To the Editor:
Allogeneic hematopoietic stem cell transplantation (HSCT) recipients are at a high risk of severe disease from SARS-CoV-2 infection with a mortality rate ranging from 22 to 25% [1, 2]. Therefore, the European Bone Marrow Transplantation guidelines recommend that HSCT be deferred until a patient is asymptomatic and has two negative polymerase chain reaction (PCR) tests [3, 4].
A 63-year-old female was diagnosed with very high-risk myelodysplastic syndrome according to the Revised International Prognostic Scoring System (pancytopenia, bone marrow blasts 10%, trisomy 8). Her past medical history was negative and had not been vaccinated for SARS-CoV-2. She achieved a partial response after five azacytidine cycles and was considered eligible for an allogeneic peripheral blood stem cell (PBSC) transplantation from her HLA-identical 57-year-old brother. Although at admission her PCR SARS-CoV-2 nasopharyngeal swab was negative, on day –2, after having completed the myeloablative preparative regimen, her monitoring swab was positive and confirmed by a second swab test. She was asymptomatic, had had no contact with confirmed SARS-CoV-2 cases, nor had she traveled to high-risk areas. Moreover, no visitors could enter the HSCT unit since November 2021. At PBSC collection, the donor, who was fully vaccinated, had a positive SARS-CoV-2 swab. The Omicron variant was identified by next-generation sequencing in both the patient’s and donor’s nasopharyngeal swabs, while only the recipient had viral RNA load in plasma. However, PBSC infusion could not be postponed as the myeloablative regimen had already been completed. The recipient was administered unmanipulated PBSC containing 5.93 × 106/kg CD34+ cells and 2.64 × 108/kg CD3+ cells on day 0. GVHD prophylaxis included cyclosporine A and a short-course methotrexate, with no ex vivo T-cell depletion.
A brief episode of desaturation occurred on day +1. The patient had neither fever nor cough and negative inflammation markers. No opacity or interstitial injury was evidenced at chest radiography and lung echography. High-flow nasal cannula therapy was discontinued two days later as arterial blood gas analysis evidenced normal oxygen saturation in room air. Although this post-transplant episode ran a mild course, it was not possible to exclude a correlation with SARS-CoV-2 as the patient’s swabs continued to be positive. Consequently, she was administered low-dose steroid (i.v. methylprednisolone at 0.5 mg/kg/day for 5 days) and remdesivir at 100 mg per day, for 5 days. Severe thrombocytopenia excluded antithrombotic prophylaxis.
Neutrophil engraftment was achieved on day +21 and complete donor chimerism was documented by fluorescent in situ hybridization on unfractionated peripheral blood cells (99.6%), CD3+ T cells (98.2%), and CD19+ B cells (99.6%) on day +30. She was discharged on self-isolation on day +31 and finally cleared the virus on day +46.
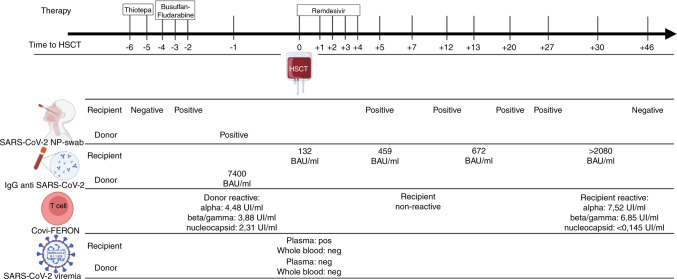
The recipient’s IgG anti-spike protein (anti-S) titer was low at day 0, before PBSC infusion (132 BAU/ml, LIAISON® SARS-CoV-2 S1/S2 IgG indirect chemiluminescent immunoassay – DiaSorin, Saluggia, Italy) and showed an upward trend until day +30 (IgG anti-S >2080 BAU/ml). Noteworthy, serum IgG rose on day +5 (439 BAU/ml), supporting the passive transfer of donor humoral immunity through the stem cell product.
The T-cell response to SARS-CoV-2 was assessed by interferon-γ release fluorescence immunoassay Covi-FERON FIA (SD BIOSENSOR, ReLab, Republic of Korea). Briefly, heparinized whole blood was incubated in test tubes for 16–24 h at 37 °C, after which, plasma was harvested and tested for the presence of interferon-γ produced in response to the SARS-CoV-2 spike protein derived from Alpha, Beta, and Gamma variants (B.1.1.7, B.1.351, and P.1, respectively) and the Nucleocapsid protein (antigen N). The recipient’s Covi-FERON test was non-reactive on day +7, whilst the donor had had a strong response to all antigens (day –1). Although the recipient’s CD4+ and CD8+ counts were low on day +30 (178 and 11, respectively), there was a significant T-lymphocyte response to the spike proteins but not to the nucleocapsid (Fig. 1).
Fig. 1. Timeline of SARS-CoV-2 testing, antiviral treatment, and transplant procedure.
The top row shows the days before and after hematopoietic stem cell transplantation (HSCT) when the conditioning regimen and antiviral therapy were administered. Below, the results of the recipient and donor SARS-CoV-2 tests are reported: SARS-CoV-2 nasopharyngeal swab (SARS-CoV-2 NP-swab); immunoglobulins G anti-SARS-CoV-2 levels (IgG); SARS-CoV-2 interferon-γ release assay (Covi-FERON) tested to the Alpha spike protein, Beta and Gamma spike proteins, and nucleocapsid antigen; B.1.1.529 SARS-CoV-2 viremia (Omicron variant).
To the best of our knowledge, this is the first reported case of a SARS-CoV-2 infection in an allogeneic HSCT recipient, whose donor was also SARS-CoV-2 positive.
Few reports have described HSCT patients with concomitant COVID-19 disease in the pre-engraftment and early post-engraftment phases. Some studies reported that HSCT recipients with COVID-19 disease have a poor outcome, but they were carried out during the period 2020-2021, when the predominant variants were Alpha and Delta [1, 2, 5]. Although the mutations identified in the Omicron spike protein (about 37) have led to serious concerns over antibody responses [6, 7], it seems that most T-cell immune responses are retained, potentially underpinning protection from a severe disease [6, 8, 9]. A recent study on vaccinated subjects evidenced a significant decrease in memory B cells and neutralizing antibodies at 6 months, with up to 42% less for the Omicron variant than others, whilst, there was an >80% average functional preservation of T-cell responses [8]. Therefore, it may be hypothesized that the myeloablative preparative regimen inhibited recipient T-cell response, at least during the early post-transplant phase. Nevertheless, the expansion of reactive T lymphocytes might well have contributed to minimizing the disease severity at one post-HSCT month. Although the interferon-γ response was evaluated to the Alpha, Beta, and Gamma variants, it may be assumed that those T-cell clones were also responsive to Omicron, in agreement with Tarke et al. [8]. The recipient had a high response to all variant spike proteins except the nucleocapsid antigen, confirming the preferential targeting of the spike protein [10].
Notwithstanding the low number of B lymphocytes, the serological assay revealed a concomitant IgG antibody rise that peaked at 2080 BAU/ml, on day +30. The recipient’s IgG anti-S protein was first detected before transplant and rose on day +5, supporting the passive transfer of donor humoral immunity through the stem cell product. It is conceivable that the remarkable immune response might have been induced by the expansion of donor lymphocytes as demonstrated by the chimerism analysis. Both T cell and humoral immunity might have contributed to limiting the extent of the disease. Indeed, although antibodies are responsible for viral neutralization, adaptive immunity is necessary for viral clearance [11].
Whether or not the absence of a SARS-CoV-2 hyperinflammatory syndrome was due to low lymphocyte and neutrophil counts and the concomitant administration of immunosuppressive agents, i.e., cyclosporine A, remains to be clarified. Although data supporting the efficacy of this antiviral agent in the immunocompromised host are scanty, treatment with remdesivir might have played a role in making the clinical course mild [2, 5].
In contrast with other authors who observed SARS-CoV-2 RNA more frequently in whole blood than in plasma, we detected SARS-CoV-2 RNA only in our recipient’s plasma [12]. However, this may be related to the differences in the viral-load kinetics in the two blood compartments analyzed, in line with other viruses in HSCT recipients [13].
The case herein reported suggests that the variant of concern, Omicron, has been associated with a mild clinical outcome even in patients at high risk of severe disease, such as HSCT recipients. However, further studies are required to clarify whether this is due to an intrinsic attenuated virulence or the capacity Omicron has to elicit a robust response of both adaptive and humoral immunity. We believe that, in our case, the potential role of humoral and cell-mediated immunity transferred from the donor should not be underestimated. Moreover, there might have been a different modulation of the immune response should the donor have been haploidentical or unrelated, or in the presence of a GVHD prophylaxis including in vivo/ex vivo T-cell depletion. Hopefully, further research will provide data able to optimize the donor selection process and clarify the impact SARS-CoV-2 infection has on immune reconstitution.
Supplementary information
Methods of viral sequencing, immune responses evaluation and analysis of chimerism
Acknowledgements
The authors would like to thank Barbara Wade, contract professor at the University of Torino, for her linguistic advice.
Author contributions
AB designed and wrote the first draft of the paper, interpreted the results, and provided clinical care for the patient described. GG designed and wrote the first draft of the paper, interpreted the results, provided clinical care for the patient described, updated the reference lists, and created the figure. FS, CC, RC, and FDR conducted the microbiological analysis and wrote the respective part of the paper. LG, ID, and CMD provided clinical care for the patient described. TF and CP conducted the stem cell collection. MR performed chimerism analysis. All authors were involved in the revision of the manuscript and approved the final version.
Data availability
All data generated or analyzed during this study are included in this article.
Competing interests
AB has received honoraria from Gilead Sciences, Merck, Pfizer Pharmaceuticals, and Jazz Pharmaceuticals; he has been a speaker for Gilead Sciences, Merck, Pfizer Pharmaceuticals, DiaSorin, GSK, Basilea, and Novartis; he has been part of an Advisory Board of Takeda, GILEAD and Pfizer. FDR has been a speaker for GSK and Gilead; he has been part of an Advisory Board of GSK and Gilead. All other authors have no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Alessandro Busca, Giulia Gabrielli.
Supplementary information
The online version contains supplementary material available at 10.1038/s41409-022-01763-x.
References
- 1.Ljungman P, de la Camara R, Mikulska M, Tridello G, Aguado B, Zahrani MA, et al. COVID-19 and stem cell transplantation; results from an EBMT and GETH multicenter prospective survey. Leukemia. 2021;35:2885–94. doi: 10.1038/s41375-021-01302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma A, Bhatt NS, St Martin A, Abid MB, Bloomquist J, Chemaly RF, et al. Clinical characteristics and outcomes of COVID-19 in haematopoietic stem-cell transplantation recipients: an observational cohort study. Lancet Haematol. 2021;8:e185–93. doi: 10.1016/S2352-3026(20)30429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.EBMT COVID-19 guidelines v. 16.03.pdf. https://www.ebmt.org/sites/default/files/2021-06/EBMT%20COVID-19%20guidelines%20v.%2016.03.pdf. Accessed Feb 7 2022.
- 4.World Health Organization. Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. 2021. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern.
- 5.Mushtaq MU, Shahzad M, Chaudhary SG, Luder M, Ahmed N, Abdelhakim H, et al. Impact of SARS-CoV-2 in hematopoietic stem cell transplantation and chimeric antigen receptor T cell therapy recipients. Transplant Cell Ther. 2021;27:796.e1–796.e7. doi: 10.1016/j.jtct.2021.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.GeurtsvanKessel CH, Geers D, Schmitz KS, Mykytyn AZ, Lamers MM, Bogers S, et al. Divergent SARS-CoV-2 Omicron–reactive T and B cell responses in COVID-19 vaccine recipients. Sci Immunol. 2022;7:eabo2202. [DOI] [PMC free article] [PubMed]
- 7.Guo H, Gao Y, Li T, Li T, Lu Y, Zheng L, et al. Structures of Omicron spike complexes and implications for neutralizing antibody development. Cell Rep. 2022;39:110770. doi: 10.1016/j.celrep.2022.110770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tarke A, Coelho CH, Zhang Z, Dan JM, Yu ED, Methot N, et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell. 2022;185:847–59.e11. [DOI] [PMC free article] [PubMed]
- 9.Gao Y, Cai C, Grifoni A, Müller TR, Niessl J, Olofsson A, et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat Med. 2022;28:472–6. doi: 10.1038/s41591-022-01700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandberg JT, Varnaitė R, Christ W, Chen P, Muvva JR, Maleki KT, et al. Longitudinal characterization of humoral and cellular immunity in hospitalized COVID-19 patients reveal immune persistence up to 9 months after infection. Immunology. 2021. 10.1101/2021.03.17.435581.
- 11.Tan AT, Linster M, Tan CW, Le Bert N, Chia WN, Kunasegaran K, et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021;34:108728. doi: 10.1016/j.celrep.2021.108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W, Du RH, Li B, Zheng XS, Yang XL, Hu B, et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9:386–9. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazzarotto T, Chiereghin A, Piralla A, Gibertoni D, Piccirilli G, Turello G, et al. Kinetics of cytomegalovirus and Epstein-Barr virus DNA in whole blood and plasma of kidney transplant recipients: implications on management strategies. PLoS ONE. 2020;15:e0238062. doi: 10.1371/journal.pone.0238062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methods of viral sequencing, immune responses evaluation and analysis of chimerism
Data Availability Statement
All data generated or analyzed during this study are included in this article.