Abstract
Integral membrane proteins historically have been challenging targets for biophysical research due to their low solubility in aqueous solution. Their importance for chemical and electrical signaling between cells, however, makes them fascinating targets for investigators interested in the regulation of cellular and physiological processes. Since membrane proteins shunt the barrier imposed by the cell membrane, they also serve as entry points for drugs, adding pharmaceutical research and development to the interests. In recent years, detailed understanding of membrane protein function has significantly increased due to high-resolution structural information obtained from single-particle cryoEM, X-ray crystallography, and NMR. In order to further advance our mechanistic understanding on membrane proteins as well as foster drug development, it is crucial to generate more biophysical and functional data on these proteins under defined conditions. To that end, different techniques have been developed to stabilize integral membrane proteins in native-like environments that allow both structural and biophysical investigations – amphipols, lipid bicelles, and lipid nanodiscs. In this chapter, we provide detailed protocols for the reconstitution of membrane proteins according to these three techniques. We also outline some of the possible applications of each technique and discuss their advantages and possible caveats.
Keywords: Membrane proteins, reconstitution, amphipol, bicelles, nanodisc, membrane scaffold, membrane protein biophysics, lipids
1. Introduction
Membrane proteins constitute about 30 % of the proteome[1], are the connection between the inside and the outside of cells, and are entry points for pathogens and pharmaceuticals. It is thus of extreme importance to understand in molecular detail how specific proteins in the cell membrane work. In recent years, the number of high-resolution structures of membrane proteins has significantly increased mostly due to the developments in single-particle cryoEM[2, 3], novel techniques in x-ray crystallography (lipidic cubic phase)[4] and continuously improving NMR techniques[5, 6]. Drawing conclusions from structural data and understanding molecular mechanisms that govern the function and regulation of these proteins is only possible if functional and biophysical data of the same proteins under comparable conditions are available. Previously, these data have been obtained for purified proteins in detergent. However, with more complex systems under investigation, the environment of the protein becomes increasingly important, and several techniques have been developed to provide a more native-like environment for integral membrane proteins, helping their stability, structural integrity, as well as their function and regulation under purified and defined conditions (Figure 1)[7]. Although many detergents were deemed acceptable for many biophysical studies, they nevertheless display a non-native environment for membrane proteins. Their amphipathic character (Figure 2 A and B) leads to the formation of large micelles around protein molecules in order to preserve their structure even after extraction from cellular membranes. However, the presence of detergents can alter protein function and biophysical characteristics[8–12], significantly alters the surface characteristics of aqueous buffers, and, in some cases, even influences the function of extra-membraneous protein domains[13].
Figure 1: Reconstitution of purified membrane proteins into native-like environments.
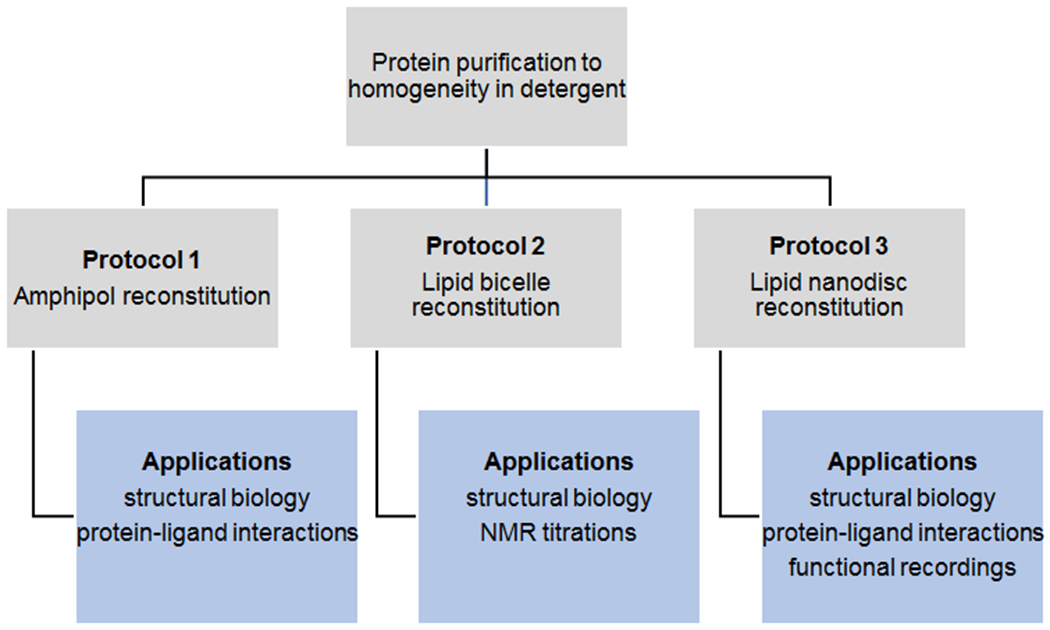
Outline of the possibilities to transfer purified, integral membrane proteins solubilized in detergent into more native-like environments. For each platform exemplary applications are listed.
Figure 2: Characteristics of detergents, amphipols and bicelles.
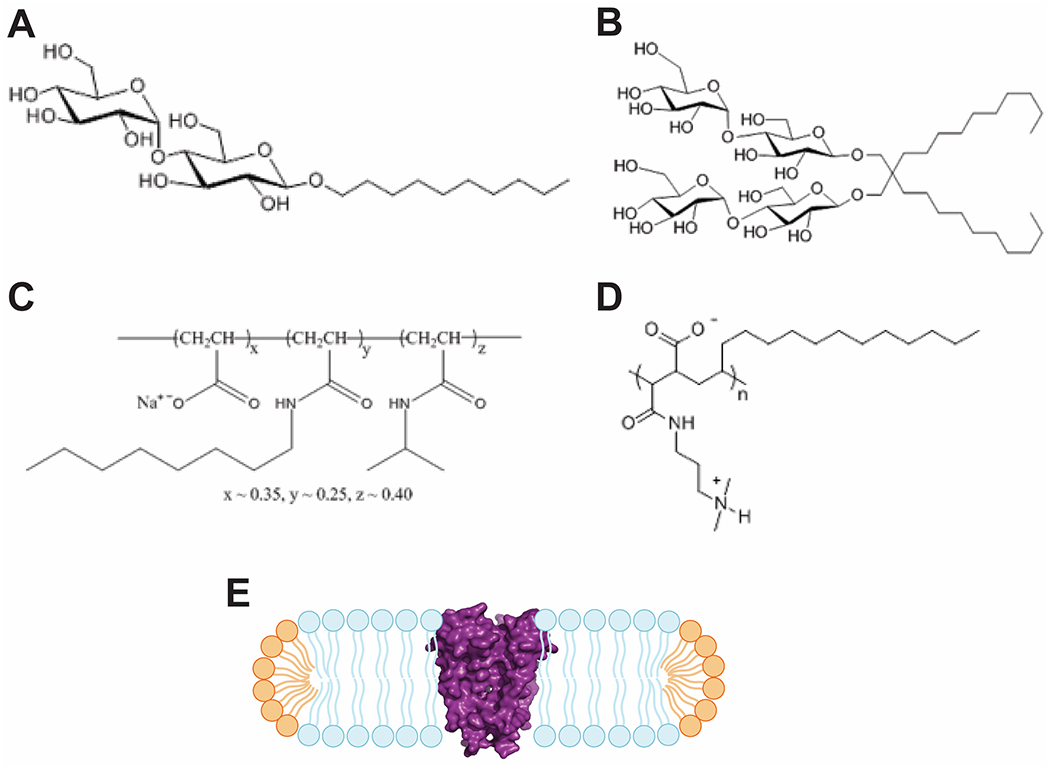
Chemical characteristics of hydrophilic headgroups and hydrophobic tails are present in detergent as shown for (A) n-Decyl-α-D-Maltopyranoside - DM, (B) Lauryl Maltose Neopentyl Glycol - LMNG, and amphipols as shown for (C) amphipol A8-35, and (D) PMAL-C12. Chemical structures were adapted from www.anatrace.com. (E) Cartoon of a lipid bicelle with a protein (KcsA ion channel, PDB: 1BL8) incorporated into a lipid bicelle. Long-chain lipids in blue, short-chain lipids in orange, KcsA in magenta surface representation
To that end, multiple reconstitution platforms for membrane proteins have been developed. Here, we describe three of them - amphipols, lipid bicelles, and lipid nanodiscs - together with their advantages and caveats, indicating that the best solution for individual experimental needs has to be determined on a case-by-case basis (Figure 1)[14].
Amphipols are amphipathic polymers that, in their chemical characteristics, resemble detergents, which combine a hydrophilic head group with a hydrophobic tail (Figure 2A–D)[15–17]. However, amphipols can stabilize membrane proteins more efficiently by interacting more strongly with the protein compared to detergents that form a lose micelle where single detergent molecules are in constant and rapid exchange with other detergent molecules in solution[18]. This is mostly due to the polymeric base structure of amphipols[19], which enables multiple hydrophobic chains of the same molecule to interact with the same protein (Figure 2C and D). Furthermore, due to the very slow dissociation of the protein-amphipol complex[20], free amphipols can and should be removed from solution after reconstitution. The downsides of the approach are: 1) the protein reconstituted into amphipol is no longer accessible for ulterior functional studies, such as lipid bilayer recordings, as it is virtually impossible to remove these polymers from the reconstituted protein, 2) amphipol-reconstituted proteins may adopt different structural and functional characteristics compared to the protein in native membranes[21]. Nevertheless, amphipols have been proven to be excellent environments for protein-ligand interactions[22–25] as well as structural studies by crystallography[26] and cryoEM[27–30].
Lipid bicelles closely resemble mixed detergent-lipid-micelles and are more native-like than amphipols as they provide a lipid environment for membrane proteins (Figure 2E)[32]. The principle of lipid bicelles relies on the fact that lipid molecules in aqueous solution spontaneously assemble in bilayers (mostly in the form of liposomes) to separate their hydrophobic tails from water. By optimizing the molar ratio of long-chain lipids to short-chain lipids (a parameter usually designated as q), it is possible to induce the assembly of small patches of lipid bilayers with the protein incorporated. The long-chain lipids form the bilayer which is stabilized by a belt of short-chain lipids, which allow more curvature and protect the hydrophobic core of the bicelle (Figure 2E)[33]. Lipid bicelles can be used for 3D-crystallography[34–37] and are an outstanding tool for analyzing membrane proteins by NMR[38–43].
A more complex, but at the same time arguably a more native-like environment for membrane proteins, are lipid nanodiscs[44–48]. The detergent-free lipid bilayer is surrounded by a membrane-scaffolding protein (MSP) and the protein of interest is inserted inside the disc (Figure 3).
Figure 3: Structural features of nanodiscs.
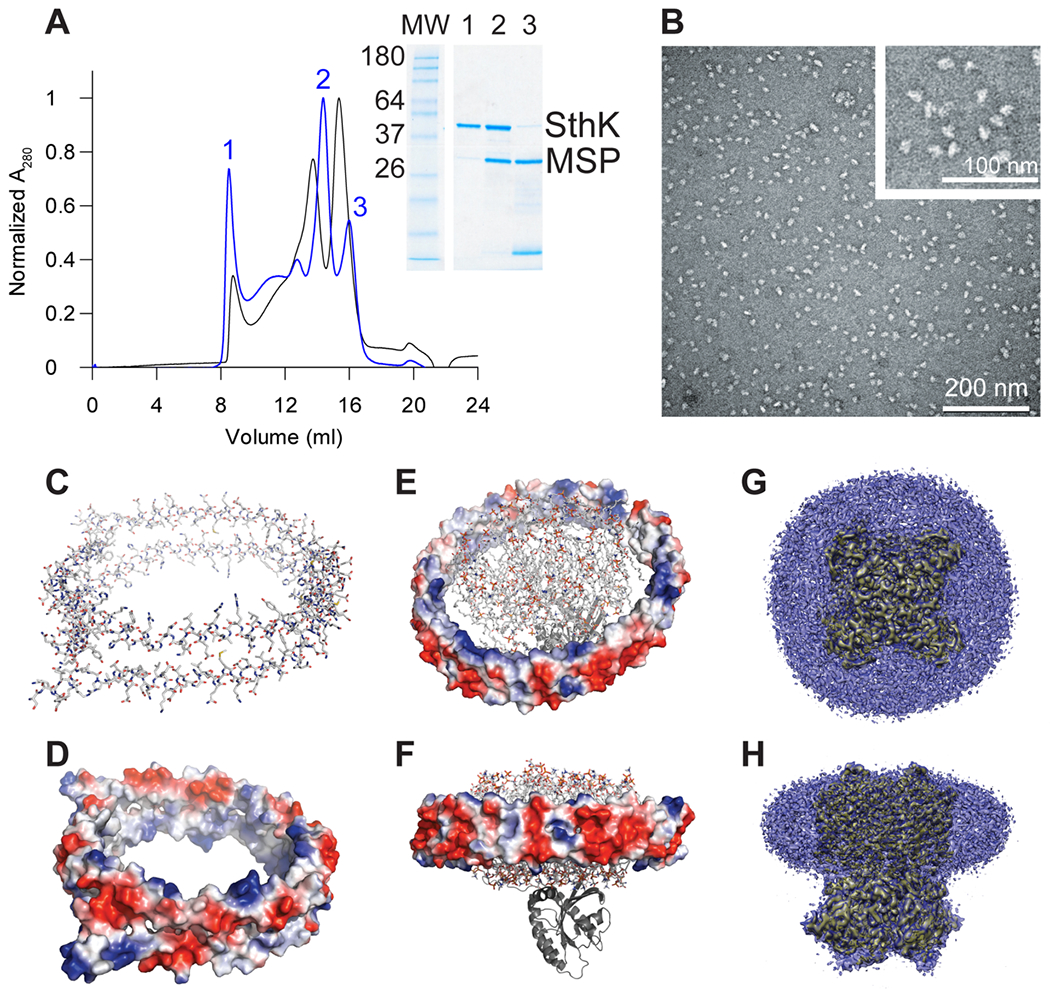
(A) Gel filtration profiles (Superose 6 16/600) used to assess the reconstitution quality of the cyclic nucleotide-gated K+ channel SthK into nanodiscs formed with different MSPs. Molar ratios of SthK:MSP2N2:POPG 1:1:125 (black) and SthK:MSP1E3:POPG 1:1:75 (blue) are shown. The bigger MSP (MSP2N2) shows an increased peak for empty nanodiscs. For the smaller MSP1E3, the SDS-PAGE clearly resolves the assembly of nanodiscs with aggregated SthK in the void peak (1), a peak for SthK inserted in MSP1E3 in nanodiscs (2) and empty nanodiscs (3). (B) Uranyl-acetate negative stain EM image of SthK in MSP1E3 (peak 2 in panel (A)) recorded on a JEM-1400 with 100 kV and a magnification of 150000. (C) Apolipoprotein A-I (PDB: 2N5E[49]) is shown in stick representation (colored by atom) and (D) colored by the surface electrostatics. (E) top view and (F) side view of the GTPase K-RAS4B tethered to an apolipoprotein A-I nanodisc (PDB: 2MSD[50]) with the nanodisc colored by surface electrostatics, lipids shown in stick representation and the GTPase as cartoon. (G) Top view and (H) side view of the density from single particle cryoEM of a ligand-gated ion channel (yellow, EMDB: 7484) incorporated into MSP1E3 nanodiscs (blue)[51]. Panels C-F were prepared using Pymol (www.pymol.org), panels (G) and (H) were prepared using UCSF Chimera[52].
The MSP is a truncated form of apolipoprotein A-I. A multitude of MSPs is used to form nanodiscs[44, 45, 53], and they all share common characteristics – two monomers of MSP assemble to form a ring structure (Figure 3C)[49, 54–56]. The two subunits are held together by up to 28 inter-subunit salt bridges, and the final ring structure creates an amphipathic environment (Figure 3D–F)[57]. The outer surface of the nanodisc is fully accessible to aqueous solution, and thus is highly charged to ensure solubility. The inner surface of the ring, however, has to accommodate the lipid bilayer and accordingly carries charges along the top and bottom rims to mediate interaction with the solvent and the charged headgroups of the lipids, while in the core predominantly hydrophobic residues are exposed to provide an amenable environment for the acyl chains of the lipids[57, 58]. The high content of charged residues and the necessity of salt-bridges to stabilize the double-layered ring structure make MSP and in turn nanodiscs strongly pH-dependent assemblies. The diameter of the nanodisc can be varied by either varying the protein:MSP:lipid ratio or by selecting MSP of different chain lengths in order to accommodate the protein of interest (Figure 3E–H)[44, 51, 54, 59–62]. Nanodiscs can be assembled from various lipid types. Therefore, they are able to meet very different requirements of the protein incorporated as well as the intended applications. However, it is important to optimize the protein:MSP:lipid ratio in order to maximize the amount of nanodiscs with protein incorporated or obtain the desired nanodisc sizes (Figure 3A and B). Recently, it was shown that, once assembled, nanodiscs can tolerate significant distortion of the enclosed membrane[61, 63].
In the following sections we will provide detailed protocols for the reconstitution of integral membrane proteins into the three systems.
2. Materials
All three protocols presented here start from pure, homogenous protein in detergent.
2.1. Amphipol reconstitution
Amphipol stock solution (100 mg/ml in ddH2O)
Detergent removal column (Pierce, ThermoScientific)
Superdex200 10/300 gel filtration column (GE Lifesciences)
2.2. Bicelle reconstitution
Lipid powder (Avanti Polar Lipids)
n-Octyl-β-D-Glucopyranoside (β-OG)
BioBeads (SM-2, BioRad)
Water bath sonicator
2.3. Nanodisc reconstitution
Lipid stock solution
Membrane-scaffolding protein (MSP)
BioBeads (SM-2, BioRad)
Spin-X column
Superose6 10/300 gel filtration column (GE Lifesciences)
3. Methods
3.1. Amphipol reconstitution
The reconstitution of membrane proteins into amphipols is the easiest of the three methods presented here and can be incorporated into any regular protein purification protocol.
Prepare amphipol stock solution ahead of time by dissolving 100 mg of amphipol in 750 μl ddH2O, yielding 1 ml of 100 mg/ml. Rotate the solution at 4 °C overnight to fully hydrate the amphipol. Prepare 100 μl aliquots and store at −20 °C until needed.
Purify the protein of interest to homogeneity. Determine the protein concentration by absorbance and concentrate to 10 mg/ml or higher if the protein is stable at high concentrations (see Note 1).
Mix protein in detergent with amphipol in ddH2O and incubate under gentle agitation at 4 °C for 2 h (see Note 2). Commonly used protein:amphipol ratios are between 1:1 and 1:3 (w/w).
Prepare detergent removal columns by washing with 3 column volumes of ddH2O followed by 3 column volumes of protein purification buffer without detergent.
Apply the protein-amphipol mix to the pre-equilibrated detergent removal column and collect fractions of 500 μl.
Check fractions for their protein content by absorbance. Pool protein-containing fractions and concentrate to 10 mg/ml.
Further purify the protein in amphipol by gel filtration in the protein purification buffer without detergent. The reconstituted protein should elute from the gel filtration as a single, symmetric peak at a volume similar to that of the protein in detergent.
3.2. Lipid-bicelle reconstitution via liposomes
Lipid bicelles provide a more native-like environment for membrane proteins than amphipols. The reconstitution protocol for lipid bicelles is more complex and needs some optimization for each protein. The protein should be purified to homogeneity with gel filtration as last purification step since the bicelles are not further purified after reconstitution.
Hydrate 20 mg/ml of long-chain lipid (usually at least tetradecyl tails) in reconstitution buffer (20 mM potassium phosphate, 20 mM NaCl, pH 7) for at least 2 h at room temperature.
Sonicate the hydrated lipids in a water bath sonicator for 1 – 2 min.
Add β-OG from a 10 % stock to reach a final concentration of 0.5 % β-OG and incubate the lipid-detergent mix for 30 min at room temperature under constant agitation. Although β-OG is most widely used for bicelle reconstitutions, other detergents can be used provided that they are compatible with the structural integrity of the protein of interest.
Add your protein of interest to the lipid-detergent mix in a molar ratio of 1:100 protein:lipid, mix and incubate slightly above the phase transition temperature of the lipids for 1 h to avoid the lipids from entering the gel phase and to provide enough time for mixed micelles to be formed.
Remove detergent by adding freshly prepared BioBeads (30 mg BioBeads per mg detergent, see Note 1) and incubate for 2 h under gentle agitation.
Transfer the solution to a new reaction tube, add fresh BioBeads and incubate overnight under gentle agitation.
Change BioBeads one more time and incubate for 2 h before transferring the liposome solution to a fresh reaction tube. Make sure to avoid transferring any BioBeads.
Spin down the liposomes at 40000 rpm for 1 h, 4 °C.
Resuspend the pellet into the final buffer of your experiment containing short-chain lipids (usually hexyl tail-lipids, see Note 2) to reach the desired protein concentration. Most prominently, lipid bicelles are used for NMR applications with a protein concentration of 0.5-1 mM.
Perform five freeze/thaw cycles to homogenize the bicelles. Freezing is best executed in liquid N2. To thaw the bicelles, incubate the reaction tube at room temperature.
3.3. Lipid-nanodisc reconstitution
The most complex protocol to reconstitute membrane proteins for functional and biophysical assays is the incorporation of proteins into lipid-nanodiscs. These particles, however, represent an environment that is very close to the conditions in a cellular membrane and can be tailored to specific experimental setups.
For an optimal reconstitution, different protein:MSP:lipid ratios need to be screened to obtain nanodiscs with a single protein incorporated. Sub-optimal reconstitution conditions can lead to aggregation, empty nanodiscs, or nanodiscs containing several proteins per disc (Figure 3A and B)[64, 65]. In general, the protein:MSP molar ratio should be 1:2 for purified membrane protein[55], however, molar ratios between 1 and 25 have been applied in practice[51, 66–70]. The optimal protein:MSP:lipid ratio can be determined by systematically testing different conditions in small-scale reconstitutions and monitoring the sample quality by size-exclusion chromatography followed by SDS-PAGE and negative stain EM (Figure 3A–B).
3.3.1. Lipid preparation
Transfer the appropriate amount of lipids from stock solutions (typically 10 or 25 mg/ml in chloroform) to a glass tube.
Dry lipids to a thin film under a constant nitrogen gas stream. Remove residual organic solvent by rinsing in one volume of pentane, and dry again under the gas stream. Alternatively, the dried lipids can be placed in a vacuum desiccator over night for complete removal of the solvent.
Add buffer to the dried lipid film to obtain a 20 mM lipid stock. Gradually add the detergent suitable for the membrane protein while sonicating in a water bath sonicator until the lipid solution is clear. The typical final concentration of detergent is at least twice the CMC. The lipids can also be dissolved by several freeze/thaw cycles.
After solubilization lipid stock solutions can be stored at −80 °C for future use.
3.3.2. Bio-Beads preparation
Wash the Bio-Beads with two volumes of methanol, followed by extensively washing with ddH2O to remove the organic solvent.
Wash the Bio-Beads with three volumes of purification buffer without detergent.
Bio-Beads can be used freshly or stored at 4 °C in ddH20 and 0.01 % NaN3 for up to three months[54].
3.3.3. Nanodisc assembly
MSP protein can be prepared in lab according to published protocols [71].
Add lipid, MSP, and the membrane protein in an Eppendorf tube at the optimized ratio. The sample volume is dependent on the application and the desired yield of nanodiscs.
Incubate the reconstitution mixture by gently inverting for 1-2 h at a temperature slightly above the phase-transition temperature of the lipid. In case of using a mix of lipids, the experimental temperature should be optimized according to the phase transition temperature of the main lipid components[72].
Start detergent removal to initiate the reconstitution by adding BioBeads (20 mg per 100 μl sample), and gently invert for ~2 h.
Transfer the reconstitution mixture to a new tube. Add fresh, pre-equilibrated BioBeads to the reconstitution mixture (20 mg per 100 μl). Gently invert overnight.
Transfer the reconstitution mixture to a new tube. Dilute the sample with sample buffer to the volume appropriate for gel filtration (typically 500 μl final volume).
Filter the sample through a 0.22 μm Spin-X centrifugation tube filter.
Apply the sample to a Superose 6 16/600 gel filtration column pre-equilibrated in sample buffer. Collect the peak fraction corresponding to the assembled nanodiscs and check for protein and MSP content by SDS-PAGE (Figure 3A).
The different fractions can be finally checked by negative-stain EM for evidence of nanodisc formation, presence of protein in nanodiscs, and absence of liposomes (Figure 3B).
Acknowledgments
This work was funded by the National Institutes of Health (GM088352 and GM124451 to CN) and the American Heart Association (18POST33960309 to PS). We thank J. Rheinberger for providing the gel filtration profiles and the negative stain EM image in Fig. 3A and B.
Amphipol reconstitution:
Protein should be purified to homogeneity (usually after gel filtration chromatography) to avoid reconstitution of bad particles. However, if the protein of interest displays low stability in detergent, amphipol exchange can also be performed right after affinity purification as long as the protein appears clean on SDS-PAGE at this stage.
Due to the free carboxylic groups, amphipols are highly water soluble at neutral pH values. The solubility is decreased at acidic pH as well as at high salt concentrations, which might lead to aggregation of amphipols.
Bicelle reconstitution:
To freshly prepare BioBeads, weigh in BioBeads, wash with at least two volumes of methanol for 10 min to remove air followed by at least 4 volumes of ddH2O. Decant the water and wash with 2 volumes of reconstitution buffer.
The concentration of short-chain lipids is given by the q value that needs to be determined for each protein and adjusted to the experimental needs. The q value is defined as the molar ratio of long-chain:short-chain lipids, and it determines the size and order parameter of the bicelles. For solid state NMR, q values of 3 – 6 are used, whereas q values of 0.15 – 0.5 will yield fast-tumbling bicelles suitable for solution NMR. For more details see [40] and [73].
Nanodisc reconstitution:
The size of MSP determines to some extent the diameter of nanodiscs and thus the number of enclosed lipid molecules [54, 59]. The selection of MSP is mainly dependent on the size of the membrane protein. Ideally, the nanodiscs should be large enough to accommodate the protein and at least two layers of phospholipids to mimic the physiological environment[55, 72]. If the assembled nanodiscs are too large it may result in floating of the protein within the discs, which may obscure structural data processing and interpretation[74]. Additionally, there is an increased chance of reconstituting more than one protein per nanodisc[65].
A variety of MSP constructs with various numbers of amphipathic helices are available to generate nanodiscs with diameters between 9.8 and 17 nm[44, 54, 70]. In addition, truncated versions of MSP have been designed, allowing the preparation of nanodiscs with diameters ranging from 6 to 8 nm[72].
Circular MSP (cMSP) has been developed to improve the homogeneity of nanodiscs, which are especially crucial for structural studies. cMSP is a variant of MSP with its N- and C-termini covalently linked by sortase A[75, 76]. The nanodisc sizes produced by this technique range from 8.5 up to 80 nm in diameter. Alternatively, cMSP generated from DnaE split intein can be used for smaller nanodiscs (7-26 nm in diameter)[77].
Based on our experience, well-defined, published reconstitution ratios can serve as a starting point for optimization, especially those from membrane proteins of a similar size with the protein of interest, as well as similar MSP and lipid types.
5 References
- [1].Fagerberg L, Jonasson K, von Heijne G, Uhlén M, Berglund L, Prediction of the human membrane proteome, Proteomics 10(6) (2010) 1141–9. [DOI] [PubMed] [Google Scholar]
- [2].Kühlbrandt W, Cryo-EM enters a new era, Elife 3 (2014) e03678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cheng Y, Single-Particle Cryo-EM at Crystallographic Resolution, Cell 161(3) (2015) 450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cherezov V, Lipidic cubic phase technologies for membrane protein structural studies, Curr Opin Struct Biol 21(4) (2011) 559–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Liang B, Tamm LK, NMR as a tool to investigate the structure, dynamics and function of membrane proteins, Nat Struct Mol Biol 23(6) (2016) 468–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Warschawski DE, Arnold AA, Beaugrand M, Gravel A, Chartrand É, Marcotte I, Choosing membrane mimetics for NMR structural studies of transmembrane proteins, Biochim Biophys Acta 1808(8) (2011) 1957–74. [DOI] [PubMed] [Google Scholar]
- [7].Hunte C, Richers S, Lipids and membrane protein structures, Curr Opin Struct Biol 18(4) (2008) 406–11. [DOI] [PubMed] [Google Scholar]
- [8].McCoy JG, Rusinova R, Kim DM, Kowal J, Banerjee S, Jaramillo Cartagena A, Thompson AN, Kolmakova-Partensky L, Stahlberg H, Andersen OS, Nimigean CM, A KcsA/MloK1 chimeric ion channel has lipid-dependent ligand-binding energetics, J Biol Chem 289(14) (2014) 9535–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Quick M, Winther AM, Shi L, Nissen P, Weinstein H, Javitch JA, Binding of an octylglucoside detergent molecule in the second substrate (S2) site of LeuT establishes an inhibitor-bound conformation, Proc Natl Acad Sci U S A 106(14) (2009) 5563–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Quick M, Shi L, Zehnpfennig B, Weinstein H, Javitch JA, Experimental conditions can obscure the second high-affinity site in LeuT, Nat Struct Mol Biol 19(2) (2012) 207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dowhan W, Molecular basis for membrane phospholipid diversity: why are there so many lipids?, Annu Rev Biochem 66 (1997) 199–232. [DOI] [PubMed] [Google Scholar]
- [12].Yu CA, Yu L, Structural role of phospholipids in ubiquinol-cytochrome c reductase, Biochemistry 19(25) (1980) 5715–20. [DOI] [PubMed] [Google Scholar]
- [13].Seddon AM, Curnow P, Booth PJ, Membrane proteins, lipids and detergents: not just a soap opera, Biochim Biophys Acta 1666(1-2) (2004) 105–17. [DOI] [PubMed] [Google Scholar]
- [14].Raschle T, Hiller S, Etzkorn M, Wagner G, Nonmicellar systems for solution NMR spectroscopy of membrane proteins, Curr Opin Struct Biol 20(4) (2010) 471–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Popot JL, Althoff T, Bagnard D, Banères JL, Bazzacco P, Billon-Denis E, Catoire LJ, Champeil P, Charvolin D, Cocco MJ, Crémel G, Dahmane T, de la Maza LM, Ebel C, Gabel F, Giusti F, Gohon Y, Goormaghtigh E, Guittet E, Kleinschmidt JH, Kühlbrandt W, Le Bon C, Martinez KL, Picard M, Pucci B, Sachs JN, Tribet C, van Heijenoort C, Wien F, Zito F, Zoonens M, Amphipols from A to Z, Annu Rev Biophys 40 (2011) 379–408. [DOI] [PubMed] [Google Scholar]
- [16].Tribet C, Audebert R, Popot JL, Amphipols: polymers that keep membrane proteins soluble in aqueous solutions, Proc Natl Acad Sci U S A 93(26) (1996) 15047–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tehei M, Perlmutter JD, Giusti F, Sachs JN, Zaccai G, Popot JL, Thermal fluctuations in amphipol A8-35 particles: a neutron scattering and molecular dynamics study, J Membr Biol 247(9-10) (2014) 897–908. [DOI] [PubMed] [Google Scholar]
- [18].Le Bon C, Marconnet A, Masscheleyn S, Popot JL, Zoonens M, Folding and stabilizing membrane proteins in amphipol A8-35, Methods 147 (2018) 95–105. [DOI] [PubMed] [Google Scholar]
- [19].Gohon Y, Giusti F, Prata C, Charvolin D, Timmins P, Ebel C, Tribet C, Popot JL, Well-defined nanoparticles formed by hydrophobic assembly of a short and polydisperse random terpolymer, amphipol A8-35, Langmuir 22(3) (2006) 1281–90. [DOI] [PubMed] [Google Scholar]
- [20].Perlmutter JD, Drasler WJ, Xie W, Gao J, Popot JL, Sachs JN, All-atom and coarse-grained molecular dynamics simulations of a membrane protein stabilizing polymer, Langmuir 27(17) (2011) 10523–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Barrera NP, Zhou M, Robinson CV, The role of lipids in defining membrane protein interactions: insights from mass spectrometry, Trends Cell Biol 23(1) (2013) 1–8. [DOI] [PubMed] [Google Scholar]
- [22].Schmidpeter PAM, Gao X, Uphadyay V, Rheinberger J, Nimigean CM, Ligand binding and activation properties of the purified bacterial cyclic nucleotide-gated channel SthK, J Gen Physiol 150(6) (2018) 821–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Martinez KL, Gohon Y, Corringer PJ, Tribet C, Mérola F, Changeux JP, Popot JL, Allosteric transitions of Torpedo acetylcholine receptor in lipids, detergent and amphipols: molecular interactions vs. physical constraints, FEBS Lett 528(1-3) (2002) 251–6. [DOI] [PubMed] [Google Scholar]
- [24].Catoire LJ, Damian M, Giusti F, Martin A, van Heijenoort C, Popot JL, Guittet E, Banères JL, Structure of a GPCR ligand in its receptor-bound state: leukotriene B4 adopts a highly constrained conformation when associated to human BLT2, J Am Chem Soc 132(26) (2010) 9049–57. [DOI] [PubMed] [Google Scholar]
- [25].Charvolin D, Perez JB, Rouvière F, Giusti F, Bazzacco P, Abdine A, Rappaport F, Martinez KL, Popot JL, The use of amphipols as universal molecular adapters to immobilize membrane proteins onto solid supports, Proc Natl Acad Sci U S A 106(2) (2009) 405–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Polovinkin V, Gushchin I, Sintsov M, Round E, Balandin T, Chervakov P, Shevchenko V, Utrobin P, Popov A, Borshchevskiy V, Mishin A, Kuklin A, Willbold D, Chupin V, Popot JL, Gordeliy V, High-resolution structure of a membrane protein transferred from amphipol to a lipidic mesophase, J Membr Biol 247(9-10) (2014) 997–1004. [DOI] [PubMed] [Google Scholar]
- [27].van Pee K, Neuhaus A, D’Imprima E, Mills DJ, Kühlbrandt W, Yildiz Ö, CryoEM structures of membrane pore and prepore complex reveal cytolytic mechanism of Pneumolysin, Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liao M, Cao E, Julius D, Cheng Y, Structure of the TRPV1 ion channel determined by electron cryo-microscopy, Nature 504(7478) (2013) 107–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hirschi M, Herzik MA, Wie J, Suo Y, Borschel WF, Ren D, Lander GC, Lee SY, Cryo-electron microscopy structure of the lysosomal calcium-permeable channel TRPML3, Nature 550(7676) (2017) 411–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Li M, Zhou X, Wang S, Michailidis I, Gong Y, Su D, Li H, Li X, Yang J, Structure of a eukaryotic cyclic-nucleotide-gated channel, Nature 542(7639) (2017) 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R, The structure of the potassium channel: molecular basis of K+ conduction and selectivity, Science 280(5360) (1998) 69–77. [DOI] [PubMed] [Google Scholar]
- [32].Sanders CR, Landis GC, Reconstitution of membrane proteins into lipid-rich bilayered mixed micelles for NMR studies, Biochemistry 34(12) (1995) 4030–40. [DOI] [PubMed] [Google Scholar]
- [33].Sanders CR, Schwonek JP, Characterization of magnetically orientable bilayers in mixtures of dihexanoylphosphatidylcholine and dimyristoylphosphatidylcholine by solid-state NMR, Biochemistry 31(37) (1992) 8898–905. [DOI] [PubMed] [Google Scholar]
- [34].Ujwal R, Bowie JU, Crystallizing membrane proteins using lipidic bicelles, Methods 55(4) (2011) 337–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK, Crystal structure of the human beta2 adrenergic G-protein-coupled receptor, Nature 450(7168) (2007) 383–7. [DOI] [PubMed] [Google Scholar]
- [36].Ujwal R, Cascio D, Colletier JP, Faham S, Zhang J, Toro L, Ping P, Abramson J, The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating, Proc Natl Acad Sci U S A 105(46) (2008) 17742–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Faham S, Bowie JU, Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure, J Mol Biol 316(1) (2002) 1–6. [DOI] [PubMed] [Google Scholar]
- [38].Kim DM, Dikiy I, Upadhyay V, Posson DJ, Eliezer D, Nimigean CM, Conformational heterogeneity in closed and open states of the KcsA potassium channel in lipid bicelles, J Gen Physiol 148(2) (2016) 119–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dürr UH, Gildenberg M, Ramamoorthy A, The magic of bicelles lights up membrane protein structure, Chem Rev 112(11) (2012) 6054–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].De Angelis AA, Opella SJ, Bicelle samples for solid-state NMR of membrane proteins, Nat Protoc 2(10) (2007) 2332–8. [DOI] [PubMed] [Google Scholar]
- [41].Opella SJ, Marassi FM, Structure determination of membrane proteins by NMR spectroscopy, Chem Rev 104(8) (2004) 3587–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Morrison EA, Henzler-Wildman KA, Reconstitution of integral membrane proteins into isotropic bicelles with improved sample stability and expanded lipid composition profile, Biochim Biophys Acta 1818(3) (2012) 814–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bibow S, Hiller S, A guide to quantifying membrane protein dynamics in lipids and other native-like environments by solution-state NMR spectroscopy, FEBS J (2018). [DOI] [PubMed] [Google Scholar]
- [44].Denisov IG, Grinkova YV, Lazarides AA, Sligar SG, Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size, J Am Chem Soc 126(11) (2004) 3477–87. [DOI] [PubMed] [Google Scholar]
- [45].Banerjee S, Huber T, Sakmar TP, Rapid incorporation of functional rhodopsin into nanoscale apolipoprotein bound bilayer (NABB) particles, J Mol Biol 377(4) (2008) 1067–81. [DOI] [PubMed] [Google Scholar]
- [46].Jonas A, Kézdy KE, Wald JH, Defined apolipoprotein A-I conformations in reconstituted high density lipoprotein discs, J Biol Chem 264(9) (1989) 4818–24. [PubMed] [Google Scholar]
- [47].Davidson WS, Thompson TB, The structure of apolipoprotein A-I in high density lipoproteins, J Biol Chem 282(31) (2007) 22249–53. [DOI] [PubMed] [Google Scholar]
- [48].Leitz AJ, Bayburt TH, Barnakov AN, Springer BA, Sligar SG, Functional reconstitution of Beta2-adrenergic receptors utilizing self-assembling Nanodisc technology, Biotechniques 40(5) (2006) 601–2, 604, 606, passim. [DOI] [PubMed] [Google Scholar]
- [49].Bibow S, Polyhach Y, Eichmann C, Chi CN, Kowal J, Albiez S, McLeod RA, Stahlberg H, Jeschke G, Güntert P, Riek R, Solution structure of discoidal high-density lipoprotein particles with a shortened apolipoprotein A-I, Nat Struct Mol Biol 24(2) (2017) 187–193. [DOI] [PubMed] [Google Scholar]
- [50].Mazhab-Jafari MT, Marshall CB, Smith MJ, Gasmi-Seabrook GM, Stathopulos PB, Inagaki F, Kay LE, Neel BG, Ikura M, Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site, Proc Natl Acad Sci U S A 112(21) (2015) 6625–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rheinberger J, Gao X, Schmidpeter PA, Nimigean CM, Ligand discrimination and gating in cyclic nucleotide-gated ion channels from apo and partial agonist-bound cryo-EM structures, Elife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, UCSF Chimera--a visualization system for exploratory research and analysis, J Comput Chem 25(13) (2004) 1605–12. [DOI] [PubMed] [Google Scholar]
- [53].Grinkova YV, Denisov IG, Sligar SG, Engineering extended membrane scaffold proteins for self-assembly of soluble nanoscale lipid bilayers, Protein Eng Des Sel 23(11) (2010) 843–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG, Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs, Methods Enzymol 464 (2009) 211–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Efremov RG, Gatsogiannis C, Raunser S, Lipid Nanodiscs as a Tool for High-Resolution Structure Determination of Membrane Proteins by Single-Particle Cryo-EM, Methods Enzymol 594 (2017) 1–30. [DOI] [PubMed] [Google Scholar]
- [56].McLean MA, Gregory MC, Sligar SG, Nanodiscs: A Controlled Bilayer Surface for the Study of Membrane Proteins, Annu Rev Biophys (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Morgan CR, Hebling CM, Rand KD, Stafford DW, Jorgenson JW, Engen JR, Conformational transitions in the membrane scaffold protein of phospholipid bilayer nanodiscs, Mol Cell Proteomics 10(9) (2011) M111.010876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Schuler MA, Denisov IG, Sligar SG, Nanodiscs as a new tool to examine lipid-protein interactions, Methods Mol Biol 974 (2013) 415–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bayburt TH, Sligar SG, Membrane protein assembly into Nanodiscs, FEBS Lett 584(9) (2010) 1721–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Koehl A, Hu H, Feng D, Sun B, Zhang Y, Robertson MJ, Chu M, Kobilka TS, Laermans T, Steyaert J, Tarrasch J, Dutta S, Fonseca R, Weis WI, Mathiesen JM, Skiniotis G, Kobilka BK, Structural insights into the activation of metabotropic glutamate receptors, Nature 566(7742) (2019) 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kalienkova V, Clerico Mosina V, Bryner L, Oostergetel GT, Dutzler R, Paulino C, Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM, Elife 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Quentin D, Ahmad S, Shanthamoorthy P, Mougous JD, Whitney JC, Raunser S, Mechanism of loading and translocation of type VI secretion system effector Tse6, Nat Microbiol 3(10) (2018) 1142–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Falzone ME, Rheinberger J, Lee BC, Peyear T, Sasset L, Raczkowski AM, Eng ET, Di Lorenzo A, Andersen OS, Nimigean CM, Accardi A, Structural basis of Ca2+-dependent activation and lipid transport by a TMEM16 scramblase., Elife 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Bayburt TH, Sligar SG, Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers, Protein Sci 12(11) (2003) 2476–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Boldog T, Grimme S, Li M, Sligar SG, Hazelbauer GL, Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties, Proc Natl Acad Sci U S A 103(31) (2006) 11509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Li Y, Soubias O, Li J, Sun S, Randazzo PA, Byrd RA, Functional expression and characterization of human myristoylated-Arf1 in nanodisc membrane mimetics, Biochemistry (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Tsukamoto H, Szundi I, Lewis JW, Farrens DL, Kliger DS, Rhodopsin in nanodiscs has native membrane-like photointermediates, Biochemistry 50(22) (2011) 5086–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Ranaghan MJ, Schwall CT, Alder NN, Birge RR, Green proteorhodopsin reconstituted into nanoscale phospholipid bilayers (nanodiscs) as photoactive monomers, J Am Chem Soc 133(45) (2011) 18318–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Mörs K, Roos C, Scholz F, Wachtveitl J, Dötsch V, Bernhard F, Glaubitz C, Modified lipid and protein dynamics in nanodiscs, Biochim Biophys Acta 1828(4) (2013) 1222–9. [DOI] [PubMed] [Google Scholar]
- [70].Inagaki S, Ghirlando R, Grisshammer R, Biophysical characterization of membrane proteins in nanodiscs, Methods 59(3) (2013) 287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].T.H.a.G.Y.V.a.S.S.G. Bayburt, Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins, Nano Letters 2(8) (2002) 853–856. [Google Scholar]
- [72].Hagn F, Nasr ML, Wagner G, Assembly of phospholipid nanodiscs of controlled size for structural studies of membrane proteins by NMR, Nat Protoc 13(1) (2018) 79–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Mäler L, Gräslund A, Artificial membrane models for the study of macromolecular delivery, Methods Mol Biol 480 (2009) 129–39. [DOI] [PubMed] [Google Scholar]
- [74].Hagn F, Etzkorn M, Raschle T, Wagner G, Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins, J Am Chem Soc 135(5) (2013) 1919–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Nasr ML, Baptista D, Strauss M, Sun ZJ, Grigoriu S, Huser S, Plückthun A, Hagn F, Walz T, Hogle JM, Wagner G, Covalently circularized nanodiscs for studying membrane proteins and viral entry, Nat Methods 14(1) (2017) 49–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Yusuf Y, Massiot J, Chang YT, Wu PH, Yeh V, Kuo PC, Shiue J, Yu TY, Optimization of the Production of Covalently Circularized Nanodiscs and Their Characterization in Physiological Conditions, Langmuir 34(11) (2018) 3525–3532. [DOI] [PubMed] [Google Scholar]
- [77].Miehling J, Goricanec D, Hagn F, A Split-Intein-Based Method for the Efficient Production of Circularized Nanodiscs for Structural Studies of Membrane Proteins, Chembiochem 19(18) (2018) 1927–1933. [DOI] [PubMed] [Google Scholar]


