Abstract
Non-Hispanic Black (NHB) people have 2.5 fold higher risk of maternal mortality when compared no Non-Hispanic White (NHW) people. Neonates of NHB people are more likely to be born preterm and small for gestational age which may be driven by structural racism. The placenta is very sensitive to the maternal environment, and may play a critical role in the translation of environmental stressors to pregnancy outcomes. Our aim was to assess the placental miRNA expression profile in both NHB and NHW people, and the association between differentially expressed miRNAs and pregnancy outcomes. Placentas were collected from 50 NHB, and 74 NHW people with a normal singleton pregnancy undergoing elective cesarean section at term prior to the onset of labor. Placental miRNA expression was measured via whole genome small RNA-sequencing in a subset of 77 placentas. Fifteen miRNAs were more highly expressed in placentas of NHB people. Several of these miRNA were associated with cellular stress response pathways, suggesting that they may be responding to environmental stressors. Placental miR-192-5p expression was lower among NHB people, and was positively associated with neonatal adiposity, suggesting it may be sensitive to structural racism with potential impacts on fetal growth.
Keywords: placenta, miRNA, stress, racial disparities
Introduction
Despite continued advancements in medical care, rates of maternal mortality in 2019 were 2.5 times higher in non-Hispanic Black (NHB) people than non-Hispanic White (NHW) people [1]. Infants of NHB mothers are more likely to be born preterm (14.13% compared to 9.09% for NHW people in 2018) [2]. NHB people in the US have an increased risk of developing placental-mediated pregnancy complications such as fetal growth restriction [3], preeclampsia [4], and pre-term birth compared to their NHW counterparts [5]. Factors driving these disparities in maternal and neonatal health are complex and multifactorial [6]. There is growing consensus and appreciation for the enormous impact of historic and ongoing structural racism (e.g. implicit bias by providers, health insurance coverage, access to care) underlying these health disparities. This translates, from a biological perspective, to chronically overwhelmed and overburdened physiological systems (e.g. increased cortisol and allostatic load) designed to compensate for environmental stressors [3, 5–7]. The placenta, as the maternal-fetal interface, plays a critical role in mediating the impact of maternal exposures on fetal growth and development, and its contribution to health disparities is a relatively unexplored area.
The placenta is exquisitely sensitive to the maternal environment, and events in early pregnancy, or even the pre-conception period, can negatively impact placental development and function, leading to poor outcomes [8]. Interestingly, some studies have reported differences in placental pathology between maternal racial groups [9]. Assibey-Mensah et al. showed that NHB had an increased risk of maternal vascular malperfusion, low placental weight, and decidual vasculopathy compared with NHW people after adjusting for maternal age, education, primiparity, and pre-pregnancy BMI [10]. In addition, Chen et al. and Matoba et.al found higher prevalence of chronic inflammatory lesions across a range of gestational ages in placentas of NHB people [9, 11]. Placental telomere length is shorter in NHB people as compared to NHW people [12]; this indicator of greater cellular stress and aging is consistent with reports that NHB people experience “weathering” or enhanced biological aging due to the chronic stress of racism [13]. Altogether, these studies suggest the placenta may play a critical role in the translation of environmental stressors to pregnancy outcomes, though the underlying mechanisms are poorly understood.
Placental microRNA – short, non-coding RNAs which alter post-transcriptional gene translation – are dynamically expressed, and guide the surveillance and response to DNA damage [14]. Placental miRNA may be involved in maternal adaptations to pregnancy [15, 16] and fetal growth outcomes. In other tissues, miRNA have been shown to be responsive to environmental stressors (e.g. pesticides, psychosocial stress, noise pollution [17]), though their role in racial health disparities has not been fully investigated [5, 18]. In this study, we utilized a large pregnancy cohort to examine placental miRNA expression profiles of NHB and NHW people, and the association between differentially expressed miRNA and neonatal adiposity.
Materials and Methods
Samples were collected from healthy people, recruited at 37 to 41 weeks of pregnancy and delivered by elective cesarean section following uncomplicated pregnancies at Metro Health Medical Center (Cleveland, OH). We selected samples based on self-identified racial data from non-Hispanic Black (NHB, n=50), and non-Hispanic White (NHW, n=74) people. Subjects with fetal anomalies, multiple gestations, preeclampsia, diabetes (pre-existing and gestational), or other comorbid disease were excluded. Multiple full-thickness placenta tissue biopsies were collected immediately after delivery from different cotyledons excluding decidual layer while avoiding any macroscopic lesions. Multiple villous samples were further dissected into small pieces, blotted for removal of blood and separately snap frozen in liquid nitrogen within 5 min of biopsy.
The study was conducted according to the guidelines in the Declaration of Helsinki. Written and informed consent was obtained prior to participation, and the analyses presented here were approved by the Institutional Review Board of Tufts Medical Center (IRB#12842) and Metro Health Medical Center (IRB #1300650).
Placental RNA isolation
Total placental RNA was extracted by homogenization of ~50 mg placental biopsy tissue in TRIzol reagent (Invitrogen), following the manufacturer’s guidelines. RNA quantification and integrity were measured using an Agilent 2100 Bioanalyzer (Agilent). All samples had an RNA integrity number >7.
Placental miRNA expression analysis by miRNA sequencing
A subset of 80 placentas were selected for small RNA sequencing considering maternal pre-pregnancy Body Mass Index (PP-BMI; lean (PP-BMI 18.5–24.9 kg/m2) and obese (PP-BMI >30kg/m2)), maternal race and smoking status, and offspring sex and adiposity at birth (high (%body fat: 12–20%) and low (%body fat: 4–10%)) [19]. Gel isolation of small RNA, library construction, and sequencing were conducted by Genome Quebec (Montreal, Quebec, Canada). After removing the adapters, all sequencing libraries were aligned to the GRCh38 human reference genome using bowtie aligner[20]. Aligned reads were intersected with mature miRNA coordinates based on miRBase (v21) (http://www.mirbase.org/). We normalize libraries using the median of ratios normalization method implemented in the R package DESeq[21]. The negative binomial test implemented in DESeq2 was used to identify miRNAs differentially expressed. Three samples were removed due to QC failure. Demographic information for the remaining samples (N=77) is presented in Supplementary Table 1. P-values of the comparisons between two groups were calculated while controlling for maternal obesity, neonatal adiposity and sex. To control the false discovery rate (FDR), Benjamini-Hochberg’s method[22] was used to adjust the p-values for multiple comparisons. Tests were considered significant if the adjusted p-value <=0.05.
Placenta cDNA synthesis and quantitative PCR
We performed reverse transcription (RT) and quantitative PCR (RT-qPCR) on 124 placenta samples with miRScript PCR system (Cat#218161 Qiagen) and ViiA 7 Real Time PCR system according to the manufacturer’s protocols. Dissociation curves were run on all reactions, and samples were normalized to the expression of the geometric mean between two housekeeping small non-coding RNAs (snoR95, snoR68) as an endogenous control best-keeper (bs). Fold increase relative to a calibrator sample was determined by using the 2−ΔΔCt method [23].
Statistical analysis
All data were presented as mean ± standard deviation. Differences in clinical characteristics between NHB and NHW people (Table 1) were analyzed using Student’s t-test. Frequency data was analyzed by Fisher’s exact test. miRNA expression results (via RT-qPCR) were expressed in arbitrary units and normalized by log-2 transformation before analysis. We used four-way ANOVA to evaluate the effects of maternal race, obesity, age, and neonatal sex on placental miRNA expression.
Table 1.
Maternal and neonatal characteristics of study population
| Maternal | NHB (N=50) | NHW (N=74) | P-value |
|---|---|---|---|
| Age (years) | 26.4 ± 6 | 30.3 ± 6 | <0.0001 |
| Pre-gravid BMI (Lean/Obese) β | 18/32 | 48/26 | 0.001 |
| Net Gestational Weight gain (kg) | 9.9 ± 8 | 12.3 ± 7 | 0.09 |
| Gestational Weight gain (kg) | 13.6 ± 9 | 16.4 ± 8 | 0.06 |
| IOM (Inadequte/Excessive)% β | 24/54 | 12/49 | 0.17 |
| Blood Pressure S/D meanβ | 118/70 | 116/68 | 0.95 |
| Smoking (N/Y)β | 37/11 | 58/16 | 0.10 |
| Neonatal | |||
| Gestational Age (wks) | 39 ± 0.7 | 39 ± 0.5 | 0.62 |
| Placenta weight (g) | 627 ± 148 | 668 ± 193 | 0.18 |
| Birth weight (kg) | 3.1 ± 0.5 | 3.3 ± 0.6 | 0.08 |
| Neonatal Adiposity (High/Low adiposity) β | 24/26 | 38/36 | 0.85 |
| Placenta Efficiency (g/g)γ | 5.2 ± 0.9 | 5.1 ± 0.9 | 0.74 |
| Length (cm) | 48 ± 3 | 49 ± 2 | 0.06 |
| Sex (M/F) β | 32/16 | 35/39 | 0.05 |
Values are mean ± standard deviation. P-values for differences between NHB and NHW people calculated by Student’s t-test or βFisher’s exact test.
Placenta efficiency: birth weight (g)/placental weight (g)
Binomial logistic regression was used to evaluate the relationship between neonatal adiposity as a marker of fetal nutrient delivery and placental miRNA expression (via RT-qPCR). Neonatal adiposity was the binary dependent variable in this analysis with categories of high/low adiposity in all participants. We adjusted for maternal age, gestational age, neonatal sex, maternal obesity, and the interaction between placental miRNAs and race. Statistical analyses were performed with R studio, Boston, MA, version 3.6.2 [24]. P-value ≤0.05 were considered significant.
Target identification and selection
We performed gene target analysis with the placental miRNAs found to be differently expressed between NHB and NHW people in the RNA-sequencing data. We identified experimentally validated target genes for the selected miRNAs using miRNet 2.0 [25]. For this analysis we used two validated databases: miRTarBase v8.0 and TarBase v8.0[26, 27]. We ran two analyses: one using the placental miRNAs found to be higher in NHB people, and the second using the miRNAs found to be higher in NHW people.
Network creation and customization
Nodes can be identified based on their position within the network. The degree of a node is the number of connections it has to other nodes. Nodes with higher node degree and betweenness act as hubs in a network. The network analyses using miRNet 2.0 were filtered by two centrality measures: degree centrality >3, and betweenness centrality >3 [25].. We did this to reduce orphan miRNAs, and improve the identification of important nodes in the network analysis. We assessed cell pathway analysis for the genes targeted by our miRNAs of interest using Reactome v.3.7, database release 77[28]. Reactome performs a pathway over-representation analysis to calculate the P-value. P-value were adjusted using the Benjamini-Hochberg’s method[22]. Adjusted p-value <0.05 was considered significant.
Results
Identification of differentially expressed miRNA between placentas of NHW vs NHB people
The demographic data from the larger cohort are summarized in (Table 1). We found that NHW people were older and leaner when compared to NHB people (P-value <0.0001). Using small RNA-sequencing we detected a total of 1677 miRNAs expressed in 77 placentas (a miRNA is considered expressed if a total of 10 or more copies were detected in the 77 samples, demographic data for this subset of the cohort in Supplementary Table 1). Out of the total, 44 miRNAs were differently expressed between placentas of NHW and NHB people according to a cut off adjusted p-value <0.05. Fifteen of these miRNAs were higher in placentas of NHB people, and 29 were higher in placentas of NHW people (Supplementary Table 2).
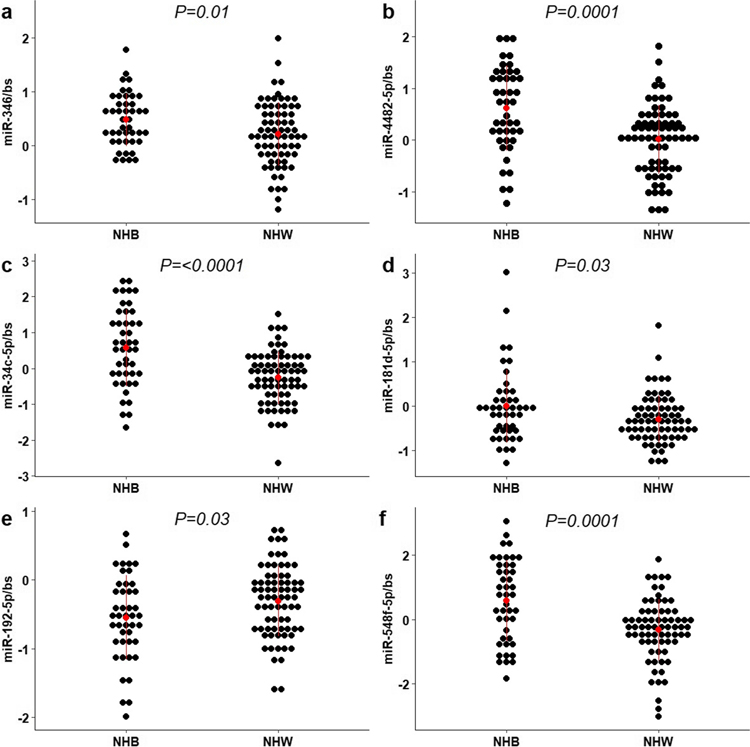
We randomly selected eight miRNAs to explore further using RT-qPCR in the larger cohort (N=124); five found to be higher in NHB placentas (hsa-miR-346, hsa-miR-4482-5p, hsa-miR-4482-3p, hsa-miR-34c-5p, hsa-miR-181d-5p), and three found to be higher in NHW placentas (hsa-miR-548f-5p, hsa-miR-192-5p, hsa-miR-505-3p) based on RNA-seq. In Fig. 1, we show that four of the five miRNAs found to be higher in placentas of NHB people measured by RT-qPCR were consistent with the RNA-sequencing results. However, for the three miRNAs found to be higher in the NHW placentas, only hsa-miR-192-5p was consistent with the RNA-sequencing results (P-value=0.03). Hsa-miR-548f-5p was lower in NHW placentas (P-value=0.0001) by RT-qPCR in the larger cohort. Placental hsa-miR-346, miR-4482-5p, miR-34c-5p, miR-181d-5p, miR-192-5p, and miR-548f-5p continued to be significantly different between NHB and NHW after adjusting for maternal obesity, maternal age, and neonatal sex.
Fig 1.
Expression of miRNAs in placentas of non-Hispanic Black (NHB) and non-Hispanic white (NHW) people. The expression levels of mature miR-346 (a), miR-4482-5p (b), miR-34c-5p (c), miR-181d-5p (d), miR-192-5p (e), and miR-548f-5p (f) was measured in 124 placentas via RT-qPCR. Red dots and lines represent means ± SD. Bs= bestkeeper (geometric mean between two housekeeping small non-coding RNA (snoR95, SNOR68) as an endogenous control). The P-value of the two-tailed t-test analysis are shown.
We assessed the association of these placental miRNAs with neonatal adiposity (high/low adiposity) among all participants (N=124). We found that increased placental miR-192-5p expression is associated with a lower probability of having a low adiposity neonate (odds ratio [OR], 0.42 [95% CI, 0.20 to 0.82]); even after adjustment for maternal age, maternal obesity, gestational age, neonatal sex, and race ([OR], 0.20 [95% CI, 0.04 to 0.64]). We did not detect an interaction between placental miR-192-5p expression and race in this model. We were not able to detect a significant association between other selected placental miRNA and neonatal adiposity.
Target analysis
Placental miRNAs with higher expression in NHB people:
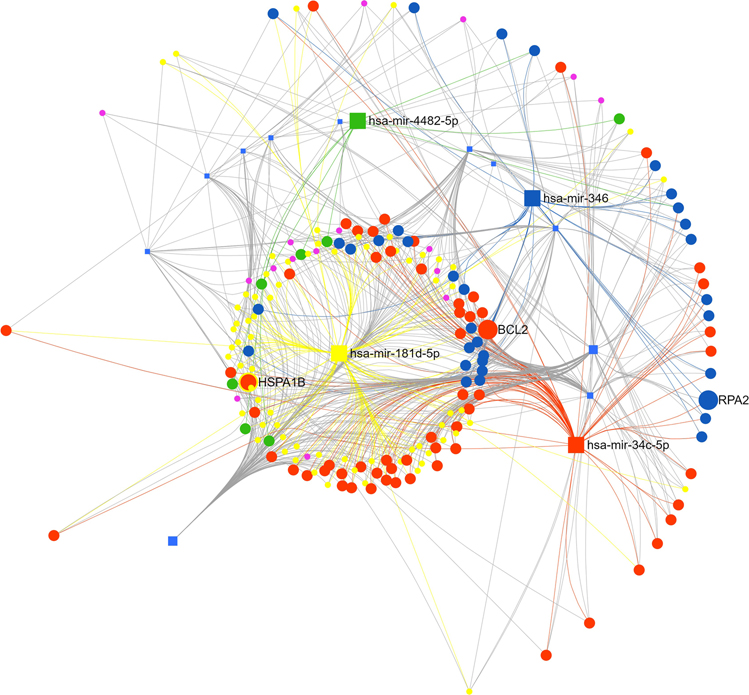
we uploaded the 15 miRNAs found to be higher in NHB people based on the RNA-sequencing data into miRNet 2.0. In the network builder we kept targets validated with CLASH, CLEAR-CLIP, Luciferase, HITS-CLIP microarray, PAR-CLIP, and Chimeric fragments methods. After the degree and betweenness centrality measures, 186 genes and 809 edges remained in the network analysis (Fig. 2), (Supplementary Table 3).
Fig 2.
Function and Network analysis of placental miRNAs highly expressed in non-Hispanic Black people. Squares represent miRNAs measured by RT-qPCR – yellow (miR-181d-5p), orange (miR-34c-5p), green (miR-4482-5p), and blue (miR-346). Connected circles represent genes targeted by these miRNAs, color and size of the circle represents the number and miRNAs targeting these genes Analysis done with miRNET 2.0.
Function and Network analysis
For the network analysis we only used genes targeted by the placental miRNAs that we validated using RT-qPCR in the larger cohort. Thus, the number of gene targets to be analyzed was reduced to 172 genes. Only 74 of the genes had a betweenness higher than 28; the rest had a degree of 1 and a betweenness of 0, so we eliminated them from the analysis. Using Reactome for cell signaling enrichment analysis we found 48 genes to interact with 102 cell signaling pathways (P value <0.05; data shown in Supplementary Table 4). Twenty-four of the gene entities were not identified by Reactome. The most significant pathways were ‘Attenuation phase’ (Adj. P-value=<0.0001), ‘HSF1-dependent transactivation’ (Adj. P-value =<0.0001), ‘HSF1-Activation’ (Adj. P-value =<0.0001), ‘Regulation of HSF1-mediated heat shock response’ (Adj. P-value =<0.0001), and ‘Cellular response to heat stress’ (Adj. P-value=0.0001) Table 2.
Table 2.
Cell signaling pathways targeted by miRNAs highly expressed in placentas of non-Hispanic Black people (Reactome 2020 Database)
| Pathway name | P-value | Adj. P-value | Genes |
|---|---|---|---|
| Attenuation phase | 3.94E-08 | 1.88E-05 | HSPA1B |
| HSF1-dependent transactivation | 1.81E-07 | 4.31E-05 | HSPA1B |
| HSF1 activation | 5.57E-07 | 8.81E-05 | RPA2;HSPA1B |
| Regulation of HSF1-mediated heat shock response | 1.12E-06 | 1.34E-04 | RPA2;HSPA1B |
| Cellular response to heat stress | 4.13E-06 | 3.93E-04 | RPA2;HSPA1B |
| Gene expression (Transcription) | 8.80E-06 | 6.95E-04 | TCF7L2;BTG2;CBX6;RPA2;BAZ2A;DICER1;SOD2;YY1;NR6A1;CCND2;CDK6;NFIA;RRAGD;CDC27;SRSF2;E2F5;ZNF664;SRSF7;E2F7;DCP2;TNRC6B |
| RNA Polymerase II Transcription | 1.14E-04 | 0.007 | TCF7L2;BTG2;CBX6;RPA2;SOD2;YY1;NR6A1;CCND2;CDK6;RRAGD;CDC27;SRSF2;E2F5;ZNF664;SRSF7;E2F7;DCP2;TNRC6B |
| Generic Transcription Pathway | 2.24E-04 | 0.013 | TCF7L2;BTG2;CBX6;RPA2;SOD2;YY1;NR6A1;CCND2;CDK6;RRAGD;CDC27;E2F5;ZNF664;E2F7;DCP2;TNRC6B |
| Nuclear Receptor transcription pathway | 3.11E-04 | 0.016 | NR6A1 |
| UCH proteinases | 5.61E-04 | 0.025 | YY1;UCHL1;INO80D;MBD6;TGFBR2 |
| Cellular responses to stress | 5.95E-04 | 0.025 | CBX6;CDK6;RRAGD;CDC27;RPA2;RPL37;SOD2;HSPA1B;DCP2;TNRC6B |
| Cellular responses to stimuli | 7.28E-04 | 0.028 | CBX6;CDK6;RRAGD;CDC27;RPA2;RPL37;SOD2;HSPA1B;DCP2;TNRC6B |
| TP53 Regulates Transcription of Cell Cycle Genes | 0.001 | 0.037 | BTG2;E2F7 |
| Interleukin-7 signaling | 0.001 | 0.042 | PIK3R3;BRWD1 |
| Regulation of RUNX1 Expression and Activity | 0.001 | 0.042 | CCND2;CDK6;TNRC6B |
Discussion
The placenta is a master regulator of fetal growth, as the principal metabolic, respiratory, and endocrine organ of the fetus; it is also a key route by which environmental exposures are transmitted from the mother to the offspring (5). In this study, we found that placental expression of 15 miRNAs were higher in NHB people compared to NHW people. We validated the expression of four of these 15 miRNAs in a larger cohort, and found the same differences in expression, even after adjusting for maternal pre-pregnancy body mass index (BMI), maternal age, neonatal adiposity and sex. Predicted gene targets of these placental miRNAs found to be higher in NHB people were related to cellular stress response pathways. One miRNA found to be higher in placentas of NHW people, miR-192-5p, was associated with lower probability of having a neonate with low adiposity. Identification of placental miRNAs targeting pathways potentially regulating response to environmental factors known to increase the risk of preterm birth and low birthweight in NHB people, provides an opportunity for early intervention using miRNAs as early biomarkers of exposure.
MiRNAs are important gene expression regulators that may mediate gene-environment interactions [29]. In this study we identified and further validated in a larger cohort, that miR-346, miR-4482-5p, miR-34c-5p, and miR-181d-5p are differentially expressed between placentas of NHB and NHW people. The expression of some of these miRNAs, such as the miR-34 family, have been shown to be altered by stress-induced modifications which affect the regulation of the hypothalamic-pituitary-adrenal (HPA) axis, the principal neuroendocrine stress response system [30]. Moreover, miR-34 and miR-181 families play a role in the cellular response to oxidative stress, mitochondrial function, redox state, and inflammatory pathways [17, 31]. MiR-34c and miR-181d target Bcl2, which controls mitochondrial outer membrane permeability and calcium homeostasis in many tissues [17, 32]. Cumulative maternal stress and trauma has been shown to be associated with a greater placental mitochondria mutational load, particularly among NHB people [33]. Based on these data, these miRNA may be part of the response to maternal chronic stress, however further studies need to be done to better understand the clinical implications for both the mother and the fetus.
We found that miR-192-5p had on average lower expression in placentas of NHB vs NHW people. Additionally, high placental expression of miR-192-5p was associated with a lower risk of giving birth to a low adiposity infant. This miRNA targets DNA damage response/repair, cell differentiation, circadian rhythm, adipocyte differentiation and lipid metabolism pathways[34]. To the best of our knowledge, ours is the first study to show an association between miR-192-5p expression in the placenta and fetal growth outcomes. Lampl et al. demonstrated that fetal fat accumulation is 5.8 times greater for NHW than for NHB people in late gestation [35]. Whether this is related to the differential expression of placental miRNAs such as the adiposity associated miR-192-5p, should be explored in future studies. As shown in Fig. 2, miRNAs can have multiple gene targets, and the specific genes targeted may be organ-specific. Future studies should investigate how miR-192-5p affects placental growth and lipid metabolism to understand the mechanisms underlying its association with neonatal fat accrual.
Our pathway network analysis showed that placental miRNAs upregulated in NHB people target genes (HSPA1B; RPA2) related to the attenuation phase, heat shock factor (HSF)1-dependent transactivation, regulation of HSF1-mediated heat shock response, and cellular responses to stress. The heat shock response (HSR) is an ordered response to diverse environmental and physiological stressors, including acute and chronic conditions such as elevated temperatures, heavy metals, chemical toxicants, and oxidative stress [36]. A hallmark of the cell stress response representing one of its first identified features is the induction of heat shock proteins, many of which function as molecular chaperones. In combination with the DNA repair machinery, molecular chaperones provide a rapid and direct mechanism of cellular defense against stress-induced damage. These proteins are extensively utilized as bio indicators of environmental stress in many different types of organisms[37, 38], which now may include the placenta.
NHB have higher rates of morbidity and mortality than NHW for almost all health outcomes in the United States. The weathering hypothesis is proposed to explain racial health disparities. It states that chronic exposure to social and economic disadvantage leads to acceleration of normal aging and earlier decline in physical health outcomes and could partially explain racial disparities in a wide array of health conditions, including low birth weight [39, 40]. In our cohort, NHB people were younger than the NHW people, which may have blunted the impacts of chronic stress. Though all were healthy with uncomplicated pregnancies, NHB people had a non-statistically significant trend towards lighter offspring and smaller placentas. With this in mind, we speculate that placental miRNA differentially expressed between NHB and NHW people may be sensitive to the maternal effects of chronic stress, and among uncomplicated pregnancies, these differences in expression may represent a beneficial reaction that counteracts/modulates the impact of environmental stress on placental pathways, preserving normal fetal growth. Future studies should investigate the regulation of these miRNAs in placentas of people with pregnancies complicated by preeclampsia, pre-term birth or intrauterine growth restriction.
A key strength of this study is that we used an unbiased method to identify miRNA candidates (small RNA-seq) and validated selected miRNA in a larger cohort via RT-qPCR, the gold standard for expression analysis. We used miRNet.2.0 a method based on experimentally validated miRNA-target interactions, to identify potential gene targets of our miRNA of interest. An important limitation of this study is the lack of information on social determinants of health (e.g. income, education, and insurance status), stress exposure, paternal race/ethnicity, and reliance on limited categories for participants to self-identify their race/ethnicity as a marker for exposure to structural racism. Additionally, it would provide more context if we were able to conduct key informant interviews with NHB and NHW pregnant patients to further examine their cultural, social, and physical environment, and capture negative outcomes in both mothers and neonates.
In conclusion, we show that placental miRNA expression patterns differing between NHB and NHW people are involved in cellular stress responses, suggesting that these miRNA may be responding to environmental stressors. We speculate that these stressors are associated with the experience of systemic racism. Though lower on average among NHB people, placental miR-192-5p was positively associated with neonatal adiposity among all pregnant participants, suggesting it may be sensitive to the experience of structural racism with potential impacts on fetal nutrition and growth. Future studies should investigate the expression of these stress-sensitive miRNA among complicated pregnancies to understand better their involvement in pregnancies where maternal and/or fetal outcomes are at risk.
Supplementary Material
Acknowledgements:
We would like to thank Dr. Mary Haghiac and Ms. Judi Minium for contributing to recruitment, sample and data collection.
Funding:
This research was supported by the National Institute of Health grant R01HD091735.
Footnotes
Competing Interests The authors declare no competing interests to disclose
Ethics Approval All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. This study was approved by the Institutional Review Board of Tufts Medical Center (IRB #12842) and Metro Health Medical Center (IRB #1300650).
Consent to Participate Informed consent was obtained from all individual participants included in the study
Publisher's Disclaimer: This AM is a PDF file of the manuscript accepted for publication after peer review, when applicable, but does not reflect post-acceptance improvements, or any corrections. Use of this AM is subject to the publisher’s embargo period and AM terms of use. Under no circumstances may this AM be shared or distributed under a Creative Commons or other form of open access license, nor may it be reformatted or enhanced, whether by the Author or third parties. See here for Springer Nature’s terms of use for AM versions of subscription articles: https://www.springernature.com/gp/open-research/policies/accepted-manuscript-terms
Reference
- 1.Hoyert DL (2020). Maternal mortality rates in the United States, 2019. Retrieved from https://stacks.cdc.gov/view/cdc/103855
- 2.Martin JA (2019). Births in the United States, 2018, (346), 8. [PubMed] [Google Scholar]
- 3.Bryant AS, Worjoloh A, Caughey AB, & Washington AE (2010). Racial/ethnic disparities in obstetric outcomes and care: prevalence and determinants. American Journal of Obstetrics and Gynecology, 202(4), 335–343. 10.1016/j.ajog.2009.10.864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka M, Jaamaa G, Kaiser M, Hills E, Soim A, Zhu M, … McNutt L-A (2007). Racial Disparity in Hypertensive Disorders of Pregnancy in New York State: A 10-Year Longitudinal Population-Based Study. American Journal of Public Health, 97(1), 163–170. 10.2105/AJPH.2005.068577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matoba N, Mestan KK, & Collins JW (2021). Understanding Racial Disparities of Preterm Birth Through the Placenta. Clinical Therapeutics, 43(2), 287–296. 10.1016/j.clinthera.2020.12.013 [DOI] [PubMed] [Google Scholar]
- 6.Pham O, Nov 10 URP, & 2020. (2020, November 10). Racial Disparities in Maternal and Infant Health: An Overview - Issue Brief. KFF. Retrieved from https://www.kff.org/report-section/racial-disparities-in-maternal-and-infant-health-an-overview-issue-brief/ [Google Scholar]
- 7.Roy-Matton N, Moutquin J-M, Brown C, Carrier N, & Bell L (2011). The Impact of Perceived Maternal Stress and Other Psychosocial Risk Factors on Pregnancy Complications. Journal of Obstetrics and Gynaecology Canada, 33(4), 344–352. 10.1016/S1701-2163(16)34852-6 [DOI] [PubMed] [Google Scholar]
- 8.Aplin JD, Myers JE, Timms K, & Westwood M (2020). Tracking placental development in health and disease. Nature Reviews Endocrinology, 16(9), 479–494. 10.1038/s41574-020-0372-6 [DOI] [PubMed] [Google Scholar]
- 9.Matoba N, Yallapragada S, Davis MM, Ernst LM, Collins JW, & Mestan KK (2019). Racial differences in placental pathology among very preterm births. Placenta, 83, 37–42. 10.1016/j.placenta.2019.06.385 [DOI] [PubMed] [Google Scholar]
- 10.Assibey-Mensah V, Parks WT, Gernand AD, & Catov JM (2018). Race and risk of maternal vascular malperfusion lesions in the placenta. Placenta, 69, 102–108. 10.1016/j.placenta.2018.07.017 [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Huang L, Zhang H, Klebanoff M, Yang Z, & Zhang J (2015). Racial disparity in placental pathology in the collaborative perinatal project. International Journal of Clinical and Experimental Pathology, 8(11), 15042–15054. [PMC free article] [PubMed] [Google Scholar]
- 12.Jones CW, Gambala C, Esteves KC, Wallace M, Schlesinger R, O’Quinn M, … Drury SS (2017). Differences in placental telomere length suggest a link between racial disparities in birth outcomes and cellular aging. American Journal of Obstetrics and Gynecology, 216(3), 294.e1–294.e8. 10.1016/j.ajog.2016.11.1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geronimus AT (1996). Black/white differences in the relationship of maternal age to birthweight: A population-based test of the weathering hypothesis. Social Science & Medicine, 42(4), 589–597. 10.1016/0277-9536(95)00159-X [DOI] [PubMed] [Google Scholar]
- 14.Wan G, Mathur R, Hu X, Zhang X, & Lu X (2011). miRNA response to DNA damage. Trends in Biochemical Sciences, 36(9), 478–484. 10.1016/j.tibs.2011.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mouillet J-F, Ouyang Y, Coyne CB, & Sadovsky Y (2015). MicroRNAs in placental health and disease. American Journal of Obstetrics and Gynecology, 213(4), S163–S172. 10.1016/j.ajog.2015.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’tierney-Ginn PF, & Alvarado F (2021). Placental miR-3940–3p is associated with maternal insulin resistance in late pregnancy. Harvard Dataverse. 10.7910/DVN/SSSL2D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miguel V, Cui JY, Daimiel L, Espinosa-Díez C, Fernández-Hernando C, Kavanagh TJ, & Lamas S (2018). The Role of MicroRNAs in Environmental Risk Factors, Noise-Induced Hearing Loss, and Mental Stress. Antioxidants & Redox Signaling, 28(9), 773–796. 10.1089/ars.2017.7175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costello JF, & Fisher SJ (2021). The Placenta - Fast, Loose, and in Control. The New England Journal of Medicine, 385(1), 87–89. 10.1056/NEJMcibr2106321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Catalano PM, Thomas AJ, Avallone DA, & Amini SB (1995). Anthropometric estimation of neonatal body composition. American Journal of Obstetrics and Gynecology, 173(4), 1176–1181. 10.1016/0002-9378(95)91348-3 [DOI] [PubMed] [Google Scholar]
- 20.Langmead B, Trapnell C, Pop M, & Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology, 10(3), R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anders S, & Huber W (2010). Differential expression analysis for sequence count data. Genome Biology, 11(10), R106. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y, & Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological), 57(1), 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 23.Livak KJ, & Schmittgen TD (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods, 25(4), 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 24.RStudio Team. (2020). RStudio: Integrated Development for R. Boston, MA: RStudio, PBC. Retrieved from http://www.rstudio.com/ [Google Scholar]
- 25.Chang L, Zhou G, Soufan O, & Xia J (2020). miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Research, 48(W1), W244–W251. 10.1093/nar/gkaa467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang H-Y, Lin Y-C-D, Li J, Huang K-Y, Shrestha S, Hong H-C, … Huang H-D (2020). miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Research, 48(D1), D148–D154. 10.1093/nar/gkz896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, … Hatzigeorgiou AG (2018). DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA–gene interactions. Nucleic Acids Research, 46(Database issue), D239–D245. 10.1093/nar/gkx1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fabregat A, Sidiropoulos K, Viteri G, Marin-Garcia P, Ping P, Stein L, … Hermjakob H (2018). Reactome diagram viewer: data structures and strategies to boost performance. Bioinformatics (Oxford, England), 34(7), 1208–1214. 10.1093/bioinformatics/btx752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q, Kappil MA, Li A, Dassanayake PS, Darrah TH, Friedman AE, … Chen J (2015). Exploring the associations between microRNA expression profiles and environmental pollutants in human placenta from the National Children’s Study (NCS). Epigenetics, 10(9), 793–802. 10.1080/15592294.2015.1066960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hollins SL, & Cairns MJ (2016). MicroRNA: Small RNA mediators of the brains genomic response to environmental stress. Progress in Neurobiology, 143, 61–81. 10.1016/j.pneurobio.2016.06.005 [DOI] [PubMed] [Google Scholar]
- 31.Li L, & Stary CM (2016). Targeting Glial Mitochondrial Function for Protection from Cerebral Ischemia: Relevance, Mechanisms, and the Role of MicroRNAs. Oxidative Medicine and Cellular Longevity, 2016, 6032306. 10.1155/2016/6032306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X-F, Shi Z-M, Wang X-R, Cao L, Wang Y-Y, Zhang J-X, … You Y-P (2012). MiR-181d acts as a tumor suppressor in glioma by targeting K-ras and Bcl-2. Journal of Cancer Research and Clinical Oncology, 138(4), 573–584. 10.1007/s00432-011-1114-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brunst KJ, Zhang L, Zhang X, Baccarelli AA, Bloomquist T, & Wright RJ (2021). Associations Between Maternal Lifetime Stress and Placental Mitochondrial DNA Mutations in an Urban Multiethnic Cohort. Biological Psychiatry, 89(6), 570–578. 10.1016/j.biopsych.2020.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren F, Yao Y, Cai X, & Fang G (2021). Emerging Role of miR-192-5p in Human Diseases. Frontiers in Pharmacology, 12, 614068. 10.3389/fphar.2021.614068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lampl M, Lee W, Koo W, Frongillo EA, Barker DJP, & Romero R (2012). Ethnic differences in the accumulation of fat and lean mass in late gestation. American Journal of Human Biology: The Official Journal of the Human Biology Council, 24(5), 640–647. 10.1002/ajhb.22285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westerheide SD, & Morimoto RI (2005). Heat Shock Response Modulators as Therapeutic Tools for Diseases of Protein Conformation*. Journal of Biological Chemistry, 280(39), 33097–33100. 10.1074/jbc.R500010200 [DOI] [PubMed] [Google Scholar]
- 37.Lewis S, Handy RD, Cordi B, Billinghurst Z, & Depledge MH (1999). Stress proteins (HSP’s): Methods of Detection and Their Use as an Environmental Biomarker. Ecotoxicology, 8(5), 351–368. 10.1023/A:1008982421299 [DOI] [Google Scholar]
- 38.Stress proteins: the biological functions in virus infection, present and challenges for target-based antiviral drug development | Signal Transduction and Targeted Therapy. (n.d.). Retrieved November 16, 2021, from https://www-nature-com.ezproxy.library.tufts.edu/articles/s41392-020-00233-4 [DOI] [PMC free article] [PubMed]
- 39.Geronimus AT, Hicken M, Keene D, & Bound J (2006). “Weathering” and Age Patterns of Allostatic Load Scores Among Blacks and Whites in the United States. American Journal of Public Health, 96(5), 826–833. 10.2105/AJPH.2004.060749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forde AT, Crookes DM, Suglia SF, & Demmer RT (2019). The weathering hypothesis as an explanation for racial disparities in health: a systematic review. Annals of Epidemiology, 33, 1–18.e3. 10.1016/j.annepidem.2019.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.