Abstract
Background
Hydrangea macrophylla var. Maculata ‘Yinbianxiuqiu’ (YB) is an excellent plant species with beautiful flowers and leaves with silvery white edges. However, there are few reports on its leaf color characteristics and color formation mechanism.
Results
The present study compared the phenotypic, physiological and transcriptomic differences between YB and a full-green leaf mutant (YM) obtained from YB. The results showed that YB and YM had similar genetic backgrounds, but photosynthesis was reduced in YB. The contents of pigments were significantly decreased at the edges of YB leaves compared to YM leaves. The ultrastructure of chloroplasts in the YB leaves was irregular. Transcriptome profiling identified 7,023 differentially expressed genes between YB and YM. The expression levels of genes involved in photosynthesis, chloroplast development and division were different between YB and YM. Quantitative real-time PCR showed that the expression trends were generally consistent with the transcriptome data.
Conclusions
Taken together, the formation of the silvery white leaf color of H. macrophylla var. maculata was primarily due to the abnormal development of chloroplasts. This study facilitates the molecular function analysis of key genes involved in chloroplast development and provides new insights into the molecular mechanisms involved in leaf coloration in H. macrophylla.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-022-03727-1.
Keywords: Hydrangea macrophylla, Leaf color, Chlorophyll, Transcriptome, Chloroplast development
Background
Hydrangea (Hydrangea macrophylla (Thunb.) Ser.) is a deciduous species belonging to the family Saxifragaceae. Hydrangea is a popular ornamental plant species in Asia, America and Europe [1, 2]. The sepal color of hydrangea plants changes from white to pink, red, purple and blue when cultured in soils with different pH values and Al3+ contents [3]. Famous for its charming large and multicolored flowers, hydrangea is extensively used as potted, bouquet and landscape plants.
Chloroplasts produce carotenoids and chlorophyll, and chlorophyll is the main pigment component in green leaves [4]. Many studies focused on the biosynthesis and degradation of chlorophyll. For example, the chlorophyll biosynthesis pathway in Arabidopsis thaliana starts from glutamyl-tRNA to chlorophylls a and b, and 27 genes encoding 15 enzymes for all 15 steps were identified [5]. Four enzymes are involved in the chlorophyll degradation pathway from chlorophyll b to nonfluorescent chlorophyll catabolites [6]. The silencing of HrHEMA (glutamyl-tRNA reductase) and HrCAO (chlorophyllide an oxygenase) genes significantly affected the structure of the chloroplast and resulted in a change in leaf color [7]. Virus-induced gene silencing of CHLI (magnesium-chelatase I subunit) reduced chlorophyll content and altered chloroplast function, which led to abnormal chloroplast structure in peas [8]. Impaired function of NYC1 (NON-YELLOW COLORING1) or NOL (NYC1-like) resulted in a stay-green phenotype in rice [9, 10].
As the site of photosynthesis, chloroplasts consist of the chloroplast membrane, thylakoid and matrix [11]. The number and distribution of chloroplasts in the tissue primarily influence leaf color. Previous studies showed that the altered expression of genes related to chloroplasts affected the biogenesis of chloroplasts [4, 7, 12]. The disruption of chloroplast assembly may lead to abnormal leaf color [13–15]. Variation in leaf color is one of the most common phenomena in higher plant species. Leaf color mutations were identified in various green plants, such as cotton [16], wheat [17], Anthurium andraeanum [13], birch [18] and Hosta plantaginea [7]. Leaf mutants help reveal the molecular mechanisms of leaf color formation and have received increasing attention. Changes in the expression levels of key genes involved in chloroplast development and division generally result in leaf color mutations [19]. For example, maize pentatricopeptide repeat 4 (PPR4) is necessary for the normal development of chloroplasts by associating with plastid rps12 pre-mRNA and splicing in trans [20]. The Golden 2-like (GLK) genes play a positive role in the regulation of chloroplast development [21], and Arabidopsis glk1-glk2 double mutants showed a pale green phenotype with a lack of chloroplast thylakoid membranes and grana [22, 23]. The gene family Accumulation and Replication of Chloroplasts (ARC), ARC3, ARC5 and ARC6 regulate the division of chloroplasts [24–26]. OscpSRP43 (chloroplast signal recognition particle 43) is required for the normal development of chloroplasts in rice, and the color mutation exhibited a distinct yellow-green leaf phenotype with impaired chloroplasts [27].
H. macrophylla var. maculata ‘Yinbianxiuqiu’ (YB) is an excellent variety with beautiful flowers and leaves with silvery white edges. A mutant with full-green leaves (YM) was obtained from one YB plant. YM and YB differed only in leaf color. However, the mechanism leading to leaf color variation in YB is not clear. Therefore, the present study compared the phenotypic and physiological characteristics of YB and YM. Leaf transcriptomes from YB and YM plants were sequenced, and genes involved in photosynthesis, chloroplast development and division and chlorophyll biosynthesis and degradation were identified. The expression of genes involved in chloroplast development and division was validated using quantitative real-time PCR (qRT–PCR). The present study elucidated the molecular mechanisms that regulate leaf color formation in H. macrophylla to provide a foundation for breeding hydrangea varieties with ornamental leaves.
Results
Phenotypic and sequence-related amplified polymorphism (SRAP) analyses
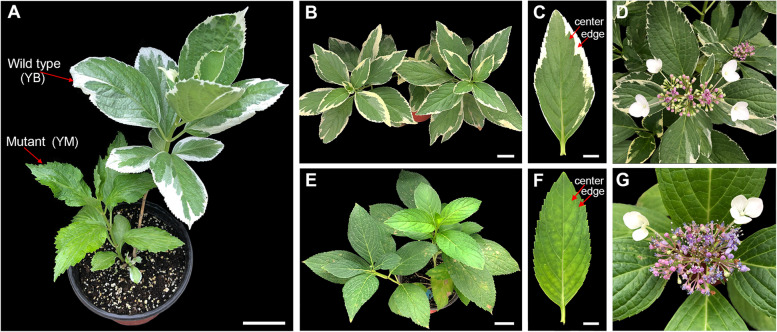
A full-green leaf mutant (YM) was obtained from YB (Fig. 1A). After years of vegetative propagation, the full-green leaves of YM were stable and bloomed normally (Fig. 1E, F, G).
Fig. 1.
Phenotypes of H. macrophylla var. maculata (YB) and full-green leaf mutant (YM). (A) YM was a bud mutation derived from YB. Bar: 5 cm. (B) YB plant. Bar: 5 cm. (C) Leaf of YB. Bar: 1 cm. (D) Flower of YB. (E) YM plant. Bar: 5 cm. (F) Leaf of YM. Bar: 1 cm. (G) Flower of YM. center: central position of leaf; edge: edge position of leaf
A total of 136 SRAP primers amplified 1,519 fragments from YB and 1,518 fragments from YM (Fig. S1). Most primer pairs amplified the same fragments from YB and YM, except three primer pairs, M6/E18, M13/E16 and M16/E11. Primer pair M6/E18 amplified three fragments in YB and two in YM. Primer pair M13/E16 amplified ten fragments in YB and nine in YM, and primer pair M16/E11 amplified eight fragments in YB and nine in YM. These results indicated that YB and YM had a similar genetic background.
Photosynthesis and photosynthetic pigments content
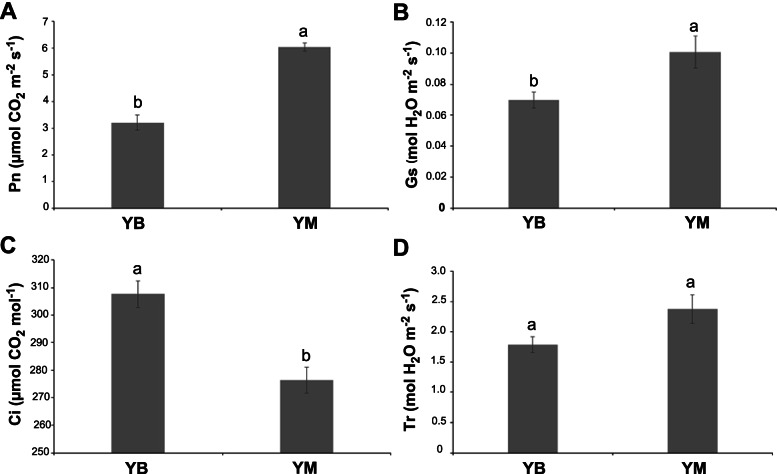
The net photosynthetic rate (Pn) and stomatal conductance (Gs) values of YB and YM varied significantly, and both values were higher in YM (Fig. 2A, B). The intercellular CO2 concentration (Ci) values also varied significantly between YB and YM, and the Ci value of YB was higher than YM (Fig. 2C). However, the transpiration rate (Tr) values showed no significant variation between YB and YM (Fig. 2D). These results suggested that the plant photosynthesis capacity differed between YB and YM, and YM capacity was higher than YB.
Fig. 2.
Photosynthetic parameters of H. macrophylla var. maculata (YB) and full-green leaf mutant (YM). (A) Net photosynthetic rate (Pn). (B) Stomatal conductance (Gs). (C) Intercellular CO2 concentration (Ci). (D) Transpiration rate (Tr)
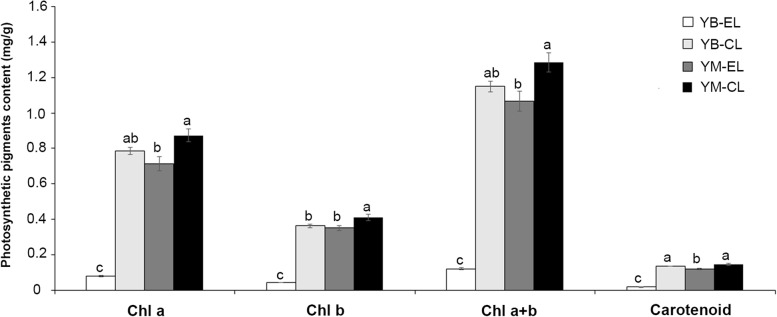
The chlorophyll and carotenoid contents were significantly different between YB and YM (Fig. 3). The highest contents of chlorophyll a, chlorophyll b and chlorophyll a + b were detected in the central position of the YM leaf, and the lowest contents were detected in the edge of the YB leaf. The highest carotenoid contents were measured in the central position of YB and YM leaves, and the lowest carotenoid contents were measured in the edges of YB leaves. These results indicated that the abnormal leaf color of YB closely correlated with the change in pigment contents.
Fig. 3.
The content of photosynthetic pigments in H. macrophylla var. maculata (YB) and full-green leaf mutant (YM). EL: edged leaf; CL: central leaf
Chloroplast ultrastructure
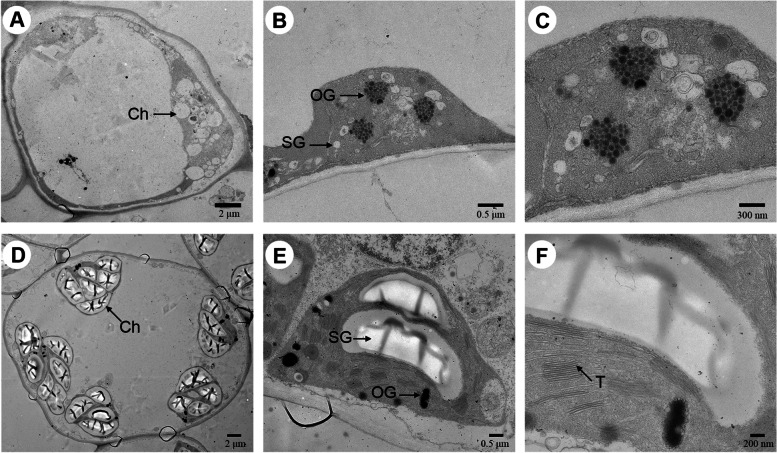
The chloroplast ultrastructure was analyzed to verify the abnormal development of chloroplasts in the mesophyll cells of YB. The shape, size and number of chloroplasts were obviously different between YB and YM (Fig. 4). Chloroplasts had an intact stromal thylakoid structure in YM (Fig. 4E, F). However, the chloroplasts in YB had no inner member structures (Fig. 4B, C). The shape of the chloroplasts was elliptical or ovoid in YM (Fig. 4D) but swollen oblate or spheroidal in YB (Fig. 4A). Chloroplasts in the mesophyll cells of YM contained small starch granules and a few osmiophilic globules (Fig. 4E, F), but large starch granules and many osmiophilic globules were observed in YB (Fig. 4B, C). These results showed that the ultrastructure of chloroplasts in the YB leaves was irregular, which confirmed the abnormal chloroplast development.
Fig. 4.
Chloroplast ultrastructure of H. macrophylla var. maculata (YB) and full-green leaf mutant (YM). (A-C) Chloroplast ultrastructure of YB. In (A), bar: 2 μm; (B), bar: 0.5 μm; (C), bar: 300 nm. (D-F) Chloroplast ultrastructure of YM. In (D), bar: 2 μm; (E), bar: 0.5 μm; (F), bar: 200 nm. Ch: chloroplast; OG: osmiophilic globules; SG: starch grains; T: thylakoid
Illumina sequencing and assembly
Based on the above phenotypic and physiological characteristics, we speculated that the expression patterns of genes involved in chloroplast development and division and pigment metabolism were altered in YM plants. To test this hypothesis, the leaf edges of YB and YM were collected and sequenced to examine the mechanism of leaf color formation. The total number of raw reads per library ranged from 47,956,778 to 64,679,848, and the total number of clean reads ranged from 47,950,946 to 64,664,862 (Table S1). The proportion of clean reads and clean data was > 99.97% in each library (Table S1). A total of 123,122 unigenes with an N50 length of 1,235 bp were obtained from the H. macrophylla transcriptome via de novo assembly (Table 1). The unigenes had an average length of 778 bp, a median length of 453 bp, a minimum length of 201 bp, a maximum length of 15,890 bp, and a total length of 95,817,440 bp. The GC content of unigenes was 38.98% (Table 1). The length distribution of unigenes is shown in Fig. S2, and 17,998 unigenes had lengths over 1,000 bp.
Table 1.
Statistics of unigene sequences in the H. macrophylla transcriptome
| Item | Total length (bp) | Total number | GC content (%) | N50 (bp) | N90 (bp) | Average (bp) | Median (bp) | Min (bp) | Max (bp) |
|---|---|---|---|---|---|---|---|---|---|
| Value | 95,817,440 | 123,122 | 38.98 | 1,235 | 316 | 778 | 453 | 201 | 15,890 |
Gene functional annotation
All 123,122 assembled unigenes were annotated in the NR, UniProt, GO, KEGG, eggNOG and Pfam databases (Table S2). A total of 42,572 (34.58%) unigenes were matched in at least one of the these databases. There were 40,235, 25,792 and 12,457 unigenes annotated in the UniProt, Pfam and eggNOG databases, respectively.
For the NR annotation, 40,796 unigenes had hits in the NR database (Table S2). The E-value distribution pattern showed that 46.83% of the top hits had high homology with an E-value < 1e−50 (Fig. S3A). For identification, more than 71% of the sequences had a similarity higher than 60%, and most of the annotated unigenes had identified distributions that ranged from 60 to 80% (Fig. S3B). On a species basis, the annotated sequences had identical fragments with genes from Actinidia chinensis var. chinensis (22.66%), Vitis vinifera (10.47%) and Quercus suber (3.85%) (Fig. S3C).
All of the unigenes were categorized according to the secondary classification of GO terms. A total of 29,824 unigenes were assigned to a GO term in three main GO classification categories: biological process, cellular component and molecular function (Fig. S4 and Table S2). The major classes of the biological process category were DNA metabolic process, biosynthetic process and cellular nitrogen compound metabolic process. The terms cellular component, nucleus and cytoplasm were dominant in the cellular component category. The main molecular function category terms were ion binding, molecular function and kinase activity.
KEGG pathway analysis revealed that 3,017 unigenes were assigned to 341 pathways (Tables S2 and S3). The major enrichments among metabolic pathways were carbon metabolism, biosynthesis of amino acids, ABC transporters, purine metabolism, two-component system and pyruvate metabolism. The metabolic pathways related to leaf color were porphyrin and chlorophyll metabolism (ko00860; 50 unigenes), photosynthesis (ko00195, 23 unigenes), carotenoid biosynthesis (ko00906, 11 unigenes), flavonoid biosynthesis (ko00941, 5 unigenes), and flavone and flavonol biosynthesis (ko00944, 3 unigenes).
Differentially expressed genes (DEGs) and functional analysis
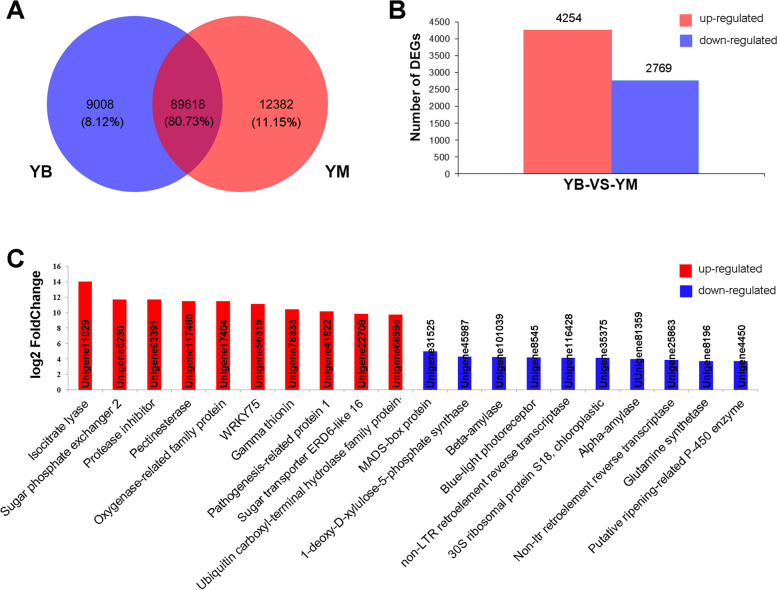
Fragments per kilobase per million (FPKM) was used to estimate the expression levels of genes. As a result, 98,626 and 102,000 unigenes were identified in the YB and YM libraries, respectively (Table S4). A total of 9,008 (8.12%) and 12,382 (11.15%) unigenes were expressed specifically in the leaves of YB and YM, respectively, and 89,618 (80.73%) unigenes were co-expressed in YB and YM (Fig. 5A). To identify DEGs between YB and YM, the expression of genes in YM was set as the control. A total of 7,023 genes showed at least a two-fold change in gene expression level, including 4,254 up-regulated and 2,769 down-regulated genes (Fig. 5B and Table S5). The top 10 significantly up- and down-regulated genes whose sequences were annotated were detected between YB and YM (Fig. 5C). These genes may be involved in leaf color formation.
Fig. 5.
The number of genes and differentially expressed genes were detected in H. macrophylla var. maculata (YB) and full-green leaf mutant (YM). (A) The number of genes was detected in the YB and YM libraries. (B) Differentially expressed genes were detected between YB and YM. (C) Top 10 significantly up- and down-regulated genes whose sequences were annotated were detected between YB and YM
The functions of the DEGs were categorized according to the secondary classification of GO terms. The results showed that 203, 192 and 454 DEGs were divided into biological process, cellular component and molecular function categories, respectively (Fig. S5 and Table S6). The most enriched terms of the biological process were cell wall organization, signal transduction and lipid metabolic process. Many DEGs in the cellular component category were associated with the membrane, plasmodesma and extracellular region. The dominant molecular function category terms were hydrolase activity, iron ion binding and heme binding.
KEGG pathway analysis was performed to categorize the DEGs. A total of 82 DEGs were enriched in 21 pathways in the KEGG database (Table S7). Plant hormone signal transduction was the most enriched pathway, followed by antigen processing and presentation. There were three DEGs in the photosynthesis pathway (Table S7). Compared to YM, the expression levels of photosystem, chlorophyll a-b binding protein, ATP synthase and cytochrome genes in YB were down-regulated (Table 2). This result further confirmed that the photosynthesis of YM was higher than YB.
Table 2.
Differentially expressed genes involved in photosynthesis in the H. macrophylla transcriptome
| Gene ID | log2(YB/YM) | Annotation |
|---|---|---|
| Unigene35224 | -3.34 | Photosystem II reaction center protein K |
| Unigene21595 | -2.78 | Photosystem II protein D1 |
| Unigene83666 | -2.42 | Photosystem II CP47 reaction center protein |
| Unigene16923 | -1.88 | Photosystem II CP43 reaction center protein |
| Unigene71781 | -1.81 | Photosystem II D2 protein |
| Unigene27991 | -1.32 | Photosystem I P700 chlorophyll a apoprotein A1 family |
| Unigene105647 | -1.31 | Photosystem II type I chlorophyll a/b-binding protein |
| Unigene25859 | -1.15 | Photosystem II reaction center PsbP family protein |
| Unigene64241 | -1.05 | Photosystem I assembly protein ycf4 |
| Unigene87772 | -1.13 | Chlorophyll a-b binding protein 2 |
| Unigene30677 | -1.94 | ATP synthase subunit beta |
| Unigene78078 | -2.08 | Cytochrome f |
| Unigene116390 | -1.33 | Cytochrome b6 |
| Unigene93247 | -1.05 | Cytochrome b6-f complex iron-sulfur subunit |
Analysis of genes related to chloroplast development and division and chlorophyll biosynthesis and degradation
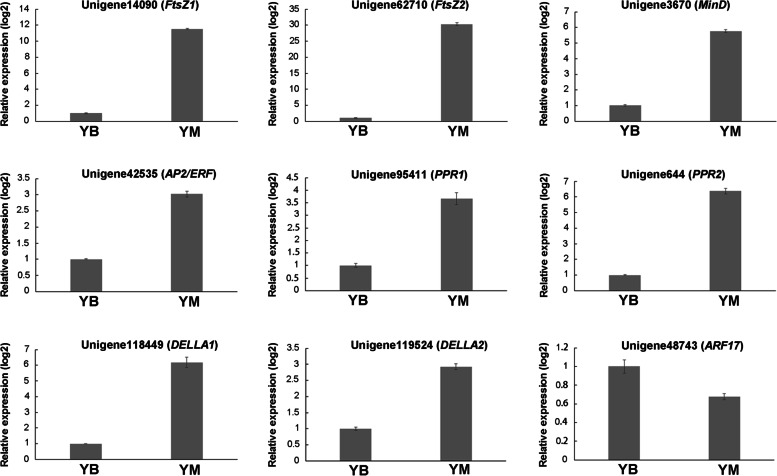
Based on the H. macrophylla transcriptome data, the unigenes involved in chloroplast development and division and chlorophyll biosynthesis and degradation were identified (Table 3). Compared to YM, the expression levels of DELLA, PPR, GLK and Thf1 (chloroplast development) and FtsZ, MinD and AP2/ERF (chloroplast division) in YB were down-regulated, and the expression levels of ARFs (chloroplast development) were up-regulated (Table 3). However, the unigenes involved in chlorophyll biosynthesis and degradation had no significant expression pattern changes between YB and YM (Table 3). These results indicated that the leaf color of YB may be caused by the expression pattern changes of chloroplast development and division genes.
Table 3.
Unigenes involved in chloroplast development and division and chlorophyll biosynthesis and degradation in the H. macrophylla transcriptome
| Function | Gene name | Gene ID | log2(YB/YM) | Annotation |
|---|---|---|---|---|
| chloroplast development | DELLA | Unigene73671 | -1.42 | DELLA protein GAI |
| Unigene118449 | -1.17 | DELLA protein GAI-like | ||
| Unigene119524 | -3.21 | GA repressor DELLA | ||
| PPR | Unigene95411 | -1.22 | pentatricopeptide repeat-containing protein At1g08070 | |
| Unigene644 | -1.04 | pentatricopeptide repeat-containing protein At2g15820 | ||
| GLK | Unigene20679 | -2.17 | transcription activator GLK1 | |
| Thf1 | Unigene88785 | -1.28 | thylakoid formation1 | |
| ARF | Unigene48743 | 1.23 | auxin response factor 17 | |
| Unigene118329 | 1.94 | Auxin response factor | ||
| Unigene122697 | 1.15 | Auxin response factor | ||
| Unigene23042 | 1.02 | Auxin response factor | ||
| Unigene32938 | 1.21 | Auxin response factor | ||
| Unigene33148 | 1.79 | Auxin response factor 17 | ||
| Unigene48743 | 1.23 | Auxin response factor 17-like | ||
| Unigene56446 | 1.25 | Auxin response factor | ||
| Unigene78971 | 1.21 | Auxin response factor 1 | ||
| chloroplast division | FtsZ | Unigene65459 | -1.70 | cell division protein FtsZ homolog 2–2, chloroplastic |
| Unigene14090 | -1.08 | tubulin beta chain | ||
| Unigene62710 | -1.09 | tubulin alpha chain | ||
| MinD | Unigene3670 | -1.31 | Adenylyl-sulfate kinase | |
| AP2/ERF | Unigene42535 | -1.43 | AP2/ERF transcription factor | |
| Unigene17785 | -1.30 | AP2/ERF transcription factor | ||
| chlorophyll biosynthesis | HEMA | Unigene94259 | 0.30 | Glutamyl-tRNA reductase |
| GSA | Unigene21371 | 0.26 | Glutamate-1-semialdehyde 2,1-aminomutase | |
| HEMB | Unigene80924 | 0.07 | Delta-aminolevulinic acid dehydratase, chloroplastic | |
| Unigene54524 | 0.52 | Delta-aminolevulinic acid dehydratase, chloroplastic | ||
| HEMC | Unigene43517 | 0.62 | Porphobilinogen deaminase | |
| Unigene80400 | -0.54 | Porphobilinogen deaminase | ||
| HEMD | Unigene42585 | -0.56 | Uroporphyrinogen-III synthase | |
| Unigene106934 | 0.75 | Uroporphyrinogen-III synthase | ||
| HEME | Unigene3363 | 0.43 | Uroporphyrinogen decarboxylase | |
| Unigene103027 | 0.30 | Uroporphyrinogen decarboxylase 1, chloroplastic | ||
| Unigene6415 | 0.41 | Uroporphyrinogen decarboxylase | ||
| HEMG | Unigene34893 | 0.19 | Protoporphyrinogen oxidase | |
| Unigene106486 | 0.63 | Protoporphyrinogen oxidase | ||
| CHLH | Unigene1894 | 0.81 | Magnesium-chelatase subunit ChlH like | |
| CHLI | Unigene48515 | 0.55 | Magnesium-chelatase subunit ChlI, chloroplastic-like | |
| CHLM | Unigene22407 | -0.59 | Mg-protoporphyrin IX methyltransferase | |
| CHLG | Unigene25797 | 0.07 | Chlorophyll synthase | |
| CAO | Unigene110336 | -0.93 | Chlorophyllide an oxygenase | |
| chlorophyll degradation | NYC1 | Unigene105397 | 0.03 | Putative chlorophyll(Ide) b reductase NYC1 |
| NOL | Unigene114249 | 0.18 | Chlorophyll b reductase NOL protein | |
| CLH1 | Unigene91850 | -0.49 | chlorophyllase-1 | |
| Unigene28136 | -0.81 | chlorophyllase-2 | ||
| PPH | Unigene14410 | 0.70 | Pheophytinase | |
| Unigene43673 | -0.03 | pheophytinase | ||
| Unigene47431 | -0.55 | pheophytinase | ||
| PAO | Unigene110336 | -0.93 | Pheophorbide an oxygenase | |
| Unigene1999 | -0.69 | Pheophorbide an oxygenase | ||
| Unigene74026 | -0.10 | Pheophorbide an oxygenase | ||
| Unigene98774 | -0.66 | Pheophorbide an oxygenase | ||
| RCCR | Unigene7212 | -0.01 | Red chlorophyll catabolite reductase |
Validation of DEGs using qRT–PCR
To verify the reliability of the transcriptome data, the expression patterns of the genes that were significantly expressed in the samples were verified using qRT–PCR (Fig. 6). The expression trends were generally consistent with the transcript abundances estimated from the RNA-Seq data, but the selected genes showed different fold-change values (Fig. 6 and Table 3). These results confirmed the reliability of the transcriptome data.
Fig. 6.
Verification of genes involved in chloroplast development and division using qRT–PCR between YB and YM
Discussion
Chlorophyll and carotenoids are the major pigments in green leaves. Previous studies demonstrated that leaf color mutants commonly contained less chlorophyll and carotenoids [13, 15, 18]. For example, the contents of pigments, including chlorophyll a, chlorophyll b, total chlorophyll and carotene, were decreased in the durum wheat mutant [28]. The present study obtained a mutant with green leaves (YM) from the YB plant. Consistent with previous reports, the pigment contents were lower in YB than YM (Fig. 3). Photosynthesis is a complex process that is easily affected by changes in pigment contents [29]. Physiological experiments showed that photosynthesis was restricted in YB (Fig. 2). Therefore, there may be fewer light-harvesting protein complexes in YB than YM, which was verified by the results that showed that the down-regulated DEGs were primarily enriched in photosynthesis (Table 2). These results revealed that the expression changes of genes involved in photosynthesis played an important role in the formation of the capacity of plant photosynthesis. Previous research reported that the expression change of genes related to photosynthesis in Brassica campestris mutants led to abnormal chloroplast development and reduced pigment content [30]. Therefore, the decreased capacity of photosynthesis in YB was likely due to the reduction of pigment contents and the low expression level of photosystem, chlorophyll a-b binding protein, ATP synthase and cytochrome genes.
Chloroplast development of higher plants requires the coordination of nuclear genes and chloroplast genes [31]. The DELLA proteins that accumulate in the nucleus are key suppressors of GA responses via inhibition of GA-regulated gene expression [32]. DELLA proteins act negatively in GA responses by interacting with diverse regulators or transcription factors [33, 34]. GA levels are reduced under light, which stimulates DELLA accumulation and abolishes negative control by DELLA targets, including PIFs that are suppressors of chloroplast development [35, 36]. Analysis of della mutants revealed the complicated regulation of chloroplast development, and it was reported that GA prevented photomorphogenesis in the dark [37]. As nuclear factors, PPR proteins are involved in the expression of chloroplast genes in many post-transcriptional processes [38, 39]. AtECB2 (a PPR gene) regulated the editing of the accD and ndhF genes in Arabidopsis early chloroplast biogenesis, and the ecb2 mutant showed a lack of thylakoid membranes with a delayed greening phenotype [40]. GhYGL1d (a PPR gene) regulated the development of thylakoids in cotton by editing the accD and ndhF genes, therefore, GhYGL1d-silenced cotton exhibited significant abnormalities in thylakoid structures compared to wild-type cotton [41]. Previous studies suggested that GLK genes were involved in regulating chloroplast development in plant species [22, 42, 43]. Members of the GLK gene family are sensitive to chloroplast retrograde signaling, and they control downstream genes for plastid retrograde signaling [44]. Compared to the wild type A. andraeanum, GLK was downregulated in the rubescent mutant [13]. The A. thaliana thylakoid formation1 (Thf1) gene controls vesicles maturation into thylakoid stacks and ultimately for leaf development, and deletion of AtThf1 leads to deficient thylakoid formation and variegated leaves [45]. Consistent with these studies, the expression levels of HmDELLA, HmPPR, HmGLK and HmThf1 in YB were lower than YM in the present study (Fig. 6 and Table 3), which indicates that these genes are related to chloroplast development.
Auxin is involved in photomorphogenesis, as proposed by the phenotype of dark-induced hypocotyl elongation in auxin-response mutants [46]. Auxin response factors (ARFs), which bind to promoters of auxin-responsive genes to regulate transcription, mediate numerous auxin responses. ARF function is suppressed by auxin/indole-3-acetic acid inducible (Aux/IAA) proteins. Auxin negatively regulates root greening via IAA14/SLR, ARF7 and ARF19 [47]. Overexpression of CYTOKININ-RESPONSIVE GATA FACTOR 1 (CGA1) and GATA, NITRATE-INDUCIBLE, CARBON-METABOLISMINVOLVED (GNC) promote the differentiation of etioplasts to chloroplasts in the light [48]. ARF2 binds to the promoters of CGA1 and GNC, suppresses their expression, and arf2 mutation promotes root greening [49]. The expression levels of HmARFs in YB were higher than YM (Fig. 6 and Table 3), which is consistent with previous reports and suggests the involvement of HmARFs in chloroplast development.
Inhibiting the expression of FtsZ genes in transgenic Arabidopsis plants significantly reduced the number of chloroplasts in mature leaves, which indicates that FtsZ genes are essential for the division of plant chloroplasts [50, 51]. The proteins AtFtsZ, AtMinD and AtMinE act in concert during chloroplast division [52]. The expression patterns of FtsZ and MinD in YB were similar to the A. andraeanum rubescent mutant [13], which indicates that HmFtsZ and HmMinD are related to chloroplast division.
Cytokinin response factor 2 (CRF2) belongs to the APETLA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) transcription factor family [53]. Overexpression of CRF2 resulted in an increased level of Plastid Division 2 (PDV2) and promoted chloroplast division [54]. The expression levels of HmAP2/ERFs (Unigene42535 and Unigene17785) in YB were lower than YM in the present study (Fig. 6 and Table 3), which indicates that HmAP2/ERF is involved in chloroplast division.
Conclusions
In conclusion, the photosynthesis and pigment contents were reduced in YB compared to YM, and the expression levels of many genes related to chloroplast development and division were changed. These results suggest that the change in gene expression patterns involved in chloroplast development and division are responsible for the abnormal ultrastructure of chloroplasts, which results in the silvery white leaf edges in YB. Our results provide a basis for further research on leaf color mutation.
Methods
Plant materials
YM was a bud mutation derived from YB (Fig. 1A), which varied only in leaf color (Fig. 1). The YB and YM were maintained in the Preservation Centre of the Hydrangea Germplasm Resources, Jiangsu Academy of Agricultural Sciences, Nanjing, China (latitude: 32°05′N, longitude: 118°08′E; 68 m above sea level) (Fig. 1B, E). The materials were propagated by cuttings. The plants were grown in a greenhouse (25 °C during the day and 15 °C at night; relative humidity of 60–70%; under natural light).
SRAP analysis
DNA was extracted from the fourth leaf of three individual YB and YM plants using a modified CTAB method [55]. The DNA was used for SRAP profiling as described by Li and Quiros [56]. A total of 136 SRAP primer pairs were used, including 24 forward and 20 reverse primers (Table S8). The pairs were M1 combined with E1 (abbreviated “M1/E1”), M1/E16, M1/E19, M2/E1, M2/E2, M2/E6, M2/E12, M2/E15, M2/E18, M3/E1, M3/E2, M3/E3, M3/E5, M3/E11, M3/E17, M4/E3, M4/E4, M4/E6, M4/E8, M4/E11, M5/E1, M5/E3, M5/E5, M5/E7, M5/E8, M6/E1, M6/E2, M6/E6, M6/E11, M6/E14, M6/E18, M6/E19, M7/E2, M7/E4, M7/E6, M7/E7, M7/E15, M7/E17, M7/E20, M8/E3, M8/E5, M8/E8, M8/E12, M8/E15, M8/E16, M8/E18, M9/E9, M9/E15, M9/E18, M10/E1, M10/E2, M10/E4, M10/E10, M11/E2, M11/E5, M11/E8, M11/E10, M11/E11, M11/E16, M12/E2, M12/E7, M12/E10, M12/E12, M12/E13, M12/E15, M12/E17, M13/E3, M13/E6, M13/E8, M13/E13, M13/E16, M14/E2, M14/E5, M14/E7, M14/E10, M14/E11, M14/E14, M14/E16, M15/E1, M15/E10, M15/E15, M15/E17, M15/E20, M16/E2, M16/E10, M16/E11, M16/E14, M16/E15, M16/E16, M16/E19, M17/E1, M17/E5, M17/E7, M17/E15, M17/E19, M18/E6, M18/E8, M18/E10, M18/E14, M18/E18, M19/E5, M19/E7, M19/E12, M19/E16, M19/E18, M19/E19, M20/E2, M20/E4, M20/E8, M20/E13, M21/E1, M21/E2, M21/E4, M21/E6, M21/E14, M21/E17, M21/E19, M22/E1, M22/E2, M22/E5, M22/E6, M22/E11, M22/E15, M22/E20, M23/E3, M23/E6, M23/E10, M23/E12, M23/E16, M23/E18, M24/E2, M24/E4, M24/E8, M24/E11, M24/E15 and M24/E16. Each 25 μl reaction mix was comprised of 2.5 μl of 10 × PCR buffer, 0.2 mM dNTPs, 1.5 mM MgCl2, 15 ng genomic DNA and 2 U of Taq polymerase (Takara, Japan). The reactions were first denatured (94 °C/5 min), followed by 5 cycles of 94 °C/1 min, 35 °C/1 min and 72 °C/2 min, followed by 35 cycles of 94 °C/1 min, 50 °C/1 min and 72 °C/2 min, with a final extension step of 72 °C/10 min. The SRAP amplicons were electrophoresed with 6% denaturing polyacrylamide gels and visualized via silver staining. Fragments in the size range of 100–500 bp were scored.
Photosynthetic parameters
Photosynthetic parameters consisting of Pn (µmol CO2 m−2 s−1), Gs (mol H2O m−2 s−1), Ci (µmol CO2 mol−1) and Tr (mol H2O m−2 s−1) were determined using an LI-6400 portable photosynthesis system (LI-COR, Lincoln, NE, USA). These data were recorded between 9:00 and 11:00 am using the first most fully expanded leaves from the apex of the shoot. The air cuvette temperature, CO2 concentration and irradiance were maintained at 30 °C, 420 µmol CO2 mol−1 and 1000 µmol m−2 s−1, respectively [57]. Ten representative plants of YB and YM were selected randomly and determined.
Chlorophyll and carotenoid content
Each leaf was cut into edged leaves (EL) and central leaves (CL) (Fig. 1C and 1F). Chlorophyll a, chlorophyll b and total carotenoid contents were measured using the methods of Zhang et al. [58]. Approximately 200 mg (fresh weight) of the fourth leaf from the stem tip was incubated in 10 mL 95% ethanol for 48 h in the dark. The absorbance of the supernatant was analyzed using spectrophotometry (UH5300, HITACHI, Tokyo, Japan) at 665, 649 and 470 nm. Total Chl (Chl a + Chl b) was also estimated. Three biological replicates were performed for each sample. Data were compared using analysis of variance (Duncan’s multiple range tests at p = 0.05) using SPSS v17.0 software (SPSS Inc.,Chicago, IL, USA).
Chloroplast ultrastructure
To observe the chloroplast ultrastructure of mesophyll cells, edge leaves of YB and YM 0.5 cm × 0.5 cm in size were immediately fixed in fresh 2.5% (v/v) glutaraldehyde (0.1 mol L−1 phosphate buffer, pH 7.2) for at least 48 h (Fig. 1C, F). The samples were immersed in 1% (v/v) osmium acid for post-fixation, embedded in resin and imaged using a transmission electron microscope (H7650, HITACHI, Tokyo, Japan).
RNA extraction and RNA-Seq analysis
The edged leaves of the fourth leaf from three individual YB and YM plants were harvested and snap frozen in liquid nitrogen. Three biological replicates were used for RNA-Seq analysis. Total RNA was extracted using RNAiso reagent (Takara, Japan) according to the manufacturer’s instructions. The quality and integrity of the total RNA were verified using a 2100 Bioanalyzer RNA Nano chip device (Agilent, Santa Clara, CA, USA). The concentration was measured using an ND-430 1000 spectrophotometer (NanoDrop, Wilmington, DE). The RNA was stored at -80 °C for subsequent use.
The mRNA of each library was sequenced on an Illumina NovaSeq 6000 platform located at Wuhan Benagen Tech Solution Co. Ltd. (Wuhan, China; http://www.benagen.com). To obtain high-quality clean reads, adapters, reads containing more than 5% poly-N and low-quality reads were removed from the raw data. The Q20, Q30 and GC contents of the clean data were calculated. De novo assembly was performed using Trinity (http://trinityrnaseq.github.io) [59]. The remaining clean reads were also spliced into unigenes by the same software. The NR (ftp://ftp.ncbi.nlm.nih.gov/blast/db), eggNOG (http://eggnogdb.embl.de/#/app/home), UniProt (http://www.uniprot.org/) [60], Pfam (v30.0) (http://pfam.xfam.org/) [61] and KEGG (v79.1) (http://www.genome.jp/kegg) [62] databases were used for blast search and annotation. All unigenes were first searched in the NR database with an E-value ≤ 10–5. Blast (v2.2.28 +) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) [63] and DIAMOND (v0.7.11) (https://github.com/bbuchfink/diamond) [64] were used for BLAST search and annotation. HMMER (v3.1) (http://hmmer.org/) was used for domain annotation [65]. ClusterProfiler (v3.6.0) (http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html) was used to obtain the GO and KEGG pathway annotations [66].
After obtaining the number of read counts of the samples, FPKM was used to estimate the expression levels of genes and compare differences in gene expression between YB and YM. DEGs were identified using an algorithm developed by Audic and Claverie [67]. The criteria applied to the thresholds for significant differences in gene expression were P-values ≤ 0.05, a false discovery rate (FDR) ≤ 0.05 and |log2Ratio|≥ 1.0.
qRT–PCR validation of DEGs
Total RNA was extracted from the edged leaf of the fourth leaf from three individual YB and YM plants using RNAiso reagent (Takara, Japan) according to the manufacturer’s recommendations. Primers were designed in Primer 5.0 software using the sequences from the transcriptome (Table S9). The H. macrophylla 18S rRNA gene was used as the reference [68]. The qRT–PCR mixtures were prepared following the instructions of the TB Green®Premin Ex Taq™ reagent kit (Takara, Japan). qRT–PCR was performed on a 7500 Real-Time PCR System (Applied Biosystems, CA, USA). The PCR cycles were first denatured (95 °C/30 s), followed by 40 cycles of 95 °C/5 s, 60 °C/34 s, and finally 1 cycle of 95 °C/15 s, 60 °C/60 s, 95 °C/15 s. Three biological replicates and three technical replicates were used for qRT–PCR analysis. Relative expression levels were calculated using the 2−△△CT method.
Supplementary Information
Additional file 1: Supplementary fig 1. SRAP profiling of H. macrophylla var. maculate (YB) and full-green leaf mutant (YM). M: Marker, 1: YB, 2: YM. M23+E3, M23+E6, M23+E10, M23+E12, M23+E16, M23+E18, M24+E2, M24+E4, M24+E8, M24+E11, and M24+E15 represented primer pairs.
Additional file 2: Supplementary fig 2. The distribution of unigenes length.
Additional file 3: Supplementary fig 3. The distribution of E-value (A), identify (B) and species (C) of H. macrophylla unigenes against non-redundant database.
Additional file 4: Supplementary fig 4. Gene ontology functional classification of H. macrophylla unigenes. Unigenes were annotated in three categories: biological process (red), cellular component (green) and molecular function (blue)
Additional file 5: Supplementary fig 5. Gene ontology functional classification of differentially expressed genes. DEGs were annotated in three categories: biological process (red), cellular component (green), and molecular function (blue).
Additional file 6: Supplementary table S1. Reading mapping summary of H. macrophylla.
Additional file 7: Supplementary table S2. The annotated genes of H. macrophylla that may be functionally classified in each corresponding database.
Additional file 8: Supplementary table S3. Pathway classification of all the unigenes in H. macrophylla.
Additional file 9: Supplementary table S4. The expression of all unigenes in H. macrophylla.
Additional file 10: Supplementary table S5. The expression of DEGs in YB-VS-YM.
Additional file 11: Supplementary table S6. The GO significant enrichment analysis of DEGs in YB-VS-YM.
Additional file 12: Supplementary table S7. Pathway classification of the DEGs in H. macrophylla.
Additional file 13: Supplementary table S8. Primer sequences used for amplification in SRAP analysis.
Additional file 14: Supplementary table S9. Primer sequences for qRT–PCR.
Acknowledgements
We are grateful for the funding support.
Abbreviations
- HEMA
Glutamyl-tRNA reductase
- CAO
Chlorophyllide an oxygenase
- CHLI
Magnesium-chelatase I subunit
- PPR
Pentatricopeptide repeat
- GLK
Golden 2-like
- SRAP
Sequence-related amplified polymorphism
- FPKM
Fragments per kilobase per million
- DEGs
Differentially expressed genes
- qRT–PCR
Quantitative real-time PCR
- ARF
Auxin response factor
Authors' contributions
X.Q. and Y.D. designed the experiments. X.Q., S.C., H.W. and H.C. performed the experiments. X.Q., J.F. and Z.Q. analyzed the data. X.Q. and Y.D. wrote and revised the manuscript. All authors read and approved the final manuscript.
Funding
The work was supported by the Foundation of Key Laboratory of Biology of Ornamental Plants in East China, National Forestry and Grassland Administration (Grant No.: KF202102), Jiangsu Agriculture Science and Technology Innovation Fund (Grant No.: CX(21)3022), and the Earmarked Fund for Jiangsu Agricultural Industry Technology System (JATS[2019]338-4).
Availability of data and materials
The raw data of transcriptome sequencing reads of H. macrophylla have been deposited in NCBI under BioProject accession number PRJNA778011 and BioSample accession numbers SAMN22898762 and SAMN22898763. The file of unigene sequences is available publicly in figshare (https://doi.org/10.6084/m9.figshare.19625256.v1).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiangyu Qi and Shuangshuang Chen are co-first authors.
References
- 1.Wu XB, Alexander LW. Genome-wide association studies for inflorescence type and remontancy in Hydrangea macrophylla. Hortic Res. 2020;7:27. doi: 10.1038/s41438-020-0255-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mcclintock EM. A monograph of the genus Hydrangea. Proc Calif Acad Sci. 1957;29:147–256. [Google Scholar]
- 3.Schreiber HD, Jones AH, Lariviere CM, Mayhew KM, Cain JB. Role of aluminum in red-to-blue color changes in Hydrangea macrophylla sepals. Biometals. 2011;24:1005–1015. doi: 10.1007/s10534-011-9458-x. [DOI] [PubMed] [Google Scholar]
- 4.Zhu T, Wang X, Xu ZM, Xu J, Li R, Liu N, et al. Screening of key genes responsible for Pennisetum setaceum “Rubrum” leaf color using transcriptome sequencing. PLoS ONE. 2020;15:e0242618. [DOI] [PMC free article] [PubMed]
- 5.Beale SI. Green genes gleaned. Trends Plant Sci. 2005;10:309–312. doi: 10.1016/j.tplants.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Hörtensteiner S. Chlorophyll degradation during senescence. Annu Rev Plant Biol. 2006;57:55–77. doi: 10.1146/annurev.arplant.57.032905.105212. [DOI] [PubMed] [Google Scholar]
- 7.Zhang JY, Sui CH, Liu HM, Chen JJ, Han ZL, Yan Q, et al. Effect of chlorophyll biosynthesis-related genes on the leaf color in Hosta (Hosta plantaginea Aschers) and tobacco (Nicotiana tabacum L.) BMC Plant Biol. 2021;21:45. doi: 10.1186/s12870-020-02805-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo T, Luo S, Araújo WL, Schlicke H, Rothbart M, Yu J, et al. Virus-induced gene silencing of pea CHLI and CHLD affects tetrapyrrole biosynthesis, chloroplast development and the primary metabolic network. Plant Physiol Bioch. 2013;65:17–26. doi: 10.1016/j.plaphy.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Thomas H, Ougham H. The stay-green trait. J Exp Bot. 2014;65:3889–3900. doi: 10.1093/jxb/eru037. [DOI] [PubMed] [Google Scholar]
- 10.Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, et al. Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell. 2007;19:1362–1375. doi: 10.1105/tpc.106.042911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bashir H, Qureshi MI, Ibrahim MM, Iqbal M. Chloroplast and photosystems: impact of cadmium and iron deficiency. Photosynthetica. 2015;53:321–335. doi: 10.1007/s11099-015-0152-z. [DOI] [Google Scholar]
- 12.Li Y, Zhang ZY, Wang P, Wang SA, Ma LL, Li LF, et al. Comprehensive transcriptome analysis discovers novel candidate genes related to leaf color in a Lagerstroemia indica yellow leaf mutant. Genes Genom. 2015;37:851–863. doi: 10.1007/s13258-015-0317-y. [DOI] [Google Scholar]
- 13.Yang YX, Chen XX, Xu B, Li YX, Ma YH, Wang GD. Phenotype and transcriptome analysis reveals chloroplast development and pigment biosynthesis together influenced the leaf color formation in mutants of Anthurium andraeanum “Sonate”. Front Plant Sci. 2015;6:139. doi: 10.3389/fpls.2015.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan YG, Zhao XX, Wang HY, Tian YY, Xiang QZ, Zhang LX. Effects of light intensity on metabolism of light-harvesting pigment and photosynthetic system in Camellia sinensis L. cultivar ‘Huangjinya’. Environ Exp Bot. 2019;166.
- 15.Zhang SZ, Wu XL, Cui J, Zhang F, Wan XQ, Liu QL, et al. Physiological and transcriptomic analysis of yellow leaf coloration in Populus deltoides Marsh. PLoS ONE. 2019;14:e0216879. [DOI] [PMC free article] [PubMed]
- 16.Karaca M, Saha S, Callahan FE, Jenkins JN, Read JJ, Percy RG. Molecular and cytological characterization of a cytoplasmic-specific mutant in pima cotton (Gossypium barbadense L.) Euphytica. 2004;139:187–197. doi: 10.1007/s10681-004-3032-3. [DOI] [Google Scholar]
- 17.Li N, Jia JZ, Xia C, Liu X, Kong XY. Characterization and mapping of novel chlorophyll deficient mutant genes in durum wheat. Breeding Sci. 2013;63:169–75. [DOI] [PMC free article] [PubMed]
- 18.Gang HX, Liu GF, Chen S, Jiang J. Physiological and transcriptome analysis of a yellow-green leaf mutant in Birch (Betula platyphylla × B. Pendula. Forests. 2019;10:120. doi: 10.3390/f10020120. [DOI] [Google Scholar]
- 19.Kato Y, Miura E, Matsushima R, Sakamoto W. White leaf sectors in yellow variegated2 are formed by viable cells with undifferentiated plastids. Plant Physiol. 2007;144:952–960. doi: 10.1104/pp.107.099002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitz-Linneweber C, Williams-Carrier RE, Williams-Voelker PM, Kroeger TS, Vichas A, Barkan A. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell. 2006;18:2650–2663. doi: 10.1105/tpc.106.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zubo YO, Blakley I, Franco-Zorrilla JM, Yamburenko M, Solano R, Kieber JJ, et al. Coordination of chloroplast development through the action of the GNC and GLK transcription factor families. Plant physiol. 2018;178:130–147. doi: 10.1104/pp.18.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waters MT, Moylan EC, Langdale JA. GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J. 2008;56:432–444. doi: 10.1111/j.1365-313X.2008.03616.x. [DOI] [PubMed] [Google Scholar]
- 23.Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA. GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 2002;31:713–727. doi: 10.1046/j.1365-313X.2002.01390.x. [DOI] [PubMed] [Google Scholar]
- 24.Gao HB, Kadirjan-Kalbach D, Froehlich JE, Osteryoung KW. ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc Natl Acad Sci USA. 2003;100:4328–4333. doi: 10.1073/pnas.0530206100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimada H, Koizumi M, Kuroki K, Mochizuki M, Fujimoto H, Ohta H, et al. ARC3, a chloroplast division factor, is a chimera of prokaryotic FtsZ and part of eukaryotic phosphatidylinositol-4-phosphate 5-kinase. Plant Cell Physiol. 2004;45:960–967. doi: 10.1093/pcp/pch130. [DOI] [PubMed] [Google Scholar]
- 26.Vitha S, Froehlich JE, Koksharova O, Pyke KA, van Erp H, Osteryoung KW. ARC6 is a J-Domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell. 2003;15:1918–1933. doi: 10.1105/tpc.013292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lv XG, Shi YF, Xu X, Wei YL, Wang HM, Zhang XB, et al. Oryza sativa chloroplast signal recognition Particle 43 (OscpSRP43) is required for chloroplast development and photosynthesis. PLoS ONE. 2015;10:e0143249. [DOI] [PMC free article] [PubMed]
- 28.Wu HY, Shi NR, An XY, Liu C, Fu HF, Cao L, et al. Candidate genes for yellow leaf color in common wheat (Triticum aestivum L.) and major related metabolic pathways according to transcriptome profiling. Int J Mol Sci. 2018;19:1594. doi: 10.3390/ijms19061594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng JS, Fan PG, Liang ZC, Wang YQ, Niu N, Li WD, et al. Accumulation of end products in source leaves affects photosynthetic rate in peach via alteration of stomatal conductance and photosynthetic efficiency. J Am Soc Hortic Sci. 2009;134:667–676. doi: 10.21273/JASHS.134.6.667. [DOI] [Google Scholar]
- 30.Nie LB, Zheng YS, Zhang LT, Wu Y, Zhu SD, Hou JF, et al. Characterization and transcriptomic analysis of a novel yellow-green leaf wucai (Brassica Campestris L.) germplasm. BMC Genomics. 2021;22:258. doi: 10.1186/s12864-021-07573-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang RJ, Zhao J, Jia M, Xu N, Liang S, Shao JX, et al. Balance between cytosolic and chloroplast translation affects leaf variegation. Plant Physiol. 2018;176:804–818. doi: 10.1104/pp.17.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun TP, Gubler F. Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol. 2004;55:197–223. doi: 10.1146/annurev.arplant.55.031903.141753. [DOI] [PubMed] [Google Scholar]
- 33.Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, et al. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007;19:3037–3057. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davière JM, de Lucas M, Prat S. Transcriptional factor interaction: a central step in DELLA function. Curr Opin Genet Dev. 2008;18:295–303. doi: 10.1016/j.gde.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 35.de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 36.Feng SH, Martinez C, Gusmaroli G, Wang Y, Zhou JL, Wang F, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pogson BJ, Ganguly D, Albrecht-Borth V. Insights into chloroplast biogenesis and development. Biochim Biophys Acta. 2015;1847:1017–1024. doi: 10.1016/j.bbabio.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 38.Zhang ZG, Cui XA, Wang YW, Wu JX, Gu XF, Lu TG. The RNA editing factor WSP1 is essential for chloroplast development in rice. Mol Plant. 2017;10:86–98. doi: 10.1016/j.molp.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 39.Du L, Zhang J, Qu SF, Zhao YY, Su BD, Lv XQ, et al. The pentratricopeptide repeat protein pigment-defective mutant2 is involved in the regulation of chloroplast development and chloroplast gene expression in Arabidopsis. Plant Cell Physiol. 2017;58:747–759. doi: 10.1093/pcp/pcx004. [DOI] [PubMed] [Google Scholar]
- 40.Cao ZL, Yu QB, Sun Y, Lu Y, Cui YL, Yang ZN. A point mutation in the pentatricopeptide repeat motif of the AtECB2 protein causes delayed chloroplast development. J Integr Plant Biol. 2011;53:258–269. doi: 10.1111/j.1744-7909.2011.01030.x. [DOI] [PubMed] [Google Scholar]
- 41.He P, Wu SY, Jiang YL, Zhang LH, Tang MJ, Xiao GH, et al. GhYGL1, a pentatricopeptide repeat protein, is required for chloroplast development in cotton. BMC Plant Biol. 2019;19:350. doi: 10.1186/s12870-019-1945-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossini L, Cribb L, Martin DJ, Langdale JA. The maize Golden2 gene defines a novel class of transcriptional regulators in plants. Plant Cell. 2001;13:1231–1244. doi: 10.1105/tpc.13.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yasumura Y, Moylan EC, Langdale JA. A conserved transcription factor mediates nuclear control of organelle biogenesis in anciently diverged land plants. Plant Cell. 2005;17:1894–1907. doi: 10.1105/tpc.105.033191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell. 2009;21:1109–1128. doi: 10.1105/tpc.108.065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Q, Sullivan RW, Kight A, Henry RL, Huang J, Jones AM, et al. Deletion of the chloroplast-localized thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol. 2004;136:3594–3604. doi: 10.1104/pp.104.049841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cortleven A, Schmülling T. Regulation of chloroplast development and function by cytokinin. J Exp Bot. 2015;66:4999–5013. doi: 10.1093/jxb/erv132. [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi K, Baba S, Obayashi T, Sato M, Toyooka K, Keranen M, et al. Regulation of root greening by light and auxin/cytokinin signaling in Arabidopsis. Plant Cell. 2012;24:1081–1095. doi: 10.1105/tpc.111.092254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiang YH, Zubo YO, Tapken W, Kim HJ, Lavanway AM, Howard L, et al. Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol. 2012;160:332–348. doi: 10.1104/pp.112.198705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richter R, Behringer C, Zourelidou M, Schwechheimer C. Convergence of auxin and gibberellin signaling on the regulation of the GATA transcription factors GNC and GNL in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2013;110:13192–13197. doi: 10.1073/pnas.1304250110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson CB, Shaik R, Abdallah R, Vitha S, Holzenburg A. FtsZ1/FtsZ2 turnover in chloroplasts and the role of ARC3. Microsc Microanal. 2015;21:313–323. doi: 10.1017/S1431927615000082. [DOI] [PubMed] [Google Scholar]
- 51.Osteryoung KW, Stokes KD, Rutherford SM, Percival AL, Lee WY. Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell. 1998;10:1991–2004. doi: 10.1105/tpc.10.12.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aldridge C, Maple J, Møller SG. The molecular biology of plastid division in higher plants. J Exp Bot. 2005;56:1061–1077. doi: 10.1093/jxb/eri118. [DOI] [PubMed] [Google Scholar]
- 53.Rashotte AM, Mason MG, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ. A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc Natl Acad Sci USA. 2006;103:11081–11085. doi: 10.1073/pnas.0602038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okazaki K, Kabeya Y, Suzuki K, Mori T, Ichikawa T, Matsui M, et al. The PLASTID DIVISION1 and 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation. Plant Cell. 2009;21:1769–1780. doi: 10.1105/tpc.109.067785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hanania U, Velcheva M, Sahar N, Perl A. An improved method for isolating high-quality DNA from Vitis vinifera nuclei. Plant Mol Biol Rep. 2004;22:173–177. doi: 10.1007/BF02772724. [DOI] [Google Scholar]
- 56.Li G, Quiros CF. Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet. 2001;103:455–461. doi: 10.1007/s001220100570. [DOI] [Google Scholar]
- 57.Deng YM, Li CC, Shao QS, Ye XQ, She JM. Differential responses of double petal and multi petal jasmine to shading: I. Photosynthetic characteristics and chloroplast ultrastructure. Plant Physiol Bioch. 2012;55:93–102. doi: 10.1016/j.plaphy.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 58.Zhang T, Qu YX, Wang HB, Wang JJ, Song AP, Hu YH, et al. The heterologous expression of a chrysanthemum TCP-P transcription factor CmTCP14 suppresses organ size and delays senescence in Arabidopsis thaliana. Plant Physiol Bioch. 2017;115:239–248. doi: 10.1016/j.plaphy.2017.03.026. [DOI] [PubMed] [Google Scholar]
- 59.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Altschul SF, Madden TL, Schffer AA, Zhang JH, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 65.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 66.Yu GC, Wang LG, Han YY, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 68.Negishi T, Oshima K, Hattori M, Yoshida K. Plasma membrane-localized Al-transporter from blue hydrangea sepals is a member of the anion permease family. Genes Cells. 2013;18:341–352. doi: 10.1111/gtc.12041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary fig 1. SRAP profiling of H. macrophylla var. maculate (YB) and full-green leaf mutant (YM). M: Marker, 1: YB, 2: YM. M23+E3, M23+E6, M23+E10, M23+E12, M23+E16, M23+E18, M24+E2, M24+E4, M24+E8, M24+E11, and M24+E15 represented primer pairs.
Additional file 2: Supplementary fig 2. The distribution of unigenes length.
Additional file 3: Supplementary fig 3. The distribution of E-value (A), identify (B) and species (C) of H. macrophylla unigenes against non-redundant database.
Additional file 4: Supplementary fig 4. Gene ontology functional classification of H. macrophylla unigenes. Unigenes were annotated in three categories: biological process (red), cellular component (green) and molecular function (blue)
Additional file 5: Supplementary fig 5. Gene ontology functional classification of differentially expressed genes. DEGs were annotated in three categories: biological process (red), cellular component (green), and molecular function (blue).
Additional file 6: Supplementary table S1. Reading mapping summary of H. macrophylla.
Additional file 7: Supplementary table S2. The annotated genes of H. macrophylla that may be functionally classified in each corresponding database.
Additional file 8: Supplementary table S3. Pathway classification of all the unigenes in H. macrophylla.
Additional file 9: Supplementary table S4. The expression of all unigenes in H. macrophylla.
Additional file 10: Supplementary table S5. The expression of DEGs in YB-VS-YM.
Additional file 11: Supplementary table S6. The GO significant enrichment analysis of DEGs in YB-VS-YM.
Additional file 12: Supplementary table S7. Pathway classification of the DEGs in H. macrophylla.
Additional file 13: Supplementary table S8. Primer sequences used for amplification in SRAP analysis.
Additional file 14: Supplementary table S9. Primer sequences for qRT–PCR.
Data Availability Statement
The raw data of transcriptome sequencing reads of H. macrophylla have been deposited in NCBI under BioProject accession number PRJNA778011 and BioSample accession numbers SAMN22898762 and SAMN22898763. The file of unigene sequences is available publicly in figshare (https://doi.org/10.6084/m9.figshare.19625256.v1).