Abstract
Granulocyte-monocyte progenitors (GMPs) differentiate into both neutrophils and monocytes. Recently, uni-potential neutrophil progenitors have been identified both in mice and humans using an array of surface markers. However, how human GMPs commit to neutrophil progenitors and the regulatory mechanisms of fate determination remain incompletely understood. In the present study, we established a human neutrophil deficiency model using the small molecule alpha-lipoic acid. Using this neutrophil deficiency model, we determined that the neutrophil progenitor commitment process from CD371+ CD115– GMPs defined by CD34 and CD15 and discovered that critical signals generated by RNA splicing and rRNA biogenesis regulate the process of early commitment for human early neutrophil progenitors derived from CD371+ CD115– GMPs. These processes were elucidated by single-cell RNA sequencing both in vitro and in vivo derived cells. Sequentially, we identified that the transcription factor ELK1 is essential for human neutrophil lineage commitment using the alpha-lipoic acid (ALA)-inducing neutrophil deficiency model. Finally, we also revealed differential roles for long-ELK1 and short-ELK1, balanced by SF3B1, in the commitment process of neutrophil progenitors. Taken together, we discovered a novel function of ALA in regulating neutrophil lineage specification and identified that the SF3B1-ELK axis regulates the commitment of human neutrophil progenitors from CD371+ CD115– GMPs.
Keywords: Alpha-lipoic acid, Neutrophils, Monocytes, ELK1, SF3B1, RNA splicing
Highlights
-
•
ALA completely blocks the differentiation of human neutrophils derived from CD34+ stem cells in ex-vivo culture.
-
•
CD34 and CD15 could be used to define the early differentiation stages of human neutrophil lineage determination.
-
•
SF3B1-ELK1 signal axis regulates human neutrophil lineage determination.
1. Introduction
Neutropenia results in serious infection, fainting, low immunity, high fever, and chills. G-CSF injection is used to stimulate emergency granulopoieses for clinical treatment of neutropenia [1]. However, the application of G-CSF carries a significant risk of severe side effects, including spleen rupture, pulmonary toxicity, and other toxic reactions, so it is not suitable for the treatment of clinical neutropenia in all cases. As alternatives to G-CSF, neutrophil or granulocyte transfusion can also be used to treat neutropenia [2]. However, large numbers of neutrophils are required for clinical treatment, and the minimum transfusion dose is 1 × 109 cells/kg [3]. Meeting the neutrophil infusion dose is particularly challenging for patients with high body weight. Therefore, studying the regulatory molecular mechanisms of neutrophil lineage determination is critical for clinical neutrophil regeneration.
Granulocyte-monocyte progenitors (GMPs) are well-known to give rise to both monocytes and neutrophils. In humans, early neutrophil progenitors have been defined as Lin– CD66b+/low CD15low CD49d+ CD11b– [4] or Lin– CD66b+ CD117+ CD71+ [5], and neutrophil committed progenitors are identified as CD66b–CD64dimCD115– in SSClow CD45dim CD34+ and CD34low/– progenitors [6], but no uni-potential neutrophil progenitors have been identified within human GMPs by single-cell liquid culture [7]. However, how human common neutrophil progenitors commit from GMPs remains unknown [8]. Single-cell RNA sequencing and mass cytometry analyses cannot answer the neutrophil lineage commitment mechanism in detail [4,5]. Previous reports have demonstrated that post-transcriptional processes regulate the myeloblast-to-monocyte differentiation transition [9], megakaryocyte and erythrocyte lineages specification [10], and normal neutrophil differentiation [11]. Recently, the small molecule thymidine has been used to reprogram mouse fibroblast cells into neutrophils [12]. These studies suggest that commitment of neutrophil progenitors could also be controlled by RNA splicing. To elucidate the detailed mechanism of neutrophil progenitor fate determination, an inducible neutrophil differentiation model is required.
It is well known that antioxidants are commonly used to regulate cell proliferation and differentiation. The natural antioxidant ALA participates in metabolic regulation as a coenzyme in the TCA cycle [13] and regulates intracellular ROS levels [14]. ALA can be synthesized through the gene LIAS or ingested from food. Previous reports show that ALA can regulate cell proliferation and apoptosis through the ROS signaling pathway [[15], [16], [17]] and inhibit cancer progression [[18], [19], [20]]. Several studies show that alpha-lipoic acid (ALA) as one of the antioxidants can inhibit the differentiation and proliferation of hematopoietic stem progenitor cells (HSPCs) and maintains the function of hematopoietic stem cells (HSCs) both in vivo and in vitro [[21], [22], [23]]. As previous report shows that reactive oxygen species (ROS) play important role in mice emergency granulopoiesis [24]. Thus, we tried to explore the function of ALA in human neutrophil differentiation. In the present study, by adding ALA to the cell culture medium, we established an inducible neutrophil differentiation model from human cord blood CD34+ HSPCs. Using this model, we determined how neutrophils differentiate from GMPs and elucidated the molecular mechanism of neutrophil progenitor commitment from GMPs. Additionally, we also confirmed that a similar commitment process of neutrophil progenitors occurs in human adult BM (bone marrow). Our findings suggested that the SF3B1-ELK1 signaling axis regulates early neutrophil commitment from CD371+ CD115– GMPs via RNA splicing.
2. Materials and methods
2.1. Cell culture
To reveal the effect of antioxidants on multiple lineage differentiation, total cord blood nucleated cells or isolated CD34+ cells were cultured in IMDM plus 10% FBS containing 8 cytokines (10 ng/mL SCF, 10 ng/mL FLT3L, 10 ng/mL TPO, 10 ng/mL IL-3, 10 ng/mL IL-6, 10 ng/mL GM-CSF, 20 ng/mL G-CSF and 2 U/mL EPO). Neutrophil and monocyte differentiation potential of indicated progenitors (CD371+ GMPs, CD371– GMPs and CD34+, CD34low or CD34– progenitors within the Lin– CD116– CD73– CD1c– CD115– CD123mid CD45RA + CD371+.) were sorted and cultured in IMDM plus 10% FBS containing 7 cytokines (10 ng/mL SCF, 10 ng/mL FLT3L 10 ng/mL TPO, 10 ng/mL IL-3, 10 ng/mL IL-6, 10 ng/mL GM-CSF and 20 ng/mL G-CSF). CD71 and GPA were used to indicate erythroid lineages, and CD15, CD66b, CD14 and CD11b were used to analyze neutrophils and monocytes.
To screen the antioxidants that affect human neutrophil differentiation, Alpha-lipoic acid (ALA) (Shanghai yuanye Bio-Technology Co., Ltd, #S26654) (25 μg/mL), l-Glutathione reduced (GSH) (MedChemExpress (MCE), #HY-D0187) (25 μg/mL), N-Acetyl-l-cysteine (NAC) (Sigma, #A9165-25G) (250 mM), N-Acetyl-d-cysteine (NAD) (MCE, #HY-136386)(250 mM), l-Ascorbic acid (MCE, # HY-B0166) (25 μg/mL), α-Vitamin E (MCE, #HY-N0683) (25 mM) and 1 mM H2O2 were added to the differentiation medium containing 7 cytokines from day0. ALA and NAC were dissolved in differentiation medium directly and stored at 4 °C for 3 weeks, while the GSG, NAD, l-Ascorbic acid and α-Vitamin E were dissolved and stored according to the manufacturer's instructions. To analyze the dose-effect of ALA on neutrophil differentiation, different doses (0.1, 0.5, 2.5, 10, 20, 50 μg/mL) of ALA were added to the 7-cytokine differentiation medium. Normally, we used 25 μg/mL ALA to analyze the effect of ALA on neutrophil differentiation for different hematopoietic progenitors.
To analyze the different effects of ALA on neutrophil differentiation derived from CD34+HSPCs, we changed normal culture medium to the ALA containing medium at day2, day4, day6, day8 and day10, or conversely, replace the ALA medium with normal medium until day14. In ex vivo culture, higher percentage of CD66b+CD15+CD14– neutrophils are detected at day14 derived from CD371+CD115– GMPs, and further culture will lead part of these cells to express CD14 (CD66b+CD15+CD14+ neutrophils, APC like TANs [25]). To avoid unnecessary misunderstandings, flow cytometry is performed on day14.
2.2. ALA treatment of mice or hematopoietic stem/progenitor cells derived from mice BM
To analyze the effect of ALA on mice neutrophil differentiation in vivo. We intraperitoneally injected 2 mg ALA dissolved in 2.5%DMSO-corn oil [15] per-mice on every other day for 2 weeks, and 1 mg/mL ALA containing water was supplied every day at the same time. Flow cytometry assay was performed on day0 (before injection), day14 and day18 to detect Ly6C+ Ly6G+ neutrophils in peripheral blood.
To analyze the effect of ALA on mice neutrophil differentiation derived from Lin(CD2,CD3,CD4,CD8,Mac1,B220,Ter119) – ckit+ Sca1+ (LSK) in ex-vivo culture, we sorted LSK for in vitro differentiation using 8 cytokines (mSCF, mTOP, mG-CSG, mGM-CSG, mIL-3, mIL-6, hFLT3L, hEPO) containing medium ±25 μg/mL ALA. Flow cytometry assay was performed to analyze Ly6C+ Ly6G+ neutrophils at day5 day7 and day9. Finally, MGG (May-Grunwald Giemsa) stain was used to analyze the cell morphology of ALA treated cultures of not.
2.3. Cell morphology assay
Total cultures or sorted cells were centrifuged onto glass slides using a Cell Cytospin machine (Cytospin 4, Thermo Fisher Scientific), and MGG (May-Grunwald Giemsa) stain was used to analyze the cell morphology as previously reported [26].
2.4. Gene-expressing plasmid and shRNA-expressing plasmid construction
The cDNA of CLEC11A (NM_002975), LRG1 (NM_052 972), PRTN3 (NM_002777), CEBPE (NM_001805), CEBPE-M (U80982.1), XRCC3, TMEM176B, PBX3, MDM1, MDB3, HMGA1, ELK1, and short-ELK1 from the dsDNA of neutrophil biased progenitors was cloned into the PCCL-MND-PGK-dsRed or PCCL-MND-PGK-EGFP lentiviral vector as previously reported [27]. Knocking down the target genes in CD34+ cells was mediated by short hairpin RNA (shRNA). All the plasmids (Plko.1-EGFP, Plko.1-PRTN3-shRNA1-EGFP, Plko.1-PRTN3-shRNA2-EGFP, Plko.1-PRTN3-shRNA3-EGFP, Plko.1-LRG1-shRNA1-EGFP, Plko.1-LRG1-shRNA2-EGFP, Plko.1-LRG1-shRNA3-EGFP, Plko.1-CLEC11A-shRNA1-EGFP, Plko.1- CLEC11A-shRNA2-EGFP and Plko.1-CLEC11A-shRNA3-EGFP were prepared by Feng Hui Sheng Wu, Co., Ltd. Three targeted sequences of each gene were inserted into the lentiviral expression vector Plko.1-EGFP-Puro (#FH1717, FENGHUISHENGWU Co., Ltd, China). In addition, the shRNA expression vectors of B3GAT3, ELK1, E2F1, HSF1, KLF16, MLLT1, MYBL2, SNRPB, SSBP4, TRIM28, and ZDHHC13 were cloned to analyze their function in neutrophil or monocyte differentiation. The target sequences are provided in Table 1.
Table 1.
Target sequences for shRNAs.
| shRNA Names | targets sequences |
|---|---|
| PRTN3_sh1 | AGCTCAATGTCACCGTGGTCA |
| PRTN3_sh2 | CCTGATCTGTGATGGCATCAT |
| PRTN3_sh3 | CCTGACTTCTTCACGCGGGTA |
| LRG1_sh1 | CATGCTGGACCTCTCCAATAA |
| LRG1_sh2 | TGGACACCCTGGTATTGAAAG |
| LRG1_sh3 | GCAATTAGAACGGCTACATCT |
| CEBPE_sh1 | GCTGGAGTACATGGCAGAGAA |
| CEBPE_sh2 | GCAAGAAGGCAGTGAACAAAG |
| CEBPE_sh3 | CCTTTGCCTACCCTCCACATA |
| CLEC11A_sh1 | CTGCCGGAACTGTTGAGGGAA |
| CLEC11A_sh2 | GTGTCCTTCTTCGCCTGGCAT |
| CLEC11A_sh3 | GCACTCCTCCTTGTTAGTGTC |
| KLF16_sh1 | ACAAGTCCTCGCACCTAAAGTCG |
| E2F1_sh1 | AGATCCCAGCCAGTCTCTACTCA |
| PBX3_sh1 | ATGGACGATCAATCCAGGATGC |
| MYBL2_sh1 | ACACAGATTCAGATGTGCCGGA |
| MLLT1_sh1 | CAAAGTAGAGGAGTCGGGGTACG |
| ZDHHC13_sh1 | CTCGCAGTGCAGGAATCACAGC |
| SNAPC4_sh1 | GCACACAGAAAGAGCAGCTGAG |
| PBX1_sh1 | TGATGAATCTCCTGCGAGAGCAA |
| SNRPB_sh1 | CAGAAAGGGAAGAGAAGCGAGT |
| B3GAT3_sh1 | CCCTTGCTGTTAGATAAGCCCA |
| SSBP4_sh1 | GTGGAAACTCGATCCCCTACTC |
| HSF1_sh1 | GCCATGAAGCATGAGAATGAGG |
| shRNA_ELK1_A | GCCTTGCGGTACTACTATGACA |
| shRNA_ELK1_L | CCAAACCTGAAATCGGAAGAGC |
| SF3B1_sh1 | GCAGATTTGCTGGATACGTGAC |
| SF3B1_sh2 | TGCTTTGATTTGGTGATGTAA |
| SF3B4_sh1 | GACCAGTAGTCAACACCCACAT |
| SF3B4_sh2 | CCCTGAGATTGATGAGAAGTT |
| SF3B5_sh1 | GCCACTTCGACCTTCTCAACTA |
| SF3B5_sh2 | TCCGCTTCAACTTGATGGAAA |
| SRSF3_sh1 | GCATCGTGATTCCTGTCCATTG |
| SRSF3_sh2 | TGGAACTGTCGAATGGTGAAA |
| SRSF11_sh1 | GAGCTTTGATAGTCGTACCAT |
2.5. Lentiviral transduction and neutrophil or monocyte differentiation potential assay
Lentiviral preparation and transduction of CD34+ HSPCs were performed as we previously described [27]. The neutrophil and monocyte differentiation potential of total transduced cells or sorted transduced cells (GFP+ CD115– CD371+ gated CD34+ CD15–, CD34mid CD15mid, CD34low/- CD15+) was analyzed by culturing in IMDM plus 10% FBS containing 7 cytokines (10 ng/mL SCF, 10 ng/mL FLT3L, 10 ng/mL TPO, 10 ng/mL IL-3, 10 ng/mL IL-6, 10 ng/mL GM-CSF and 20 ng/mL G-CSF) for 7–14 days.
2.6. Bulk cell RNA-seq
Two thousand CD371+CD115– GMP, CD34midCD115+MoP, NbP1, NbP2 and NbP3 cells derived from cultures or BM were sorted into 1.5 mL tube containing 200 μL 0.5% BSA-DPBS solution. The dsDNA and libraries were generated as previously described. And all the libraries were sequenced using the Illumina NovaSeq6000 system (Novogene Co.,Ltd.). The clean fastq data were aligned to the hg38 human genome using Hisat2. R packages of DESeq2, ggplots clusterProfiler and GSEA were used to analyze the differentially expressed genes and gene ontology enrichment and gene set enrichment.
To assay the signals changing under the overexpression of L-ELK1 and S-ELK1, we sorted two thousand NbP1 derived from L-ELK1 and S-ELK1 transduced CD34+ HSPCs to generate DNA libraries as previously described. Data analysis was performed as described above.
2.7. In vitro single-cell RNA-seq analysis
To reveal the in vitro transcriptome characteristics of neutrophil differentiation at different stages at the single cells level, we sorted the CD371+ CD115– GMPs, CD115+ CD34mid MoPs, CD115–CD34mid CD371+ early neutrophil biased progenitors (ENbPs), CD115– CD34low CD371+ late neutrophil biased progenitors (LNbPs) CD371+ CD66b–, and CD371+ CD66b+ cells for single cell RNA-Seq and the double-strand cDNA library of single cells was generated as previously reported [28]. The quality control of amplified cDNA was analyzed by RT-PCR analysis of the housekeeping gene B2M. Samples with CT values less than 25 were used for sequencing library preparation using Vazyme TruePrepTM DNA Library Prep Kit V2 from Illumina (Vazyme, TD-503). Library sequencing was performed using the Illumina NovaSeq 6000 system (Novogene Co., Ltd.).
All fastq files were mapped to the human hg38 genome by HISAT2 software, and raw count data were calculated using featureCounts software. For bulk cell RNA-seq, the DESeq2 package was used to normalize the raw data and determine the differentially expressed genes. For the single-cell RNA-seq data, all the raw count data were normalized by transforming into CPM (counts per million) values by the edgeR program. Unsupervised cluster analysis was performed to evaluate the accuracy of sorted cells. In total, 165 single cells (40 CD371+ CD115– GMPs, 24 CD115+ CD34mid MoPs, 29 CD115–CD34mid CD371+ early neutrophil biased progenitors (ENbPs), 34 CD115– CD34low CD371+ late neutrophil biased progenitors (LNbPs), 18 CD371+ CD66b–, and 20 CD371+ CD66b+ cells) were retained for further analysis. Multiple t-tests were used to identify the statistically significant differentially expressed genes for different cell groups. PCA and the UMAP method were used to analyze the single-cell RNA, and Monocle was used to perform the cell trajectory interference.
The R packages sincell, pcaMethods, uwot, gplots, clusterProfiler, and reshape2 were used to analyze the single-cell data. The DESeq2, clusterProfiler, and GSEA packages were used to analyze the differentially expressed genes, GO enrichment, and pathways for bulk cell RNA-seq data.
2.8. 10x single-cell RNA-seq of BM derived cells
To dissect the in vivo transcriptome characteristics of neutrophil lineage cells at different stages, 10 000 Lin/CD116/CD1C/CD73– CD371+ CD123+ cells derived from human adult BM of two healthy donors were sorted for droplet-based cell harvest by a 10X Genomics Chromium system. The fastq raw files were processed by Cell Ranger 4.0.4 software to obtain the gene expression matrix. Cells with more than 2500 detected genes (an average of 4500 genes were detected) were used to perform further clustering. To further define the populations, we used CD1c, CD1d, CCR6, and CLEC10A to define dendritic progenitor cells, HOXA9, HOXA10, HOXB4, and CD34 to define CD371+ GMPs, CD34, MPO, PRTN3, ELANE, and CLEC11A to define neutrophil biased progenitors, and CD115 (CSF1R), MAFB, MAFF, and CD14 to define monocyte progenitors. Finally, Monocle2 was used to determine the cell trajectory of CD371+ GMPs, neutrophil biased progenitors and monocyte progenitors. Fastq files or processed data were uploaded to the Gene Expression Omnibus public database (GSE184864).
2.9. Immunofluorescence staining
Sorted cells were centrifuged onto glass slides using a Cell Cytospin machine (Cytospin 4, Thermo Fisher Scientific), and cells were fixed using 4% paraformaldehyde in PBS pH 7.4 for 10 min at room temperature. The fixed cells were washed three times with cold PBS and then permeabilized using 0.2% Triton X-100 for 10 min at room temperature. The cells were gently washed with PBS three times. The cells were blocked with 1% BSA for 30 min and then incubated with 1:50 diluted primary antibodies (anti-ELK1 N-terminus antibody (Biorbyt, orb213892, indicates S-ELK1 and L-ELK1) and anti-ELK1 C-terminus antibodies (Affinity, AF6212, only indicates the L-ELK1)) in 1% BSA overnight at 4 °C. The cells were gently washed with cold PBS three times. The cells were incubated with goat IgG (H + L) cross-adsorbed secondary antibody Alexa Fluor ® 555 conjugate antibody (ThermoFisher, A-21432)) for 1 h at room temperature. The secondary antibody solution was decanted and washed three times for 5 min. Finally, the cells were incubated with 1 μg/mL Hoechst3342 for 1 min. The pictures were harvested by a confocal microscope LSM900 with the same parameters.
2.10. RNA-protein interaction assay
RNA pull-down and immunoblotting were performed to assay the interaction of SF3B1 and ELK1 mRNA. According to the absent sequence of S-ELK1 compared to L-ELK1, we synthesized 5′-biotin-modified RNA (20 bp) around the splicing site. RNA pulldown and immunoblotting were performed as previously described [29,30]. In brief, 1 mol of each biotin-modified RNA was added to the washed agarose beads, incubated at 4 °C for 3–4 h with 180 U/mL RNase inhibitor (Biosystems, AM2694) and resuspended in 500 μL of RNA-streptavidin interaction buffer after washing twice. Cellular nuclear proteins were extracted by a Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime), and then 100 μL was added to an agarose-streptavidin-biotin-RNA tube and incubated at 4 °C overnight under 100–150 RPM rotation. After this procedure, 15 μL SDT buffer (2% SDS, 100 mM DTT, 100 mM Tris-HCl, pH = 7.6) was added to the pellets and incubated at 95 °C for 5 min to elute the proteins after washing twice with the interaction buffer. The supernatant liquid was used as the input for the mass spectrometry assay (Performed by Aptbiotech. Ltd.) to identify the proteins binding to ELK1 biotin-RNA.
2.11. Humanized immunodeficient mouse transplantation
L-ELK1-, S-ELK1-, ELK1-shRNA-A (target to all transcripts)- and ELK1-shRNA-L (target to only the long coding protein transcript)-transduced CD34+ HSPCs were transplanted into B-NDG-SM humanized immunodeficient mice expressing human SCF and GM-CSF purchased from Beijing Biocytogen Co., Ltd. Transplantation was conducted as previously reported [21]. To improve neutrophil differentiation, 100 ng/mL human IL-3 and G-CSF were injected into mice plus cells. Flow cytometry assay was performed on day18 after transplantation. All animal experiments were approved by the Institutional Ethics Review Committee of Institute of Blood Transfusion (IERC-IBT).
2.12. Statistical analysis
The R packages of edgeR, DESeq2, clusterProfiler, pcaMethods, gplots, reshape2, ggplot2, Seurat, monocle2, GSEA, cluster, factoextra, uwot and sincell were used to perform statistical analysis of RNA-Seq data. Other significant analyses were performed using unpaired Student's t-tests (GraphPad Prism, GraphPad Software).
2.13. Data availability
The data that support the findings of this study are available from the corresponding author upon request. All RNA-Seq data are deposited in the GEO database with accession code GSE184864.
3. Results
3.1. ALA impedes human neutrophil commitment
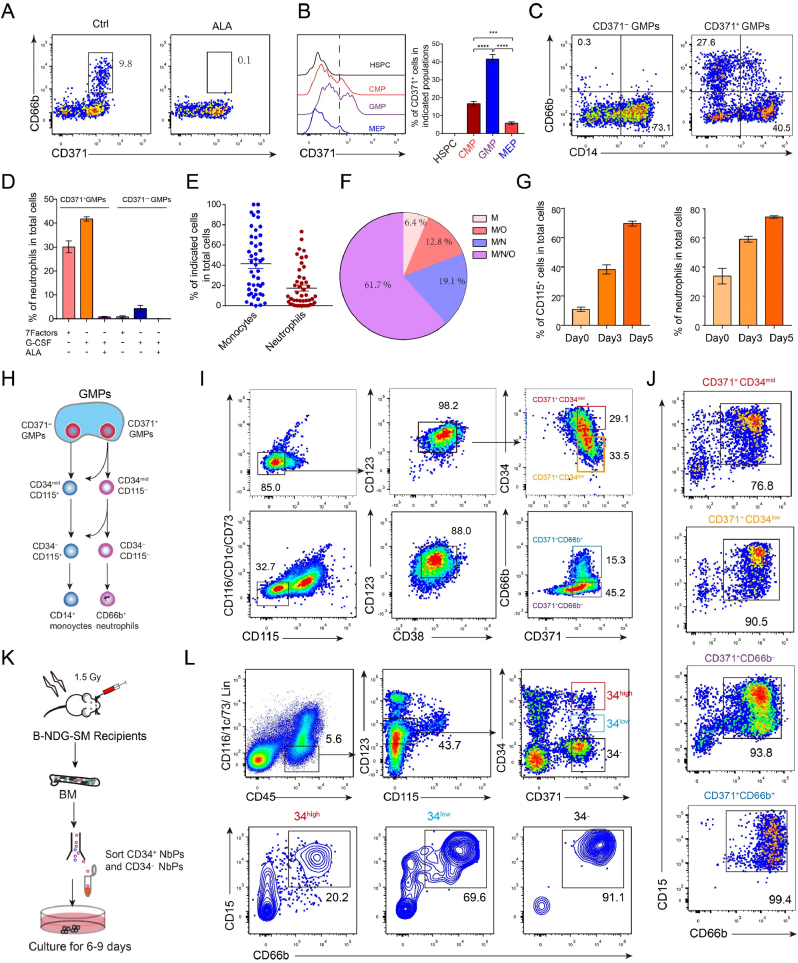
Our previous report shows that ALA is helpful to maintain the stemness of hematopoietic stem cells [21]. To determine the function of ALA on blood cell lineage differentiation, CB CD34+ hematopoietic stem/progenitor cells (HSPCs) were sorted and cultured in 10% fetal bovine serum (FBS) IMDM medium containing SCF, FLT3L, IL-3, IL-6, TPO, GM-CSF, G-CSF and EPO ± ALA. Surprisingly, ALA completely inhibited production of CD11b+ CD14– CD66b+ CD15+ neutrophils and increased CD11b+ CD14+ CD66b– CD15– monocytes production (Fig. 1. A). Morphological analysis also demonstrated that ALA blocked generation of CD11b+ CD66b+ CD15+ polymorphonuclear neutrophils (PMNs) (Fig. 1. B) We also determined the effect of ALA on CD66b+ CD14– and CD66b– CD14– populations. ALA did not block the cell proliferation of CD66b+ CD15+ neutrophils. Although ROS are reported to be related to emergency granulopoiesis [24], we contend that ROS are not sufficient to control neutrophil commitment. Thus, we postulated that ALA inhibition of neutrophil differentiation was unlikely to be related to its antioxidant function, but rather by other unknown functions. To test this hypothesis, we added 1 mM H2O2 and commonly used antioxidants that could scavenge ROS in CD34+ HSPCs (Supplementary Fig. 1. A), including N-Acetyl-l-Cysteine (NAC), N-Acetyl-d-cysteine (NAD), l-Glutathione reduced (GSH), α-Vitamin E ((+)-α-Tocopherol), and l-Ascorbic acid (Vitamin C) to the culture medium ± ALA. Neither H2O2 nor other antioxidants (GSH, NAC, NAD, α-Vitamin E and Vitamin C) treatment affected the production of neutrophils and monocytes (Fig. 1 C-D), indicating that ALA inhibited human neutrophil differentiation independently of its effects on ROS. Additionally, ALA did not block neutrophil differentiation from Lin (CD2, CD3, CD4, CD8, B220, Mac1, Ter119)– c-kit+ sca1+ mouse HSPCs (Supplementary figure 1 B-D).
Fig. 1.
ALA impedes human neutrophil commitment.
(A) Representative flow cytometry plots of CD11b+ CD66b+ CD15+ CD14– neutrophils and CD11b+ CD14+ CD66b– CD15– monocytes derived from CD34+ HSPCs of CB ± ALA. Flow cytometry was performed on day 14 of culture.
(B) MGG staining assay to determine the morphology of Ctrl cultures, ALA-treated cultures, and CD66b+ CD15+ CD14– neutrophils derived from Ctrl cultures and peripheral blood. Scale bar = 20 μm (C) Percentages of CD11b+ CD66b+ CD15+ CD14– neutrophils and CD11b+ CD14+ CD66b– CD15– monocytes derived from cord blood CD34+ hematopoietic stem/progenitor cells (HSPCs) treated with different antioxidants (Alpha-lipoic acid = ALA, l-Glutathione reduced = GSH, N-Acetyl-l-Cysteine = NAC, N-Acetyl-d-cysteine = NAD, l-Ascorbic acid = VC, α-Vitamin E = VE). Flow cytometry was performed to measure neutrophil and monocyte percentages on day 14 of culture.
(D) Statistical analysis of neutrophil percentages in total cells derived from CD34+ HSPCs ± ALA or H2O2 treatment.
(E) Statistical analysis of neutrophil percentages derived from CD34+ HSPCs with different doses of ALA (0.1, 0.5, 2.5, 10, 20, 50 μg/mL).
(F) Absolute cell number of neutrophils derived from CD34+ HSPCs under 20 μg/mL ALA treatment at different days. ALA was added to (I) and removed from (II) culture medium at day 2 (D2), day 4 (D4), day 6 (D6), day 8 (D8), and day 10 (D10). Flow cytometry was performed on day14. The Ctrl for addition of ALA was the absence of ALA, and the Ctrl for removing ALA was treatment for the duration of the experiment.
(G–H) Proportion of CD115– cells in Lin– CD38+ CD34+ CD123+ CD45RA+ GMPs and Lin– CD38+ CD34–CD123+ CD45RA+. CD34+ HSPCs were cultured ± ALA treatment. Flow cytometry analysis of CD115– cells in the indicated populations was performed on day 2 (D2), day 4 (D4), day 6 (D6), or day 8 (D8).
(I–J) Percentages of neutrophils derived from CD34+, CD34low, or CD34– cells within CD115– cells at day 5 culture with ALA treatment (J) or vehicle treatment (I). Sorted CD115– CD34+, CD115– CD34low, or CD115– CD34– cells were cultured ± ALA treatment for 6–12 days and flow cytometry assay was performed to analyze CD11b+ CD66b+ CD15+ CD14– neutrophils and CD11b+ CD14+ CD66b– CD15– monocytes.
Data of (C–I) are expressed as means ± SD. Each symbol in (C, D, G, H) represents an individual replicate, and small horizontal lines indicate the mean (±SD.). An unpaired Student's t-test (two-tailed) was performed to evaluate statistical significance. NS = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, N = 3–7.
Furthermore, we determined the dose-dependent effect of ALA on CD34+ HSPC differentiation. ALA had a dose-dependent effect on neutrophil differentiation of CD34+ HSPCs, and 20 μg/mL of ALA completely inhibited neutrophil production (Fig. 1. E). To determine the stage at which ALA blocks neutrophil differentiation, CD34+ HSPCs were sorted and cultured ± ALA treatment. ALA was added to or removed from culture medium at different time points, and flow cytometry analysis was performed on day 14. Both the addition and removal of ALA hindered neutrophil differentiation (Fig. 1. F). We thus postulated that ALA impaired the commitment of neutrophil progenitors. Given that neutrophils are mainly derived from CD115– populations [31], we analyzed the CD115– GMP-like cells and CD115– CD34– CD123mid CD45RA + CD38+ progenitors on day 2, day 4, day 6, and day 8. CD34– CD115– CD123mid CD45RA + CD38+ progenitors, but not the CD115– GMP-like cells, were significantly decreased on day 4, day 6, and day 8 with ALA treatment (Fig. 1G-H). Finally, to determine which stages does ALA block neutrophil differentiation, we sorted CD115– CD34+, CD115– CD34mid, and CD115– CD34– cells derived from Ctrl cultures for continuous differentiation culture. A higher proportion of neutrophils were detected in CD115– CD34– progenitors derived cultures compared to CD115– CD34+, CD115– CD34mid progenitors, and ALA completely blocked neutrophil differentiation of CD34high and CD34mid progenitors, but not in CD34– progenitors (Fig. 1. I). Furthermore, CD115– CD34+, CD115– CD34mid, and CD115– CD34– progenitors derived from ALA treated cultures at day5 were sorted for continuous culture. Almost no neutrophils were detected in CD34high CD34mid and CD34– progenitors with continuous ALA treatment (Fig. 1. J). A significantly lower proportion of neutrophils were detected from ALA-treated CD115– CD34mid (16.17 ± 1.2%) compared to Ctrl culture-derived CD115– CD34mid (53.4 ± 2.1%), and ALA-treated CD115– CD34– (14.7 ± 1.3%) progenitors compared to culture-derived CD115– CD34– (58.4 ± 1.8%). but not for CD115– CD34high progenitors (Fig. 1 I-J)). Together, these findings indicated that neutrophil progenitors commit to the neutrophil fate concomitant with decreasing CD34 expression, and that ALA impedes the commitment of neutrophil progenitors.
3.2. CD371+ GMPs commit to neutrophil progenitors by losing monocyte differentiation potential
In prior studies, single-cell culture of human GMPs revealed that cells differentiated from the culture were neutrophils, but that this is accompanied by a small amount of monocyte production [7], indicating that human neutrophil progenitor fate determination could occur downstream of GMP differentiation. However, the ways by which neutrophil progenitors are committed from GMPs remains unknown. Prior reports have indicated that almost all CFU-G colonies or CD66b+ granulocytes are derived from CD64mid/–CD371+ GMPs [32] excluding the CD115+ cells [6]. Therefore, to confirm that CD66b+ neutrophils were derived from CD371+ progenitor cells, we performed flow cytometry analysis to measure the neutrophils at day 7 of in vitro differentiation of CB-derived GMPs. Only CD371+ cells produced CD66b+ neutrophils and ALA blockage of CD371+ cells gave rise to CD66b+ neutrophils (Fig. 2. A). Flow cytometric analysis of CD34+ CD38− HSPCs, CD34+ CD38+ CD123mid CD45RA– CMPs (Common myeloid progenitors, CMPs), CD34+ CD38+ CD123mid CD45RA + GMPs, and CD34+ CD38+ CD123– CD45RA– MEPs (Megakaryocyte-erythroid progenitor cell, MEPs) demonstrated that GMPs contained a higher proportion of CD371+ progenitor cells (Fig. 2. B). To determine the neutrophil differentiation potential of CD371+ progenitors, we sorted CD371+ GMPs and CD371– GMPs for neutrophil differentiation, revealing that neutrophils are mainly derived from CD371+ GMPs, but not CD371– GMPs (Fig. 2. C). Furthermore, ALA completely blocked the differentiation of CD371+GMPs into neutrophils, even under treatment with G-CSF only (Fig. 2. D). To further analyze the neutrophil differentiation potential of CD371+ GMPs, we sorted CD371+ GMPs for single-cell differentiation analysis. CD371+ GMPs contained no committed neutrophil progenitors, except monocyte progenitors (Fig. 2 E-F), and CD371+ GMPs contained different neutrophil differentiation potential progenitors (Fig. 2E-F). In addition, flow cytometry analysis revealed that CD371+ GMPs contained CD115low cells (Supplementary Fig. 2. A).
Fig. 2.
Human neutrophils are derived from CD115–CD371+GMPs.
(A) Representative flow cytometry plots of CD66b+ neutrophils within CD371+ cells derived from CB CD34+ HSPCs ± ALA. Flow cytometry was performed at day 9.
(B) Flow cytometry analysis of the proportion of CD371+ progenitors in Lin– CD38– CD34+HSPCs, Lin– CD38+ CD34+ CD123mid CD45RA– CMPs (common myeloid progenitors), Lin– CD38+ CD34+ CD123mid CD45RA + GMPs and Lin– CD38+ CD34+ CD123– CD45RA– MEPs (megakaryocyte-erythroid progenitors) ± ALA.
(C) Representative flow cytometry plots of CD11b+CD66b+CD14–neutrophils and CD11b+CD66b–CD14+ monocytes derived from CD371– GMPs and CD371+ GMPs.
(D) Neutrophil production in CD371+ GMPs and CD371– GMPs under the specified treatments, including culture with seven cytokines, G-CSF only, or G-CSF and ALA. Flow cytometry assay to performed to analyze the CD11b+CD15+CD66b+CD14– neutrophils in total cells at day14 of culture.
(E) Single-cell assay for the neutrophil and monocyte differentiation potentials of CD371+ GMPs.
(F) Pie diagram illustrating percentages of progenitors with different lineages (M: monocyte, N: neutrophil, O: others).
(G) Neutrophil differentiation potential of CD115– progenitors derived from CD371+ GMPs at different time points. Percentages of CD115+ progenitors and neutrophils derived from CD115– progenitors are shown.
(H) Conjecture model diagram of neutrophil and monocyte differentiation derived from GMPs.
(I) Representative flow cytometry plots of the sorting gate for neutrophil-biased progenitors derived from CD115–CD371+ GMPs at day 3 and day 6.
(J) Flow cytometry analysis of the neutrophils from sorted CD371+ CD34high, CD371+ CD34mid (day 3), CD371+ CD66b– and CD371+CD66b+ (day 6) cells derived from CD115– CD371+ GMPs from day 6 to day 9.
(K)In vivo experimental strategy to analyze neutrophil-biased progenitors from B-NDG-SM mouse BM.
(L) Flow cytometry analysis of the neutrophil differentiation potential of sorted neutrophil-biased progenitors from B-NDG-SM mouse BM at day 6–day 9. Data in (B, D, E, G) are represented as means ± SD. Each symbol of (E) represents one cell, and small horizontal lines indicate the mean (±SEM.). An unpaired Student's t-test (two-tailed) was performed to assess statistical significance. ***P < 0.001, ***P < 0.0001.
To further explore the characteristics of neutrophil differentiation from CD371+ GMPs, we first analyzed CD115+ progenitor proportions at different time points, and subsequently analyzed the neutrophil differentiation potential from CD115+-excluded progenitor cells. The percentages of CD115+ cells increased up to 75% on day 5, and the neutrophil purity increased to 80% (Fig. 2. G). Thus, we postulated that neutrophil commitment could be stimulated by gradual loss of monocyte differentiation potential (Fig. 2. H). To test this hypothesis, we measured a set of surface markers in CD371+ CD115– CD66b– CD14– cells on day 14 to further improve neutrophil purity. CD371+ CD115– CD66b– CD14–cells expressed CD1c, CD73, and CD116, while CD371+ CD66b+ neutrophils did not express these markers (Supplementary Fig. 2. B). In addition, we also analyzed in vitro early differentiation of GMPs at day 5 and in vivo CD34+ HSPCs transplanted NSG-BM. Both in vitro and in vivo studies demonstrated that CD123+CD371+CD115–CD66b– cells contained CD116/CD1c/CD73 positive cells (Supplementary Fig. 2. C). Excluding CD116/CD1c/CD73 positive cells from the CD115– progenitors could improve neutrophil purity.
Subsequently, we sorted CD34mid CD371+, CD34low CD371+ and CD66b– CD371+, CD66b+ CD371+ cells (Fig. 2. I) within the CD115/CD116/CD73/CD1c– CD123mid CD38+ population derived from CD371+ CD115– GMPs. CD34mid CD371+ progenitors gave rise to approximately 80% neutrophils, while CD34– CD371+ CD66b– late-committed neutrophil progenitors produced more than 90% neutrophils (Fig. 2. J). In addition, we also sorted equal populations from CB-transplanted B-NDG-SM (expressing human SCF and GM-CSF) BM (Fig. 2. K). In a 6–9 days culture of the sorted populations, we found that CD34low CD371+ and CD34– CD371+ cells produced CD15+ CD66b+ neutrophils of high purity (Fig. 2 L). Conclusively, we preliminary validated the differentiation model of neutrophils, in which CD371+ CD115– GMPs gradually lose monocyte differentiation potential to become neutrophil progenitors (Fig. 2. H).
3.3. Single-cell transcriptome reveals the characteristics of neutrophil commitment
To reveal the commitment characteristics of neutrophil progenitors from a transcriptome perspective, we performed single-cell RNA-seq to evaluate transcriptomic changes. We sorted CD371+ CD34+ CD115– GMPs, CD115– CD34mid CD371+ early neutrophil biased progenitors (ENbPs), CD115– CD34low CD371+ late neutrophil biased progenitors (LNbPs), and CD34mid CD115+ monocyte progenitors (MoPs) (Fig. 2. I, Supplementary Fig. 3. A) to generate dsDNA from mRNA as previously reported [28], and constructed libraries for sequencing with 1 ng dsDNA [21,33]. We performed principal component analysis to determine the relationship of CD34mid CD115+ MoPs, CD34low CD371+ ENbPs, CD34– CD371+ LNbPs, and CD371+ CD115– GMPs (Fig. 3. A). First, we analyzed the expression pattern of previously reported neutrophil progenitor high expression markers [4]. The high expression surface markers were expressed in ENbPs, LNbPs and MoPs (Supplementary Fig. 3. B). Differential expression genes (DEGs) (p < 0.01, log2foldchange >2) analysis was performed to screen out genes uniquely expressed in neutrophils. Only a set of upregulated genes was uniquely expressed in MoPs, but not in ENbPs or LNbPs compared with CD371+ CD115– GMPs (Fig. 3. B).
Fig. 3.
Dissecting the transcriptome characterization of neutrophil differentiation from CD115–CD371+GMPs at the single-cell level.
(A) Principal component analysis (PCA) of CD115– CD371+ GMPs, CD115– CD371+ GMP-derived CD34mid ENbPs, CD34low LNbPs and CD34mid CD115+ MoPs. The CPM (count per million) values of genes expressed by CD115– CD371+ GMPs (n = 40), CD34low ENbPs (n = 29), CD34low LNbPs (n = 34) and CD34+ CD115+ MoPs (n = 24) were calculated by pcaMethod packages.
(B) Heatmaps showing the expression pattern of different populations by all DEGs (p-value < 0.01, log2 fold change >2).
(C) Heatmaps representing the differential expression of surface markers in CD371+ CD115–GMPs, MoPs, ENbPs and LNbPs.
(D) Density plots presenting the expression levels of representative differentially expressed transcription factors.
(E) GO enrichment analysis of genes upregulated in CD34mid neutrophil-biased progenitors relative to CD371+ GMPs.
(F) GSEA analysis of the active signals in CD371+ CD115–GMPs (GMP), MoPs, and ENbPs.
(G) Violin plots indicating the expression levels of granule generation-related genes in neutrophil-biased progenitors and neutrophil progenitors.
(H–I) Representative flow cytometry plots of CD15 expression on CD371– CD115– GMPs, CD371+ CD115– GMPs, Lin– CD116– CD1c– CD73– CD115– CD123mid CD371+ CD34mid ENbPs, Lin– CD116– CD1c– CD73– CD115– CD123mid CD371+ CD34low LNbPs and CD115+ CD34mid MoPs derived from CD371+ GMP cultures. The numbers indicate the percentages.
(J–K) Neutrophil differentiation potentials of CD34+ CD15– (NbP1), CD34mid CD15mid (NbP2), and CD34low/–CD15+ (NbP3) sorted from Lin/CD116/CD1c/CD73– CD115– CD123mid CD371+ cells at day 3 of CD371+ CD115– GMP culture.
(L) MGG staining assays indicating the morphologies of defined NbP1s, NbP2s, NbP3s, and neutrophils. Scale bar = 10 μm. Data in (K) are represented as means ± SD.
To further identify the characteristics of neutrophil commitment, we analyzed the expression pattern of neutrophil surface markers and transcription factors. Differentially expressed surface markers revealed that in CD115+CD34mid MoPs, the surface markers CD9, KDR, IGF2R, TLR8, CD1d, CD86, CD300A, FCGR3A(CD16) were specifically upregulated, and that FCGR1A(CD64). CSF3R, TLR2, TLR4, CXCR4, and CD84 were moderately expressed in ENbPs and LNbPs (Fig. 3. C). Furthermore, we screened out differentially (p < 0.01, log2foldchange >2) expressed transcription factors, revealing that the monocyte- or dendritic cell-related transcription factors MAFB [34,35], MAF [36], PBX1, HOXB6 [37], IRF8 [38,39] were upregulated in MoPs (Fig. 3. D. (I)), while the reported mice neutrophil master regulator factor CEBPE [40] was expressed in both MoPs and neutrophil progenitors (Fig. 3. D.(I)). Unexpectedly, only three transcription factors, ZBTB1, ATF1, and MLLT3, were differentially expressed in ENbPs and LNbPs (Fig. 3. D. (II)). However, gene ontology (GO) enrichment analysis of genes upregulated in ENbPs (p < 0.01, log2foldchange >2) relative to those in CD371+ CD115– GMPs indicated that neutrophil degranulation, neutrophil activation, and neutrophil-mediated immunity genes were upregulated in neutrophil-biased progenitors (Fig. 3. E). To analyze the signals altered by differentiation of CD371+ CD115– GMPs into MoPs and ENbPs, we performed gene set enrichment analysis (GSEA). rRNA catabolic processing-, rRNA binding-, and mRNA decay-related signals were upregulated in both MoPs and ENbPs, with more robust signal changes in MoPs relative to ENbPs. In addition, both CD371+ CD115– GMP and ENbPs highly expressed genes related to RNA splicing signaling pathways, while ENbPs had stronger mRNA splicing signals (Fig. 3. F, Supplementary Fig. 3. C).
Additionally, we sorted CD116– CD1c– CD115– CD123+ CD34– CD66b– CD371+ and CD66b+ CD371+ cells (Supplementary Fig. 3. A) from cultures of CD371+ CD115– GMPs at day 7 for single-cell RNA-seq. Among the four populations, only 75 DEGs were identified. Among DEGs, only granule-related genes and genes highly expressed in neutrophils, including LPO, MPO, LYZ [41], ELANE, PRTN3, CSTG [42], CST7 [43], and CSTA were upregulated by differentiation into neutrophils (Fig. 3. G). Taken together, these findings indicated that neutrophil-biased progenitors had a similar transcriptome pattern, suggesting that the further differentiation of neutrophil-biased progenitor cells into committed neutrophil progenitor cells could be regulated by post-transcriptional modifications.
To further identify the specific surface markers for MoPs and neutrophil-biased progenitors, we used flow cytometry to analyze the surface expression of CD15, FCGR1A (CD64), TLR2, TLR4, CXCR4, and CSF3R (CD114), which were upregulated in MoPs, ENbPs, and LNbPs relative to CD371+ CD115– GMPs (Fig. 3. C). Although both CD64 and CD15 were significantly upregulated in ENbP to LNbP (Supplementary Fig. 3. D), CD64 was expressed in both monocytes and neutrophils [44] and moderately expressed in neutrophil committed progenitors [6], so we further analyzed the expression of CD15 in the process of CD371+CD115– GMPs to LNbPs. Surprisingly, CD15 was first expressed at low levels in CD371+ CD115– GMPs, and highly expressed in LNbPs (Fig. 3 H-I). Thus, we postulated that the expression level of CD15 could be indicative of the neutrophil differentiation potential of neutrophil-biased progenitors. Sequentially, we sorted CD34+ CD15– (NbP1), CD34mid CD15mid (NbP2), and CD34low/– CD15+ (NbP3) cells gated from CD1c/CD73/CD116– CD115–CD123+CD371+ cells derived from CD371+ GMP cultures at day 3 to analyze the neutrophil differentiation potential. The neutrophil differentiation potential was enhanced, together with increased expression of CD15 and decreased expression of CD34 (Fig. 3 J-K). A morphology assay of the different stages of neutrophil differentiation also indicated a gradual change in cell morphology (Fig. 3 L). Together, these findings indicated that CD15 expression is specific to the neutrophil differentiation potential of neutrophil-biased progenitors.
3.4. Dissecting the characteristics of the human neutrophil lineage commitment process in adult bone marrow at the single-cell level
To further validate the commitment characteristics of neutrophil-biased progenitors in adult human BM, we analyzed the expression pattern of CD34 and CD15 gated from Lin/CD1c/CD116/CD73/CD115– CD371+ CD123+ CD45RA+ cells by flow cytometry. The same expression pattern of CD34 and CD15 was observed in human adult BM as in the in vitro model (Fig. 4. A). In addition, neutrophil and monocyte differentiation potential assays of CD34+ CD15– NbP1, CD34mid CD15mid NbP2 and CD34low/– CD15+ NbP3 cells indicated that human neutrophil progenitors also committed gradually with decreased CD34 and increased CD15 expression (Fig. 4. B), which was consistent with the in vitro results (Fig. 3. K).
Fig. 4.
Characteristics of neutrophil differentiation in human bone marrow at the single-cell level.
(A) Representative flow cytometry plots of neutrophil-biased progenitors in adult human BM.
(B) Neutrophil differentiation potential analyses of CD34+ CD15– NbP1, CD34mid CD15mid NbP2s and CD34low/– CD15+ NbP3s sorted from adult human BM Lin/CD116/CD1c/CD73– CD115– CD371+ CD123mid CD45RA+ cells. Flow cytometry assay to performed to analyze the CD11b+CD15+CD66b+CD14– neutrophils and CD11b+ CD66b–CD14+ monocytes in total cells at day 6 to day 10.
(C) Unsupervised cluster analysis of bulk cell RNA-seq data from neutrophil-biased progenitors and monocyte progenitors derived from adult human BM and cultures of CB CD371+ GMPs at day 3.
(D) Heat maps showing representative highly expressed genes in neutrophil-biased progenitors.
(E–F) UMAP analysis of representative highly expressed genes in CD371+ GMPs, neutrophil related progenitors (NeuP1 and NeuP2), MoP related progenitors (MoP1, and MoP2).
(G) Heatmaps presenting highly expressed genes in adult human bone marrow-derived neutrophil related progenitors (NeuP1 and NeuP2).
(H) GO enrichment analysis of DEGs in CD371+ GMPs compared to MoPs and NeuPs compared to MoPs.
(I) Single-cell trajectory analysis of neutrophil progenitors and monocyte progenitors derived from human adult BM CD371+ GMP, NeuP1, NeuP2, MoP1, MoP2 and MoP, NbP1, NbP2, NbP3, CD66b+ neutrophils derived from CB CD115– CD371+ GMPs. Data in (B) are represented as the means ± SD.
Unsupervised cluster analysis of bulk cell RNA-seq data of MoPs and neutrophil-biased progenitors both in vivo and in vitro indicated that neutrophil-biased progenitors had similar transcriptomic characteristics to those of MoPs (Fig. 4. C). We performed DEG analysis to screen a list of genes that are highly expressed in neutrophil-biased progenitors (Supplementary Fig. 4. A). We found that both in vivo- and in vitro-derived neutrophil-biased progenitors highly expressed neutrophil uni-expressed genes or granule-related genes, including MPO, LRG1 [45], PRTN3, ELANE, CTSG [42], CST7 [43], CSTA [46], and CLEC11A [47] (Fig. 4. D).
To further delineate the neutrophil commitment characteristics in adult human BM, we performed single-cell RNA-seq of sorted Lin/CD116/CD1C/CD73– CD123+ CD371+ populations (Supplementary Fig. 4. B) that contained neutrophil-biased progenitors at different stages of differentiation. The Seurat R software package was used for cluster identification (Supplementary figure 4 C-D). We first excluded populations of dendritic progenitor cells that highly expressed CD1c, CCR6 [48], CLEC10A [49], CD1E [50], CLECL1 [51], GZMB [52] (Supplementary Fig. 4. E). Subsequently, GATA2, HOXA9, HOXA10, HOXB4, MPO, PRTN3, ELANE, CLEC11A, MAFB, CD115, and CD14 were used to identify early CD371+ GMPs, neutrophil related progenitors named NeuP1 and NeuP2 (not equal to the NbP1, NbP2 and NbP3 defined by CD34 and CD15 at protein level), and monocyte progenitors named MoP1 and MoP2 (not equal to CD34mid CD115+MoPs). CD371+ GMPs exhibited high expression of the transcription factors GATA2, HOXA9, HOXA10, HOXB4, and HOXA7, and neutrophil related progenitors (NeuP1, NeuP2) exhibited high expression of CD15 (FUT4), PRTN3, MPO, ELANE, MLLT3, and CLEC11A. Monocyte progenitors highly expressed MAFF, MAFB, CD115 (CSF1R), CD14, CCL3, and CCR1 (Fig. 4 E-F). Further, monocyte progenitors also expressed surface markers that were highly expressed in MoPs in vitro, including TLR2, TLR4, IGF2R, FCGR2A, FCGR2B, CCR1, CD226, CD267, and CD86 (Supplementary Fig. 4. F, Fig. 3. C). Furthermore, neutrophil-related progenitors highly expressed DNA replication- and myeloid differentiation-related transcription factors, including TCF4, which is related to granulopoiesis [53] (Fig. 4. G, Supplementary Fig. 4. G). In addition, GO enrichment analysis revealed that DEGs in CD371+ CD115– GMP cells compared to MoPs and NeuPs compared to MoPs were also mainly related to mRNA processing and rRNA processing (Fig. 4. H), which was similar to in vitro signal changes (Fig. 3. F). Finally, a single-cell trajectory assay was performed to evaluate the relationship between CD371+ CD115– GMPs, neutrophil-related progenitors, and monocyte progenitors. Both in vivo and in vitro-derived cells shared a similar cell trajectory, and neutrophil progenitors were more similar to CD371+ CD115– GMPs (Fig. 4. I). Together, these results indicated that the transcriptomes of CD371+ CD115– GMPs, neutrophil-related progenitors, and monocyte progenitors had similar developmental characteristics both in vivo and in vitro.
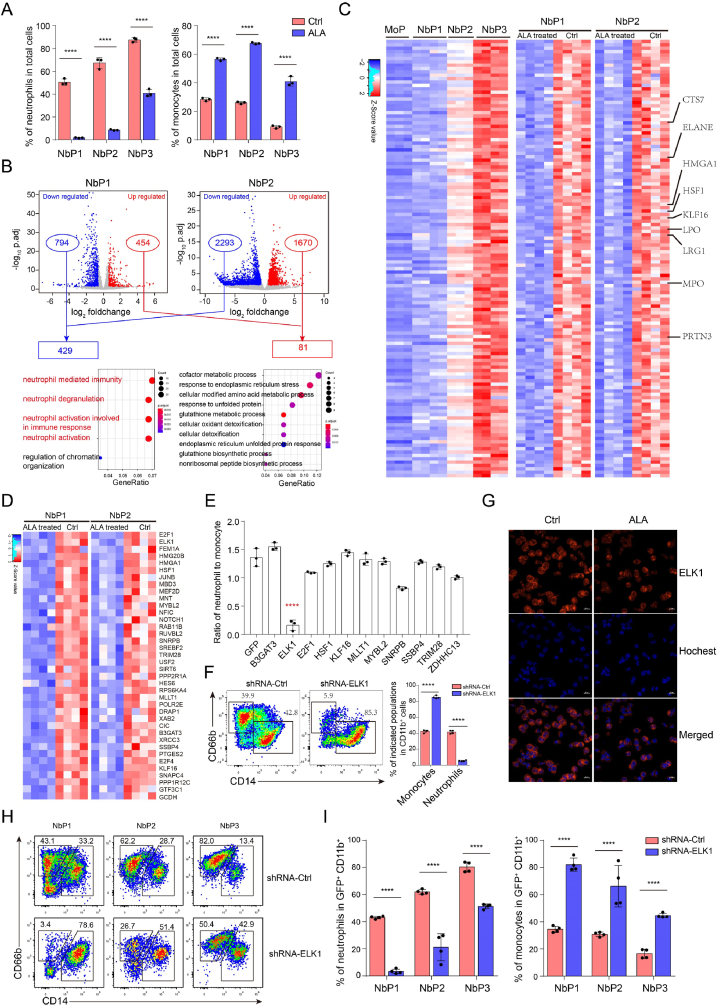
3.5. ALA targets ELK1 to regulate neutrophil differentiation
To further identify the key regulatory genes of neutrophil differentiation, we analyzed the effects of ALA on neutrophil differentiation of neutrophil-biased progenitor cells (CD34+ CD15– NbP1, CD34mid CD15mid NbP2, CD34low/– CD15+ NbP3) (Supplementary Fig. 5. A) derived from CD371+ CD115– GMPs at day 3. ALA significantly impeded neutrophil differentiation from NbP1, NbP2, and NbP3 cells, with the most potent blockage of NbP1 cells giving rise to neutrophils (Fig. 5. A). Based on this model, we performed bulk cell RNA-seq to analyze the transcriptomic changes of NbP1 and NbP2 cells ± ALA. Downregulated DEGs were 5-fold greater than upregulated DEGs (Fig. 5. B). The downregulated genes were more related to neutrophil activity (Fig. 5. B). About 120 neutrophil-specific genes were downregulated by ALA, including PRTN3 [54], LRG1 [45], CEBPE [55], LPO, MPO, ELANE [6,56], CLEC11A [57] and KLF16, CTS7, HMGA1 (Fig. 5. C, Supplementary Fig. 5. B).
Fig. 5.
ALA regulates commitment of neutrophil progenitors by targeting ELK1.
(A) Statistical analysis of neutrophils and monocytes differentiated from NbP1s, NbP2s, and NbP3s ± ALA treatment. To analyze the effect of ALA on neutrophil differentiation, sorted neutrophil biased progenitors were cultured ± ALA and flow cytometry assay to performed to analyze the CD11b+CD15+CD66b+CD14– neutrophils and CD11b+ CD66b–CD14+ monocytes at day6 to day14.
(B) Volcano plots and GO enrichment analysis of genes upregulated and downregulated by ALA.
(C) Heatmaps showing ALA-downregulated genes among neutrophil-biased progenitor-specific genes.
(D) Heatmaps presenting ALA-downregulated transcription factors in NbP1s and NbP2s.
(E) Flow cytometry analysis of neutrophils and monocytes derived from CD34+ HSPCs transduced with shRNAs targeting ALA-downregulated transcription factors. The ratio of neutrophils to monocytes is presented to analyze the neutrophil differentiation potential of CD34+ HSPCs transduced with different shRNAs.
(F) Flow cytometry plots of neutrophils and monocytes from ELK1-shRNA knockdown CD34+ progenitors. Flow cytometry analysis was performed on day 9, and neutrophils and monocytes were analyzed with CD11b+ gated cells. Representative flow cytometry plots and statistical analysis bar plots are shown.
(G) Immunofluorescence analysis of ELK1 was performed to measure ELK1 protein levels in NbP1 cells ± ALA treatment. Scale bar = 20 μm.
(H–I)In vitro flow cytometry analysis of neutrophils and monocytes from ELK1-shRNA-transduced NbP1, NbP2, and NbP3 cells. Flow cytometry analysis was performed from day 6 to day 9, and neutrophils and monocytes were analyzed within GFP+ CD11b+ gated cells. Representative flow cytometry plots (G) and statistical analysis bar plots (H) are shown.
Data in (A, E, I) are expressed as means ± SD. An unpaired Student's t-test (two-tailed) was performed to assess statistical significance. NS = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, N = 3–4.
To identify the primary neutrophil commitment regulatory genes, we first cloned cDNAs of CEBPE, LRG1, PRTN3 and CLEC11A into PCCL-desRed or PCCL-EGFP overexpression vectors [27], and the shRNA target sequences of these genes into plKO.1-GFP vectors. Both overexpression and shRNA knockdown experiments indicated that CEBPE, PRTN3, LRG1, and CLEC11A did not affect human neutrophil differentiation (Supplementary Fig. 5. C). Given that granule-related genes MPO, ELANE, PRTN3, LRG1, and CLEC11A do not affect neutrophil differentiation, we analyzed ALA-downregulated transcription factors in NbP1 and NbP2 cells. We identified 38 differentially expressed (p.adj, < 0.05; foldchange, > 1.5) transcription factors (Fig. 5. D). GO enrichment analysis demonstrated that these transcription factors were related to epigenetic remodeling events such as chromosome organization, chromatin remodeling, and heterochromatin assembly (Supplementary Fig. 5. D), including ELK1 reported to interact with CSF-1R (CD115) [58]and regulate micro-RNA expression in monocyte differentiation [59]. To determine the function of these transcription factors in neutrophil differentiation, overexpression of XRCC3, TMEM176B, PBX3, MDM1, MDB3, HMGA1 and knockdown of B3GAT3, ELK1, E2F1, HSF1, KLF16, MLLT1, MYBL2, SNRPB, SSBP4, TRIM28, and ZDHHC13 in CD34+ HSPCs was performed to analyze neutrophil differentiation potential. Overexpression of XRCC3, TMEM176B, PBX3, MDM1, MDB3, HMGA1 (Supplementary Fig. 5. E) and knocking down of B3GAT3, E2F1, HSF1, KLF16, MLLT1, MYBL2, SNRPB, SSBP4, TRIM28, and ZDHHC13 did not affect neutrophil differentiation (Fig. 5. E), while ELK1 knocking down significantly impeded neutrophil differentiation and promoted monocyte differentiation (Fig. 5 E-F). Additionally, ALA downregulated ELK1 expression at the protein level, as demonstrated by immunofluorescence analysis (Fig. 5. G).
To reveal the roles of ELK1 in early neutrophil progenitor commitment, we analyzed the neutrophil differentiation potential of ELK1-shRNA transduced NbP1, NbP2, and NbP3 cells. Knocking down ELK1 had the same effect as ALA on the commitment of neutrophil progenitors (Fig. 5. A, H–I). Taken together, these findings indicated that ALA impeded neutrophil differentiation by targeting ELK1, and that ELK1 was required for granulopoiesis.
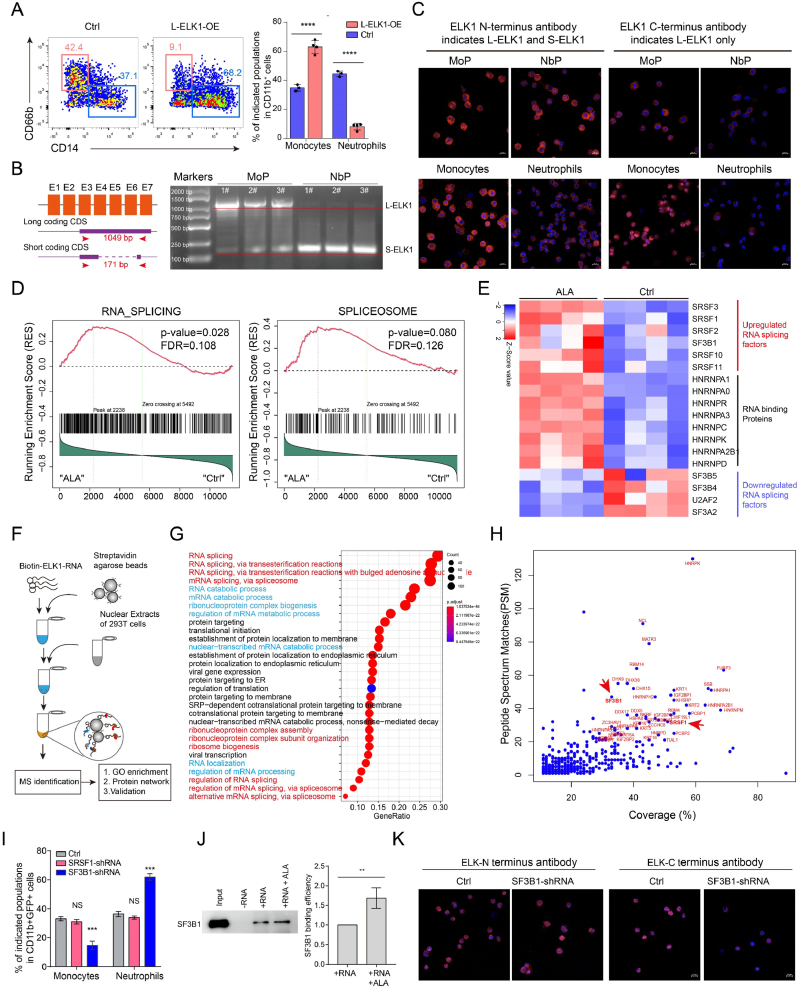
3.6. SF3B1 regulates ELK1 RNA splicing
Because ELK1 is essential for human neutrophil differentiation, we overexpressed ELK1 in CD34+ HSPCs to increase neutrophil differentiation. Unexpectedly, overexpression of the ELK1 long coding protein isoform (L-ELK1) also impeded neutrophil differentiation (Fig. 6. A). Thus, we hypothesized that different ELK1 isoforms could play different roles in balancing neutrophil and monocyte differentiation. PCR analysis of ELK1 isoforms indicated that CD34mid MoPs highly expressed theL-ELK1, while neutrophil-biased progenitors highly expressed the short coding protein isoform (S-ELK1) (Fig. 6. B). Furthermore, immunofluorescence staining for L-ELK1 indicated by the ELK1 C- terminus antibody and S-ELK1 (absent the C- terminus domain) indicated by ELK1 N- terminus antibody (measure both L-ELK1 and S-ELK1) deduct C- terminus antibody (measure L-ELK1 only) showed that monocyte progenitors and monocytes highly expressed L-ELK1, while neutrophil progenitors and neutrophils highly expressed S-ELK1 (Fig. 6. C). Further analysis demonstrated that ALA downregulated the expression of S-ELK1 but not L-ELK1 (Supplementary Fig. 6. A).
Fig. 6.
SF3B1 regulates ELK1 mRNA splicing.
(A) Flow cytometry analysis of neutrophils and monocytes derived from L-ELK1-overexpressing CD115- CD34+ progenitors. Flow cytometry analysis was performed on day 9.
(B). Identification of different ELK1 isoforms in MoPs and NbPs by PCR. Ten nanograms of dsDNA generated from MoPs or neutrophil-biased progenitors (NbPs) was used for PCR.
(C) Immunofluorescence for ELK1 N-terminus and ELK1 C terminus antibodies were performed to assess L-ELK1 and S-ELK1 expression levels in MoPs and neutrophil biased progenitor (NbP2, NbP3) cells. For the absence of C-terminus domain of S-ELK1, the C-terminus antibody indicates L-ELK1 only and the ELK1 N-terminus antibody indicates both L-ELK1 and S-ELK1.
(D) GSEA was performed to analyze the effect of ALA on RNA splicing-related signal changes in NbP1.
(E) Heatmaps present ALA-upregulated and ALA-downregulated RNA splicing-related genes.
(F) Flow diagram for mass spectrometry (MS) analysis of ELK1 RNA pull-down proteins.
(G) GO enrichment analysis of ELK1 RNA pull-down proteins. Proteins with coverage >10% (391 proteins) were subjected to GO enrichment analysis.
(H) Scatter plots showing the relationship of peptide-spectral matches (PSM) and protein coverage. Proteins with coverage >30% and PSM >20 are denoted by red gene names.
(I) Statistical analysis of the proportions of neutrophils and monocytes derived from SRSF1 shRNA- and SF3B1 shRNA-transduced CD371+ CD115- GMPs. Flow cytometry analysis was performed on day 14 of differentiation.
(J) RNA pull-down and immunoblotting were performed to measure the interaction of ELK1 RNA and SF3B1 under ALA treatment.
(K) Immunofluorescence staining for ELK1 N-terminus and ELK1 C- terminus antibodies were performed to assess L-ELK1 and S-ELK1 expression levels in SF3B1-shRNA transduced NbP1 cells and Ctrl NbP1 cells. For the absence of C-terminus domain of S-ELK1, the C- terminus antibody indicates L-ELK1 only and the ELK1 N-terminus antibody indicates L-ELK1 and S-ELK1 Data in (A, I) are represented as the means ± SD. An unpaired Student's t-test (two-tailed) was performed to determine statistical significance. NS = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, N = 3–7. (C, J) Scale bar = 20 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Because single-cell transcriptome findings demonstrated that ENbPs had stronger RNA splicing signals (Fig. 3. E), we further analyzed the changes in ELK1 RNA splicing ± ALA. The results indicated that ALA significantly changed RNA splicing and spliceosome-related signals (Fig. 6. D). DEG analysis revealed that ALA-upregulated splicing factors such as SRSF3, SRSF11, SF3B1, and downregulated SF3B4 and SF3B5, which were differentially expressed in MoPs and neutrophil-biased progenitors (Fig. 6. E, Supplementary Fig. 6. B). Furthermore, mass spectrometry analysis of ELK1 RNA pull-down proteins demonstrated that most of the pulled-down proteins were related to RNA splicing and RNA binding (Fig. 6 F-G). Further, the ALA-upregulated splicing factors SF3B1 and SRSF1 accounted for a high proportion of the identified ELK RNA-binding proteins (Fig. 6. H). Thus, we determined the functions of SRSF1 and SF3B1 in neutrophil differentiation. SF3B1 knockdown significantly increased the production of neutrophils and decreased monocyte percentages (Fig. 6. I, Supplementary Fig. 6. C).
We postulated that ELK1 isoform expression could be controlled by SF3B1. We analyzed the RNA-protein interaction of ELK1 RNA and SF3B1 ± ALA. These findings demonstrated that SF3B1 bound the splicing site of ELK1 mRNA, and that treatment with ALA increased the binding of SF3B1 to ELK1 mRNA (Fig. 6. J). In addition, SF3B1 knockdown partially impaired the effects of ALA on neutrophil and monocyte differentiation (Supplementary figure 6 C-D). Immunofluorescence also indicated that SF3B1 downregulated L-ELK1 and upregulated S-ELK1 (Fig. 6. K). Taken together, these findings demonstrated that SF3B1 regulated neutrophil and monocyte differentiation from CD371+ CD115– GMPs by regulating expression of S-ELK1 and L-ELK1.
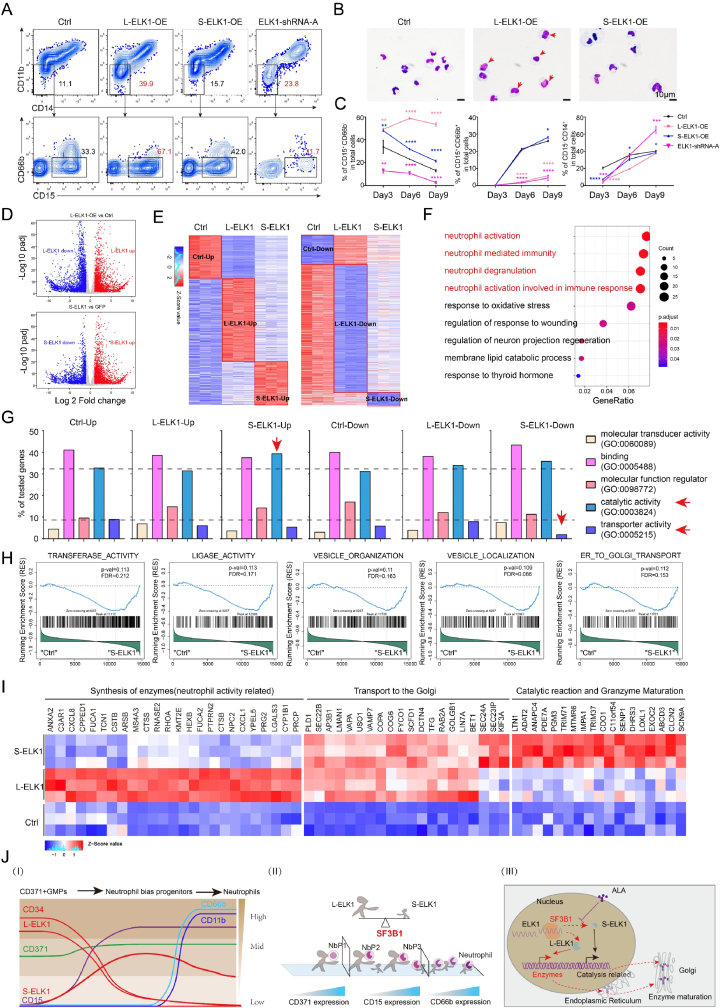
3.7. L-ELK1 and S-ELK1 have differential roles in human neutrophil differentiation
To analyze the effects of L-ELK1 and S-ELK1 on neutrophil and monocyte differentiation derived from CD34+ HSPCs, we sorted Ctrl (GFP control virus), L-ELK1, S-ELK1 and ELK1-shRNA-A and ELK1-shRNA-L transduced CD34+ HSPCs to perform in vitro differentiation culture or transplant into NDG-SM mice. Both in vivo and in vitro data demonstrated that L-ELK1 overexpression impaired neutrophil differentiation, while knockdown of only L-ELK1 enhanced neutrophil differentiation (Supplementary figure 7 A-B). However, flow cytometric analysis of NbP1 cells overexpressing of L-ELK1 and S-ELK1 or knocking down of ELK1 demonstrated that L-ELK1 inhibited CD15+CD66b+ expression but did not block expression of CD11b– CD14– CD66b– CD15+, a characteristic of NbP1s (Fig. 7. A). Thus, we postulated that L-ELK1 was required for early commitment of neutrophil progenitors (NbP1, NbP2, and NbP3), but not for committed neutrophil progenitors (CD15+ CD66b+). Giemsa staining also revealed that overexpression of L-ELK1 suppressed most of the cells at the CD115– CD34– CD11b– CD15+ NbP3 stage (Fig. 7. B). Subsequently, we sorted CD115– CD34+ CD371+ CD15– NbP1s from L-ELK1-OE-, S-ELK1-OE-, and ELK1-shRNA-A-transduced progenitors at day 3 for further differentiation analyses. Both L-ELK1 and S-ELK1 overexpression upregulated CD15, while ELK1 knockdown downregulated CD15 at an early stage (Fig. 7. C). Overexpression of L-ELK1 or knockdown of ELK1 significantly decreased CD66b expression. In addition, overexpression of L-ELK1 and S-ELK1 did not block CD14+ monocyte differentiation, while ELK1 knockdown enhanced monocyte differentiation (Fig. 7. C). This indicated that L-ELK1 was required for early neutrophil progenitor commitment and was gradually downregulated with commitment.
Fig. 7.
L-ELK1 and S-ELK1 play different roles in neutrophil commitment.
(A) Representative flow cytometry plots of CD15+ CD66b+ neutrophils and CD15+ CD66b− neutrophil progenitors derived from Ctrl (GFP empty vector), L-ELK1 overexpression (L-ELK1-OE), S-ELK1 overexpression (S-ELK1-OE), and ELK1-shRNA-A viruses transduced NbP1 progenitors. Flow cytometry was performed on day 9 of culture.
(B) Giemsa staining of the morphology of CD14– CD15+ cells derived from L-ELK1- and S-ELK1-overexpressing NbP1 progenitors on day 9. Arrows indicate immature neutrophil progenitors. Scale bar = 10 μm (C) Regular analysis of CD15– CD66b–, CD15+ CD66b+, and CD15– CD14+ expression percentages derived from Ctrl (GFP empty vector), L-ELK1 overexpression, S-ELK1 overexpression, and ELK1-shRNA-A knockdown viruses transduced NbP1 progenitors.
(D) Volcano plots were used to analyze differential expression of genes in Ctrl, L-ELK1- and S-ELK1-overexpressing NbP1 progenitors.
(E) Heatmaps presenting upregulated and downregulated gene sets in Ctrl-, L-ELK1-, and S-ELK1-overexpressing NbP1 progenitors.
(F) GO enrichment analysis of the L-ELK1-specific upregulated genes.
(G) Gene ontology category analysis of specific upregulated and downregulated gene sets in Ctrl-, L-ELK1- and S-ELK1-overexpressing NbP1 progenitors. Gene ontology category analysis was performed using http://www.pantherdb.org/.
(H) GSEA was used to analyze signal changes in S-ELK1-overexpressing NbP1 progenitors.
(I) Heatmaps present the L-ELK1- and S-ELK1-upregulated genes related to enzyme synthesis, endoplasmic reticulum to Golgi transport, and enzyme catalytic reaction signals.
(J) Diagram of the marker-changing model in CD371+ CD115– GMPs differentiating into neutrophil progenitors (I), regulation of the SF3B1-ELK1 signaling axis (II), and the working model of the roles of L-ELK1 and S-ELK1 in the different differentiation stages of neutrophils (III).
To further identify the key signals altered by L-ELK1 or S-ELK1 in neutrophil differentiation, we performed RNA-seq analysis of L-ELK1- and S-ELK1-transduced CD34+ CD371+ CD115– CD123+ CD38+ progenitors (CD371+ CD115– GMPs). DEG (Fig. 7. D) (p.adj, < 0.05; log2 fold change, > 1) analysis revealed that over 75% of DEGs were upregulated or downregulated in both L-ELK1- and S-ELK1-transduced CD371+ CD115– GMPs (Supplementary Fig. 7. C). Because L-ELK1 and S-ELK1 have differential roles in regulating neutrophil or monocyte differentiation, we further analyzed DEGs in L-ELK1, S-ELK1 or Ctrl cells (Fig. 7. E). Surprisingly, GO enrichment analysis revealed that genes specifically upregulated by L-ELK1 were significantly enriched for neutrophil activation, neutrophil-mediated immunity, and neutrophil degranulation (Fig. 7. F). This trend was not observed for S-ELK1 DEGs. GO function classification revealed that S-ELK1 upregulated catalytic activity-related genes and downregulated transporter activity-related genes (Fig. 7. G). GSEA also demonstrated that S-ELK1 significantly changed genes related to catalytic processing, ligase activity, and ER-to-Golgi transport (Fig. 7. H). Further DEG analysis revealed that L-ELK1 increased genes related to enzyme synthesis (CTSS, CTSB, CSTB, HEXB, FUCA1 PRCP, etc.) and endoplasmic reticulum to Golgi transport (COPA, COG6, GOLGB1, BET1, LMAN1, USO1, VAPA, BET1, etc.), while S-ELK1 upregulated additional genes related to endoplasmic reticulum to Golgi transport (SEC24A, SEC23IP and KIF3A), catalytic reactions, and enzyme maturation-related genes (LTN1, TRIM37, TRIM71, and LOXL1) (Fig. 7. I).
In summary, the expression of the surface marker CD15 can be used to define the degree of neutrophil progenitor fate determination (Fig. 7. J. (I)). Further, the splicing factor SF3B1 directly regulates mRNA processing of ELK1 to produce L-ELK1 and S-ELK1, and S-ELK1 was upregulated to replace L-ELK1, pushing uncommitted neutrophil-biased progenitors into neutrophil progenitors in the differentiation of CD371+ CD115– GMPs into neutrophil progenitors (Fig. 7. J. (II)-(III)). Taken together, the results indicate that ELK1 is an effective regulatory gene for neutrophil fate determination, and its function is regulated by multiple factors, including the splicing factor SF3B1.
4. Discussion
Neutrophils are well-known to be derived from GMPs. Although mouse neutrophil progenitors have been found in GMPs [4] and the absence of CEBPE causes neutrophil deficiency in vivo [40], neither CEBPE mutation [55] nor knockdown results in the disappearance of human neutrophils. Additionally, intravenous immunoglobulin regulates the survival of human but not mouse neutrophils [60]. All these studies show that there exists a difference in the differentiation and function of human and mouse neutrophils. Recently, neutrophil uni-potential progenitors have been identified in fetal bone marrow as Lin− CD15low/+ CD66b+ SSClow CD49d+ [4] or as Lin– CD66b+ in adult bone marrow [5]. Excluding CD371highCD64high [32]and CD64high CD115+ cells [6] from the GMPs would improve the percentage of progenitors holding neutrophil potential. Here we established an inducible neutrophil differentiation by ALA and dissected the process of neutrophil lineage differentiation from cord blood CD371+CD115– GMPs (contains both the Lin– CD64dim CD115– CD34+ CD45RA+ NCPs [6] and GMDP [61]). Although MPO, ELANE, PRTN3 and ELANE are upregulated in the reported NCPs [6] and NbPs (Supplementary Fig. 8), these genes are not the neutrophil lineage early cell fate decision genes.
Post-transcriptional modification is widely reported to influence cell metabolism and cell fate [62], such as the mitochondrial metabolism drives erythropoiesis [63] and post-transcriptional networks regulating the transition of myeloblasts to monocytes [9]. In addition, alternative splicing has been reported to play important roles in regulating erythropoiesis [10], granulopoiesis [11], and monocyte-to-macrophage differentiation [64]. In our study, single-cell transcriptome analysis revealed that the transcriptomes of ENbPs and GMPs are more similar than that of MoPs. Most DEGs in ENbPs relative to those in GMPs were RNA binding and RNA splicing-related genes, suggesting that neutrophil maturation could be controlled by post-transcriptional, post-translational, or metabolic regulation. Using the ALA-induced neutrophil deficiency model, we found that ALA significantly changed RNA splicing signals. Based on this analysis, we demonstrated that the critical signal in neutrophil commitment is SF3B1-regulated splicing of ELK1 isoforms to balance neutrophil and monocyte differentiation, with the short isoform regulating neutrophil differentiation and the long isoform regulating monocyte differentiation.
Prior reports have demonstrated that different isoforms of ELK1 have opposite roles in neuronal differentiation [65]. The present study identified differential roles for L-ELK1 and S-ELK1 in the neutrophil commitment. Both L-ELK1 and S-ELK1 played important roles in the commitment of neutrophil progenitors. Normally, ELK1 is required for GMPs to differentiate into neutrophil biased progenitors. Overexpression of L-ELK1 or S-ELK1 increases the production of CD15+ CD66b– late neutrophil-biased progenitors derived from CD371+ CD15– NbP1 cells. However, overexpression of L-ELK1 impeded differentiation of CD115– CD15+ neutrophil-biased progenitors into CD15+ CD66b+ neutrophil progenitors, while knocking down L-ELK1 enhanced differentiation of CD115– CD15+ neutrophil-biased progenitors into CD15+ CD66b+ neutrophil progenitors.
Numerous prior studies have demonstrated that SF3B1 has critical functions in hematopoiesis, such as myeloid differentiation [66] and erythropoiesis [67], and SF3B1 mutations are commonly detected in MDS (myelodysplastic syndromes) [68]. However, the function of SF3B1 in neutrophil lineage commitment has not been reported. In the present study, we identified that SF3B1 directly binds ELK1 RNA to regulate the generation of L-EKL1 and S-ELK1, balancing neutrophil and monocyte differentiation. SF3B1 knockdown down-regulates L-ELK1 and up-regulates S-ELK1 to enhance neutrophil differentiation. Thus, ELK1 is a critical SF3B1 target for regulation of myeloid differentiation in GMPs.
As previous reports show that upregulation of ELK1 promotes the progression of cervical cancer [69], pancreatic cancer [70], thyroid cancer [71], colorectal cancer [72] and breast cancer [73], and suppression of ELK1 help to inhibit thyroid cancer [71], pancreatic cancer [74], breast cancer [75] and cervical cancer [76]. ALA has been applied to cancer therapy in multiple cancers [77], but it's unclear how ALA suppress cancer progression. Our founding might help to explain the mechanism of cancer therapy using ALA. And ALA could use to inhibit the progression of the cancers promoted by upregulations of ELK1. In addition, ALA could completely inhibit the neutrophil differentiation derived from hematogenic stem cells, ALA would be applied to inhibit the inflammation in many cases, such as serious inflammation caused by COVID-19.
In summary, the present study provides new insights into the regulatory mechanisms of human neutrophil lineage commitment at post-transcriptional modification by ALA, which can potentially be translated to targeted drug development for MDS, neutrophil regeneration and cancer therapy.
5. Conclusions
Overall, we established an inducible neutrophil differentiation model by using an antioxidant ALA and defined the neutrophil progenitor commitment process from CD371+ CD115– GMPs by CD34 and CD15. Moreover, we confirmed that the neutrophil transcriptomes of CD371+ CD115– GMPs, neutrophil-biased progenitors, and monocyte progenitors had similar developmental characteristics both in vivo and in vitro. Sequentially, we discovered that ELK1 was essential for human neutrophil lineage determination, SF3B1 regulated RNA splicing of L-ELK1 and L-ELK1 increased genes related to enzyme synthesis and endoplasmic reticulum to Golgi transport while S-ELK1 upregulated additional genes related to endoplasmic reticulum to Golgi transport catalytic reactions, and enzyme maturation-related genes.
Author contributions
Yong Dong conceived, initialed, and designed this project. Yong Dong and Yimeng Zhang collected all the data. Yong Dong analyzed and interpreted all the data analysis. Yongping Zhang collected the human BM. Zhenyang Lai and Qiang Chen provided cord blood. Xu Pan and Yijin Chen performed cell sorting, Ju Bai, Ya Zhou, Qiongxiu Zhou, Yonggang Zhang provided help in this research. Yong Dong, Yimeng Zhang and Feng Ma discussed the data and wrote the manuscript. Feng Ma approved the manuscript.
Funding
This research is supported by the China Postdoctoral Fund Program (grant number 2018M641265), the CAMS Initiatives for Innovative Medicine (2019-I2M-1–006, 2017-I2M−2005, 2019-RC-HL-016), the grant from the National Natural Science Foundation of China (grant number 82000119, 82170121), and the Disciplinary Construction Innovation Team Foundation of Chengdu Medical College (grant number CMC-XK-2102).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank professor Jinyong Wang for providing PCCL, PLP1 and PLP2 overexpression vectors.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102392.
Contributor Information
Yong Dong, Email: dong_yong@ibt.pumc.edu.cn.
Feng Ma, Email: mafeng@ibt.pumc.edu.cn.
Abbreviations
- ALA
Alpha lipoic acid
- HSC
Hematopoietic stem cell
- HPSC
Hematopoietic stem and progenitor cell
- GMP
Granulocyte-monocyte progenitor
- CMP
Common myeloid progenitor
- MEP
Megakaryocyte-erythrocyte progenitor
- NbP
Neutrophil-biased progenitor
- ENbP
Early neutrophil-biased progenitor
- LNbP
Late neutrophil-biased progenitor
- ELK1
ETS domain-containing protein Elk-1
- SF3B1
Splicing factor 3b, subunit 1
- SCF
Stem cell factor
- IL-3
Interleukin-3
- IL-6
Interleukin-6
- TPO
Thrombopoietin
- EPO
Erythropoietin
- FLT3L
FMS-like tyrosine kinase-3 ligand
- G-CSF
Granulocyte colony stimulating factor
- GM-CSF
Granulocyte-macrophage colony stimulating factor
- ROS
Reactive oxygen species
- LSK
Lin (CD2, CD3, CD4, CD8, CD11b, B220, Ter19, Mac1)- Sca1+ ckit+
- BM
Bone marrow
- MDS
Myelodysplastic syndromes
- NAC
N-Acetyl-L-cysteine
- NAD
N-Acetyl-d-cysteine
- VC
l-Ascorbic acid (Vitamin C)
- VE
α-Vitamin E
- GSH
l-Glutathione reduced
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Dale D.C. How I diagnose and treat neutropenia. Curr. Opin. Hematol. 2016;23(1):1–4. doi: 10.1097/MOH.0000000000000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marfin A.A., Price T.H. Granulocyte transfusion therapy. J. Intensive Care Med. 2015;30(2):79–88. doi: 10.1177/0885066613498045. [DOI] [PubMed] [Google Scholar]
- 3.Cancelas J.A. Granulocyte transfusion: questions remain. Blood. 2015;126(18):2082–2083. doi: 10.1182/blood-2015-09-669085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwok I., et al. Immunity; 2020. Combinatorial Single-Cell Analyses of Granulocyte-Monocyte Progenitor Heterogeneity Reveals an Early Uni-Potent Neutrophil Progenitor. [DOI] [PubMed] [Google Scholar]
- 5.Dinh H.Q., et al. Coexpression of CD71 and CD117 identifies an early unipotent neutrophil progenitor population in human bone marrow. Immunity. 2020;53(2):319–334 e6. doi: 10.1016/j.immuni.2020.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calzetti F., et al. CD66b(-)CD64(dim)CD115(-) cells in the human bone marrow represent neutrophil-committed progenitors. Nat. Immunol. 2022;23(5):679–691. doi: 10.1038/s41590-022-01189-z. [DOI] [PubMed] [Google Scholar]
- 7.Drissen R., et al. Identification of two distinct pathways of human myelopoiesis. Sci Immunol. 2019;4(35) doi: 10.1126/sciimmunol.aau7148. [DOI] [PubMed] [Google Scholar]
- 8.Ng L.G., Ostuni R., Hidalgo A. Heterogeneity of neutrophils. Nat. Rev. Immunol. 2019;19(4):255–265. doi: 10.1038/s41577-019-0141-8. [DOI] [PubMed] [Google Scholar]
- 9.Fontemaggi G., et al. Identification of post-transcriptional regulatory networks during myeloblast-to-monocyte differentiation transition. RNA Biol. 2015;12(7):690–700. doi: 10.1080/15476286.2015.1044194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards C.R., et al. A dynamic intron retention program in the mammalian megakaryocyte and erythrocyte lineages. Blood. 2016;127(17):e24–e34. doi: 10.1182/blood-2016-01-692764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong J.J., et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154(3):583–595. doi: 10.1016/j.cell.2013.06.052. [DOI] [PubMed] [Google Scholar]
- 12.Ye B., et al. Cell Mol Immunol; 2022. Induction of Functional Neutrophils from Mouse Fibroblasts by Thymidine through Enhancement of Tet3 Activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Solmonson A., DeBerardinis R.J. Lipoic acid metabolism and mitochondrial redox regulation. J. Biol. Chem. 2018;293(20):7522–7530. doi: 10.1074/jbc.TM117.000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moini H., Packer L., Saris N.E. Antioxidant and prooxidant activities of alpha-lipoic acid and dihydrolipoic acid. Toxicol. Appl. Pharmacol. 2002;182(1):84–90. doi: 10.1006/taap.2002.9437. [DOI] [PubMed] [Google Scholar]
- 15.Xia D., et al. Alpha lipoic acid inhibits oxidative stress-induced apoptosis by modulating of Nrf2 signalling pathway after traumatic brain injury. J. Cell Mol. Med. 2019;23(6):4088–4096. doi: 10.1111/jcmm.14296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rahimifard M., et al. Multiple protective mechanisms of alpha-lipoic acid in oxidation, apoptosis and inflammation against hydrogen peroxide induced toxicity in human lymphocytes. Mol. Cell. Biochem. 2015;403(1–2):179–186. doi: 10.1007/s11010-015-2348-8. [DOI] [PubMed] [Google Scholar]
- 17.Li Y., et al. Alpha lipoic acid protects lens from H(2)O(2)-induced cataract by inhibiting apoptosis of lens epithelial cells and inducing activation of anti-oxidative enzymes. Asian Pac. J. Trop. Med. 2013;6(7):548–551. doi: 10.1016/S1995-7645(13)60094-2. [DOI] [PubMed] [Google Scholar]
- 18.Tripathy J., et al. alpha-Lipoic acid inhibits the migration and invasion of breast cancer cells through inhibition of TGFbeta signaling. Life Sci. 2018;207:15–22. doi: 10.1016/j.lfs.2018.05.039. [DOI] [PubMed] [Google Scholar]
- 19.Peng P., et al. Alpha-lipoic acid inhibits lung cancer growth via mTOR-mediated autophagy inhibition. FEBS Open Bio. 2020;10(4):607–618. doi: 10.1002/2211-5463.12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeon M.J., et al. Alpha lipoic acid inhibits proliferation and epithelial mesenchymal transition of thyroid cancer cells. Mol. Cell. Endocrinol. 2016;419:113–123. doi: 10.1016/j.mce.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Dong Y., et al. Alpha lipoic acid promotes development of hematopoietic progenitors derived from human embryonic stem cells by antagonizing ROS signals. J. Leukoc. Biol. 2020 doi: 10.1002/JLB.1A0520-179R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wambi C., et al. Dietary antioxidants protect hematopoietic cells and improve animal survival after total-body irradiation. Radiat. Res. 2008;169(4):384–396. doi: 10.1667/RR1204.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramakrishnan N., Wolfe W.W., Catravas G.N. Radioprotection of hematopoietic tissues in mice by lipoic acid. Radiat. Res. 1992;130(3):360–365. [PubMed] [Google Scholar]
- 24.Kwak H.J., et al. Myeloid cell-derived reactive oxygen species externally regulate the proliferation of myeloid progenitors in emergency granulopoiesis. Immunity. 2015;42(1):159–171. doi: 10.1016/j.immuni.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saha S., Biswas S.K. Tumor-Associated neutrophils show phenotypic and functional divergence in human lung cancer. Cancer Cell. 2016;30(1):11–13. doi: 10.1016/j.ccell.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 26.Mao B., et al. Early development of definitive erythroblasts from human pluripotent stem cells defined by expression of glycophorin A/CD235a, CD34, and CD36. Stem Cell Rep. 2016;7(5):869–883. doi: 10.1016/j.stemcr.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang T., et al. Trim27 confers myeloid hematopoiesis competitiveness by up-regulating myeloid master genes. J. Leukoc. Biol. 2018;104(4):799–809. doi: 10.1002/JLB.1A1217-480R. [DOI] [PubMed] [Google Scholar]
- 28.Tang F., et al. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat. Protoc. 2010;5(3):516–535. doi: 10.1038/nprot.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng N., et al. Small protein hidden in lncRNA LOC90024 promotes "cancerous" RNA splicing and tumorigenesis. Adv. Sci. 2020;7(10) doi: 10.1002/advs.201903233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou S., Shi L., Lei H. Biotin-streptavidin affinity purification of RNA-protein complexes assembled in vitro. Methods Mol. Biol. 2016;1421:23–34. doi: 10.1007/978-1-4939-3591-8_3. [DOI] [PubMed] [Google Scholar]
- 31.Olweus J., Thompson P.A., Lund-Johansen F. Granulocytic and monocytic differentiation of CD34hi cells is associated with distinct changes in the expression of the PU.1- regulated molecules, CD64 and macrophage colony-stimulating factor receptor. Blood. 1996;88(10):3741–3754. [PubMed] [Google Scholar]
- 32.Kawamura S., et al. Identification of a human clonogenic progenitor with strict monocyte differentiation potential: a counterpart of mouse cMoPs. Immunity. 2017;46(5):835–848 e4. doi: 10.1016/j.immuni.2017.04.019. [DOI] [PubMed] [Google Scholar]
- 33.Zhang M., et al. Transcription factor Hoxb5 reprograms B cells into functional T lymphocytes. Nat. Immunol. 2018;19(3):279–290. doi: 10.1038/s41590-018-0046-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gemelli C., et al. Virally mediated MafB transduction induces the monocyte commitment of human CD34+ hematopoietic stem/progenitor cells. Cell Death Differ. 2006;13(10):1686–1696. doi: 10.1038/sj.cdd.4401860. [DOI] [PubMed] [Google Scholar]
- 35.Kelly Louise M., Isabelle Lafon U.E., Sieweke1 Michael H., Graf2 Thomas. 2000. MafB Is an Inducer of Monocytic Differentiation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hegde S.P., et al. c-Maf induces monocytic differentiation and apoptosis in bipotent myeloid progenitors. Blood. 1999;94(5):1578–1589. [PubMed] [Google Scholar]
- 37.Fischbach N.A., et al. HOXB6 overexpression in murine bone marrow immortalizes a myelomonocytic precursor in vitro and causes hematopoietic stem cell expansion and acute myeloid leukemia in vivo. Blood. 2005;105(4):1456–1466. doi: 10.1182/blood-2004-04-1583. [DOI] [PubMed] [Google Scholar]
- 38.Cytlak U., et al. Differential IRF8 transcription factor requirement defines two pathways of dendritic cell development in humans. Immunity. 2020;53(2):353–370. doi: 10.1016/j.immuni.2020.07.003. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurotaki D., et al. IRF8 inhibits C/EBPalpha activity to restrain mononuclear phagocyte progenitors from differentiating into neutrophils. Nat. Commun. 2014;5:4978. doi: 10.1038/ncomms5978. [DOI] [PubMed] [Google Scholar]
- 40.Evrard M., et al. Developmental analysis of bone marrow neutrophils reveals populations specialized in expansion, trafficking, and effector functions. Immunity. 2018;48(2):364–379 e8. doi: 10.1016/j.immuni.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 41.Grigorieva D.V., et al. Myeloperoxidase stimulates neutrophil degranulation. Bull. Exp. Biol. Med. 2016;161(4):495–500. doi: 10.1007/s10517-016-3446-7. [DOI] [PubMed] [Google Scholar]
- 42.Ahmad J., et al. Physics-driven identification of clinically approved and investigation drugs against human neutrophil serine protease 4 (NSP4): a virtual drug repurposing study. J. Mol. Graph. Model. 2020;101 doi: 10.1016/j.jmgm.2020.107744. [DOI] [PubMed] [Google Scholar]
- 43.Sawyer A.J., et al. Transcriptomic profiling identifies neutrophil-specific upregulation of cystatin F as a marker of acute inflammation in humans. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.634119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gatti A., et al. Quantification of neutrophil and monocyte CD64 expression: a predictive biomarker for active tuberculosis. Int. J. Tubercul. Lung Dis. 2020;24(2):196–201. doi: 10.5588/ijtld.19.0147. [DOI] [PubMed] [Google Scholar]
- 45.Druhan L.J., et al. Leucine rich alpha-2 glycoprotein: a novel neutrophil granule protein and modulator of myelopoiesis. PLoS One. 2017;12(1) doi: 10.1371/journal.pone.0170261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barrett M. Immunolocalization of human cystatins in neutrophils and lymphocytes. Histochemistry. 1984;80:373–377. doi: 10.1007/BF00495420. [DOI] [PubMed] [Google Scholar]
- 47.Perrina Christophe, et al. Expression of LSLCL, a new C-type lectin, is closely restricted, in bone marrow, to immature neutrophils. C R Acad. Sci. III. 2001;324:1125–1132. doi: 10.1016/s0764-4469(01)01392-0. [DOI] [PubMed] [Google Scholar]
- 48.Greaves David R., et al. CCR6, a CC chemokine receptor that interacts with macrophage inflammatory protein 3a and is highly expressed in human dendritic cells. J. Exp. Med. 1997;186(6):837–844. doi: 10.1084/jem.186.6.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heger L., et al. CLEC10A is a specific marker for human CD1c+ dendritic cells and enhances their toll-like receptor 7/8-induced cytokine secretion. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Angénieux C., et al. The Cellular Pathway of CD1e in Immature and Maturing Dendritic Cells. Traffic. 2005;6(4):286–302. doi: 10.1111/j.1600-0854.2005.00272.x. [DOI] [PubMed] [Google Scholar]
- 51.Ryan E.J., et al. Ligation of dendritic cell–associated lectin–1 induces partial maturation of human monocyte derived dendritic cells. Hum. Immunol. 2009;70(1):1–5. doi: 10.1016/j.humimm.2008.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jahrsdörfer B., et al. Granzyme B produced by human plasmacytoid dendritic cells suppresses T-cell expansion. Blood. 2010;115(6):1156–1165. doi: 10.1182/blood-2009-07-235382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Danek Petr, et al. blood; 2020. beta-Catenin-TCF/LEF Signaling Promotes Steady-State and Emergency Granulopoiesis via G-CSF Receptor Upregulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J., et al. Histone modification signature at myeloperoxidase and proteinase 3 in patients with anti-neutrophil cytoplasmic autoantibody-associated vasculitis. Clin. Epigenet. 2016;8:85. doi: 10.1186/s13148-016-0251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Serwas N.K., et al. CEBPE-mutant specific granule deficiency correlates with aberrant granule organization and substantial proteome alterations in neutrophils. Front. Immunol. 2018;9:588. doi: 10.3389/fimmu.2018.00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garg B., et al. Inducible expression of a disease-associated ELANE mutation impairs granulocytic differentiation, without eliciting an unfolded protein response. J. Biol. Chem. 2020 doi: 10.1074/jbc.RA120.012366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perrin C., et al. Expression of LSLCL, a new C-type lectin, is closely restricted, in bone marrow, to immature neutrophils. C R Acad. Sci. III. 2001;324(12):1125–1132. doi: 10.1016/s0764-4469(01)01392-0. [DOI] [PubMed] [Google Scholar]
- 58.Bencheikh L., et al. Dynamic gene regulation by nuclear colony-stimulating factor 1 receptor in human monocytes and macrophages. Nat. Commun. 2019;10(1):1935. doi: 10.1038/s41467-019-09970-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmeier S., et al. Deciphering the transcriptional circuitry of microRNA genes expressed during human monocytic differentiation. BMC Genom. 2009;10:595. doi: 10.1186/1471-2164-10-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schneider C., et al. IVIG regulates the survival of human but not mouse neutrophils. Sci. Rep. 2017;7(1):1296. doi: 10.1038/s41598-017-01404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee J., et al. Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow. J. Exp. Med. 2015;212(3):385–399. doi: 10.1084/jem.20141442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tarazona O.A., Pourquie O. Exploring the influence of cell metabolism on cell fate through protein post-translational modifications. Dev. Cell. 2020;54(2):282–292. doi: 10.1016/j.devcel.2020.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rossmann M.P., et al. Cell-specific transcriptional control of mitochondrial metabolism by TIF1gamma drives erythropoiesis. Science. 2021;372(6543):716–721. doi: 10.1126/science.aaz2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu H., et al. Alternative splicing analysis in human monocytes and macrophages reveals MBNL1 as major regulator. Nucleic Acids Res. 2018;46(12):6069–6086. doi: 10.1093/nar/gky401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vanhoutte P., et al. Opposing roles of Elk-1 and its brain-specific isoform, short Elk-1, in nerve growth factor-induced PC12 differentiation. J. Biol. Chem. 2001;276(7):5189–5196. doi: 10.1074/jbc.M006678200. [DOI] [PubMed] [Google Scholar]
- 66.Mian S.A., et al. SF3B1 mutant MDS-initiating cells may arise from the haematopoietic stem cell compartment. Nat. Commun. 2015;6(1) doi: 10.1038/ncomms10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang Y., et al. SF3B1 deficiency impairs human erythropoiesis via activation of p53 pathway: implications for understanding of ineffective erythropoiesis in MDS. J. Hematol. Oncol. 2018;11(1) doi: 10.1186/s13045-018-0558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luca Malcovati K.S., et al. SF3B1-mutant MDS as a distinct disease subtype: a proposal from the International Working Group for the Prognosis of MDS. Blood. 2020;136(2):157–170. doi: 10.1182/blood.2020004850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tang Q., et al. Circular RNA hsa_circ_0000515 acts as a miR-326 sponge to promote cervical cancer progression through up-regulation of ELK1. Aging (Albany NY) 2019;11(22):9982–9999. doi: 10.18632/aging.102356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yan Q., et al. ELK1 enhances pancreatic cancer progression via LGMN and correlates with poor prognosis. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.764900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kong Y., et al. Suppression of Elk1 inhibits thyroid cancer progression by mediating PTEN expression. Oncol. Rep. 2018;40(3):1769–1776. doi: 10.3892/or.2018.6554. [DOI] [PubMed] [Google Scholar]
- 72.Ma J., et al. c-KIT-ERK1/2 signaling activated ELK1 and upregulated carcinoembryonic antigen expression to promote colorectal cancer progression. Cancer Sci. 2021;112(2):655–667. doi: 10.1111/cas.14750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang S., et al. ELK1-induced up-regulation of KIF26B promotes cell cycle progression in breast cancer. Med. Oncol. 2021;39(1):15. doi: 10.1007/s12032-021-01607-6. [DOI] [PubMed] [Google Scholar]
- 74.Li S., et al. miR-597-5p inhibits cell growth and promotes cell apoptosis by targeting ELK1 in pancreatic cancer. Hum. Cell. 2020;33(4):1165–1175. doi: 10.1007/s13577-020-00395-x. [DOI] [PubMed] [Google Scholar]
- 75.Ahmad A., et al. Tumor-suppressive miRNA-135a inhibits breast cancer cell proliferation by targeting ELK1 and ELK3 oncogenes. Gene. Genomic. 2018;40(3):243–251. doi: 10.1007/s13258-017-0624-6. [DOI] [PubMed] [Google Scholar]
- 76.Zhang W., Zhang S. Downregulation of circRNA_0000285 suppresses cervical cancer development by regulating miR197-3p-ELK1 Axis. Cancer Manag. Res. 2020;12:8663–8674. doi: 10.2147/CMAR.S253174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Novotny L., Rauko P., Cojocel C. alpha-Lipoic acid: the potential for use in cancer therapy. Neoplasma. 2008;55(2):81–86. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request. All RNA-Seq data are deposited in the GEO database with accession code GSE184864.
All data generated or analyzed during this study are included in this published article.