Abstract
The Sigma 1 receptor (SIGMAR1) is a transmembrane protein located in the mitochondria-associated endoplasmic reticulum membrane, and plays an important role in cell survival as a pluripotent modulator of a variety of signaling pathways related to neurodegeneration. Though SIGMAR1 is a potential target for neurodegenerative diseases, the specific role of SIGMAR1 in different tissue and cell types remains unclear. Here we reported the generation of Sigmar1 conditional knockout (Sigmar1loxP) mice using CRISPR-Cas9 method to insert loxP sites into the 5’- and 3’- untranslated regions of Sigmar1. We showed that the insertion of loxP sequences did not affect the expression of Sigmar1 and that Sigmar1loxP/loxP mice exhibited no detectable visual defects compared to wild type mice at the early adult stage. By crossing Sigmar1loxP mice with retina-specific Six3-Cre and ubiquitous CMV-Cre mice, we confirmed the deletion of Sigmar1 coding regions of exons 1-4, and the retina-specific and global loss of SIGMAR1 expression, respectively. Thus, Sigmar1loxP mice provide a valuable tool for unraveling the tissue and cell type specific role of Sigmar1.
Keywords: Sigma receptor, sconditional knockout, CRISPR-Cas9, retina
INTRODUCTION
The Sigma 1 receptor (SIGMAR1) is a highly conserved protein of 223 amino acids and shares no homology with any other known human proteins (Boehning et al., 2003; Delprat, Crouzier, Su, & Maurice, 2020; Schmidt et al., 2016). SIGMAR1 is an endoplasmic reticulum (ER) protein that resides at the mitochondria-associated endoplasmic reticulum membrane (MAM) and acts as a chaperone to modulate the activity of various ion channels and signaling molecules via translocation and protein-protein interactions (Hanner et al., 1996; Hayashi & Su, 2007; Matsumoto, Bowen, & Su, 2007; L. Nguyen et al., 2015; Smith & Su, 2017; Su, Hayashi, Maurice, Buch, & Ruoho, 2010). SIGMAR1 exhibits a widespread expression pattern including the central and the peripheral nervous systems (Hayashi & Su, 2004). SIGMAR1 has proven to be relevant to several neurodegenerative diseases, such as Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), amyotrophic lateral sclerosis (ALS), and retinal degeneration (Al-Saif, Al-Mohanna, & Bohlega, 2011; Hedskog et al., 2013; Jansen, Faull, Storey, & Leslie, 1993; Luty et al., 2010; Matsumoto et al., 2007; Maurice & Goguadze, 2017; M. Mishina et al., 2005; Masahiro Mishina et al., 2008; Linda Nguyen, Lucke-Wold, Mookerjee, Kaushal, & Matsumoto, 2017; L. Nguyen et al., 2015; Prause et al., 2013; Sylvia B Smith et al., 2018; Wu et al., 2021). SIGMAR1 has gained considerable attention for its potential to slow the progression of neuron death or reverse an existing pathology of neurodegenerative diseases (Hedskog et al., 2013; Hyrskyluoto et al., 2013; Mori, Hayashi, & Su, 2012; L. Nguyen et al., 2015; Sylvia B Smith et al., 2018; Weng, Tsai, & Su, 2017; Wu et al., 2021). Pharmacological studies using SIGMAR1 agonists or antagonists have shown that SIGMAR1 plays a neuroprotective role by modulating calcium homeostasis, attenuating reactive oxygen species production, regulating ER and mitochondrial function, and glial activities (Chu & Ruoho, 2016; Hayashi & Su, 2004; Maurice & Su, 2009; Linda Nguyen, Kaushal, Robson, & Matsumoto, 2014; L. Nguyen et al., 2015; Wang, Saul, Roon, & Smith, 2016). Though significant progress has been made in exploring the role of SIGMAR1, its broad expression pattern in diverse cell types makes it challenging to assess the precise function of SIGMAR1 in individual tissues and cell types in the absence of a Sigmar1 conditional knockout allele.
In this study, we generated the Sigmar1 conditional knockout mouse line (Sigmar1loxP) using the CRISPR-Cas9 gene targeting method to flox Sigmar1 exons 1-4. Heterozygous and homozygous Sigmar1loxP mice were viable, fertile, aphenotypic, and display normal visual function through early adulthood. By crossing Sigmar1loxP mice with retina-specific Six3-Cre or ubiquitous CMV-Cre lines, we demonstrated that Sigmar1 could be successfully deleted by Cre recombinase tissue-specifically or ubiquitously. Thus, this Sigmar1 conditional knockout mouse line offers a useful tool for studying the role of Sigmar1 spatiotemporally.
MATERIALS AND METHODS
Animals
To generate the Sigmar1 conditional knockout allele, we used the CRISPR-Cas9 genome editing method to insert two loxP sites to flank the entire Sigmar1 coding region in exon 1-4. Two single guide RNAs (sgRNAs) separately targeting within the 5’ untranslated region (UTR) and 3’ UTR of Sigmar1 were co-injected with two single-stranded oligonucleotide (ssODN) repair templates containing loxP sites. The sgRNA-targeted sequences in 5’ and 3’ UTRs were 5’- TAAGACCGAACTAGGGCCAC AGG -3’ (with protospacer adjacent motif (PAM) site AGG) and 5’-TCTGTGGATGGACAGGAGCG CGG-3’ (with PAM site CGG), respectively. In the 5’ ssODN repair template, a loxP consensus site replaced the PAM site and base 19-20 (AC AGG) and was flanked by 57 bp 5’ homology arm and 60 bp 3’ homology arm. In the 3’ ssODN template, PAM site and base 18-20 (GCG CGG) were replaced by a loxP sequence that was flanked by 66 bp 5’ and 58 bp 3’ homology arms. The sgRNAs and ssODNs were co-injected into approximately 170 C57BL/6J pronuclear-stage zygotes. After transfer into pseudopregnant Swiss webster recipient female mice, 14 pups were obtained and were PCR genotyped to verify the correct insertion of 5’ and 3’ loxP sites. The primer sets to verify 5’ loxP site are 5’-GGTGCTAGGTGACCCGGACGT-3’ and 5’-CACCCGCATACTGTCGAGCAAGCT-3’, with PCR products of 305 bp for Sigmar1 wild type and 335 bp for Sigmar1loxP. To verify the insertion of 3’ loxP site, primer sets 5’-GCCTCCGGCTTGAGCTTACCA-3’ and 5’-GCTTCCCAAGAGCTGTGTCTGGA-3’ were used, with PCR products of 222 bp for Sigmar1 wild type and 255 bp for Sigmar1loxP. One potential male founder who carried both the 5’ and 3’ loxP sites was further verified using long-range PCR (LRPCR) with external primer set 5’-GAGTCGTGAACCCACAGGATCCA-3’ and 5’-GAGAGATGGATGTGGTCCTGAAGCT-3’ to confirm that two loxP sites were inserted in the same Sigmar1 allele. The identified founder was bred with C57BL/6J and progeny were screened to determine the germline transmission by Sanger sequencing of the 5’ and 3’ loxP sites from genomic DNA isolated from the founder and progeny mice. All animal procedures used in this study were approved by Institutional Animal Care and Use Committee (IACUC) at Augusta University. The corresponding author will make this mouse strain available to the research community upon acceptance of this manuscript.
Western blot
Western blot was performed as previously described (Dong et al., 2020). Briefly, tissue samples were collected and lysed in the RIPA buffer (ThermoFisher Scientific, Waltham, MA, #89900) with a protease inhibitor cocktail (Roche, #04693124001). The cell lysate was centrifuged for 15 minutes at 12,000 rpm at 4°C. The supernatant was suspended with 2X Laemmli Sample Buffer (Bio-Rad, Hercules, CA #1610737) and boiled for 5 minutes. The sample was loaded onto 20% SDS-PAGE gel (Bio-Rad), and electrophoresed for 2 hours at 100 V. Proteins were then transferred onto the PVDF membrane using Mini Trans-Blot electrophoretic transfer cell (Bio-Rad). Next, the membrane was blocked with the blocking buffer (5% milk in TBST) for 1 hour at room temperature (RT) and incubated with anti-SIGMAR1 (Y. Ha et al., 2014; Ola et al., 2002) (1:1,000) at 4°C overnight. Then, the membrane was washed three times with TBST for 5 minutes each, and incubated with VeriBlot for IP Detection Reagent (1:1,000, Abcam #ab131366) for 1 hour at RT. After the membrane was washed four times with TBST for 5 minutes each, detection was performed using a ImageQuant LAS4000 biomolecular imager (GE Healthcare, Chicago, IL).
Optical Coherence Tomography
To assess retinal architecture in situ, spectral domain- optical coherence tomography (SD-OCT) was performed. Mice were anesthetized using intraperitoneal injection (i.p.) of rodent anesthesia cocktail (Ketamine 85mg/cc, Xylazine 10 mg/cc) at a dosage of 1 μl/g body weight. Anesthetized mice were wrapped in soft tissue paper and placed within the mouse holding cassette, pupils were dilated with 1% tropicamide. 0.3% hypromellose eye gel (Genteal, Alcon Corp., Fort Worth, TX) was used throughout the procedure to maintain corneal moisture and clarity. Additionally, 0.4% polyethylene glycol 400 (Systane Ultra, Alcon Corp) was applied liberally to maintain corneal moisture during imaging. The SD-OCT images were obtained using the Bioptigen Spectral Domain Ophthalmic Imaging System (SDOIS; Bioptigen Envisu R2200, North Carolina) equipped with the rodent alignment stage and mouse retina probe using proprietary InVivoVue™ Diver 2.4 software (Bioptigen). The Three repeated volume intensity projections, centered on the optic nerve head, were acquired for each eye. The images consisted of 100 averaged B-scans. Images were acquired within 20 minutes after anesthesia administration to ensure that all three scans were acquired rapidly to avoid the development of reversible cataracts.
Optomotor response
Visual acuity was assessed as the optomotor response (OMR) using the OptoMotry device from Cerebral Mechanics (Medicine Hat, Alberta, Canada). OptoMotry is a non-invasive behavioral test that measures visual acuity in mice and rats (Prusky, Alam, Beekman, & Douglas, 2004). This test causes minimal stress to the rodents. Briefly, a single mouse was placed on a platform within the OptoMotry device. Video monitors surrounding the rodent displayed gratings that moved around the rodent, periodically reversing direction. These stimuli evoked optomotor reflexes, causing the rodent to turn its head if the grating was visible. A camera above the rodent was used to detect these responses, and to determine the spatial frequency and/or contrast thresholds needed to generate a response.
Electroretinogram
To assess electrophysiological responses of the retina, electroretinography was performed to assess scotopic (rod) and photopic (cone) responses. Mice were dark-adapted overnight (>12 hours), and anesthetized as described above for the OCT test, the pupils were dilated using 0.5% tropicamide and 2.5% phenylephrine, 0.3% hypromellose (GenTeal, Alcon Corp.). The gel was applied to the ERG probes as well as the cornea, which provided a lubricating cushion between cornea and probe. The Celeris Ophthalmic Electrophysiology System, a high throughput testing system, manufactured by Diagnosys LLC (Lowell, MA) was used for testing. For scotopic testing, a series of full-field light flashes were presented via the probes and presented from dim to bright. After dark-adapted testing, animals were light-adapted for 5 minutes. To record cone isolated (photopic) responses, a series of full-field flashes with increasing intensity was presented in the presence of a background light. After the recordings, the probes were removed and the animal received an additional 0.3% hypromellose ointment, which keeps the cornea lubricated and prevents corneal lesions. The mouse was on a 37° heated platform during testing and was returned to the laboratory animal facility upon full recovery.
RESULTS AND DISCUSSION
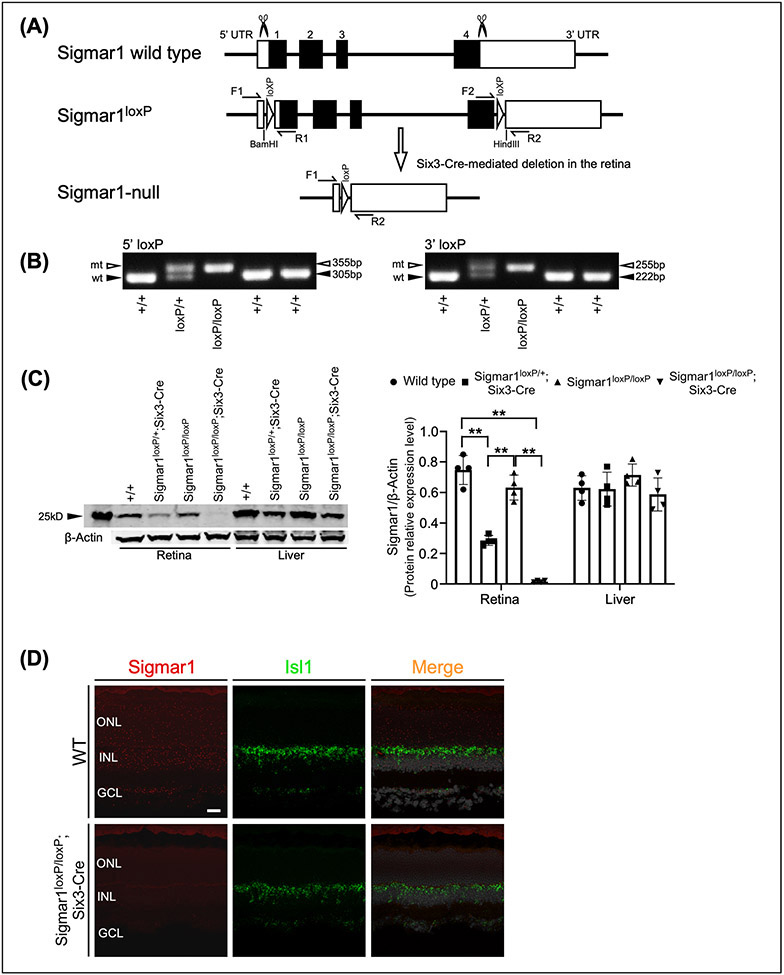
To generate the SIGMAR1 conditional knockout mouse, we used CRISPR-Cas9 gene targeting to insert two syntropic loxP sites that flank exon 1-4 of SIGMAR1 to obtain the conditional knockout allele Sigmar1loxP (Fig. 1A). Correct insertion of 5’ and 3’ loxP sites was verified by Sanger sequencing and PCR genotyping with primers flanking the individual loxP sites (Fig. 1B). Mice heterozygous and homozygous for the conditional Sigmar1 allele, Sigmar1loxP/+ and Sigmar1loxP/loxP, were viable and fertile, and did not display any gross abnormalities through adulthood.
Figure 1.
Generation of Sigmar1loxP allele and retinal specific deletion of Sigmar1. (A) CRISPR-Cas9 targeting strategy to generate the Sigmar1loxP conditional knockout allele. (B) PCR genotyping used to identify 5’ Sigmar1 wild-type (305 bp) and mutant (355 bp) alleles, and 3’ Sigmar1 wild-type (222 bp) and mutant (255 bp) alleles. (C) Breeding strategy to generate Sigmar1 heterozygous (Sigmar1loxP/+; Six3-Cre) and homozygous (Sigmar11loxP/loxP; Six3-Cre) mice. (D) Western blot analysis of SIGMAR1 expression in retina and liver tissues from wild type, Sigmar1 heterozygous and homozygous mice.
To test whether tissue-specific Cre-mediated excision of exon1-4 could occur at the Sigmar1loxP allele, we crossed the Sigmar1loxP mice with the Six3-Cre mouse line (JAX stock No. 019775), which express Cre recombinase in the retina and forebrain (Furuta, Lagutin, Hogan, & Oliver, 2000) (Fig. 1C). Retina and liver tissues from wild type (Sigmar1+/+), heterozygous (Sigmar1loxP/+; Six3-Cre) and homozygous (Sigmar11loxP/loxP; Six3-Cre) mice were collected at postnatal day 30 (P30). Western blot analysis showed that compared to Sigmar1+/+, the expression of SIGMAR1 was significantly decreased in Sigmar1loxP/+; Six3-Cre retinas and was absent in Sigmar1loxP/loxP; Six3-Cre retinas, but was not affected in liver tissues (Fig. 1D). The absence of SIGMAR1 protein specifically in Sigmar1loxP/loxP; Six3-Cre retinas confirms the Cre-mediated tissue-specific deletion of Sigmar1.
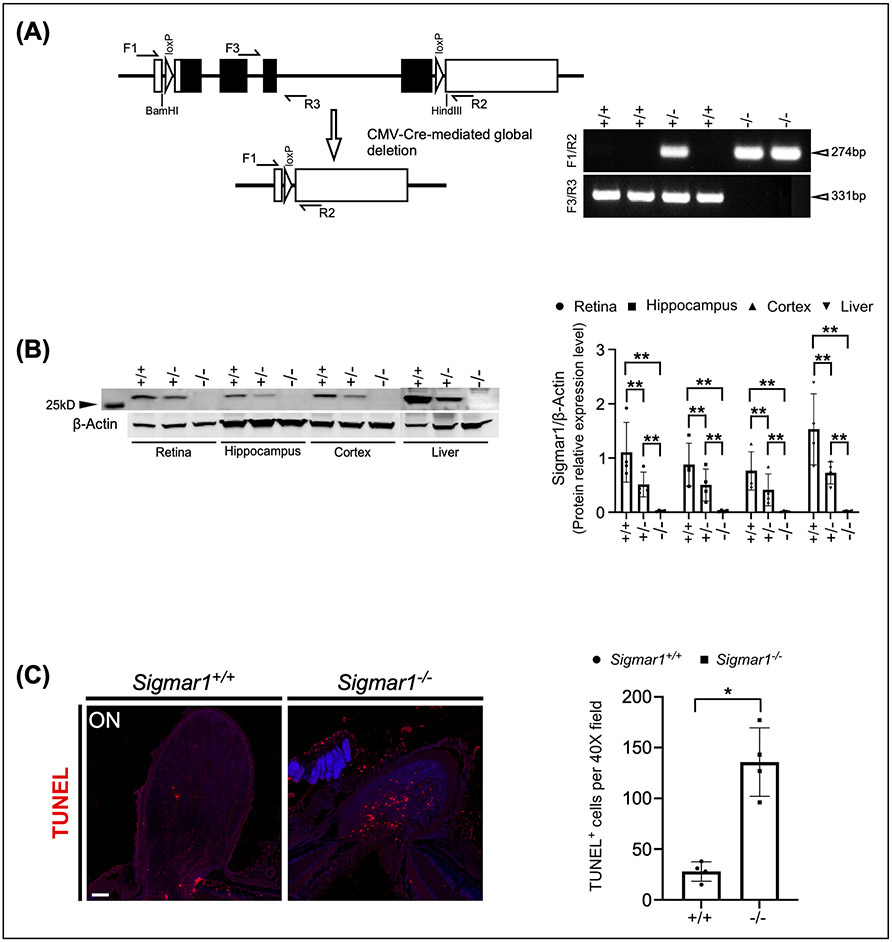
In addition, we crossed the Sigmar1loxP mice with the ubiquitous Cre-expressing transgenic mice CMV-Cre (JAX stock No. 006054), in which Cre expression is under the control of a human cytomegalovirus minimal promoter and is expressed in all tissues (Schwenk, Baron, & Rajewsky, 1995) (Fig. 2A). We used primer sets flanking the two loxP sites (primer F1/R2) and the exon 3 (primer F3/R3) to verify the deletion of Sigmar1 (Fig. 2B). Mouse tissues of retina, hippocampus, cortex and liver were collected from Sigmar1+/+, Sigmar1loxP/+; CMV-Cre (hereafter Sigmar1+/−) and Sigmar1loxP/loxP; CMV-Cre (hereafter Sigmar1−/−) mice at P30. Western blot analysis revealed that the expression of SIGMAR1 was significantly reduced in all tested tissues of Sigmar1+/− mice and vanished in Sigmar1−/− mice (Fig. 2C), confirming the global knockout of Sigmar1 by CMV-Cre.
Figure 2.
Global knockout of Sigmar1 by CMV-Cre. (A) Breeding strategy to generate Sigmar1 global knockout (Sigmar1−/−) mice. (B) Cre-mediated removal of exon 1-4 of Sigmar1 and PCR genotyping of Sigmar1 wild-type (331 bp) and mutant (274 bp) alleles. (C) Western blot analysis of SIGMAR1 expression in retina, hippocampus, cortex and liver tissues from wild type, Sigmar1+/− and Sigmar1−/− mice.
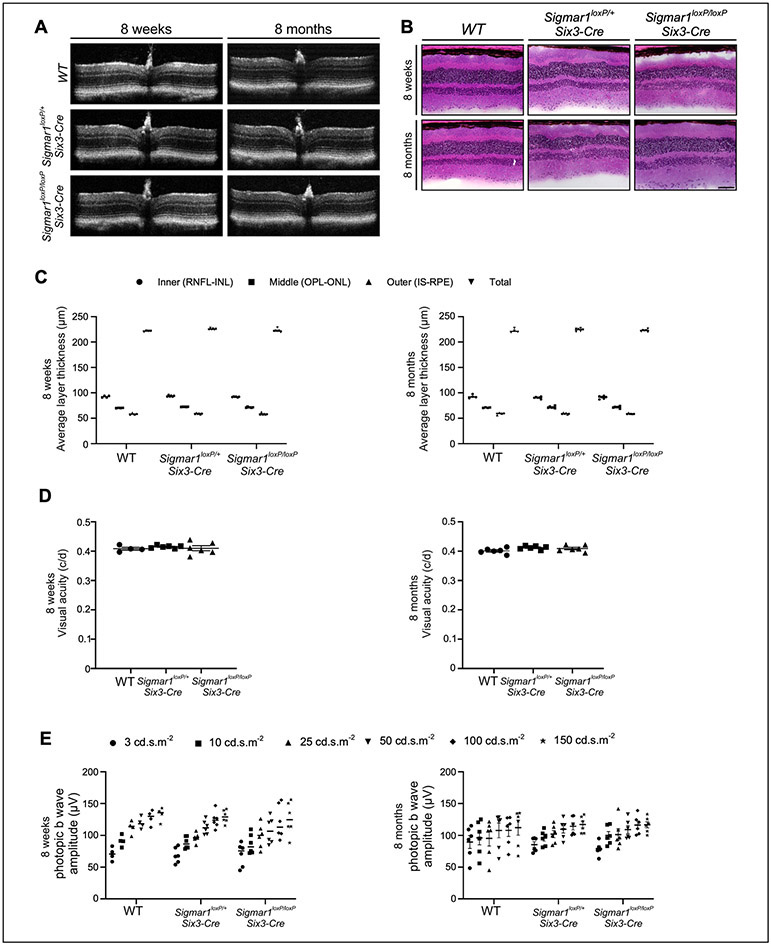
Previously studies have shown that SIGMAR1 is expressed broadly in the retina and other ocular tissues (S. B. Smith et al., 2018). Mice null for Sigmar1 initially exhibit comparable morphologic appearance and visual functions to wild type mice but develop the late-onset functional abnormalities starting from 12-month age (Yonju Ha et al., 2011). To evaluate the visual function of Sigmar1 conditional knockout mice, we assessed retinal architecture by optical coherence tomography (OCT), visual acuity by evaluating the optomotor response (OMR), and retinal electrophysiological response by electroretinogram (ERG) analyses of wild type, Sigmar1loxP/+; Six3-Cre, and Sigmar11loxP/loxP; Six3-Cre mice at P56 (Fig. 3). As expected, the morphologic structure observed by OCT (Fig. 3A and B), the visual acuity measured by OMR (Fig. 3C), and cone responses reflected as the ERG b-wave amplitude (Fig. 3D) of both Sigmar1loxP/+; Six3-Cre, and Sigmar11loxP/loxP; Six3-Cre mice were comparable to the wild type controls, suggesting normal visual function of adult Sigmar1loxP/+; Six3-Cre, and Sigmar11loxP/loxP; Six3-Cre mice, at least through P56 days.
Figure 3.
Normal visual function of Sigmar1 conditional knockout mice at the early adult stage. (A) Representative optical coherence tomography (OCT) B-scan image, (B) Average thickness of retinal layers obtained from OCT images and quantified using DIVERS software including total retinal thickness (total), inner retina (RNFL, inner plexiform layer, INL); middle retina (OPL & ONL), outer retina (IS, outer segments, RPE). (C) Visual acuity measured by OMR and expressed as cycles/degree (c/d) and (D) Photopic b-wave amplitudes measured by ERG as responses to increasing light intensities expressed as candela-seconds per meter squared (cd.s.m2) were compared in wild type, Sigmar1loxP/+; Six3-Cre, and Sigmar1loxP/loxP; Six3-Cre mice at P56. RNFL: retinal nerve fiber layer; INL: inner nuclear layer; OPL: outer plexiform layer; ONL: outer nuclear layer; IS: inner segment; RPE: retinal pigment epithelium.
In conclusion, we generated a novel conditional knockout mouse model of Sigmar1 using CRISPR-Cas9 genome editing, which will serve as an invaluable tool to elucidate the role of Sigmar1 in tissue and cell type-specific manner.
Acknowledgment
Sigmar1loxPconditional knockout mice were generated and funded by the resource sharing program at Transgenic and Genome Editing Core of Augusta University Medical College of Medicine. Sigmar1loxP characterization study reported in this publication was supported by the National Eye Institute grants (R01EY026614, R01EY028103 and P30EY031631).
References
- Al-Saif A, Al-Mohanna F, & Bohlega S (2011). A mutation in sigma-1 receptor causes juvenile amyotrophic lateral sclerosis. Annals of neurology, 70(6), 913–919. [DOI] [PubMed] [Google Scholar]
- Boehning D, Patterson RL, Sedaghat L, Glebova NO, Kurosaki T, & Snyder SH (2003). Cytochrome c binds to inositol (1, 4, 5) trisphosphate receptors, amplifying calcium-dependent apoptosis. Nature cell biology, 5(12), 1051–1061. [DOI] [PubMed] [Google Scholar]
- Chu UB, & Ruoho AE (2016). Biochemical pharmacology of the sigma-1 receptor. Molecular pharmacology, 89(1), 142–153. [DOI] [PubMed] [Google Scholar]
- Delprat B, Crouzier L, Su T-P, & Maurice T (2020). At the crossing of ER stress and MAMs: a key role of sigma-1 receptor? Calcium Signaling, 699–718. [DOI] [PubMed] [Google Scholar]
- Dong X, Yang H, Zhou X, Xie X, Yu D, Guo L, Gan L (2020). LIM-homeodomain transcription factor LHX4 is required for the differentiation of retinal rod bipolar cells and OFF-cone bipolar subtypes. Cell Rep, 32(11), 108144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y, Lagutin O, Hogan BL, & Oliver GC (2000). Retina- and ventral forebrain-specific Cre recombinase activity in transgenic mice. Genesis, 26(2), 130–132. [PubMed] [Google Scholar]
- Ha Y, Saul A, Tawfik A, Williams C, Bollinger K, Smith R, Smith SB (2011). Late-onset inner retinal dysfunction in mice lacking sigma receptor 1 (σR1). Investigative ophthalmology & visual science, 52(10), 7749–7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha Y, Shanmugam AK, Markand S, Zorrilla E, Ganapathy V, & Smith SB (2014). Sigma receptor 1 modulates ER stress and Bcl2 in murine retina. Cell Tissue Res, 356(1), 15–27. doi: 10.1007/s00441-013-1774-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanner M, Moebius FF, Flandorfer A, Knaus H-G, Striessnig J, Kempner E, & Glossmann H (1996). Purification, molecular cloning, and expression of the mammalian sigma1-binding site. Proceedings of the National Academy of Sciences, 93(15), 8072–8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, & Su T-P (2004). σ-1 receptor ligands. CNS drugs, 18(5), 269–284. [DOI] [PubMed] [Google Scholar]
- Hayashi T, & Su T-P (2007). Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca2+ signaling and cell survival. Cell, 131(3), 596–610. [DOI] [PubMed] [Google Scholar]
- Hedskog L, Pinho CM, Filadi R, Rönnbäck A, Hertwig L, Wiehager B, … Westerlund M (2013). Modulation of the endoplasmic reticulum–mitochondria interface in Alzheimer’s disease and related models. Proceedings of the National Academy of Sciences, 110(19), 7916–7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrskyluoto A, Pulli I, Törnqvist K, Ho TH, Korhonen L, & Lindholm D (2013). Sigma-1 receptor agonist PRE084 is protective against mutant huntingtin-induced cell degeneration: involvement of calpastatin and the NF-κ B pathway. Cell death & disease, 4(5), e646–e646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen K, Faull R, Storey P, & Leslie R (1993). Loss of sigma binding sites in the CA1 area of the anterior hippocampus in Alzheimer's disease correlates with CA1 pyramidal cell loss. Brain research, 623(2), 299–302. [DOI] [PubMed] [Google Scholar]
- Luty AA, Kwok JB, Dobson-Stone C, Loy CT, Coupland KG, Karlström H, Barcikowska M (2010). Sigma nonopioid intracellular receptor 1 mutations cause frontotemporal lobar degeneration–motor neuron disease. Annals of neurology, 68(5), 639–649. [DOI] [PubMed] [Google Scholar]
- Matsumoto RR, Bowen WD, & Su TP (2007). Sigma receptors: Chemistry, Cell Biology and Clinical Implications: Springer. [Google Scholar]
- Maurice T, & Goguadze N (2017). Sigma-1 (σ 1) receptor in memory and neurodegenerative diseases. Sigma Proteins: Evolution of the Concept of Sigma Receptors, 81–108. [DOI] [PubMed] [Google Scholar]
- Maurice T, & Su T-P (2009). The pharmacology of sigma-1 receptors. Pharmacology & therapeutics, 124(2), 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina M, Ishiwata K, Ishii K, Kitamura S, Kimura Y, Kawamura K, … Katayama Y (2005). Function of sigma1 receptors in Parkinson's disease. Acta Neurol Scand, 112(2), 103–107. doi: 10.1111/j.1600-0404.2005.00432.x [DOI] [PubMed] [Google Scholar]
- Mishina M, Ohyama M, Ishii K, Kitamura S, Kimura Y, Oda K. -i., …Katayama Y (2008). Low density of sigma 1 receptors in early Alzheimer’s disease. Annals of nuclear medicine, 22(3), 151–156. [DOI] [PubMed] [Google Scholar]
- Mori T, Hayashi T, & Su T-P (2012). Compromising σ-1 receptors at the endoplasmic reticulum render cytotoxicity to physiologically relevant concentrations of dopamine in a nuclear factor-κB/Bcl-2-dependent mechanism: potential relevance to Parkinson's disease. Journal of Pharmacology and Experimental Therapeutics, 341(3), 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L, Kaushal N, Robson MJ, & Matsumoto RR (2014). Sigma receptors as potential therapeutic targets for neuroprotection. European journal of pharmacology, 743, 42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L, Lucke-Wold BP, Mookerjee S, Kaushal N, & Matsumoto RR (2017). Sigma-1 receptors and neurodegenerative diseases: towards a hypothesis of sigma-1 receptors as amplifiers of neurodegeneration and neuroprotection. Sigma Receptors: Their Role in Disease and as Therapeutic Targets, 133–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L, Lucke-Wold BP, Mookerjee SA, Cavendish JZ, Robson MJ, Scandinaro AL, & Matsumoto RR (2015). Role of sigma-1 receptors in neurodegenerative diseases. J Pharmacol Sci, 127(1), 17–29. doi: 10.1016/j.jphs.2014.12.005 [DOI] [PubMed] [Google Scholar]
- Ola MS, Moore P, Maddox D, El-Sherbeny A, Huang W, Roon P, Smith SB (2002). Analysis of sigma receptor (sigmaR1) expression in retinal ganglion cells cultured under hyperglycemic conditions and in diabetic mice. Brain Res Mol Brain Res, 107(2), 97–107. doi: 10.1016/s0169-328x(02)00444-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prause J, Goswami A, Katona I, Roos A, Schnizler M, Bushuven E, … Beyer C (2013). Altered localization, abnormal modification and loss of function of Sigma receptor-1 in amyotrophic lateral sclerosis. Hum Mol Genet, 22(8), 1581–1600. [DOI] [PubMed] [Google Scholar]
- Prusky GT, Alam NM, Beekman S, & Douglas RM (2004). Rapid quantification of adult and developing mouse spatial vision using a virtual optomotor system. Invest Ophthalmol Vis Sci, 45(12), 4611–4616. doi: 10.1167/iovs.04-0541 [DOI] [PubMed] [Google Scholar]
- Schmidt HR, Zheng S, Gurpinar E, Koehl A, Manglik A, & Kruse AC (2016). Crystal structure of the human σ 1 receptor. Nature, 532(7600), 527–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F, Baron U, & Rajewsky K (1995). A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res, 23(24), 5080–5081. doi: 10.1093/nar/23.24.5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SB, & Su T-P (2017). Sigma receptors: Their role in disease and as therapeutic targets: Springer. [DOI] [PubMed] [Google Scholar]
- Smith SB, Wang J, Cui X, Mysona BA, Zhao J, & Bollinger KE (2018). Sigma 1 receptor: a novel therapeutic target in retinal disease. Prog Retin Eye Res, 67, 130–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SB, Wang J, Cui X, Mysona BA, Zhao J, & Bollinger KE (2018). Sigma 1 receptor: A novel therapeutic target in retinal disease. Prog Retin Eye Res, 67, 130–149. doi: 10.1016/j.preteyeres.2018.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T-P, Hayashi T, Maurice T, Buch S, & Ruoho AE (2010). The sigma-1 receptor chaperone as an inter-organelle signaling modulator. Trends in pharmacological sciences, 31(12), 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Saul A, Roon P, & Smith SB (2016). Activation of the molecular chaperone, sigma 1 receptor, preserves cone function in a murine model of inherited retinal degeneration. Proc Natl Acad Sci U S A, 113(26), E3764–3772. doi: 10.1073/pnas.1521749113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng T-Y, Tsai S-YA, & Su T-P (2017). Roles of sigma-1 receptors on mitochondrial functions relevant to neurodegenerative diseases. Journal of biomedical science, 24(1), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu NH, Ye Y, Wan BB, Yu YD, Liu C, & Chen QJ (2021). Emerging Benefits: Pathophysiological Functions and Target Drugs of the Sigma-1 Receptor in Neurodegenerative Diseases. Mol Neurobiol. doi: 10.1007/s12035-021-02524-5 [DOI] [PubMed] [Google Scholar]