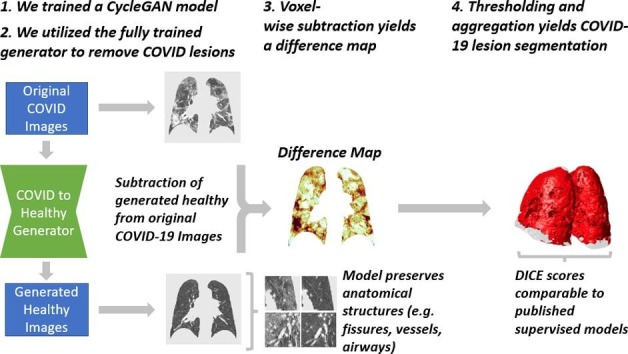
Graphical abstract
Keywords: CycleGAN, COVID-19, CT, Lesion segmentation
Abstract
Background
Lesion segmentation is a critical step in medical image analysis, and methods to identify pathology without time-intensive manual labeling of data are of utmost importance during a pandemic and in resource-constrained healthcare settings. Here, we describe a method for fully automated segmentation and quantification of pathological COVID-19 lung tissue on chest Computed Tomography (CT) scans without the need for manually segmented training data.
Methods
We trained a cycle-consistent generative adversarial network (CycleGAN) to convert images of COVID-19 scans into their generated healthy equivalents. Subtraction of the generated healthy images from their corresponding original CT scans yielded maps of pathological tissue, without background lung parenchyma, fissures, airways, or vessels. We then used these maps to construct three-dimensional lesion segmentations. Using a validation dataset, Dice scores were computed for our lesion segmentations and other published segmentation networks using ground truth segmentations reviewed by radiologists.
Results
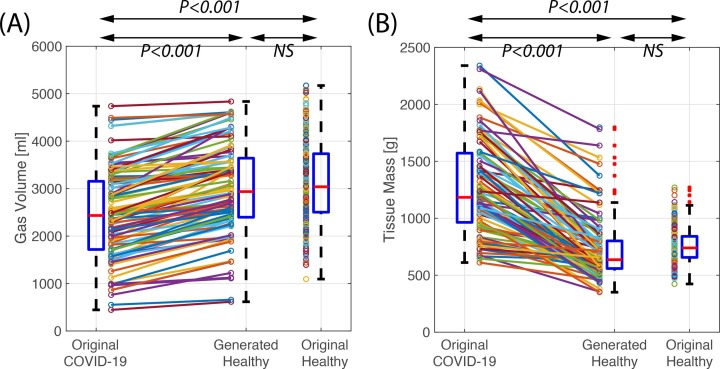
The COVID-to-Healthy generator eliminated high Hounsfield unit (HU) voxels within pulmonary lesions and replaced them with lower HU voxels. The generator did not distort normal anatomy such as vessels, airways, or fissures. The generated healthy images had higher gas content (2.45 ± 0.93 vs 3.01 ± 0.84 L, P < 0.001) and lower tissue density (1.27 ± 0.40 vs 0.73 ± 0.29 Kg, P < 0.001) than their corresponding original COVID-19 images, and they were not significantly different from those of the healthy images (P < 0.001). Using the validation dataset, lesion segmentations scored an average Dice score of 55.9, comparable to other weakly supervised networks that do require manual segmentations.
Conclusion
Our CycleGAN model successfully segmented pulmonary lesions in mild and severe COVID-19 cases. Our model’s performance was comparable to other published models; however, our model is unique in its ability to segment lesions without the need for manual segmentations.
1. Introduction
The Coronavirus Disease 2019 (COVID-19) pandemic has caused hundreds of millions of confirmed infections worldwide as of 2022 [1]. Of significant clinical consequence was the high case-fatality rate during the beginning of the pandemic, peaking in May of 2020 and has decreased worldwide since. Similarly, within Italy’s initial outbreak the case fatality rate was nearly 5 times greater than that at the time of this publication [2]. During this time in Italy, the Lombardy region hospitals bore the greatest number of COVID-19 cases, attributed deaths, and the highest case fatality rate compared to all other regions. COVID-19 outbreaks risk overwhelming healthcare resources and remain a threat to under vaccinated and resource-poor nations, especially as new variants emerge.
Characterizing the radiologic findings of lung injury can help the clinical evaluation of COVID-19 [3], [4], [5], [6], [7], [8], [9] and in fact, the Fleischner Society recommends chest computed tomography (CT) in patients with - or at risk of - worsening respiratory status [10]. Clinical studies correlated density of opacities and greater total lung involvement on chest CT with worse clinical outcomes, including intensive care admission, greater respiratory support requirements, organ failure, and death [11], [12]. These findings suggest a role of quantitative CT characterization of COVID-19 patients to inform prognostication and treatment plans. Unsupervised machine learning applied to medical imaging has the potential to aid in the diagnosis, prognosis, and characterization of COVID-19 lung findings. These approaches seek to improve the efficiency of imaging analysis workflows and offer clinicians a valuable tool during outbreak conditions with high patient volumes.
Many machine learning techniques have been proposed for COVID-19 imaging analysis. A key step in CT analysis is the lesion segmentation: separation of pathologic tissue from the surrounding non-pathologic tissue. There are a broad range of segmentation approaches. Some techniques have focused on preprocessing to exaggerate the distinction between the target lesion tissue and the surrounding tissue, which then aids in Hounsfield unit (HU) thresholding-based approaches. One such study, Oulefki et al, uses predefined filters to improve segmentation performance via a previously published multilevel thresholding process [13]. Most techniques, however, have made use of deep neural networks to achieve accurate segmentations. Supervised networks are quite common, despite the labor-intensive manual lesion segmentations required to train such models. These supervised approaches have recently seen considerable improvement. He et al introduced a supervised evolution-based adversarial network that outperformed multiple existing supervised networks [14]. Mu et al also introduced a novel supervised approach to COVID-19 lesion segmentation that segments on multiple topographic scales, each segmentation improving on the previous resulting in “polished” segmentations [15]. A major disadvantage of these supervised approaches is that they involve manually segmenting CT scans, which is expensive, time-consuming, prone to observer bias, and requires radiology experts [16], [17]. Moreover, the reliance on labeled images limits the scale at which training data can be generated. This prevents the use of large imaging data sources such as aggregated repositories of health records, which could further enhance machine learning-based methods. An unsupervised learning approach is particularly useful for lesion segmentation, often the first step in quantitative image analysis, as it eliminates the need for manual annotation of training data lesions.
Several strategies have been proposed for weakly (minimally) supervised lesion segmentation. CoSinGan used a multi-scale architecture with conditional GANs to achieve plausible lesion segmentation performance with only two manually labeled images [18]. Laradji et al’s Active Learning allows for weakly supervised learning by working with human annotations to label regions of high interest, and allowing point-level labeling rather than pixel-level labelling [19]. Similarly, Liu et al allow an annotator to “scribble” rough hand-drawn segmentations, which the network then refines [20], [21]. Still, CoSinGan, Active Learning and Liu et al’s “scribble” approach still require some human annotation for the generation of each segmentation. Xu et al. proposed GASNet, which requires as little as one manual segmentation to achieve good performance [20]. GASNet makes use of a CycleGAN-like network that incorporates a supervised “Segmenter” network to perform lesion segmentation. Additionally, Yao et al. proposed a network that randomly generates fake lesions to superimpose on normal CT images. The resulting NormNet is able to recognize out-of-distribution voxels, which correspond to COVID-19 lesions [22]. While these approaches perform strongly, they have certain limitations. GASNet requires some manual segmentation, and NormNet generates synthetic lesions to augment U-net training, rather than using an unsupervised method that incorporates real lesions. Our proposed model is unique as it is fully unsupervised, requires no manual annotations to learn lesion segmentation, and trains on real COVID-19 lesions.
GANs are useful for unsupervised training because they can learn the data distribution of a given domain without requiring a human to explicitly annotate domain-specific features [23]. In particular, conditional GANs, which are capable of image-to-image translation, have become popular in the medical imaging research community [24]. These networks have proven useful in a wide range of tasks, including denoising, cross-modality image synthesis, and data augmentation [24]. Recent work in brain MRI and liver CT also showed that cycle-consistent GANs, given images from both a normal and pathological population, can be used to create a “normal” looking version of pathological tissue [25], yielding an “abnormality map”.
CycleGANs [26] are a particular class of GAN that allows for images to be converted from one domain into another. Critically, there does not need to be direct correspondence between the two sets of images (i.e. the images can be “unpaired”), and they do not require explicit annotation of the features that differ between the two domains. In this paper, we leverage these advantages to create an algorithm capable of recognizing and removing lesioned tissue.
Here we train a CycleGAN on a dataset of healthy and COVID-19 positive chest CT scans to create a model that recognizes and removes pulmonary lesions. By subtracting these synthetically produced healthy equivalents, we are left with a map of pathological tissue, which can then be thresholded to yield a lesion segmentation.
2. Materials and methods
2.1. Definitions
In order to clarify terminology we use repeatedly, here we provide a list of terms along with their definition in the context of this paper.
Domain: a group of images that share some common characteristic, in this case the presence of COVID-19 or the lack of pathology. These are referred to as the COVID domain and the healthy domain, respectively. In other CycleGAN implementations, the network may learn to convert images of apples into images of oranges and vice versa. In this case, the network learns to convert images from the COVID-19 domain into the healthy domain and vice versa.
COVID-to-Healthy generator: a neural network that learns to convert images from the COVID domain to the healthy domain. One of four networks that comprise the CycleGAN.
Healthy-to-COVID generator: a neural network that learns to convert images from the healthy domain to the COVID domain. One of four networks that comprise the CycleGAN.
COVID discriminator: a neural network that learns to differentiate real images from the COVID domain from those made by the Healthy-to-COVID generator.
Healthy discriminator: a neural network that learns to differentiate real images from the healthy domain from those made by the COVID-to-Healthy generator.
Original COVID-19 images: 153 chest CT scans obtained from Italian hospitals of patients who tested positive for COVID-19 via RT-PCR.
Original healthy images: 356 chest CT scans obtained from the COPDGene dataset with <15% abnormal aeration of the lung. We recognize that these images do not represent truly healthy lungs, however, they do appear grossly normal upon inspection. Furthermore, there is minimal pathology visible on the scans, and no obvious emphysema or bronchitis.
Generated healthy images: 153 original COVID-19 images after being passed through the COVID-to-Healthy generator.
Original healthy images passed through the COVID-to-Healthy generator: The 356 COPDGene images after being processed by the COVID-to-Healthy generator. This was done for the purpose of studying the effects of the COVID-to-Healthy generator on healthy images.
2.2. Dataset
The original COVID-19 images group consisted of chest CT scans from 153 unique patients with SARS-CoV-2 infections confirmed by nucleic acid amplification tests from two Italian hospitals. The original healthy group consisted of 356 inspiratory chest CT scans, all from unique patients. All original healthy images came from the COPDGene dataset and had <15% of lung tissue with high attenuation (HU 〈−950) [27]. Finally, the publicly available Coronacases dataset was used as an external validation set [28]. The validation dataset contains 10 complete chest CT scans of patients with confirmed COVID-19, consisting of 2,506 individual coronal slices used for validation testing, as well as the corresponding lung segmentations and radiologist-reviewed lesion segmentations. As a validation set, these images were never used in the training of the network. These 10 scans represent a subset of the total linked dataset, as not all images had thin axial slices and thus sufficient coronal resolution to be used in this study. Furthermore, the data used in the regular dataset of 153 COVID-19 images and 356 healthy images were all thin-slice CT scans with high resolution in the coronal dimension. All methods and data use performed in this study were in accordance with the guidelines and regulations set forth by COPDGene, CoronaCases, and the Institutional Review Boards at the University of Milano-Bicocca, the Hospital of San Gerardo, and the University of Pennsylvania. All data was anonymized and considered non-patient data.
2.3. Data preprocessing and network training
Chest CT scans from COVID-19 and healthy images were sliced coronally, resampled from various dimensions to 256 × 256 pixels, and restricted to a range of −1150 to 350 Hounsfield units (HU). Slices not including any lung tissue were removed. The model was then trained on the original COVID-19 images as well as the original healthy images for 40,000 iterations at a learning rate of 0.0005 with batch size of 1 on an NVIDIA Tesla T4 GPU. The training was repeated at a learning rate of 0.005 and with various network architectures. Following training, all images from the COVID-19 dataset were converted to their generated healthy equivalents. Once an image in the COVID group was converted to the healthy domain, each voxel in the generated healthy image was subtracted from the original to yield a lesion map. For the purposes of displaying and analyzing results, a whole-lung segmentation mask of the original image was generated using a previously trained network [29], and regions outside the mask were set to zero. Every 1000 iterations, the validation set was tested which produced lesion segmentations that were compared to the radiologist-reviewed segmentations and Dice coefficients of similarity were recorded. Alternative learning rates and architectures were tested to determine their effect on model performance.
Outside of the validation set, cases were sorted by mean lung density inside the whole-lung segmentation of the original image as a proxy for severity, and representative images were chosen. Healthy cases were run through the COVID-to-Healthy generator to determine whether the model introduces undesired transformation in healthy tissue. To obtain a lesion segmentation mask, threshold cutoffs from 10 to 500 HU were tried, and 200 units of HU difference was determined to include unhealthy tissue while still removing most noise. Therefore, all voxels above a difference of 200 HU were labeled as lesions. Frequency distributions of HU densities in COVID and COVID-to-Healthy GAN generated images and used to calculate metrics of whole-lung gas volume and tissue weight using a previously established methodology [30]. Paired T-tests were performed on lung gas volumes and lung masses of original COVID images vs. generated healthy images, as well as generated healthy images vs. original healthy images and original COVID images vs. original healthy images. In order to determine whether the network preserved the pulmonary gravitational gradient, all segmented lungs from generated healthy images and original healthy images were sliced into six equally spaced regions along the anterior-posterior axis.
2.4. Implementation
The network was trained on a virtual machine instance created via Google Cloud Platform. The instance, n1-highmem-8, was equipped with 8 virtual CPUs, 400 GB of disc storage, and 52 GB of RAM. An NVIDIA Tesla T4 graphics card was attached to the instance. A boot disc with Pytorch version 1.4 and CUDA version 10.1.243 preinstalled was selected. An XFCE desktop environment was installed and managed via Google Remote Desktop to facilitate development and visual inspection of network results. Development was primarily conducted using Jupyter Notebooks. The official CycleGAN implementation (https://github.com/junyanz/pytorch-CycleGAN-and-pix2pix) [26] was cloned from GitHub and functions to allow for intermittent comparison of network output vs CoronaCases ground truth were added. To facilitate this, functions to allow for voxel-wise subtraction, thresholding, and 3D image reassembly were written. Finally, the network architecture was modified to allow for testing of different-sized networks. Beyond this, all network customizations were made using built-in options available in the public implementation.
3. Theory
3.1. Network architecture
Unpaired image-to-image translation (Fig. 1 ) was accomplished via a modified version of a publicly available 2-dimensional CycleGAN implementation (Fig. 2 ) [26]. All networks used an Adam optimizer with a batch size of 1. Adversarial loss was calculated using L2 loss, while cycle-consistency and identity loss used L1 [31]. Generators with 6, 9, and 18 ResBlocks were trained, and the version with 9 ResBlocks was determined to have the best combination of speed and accuracy (Supplementary Figure 1). From these data the model performed optimally with 9 Resnet blocks, 40,000 training iterations, and a learning rate of 0.0005 (Supplementary Figure 2). As such, the 9-ResBlock model with these hyperparameters was selected for further use. The model’s performance with and without the validation set included in the training data set is shown in Supplementary Figure 3.
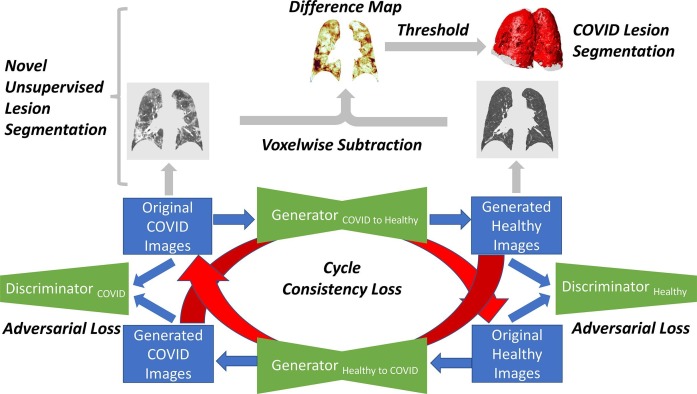
Fig. 1.
Diagram of the CycleGAN training loop and the novel process for generating unsupervised COVID-19 lesion segmentations. The model is composed of four networks: two generators and two discriminators. Beginning with an original image from the COVID domain, the COVID-to-Healthy generator converts it into the healthy domain. The image is then converted back into the COVID domain by the healthy-to-COVID generator, and it is compared to the original COVID image to calculate cycle consistency loss. Along the way, the original healthy and generated healthy images are given as input to the healthy discriminator, which attempts to correctly classify the image as original or generated and subsequently calculate adversarial loss. This process is then repeated with a real image from the healthy domain. After training, the COVID-to-Healthy generator is used to convert unseen COVID images to generated healthy equivalents. These are then subtracted from the original lesioned tissue to create a difference map, which can then be thresholded to produce a lesion segmentation.
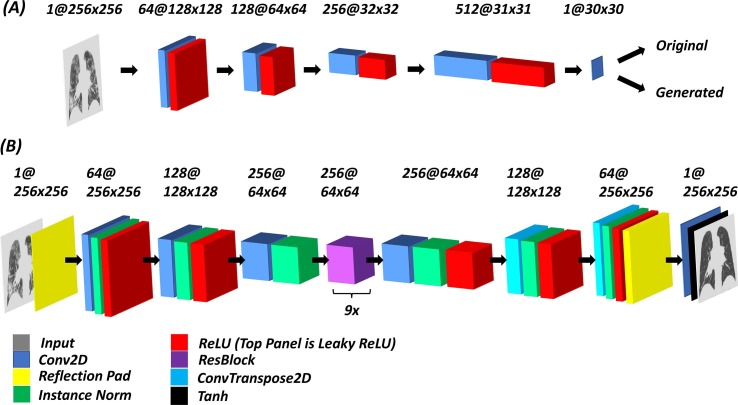
Fig. 2.
General network architecture of the COVID and Healthy discriminators (A) and the COVID-to-Healthy and Healthy-to-COVID generators (B). Tensor dimensions are given above their respective representations, with the number of filters preceding the @ sign, and spatial dimensions following it. Sample images show inputs and outputs from COVID-19 discriminator and COVID-19-to-Healthy generator.
3.2. Loss function
The loss function of a CycleGAN involves two primary types of loss terms: adversarial loss, which is dependent on the discriminator’s evaluation of the generated images as compared to the originals, and cycle-consistency loss, which is dependent on the generator’s ability to reconstruct the original image after processing by both generators [26]. Some CycleGANs, including ours, also make use of identity loss, which encourages a generator to not change an image that is already in that generator’s output distribution [32]. Adversarial loss, also called GAN loss, is commonly used in other generative adversarial networks. For a given domain, the generator tries to minimize the GAN loss by generating realistic outputs that will fool the discriminator, while the discriminator tries to maximize it by correctly classifying real images as real and generated images as generated. Note that adversarial losses use L2 loss, while cycle-consistency and identity losses use L1. Mathematically, adversarial loss for COVID discriminator Dc and healthy-to-COVID generator GH2C is as follows:
| (1) |
Similarly, the adversarial loss for healthy discriminator DH and COVID-to-Healthy generator GC2H is given by:
| (2) |
Cycle-consistency loss represents the intuition that putting a COVID image through the COVID-to-Healthy generator and then putting the resulting image through the healthy-to-COVID generator should result in the recreation of the original image. The difference between this recreated image and the original gives the cycle-consistency loss. Cycle-consistency loss for generators GH2C and GC2H is as follows:
| (3) |
We also make use of an identity loss, which helps preserve the true HU values of each region. It does this by encouraging a generator to not change an image that is already in that generator’s output distribution. Identity loss for GH2C and GC2H is given by:
| (4) |
Different types of loss are given different weight in the combined loss function. The weights for cycle consistency loss and identity loss were 10 and 5, respectively. These numbers were based off recommendations in the original CycleGAN paper. The combined loss function is thus given as:
| (5) |
4. Calculations
4.1. Evaluation metrics
Hounsfield Units: Hounsfield units (HU) are a commonly used measure of radiodensity in CT scans. HU represents the radiodensity on a scale defined by the radiodensities (μ) of air and water. A Hounsfield unit is defined as:
| (6) |
Tissue Mass: Tissue mass was calculated using a previous method [30]. The lungs are modeled as a mixture of tissues (density equal to that of water, 1 g/cm3) and air (density of 0 g/cm3). The percent of the lung that corresponds to tissue is called the tissue fraction (Ft), thus the air fraction is given by (1- Ft). Given that HU are measured on a scale from −1000 (air) to 0 (water and tissue), we can derive the tissue fraction by dividing the mean lung HU value by −1000 (i.e. a lung with mean HU of −500 will be 50% air and 50% tissue, −500/-1000 = 0.5). Once we have the tissue fraction, we can obtain the tissue mass by multiplying by the total lung volume and the density of tissue (defined as 1 g/cm3):
| (7) |
Tissue Volume: Tissue volume was calculated using a similar equation [30], but with the tissue fraction replaced by the air fraction, and without the density term:
| (8) |
Dice Score: For two volumes X and Y, the Dice score between them is:
| (9) |
4.2. Lesion segmentation operations
To generate segmentations, we first train a CycleGAN as described above. Once training is complete, we generate a healthy equivalent for each COVID image using the COVID-to-Healthy generator. This healthy equivalent is then subtracted from the original to yield a difference map, represented as :
| (10) |
These difference maps represent the degree of aberrant radiodensity present in lesioned tissue as compared to a healthy equivalent. We threshold them such that each voxel at or >200 HU is included in the segmentation. This voxelwise binary classification results in a lesion mask M, thus the value of each voxel in difference map determines the value of corresponding mask voxel :
| (11) |
5. Results and discussion
5.1. COVID-19 lesion segmentation
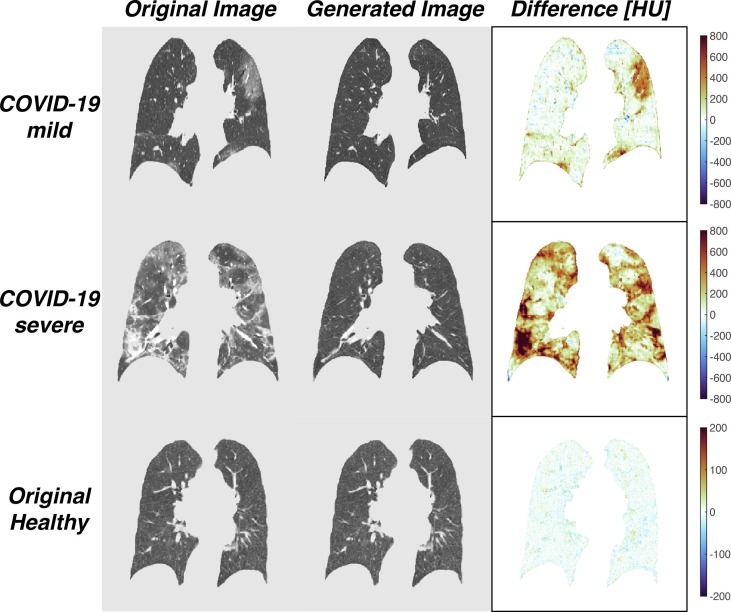
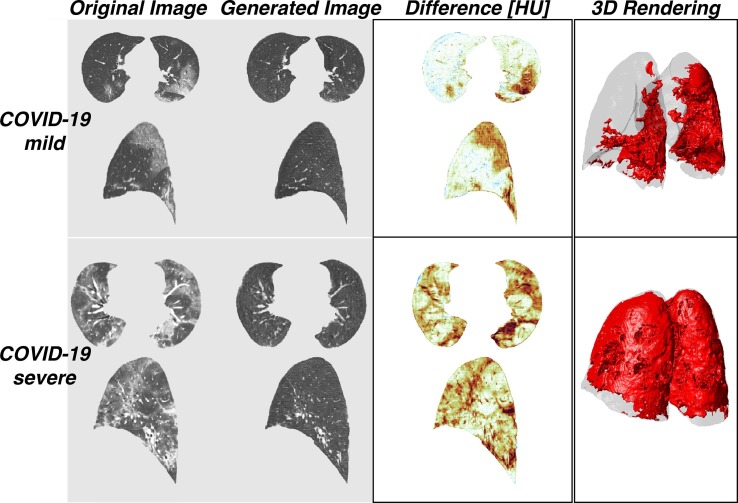
After model training was completed, original COVID-19 images were used as inputs to the COVID-to-Healthy generator, resulting in generated healthy images. Difference maps were created by subtracting the HU value for each corresponding voxel in the generated image from that of the original image (Fig. 3 top rows). In order to determine whether the COVID-to-Healthy generator altered scans with normal radiological features, healthy images from the COPDGene data set were used as inputs to the COVID-to-Healthy generator. A representative example shows the results are nearly superimposable with the originals, as demonstrated by the difference map (Fig. 3 , bottom row).
Fig. 3.
Representative original images from mild COVID-19, severe COVID-19, and healthy cases (left) and their corresponding generated images (middle). Subtraction of the generated image from the original image results in the difference map (right).
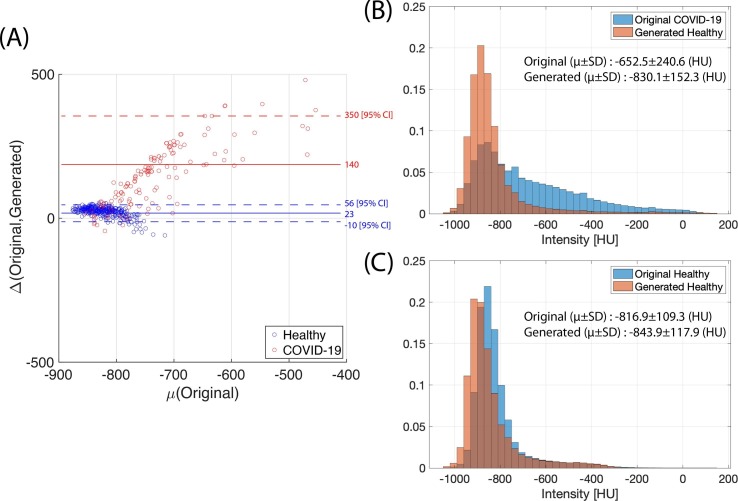
Generated healthy images had lower mean HU than their corresponding original COVID-19 images (−830.1 ± 152.3 HU, 95% CI for SEM [−842.4, −817.8] vs −652.5 ± 240, 95% CI for SEM [−690.7, −614.2], P < 0.001 via paired t-test ( Fig. 4 B, Table 2 ). There was no significant difference between the mean HU of the original healthy images and the generated healthy images (P > 0.05). Original COVID-19 images showed a significantly greater decrease in mean HU following input to the COVID-to-Healthy generator than the original healthy images (mean ΔHU 140, 95% CI [123.02, 156.98] and 23, 95% CI [21.25, 24.75]) for original COVID and original healthy, respectively, P < 0.001 via paired t-test, Fig. 4 A). When the original COVID-19 images were converted into the healthy domain, voxels in the higher ranges of HU were predominantly converted to lower HU ( Fig. 4 B). While the original healthy images did have voxels in high HU ranges when they were run through the COVID-to-Healthy generator, these voxels were not preferentially substituted, and there is no discernable pattern in their substitution ( Fig. 4 C). The model successfully replaced high HU voxels in pulmonary lesions with voxels in the range of normal healthy lung parenchyma without altering normally present high HU voxels found in vessels, or fissures. Furthermore, the generator functioned consistently among the coronal slices resulting in normal-appearing sagittal and axial views; inconsistency between coronal slices would result in a “choppy” appearance when reconstructed sagittally or axially (Fig. 5 , left). The final step in our pipeline - thresholding based on difference >200 - resulted in 3D segmentation maps (Fig. 5 , right).
Fig. 4.
(A) Scatter plot of original mean lung attenuation versus difference of means between original COVID-19 and the generated healthy images (Red) and between the original healthy and original healthy run through the COVID-Healthy generator (Blue). Dotted lines represent 95% confidence intervals. (B) Histogram of attenuation including all in-lung voxels of 153 original COVID-19 images and their corresponding generated healthy images. (C) Histogram of attenuation including all in-lung voxels of 356 original healthy images and their corresponding generated images, created by passing original healthy images through the COVID-to-Healthy generator.
Table 2.
Summary of Computed Tomography-Derived Physiology among the Original Healthy, Original COVID-19, and Generated Healthy Images. CT density is displayed in Hounsfield units (HU). All measures are averages followed by their standard deviation. P values result from two-tailed paired t tests.
| Images | Average CT density (HU) | Gas volume | Tissue mass |
|---|---|---|---|
| Original COVID-19 | −652.5 ± 240.6** | 2.45 ± 0.93** | 1.27 ± 0.40** |
| Generated Healthy | −830.1 ± 152.3 | 3.01 ± 0.84 | 0.73 ± 0.29 |
| Original Healthy | −816.9 ± 109.3 | 3.12 ± 0.83 | 0.75 ± 0.15 |
**P < 0.001 compared to Generated Healthy group.
Fig. 5.
Axial and sagittal views of the two original COVID-19 images from Fig. 1 (Left), their corresponding COVID-to-Healthy generated images, and the resulting difference map (Middle). 3D rendering of lesion segmentation mask (Right). Difference maps were thresholded to 200 Hounsfield units of difference, and all voxels above the threshold were included in the mask.
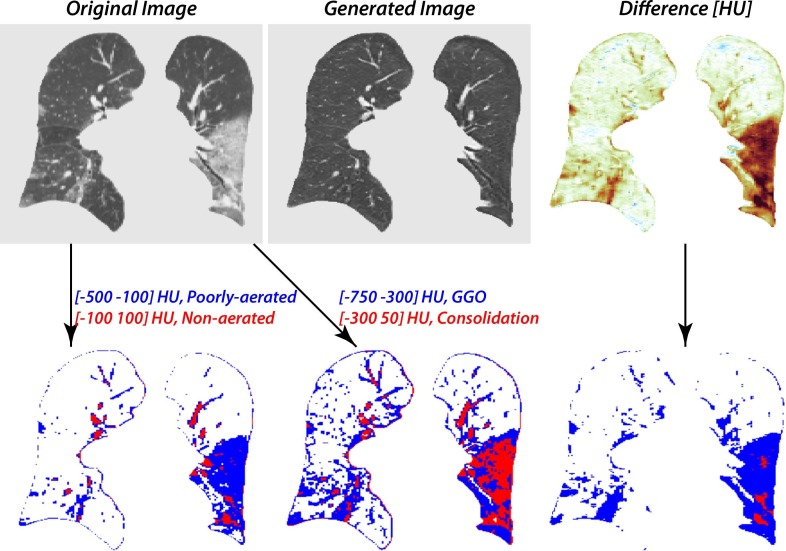
Our method was compared to the traditional thresholding techniques using HU cut offs published for ARDS and COVID-19 ( Fig. 6 ). Our model was able to differentiate between lesioned tissue and blood vessels of similarly high radiodensity, a feat well outside the capabilities of thresholding. Previous thresholding guidelines for ARDS have used a range of −500 to −100 HU to segment poorly aerated regions and −100 to 100 for loss of aeration [31], [33]. However, the traditional Hounsfield unit range for poorly aerated lung tissue does not include all ground-glass regions. Recent work suggested that ranges for COVID-19 patients be adjusted to −750 to −300 for ground glass opacities (GGO) and −300 to 50 for consolidation, in order to include more unhealthy tissue and lessen the amount of pulmonary vasculature included in a given segmentation [31]. Even with these adjustments, thresholding still results in considerable erroneous inclusion of proximal and prominent distal pulmonary vasculature. In contrast, our model has the ability to separate GGO, atelectasis, and consolidation from healthy tissue. Rather than simply lowering the attenuation globally, our network distinguishes between healthy and unhealthy regions of tissue, and selectively modifies the latter.
Fig. 6.
Comparison of thresholding-based segmentation and our method. ARDS guidelines for lesion segmentation define HU between −500 and −100 as poorly aerated and −100 to 100 as loss of aeration. COVID-19 threshold guidelines define HU between −750 and −300 as ground glass opacities and between −300 and 50 as consolidation.
Taken together, these data indicate that the COVID-to-Healthy generator distinguishes between diseased lung tissue and normal lung tissue. We observed that lesions were removed from original COVID-19 images and that the generator made nearly no changes to the original healthy images. Furthermore, the COVID-to-Healthy generator was able to discern between the abnormal tissue and the normal anatomic features within the COVID-19 scans as only the lesions were removed, without affecting the vessels, airways, and fissures regardless of their HU ranges. Our model removes lesions from COVID-19 scans generating an anatomically healthy image that can be subtracted from the original yielding a lesion segmentation.
5.2. Preservation of normal physiology in generated healthy images
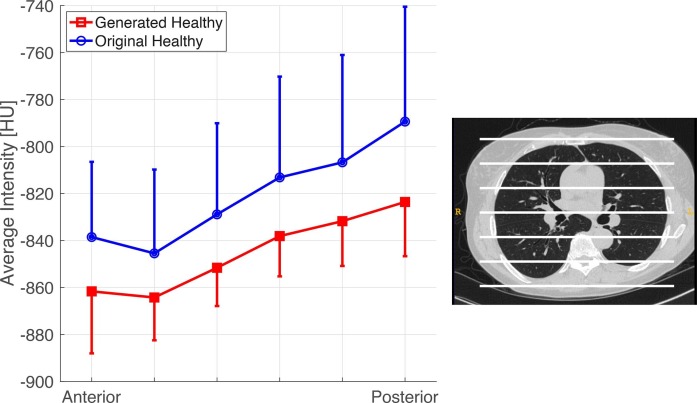
After the original COVID-19 images were processed through the COVID-to-Healthy generator, the generated healthy images showed an increased gas volume (2.45 ± 0.93 vs 3.01 ± 0.84 L, P < 0.001) and decreased lung tissue mass (1.27 ± 0.40 vs 0.73 ± 0.29 Kg, P < 0.001), indicating the physiologic effects of lesion removal Fig. 7 A and B). The generated healthy lungs had no difference in gas volume (3.01 ± 0.84 vs 3.12 ± 0.83 L, P > 0.05) Fig. 7 A) or mass (0.73 ± 0.29 vs 0.75 ± 0.15 Kg, P > 0.05) Fig. 7 B) compared to the original healthy images (Table 2). Upon averaging the HU among 6 equidistant regions along the anterior to posterior axis, the physiologic gravitational tissue gradient is preserved between original healthy images and generated healthy images (P > 0.05 in all regions, Fig. 8 ). This supports the notion that generated healthy images contain the same gravitational gradient of tissue density seen in normal healthy supinated lungs. Thus, our model substitutes diseased tissue with healthy tissue in a physiologically realistic manner.
Fig. 7.
Lung gas volumes (A) and lung masses (B) of 153 COVID-19 images in their original and generated healthy images compared with the 356 original healthy group images.
Fig. 8.
Mean Hounsfield attenuations of six equally spaced regions of lung along the anterior-posterior axis of the generated healthy images and original healthy images. Intensities reflect the mean HU value of each equidistant section across all available CT scans and the corresponding 95% confidence intervals are represented by the bars. (n = 153 for generated healthy, n = 356 for original healthy).
5.3. Validation of COVID-19 lesion segmentations
We sought to objectively evaluate our lesion segmentations using a validation dataset. We tested our model on 10 CoronaCases COVID-19 images with which our model was not trained. We compared our lesion segmentation to the radiologist reviewed lesion segmentations of CoronaCases yielding an average Dice score of 55.9 (Table 1 ). Our model had comparable performance to three other models, however ours is the only model that trains on real data without the need for manual segmentation.
Table 1.
Comparison of Dice scores for lesion segmentation between various weakly supervised networks and ours. Note that our evaluation only used 10 of 20 images in the CoronaCases dataset due to lack of sufficient coronal resolution on the other 10. Other networks in this chart trained on axial images and were therefore able to make use of all 20.
| Method | Number of manual segmentations used in training | Dice score |
|---|---|---|
| ActiveLearning | 16 | 52.4 |
| CoSinGan | 2 | 57.8 |
| GASNet | 1 | 70.3 |
| Ours | 0 | 55.9 |
Semi-supervised approaches to lesion segmentation, such as GASNet, achieve good results, but still require some level of manual segmentation. This still requires resources and limits their ability to be translated into other problem domains. Other approaches to lesion segmentation use convolutional neural networks, namely U-Net and U-Net-like architectures, to identify COVID-19 lesions [16]. While effective, these methods require extensive manually labeled training data, and often perform suboptimally on images that are unlike those contained in the original training set. Their performance is thus restricted by the amount and diversity of training data available, which is in turn restricted by the labor required to produce manual annotations. Our model has no such labor requirement, which removes a critical barrier to increased training set sizes and performed similarly to them on the validation dataset. Furthermore, inclusion of the validation dataset into the training dataset did not improve our model’s performance, underlining how our unsupervised model is able to generalize it’s COVID-to-Healthy generator network to succeed on images it has not seen before (Supplementary Figure 3).
5.4. Future directions and limitations of our approach
Beyond lesion segmentation and quantifying characteristics such as lung mass and volume, we believe that our healthy-to-COVID generator can be used to create synthetic training data. These can then be used along with the original segmentation mask to train segmentation models using supervised methods. This would be further augmented by the use of network structures that allow for multimodal outputs, such as MUNIT [29] or U-GAT-IT [34].
Our approach has several limitations. On rare occasions, our COVID-to-Healthy generator generates false positive healthy lung tissue in areas such as the chest wall or large airways. Although this does not impede lesion segmentation, which uses a separately generated whole-lung segmentation, it does prevent our current model from being used for thresholding-based whole-lung segmentation. Although these issues improve with longer training times, another issue arises after approximately 50,000 training iterations: the model learns to “cheat” the cycle-consistency constraint. This issue has been addressed in previous literature, and a solution was presented [35]. Future work could implement a version of this solution that is customized for COVID-19 pathology recognition wherein generators are trained separately to prevent cooperative “cheating”. We believe that once this limitation is overcome, errors will be further reduced by training on a larger data set and extending our current 2D CycleGAN to 3D. A further limitation was our use of a 2-dimensional model, which was chosen because CycleGANs are very memory-intensive, and thus difficult to fit on a single GPU if training in 3D. Although the model showed good spatial consistency of healthy features between slices, there were severe cases where pathological tissue was removed unevenly between slices. Many of the COVID-19 scans were taken with contrast, and as a result the network tends to remove contrast in images where it is present. This occasionally results in changes in intensity on pulmonary blood vessels, yielding sub-optimal difference masks. In the future, we plan on training contrast and non-contrast groups separately in order to avoid this issue. An additional limitation to this approach is that healthy training data was a subset of the COPDGene dataset, specifically patients that had <15% of lung tissue with abnormal aeration. Patients do not need a diagnosis of COPD in order to be enrolled in the study, but they do need to have certain risk factors, such as a history of smoking. Thus, it is possible that our healthy dataset does contain pathologies that went undetected by our review, despite having mostly healthy lung tissue. Finally, our validation set contained only 10 complete COVID-19 CT images. It reflects a range of disease severity, and contains a total of 2506 coronal slices, but is still small for a robust test of a deep learning model’s performance. While other publications similarly use small validation sets, a larger sample would improve our ability to evaluate and compare the performance of our unsupervised model to that of competing models.
6. Conclusion
We presented a CycleGAN-based model for unsupervised COVID-19 lesion segmentation. Our CycleGAN model successfully segmented pulmonary lesions in mild and severe COVID-19 cases. Our model’s performance was comparable to other published models when tested on a validation dataset. Our model, at the time of writing, is the first approach that describes an unsupervised COVID-19 lesion segmentation process that trains on real human data. The lack of manually labeled data represents a key bottleneck in COVID-19 research, and our approach circumvents this problem. This is of particular importance given the clinical burden placed on much of the expert population by the present COVID-19 pandemic.
Funding
This work was supported by the National Institutes of Health 1R01HL137389-01A1: “An integrated approach to predict and improve the outcomes of lung injury.”
CRediT authorship contribution statement
Marc Connell: Conceptualization, Formal Analysis, Writing – original draft, Writing – review & editing. Yi Xin: Conceptualization, Formal Analysis, Writing – original draft, Writing – review & editing. Sarah E. Gerard: Conceptualization, Formal Analysis. Jacob Herrmann: Conceptualization, Formal Analysis. Parth K. Shah: Conceptualization, Writing – original draft, Writing – review & editing. Kevin T. Martin: Conceptualization, Writing – original draft, Writing – review & editing. Emanuele Rezoagli: Data curation, Writing – review & editing. Davide Ippolito: Data curation, Writing – review & editing. Jennia Rajaei: Writing – review & editing. Ryan Baron: Data curation, Writing – review & editing. Paolo Delvecchio: Formal analysis. Shiraz Humayun: Formal analysis. Rahim R. Rizi: Writing – review & editing. Giacomo Bellani: Data curation, Writing – review & editing. Maurizio Cereda: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ymeth.2022.07.007.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Modi C., Böhm V., Ferraro S., Stein G., Seljak U. Estimating COVID-19 mortality in Italy early in the COVID-19 pandemic. Nat. Commun. 2021;12:2729. doi: 10.1038/s41467-021-22944-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernheim A., Mei X., Huang M., Yang Y., Fayad Z.A., Zhang N., Diao K., Lin B., Zhu X., Li K., Li S., Shan H., Jacobi A., Chung M. Chest CT findings in Coronavirus Disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020:200463. doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., Cui J., Xu W., Yang Y., Fayad Z.A., Jacobi A., Li K., Li S., Shan H. CT imaging features of 2019 Novel Coronavirus (2019-nCoV) Radiology. 2020;295:202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng M.-Y., Lee E.Y.P., Yang J., Yang F., Li X., Wang H., Lui M.M., Lo C.-S.-Y., Leung B., Khong P.-L., Hui C.-K.-M., Yuen K., Kuo M.D. Imaging profile of the COVID-19 infection: radiologic findings and literature review, radiology: cardiothoracic. Imaging. 2020;2:e200034. doi: 10.1148/ryct.2020200034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan F., Ye T., Sun P., Gui S., Liang B., Li L., Zheng D., Wang J., Hesketh R.L., Yang L., Zheng C. Time course of lung changes on chest CT during recovery from 2019 novel Coronavirus (COVID-19) pneumonia. Radiology. 2020:200370. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C. Articles Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect. Dis. 2020;20(4):425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song F., Shi N., Shan F., Zhang Z., Shen J., Lu H., Ling Y., Jiang Y., Shi Y. Emerging 2019 novel Coronavirus (2019-nCoV) pneumonia. Radiology. 2020;295:210–217. doi: 10.1148/radiol.2020200274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Z., McGoogan J.M. Characteristics of and important lessons from the Coronavirus Disease 2019 (COVID-19) outbreak in china: summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 10.Rubin G.D., Ryerson C.J., Haramati L.B., Sverzellati N., Kanne J.P., Raoof S., Schluger N.W., Volpi A., Yim J.-J., Martin I.B.K., Anderson D.J., Kong C., Altes T., Bush A., Desai S.R., Goldin O., Goo J.M., Humbert M., Inoue Y., Kauczor H.-U., Luo F., Mazzone P.J., Prokop M., Remy-Jardin M., Richeldi L., Schaefer-Prokop C.M., Tomiyama N., Wells A.U., Leung A.N. The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the Fleischner society. Radiology. 2020;296(1):172–180. doi: 10.1148/radiol.2020201365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oulefki A., Agaian S., Trongtirakul T., Kassah Laouar A. Automatic COVID-19 lung infected region segmentation and measurement using CT-scans images. Pattern Recognit. 2021;114 doi: 10.1016/j.patcog.2020.107747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He J., Zhu Q., Zhang K., Yu P., Tang J. An evolvable adversarial network with gradient penalty for COVID-19 infection segmentation. Appl. Soft Comput. 2021;113 doi: 10.1016/j.asoc.2021.107947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mu N., Wang H., Zhang Y., Jiang J., Tang J. Progressive global perception and local polishing network for lung infection segmentation of COVID-19 CT images. Pattern Recogn. 2021;120 doi: 10.1016/j.patcog.2021.108168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.F. Shi, J. Wang, J. Shi, Z. Wu, Q. Wang, Z. Tang, K. He, Y. Shi, D. Shen, Review of Artificial Intelligence Techniques in Imaging Data Acquisition, Segmentation and Diagnosis for COVID-19, (2020). https://arxiv.org/abs/2004.02731. [DOI] [PubMed]
- 17.Cao Y., Xu Z., Feng J., Jin C., Han X., Wu H., Shi H. Longitudinal assessment of COVID-19 using a deep learning–based quantitative CT pipeline: illustration of two cases. Radiol.: Cardiothoracic Imaging. 2020;2:e200082. doi: 10.1148/ryct.2020200082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang P., Zhong Y., Deng Y., Tang X., Li X. CoSinGAN: learning COVID-19 infection segmentation from a single radiological image. Diagnostics (Basel) 2020;10:E901. doi: 10.3390/diagnostics10110901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.I. Laradji, P. Rodriguez, F. Branchaud-Charron, K. Lensink, P. Atighehchian, W. Parker, D. Vazquez, D. Nowrouzezahrai, A Weakly Supervised Region-Based Active Learning Method for COVID-19 Segmentation in CT Images, ArXiv:2007.07012 [Cs, Eess]. (2020). http://arxiv.org/abs/2007.07012 (accessed October 29, 2021).
- 20.Z. Xu, Y. Cao, C. Jin, G. Shao, X. Liu, J. Zhou, H. Shi, J. Feng, GASNet: Weakly-supervised Framework for COVID-19 Lesion Segmentation, ArXiv:2010.09456 [Cs, Eess]. (2020). http://arxiv.org/abs/2010.09456 (accessed October 29, 2021).
- 21.Liu X., Yuan Q., Gao Y., He K., Wang S., Tang X., Tang J., Shen D. Weakly Supervised Segmentation of COVID19 Infection with Scribble Annotation on CT Images. Pattern Recogn. 2022;122 doi: 10.1016/j.patcog.2021.108341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Q. Yao, L. Xiao, P. Liu, S.K. Zhou, Label-Free Segmentation of COVID-19 Lesions in Lung CT, ArXiv:2009.06456 [Cs, Eess]. (2021). http://arxiv.org/abs/2009.06456 (accessed April 27, 2022). [DOI] [PMC free article] [PubMed]
- 23.I.J. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Warde-Farley, S. Ozair, A. Courville, Y. Bengio, Generative Adversarial Networks, (2014). https://arxiv.org/abs/1406.2661.
- 24.Yi X., Walia E., Babyn P. Generative adversarial network in medical imaging: a review. Med. Image Anal. 2019;58 doi: 10.1016/j.media.2019.101552. [DOI] [PubMed] [Google Scholar]
- 25.Sun L., Wang J., Huang Y., Ding X., Greenspan H., Paisley J. An adversarial learning approach to medical image synthesis for lesion detection. IEEE J. Biomed. Health Inform. 2020;24(8):2303–2314. doi: 10.1109/JBHI.2020.2964016. [DOI] [PubMed] [Google Scholar]
- 26.J.-Y. Zhu, T. Park, P. Isola, A.A. Efros, Unpaired image-to-image translation using cycle-consistent adversarial networks, ArXiv:1703.10593 [Cs]. (2018). http://arxiv.org/abs/1703.10593 (accessed May 12, 2020).
- 27.Regan E.A., Hokanson J.E., Murphy J.R., Make B., Lynch D.A., Beaty T.H., Curran-Everett D., Silverman E.K., Crapo J.D. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.M. Jun, G. Cheng, W. Yixin, A. Xingle, G. Jiantao, Y. Ziqi, Z. Minqing, L. Xin, D. Xueyuan, C. Shucheng, W. Hao, M. Sen, Y. Xiaoyu, N. Ziwei, L. Chen, T. Lu, Z. Yuntao, Z. Qiongjie, D. Guoqiang, H. Jian, COVID-19 CT Lung and Infection Segmentation Dataset, (2020). https://doi.org/10.5281/zenodo.3757476.
- 29.X. Huang, M.-Y. Liu, S. Belongie, J. Kautz, Multimodal Unsupervised Image-to-Image Translation, ArXiv:1804.04732 [Cs, Stat]. (2018). http://arxiv.org/abs/1804.04732 (accessed May 11, 2020).
- 30.Xin Y., Cereda M., Hamedani H., Pourfathi M., Siddiqui S., Meeder N., Kadlecek S., Duncan I., Profka H., Rajaei J., Tustison N.J., Gee J.C., Kavanagh B.P., Rizi R.R. Unstable inflation causing injury. Insight from prone position and paired computed tomography scans. Am. J. Respiratory Crit. Care Med. 2018;198:197–207. doi: 10.1164/rccm.201708-1728OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vieira S.R., Puybasset L., Richecoeur J., Lu Q., Cluzel P., Gusman P.B., Coriat P., Rouby J.J. A lung computed tomographic assessment of positive end-expiratory pressure-induced lung overdistension. Am. J. Respiratory Crit. Care Med. 1998;158:1571–1577. doi: 10.1164/ajrccm.158.5.9802101. [DOI] [PubMed] [Google Scholar]
- 32.Y. Taigman, A. Polyak, L. Wolf, Unsupervised cross-domain image generation, ArXiv:1611.02200 [Cs]. (2016). http://arxiv.org/abs/1611.02200 (accessed April 25, 2022).
- 33.Gattinoni L., Pesenti A., Avalli L., Rossi F., Bombino M. Pressure-volume curve of total respiratory system in acute respiratory failure. Computed tomographic scan study. Am. Rev. Respir. Dis. 1987;136:730–736. doi: 10.1164/ajrccm/136.3.730. [DOI] [PubMed] [Google Scholar]
- 34.J. Kim, M. Kim, H. Kang, K. Lee, U-GAT-IT: Unsupervised generative attentional networks with adaptive layer-instance normalization for image-to-image translation, ArXiv:1907.10830 [Cs, Eess]. (2020). http://arxiv.org/abs/1907.10830 (accessed May 12, 2020).
- 35.A.W. Harley, S.-E. Wei, J. Saragih, K. Fragkiadaki, Image disentanglement and uncooperative re-entanglement for high-fidelity image-to-image translation, ArXiv:1901.03628 [Cs]. (2019). http://arxiv.org/abs/1901.03628 (accessed May 11, 2020).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.