Abstract
The global scourge of COVID-19 is a serious threat to public health, but effective therapies remain very limited for this disease. Therefore, the discovery of novel antiviral agents is urgently needed to fight against COVID-19. In the lifecycle of SARS-CoV-2, the causing pathogen of COVID-19, papain-like protease (PLpro) is responsible for the cleavage of polyprotein into functional units as well as immune evasion of vaccines. Hence, PLpro has been regarded as an attractive target to develop antiviral agents. Herein, we first developed a robust and simple sandwich-like fluorescence polarization (FP) screening assay for the discovery of PLpro inhibitors, and identified anacardic acid as a novel competitive inhibitor against PLpro in vitro with an IC50 value of 24.26 ± 0.4 μM. This reliable FP screening assay could provide a prospective avenue for rapid discovery of antiviral agents targeting PLpro in a large-scale screening.
Keywords: SARS-CoV-2, Papain-like protease inhibitors, Fluorescence polarization, High-throughput screening, Anacardic acid
Graphical abstract
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the cause of the ongoing pandemic of coronavirus disease 2019 (COVID-19), and its infection has led to a great threat to global public health. Recently, the emergence of omicron variant has brought a new challenge for ending the COVID-19 pandemic due to its increased transmissibility and immune evasion from approved vaccines (Cao et al., 2022; Tian et al., 2022). Therefore, highly effective antiviral agents are urgently required to fight a global pandemic in the future (Case et al., 2021).
SARS-CoV-2 is an enveloped, positive-sense, single-stranded RNA virus and belongs to the β-coronavirus genera. Its large viral RNA genome shares 79% sequence identity with SARS-CoV. SARS-CoV-2 virus mainly depends on their spike proteins for binding to the host cell surface receptor, angiotensin-converting enzyme 2 (ACE2), to enter into host cells (Jackson et al., 2022; Medina-Enríquez et al., 2020). After virus entry, ORF1a and ORF1ab are translated to produce two polyproteins pp1a and pp1ab, which are further cleaved by two cysteine proteases, the main protease (Mpro, 3CLpro) and the papain-like protease (PLpro), to yield 16 non-structural proteins (nsp) for RNA transcription and virus replication. During the proteolytic process, PLpro is responsible for releasing nsp 1-3. More importantly, PLpro regulates virus evasion of the host innate immune response by removing the ubiquitin (Ub) and interferon-stimulated gene 15 (ISG15) from host cellular proteins, making it a promising target for antiviral agent discovery (McClain and Vabret, 2020; Tan et al., 2022; Yuan et al., 2022).
Recently, a co-packaged combination of nirmatrelvir (PF-07321332) and ritonavir named paxlovid by Pfizer has received its first conditional authorization for the treatment of COVID-19 patients in the United States. This milestone has greatly encouraged researchers around the world to develop more potent antiviral agents targeting Mpro for the treatment of COVID-19 patients (Lamb, 2022; Owen et al., 2021). Although an intensive hunt for Mpro inhibitors has been undertaken, only scarce information is available for the discovery of PLpro inhibitors. At present, the fluorescence resonance energy transfer (FRET) and cell-based assays have been widely used for high-throughput screening (HTS) of PLpro inhibitors (Ma et al., 2021, 2022; Shan et al., 2021; Zhao et al., 2021). However, these screening assays are frequently associated with unstable enzymatic kinetics, false positives, expensive screening cost, tedious cell culture, long screening cycles, and poor repeatability. Hence, development of a simple, robust, and affordable biochemical assay for rapid screening of novel PLpro inhibitors is highly needed.
Here, we first developed a novel sandwich-like fluorescence polarization (FP) screening assay for the discovery of PLpro inhibitors. With this assay, we identified anacardic acid as a competitive hit compound targeting PLpro in vitro. Therefore, this robust, simple, and reliable FP screening assay will be vital for rapid screening of antiviral agents targeting PLpro, and could provide a prospective avenue for the discovery of viral protease inhibitors.
2. Materials and methods
2.1. Chemical reagents
The natural product library, GRL0617, PF-07321332 (nirmatrelvir), and anacardic acid were purchased from TargetMol (Shanghai, China). All tested compounds were dissolved in DMSO at a final concentration of 20 mM, and stored at −20 °C before use. The FRET substrate (Dabcyl-FTLKGGAPTKVTE-Edans; λex/λem = 340/500 nm), FP probe (FITC-FTLKGGAPTKVTK-Biotin; λex/λem = 485/535 nm), FITC-FTLKGG peptide, and the fluorogenic peptide Z-RLRGG-AMC (λex/λem = 340/460 nm) were purchased from GL Biochem (Shanghai, China), respectively.
2.2. Expression, purification, and enzymatic activity analysis of SARS-CoV-2 PLpro
The detailed expression and purification procedures of PLpro were performed as a previous study (Shin et al., 2020). Briefly, a codon-optimized PLpro gene (GenBank: NC_045512.2) was cloned into a pET-28a vector with Nco I and Xho I to carry an extra C-terminal 6 × His-tag. Protein production was induced by addition of 1 mM isopropyl-β-d-thiogalactoside (IPTG) and zinc chloride (ZnCl2) solution at 16 °C. After 12 h incubation, the E. coli Rosetta (DE3) cells were harvested, and then the cell pellets were lysed by an ultrasonic cell disruptor (SCIENTZ, Ningbo, China). Subsequently, the supernatant was purified by a HisTrap™ chelating column (Cytiva, Uppsala, Sweden), and the purity of purified PLpro was analyzed by SDS-PAGE. After dialysis, the quantitative PLpro solution was stored at −80 °C before use.
According to the published FRET protocol (Gao et al., 2021), the fluorogenic peptide (Dabcyl-FTLKGGAPTKVTE-Edans) was used as PLpro hydrolysis substrate. Each reaction mixture contained 2 μM PLpro and the indicated amount of FRET substrate in HEPES buffer (10 mM HEPES, 200 mM NaCl, 1 mM EDTA, 1 mM DTT, pH7.0), and the relative fluorescence unit (RFU) value was recorded every second for 3 min by a microplate reader (BioTek, Winooski, USA) with an excitation wavelength of 340 nm and emission wavelength of 500 nm at room temperature (RT). The initial velocity (V = ΔRFU/s) was calculated for the first 30 s by a slope of a linear regression, and further plotted against the FRET substrate concentrations to yield a Michaelis–Menten equation for the determination of enzyme kinetic parameters.
2.3. Establishment of a novel sandwich-like FP screening assay
The experiments were performed based on our previous publication (Yan et al., 2021). To determine the optimal amount of FP probe in this FP screening assay, the FP probe (2 mM) was diluted to the fixed concentrations ranking from 1.25 to 100 nM in HEPES buffer, and 60 μL dilution was pipetted into a black 96-well microplate (Corning, Product code: 3686, Kennebunk, USA) in triplicate. After incubation for 10 min at RT, the millipolarization unit (mP) value was recorded by a microplate reader with the excitation at 485 nm and emission at 535 nm. The optimal amount of FP probe in this FP screening assay should be selected as a minimum concentration that produces the lowest background signal and stable mP value.
To find an optimal working concentration of avidin (Aladdin, Shanghai, China) in this FP screening assay, the reaction mixture containing 20 nM FP probe (30 μL/well) and avidin dilution (2.5~160 nM, 30 μL/well) was pipetted into a black 96-well microplate in triplicate. After incubation for 5 min at RT, the mP value was measured as described above.
To determine the most reasonable duration of PLpro hydrolysis, the reaction mixture containing 20 nM FP probe (30 μL/well) and the indicated concentrations of PLpro dilution (0~6 μM, 20 μL/well) was incubated for 5, 10, 20, 30, 40, or 50 min at RT. After quenching by avidin (240 nM, 10 μL/well) for additional 5 min at RT, the mP value was separately recorded by a microplate reader.
In addition, the reaction mixture described above was also incubated at 4, 20, 25, 30, or 37 °C for 30 min. After addition of avidin, the mP value was monitored separately.
For DMSO tolerance test, the reaction mixture mentioned above was incubated for 30 min at RT in the absence or presence of DMSO at the indicated concentrations, and the mP value was monitored as described above. All mentioned proteolytic reaction curves of PLpro in these experiments were plotted using GraphPad Prism 8.0, and the half concentration of maximal effect (EC50) and dynamic range (ΔmP) values were compared.
As mentioned above, the reaction mixture was incubated at RT for 30 min, and the mP value was further recorded after avidin addition. Based on the proteolytic trajectory of PLpro, an optimal working concentration should be the minimum amount of PLpro that can adequately cleave the FP probe into a small FITC-FTLKGG fragment in this FP screening assay.
To test the assay reliability, the inhibition of PLpro activity by GRL0617 was assessed. In brief, 30 μL reaction mixture containing 1 μM PLpro and 8 two-fold dilutions of GRL0617 (initial concentration: 80 μM) was incubated for 30 min at RT in a black 96-well microplate. The reaction mixture was further incubated for 30 min at RT after addition of 30 nM FP probe (20 μL/well). After quenching by 240 nM avidin (10 μL/well) for additional 5 min, the mP value was monitored as described above. In this test, the wells containing FP probe/avidin binding complex and FITC-FTLKGG peptide were used as positive and negative controls, respectively. The following equation was used to calculate the half-maximum inhibitory concentration (IC50) value of GRL0617.
| (Eq. 1) |
where μHit, μN and μP represent the average mP values of the testing compound, negative, and positive controls, respectively.
For the determination of Z factor, the wells containing 50 μM GRL0617 or DMSO were served as positive and negative controls, respectively. For a better statistical analysis, each control covered a separate 96-well microplate. As mentioned above, the mP value was recorded using a microplate reader for the calculation of Z factor (Chen et al., 2021a; Yan et al., 2021, 2022a).
2.4. The primary screening of a natural product library and evaluation of the inhibitory activity of anacardic acid against SARS-CoV-2 PLpro in the FP screening assay
In a pilot screening, 1 μL natural product (2 mg/mL in DMSO) was mixed with 29 μL sample containing 1 μM PLpro in HEPES buffer in a black 96-well microplate, and the reaction mixture was further incubated for 30 min at RT. After addition of 20 μL FP probe (30 nM), the mixture was incubated for 30 min at RT. After quenching by 10 μL avidin (240 nM) for additional 5 min, the mP values were recorded by a microplate reader. In each assay plate, GRL0617 (50 μM), DMSO, and FITC-FTLKGG peptide (10 nM) were served as a positive, negative, and background controls, respectively. The inhibition of PLpro activity by a natural product was calculated using Eq. (1), and the candidate compounds (>50% inhibition) were repeated in the second screening.
As described in GRL0617 inhibitory potency test, the inhibition of PLpro activity by anacardic acid was also evaluated by this FP screening assay. According to the plotted dose-response curve, an IC50 value was calculated.
2.5. Evaluation of the inhibitory activity of anacardic acid against SARS-CoV-2 cysteine proteases by the FRET assay
To confirm the possible inhibitory activity of anacardic acid, its inhibition of PLpro activity was evaluated by the FRET assay (Ma et al., 2022). In brief, 25 μL PLpro solution (1 μM) was preincubated with the indicated concentrations of anacardic acid (initial concentration: 160 μM, 8 two-fold dilutions) for 30 min at RT in HEPES buffer before addition of 25 μL FRET substrate (20 μM) to initiate the reaction. GRL0617 (50 μM) and DMSO were used as the positive and negative controls, respectively. The RFU value was monitored as described previously. According to the initial velocity, the inhibitory activity was calculated using the following equation.
| (Eq. 2) |
For the inhibition of Mpro activity by anacardic acid, the FRET assay was carried out as previously described (Yan et al., 2021, 2022a). PF-07321332 was used as a positive control.
2.6. Inhibition of PLpro isopeptidase activity by anacardic acid in vitro
The enzymatic assay was performed as previously described with slight modification (Chen et al., 2021b). Briefly, 25 μL of 1 μM PLpro solution was added to the solution with indicated concentrations of anacardic acid, and the mixture was incubated at RT for 30 min. The reaction was initiated by adding 25 μL of 40 μM fluorogenic peptide (Z-RLRGG-AMC). Later, the RFU signal was measured every second for 3 min by a microplate reader with the excitation wavelength of 340 nm and emission wavelength of 460 nm. The inhibition of PLpro isopeptidase activity by anacardic acid was assessed using Eq. (2). The final concentration of testing compound was 80 μM, and 8 two-fold dilutions was prepared. GRL0617 served as a positive control.
2.7. Inhibition mechanism of anacardic acid against SARS-CoV-2 PLpro in vitro
To determine the inhibition mechanism of anacardic acid against PLpro in vitro, 25 μL reaction mixtures containing PLpro (1 μM) and anacardic acid (0, 5, 10, 20 μM) were separately mixed with 25 μL FRET substrate (10, 20, 30, 40 μM), and the initial velocity was measured as described in the FRET assay. The inhibition mechanism of anacardic acid against PLpro in vitro was inferred using the Lineweaver-Burk (LB) plot and an inhibitory constant (K i) value was generated by its secondary plot.
3. Results
3.1. Production and characterization of SARS-CoV-2 PLpro
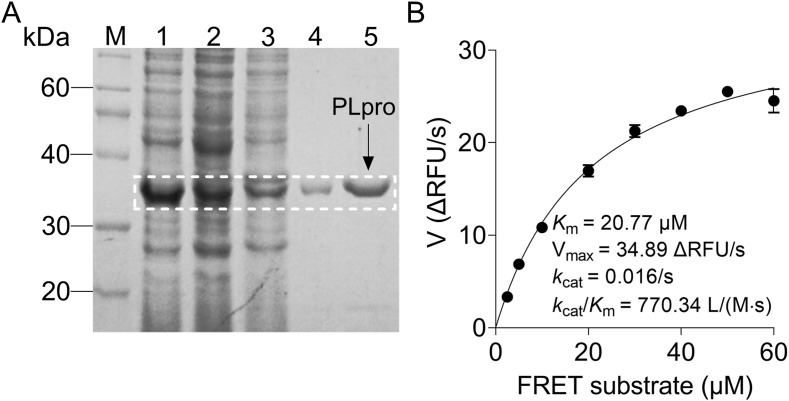
A codon-optimized SARS-CoV-2 PLpro gene was cloned into a pET-28a vector, and the recombinant PLpro proteins were expressed in E. coli Rosetta (DE3) cells with an additional C-terminal 6 × His-tag. After preparation of crude extract by salting out method, PLpro proteins were purified by a HisTrap™ chelating column (Fig. 1 A). To better characterize the enzymatic activity of purified PLpro, a fluorescently labeled FRET substrate, Dabcyl-FTLKGGAPTKVTE-Edans, was used as PLpro hydrolysis substrate in the FRET assay. Importantly, this peptide sequence was derived from the proteolytic processing site between nsp 2 and nsp 3 of pp1a polyprotein (Gao et al., 2021). Michaelis–Menten equation gave the best-fit values of K m and catalytic efficiency (k cat/K m) as 20.77 μM and 770.34 L/(M·s), respectively (Fig. 1B). These results indicate that the purified PLpro proteins exhibit an expected enzymatic activity.
Fig. 1.
Production and characterization of SARS-CoV-2 papain-like protease (PLpro). (A) Bacterial expression and purification of PLpro. After IPTG induction, E. coli cells were harvested and resuspended in the lysis buffer for sonication. After removing the cell debris by centrifugation, the supernatant was purified by a HisTrap™ chelating column, and the purity of purified PLpro was further analyzed by SDS-PAGE. The purified PLpro band in the stained gel is marked by a box with dashed white line. M: Protein marker; 1: Total cell proteins; 2: Supernatant; 3: Crude extract by salting out method; 4–5: Purified PLpro proteins (36 kDa). (B) Determination of the enzyme kinetics parameters of purified PLpro by the FRET assay. According to the initial velocity (V) in the FRET assay, the Km, Vmax, kcat and kcat/Km values were calculated using a Michaelis-Menten equation. All experiments were carried out in triplicate.
3.2. Basic principle of a sandwich-like FP screening assay for the discovery of SARS-CoV-2 PLpro inhibitors
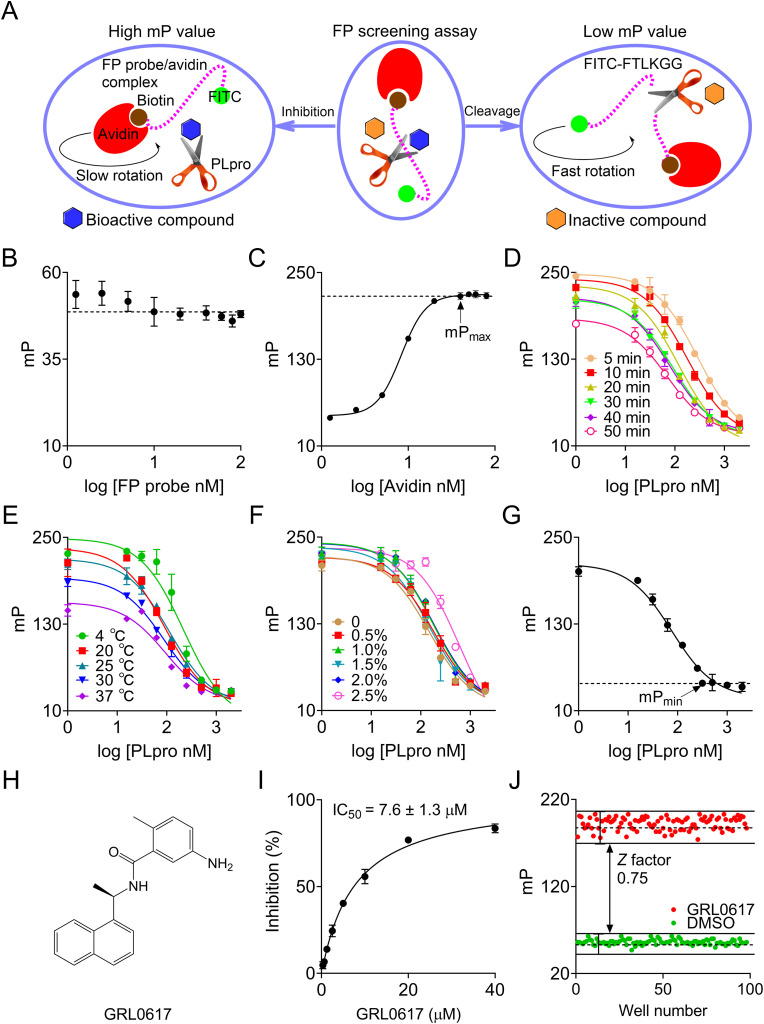
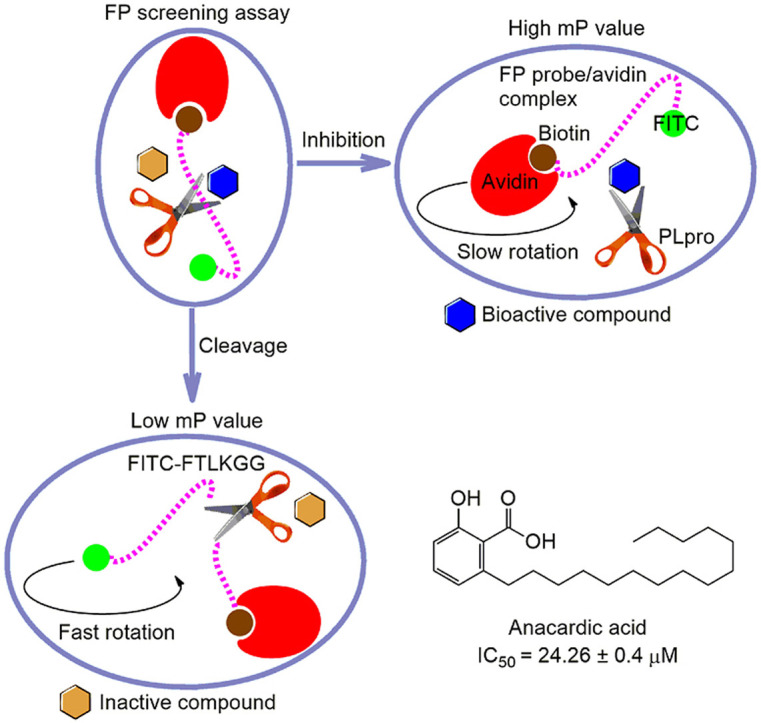
To further improve the existing HTS assay, we developed a simple and robust sandwich-like FP screening assay for the discovery of PLpro inhibitors. This FP approach is based on the positive correlation change between mP value and apparent molecular weight of a fluorescent moiety in the screening solution (Hall et al., 2016). In this FP screening assay, the synthetic peptide with a similar sequence in the FRET assay was conjugated with a fluorescein isocyanate (FITC) fluorophore and biotin to generate the FP probe FITC-FTLKGGAPTKVTK-Biotin. As described in our previous studies (Yan et al., 2021, 2022b), PLpro cleaves the FP probe to release a small fluorescent molecule FITC-FTLKGG, resulting in a low mP value due to a fast rotation. In contrast, when a bioactive compound inhibits the PLpro activity, the binding of this intact FP probe to avidin leads to formation of a large binding complex, resulting in a high mP value due to a slow rotation (Fig. 2 A). Therefore, candidate compounds from large compound libraries could be rapid identified by monitoring the change of mP value. This FP screening system mainly includes a natural product, active PLpro, FP probe, and avidin, which looks like a sandwich.
Fig. 2.
Optimization and quality assessment of a novel sandwich-like FP screening assay for the discovery of SARS-CoV-2 PLpro inhibitors. (A) The sketch map of the developed sandwich-like FP screening assay. As previously described (Yan et al., 2021), incubation of FP probe (dashed purple line) with PLpro (the scissors) and subsequent addition of avidin (red crescent) will produce a FP signal that is proportional to the relative amount of cleaved and uncleaved FP probe. In the presence of a bioactive compound (blue hexagon) that inhibits PLpro activity, the uncleaved FP probe produces a high mP value upon binding to avidin, whereas in the presence of an inactive compound (yellow hexagon), the cleaved fragment FITC-FTLKGG produces a low mP value because of the failure for avidin binding. (B) Determination of the optimal working concentration of FP probe in the FP screening assay. The dashed line represents the average of mP values in the indicated FP probe concentrations. (C) Saturation binding assay between FP probe and avidin for the determination of the optimal concentration of avidin used in the FP screening assay. The optimal avidin concentration should be the minimum amount that reaches the plateau of this saturation binding curve. (D) The reaction course trajectories of PLpro in the FP screening assay at the indicated time points. (E) The reaction course trajectories of PLpro in the FP screening assay at the indicated temperatures. (F) DMSO tolerance assay. The proteolytic reaction of PLpro was performed in the presence of the indicated DMSO concentrations, and the mP values were compared. (G) Determination of the optimal concentration of PLpro in the FP screening assay. For a high sensitivity of screening assay, the optimal working concentration of used PLpro should be the minimum amount that could reach the reaction end. (H) The chemical structure of GRL0617. (I) Dose-response curve of GRL0617 in the FP screening assay. (J) Determination of Z factor in the FP screening assay. GRL0617 and DMSO were used as positive and negative controls, respectively. The Z factor of this screening assay was 0.75. All mentioned biochemical assays were performed in triplicate.
3.3. Development of a simple and reliable sandwich-like FP screening assay
As previously noted, the working concentration of FP probe can affect the assay background and screening cost. To develop a sensitive and economical screening assay, the mP values for different working concentrations of FP probe were determined. It was obvious that the mP value became stable when the concentration of FP probe was increased from 5 to 100 nM (Fig. 2B). Considering both stability of mP value and importance of low background, 10 nM FP probe was used as the optimal working concentration in this FP screening assay. Subsequently, the titration of 10 nM FP probe with avidin was carried out. Based on the avidin titration curve in Fig. 2C, the maximum mP (mPmax) value achieved when the concentration of avidin was more than 40 nM. Given that avidin molecules form tetramers, this represented a 16-fold molar excess of avidin in binding equivalence when 40 nM avidin was used to quench the reaction with 10 nM FP probe. Next, we recorded the PLpro proteolytic reaction curves. The obvious overlap of the curves from 30 min to 40 min reactions, the minimum EC50, and highest ΔmP values from this figure indicate that the endpoint was reached within 30 min (Fig. 2D). Hence, the incubation duration was defined in 30 min in this FP screening assay. Moreover, the proteolytic reaction curves were stable after incubation at 20 °C, 25 °C, and 30 °C for 30 min, resulting in highly overlapped trajectories (Fig. 2E). It should be noted that the actual working concentration of DMSO used in this assay must be less than 2% to achieve a reliable screening for natural products (Fig. 2F). More importantly, the minimum mP (mPmin) value of the proteolytic reaction curve appeared in the presence of 0.5 μM PLpro, indicating the endpoint of this reaction at RT (Fig. 2G). All these results strongly suggest that this FP screening assay is significantly stable and sensitive, and the reaction mixture containing 0.5 μM PLpro and 10 nM FP probe should be incubated for 30 min at RT for natural product screening in this FP assay.
As a PLpro inhibitor, GRL0617 has been widely used as a positive control for the quality assessment of the assays used to examine PLpro inhibition (Ma et al., 2022; Shan et al., 2021; Zhao et al., 2021). As expected, GRL0617 exhibited an IC50 value of 7.6 ± 1.3 μM in this FP screening assay (Fig. 2H and I), and this result was consistent with a published study (Shan et al., 2021). Furthermore, as shown in Fig. 2J, the Z factor of this FP screening assay reached 0.75, and met the HTS requirement of Z>0.5, indicating that this assay is robust and amenable to a HTS pipeline.
3.4. Main procedure for a large-scale screening
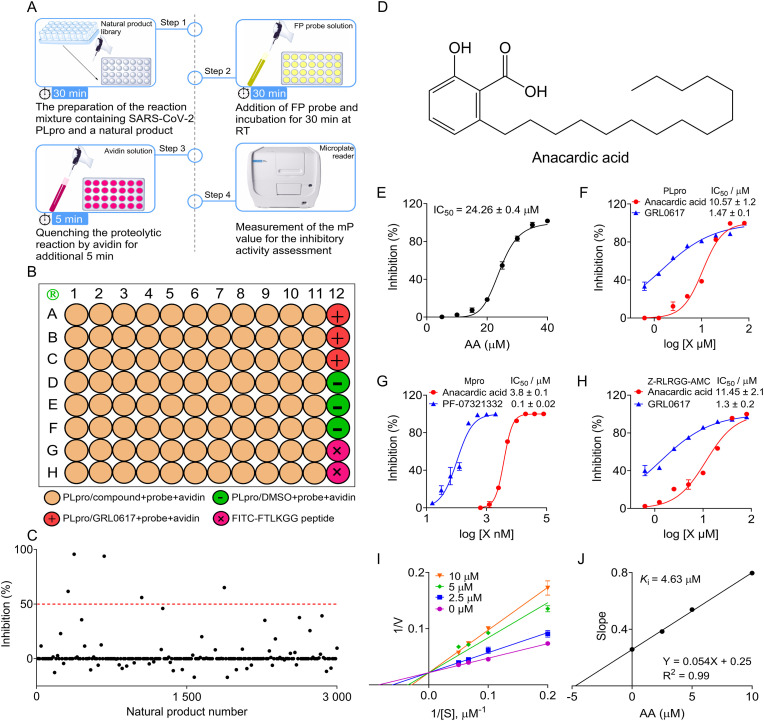
The operation of the FP screening assay was divided into 4 steps for laboratory use in a large-scale screening. As shown in Fig. 3 A, this entire procedure took about 70 min for each round of screening. First, the 30 μL reaction mixture containing 1 μM PLpro (29 μL/well) and a natural product (2 mg/mL, 1 μL/well) was incubated for 30 min at RT. Subsequently, 20 μL FP probe solution (30 nM) was added and further incubated for 30 min at RT. Later, 10 μL avidin solution (240 nM) was used to quench the reaction for additional 5 min, and the mP value was measured by a microplate reader. For a rapid and reliable screening of natural products, each screening microplate should be arranged as shown in Fig. 3B, and the positive (GRL0617), negative (DMSO), and background controls should be included in each screening cycle.
Fig. 3.
The inhibitory activity and mechanism of a natural product anacardic acid (AA) against SARS-CoV-2 PLpro in vitro. (A) The 4-step procedure of the FP screening assay for rapid screening of PLpro inhibitors. Step 1: 29 μL PLpro solution (1 μM) was incubated with 1 μL sample of a natural product (2 mg/mL) for 30 min at RT in a black 96-well microplate. Step 2: 20 μL FP probe solution (30 nM) was added and incubated for 30 min at RT. Step 3: The reaction was quenched by adding 10 μL avidin solution (240 nM) followed by 5 min incubation. Step 4: The mP value was measured by a microplate reader (BioTek). (B) The natural product layout in a black 96-well microplate for a large-scale screening. The positive (GRL0617), negative, and background wells were highlighted. (C) The illustration for the primary screening cycle of a natural product library using the FP screening assay. The red dotted line indicates a baseline (equivalent to 50% inhibition) in the primary screening cycle, and 5 candidate compounds were identified. (D) The chemical structure of anacardic acid. (E) Inhibitory effect of anacardic acid on the PLpro activity in the FP screening assay. (F) Inhibitory effect of anacardic acid on the PLpro activity in the FRET screening assay. GRL0617 was used as a positive control. (G) Inhibitory effect of anacardic acid on the Mpro activity in the FRET screening assay. PF-07321332 served as a positive control. (H) Inhibitory effect of anacardic acid on the PLpro isopeptidase activity. The fluorogenic peptide Z-RLRGG-AMC was used as the substrate for inhibition of PLpro isopeptidase activity by anacardic acid in vitro. GRL0617 served as a positive control. (I) The Lineweaver-Burk double-reciprocal plots for inhibition of PLpro activity by anacardic acid in vitro for the FRET substrate. (J) The secondary plots for a Ki value.
3.5. Screening and validation anacardic acid as a competitive inhibitor against SARS-CoV-2 PLpro in vitro
As a natural medicine research group, we have a long-term interest in identifying more potent bioactive compounds from traditional Chinese herbal medicine as novel antiviral agents. Therefore, we performed a screening with a natural product library (3 000 compounds) for novel PLpro inhibitors using this FP screening assay. Interestingly, 5 compounds exhibited IC50 values of >50% in a pilot screening (Fig. 3C). During the second screening cycle, a natural product anacardic acid exhibited an expected dose-response curve, and was a novel PLpro inhibitor with an IC50 value of 24.26 ± 0.4 μM (Fig. 3D and E). Importantly, anacardic acid also exhibited a strong inhibition of PLpro activity with an IC50 value of 10.57 ± 1.2 μM using the FRET assay (Fig. 3F). Interestingly, anacardic acid still dose-dependently inhibited Mpro activity in vitro using the FRET assay with an IC50 value of 3.8 ± 0.1 μM (Fig. 3G), indicating that anacardic acid is a dual inhibitor targeting SARS-CoV-2 cysteine proteases. Moreover, the synthetic peptide Z-RLRGG-AMC consists of the Ub and ISG15 C-terminal sequences and is a fluorogenic substrate for PLpro, deubiquitinating enzyme isopeptidase T (IPaseT) and other ubiquitin C-terminal hydrolases (Shan et al., 2021). We used this peptide as the PLpro substrate to mimic the in vitro deubiquitylation and deISGylation processes. Our result demonstrated that anacardic acid exhibited inhibition of PLpro isopeptidase activity in vitro with an IC50 value of 11.45 ± 2.1 μM (Fig. 3H). Subsequent LB plot graphical method yielded an obvious character of a series of straight lines with the same y-axis intercept in the presence of anacardic acid, suggesting that anacardic acid is a novel competitive inhibitor against PLpro in vitro with a K i value of 4.63 μM (Fig. 3I and J).
4. Discussion
Currently, the ongoing COVID-19 pandemic caused by SARS-CoV-2 continues to be a serious threat to global public health, but there are still few antiviral agents available for the treatment of COVID-19 patients. In the lifecycle of SARS-CoV-2, the conserved PLpro is responsible for viral polyprotein cleavage and immune escape (McClain and Vabret, 2020). Importantly, genetic reshuffling and mutations are often absent in viral proteases (Sacco et al., 2022). Therefore, PLpro represents a promising drug target to generate more potent and broad-spectrum antiviral agents against COVID-19. The discovery of novel PLpro inhibitors with diverse chemical scaffolds using a rapid and robust screening assay is of the highest significance.
HTS technique has been regarded as an efficient approach in drug discovery field (Blay et al., 2020). However, its success rate depends not only on the capacity and diversity of screened chemical libraries, but also on the robustness, convenience, and cost efficiency of the HTS assays (Shan et al., 2021). As a homogenous biochemical assay, our research group has successfully utilized FP technique for screening of protein-protein interaction antagonists and Mpro inhibitors (Chen et al., 2017, 2021a; Li et al., 2022; Yan et al., 2021, 2022b). At present, the discovery of PLpro inhibitors mainly relies on FRET and cell-based screening assays, but these approaches often face more obstacles in a large-scale screening. In this study, we first applied FP technique to develop an optimized sandwich-like FP screening assay for the discovery of PLpro inhibitors, and this FP screening assay showed a high robustness, convenience, and cheapness in a HTS pipeline. In contrast to the published biochemical assays, the 4-step screening procedure only takes 70 min for each screening cycle. As such, this FP screening assay is very simple and rapid for laboratory use. Moreover, only a trace peptide substrate (FP probe, 10 nM/well) is used in this FP screening assay, and the monitoring indicator of mP value is rather stable compared with the RFU signal. Therefore, this FP screening assay is featured by simple operation, greater efficiency, high reproducibility, and most affordable cost, which is appropriate for a HTS pipeline. In addition, because the emission wavelength of FITC was 535 nm in this FP screening assay, the interference by the autofluorescence of natural products on the reliability of the screening assay is minimal. For this reason, this FP screening assay is expected to generate reliable results and will have broad applications for the screening and evaluation of antiviral agents targeting PLpro.
Through a pilot screening of a natural product library, anncardic acid was identified as a competitive PLpro inhibitor in vitro with a K i value of 4.63 μM. Anacardic acid is the main component of extracts from Ginkgo biloba and Rhus verniciflua, which has a wide range of pharmacological activities such as antibacterial, anticancer, and antiviral (Hundt et al., 2015; Park et al., 2018; Saedtler et al., 2020). Recent studies have confirmed that the EC50 value of antiviral potency of anacardic acid is 9.0 ± 2.5 μM in live SARS-CoV-2 virus by targeting Mpro (Chen et al., 2021b; Hicks et al., 2022; Li et al., 2022; Xiong et al., 2021; Yan et al., 2022b). Our study demonstrated that anacardic acid is likely a novel competitive inhibitor targeting both PLpro and Mpro of SARS-CoV-2, and more in vivo experiments for its antiviral mechanism will be explored in the future. However, anacardic acid shows cytotoxicity, and further chemical modification and structural optimization are needed to improve its target specificity and reduce its cytotoxicity.
5. Conclusion
Overall, we first developed a novel sandwich-like FP screening assay for rapid screening of PLpro inhibitors. With this assay, we further identified anacardic acid as a novel competitive inhibitor against PLpro in vitro from a natural product library. This robust and simple FP screening assay will facilitate the rapid screening of antiviral agents targeting PLpro, and could shed light on a promising avenue for the discovery of viral protease inhibitors.
Declaration of competing interest
The authors declare no competing interests.
Funding
This work was supported by National Natural Science Foundation of China (No. 81703546); Natural Science Foundation of Anhui Province, China (No. 1808085QH265); University Natural Science Research Project of Anhui Province, China (Nos. KJ2019ZD30, KJ2021A0839, YJS20210549); and Key Technologies Research and Development Program of Anhui Province, China (No. 202004a07020041).
CRediT authorship contribution statement
Haohao Yan: Methodology, Data curation, Writing – original draft, Preparation. Zhicheng Liu: Methodology, Data curation, Writing – original draft, Preparation. Gangan Yan: Methodology, Data curation, Writing – original draft, Preparation, Funding acquisition, Validation. Xiaoli Liu: Investigation, Data curation, Visualization, Validation. Xiaoping Liu: Resources, Validation, Supervision, Funding acquisition. Yanchang Wang: Conceptualization, Methodology, Supervision, Writing – review & editing. Yunyu Chen: Conceptualization, Methodology, Supervision, Resources, Project administration, Funding acquisition, Writing – review & editing, All authors read and approved the final manuscript.
Acknowledgements
We are grateful to Mrs. Jingzhe Yang (BioTek Shanghai Representative Office, Shanghai, China) for her kind technical assistance on the development of fluorescence polarization screening assay.
Data availability
Data will be made available on request.
References
- Blay V., Tolani B., Ho S.P., Arkin M.R. High-Throughput Screening: today's biochemical and cell-based approaches. Drug Discov. Today. 2020;25:1807–1821. doi: 10.1016/j.drudis.2020.07.024. [DOI] [PubMed] [Google Scholar]
- Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., Wang J., Wang Y., Niu X., Yang S., Liang H., Sun H., Li T., Yu Y., Cui Q., Liu S., Yang X., Du S., Zhang Z., Hao X., Shao F., Jin R., Wang X., Xiao J., Wang Y., Xie X.S. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Winkler E.S., Errico J.M., Diamond M.S. On the road to ending the COVID-19 pandemic: are we there yet? Virology. 2021;557:70–85. doi: 10.1016/j.virol.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Fu Z., Li D., Yue Y., Liu X. Optimizations of a novel fluorescence polarization-based high-throughput screening assay for β-catenin/LEF1 interaction inhibitors. Anal. Biochem. 2021;612 doi: 10.1016/j.ab.2020.113966. [DOI] [PubMed] [Google Scholar]
- Chen Y., Zhang J., Li D., Jiang J., Wang Y., Si S. Identification of a novel Polo-like kinase 1 inhibitor that specifically blocks the functions of Polo-Box domain. Oncotarget. 2017;8:1234–1246. doi: 10.18632/oncotarget.13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Cui Q., Cooper L., Zhang P., Lee H., Chen Z., Wang Y., Liu X., Rong L., Du R. Ginkgolic acid and anacardic acid are specific covalent inhibitors of SARS-CoV-2 cysteine proteases. Cell Biosci. 2021;11:45. doi: 10.1186/s13578-021-00564-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Qin B., Chen P., Zhu K., Hou P., Wojdyla J.A., Wang M., Cui S. Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm. Sin. B. 2021;11:237–245. doi: 10.1016/j.apsb.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M.D., Yasgar A., Peryea T., Braisted J.C., Jadhav A., Simeonov A., Coussens N.P. Fluorescence polarization assays in high-throughput screening and drug discovery: a review. Methods Appl. Fluoresc. 2016;4 doi: 10.1088/2050-6120/4/2/022001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks E.G., Kandel S.E., Lampe J.N. Identification of Aloe-derived natural products as prospective lead scaffolds for SARS-CoV-2 main protease (Mpro) inhibitors. Bioorg. Med. Chem. Lett. 2022;66 doi: 10.1016/j.bmcl.2022.128732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundt J., Li Z., Liu Q. The inhibitory effects of anacardic acid on hepatitis C virus life cycle. PLoS One. 2015;10 doi: 10.1371/journal.pone.0117514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C.B., Farzan M., Chen B., Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022;23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb Y.N. Nirmatrelvir plus ritonavir: first approval. Drugs. 2022;82:585–591. doi: 10.1007/s40265-022-01692-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Yan G., Zhou W., Si S., Liu X., Zhang J., Li Y., Chen Y. Ginkgolic acid and anacardic acid are reversible inhibitors of SARS-CoV-2 3-chymotrypsin-like protease. Cell Biosci. 2022;12:65. doi: 10.1186/s13578-022-00806-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Hu Y., Wang Y., Choza J., Wang J. Drug-repurposing screening identified tropifexor as a SARS-CoV-2 papain-like protease inhibitor. ACS Infect. Dis. 2022;8:1022–1030. doi: 10.1021/acsinfecdis.1c00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Sacco M.D., Xia Z., Lambrinidis G., Townsend J.A., Hu Y., Meng X., Szeto T., Ba M., Zhang X., Gongora M., Zhang F., Marty M.T., Xiang Y., Kolocouris A., Chen Y., Wang J. Discovery of SARS-CoV-2 papain-like protease inhibitors through a combination of high-throughput screening and a FlipGFP-based reporter assay. ACS Cent. Sci. 2021;7:1245–1260. doi: 10.1021/acscentsci.1c00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain C.B., Vabret N. SARS-CoV-2: the many pros of targeting PLpro. Signal Transduct. Targeted Ther. 2020;5:223. doi: 10.1038/s41392-020-00335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Enríquez M.M., Lopez-León S., Carlos-Escalante J.A., Aponte-Torres Z., Cuapio A., Wegman-Ostrosky T. ACE2: the molecular doorway to SARS-CoV-2. Cell Biosci. 2020;10 doi: 10.1186/s13578-020-00519-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., Dantonio A., Di L., Eng H., Ferre R., Gajiwala K.S., Gibson S.A., Greasley S.E., Hurst B.L., Kadar E.P., Kalgutkar A.S., Lee J.C., Lee J., Liu W., Mason S.W., Noell S., Novak J.J., Obach R.S., Ogilvie K., Patel N.C., Pettersson M., Rai D.K., Reese M.R., Sammons M.F., Sathish J.G., Singh R.S.P., Steppan C.M., Stewart A.E., Tuttle J.B., Updyke L., Verhoest P.R., Wei L., Yang Q., Zhu Y. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374:1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- Park M., Upton D., Blackmon M., Dixon V., Craver S., Neal D., Perkins D. Anacardic acid inhibits pancreatic cancer cell growth, and potentiates chemotherapeutic effect by Chmp1A-ATM-p53 signaling pathway. BMC Compl. Alternative Med. 2018;18:71. doi: 10.1186/s12906-018-2139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco M.D., Hu Y., Gongora M.V., Meilleur F., Kemp M.T., Zhang X., Wang J., Chen Y. The P132H mutation in the main protease of Omicron SARS-CoV-2 decreases thermal stability without compromising catalysis or small-molecule drug inhibition. Cell Res. 2022;32:498–500. doi: 10.1038/s41422-022-00640-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saedtler M., Fortig N., Ohlsen K., Faber F., Masota N., Kowalick K., Holzgrabe U., Meinel L. Antibacterial anacardic acid derivatives. ACS Infect. Dis. 2020;6:1674–1685. doi: 10.1021/acsinfecdis.9b00378. [DOI] [PubMed] [Google Scholar]
- Shan H., Liu J., Shen J., Dai J., Xu G., Lu K., Han C., Wang Y., Xu X., Tong Y., Xiang H., Ai Z., Zhuang G., Hu J., Zhang Z., Li Y., Pan L., Tan L. Development of potent and selective inhibitors targeting the papain-like protease of SARS-CoV-2. Cell Chem. Biol. 2021;28:855–865. doi: 10.1016/j.chembiol.2021.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., Schulz L., Widera M., Mehdipour A.R., Tascher G., Geurink P.P., Wilhelm A., van der Heden van Noort G.J., Ovaa H., Muller S., Knobeloch K.P., Rajalingam K., Schulman B.A., Cinatl J., Hummer G., Ciesek S., Dikic I. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H., Hu Y., Jadhav P., Tan B., Wang J. Progress and challenges in targeting the SARS-CoV-2 papain-like protease. J. Med. Chem. 2022;65:7564–7580. doi: 10.1021/acs.jmedchem.2c00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., Sun Y., Xu H., Ye Q. The emergence and epidemic characteristics of the highly mutated SARS-CoV-2 Omicron variant. J. Med. Virol. 2022;94:2376–2383. doi: 10.1002/jmv.27643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Zhu G.H., Wang H.N., Hu Q., Chen L.L., Guan X.Q., Li H.L., Chen H.Z., Tang H., Ge G.B. Discovery of naturally occurring inhibitors against SARS-CoV-2 3CLpro from Ginkgo biloba leaves via large-scale screening. Fitoterapia. 2021;152 doi: 10.1016/j.fitote.2021.104909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G., Li D., Lin Y., Fu Z., Qi H., Liu X., Zhang J., Si S., Chen Y. Development of a simple and miniaturized sandwich-like fluorescence polarization assay for rapid screening of SARS-CoV-2 main protease inhibitors. Cell Biosci. 2021;11:199. doi: 10.1186/s13578-021-00720-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G., Li D., Qi H., Fu Z., Liu X., Zhang J., Chen Y. Discovery of SARS-CoV-2 main protease inhibitors using an optimized FRET-based high-throughput screening assay. Chin. J. Biotechnol. 2022;38:2236–2249. doi: 10.13345/j.cjb.210657. [DOI] [PubMed] [Google Scholar]
- Yan H., Yan G., Qi H., Liu Z., Liu X., Liu X., Li N., Chen Y. Identifying SARS-CoV-2 main protease inhibitors by a novel sandwich-like fluorescence polarization screening assay. Chin. J. Biotechnol. 2022;38:2352–2364. doi: 10.13345/j.cjb.210949. [DOI] [PubMed] [Google Scholar]
- Yuan S., Gao X., Tang K., Cai J.P., Hu M., Luo P., Wen L., Ye Z.W., Luo C., Tsang J.O., Chan C.C., Huang Y., Cao J., Liang R., Qin Z., Qin B., Yin F., Chu H., Jin D.Y., Sun R., Chan J.F., Cui S., Yuen K.Y. Targeting papain-like protease for broad-spectrum coronavirus inhibition. Protein Cell. 2022;13:940–953. doi: 10.1007/s13238-022-00909-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Du X., Duan Y., Pan X., Sun Y., You T., Han L., Jin Z., Shang W., Yu J., Guo H., Liu Q., Wu Y., Peng C., Wang J., Zhu C., Yang X., Yang K., Lei Y., Guddat L.W., Xu W., Xiao G., Sun L., Zhang L., Rao Z., Yang H. High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors. Protein Cell. 2021;12:877–888. doi: 10.1007/s13238-021-00836-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.