Abstract
The stochasticity of molecular motion results in the existence of multiple kinetically relevant pathways in many biomolecular mechanisms. Because it is highly demanding to characterize them for complex systems, mechanisms are often described with a single-pathway perspective. However, kinetic network analysis and sub-ensemble experimental insight are increasingly demonstrating not only the existence of competing pathways, but also the importance of kinetic selection in biology. This review focuses on advances in multiscale kinetic analysis of proteins, which connects molecular level information from simulations to macroscopic data to characterize mechanistic reaction networks and the reactive flux through them. We describe a range of methods used and highlight several examples where kinetic modeling has revealed functional importance of pathway heterogeneity.
Keywords: kinetic network analysis, multiscale kinetic modeling, kinetic selection, kinetic proofreading, multiscale modeling
INTRODUCTION
Protein mechanisms are often described as a series of transitions between intermediates (Figure 1). This single-pathway perspective can be helpful for envisioning how a process might proceed. However, it can also be qualitatively wrong, missing the mechanistic heterogeneity exemplified in single-molecule experiments [2]. The relevance of competing pathways is obvious in heterogeneous catalysis, where percent yields quantify the dominance of particular pathways in a reaction network. While the same principles hold for biomolecular transformations [3], pathway heterogeneity has been especially challenging to discern because it requires identifying not only all relevant intermediates, but also the transition rates between them and reactive flux through them under a given set of conditions. Kinetic network analysis aims to do just this, and its multiscale application to proteins, wherein molecular-level information is combined with experimental restraints, is the focus of this review.
Figure 1.
Single pathway mechanism proposed for Cl-/H+ antiporter [1].
We open with several biological contexts in which mechanistic heterogeneity has been demonstrated. This discussion is far from exhaustive, but intended to provide key examples of cases where kinetic selection is functionally important. We then summarize several methodological approaches to kinetic analysis to provide context for our efforts to advance multiscale kinetic modeling (MKM). Despite fruitful methodological overlap [4], we retain a molecular perspective for space reasons and refer the reader to recent reviews and method development in the network modeling central to systems biology [5–8]. We close with the insights gained on secondary active ion transport and a perspective on future directions for multiscale kinetic analysis of proteins.
KINETIC SELECTION IN BIOLOGY
The importance of kinetic selection in biology was poignantly brought to light by the independent work of Hopfield and Ninio circa 1974 [9, 10], who proposed the concepts of kinetic proofreading (or kinetic amplification) during replication and translation. Their work revealed how substrate specificity can be increased by introducing a delay following binding (e.g., tRNA-ribosome association followed by slow ATP hydrolysis), such that incorrect substrates have time to dissociate prior to product formation. In this case, kinetic selection between competing pathways, one with the correct substrate and one with the incorrect substrate, is driven by phosphate hydrolysis. This results in a lower error rate than would be possible based on relative substrate binding affinities alone. Hopfield noted then that, ‘Understanding the meaning of biosynthetic pathways in such cases will involve the nuances of minor pathways, competitive rates, and side reactions.’ [10]. Kinetic proofreading has since been demonstrated in other processes, including transcription [11], signal transduction [12, 13] and pathogen recognition [14]. A fascinating recent extension to the concept of kinetic proofreading is the replacement of slow enzymatic steps with slow diffusion such that spatial localization affects the same substrate discrimination [15]•. Additionally, the concept that relative steady state fluxes are kinetically controlled has been proposed more broadly based on flux ratios that are not altered by the thermodynamic stability of intermediates in the absence of altered rates[16].
Branched pathways have also been demonstrated in motor proteins. In 2007 Liepelt and Lipowsky proposed a network theory to describe the chemomechanical coupling (i.e., coupling between ATP hydrolysis and mechanical motion) of the molecular motor kinesin [17]. Their work and subsequent work on other motor proteins detail the functional relevance of branched pathways that dominate under specific conditions. For example, myosin V was shown to proceed with both ATP-dependent force-generating mechanical steps and ATP-independent high-speed backward steps induced by superstall loading, consistent with single-molecule observations under varying ATP/ADP concentrations [18]•. F1-ATPase was shown to function with a forward rotary motor, driven by ATP hydrolysis, and reverse ATP synthesis, driven by forced rotation, revealing that high torque conditions can induce mechanical slip (rotation) without coupled chemistry (ATP synthesis) [19]•.
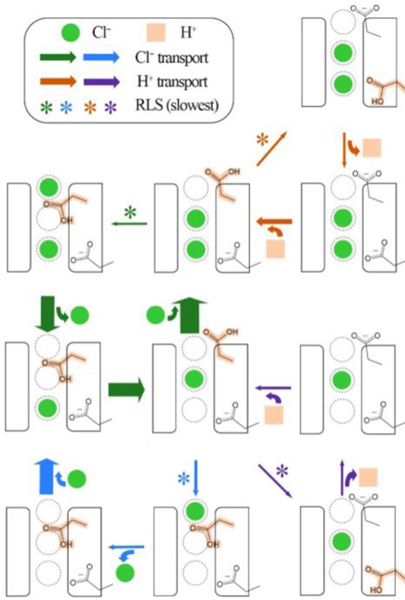
The role of kinetic selection is also increasingly apparent in membrane transporters, where inconsistent ratios of transported substrates can only be explained by competing transport pathways. Our work, summarized below, demonstrates how multiple pathways (Figure 2) explain the non-integer stoichiometry of ion exchange (2.2 Cl− to 1 H+) in Cl−/H+ antiporters [20]. Variable coupling under different conditions has been demonstrated in multiple transporters[21–23]••, perhaps most strikingly in the multidrug efflux pump EmrE [24]••. This mechanistic heterogeneity can confer biological advantage, such as toxin discrimination in symporters [25, 26]••. It is likely that mechanistic heterogeneity from kinetic selection is much more widespread than the examples given above. However, discerning this requires a more detailed and nuanced understanding of biomolecular processes than has traditionally been achievable. The methodological advances described next are getting us closer to this goal.
Figure 2.
Competing ion transport pathways in ClC-ec1.
METHODOLOGICAL APPROACHES
Kinetic modeling is carried out in a vast array of research fields, each with its own nuances and domain-specific challenges. The most common approaches are phenomenological kinetic models employing physics-based theoretic analyses. These ‘top-down’ approaches fit unknown transition rates to known macroscopic data. Because the kinetic solution space (i.e., all sets of transition rates that fit the few known observables) is generally vast, it has traditionally been essential to simplify the reaction network. Consequently, a central challenge has been making sense of the wide range of possible solutions and understanding solution dependence on the selected network representation [18]•. Despite these challenges, most of the insights described above have used this top-down approach. Valuable trends can be extracted, and the models improved with increasing experimental data. Moreover, advances in sampling are enabling more systematic exploration of solution space leading to increased network complexity and the generation of solution sets that can be experimentally tested [26]••.
In contrast, Markov state models (MSMs) can be built directly from molecular-level data and have become a powerful approach for understanding biomolecular processes [27, 28]. They are most often used to characterize molecular dynamics (MD) simulations, coarse-graining the captured phase space into metastable intermediates (states) and quantifying the transition probabilities between them. Configurations must be sampled beyond the state correlation times (lag times) to achieve Markovian transitions, which can create MSMs with a large number of microstates, each with sufficiently fast relaxation dynamics, and complex resulting networks. The benefit of this is that the multiplicity of pathways is retained; the challenge is in adequately sampling and reducing phase space to resolve them. MSM-associated methods to address these challenges are rapidly advancing (see, e.g., [29]•), lending hope to our future prospects for rigorous kinetic modeling of complex systems (see Conclusions). Already, MSM is revealing novel mechanistic insight from kinetic modeling. As just one example, sequence changes in myosin motor domains were shown to shift pathway flux to explain ADP release rates [30].
Finally, microkinetic modeling must be mentioned. Although applied in heterogeneous catalysis, the advanced methodolgy [31] may lead to promising new avenues for biomolecular kinetic modeling. In ‘bottom-up’ approaches, rates for chemical reactions are calculated with ab initio methods and then factored into a kinetic master equation. Consistent with our findings [20], pure bottom-up models generally do not produce physically meaningful solutions and must be combined with a top-down optimization of rate coefficients. Increasingly sophisticated methods are being developed for parameter refinement, including Bayesian and other stochastic optimization methods [32, 33]. Solution refinement can be further improved with sensitivity analysis and the removal of transitions that have little impact on macroscopic flow rates [31]. Correlative global sensitivity analysis additionally accounts for co-variance of parameters [34].
MULTISCALE KINETIC MODELING
Similar to some MSMs [11] and to microkinetic modeling, the MKM we have been developing also combines bottom-up quantification of transition rates with top-down refinement based on experimental data. Distinct from these approaches has been the need for enhanced free energy sampling, sometimes at multiscale resolution, to obtain calculated transition rates [35–37] and the refinement of all calculated or measured rates within estimated error [20]. Additional differences from microkinetic modeling that motivated our efforts include cyclic mechanisms as opposed to a series of chemical transformations; the condensed phase medium (often including spatial relevance such as membrane transport) instead of gas-phase surface adsorption; diverse transitions (e.g., enzymatic reactions, noncovalent association, ion transport) often coupled with intermediate-distinguishing hydration and conformational changes as opposed to chemical reactions; and diverse experimental conditions beyond concentrations and temperature, such as localization, orientation, pH, ion gradients, transmembrane voltage and more.
MKM can be broken down into 6 steps, each of which can be a formidable challenge:
Intermediate Determination: This can be the crux of a model as the fidelity of the solution depends on inclusion of all relevant intermediates, which requires extensive system characterization. In principle, any state separated from others by rate-influencing barriers that contributes to the total flux should be included. In practice, fast transitioning states can be grouped together (see Network Reduction).
Rate Determination: Initial rate coefficients can be measured in experiment, as nicely demonstrated on EmrE [24]••, or calculated in silico. This significantly reduces the kinetic solution space compared to top-down approaches. Depending on the transition, this step alone can require multiscale treatment (e.g., QM/MM for an enzymatic reaction or reactive MD for charge transport [37]).
Kinetic Modeling: Obtained rate coefficients are mapped into a kinetic master equation, a set of differential equations describing the conversion between intermediates over time that can be solved for the steady-state solution or for the evolution of populations over time given initial concentrations [38].
Solution Refinement: Given the exponential in the Eyring-Polanyi relationship (ki =Aie−βΔG‡), even small errors in calculated ΔG‡ values result in large rate deviations. Solutions consistent with established data thus require parameter refinement [32, 33] and can be improved by focusing on the important transitions with sensitivity analysis [31].
Network Reduction: Kinetic networks can be large, complex and sparse. Reducing their dimensionality by lumping states together can alleviate numerical challenges and reveal important pathways. The simplest approach is to combine states separated by fast transitions with a local equilibrium approximation. More rigorous approaches utilize methods like the graph transformation algorithm, which computes mean first passage times [39], to group states and reweight rates while retaining the network dynamic properties [40, 41].
Network Analysis: Once a solution is obtained, the network must be analyzed by quantifying dominant flow cycles, identifying rate-influencing steps, tracking time-dependent populations, and testing condition-dependent behavior. Again, there are domain-specific challenges. Since many biomolecular mechanisms must be described in a cyclic manner, we used Markov cycle theory [42] to develop a method that quantifies the flux through cyclic pathways in a closed network [43].
CHLORIDE CHANNEL ANTIPORTERS
Our work in this field was motivated by a long-standing effort to characterize the coupled Cl−/H+ exchange mechanism in the chloride channel, ClC-ec1. Different multiscale approaches, including QM/MM, multiscale reactive MD, polarizable MD, and Brownian dynamics simulations, were employed to determine free energy profiles for each involved transition [35–37, 44]. Even with these, a full mechanistic description was undecipherable without a kinetic model. The above methodology was developed for this purpose. The calculated rate coefficients were refined with macroscopic restraints (experimental Cl– transport rates and Cl−:H+ exchange ratios [45, 46]) in a kinetic master equation to generate physically meaningful solutions [44].
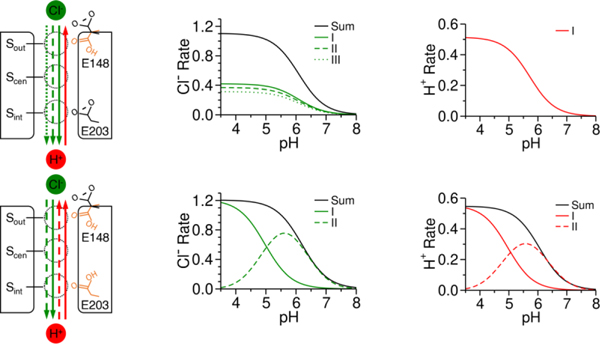
In contrast to the many single-pathway Cl−/H+ exchange mechanisms proposed (e.g., Figure 1), our work revealed multiple pathways contribute to the flux of each ion (Figure 2). This explains how the exact exchange stoichiometry is consistently non-integer, 2.2 Cl− to every 1 H+, for the wildtype system. This facet alone is indicative of pathway heterogeneity. The kinetic solutions also resolve how the upper glutamic acid, E148, couples ion exchange through protonation-dependent blockage of Cl− transport and anion-dependent release of the excess proton. We next probed how flux varies with pH [47], a reactant concentration in this case. Similar to the findings for molecular motors, the mechanism changes under different reaction conditions as the dominance of various pathways shifts (Figure 3) [47]. Interestingly, the lower glutamic acid, which has had questionable importance in the transport cycle to date, determines the distribution between various pathways. Increased back flux through this residue enables stabilization of the ion exchange rates and stoichiometry at lower pH values. This could explain the importance of a lower glutamic acid in the evolution of ClC antiporters.
Figure 3.
Shift in dominant pathways controlled by lower glutamic acid protonation state
CONCLUSIONS/PERSPECTIVE
The examples above highlight an increasing recognition of the fascinating and important role pathway heterogeneity can play in biology. Despite this, it remains a significant challenge to characterize kinetic networks for most complex biomolecular processes. With continued method development and increasing sub-ensemble experimental data, we are optimistic that kinetic analysis will someday be a standard tool used to query any biomolecular mechanism. The rapid evolution of kinetic analysis methods is encouraging in this sense. From phase space exploration with improved adaptive sampling [48–50] to feature extraction and phase space reduction with machine-learned and nonlinear collective variable discovery [51], each of the sub-domains discussed herein is quickly improving. Combining these rate-determining methods with effective kinetic solution space sampling [26]•• could offer a promising paradigm. The impressive experimental characterization for systems like EmrE additionally offers system-specific insight and an essential training ground for method development and benchmarking [24]••. Collectively, the field is progressing toward the ability to understand when and how kinetic selection influences any mechanism. This, in turn, may give us new ways to control biological systems and affect changes in mechanistic outcomes.
ACKNOWLEDGMENTS
The author gratefully acknowledges support from the National Institute of General Medicine of the National Institutes of Health under award number R35GM143117 and computational support from the Extreme Science and Engineering Discovery Environment supported by the National Science Foundation (Grant No. ACI-1548562) under allocation MCB200018 as well as the Center for High Performance Computing at the University of Utah.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES CITED
- 1.Park E, and MacKinnon R.(2018). Structure of the CLC-1 chloride channel from Homo sapiens. Elife 7, e36629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elf J, and Barkefors I.(2019). Single-Molecule Kinetics in Living Cells. Annu Rev Biochem 88, 635–659. [DOI] [PubMed] [Google Scholar]
- 3.Hill TL (2004). Free Energy Transduction and Biochemical Cycle Kinetics, (Courier Corporation; ). [Google Scholar]
- 4.Stein M, Gabdoulline RR, and Wade RC (2007). Bridging from molecular simulation to biochemical networks. Curr Opin Struct Biol 17, 166–172. [DOI] [PubMed] [Google Scholar]
- 5.Loskot P, Atitey K, and Mihaylova L.(2019). Comprehensive Review of Models and Methods for Inferences in Bio-Chemical Reaction Networks. Front Genet 10, 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katsoulakis MA, and Vilanova P.(2020). Data-driven, variational model reduction of high-dimensional reaction networks. J Comput Phys 401. [Google Scholar]
- 7.Warne DJ, Baker RE, and Simpson MJ (2019). Simulation and inference algorithms for stochastic biochemical reaction networks: from basic concepts to state-of-the-art. J R Soc Interface 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.uken A, Wendering P, Langary D, and Nikoloski Z.(2021). A structural property for reduction of biochemical networks. Sci Rep-Uk 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ninio J.(1975). Kinetic amplification of enzyme discrimination. Biochimie 57, 587–595. [DOI] [PubMed] [Google Scholar]
- 10.Hopfield JJ (1974). Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci USA 71, 4135–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unarta IC, Zhu L, Tse CKM, Cheung PP, Yu J, and Huang X.(2018). Molecular mechanisms of RNA polymerase II transcription elongation elucidated by kinetic network models. Curr Opin Struct Biol 49, 54–62. [DOI] [PubMed] [Google Scholar]
- 12.Swain PS, and Siggia ED (2002). The role of proofreading in signal transduction specificity. Biophys J 82, 2928–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo WL, and Weiss A.(2021). Adapting T Cell Receptor Ligand Discrimination Capability via LAT. Front Immunol 12, 673196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yousefi OS, Gunther M, Horner M, Chalupsky J, Wess M, Brandl SM, Smith RW, Fleck C, Kunkel T, Zurbriggen MD, et al. (2019). Optogenetic control shows that kinetic proofreading regulates the activity of the T cell receptor. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Galstyan V, Husain K, Xiao F, Murugan A, and Phillips R.(2020). Proofreading through spatial gradients. Elife 9. •Extension of the concept of kinetic proofreading to cases in which substrate binding and product formation are separated spatially such that the diffusion between locations acts as the slow step. This increases substrate specificity without the requirement of free energy dissipation (e.g., ATP hydrolysis), and could be at play in the many processes with controlled concentration gradients.
- 16.Mallory JD, Kolomeisky AB, and Igoshin OA (2020). Kinetic control of stationary flux ratios for a wide range of biochemical processes. P Natl Acad Sci USA 117, 8884–8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liepelt S, and Lipowsky R.(2007). Kinesin’s network of chemomechanical motor cycles. Phys Rev Lett 98, 258102. [DOI] [PubMed] [Google Scholar]
- 18. Sumi T.(2017). Myosin V: Chemomechanical-coupling ratchet with load-induced mechanical slip. Sci Rep 7, 13489. •This work nicely builds on many single-cycle and simplified branched-cycle networks built for myosin V, but intentionally retains as many cycles as possible. By including mechanical transitions not coupled to chemical transitions the author shows how additional network complexity explains the varying behavior under different ATP/ADP concentrations observed in single-molecule experiments.
- 19. Sumi T, and Klumpp S.(2019). Is F1-ATPase a Rotary Motor with Nearly 100% Efficiency? Quantitative Analysis of Chemomechanical Coupling and Mechanical Slip. Nano Lett 19, 3370–3378. •Chemomechanical network modeling of F1-ATPase resulting in quantitative models describing the forward rotary motor driven by ATP hydrolysis as well as reverse ATP synthesis driven by forced rotation. The authors propose that slippage, rotation without coupled synthesis that irreversibly dissipates heat and reduces the free energy transduction efficiency, is inherent in biological nanomachines because they are easily deformed. This is an interesting alternative to kinetic selection being an evolutionary consequence of improved performance.
- 20.Mayes HB, Lee S, White AD, Voth GA, and Swanson JMJ (2018). Multiscale Kinetic Modeling Reveals an Ensemble of Cl(−)/H(+) Exchange Pathways in ClC-ec1 Antiporter. J Am Chem Soc 140, 1793–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Henderson RK, Fendler K, and Poolman B.(2019). Coupling efficiency of secondary active transporters. Curr Opin Biotechnol 58, 62–71. ••Review of imperfect coupling in secondary active transporters, including altered coupling due to reaction conditions, mutations, and across family members. Multiple examples highlight the existence of leak pathways and demonstrate that conditions can even change an active symporter into a passive uniporter. Authors suggest that imperfect coupling can be beneficial, e.g., leading to toxicity control or faster transport.
- 22.Bazzone A, Zabadne AJ, Salisowski A, Madej MG, and Fendler K.(2017). A Loose Relationship: Incomplete H(+)/Sugar Coupling in the MFS Sugar Transporter GlcP. Biophys J 113, 2736–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kettner C, Bertl A, Obermeyer G, Slayman C, and Bihler H.(2003). Electrophysiological analysis of the yeast V-type proton pump: variable coupling ratio and proton shunt. Biophys J 85, 3730–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hussey GA, Thomas NE, and Henzler-Wildman KA (2020). Highly coupled transport can be achieved in free-exchange transport models. J Gen Physiol 152. ••Based on extensive NMR characterization of the proton-coupled multi-drug efflux pump, EmrE, this work presents a bottom-up, mass-action kinetic network model that captures a range of behaviors. Some solutions demonstrate robust coupling across a range of conditions while others switch from symport to antiport, exemplifying how coupled transport properties can vary significantly depending on the network flux. The experimental characterization of EmrE is exemplary, offering a promsing testing ground for kinetic analysis method development.
- 25.Bisignano P, Lee MA, George A, Zuckerman DM, Grabe M, and Rosenberg JM (2020). A kinetic mechanism for enhanced selectivity of membrane transport. Plos Comput Biol 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. George A, Bisignano P, Rosenberg JM, Grabe M, and Zuckerman DM (2020). A systems-biology approach to molecular machines: Exploration of alternative transporter mechanisms. Plos Comput Biol 16, e1007884. ••Motivated by the observation of an alternative leak pathway in simulations of a sodium/glucose transporter, these back to back publications reveal how symporters can also perform proofreading by leaking incorrect substrates (e.g., toxins) more frequently than the correct substrates, thereby increasing substrate-fidelity at the cost of some proportion of futile transport cycles. This insight is found with a Metropolis Markov chain Monte Carlo sampling methodology that enables more thorough exploration of the kinetic solution space based on both equilibrium and non-equilibrium constraints and produces thermodynamically consistent solution sets for further testing.
- 27.Noe F, and Rosta E.(2019). Markov Models of Molecular Kinetics. J Chem Phys 151, 190401. [DOI] [PubMed] [Google Scholar]
- 28.Husic BE, and Pande VS (2018). Markov State Models: From an Art to a Science. J Am Chem Soc 140, 2386–2396. [DOI] [PubMed] [Google Scholar]
- 29. Cao S, Montoya-Castillo A, Wang W, Markland TE, and Huang X.(2020). On the advantages of exploiting memory in Markov state models for biomolecular dynamics. J Chem Phys 153, 014105. •The Markovian requirement in MSM results in complex networks with many states, which can obscur biological insight into the relevant transitions. This work introduces quasi MSMs, which include non-Markovian dynamics with time-dependent memory kernels and thereby capture the relevant dynamics more effeiciently.
- 30.Porter JR, Meller A, Zimmerman MI, Greenberg MJ, and Bowman GR (2020). Conformational distributions of isolated myosin motor domains encode their mechanochemical properties. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wittreich GR, Alexopoulos K, and Vlachos DG (2020). Microkinetic Modeling of Surface Catalysis, (Spinger Cham; ). [Google Scholar]
- 32.Matera S, Schneider WF, Heyden A, and Savara A.(2019). Progress in Accurate Chemical Kinetic Modeling, Simulations, and Parameter Estimation for Heterogeneous Catalysis. ACS Catal. 9, 6624–6647. [Google Scholar]
- 33.Ashraf C, and Pfaendtner J.(2020). Assessing the Performance of Various Stochastic Optimization Methods on Chemical Kinetic Modeling of Combustion. Ind Eng Chem Res 59, 19212–19225. [Google Scholar]
- 34.Sutton JE, Guo W, Katsoulakis MA, and Vlachos DG (2016). Effects of correlated parameters and uncertainty in electronic-structure-based chemical kinetic modelling. Nat Chem 8, 331–337. [DOI] [PubMed] [Google Scholar]
- 35.Lee S, Liang R, Voth GA, and Swanson JMJ (2016). Computationally Efficient Multiscale Reactive Molecular Dynamics to Describe Amino Acid Deprotonation in Proteins. J Chem Theory Comput 12, 879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S, Mayes HB, Swanson JM, and Voth GA (2016). The Origin of Coupled Chloride and Proton Transport in a Cl−/H+ Antiporter. J. Am. Chem. Soc 138, 14923–14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee S, Swanson JMJ, and Voth GA (2016). Multiscale Simulations Reveal Key Aspects of the Proton Transport Mechanism in the ClC-ec1 Antiporter. Biophys. J 110, 1334–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker G, and Lagana A.(2018). Chemical Reactions: Basic Theory and Computing, Volume 1, (Springer International Publishing; ). [Google Scholar]
- 39.Kannan D, Sharpe DJ, Swinburne TD, and Wales DJ (2020). Optimal dimensionality reduction of Markov chains using graph transformation. J Chem Phys 153, 244108. [DOI] [PubMed] [Google Scholar]
- 40.Hummer G, and Szabo A.(2015). Optimal Dimensionality Reduction of Multistate Kinetic and Markov-State Models. J Phys Chem B 119, 9029–9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kells A, Koskin V, Rosta E, and Annibale A.(2020). Correlation functions, mean first passage times, and the Kemeny constant. Journal of Chemical Physics 152. [DOI] [PubMed] [Google Scholar]
- 42.Kalpazidou SL (2007). Cycle representations of Markov processes, (New York, NY: Springer; ). [Google Scholar]
- 43.Bernardi A, and Swanson JMJ (2021). CycFlowDec: A Python module for decomposing flow networks using simple cycles. SoftwareX 14, 100676–100671-100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayes HB, Lee S, White AD, Voth GA, and Swanson JMJ (2018). Multiscale Kinetic Modeling Reveals an Ensemble of Cl−/H+ Exchange Pathways in ClC-ec1 Antiporter. J. Am. Chem. Soc 140, 1793–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim H-H, and Miller C.(2009). Intracellular Proton-Transfer Mutants in a CLC Cl−/H+ Exchanger. J. Gen. Physiol 133, 131–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Picollo A, Xu Y, Johner N, Bernèche S, and Accardi A.(2012). Synergistic Substrate Binding Determines the Stoichiometry of Transport of a Prokaryotic H+/Cl− Exchanger. Nat Struct Mol Biol 19, 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yue Z, Bernardi A, Li C, Mironenko AV, and Swanson JMJ (2021). Toward a Multipathway Perspective: pH-Dependent Kinetic Selection of Competing Pathways and the Role of the Internal Glutamate in Cl(−)/H(+) Antiporters. J Phys Chem B 125, 7975–7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zimmerman MI, Porter JR, Sun X, Silva RR, and Bowman GR (2018). Choice of Adaptive Sampling Strategy Impacts State Discovery, Transition Probabilities, and the Apparent Mechanism of Conformational Changes. J Chem Theory Comput 14, 5459–5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuckerman DM, and Chong LT (2017). Weighted Ensemble Simulation: Review of Methodology, Applications, and Software. Annu Rev Biophys 46, 43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hruska E, Abella JR, Nuske F, Kavraki LE, and Clementi C.(2018). Quantitative comparison of adaptive sampling methods for protein dynamics. J Chem Phys 149, 244119. [DOI] [PubMed] [Google Scholar]
- 51.Sidky H, Chen W, and Ferguson AL (2020). Machine learning for collective variable discovery and enhanced sampling in biomolecular simulation. Mol Phys 118. [Google Scholar]