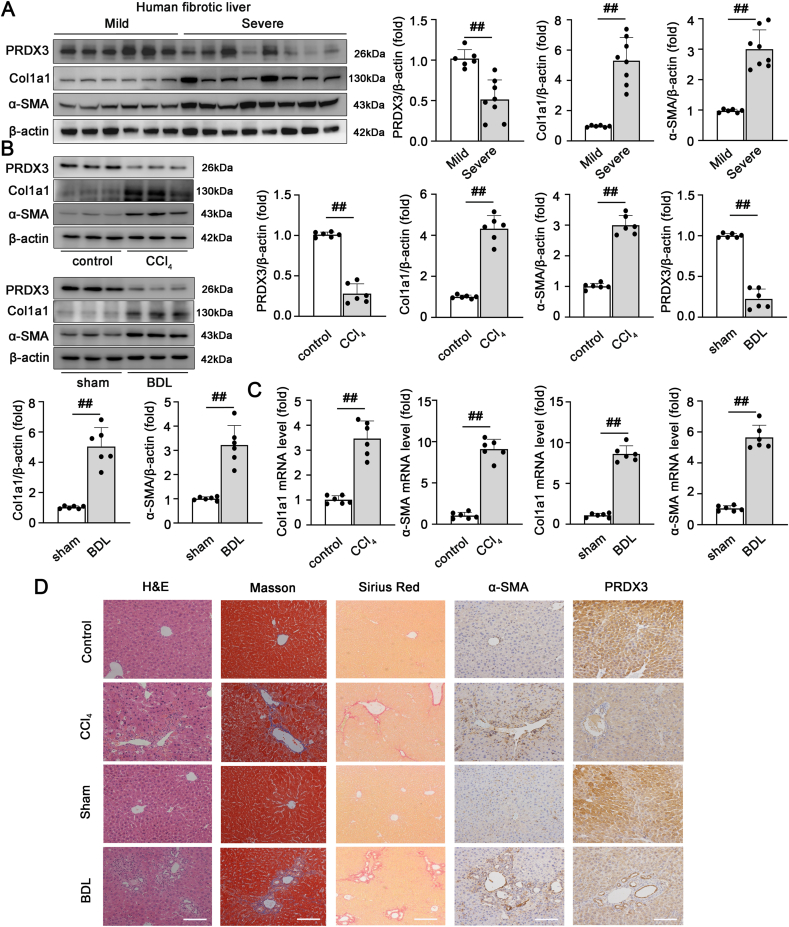
We initially measured the expression of PRDX3 in liver tissues from patients with chronic fibrosis to explore the potential role of PRDX3 in clinical liver fibrosis. The PRDX3 protein levels were markedly lower in patients with severe fibrosis than in patients with mild fibrosis. Furthermore, the accumulation of collagen and α-SMA was detected in fibrotic livers (Fig. 1A). These results indicate that PRDX3 downregulation is involved in human liver fibrosis.
Fig. 1.
PRDX3 expression is decreased in fibrotic liver tissues from humans and mice. (A) The PRDX3, Col1a1, and α-SMA protein levels in liver tissues from patients with mild fibrosis (n = 6) or severe fibrosis (n = 8) were analyzed. (B–D) Mice were injected with CCl4 or subjected to BDL. (B) Liver PRDX3, Col1a1, α-SMA proteins, n = 6. (C) Col1a1 and α-SMA mRNA levels, n = 6. (D) H&E, Masson, Sirius Red staining, n = 6, and IHC staining for PRDX3 and α-SMA, n = 3. Scale bar: 200 μm ##P < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We then used fibrotic animal models to further determine the correlation between PRDX3 expression and liver fibrosis. Mice treated with CCl4 exhibited substantially lower PRDX3 protein and mRNA levels, and these decreases were accompanied by increased expression of the fibrosis markers Col1a1 and α-SMA. Similar changes were observed in mice with bile duct ligation (BDL) (Fig. 1B and C; Figs. S1A and B). Additionally, immunohistochemical (IHC) staining revealed that the decreased PRDX3 expression induced by CCl4 in mice was inversely correlated with α-SMA expression in the fibrotic liver (Fig. 1D). These data showed that PRDX3 expression is negatively correlated with the severity of liver fibrosis.
-
2.
PRDX3 downregulation exacerbates liver fibrosis.
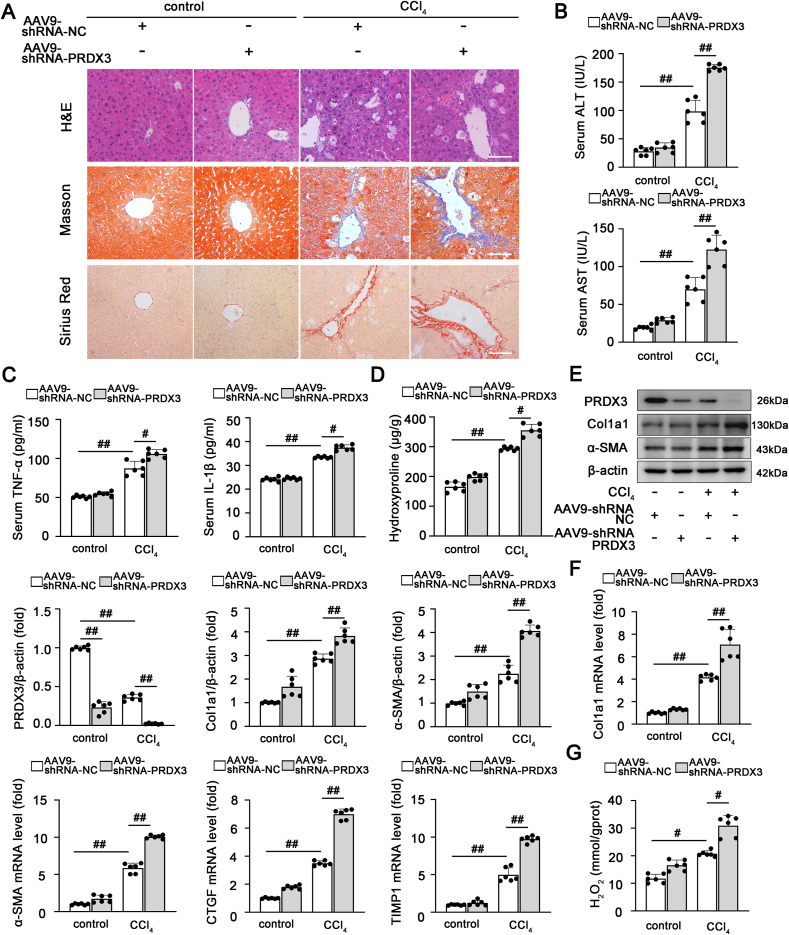
To obtain further insights into the function of PRDX3 in liver fibrosis, the adeno-associated virus 9 (AAV9)-shRNA-PRDX3 vector (PRDX3 shRNA) was injected into mice via the tail vein. As shown in Fig. 2A–C, PRDX3 knockdown aggravated liver injury, as evidenced by H&E staining, and increased the levels of AST, ALT, TNF-α and IL-1β. Consistent with these findings, significant induction of fibrosis was detected in PRDX3 shRNA-injected mice, as evidenced by stronger Sirius Red and Masson staining, a higher hydroxyproline content, and increased expression levels of fibrogenic and HSC activation-related genes (Col1a1, α-SMA, CTGF and TIMP1) (Fig. 2A, D-F). Furthermore, PRDX3 knockdown triggered a substantial increase in the H2O2 content, which indicated that PRDX3 silencing induced intracellular ROS accumulation during liver fibrosis (Fig. 2G). Based on these results. PRDX3 knockdown aggravates HSC activation and liver fibrosis in vivo.
-
3.
The HSC-specific overexpression of PRDX3 attenuates liver fibrosis.
Fig. 2.
PRDX3 downregulation exacerbates liver fibrosis. PRDX3 silencing was induced by the injection of AAV9 into mice exposed to CCl4. (A) H&E, Masson, Sirius Red staining, n = 6. Scale bar: 200 μm. (B) ALT and AST activities, n = 6. (C) TNF-α and IL-1β levels, n = 6. (D) Hydroxyproline content, n = 6. (E) Liver PRDX3, Col1a1, and α-SMA proteins, n = 6. (F) Col1a1, α-SMA, TIMP1, CTGF mRNA levels, n = 6. (G) H2O2 content, n = 6. #P < 0.05, ##P < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
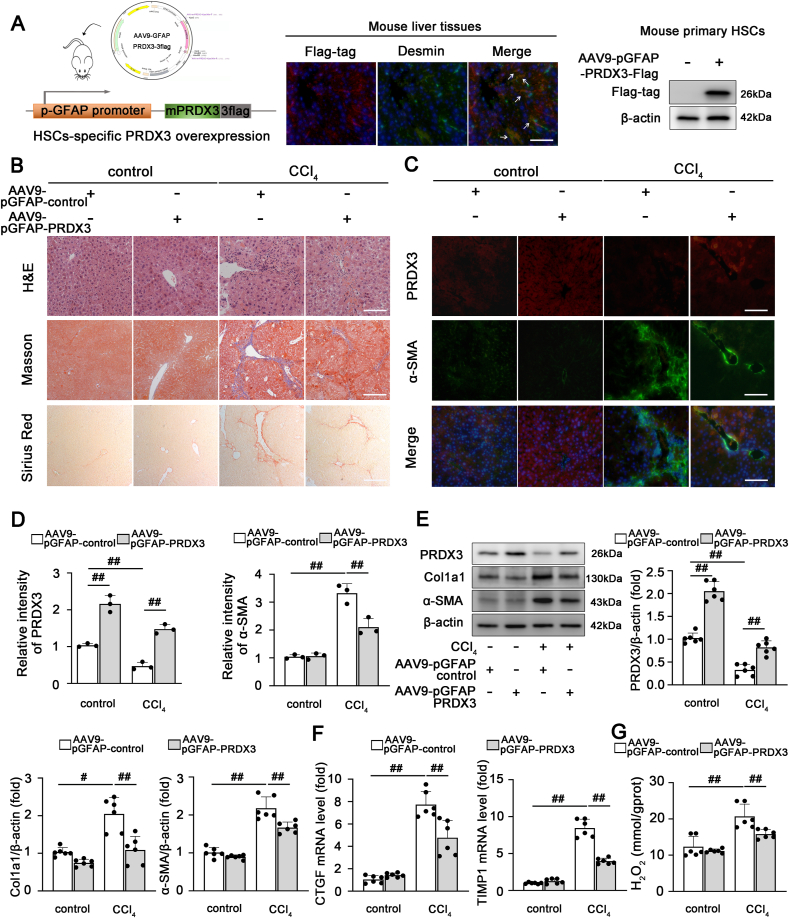
Because HSC activation acts as a crucial contributor to liver fibrosis, we hypothesized that therapies targeting PRDX3 in HSCs may protect against liver fibrosis. Similar to previous studies [21,22], the liver-specific gene delivery of AAV9 containing the promoter of glial fibrillary acidic protein (pGFAP) was employed for targeted PRDX3 overexpression in HSCs in mice to validate this hypothesis. As shown in Fig. 3A and Fig. S1C, the successful delivery of exogenous flag-tagged PRDX3 into HSCs was confirmed by costaining of Desmin-positive cells and western blotting analysis of isolated primary HSCs. As expected, PRDX3 was specifically overexpressed in HSCs (Fig. 3A; Fig. S1D), which significantly alleviated liver injury and fibrosis, as indicated by improvements in liver H&E, Masson, Sirius Red, and α-SMA-PRDX3 staining and the levels of fibrogenic and HSC activation-related markers (Fig. 3B–F; Figs. S1E–G). Additionally, a decreased H2O2 content was observed in AAV9-pGFAP-PRDX3-treated mice, which suggested that PRDX3 overexpression reduced intracellular ROS accumulation in liver fibrosis (Fig. 3G). Thus, HSC-specific PRDX3 overexpression substantially alleviates liver fibrosis in mice.
-
4.
PRDX3 inhibits HSC activation in vitro.
Fig. 3.
The HSC-specific overexpression of PRDX3 attenuates liver fibrosis in mice. The HSC-specific overexpression of PRDX3 was induced by the injection of AAV9-pGFAP into mice exposed to CCl4. (A) Schematic diagram showing the AAV9-pGFAP-PRDX3 treatment of mice (left panel). Dual immunofluorescence staining for the Flag-tagged PRDX3 and Desmin was performed, n = 3. Scale bar: 200 μm (middle panel). Immunoblots of Flag-tagged PRDX3 in primary HSCs isolated from mice after AAV injection, n = 6 (right panel). (B) H&E, Masson, Sirius Red staining, n = 6. Scale bar: 200 μm. (C, D) Dual immunofluorescence for PRDX3 and α-SMA. Representative images and a quantitative analysis are shown, n = 3. Scale bar: 200 μm. (E) Liver PRDX3, Col1a1, α-SMA proteins, n = 6. (F) CTGF, TIMP1 mRNA levels, n = 6. (G) H2O2 content, n = 6. #P < 0.05, ##P < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Because TGF-β1-induced HSC activation plays an indispensable role in liver fibrogenesis [23], we subsequently explored the potential function of PRDX3 in human LX-2 cells and mouse primary HSCs following TGF-β1 treatment. PRDX3 overexpression markedly reduced hepatic fibrogenesis and HSC activation (Figs. S2A–D). Moreover, immunohistofluorescence (IHF) staining revealed that PRDX3 colocalized with α-SMA-positive cells and that PRDX3 overexpression abolished the decrease in PRDX3 expression and increase in α-SMA expression induced by TGF-β1 (Fig. S2E). In contrast, PRDX3 knockdown amplified the TGF-β1-induced expression of genes related to HSC activation and fibrosis in both LX-2 cells and primary HSCs (Fig. S3). These results provide evidence showing that PRDX3 inhibits HSC activation and liver fibrosis in vitro.
-
5.
PRDX3 suppresses HSC activation by regulating mitochondrial ROS/TGF-β1/Smad2/3 pathway.
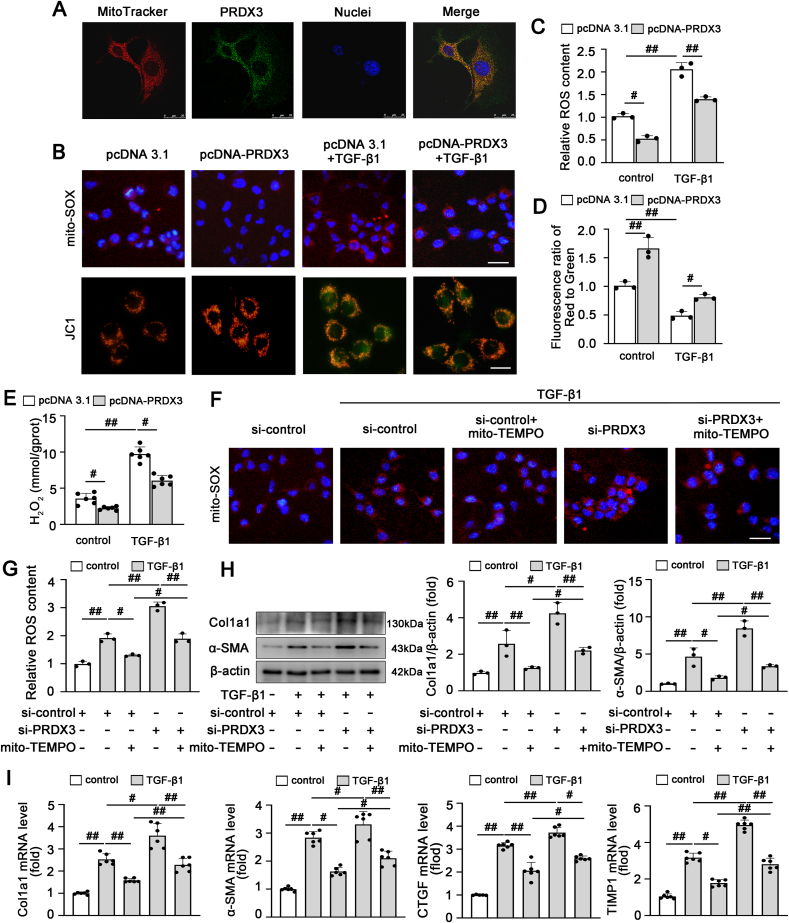
We further examined whether PRDX3-mediated mitochondrial ROS are involved in HSC activation. As shown in Fig. 4A, PRDX3 was localized mainly in mitochondria in LX-2 cells. The mitochondrial ROS level, mitochondrial membrane potential and H2O2 content were increased after treatment with TGF-β1 and significantly decreased by PRDX3 overexpression (Fig. 4B–E). In addition, PRDX3 overexpression increased SOD activity and SOD2 expression (Fig. S4). In contrast, PRDX3 silencing resulted in robust ROS generation, which was blunted by treatment with mito-TEMPO, a specific mitochondria-targeted ROS scavenger (Fig. 4F and G). Moreover, the exacerbation of hepatic fibrogenesis and HSC activation induced by PRDX3 knockdown were markedly attenuated by mito-TEMPO (Fig. 4H and I). Collectively, these results indicated that the underlying mechanism through which PRDX3 reduces HSC activation in liver fibrosis may be mediated mainly by the inhibition of mitochondrial ROS production.
Fig. 4.
PRDX3 suppresses HSC activation by regulating mitochondrial ROS production. (A) Colocalization of PRDX3 and mitochondria (MitoTracker) in LX-2 cells, n = 3. Scale bar: 25 μm. (B–E) LX-2 cells were transfected with pcDNA-PRDX3 and then treated with TGF-β1. (B–D) Images of JC-1- and MitoSOX-stained cells, n = 3. Scale bar: 200 μm. (E) H2O2 content, n = 6. (F–I) LX-2 cells were transfected with PRDX3 siRNA and incubated with mito-TEMPO under TGF-β1 treatment. (F, G) Images of MitoSOX-stained cells, n = 3. (H) Col1a1 and α-SMA proteins, n = 3. (I) Col1a1, α-SMA, CTGF, TIMP1 mRNA levels, n = 6. #P < 0.05, ##P < 0.01.
Accumulating evidence indicates the involvement of mitochondrial ROS in liver fibrogenesis through a mechanism mediated by the TGF-β1/Smad2/3 pathway [23]. We then explored whether the PRDX3-mediated regulation of mitochondrial ROS production is associated with the TGF-β1/Smad2/3 pathway in HSCs. As shown in Figs. S5A–B, the HSC-specific overexpression of PRDX3 substantially decreased the levels of p-Smad2 and p-Smad3 both in vivo and in vitro. Moreover, PRDX3 knockdown increased the levels of HSC activation-related and fibrogenic genes, and these increases were considerably reduced by treatment with the TGF-β1 inhibitor SB431542 (Figs. S5C and D). Thus, these data implied that the mitochondrial ROS scavenger PRDX3 attenuates HSC activation and liver fibrosis by inhibiting the TGF-β1/Smad2/3 pathway.
-
6.
The m6A modification regulates PRDX3 expression and affects its function in liver fibrosis.
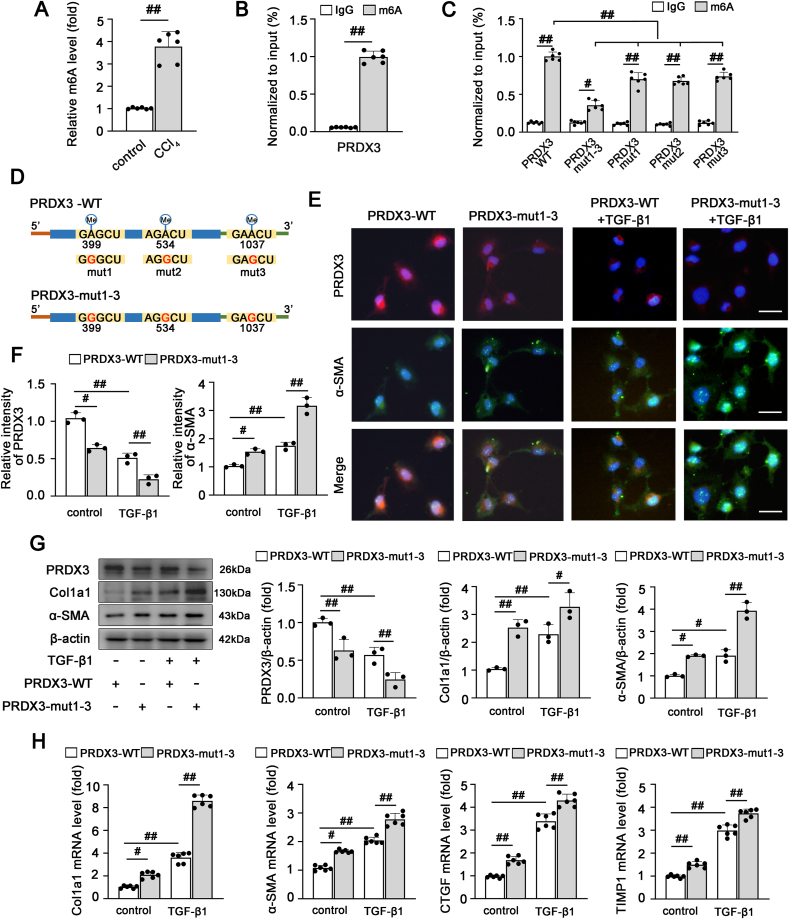
Because PRDX3 is a crucial regulator of liver fibrosis, we further explored the potential mechanisms upstream of PRDX3. Recent findings have revealed that m6A is the most prevalent internal modification of mRNA and affects mRNA export, stability and translation [13]. Sequence-based RNA adenosine methylation site predictor (SRAMP) (http://www.cuilab.cn/sramp) [24] predicted that several potential m6A sites are distributed in PRDX3 mRNA; thus, we speculated that the regulation of PRDX3 in liver fibrosis depends on m6A modification. First, we examined the m6A levels in liver fibrosis. As shown in Fig. 5A, an elevated m6A level in the total RNA population was observed in liver fibrosis. Subsequently, MeRIP-qRT–PCR with an m6A-specific antibody demonstrated that PRDX3 mRNA was significantly enriched in LX-2 cells (Fig. 5B), which suggested that the effect of PRDX3 on liver fibrosis may be related to its m6A modification.
Fig. 5.
The m6A modification regulates PRDX3 expression and affects its function in liver fibrosis. (A) The total m6A levels in control mice and mice with CCl4-induced liver fibrosis were determined using an m6A methylation quantification kit, n = 6. (B) The m6A enrichment of PRDX3 mRNA in LX-2 cells was validated by MeRIP-qRT–PCR, n = 6. (C) LX-2 cells were transfected with the indicated gene expression plasmids, and the m6A level in PRDX3 was then determined, n = 6. (D) Schematic representation of putative m6A modification sites in the PRDX3 mRNA sequence. (E–H) LX-2 cells were transfected with pcDNA-PRDX3-WT or pcDNA-PRDX3-mut1-3 and then exposed to TGF-β1. (E, F) Dual immunofluorescence for PRDX3 and α-SMA. Representative images and a quantitative analysis are shown, n = 3. Scale bar: 200 μm. (G) PRDX3, Col1a1, α-SMA proteins, n = 3. (H) Col1a1, α-SMA, TIMP1, CTGF mRNA levels, n = 6. #P < 0.05, ##P < 0.01.
Moreover, to further investigate the function of m6A motifs in the modulation of PRDX3, three m6A sites in PRDX3 predicted with high confidence by SRAMP [24] were replaced by adenine-guanine (A-G) mutations as follows: PRDX3-mut1 (A399G), PRDX3-mut2 (A534G), PRDX3-mut3 (A1037G), and PRDX3-mut1-3 (A399G, A534G, A1037G). These mutants were transfected into LX-2 cells. The m6A levels in PRDX3 mutants were clearly lower than those in wild-type PRDX3 (PRDX3-WT) (Fig. 5C and D). More importantly, the degree of decrease in the m6A level of PRDX3 mut1-3, which contains three m6A site mutations, was greater than that of the mRNA with only one mutation of a potential m6A site, suggesting that the cooperation of three m6A sites was the key mechanism of PRDX3 m6A modification (Fig. 5C). Furthermore, the PRDX3 protein levels were decreased considerably upon mutation of the m6A motifs (Fig. 5E–G), which indicated that the m6A modification of PRDX3 may regulate PRDX3 protein expression in liver fibrosis.
We then explored whether the m6A modification of PRDX3 further modulated its protective effect on liver fibrosis. As expected, PRDX3 m6A site mutations markedly induced HSC activation and hepatic fibrogenesis compared with the results obtained with PRDX3-WT (Fig. 5E–H). Based on these data, m6A modification may be involved in the posttranscriptional modulation of PRDX3 and alter its expression and function in liver fibrosis.
-
7.
PRDX3 mRNA translation is regulated by YTHDF3 in an m6A-dependent manner
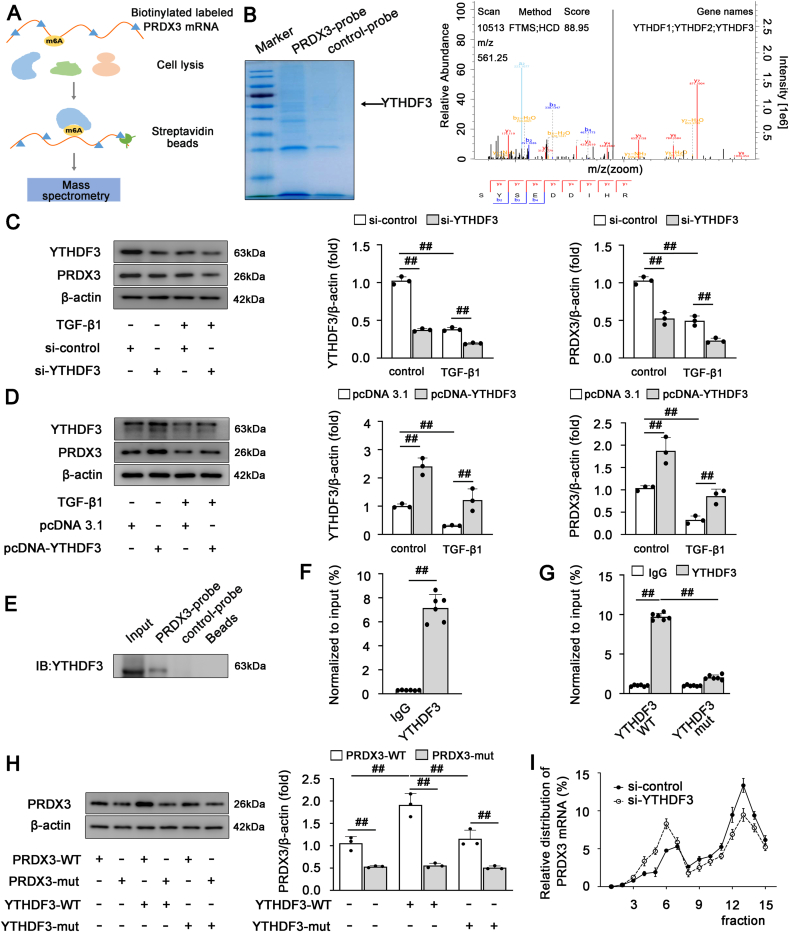
Previous studies have shown that m6A modification mediates multiple layers of posttranscriptional gene control through a mechanism that mainly depends on the specific m6A reader [15]. We then sought to identify the key m6A reader that regulates PRDX3 expression via m6A modification in liver fibrosis. RNA pull-down combined with mass spectrometry was performed to identify PRDX3 mRNA-interacting proteins in LX-2 cells (Fig. 6A). In the mass spectrometry results, among the m6A reader proteins, the peptides of YTH domain family proteins and Insulin-Like Growth Factor 2 mRNA-Binding Proteins (IGF2BP), including YTHDF1-3 and IGF2BP2-3, were found to specifically bind to PRDX3 mRNA with signal intensity (Fig. 6B, Supplementary Material 2). The YTHDF1-3 expression were clearly downregulated in the progression of liver fibrosis, while the expression of IGF2BP2-3 were not significantly different, indicating that YTHDF1-3 may be the potential m6A readers affecting PRDX3 mRNA during liver fibrosis (Figs. S6A–C). Moreover, the knockdown of YTHDF3 but not YTHDF1/2 suppressed PRDX3 protein expression (Fig. 6C; Figs. S6D and E). In contrast, YTHDF3 overexpression significantly increased the PRDX3 protein expression (Fig. 6D). The specific interaction between YTHDF3 and PRDX3 was further determined by performing YTHDF3 pull down with a biotinylated PRDX3 mRNA (Fig. 6E). Moreover, RIP assays showed that PRDX3 mRNA was enriched in YTHDF3-containing immunoprecipitates compared with IgG immunoprecipitates (Fig. 6F). These data revealed that YTHDF3 specifically binds to PRDX3 and regulates its expression in LX-2 cells.
Fig. 6.
PRDX3 mRNA translation is regulated by YTHDF3 in an m6A-dependent manner. (A) Schematic representation of the procedure for the RNA pull-down and mass spectrometry analysis aiming to screen PRDX3 mRNA-interacting proteins. (B) Coomassie Brilliant Blue staining (left panel). Peptide spectrum of YTHDFs generated by LC–MS/MS (right panel). (C, D) LX-2 cells were transfected with YTHDF3 siRNA or pcDNA-YTHDF3 and then exposed to TGF-β1. YTHDF3 and PRDX3 proteins, n = 3. (E) YTHDF3 was pulled down and enriched with biotinylated PRDX3 mRNA in LX-2 cells. (F, G) The interaction between YTHDF3 and PRDX3 mRNA in LX-2 cells was detected by YTHDF3 RIP followed by qRT–PCR, n = 6. (H) LX-2 cells were cotransfected with pcDNA3.1, pcDNA-YTHDF3-WT or pcDNA-YTHDF3-mut and pcDNA-PRDX3-WT or pcDNA-PRDX3-mut1-3. PRDX3 protein, n = 3. (I) LX-2 cells were transfected with YTHDF3 siRNA, and a qRT–PCR analysis of PRDX3 transcripts in different polysome fractions was performed, n = 6. ##P < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Because YTHDF3 binds to m6A sites via its m6A-binding pockets (W438 and W492) in the YTH domain, the W438 and W492 mutations abolish the mRNA-binding capacity of YTHDF3 [25] (Fig. S6F). Thus, LX-2 cells were transfected with plasmids expressing YTHDF3-WT or YTHDF3-mut with the W438A and W492A mutations. The RIP assay showed that PRDX3 mRNA was immunoprecipitated effectively in cells transfected with YTHDF3-WT, whereas the interaction between YTHDF3-mut (m6A binding-defective YTHDF3) and PRDX3 was significantly reduced (Fig. 6G). Furthermore, YTHDF3-WT but not YTHDF3-mut increased the expression of PRDX3. Additionally, PRDX3 with mutation of its m6A sites (PRDX3-mut) did not exhibit a significantly increased protein level in the presence of YTHDF3-WT overexpression (Fig. 6H), which indicated that the YTHDF3-mediated regulation of PRDX3 expression was dependent on m6A.
Because YTHDF3 increases the translational efficiency of m6A-modified mRNAs [26], we subsequently sought to determine whether YTHDF3 regulates the translation of PRDX3. Indeed, YTHDF3 knockdown inhibited PRDX3 translation, and this finding was verified by the moderate shift in PRDX3 mRNA from the polysome to the subpolysome fraction without a change in its mRNA level or mRNA stability (Fig. 6I; Figs. S6G and H). Collectively, these results revealed that PRDX3 mRNA translation is likely regulated by YTHDF3 in an m6A-dependent manner in liver fibrosis.
-
8.
PRDX3 knockdown suppresses the protective effect of HSC-specific YTHDF3 overexpression on liver fibrosis in mice.
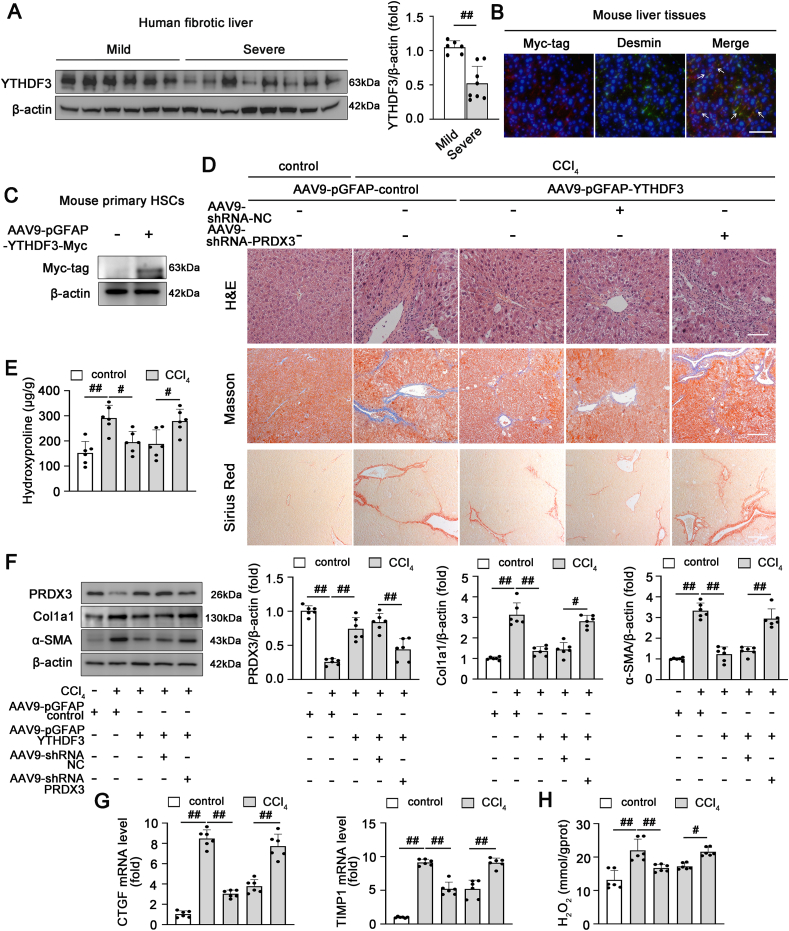
According to recent studies, YTHDF3 expression is significantly changed in hepatic steatosis and HCC, which indicates that YTHDF3 is likely to be closely associated with liver diseases [27,28]. We observed a marked decrease in the YTHDF3 protein level in fibrotic livers from both humans and mice, which suggested that YTHDF3 expression was negatively correlated with the progression of liver fibrosis (Fig. 7A; Figs. S6A and B). Furthermore, based on the aforementioned findings regarding the regulatory effect of YTHDF3 on PRDX3, we further explored whether YTHDF3 functions in liver fibrosis by modulating PRDX3 expression. To this end, AAV9-pGFAP-YTHDF3 was used to achieve the HSC-specific overexpression of YTHDF3 accompanied by PRDX3 knockdown in mice. The specificity of YTHDF3 overexpression in HSCs was validated (Fig. 7B and C). HSC-specific YTHDF3 overexpression markedly attenuated liver injury and fibrosis, as evidenced by an improved liver histopathology and reductions in hepatic profibrogenic and HSC activation (Fig. 7D–G). Notably, YTHDF3 overexpression in HSCs significantly increased the PRDX3 protein level and reduced cellular oxidative damage, as revealed by a substantial decrease in the H2O2 content (Fig. 7F, H). However, the protective effects of AAV9-pGFAP-YTHDF3 on liver fibrosis were abrogated by PRDX3 knockdown (Fig. 7D–H). Consistent with these findings, primary HSCs and LX-2 cells transfected with pcDNA-YTHDF3 along with PRDX3 siRNA displayed the same trends observed in vivo (Fig. S7). Collectively, these data showed that HSC-specific YTHDF3 overexpression markedly attenuates CCl4-induced liver fibrosis mainly by upregulating PRDX3 expression.
Fig. 7.
PRDX3 knockdown suppresses the protective effect of HSC-specific YTHDF3 overexpression on liver fibrosis in mice. (A) The YTHDF3 protein level in liver tissues from patients with mild fibrosis (n = 6) or severe fibrosis (n = 8) was analyzed. (B–H) The HSC-specific overexpression of YTHDF3 and PRDX3 shRNA was induced via AAV9-mediated delivery into mice, and the mice were then treated with CCl4. (B) Dual immunofluorescence staining for the Myc-tagged YTHDF3 and Desmin was performed, n = 3. Scale bar: 200 μm. (C) Immunoblots of Myc-tagged YTHDF3 in primary HSCs isolated from mice after AAV injection, n = 6. (D) H&E, Masson, Sirius Red staining, n = 6. (E) Hydroxyproline content, n = 6. (F) PRDX3, Col1a1, α-SMA proteins, n = 6. (G) TIMP1 and CTGF mRNA levels, n = 6. (H) H2O2 content, n = 6. #P < 0.05, ##P < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3. Discussion
Liver fibrosis is a condition accompanied by aberrant deposition of ECM in liver tissue, and activated HSCs have been recognized as the predominant producer of ECM components [1]. Excess oxidative stress is considered the key event in HSC activation and liver fibrogenesis [4]. The present study identified the potent ROS scavenger PRDX3 as a novel regulator that inhibits activated HSCs. Furthermore, we identified YTHDF3 as an m6A reader that modulates PRDX3 expression in an m6A-dependent manner during liver fibrosis. Thus, targeting the YTHDF3-PRDX3 axis may be a promising therapeutic approach for liver fibrosis.
Mitochondrial ROS mainly include O2·− and H2O2, which are the dominant sources of intracellular oxidative stress. Most O2·− is dismutated by MnSOD to form H2O2 in mitochondria [8]. Subsequently, mitochondrial H2O2 diffuses into the cytoplasm to enhance oxidative damage, which is presumed to exert critical effects on the development of liver diseases, including hepatic steatosis, cholestatic liver injury and liver fibrosis [4,29,30]. Based on accumulating evidence, H2O2 induces ECM synthesis by directly promoting collagen gene transcription in activated HSCs [31,32], which suggests that interventions designed to reduce mitochondrial H2O2 production might be effective strategies for preventing liver fibrosis.
PRDXs are abundantly expressed in cells and have a structure that renders the cysteine (Cys) residue in the active site highly sensitive to oxidation by H2O27. Among PRDXs, PRDX3 and PRDX5 are responsible for the decomposition of mitochondrial H2O2. However, studies have suggested that the concentration of PRDX5 in mitochondria is markedly lower than that of PRDX3 [7]. Based on both its abundance and rapid reactivity with H2O2, PRDX3 removes approximately 90% of mitochondrial H2O2, which suggests that it is a potent antioxidant enzyme that protects against mitochondrial oxidative damage [7,33]. Furthermore, PRDX3 is considered a crucial regulator of mitochondrial ROS production in liver diseases [[10], [11], [12]]. Nevertheless, the mechanisms through which PRDX3 regulates mitochondrial oxidative stress during liver fibrosis remain to be fully clarified. In the current study, our data revealed that PRDX3 expression was negatively correlated with the severity of liver fibrosis. More importantly, PRDX3 silencing resulted in robust intracellular ROS generation and ECM accumulation, and both of these effects were markedly attenuated by treatment with the mitochondria-targeted ROS scavenger mito-TEMPO, which suggested that PRDX3 suppresses liver fibrosis by regulating mitochondrial ROS production. Moreover, because mitochondrial ROS activate the TGF-β1/Smad2/3 pathway and are considered a crucial trigger of the profibrogenic effects in the liver, we further identified a new function of PRDX3 in inhibiting HSC activation that is at least partially dependent on the mitochondrial ROS/TGF-β1/Smad2/3 pathway. Although our data indicated that PRDX3 knockdown may also affect SOD activity, we concluded that the effect of PRDX3 on inhibiting mitochondrial ROS production during liver fibrosis mainly depended on regulation of the mitochondrial H2O2 levels due to the direct role of PRDX3 in neutralizing mitochondrial H2O2. In addition, previous studies have revealed that PRDX3 expressed in hepatocytes or Kupffer cells exerts a protective effect on liver injury [10,12]. Because hepatocytes and Kupffer cells also play key roles in the development of liver injury and fibrogenesis [34], PRDX3 expressed in other cell types may also participate in the progression of liver fibrosis. Although PRDX3 exerts hepatoprotective effects via its antioxidant properties, it also mediates hepatic tumorigenesis by promoting HCC growth and invasion [35]. PRDX3 exerts these different regulatory effects on liver diseases, possibly because H2O2 plays a dual role in liver pathological processes, and PRDXs may also act as redox relays during redox signaling under certain conditions [[36], [37], [38]]. Thus, the precise mechanism of PRDX3 in the progression of liver fibrosis to HCC requires further exploration.
m6A is the most prevalent posttranscriptional modification of mRNA, occurring at approximately three to five sites per mRNA in mammals [39]. Recent studies have verified that the m6A modification of mRNA plays crucial roles in various biological events [13]. Specifically, the m6A modification is crucial for regulating the progression of liver diseases, such as liver cancer, nonalcoholic fatty liver disease, and liver fibrosis [20,40]. Notably, an analysis of the m6A methylation profile of fibrotic liver tissues has revealed that the differentially m6A-methylated genes are involved in the biological functions of oxidative stress and cytochrome metabolism [20]. Thus, we reasoned that the reversible m6A modification of specific mRNAs influences the progression of liver fibrosis. Interestingly, three high-confidence m6A sites have been predicted to be present in PRDX3 mRNA [24]. Furthermore, our results suggested that PRDX3 mRNA was modified by m6A and revealed that both the m6A and expression levels of PRDX3 were reduced upon mutation of the m6A sites, and these reductions further aggravated hepatic fibrogenesis and HSC activation. Thus, we concluded that m6A modification may be involved in regulating PRDX3 expression and thus affects its function in liver fibrosis.
The critical m6A readers YTHDF1-3 have been reported to specifically bind and recognize m6A-methylated sites on mRNA, which may affect the fate and function of the mRNA [15]. Indeed, in this study, RNA pull-down/mass spectrometry results showed that PRDX3 mRNA interacted with the YTHDF1-3 with signal intensity and their protein expression were clearly downregulated in the progression of liver fibrosis, indicating that YTHDF1-3 may be the potential mediators in regulating the m6A modification of PRDX3 mRNA during liver fibrosis. More importantly, PRDX3 expression was suppressed after the knockdown of YTHDF3 but not YTHDF1 or YTHDF2. The increase in PRDX3 expression induced by YTHDF3 overexpression was further abrogated in the presence of m6A binding-defective YTHDF3. Polysome profiling showed that YTHDF3 directly regulated the translation of PRDX3. Thus, we concluded that YTHDF3 induces PRDX3 translation in an m6A-dependent manner. Additionally, the HSC-specific overexpression of YTHDF3 attenuated liver fibrosis, and this attenuation was abrogated by PRDX3 knockdown, which indicated that the protective role of YTHDF3 in liver fibrosis was mainly mediated by the upregulation of PRDX3 expression. However, limitation exists in our research. For one thing, the HSC-specific knockdown of PRDX3 was not applied in the study with technical limitation. Further experiments are needed to explain the role of YTHDF3/PRDX3 in liver fibrosis. For another, our RNA pull-down/mass spectrometry results were based on the previously discovered m6A reader proteins. With the cognitive improvement of m6A regulators and the post-transcriptional modifications, other potential targets affecting PRDX3 mRNA may be found and related regulatory mechanism needs to be further studied.
Therapeutic strategies that modulate ROS levels through the antioxidant defense system have been shown to be effective for various diseases [41]. Indeed, our data showed that overexpression of the potent antioxidant PRDX3 significantly attenuated liver fibrosis by inhibiting mitochondrial ROS production. However, studies have also indicated that off-target ROS inhibition might contribute to genetic instability and mutations in cells, and the selected elimination of ROS in specific cell types may provide a more balanced approach [34,42]. Based on this information, we found that antioxidant therapy targeting mitochondrial PRDX3 in HSCs potentially represents an attractive therapeutic strategy for the treatment of liver fibrosis. Furthermore, numerous studies have suggested that the m6A modification of target genes is gradually showing increasingly more significant clinical potential [43]. As shown in the current study, the YTHDF3-mediated m6A modification of PRDX3 affects the progression of liver fibrosis. Thus, regulation of the YTHDF3/PRDX3 axis might be a potentially feasible therapeutic approach for liver fibrosis. However, YTHDF3 functions in liver fibrosis might be a global effect related with other targets, such as the fibrosis-related regulators FOXO3 and YAP [26,44], but this potential requires further exploration.
In summary, the present study verified that the HSC-specific overexpression of PRDX3 inhibits HSC activation and ameliorates liver fibrosis by suppressing mitochondrial ROS/TGF-β1/Smad2/3 signaling. We further revealed that the m6A reader YTHDF3 specifically regulates PRDX3 translation and expression and affects its function during liver fibrosis. Taken together, our results provide strong evidence showing that targeting the YTHDF3/PRDX3 axis in HSCs may be a promising therapeutic strategy for liver fibrosis.
4. Materials and methods
4.1. Human liver samples
Human fibrotic liver tissue samples were collected from tumor- and cyst-free liver tissues of patients who underwent liver resection at Shulan Hospital (Hangzhou, China). Written consent was obtained, and the procedures were approved by the human ethics committees of Shulan Hospital (Hangzhou) and were consistent with principles of the Declaration of Helsinki. Additional detailed information of the patients is provided in Supplementary Material 1.
4.2. Animal experiments
C57BL/6 mice (male, 20 ± 2 g, 6–8 weeks) were obtained from the Laboratory Animal Center of Dalian Medical University (Dalian, China). Mice were intraperitoneally injected with CCl4 (2 ml/kg) twice weekly for four weeks. Other mice were injected with olive oil and served as a control group. The bile duct ligation (BDL)-induced liver fibrosis model was established according to a previously described procedure [21]. Briefly, after laparotomy, the common bile duct was double ligated with 3–0 surgical silk and cut between the ligatures. A sham operation without ligation was performed similarly. The mice were sacrificed two weeks after the sham and BDL operations. A detailed description of the number of animals included in each group is provided in Supplementary Material 1. The animal experiments were approved by the ethics committee of Dalian Medical University and complied with the Guide for the Care and Use of Laboratory Animals.
For in vivo PRDX3 knockdown, PRDX3 shRNA was driven by the U6 promoter and packaged into AAV9 [45]. In addition, PRDX3-Flag or YTHDF3-Myc plasmids were driven by the pGFAP promoter and packaged into AAV9 for the overexpression of PRDX3 or YTHDF3 in HSCs. The mice were injected with AAV9-shRNA NC (AAV9-U6-shRNA-negative control), AAV9-shRNA-PRDX3 (AAV9-U6-shRNA-PRDX3), AAV9-pGFAP-control (AAV9-pGFAP empty vector), AAV9-pGFAP-PRDX3 or AAV9-pGFAP-YTHDF3 (1 × 1012 vg/ml in a volume of 100 μl per mouse) via the tail vein before the first CCl4 injection. The AAV9 vectors were designed and produced by Hanbio Biotechnology Co., Ltd. (Shanghai, China).
4.3. Liver histology
Liver tissues were fixed with paraformaldehyde (4%) and embedded in paraffin. The sections were subjected to Sirius Red, Masson and H&E staining. Immunohistochemical staining for PRDX3 and α-SMA was performed with anti-α-SMA and anti-PRDX3 antibodies, respectively, as described previously [46]. Images were observed with a light microscope.
4.4. Biochemical assays
The AST (Cat# C010-2-1), ALT (Cat# C009-2-1), IL-1β (Cat# H002), TNF-α (Cat# H052-1) levels in serum and the hydroxyproline content (Cat# A030) in the liver were measured using appropriate examination kits (Jiancheng Corp., Nanjing, China).
4.5. H2O2 measurement
The H2O2 levels were examined using kits (Solarbio, Beijing, China, Cat# BC3595) following the manufacturer's protocol. H2O2 reacted with molybdic acid to form a complex, which was detected at 405 nm. The extracts of liver tissues or LX-2 cells were pretreated with excess catalase to remove H2O2, and an H2O2 standard liquid was applied as a control to calculate the H2O2 concentration.
4.6. SOD activity
The SOD levels were examined using a SOD assay kit (Jiancheng, Cat# A001-3) following the manufacturer's protocols. Briefly, liver tissue or cells were centrifuged to obtain the supernatant. The activity of SOD in the supernatant was measured by detecting the water-soluble tetrazolium salt (WST) at 450 nm and reported in units/mg protein.
4.7. Cell culture and transfection
LX-2 cell line was obtained from the China Cell Culture Center. Mouse primary HSCs were extracted by digestion with collagenase/proteinase enzymes (Solarbio, Cat# C8160) as described previously [47]. The cells were cultivated in DMEM containing 10% fetal bovine serum (Gibco, USA, Cat# 11965092, Cat# 10099141) at 37 °C with 5% CO2.
siRNAs targeting PRDX3 and YTHDF3, pcDNA-PRDX3 plasmid, pcDNA-PRDX3 mutant plasmids (A399G, A534G, and A1037G), pcDNA-YTHDF3 plasmid, pcDNA-YTHDF3 mutant plasmids (W438A and W492A) or the negative controls (si-control or pcDNA 3.1) were transfected into cells. The PRDX3 mutant plasmids containing mutations at sites 399, 534 and 1037 (A to G) and YTHDF3 mutant plasmids containing mutations at amino acids 438 and 492 (W to A) were cloned into the pcDNA3.1 vector and synthesized by GenePharma (Shanghai, China). The PRDX3 siRNA sequence was as follows: sense, 5′-GCA CUC UUG UCA GAC UUA ATT-3′, and antisense, 5′-UUA AGU CUG ACA AGA GUG CTT-3’. The YTHDF3 siRNA sequence was as follows: sense, 5′-GGA CGU GUG UUU AUA AUU ATT-3′, and antisense, 5′-UAA UUA UAA ACA CAC GUC CTT-3’. The cells were then incubated with TGF-β1 (2 ng/ml, PeproTech, Cat# AF-100-21C) for different experiments.
4.8. Western blotting
Liver tissues, LX-2 cells or mouse primary HSCs were lysed, and the protein concentrations were detected. The protein level was measured by western blotting as previously described [48]. Briefly, equal amounts of protein samples were separated on SDS–PAGE gels, and the different target proteins from the same gel were transblotted onto PVDF membranes and blotted with appropriate primary antibodies and secondary antibodies. The immunoreactive bands were visualized with an enhanced chemiluminescence reagent (Beyotime, Shanghai, China, Cat# P0018S) and analyzed using Gel-Pro Analyzer. Afterward, the previous antibodies were stripped with stripping buffer (Beyotime, Cat# P0025), and the membranes were reprobed with an anti-β-actin antibody as a loading control. The relevant steps were consistent with previous studies [49,50]. The primary antibodies used are shown in Table S2.
4.9. RNA extraction and qRT–PCR
Total RNA was extracted with TRIzol reagent (TaKaRa, Japan, Cat# 9109). The cDNA templates were synthesized, and the RNA abundance was detected using a reverse transcription kit (TaKaRa, Cat# RR047A) and SYBR Green PCR kit (TaKaRa, Cat# RR820A). The target mRNA expression was normalized to β-actin. The expression levels were analyzed by calculating the ΔΔCt values. The primer sequences are shown in Table S3.
4.10. Mitochondrial ROS detection
Mitochondrial ROS production was measured by MitoSOX Red staining (Invitrogen, Carlsbad, CA, USA, Cat# M36008). Briefly, the cells were treated with MitoSOX (5 μM), and nuclei were detected by Hoechst staining (Beyotime, Cat# C1025). Images were acquired with a Nikon 80i microscope.
4.11. Mitochondrial membrane potential
The JC-1 probe was applied to determine the mitochondrial membrane potential following the manufacturer's instructions (Beyotime, Cat# C2006). Images were acquired with a Nikon 80i microscope.
4.12. Dual immunofluorescence staining
Fixed liver tissue sections or LX-2 cells were incubated overnight with the primary antibody and then incubated with the appropriate secondary antibody (1 h). Subsequently, nuclei were detected by DAPI staining (Beyotime, Cat# C1006). Immunofluorescence staining images were acquired with a Nikon 80i microscope.
4.13. m6A measurement
The total m6A levels in CCl4-induced liver fibrosis samples were measured using an EpiQuik m6A RNA Methylation Quantification Kit (EpiGentek, Farmingdale, NY, USA, Cat# P-9005). Briefly, total RNA (200 ng) was added to each well, and capture antibody solution and detection antibody solution were added following the manufacturer's protocol. The m6A levels were quantified colorimetrically by measuring the absorbance at 450 nm.
Magna MeRIP™ m6A Kit (Millipore, Bedford, MA, USA, Cat# 17–10499) was applied to measure the m6A levels of target mRNAs as previously described [51]. Briefly, total RNA from LX-2 cells was extracted and chemically fragmented into approximately 100-nt fragments. The fragmented RNA was incubated with the m6A antibody for immunoprecipitation following the manufacturer's protocol. The enrichment of m6A-containing mRNA was then analyzed by qRT–PCR.
4.14. RIP
RIP assays were detected using an RNA-Binding Protein Immunoprecipitation Kit (Millipore, Cat# 17–700). Briefly, LX-2 cells were harvested and lysed in RIP lysis buffer containing protease and RNase inhibitors. The centrifuged supernatant was then incubated with anti-YTHDF3 antibody-conjugated Protein A/G magnetic beads. After overnight incubation, the beads were washed with IP buffer and treated with proteinase K buffer for digestion. The coimmunoprecipitated RNA was then further measured by qRT–PCR.
4.15. RNA pull-down
The biotin-labeled PRDX3-mRNA complexes were incubated with streptavidin-coated magnetic beads (Invitrogen, Cat# 20164) overnight. The LX-2 cell lysates were added to the binding reaction with RNase inhibitor and protease/phosphatase inhibitor cocktail and then incubated with the beads (1 h). The coprecipitated proteins in the capture complex were measured by western blotting or mass spectrometry performed by Shanghai Bioprofile Technology (Shanghai, China).
4.16. Polysome fractionation
LX-2 cells were incubated with cycloheximide (100 μg/ml, Millipore, Cat# 239763) for 10 min and then lysed on ice with lysis buffer. After centrifugation, the supernatant was gathered, loaded onto a 10/50% (w/v) sucrose gradient and then centrifuged at 39,000 rpm in a Beckman SW-41Ti rotor (3 h, 4 °C). Samples were collected as 15 fractions from the top of the gradient and then analyzed by qRT–PCR.
4.17. Statistical analysis
The data were assessed for a normal distribution using the Shapiro–Wilk test and are presented as the means ± SDs. The results were analyzed by Student's unpaired t-test (two-group comparisons) or one-way ANOVA (multigroup comparisons) followed by Tukey's post-hoc test (GraphPad Software). P < 0.05 was considered to indicate significance.
Authors’ contributions
R.S. and J.Y. designed the study. R.S., X.T., Y.L., Z.W., Y.Z. and L.Z. performed the experiments. Y.H., D.G. and Y.W. analyzed the results. X.T., S.Z. contributed materials and gathered liver samples. J.Y., R.S. and X.T. prepared the manuscript. J.Y. provided financial support.
Ethical approval
The use of clinical samples was approved by the human ethics committees of Shulan Hospital (Hangzhou, China). The animal experiments were approved by the ethics committee of Dalian Medical University and complied with the Guide for the Care and Use of Laboratory Animals.
Financial support
This work was supported by grants from the National Natural Science Foundation of China (Nos. 82004044 and 81973381) and the Innovative Leading Talents of LiaoNing (No. XLYC1902098).
Declaration of competing interest
None of the authors have potential conflicts of interest to declare.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102378.
Contributor Information
Shusen Zheng, Email: shusenzheng@zju.edu.cn.
Jihong Yao, Email: yaojihong65@hotmail.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Bataller R., Brenner D.A. Liver fibrosis. J. Clin. Invest. 2005;115:209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J., Li Y., Liu Q., Huang Y., Li R., Wu T., et al. Sirt6 alleviated liver fibrosis by deacetylating conserved lysine 54 on Smad2 in hepatic stellate cells. Hepatology. 2021;73:1140–1157. doi: 10.1002/hep.31418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dat N.Q., Thuy L.T.T., Hieu V.N., Hai H., Hoang D.V., Thi Thanh Hai N., et al. Hexa histidine-tagged recombinant human cytoglobin deactivates hepatic stellate cells and inhibits liver fibrosis by scavenging reactive oxygen species. Hepatology. 2021;73:2527–2545. doi: 10.1002/hep.31752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez-Valle V., Chavez-Tapia N.C., Uribe M., Mendez-Sanchez N. Role of oxidative stress and molecular changes in liver fibrosis: a review. Curr. Med. Chem. 2012;19:4850–4860. doi: 10.2174/092986712803341520. [DOI] [PubMed] [Google Scholar]
- 5.Wonsey D.R., Zeller K.I., Dang C.V. The c-Myc target gene PRDX3 is required for mitochondrial homeostasis and neoplastic transformation. Proc. Natl. Acad. Sci. U. S. A. 2002;99:6649–6654. doi: 10.1073/pnas.102523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xi H., Gao Y.H., Han D.Y., Li Q.Y., Feng L.J., Zhang W., et al. Hypoxia inducible factor-1alpha suppresses Peroxiredoxin 3 expression to promote proliferation of CCRCC cells. FEBS Lett. 2014;588:3390–3394. doi: 10.1016/j.febslet.2014.07.030. [DOI] [PubMed] [Google Scholar]
- 7.Rhee S.G., Kil I.S. Multiple functions and regulation of mammalian peroxiredoxins. Annu. Rev. Biochem. 2017;86:749–775. doi: 10.1146/annurev-biochem-060815-014431. [DOI] [PubMed] [Google Scholar]
- 8.Arkat S., Umbarkar P., Singh S., Sitasawad S.L. Mitochondrial Peroxiredoxin-3 protects against hyperglycemia induced myocardial damage in Diabetic cardiomyopathy. Free Radic. Biol. Med. 2016;97:489–500. doi: 10.1016/j.freeradbiomed.2016.06.019. [DOI] [PubMed] [Google Scholar]
- 9.Hwang I., Uddin M.J., Lee G., Jiang S., Pak E.S., Ha H. Peroxiredoxin 3 deficiency accelerates chronic kidney injury in mice through interactions between macrophages and tubular epithelial cells. Free Radic. Biol. Med. 2019;131:162–172. doi: 10.1016/j.freeradbiomed.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y., Zhao Y., Wang Z., Sun R., Zou B., Li R., et al. Peroxiredoxin 3 inhibits acetaminophen-induced liver pyroptosis through the regulation of mitochondrial ROS. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.652782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Park J., Han S.J., Park I., Huu T.N., Kim J.S., et al. The critical role of redox regulation of PTEN and peroxiredoxin III in alcoholic fatty liver. Free Radic. Biol. Med. 2021;162:141–148. doi: 10.1016/j.freeradbiomed.2020.11.022. [DOI] [PubMed] [Google Scholar]
- 12.Geng Y., Wang Y., Sun R., Kang X., Zhao H., Zhu M., et al. Carnosol alleviates nonalcoholic fatty liver disease by inhibiting mitochondrial dysfunction and apoptosis through targeting of PRDX3. Toxicol. Appl. Pharmacol. 2021;432 doi: 10.1016/j.taap.2021.115758. [DOI] [PubMed] [Google Scholar]
- 13.Liu L., Wang J., Sun G., Wu Q., Ma J., Zhang X., et al. m(6)A mRNA methylation regulates CTNNB1 to promote the proliferation of hepatoblastoma. Mol. Cancer. 2019;18:188. doi: 10.1186/s12943-019-1119-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathiyalagan P., Adamiak M., Mayourian J., Sassi Y., Liang Y., Agarwal N., et al. FTO-dependent N(6)-methyladenosine regulates cardiac function during remodeling and repair. Circulation. 2019;139:518–532. doi: 10.1161/CIRCULATIONAHA.118.033794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhuang M., Li X., Zhu J., Zhang J., Niu F., Liang F., et al. The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3.1 expression. Nucleic Acids Res. 2019;47:4765–4777. doi: 10.1093/nar/gkz157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin Z., Niu Y., Wan A., Chen D., Liang H., Chen X., et al. RNA m(6) A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 2020;39 doi: 10.15252/embj.2019103181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen M., Wei L., Law C.T., Tsang F.H., Shen J., Cheng C.L., et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 18.Wang M., Yang Y., Yang J., Yang J., Han S. circ_KIAA1429 accelerates hepatocellular carcinoma advancement through the mechanism of m(6)A-YTHDF3-Zeb1. Life Sci. 2020;257 doi: 10.1016/j.lfs.2020.118082. [DOI] [PubMed] [Google Scholar]
- 19.Zhou B., Liu C., Xu L., Yuan Y., Zhao J., Zhao W., et al. N(6) -methyladenosine reader protein YT521-B homology domain-containing 2 suppresses liver steatosis by regulation of mRNA stability of lipogenic genes. Hepatology. 2021;73:91–103. doi: 10.1002/hep.31220. [DOI] [PubMed] [Google Scholar]
- 20.Cui Z., Huang N., Liu L., Li X., Li G., Chen Y., et al. Dynamic analysis of m6A methylation spectroscopy during progression and reversal of hepatic fibrosis. Epigenomics. 2020;12:1707–1723. doi: 10.2217/epi-2019-0365. [DOI] [PubMed] [Google Scholar]
- 21.Wu X., Wu X., Ma Y., Shao F., Tan Y., Tan T., et al. CUG-binding protein 1 regulates HSC activation and liver fibrogenesis. Nat. Commun. 2016;7 doi: 10.1038/ncomms13498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K., Fang S., Liu Q., Gao J., Wang X., Zhu H., et al. TGF-beta1/p65/MAT2A pathway regulates liver fibrogenesis via intracellular SAM. EBioMedicine. 2019;42:458–469. doi: 10.1016/j.ebiom.2019.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu R.M., Desai L.P. Reciprocal regulation of TGF-beta and reactive oxygen species: a perverse cycle for fibrosis. Redox Biol. 2015;6:565–577. doi: 10.1016/j.redox.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y., Zeng P., Li Y.H., Zhang Z., Cui Q. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44:e91. doi: 10.1093/nar/gkw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang G., Shi L., Ye Y., Shi H., Zeng L., Tiwary S., et al. YTHDF3 induces the translation of m(6)a-enriched gene transcripts to promote breast cancer brain metastasis. Cancer Cell. 2020;38:857–871 e857. doi: 10.1016/j.ccell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y., Wang X., Zhang X., Wang J., Ma Y., Zhang L., et al. RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc. Natl. Acad. Sci. U. S. A. 2019;116:976–981. doi: 10.1073/pnas.1812536116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J., Li Y., Yu J., Gan Z., Wei W., Wang C., et al. Resveratrol attenuates high-fat diet induced hepatic lipid homeostasis disorder and decreases m(6)A RNA methylation. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.568006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J., Sun G., Pan S., Qin M., Ouyang R., Li Z., et al. The Cancer Genome Atlas (TCGA) based m(6)A methylation-related genes predict prognosis in hepatocellular carcinoma. Bioengineered. 2020;11:759–768. doi: 10.1080/21655979.2020.1787764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kakimoto P.A., Tamaki F.K., Cardoso A.R., Marana S.R., Kowaltowski A.J. H2O2 release from the very long chain acyl-CoA dehydrogenase. Redox Biol. 2015;4:375–380. doi: 10.1016/j.redox.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L., Yang F., Jia R., Yan P., Ma L. Velvet antler polypeptide prevents the disruption of hepatic tight junctions via inhibiting oxidative stress in cholestatic mice and liver cell lines. Food Funct. 2020;11:9752–9763. doi: 10.1039/d0fo01899f. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Trevijano E.R., Iraburu M.J., Fontana L., Dominguez-Rosales J.A., Auster A., Covarrubias-Pinedo A., et al. Transforming growth factor beta1 induces the expression of alpha1(I) procollagen mRNA by a hydrogen peroxide-C/EBPbeta-dependent mechanism in rat hepatic stellate cells. Hepatology. 1999;29:960–970. doi: 10.1002/hep.510290346. [DOI] [PubMed] [Google Scholar]
- 32.Gu J., Zhuang L., Huang G.C. Melatonin prevents H2O2-induced activation of rat hepatic stellate cells. J. Pineal Res. 2006;41:275–278. doi: 10.1111/j.1600-079X.2006.00364.x. [DOI] [PubMed] [Google Scholar]
- 33.Cox A.G., Winterbourn C.C., Hampton M.B. Mitochondrial peroxiredoxin involvement in antioxidant defence and redox signalling. Biochem. J. 2009;425:313–325. doi: 10.1042/BJ20091541. [DOI] [PubMed] [Google Scholar]
- 34.Luangmonkong T., Suriguga S., Mutsaers H.A.M., Groothuis G.M.M., Olinga P., Boersema M. Targeting oxidative stress for the treatment of liver fibrosis. Rev. Physiol. Biochem. Pharmacol. 2018;175:71–102. doi: 10.1007/112_2018_10. [DOI] [PubMed] [Google Scholar]
- 35.Liu Z., Hu Y., Liang H., Sun Z., Feng S., Deng H. Silencing PRDX3 inhibits growth and promotes invasion and extracellular matrix degradation in hepatocellular carcinoma cells. J. Proteome Res. 2016;15:1506–1514. doi: 10.1021/acs.jproteome.5b01125. [DOI] [PubMed] [Google Scholar]
- 36.Mari M., Colell A., Morales A., von Montfort C., Garcia-Ruiz C., Fernandez-Checa J.C. Redox control of liver function in health and disease. Antioxidants Redox Signal. 2010;12:1295–1331. doi: 10.1089/ars.2009.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sies H. Role of metabolic H2O2 generation: redox signaling and oxidative stress. J. Biol. Chem. 2014;289:8735–8741. doi: 10.1074/jbc.R113.544635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobotta M.C., Liou W., Stocker S., Talwar D., Oehler M., Ruppert T., et al. Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat. Chem. Biol. 2015;11:64–70. doi: 10.1038/nchembio.1695. [DOI] [PubMed] [Google Scholar]
- 39.Lee M., Kim B., Kim V.N. Emerging roles of RNA modification: m(6)A and U-tail. Cell. 2014;158:980–987. doi: 10.1016/j.cell.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 40.Xu K., Sun Y., Sheng B., Zheng Y., Wu X., Xu K. Role of identified RNA N6-methyladenosine methylation in liver. Anal. Biochem. 2019;578:45–50. doi: 10.1016/j.ab.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 41.Idelchik M., Begley U., Begley T.J., Melendez J.A. Mitochondrial ROS control of cancer. Semin. Cancer Biol. 2017;47:57–66. doi: 10.1016/j.semcancer.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okon I.S., Zou M.H. Mitochondrial ROS and cancer drug resistance: implications for therapy. Pharmacol. Res. 2015;100:170–174. doi: 10.1016/j.phrs.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang X., Liu B., Nie Z., Duan L., Xiong Q., Jin Z., et al. The role of m6A modification in the biological functions and diseases. Signal Transduct. Targeted Ther. 2021;6:74. doi: 10.1038/s41392-020-00450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ni W., Yao S., Zhou Y., Liu Y., Huang P., Zhou A., et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol. Cancer. 2019;18:143. doi: 10.1186/s12943-019-1079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Y., Jiao H., Yue Y., He K., Jin Y., Zhang J., et al. Ubiquitin ligase E3 HUWE1/MULE targets transferrin receptor for degradation and suppresses ferroptosis in acute liver injury. Cell Death Differ. 2022 doi: 10.1038/s41418-022-00957-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Y., Wang Z., Feng D., Zhao H., Lin M., Hu Y., et al. p66Shc contributes to liver fibrosis through the regulation of mitochondrial reactive oxygen species. Theranostics. 2019;9:1510–1522. doi: 10.7150/thno.29620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mederacke I., Dapito D.H., Affo S., Uchinami H., Schwabe R.F. High-yield and high-purity isolation of hepatic stellate cells from normal and fibrotic mouse livers. Nat. Protoc. 2015;10:305–315. doi: 10.1038/nprot.2015.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao Y., Wang Z., Zhou J., Feng D., Li Y., Hu Y., et al. LncRNA Mical2/miR-203a-3p sponge participates in epithelial-mesenchymal transition by targeting p66Shc in liver fibrosis. Toxicol. Appl. Pharmacol. 2020;403 doi: 10.1016/j.taap.2020.115125. [DOI] [PubMed] [Google Scholar]
- 49.Zhao Q., Liu J., Deng H., Ma R., Liao J.Y., Liang H., et al. Targeting mitochondria-located circRNA SCAR alleviates NASH via reducing mROS output. Cell. 2020;183:76–93 e22. doi: 10.1016/j.cell.2020.08.009. [DOI] [PubMed] [Google Scholar]
- 50.Belcher J.D., Young M., Chen C., Nguyen J., Burhop K., Tran P., et al. MP4CO, a pegylated hemoglobin saturated with carbon monoxide, is a modulator of HO-1, inflammation, and vaso-occlusion in transgenic sickle mice. Blood. 2013;122:2757–2764. doi: 10.1182/blood-2013-02-486282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y., Wang J., Xu D., Xiang Z., Ding J., Yang X., et al. m(6)A mRNA methylation regulates testosterone synthesis through modulating autophagy in Leydig cells. Autophagy. 2021;17:457–475. doi: 10.1080/15548627.2020.1720431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.