Abstract
Recent developments in cryogenic electron microscopy (cryo-EM) led to an exponential increase in high-resolution structures of membrane proteins, and in particular ion channels. However, structures alone can only provide limited information about the workings of these proteins. In order to understand ion channel function and regulation in molecular detail, the obtained structural data need to be correlated to functional states of the same protein. Here, we describe several techniques that can be employed to study ion channel structure and function in vitro and under defined, similar conditions. Lipid nanodiscs provide a native-like environment for membrane proteins and have become a valuable tool in membrane protein structural biology and biophysics. Combined with liposome-based flux assays for the kinetic analysis of ion channel activity as well as electrophysiological recordings, researchers now have access to an array of experimental techniques allowing for detailed structure-function correlations using purified components. Two examples are presented where we put emphasis on the lipid environment and time-resolved techniques together with mutations and protein engineering to interpret structural data obtained from single particle cryo-EM on cyclic nucleotide-gated or Ca2+-gated K+ channels. Furthermore, we provide short protocols for all the assays used in our work so that others can adapt these techniques to their experimental needs. Comprehensive structure-function correlations are essential in order to pharmacologically target channelopathies.
Introduction
Ion channels have fascinated researchers for a long time due to their critical role in cellular signaling(Hodgkin and Huxley, 1939; Hodgkin et al., 1952; Bezanilla et al., 1970; Latorre and Miller, 1983; Bezanilla, 2008). With the invention of patch-clamp electrophysiology it became possible to study the mechanism and functional diversity of these proteins with unprecedented accuracy(Neher and Sakmann, 1976; Conti and Neher, 1980). This quickly led to a fundamental understanding of ion conduction and selectivity(Eisenman and Horn, 1983; Latorre and Miller, 1983; Yellen, 1987; Bezanilla, 2008), as well as the allosteric regulation of channels by stimuli such as ligands, temperature, or mechanical stress(Dittert et al., 2006; Petrov et al., 2013; Gonzalez-Perez et al., 2014). Knowledge obtained from such functional studies guided the understanding of detailed, microscopic mechanisms for many different ion channels and could explain associated channel-related diseases. However, in order to target specific ion channel malfunction pharmacologically, it is necessary also to know the exact three-dimensional structure of these different states in order to design and develop state-specific drugs. X-ray crystallography can provide very detailed snapshots of proteins, including ion channels (Doyle et al., 1998; Zhou et al., 2001), but it generally only captures a single conformation. Mutations, state-specific antibodies, and other ligands can help identify other conformations, but these approaches are laborious.
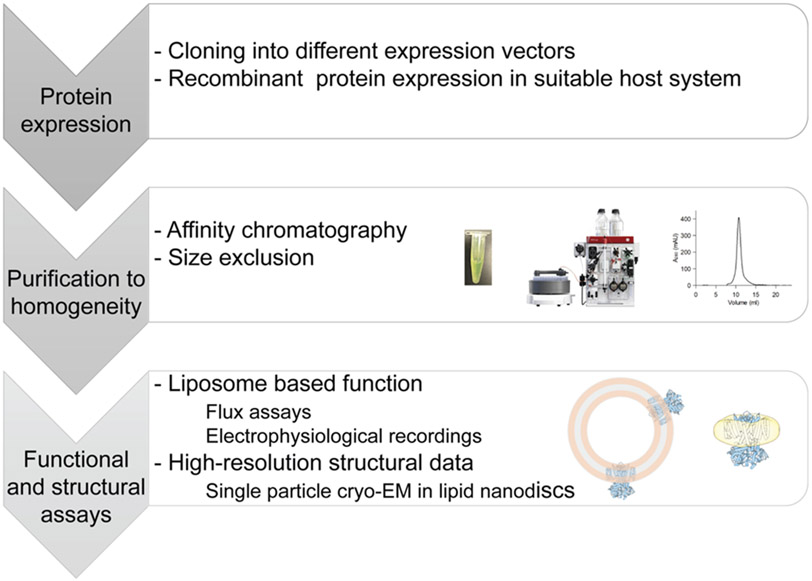
With the advent of cryo-EM, structural biology of membrane proteins (MP) and large protein complexes has been lifted to a new level(Liao et al., 2013; Kühlbrandt, 2014; Nakane et al., 2020; Yip et al., 2020). Similar to NMR and in contrast to crystallography, cryo-EM is believed to capture the conformational landscape of proteins virtually in solution, due to rapid plunge freezing of the sample. In contrast to NMR, which can only be used to solve the three-dimensional structures of small proteins, cryo-EM can resolve molecules from currently ~60 kDa(Wu and Lander, 2020) up to large, multi-protein assemblies in the MDa range. Combined with nanodisc technology(Bayburt et al., 2002; Sligar and Denisov, 2020), which provides a native-like environment for membrane proteins, it is now possible to capture multiple different conformations of a single protein in a single sample. In order to assign such co-existing conformations to functional states it is important to analyze protein function under similar conditions (Figure 1). While such combined studies are becoming more routine, it still can be challenging to correlate structural and functional data. In the following, we provide a short overview of techniques that can be applied to achieve an inclusive structure-function correlation for ion channel proteins and provide two recent examples from our research demonstrating their applicability and limitations.
Figure 1: General workflow for structure-function studies.
Optimized expression and purification of ion channel proteins is followed by reconstituting pure, homogeneous protein into liposomes or nanodiscs under identical, defined conditions in order to correlate results.
1. Nanodiscs
Nanodiscs are composed of a membrane scaffolding protein (MSP) derived from apolipoprotein that encloses a lipid bilayer in which the membrane protein under investigation is inserted (Figure 2A)(Bayburt et al., 2002; Denisov et al., 2004; Sligar and Denisov, 2020). They thus provide a more native-like environment for membrane proteins than detergents that are used to extract MPs from cellular membranes. In combination with single particle cryo-EM, nanodiscs have become an important and versatile tool for structural biology of MPs(Hagn et al., 2013; Kalienkova et al., 2020). Comprehensive summaries of available MSPs and recent cryo-EM structures employing nanodiscs are available(Denisov et al., 2004; Kalienkova et al., 2020). Nanodiscs are small bilayer patches enclosed by rings of MSP dimers. The MSPs are composed of two α-helices (one per MSP) that circularize and are held together by salt-bridge interactions. The length of these amphipathic helices is the main determinant of the diameter of the disc(Denisov et al., 2004). The outside of this ring, which faces the solution, is hydrophilic, whereas the MSP surface on the inside of the ring is hydrophobic to accommodate lipids(Bayburt et al., 2002; Denisov et al., 2004; Ritchie et al., 2009; Sligar and Denisov, 2020). Thus, once formed, nanodiscs can be treated like soluble entities.
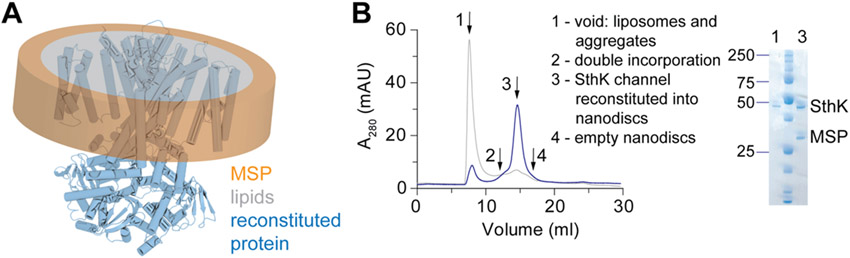
Figure 2: Ion channel reconstitution into lipid nanodiscs.
A schematic representation of a membrane protein reconstituted into lipid nanodiscs. The MSP ring is shown in orange, the lipid bilayer in grey, and the protein (from PDB-6CJU) in blue. B Gel filtration analysis (Superose 6 10/300) of small-scale reconstitutions with different MP:MSP:lipid ratios (grey and blue) for SthK is shown together with SDS-PAGE analysis of the peaks 1 and 3.
Two aspects should be kept in mind when preparing nanodiscs: During MSP purification detergents (such as Triton X-100 and cholate) need to be employed to remove contaminants from the hydrophobic surface areas of MSP. To ensure complete removal of contaminants, certain MSPs are even purified under denaturing conditions using guanidine and refolded on an IMAC column after thorough washing(Banerjee et al., 2008). Second, the assembly of two MSPs to form one nanodisc is stabilized by salt-bridge interactions between the two MSP monomers. Thus, high salt concentrations and extreme pH values are incompatible with nanodiscs.
For each membrane protein and each lipid composition the reconstitution into nanodiscs needs to be optimized(Ritchie et al., 2009) (Figure 2). An initial estimate of the MP:MSP:lipid ratio can be calculated based on the lipid mix, the theoretical diameter of the disc based on the MSP used (~ 9-17 nm), and the cross-sectional area of the MP(Ritchie et al., 2009). The reconstitution condition can then be experimentally optimized by setting up small scale reconstitutions with varying MP:MSP:lipid ratios. Most frequently, lipids are solubilized in cholate or CHAPS containing buffer. In principle, any detergent that facilitates complete solubilization of the lipids can be used, as long as the stability of the MP under investigation is not affected. Purified MP in detergent and MSP are added and the mix is incubated for about 30 min before detergent removal is initiated by addition of BioBeads. The reconstitution should be performed slightly above the phase transition temperature of the lipids used in order to prevent lipids from entering the gel phase. The quality and homogeneity of the reconstitution is then analyzed by size-exclusion chromatography and SDS-PAGE (Figure 2B).
Once the reconstitution conditions are optimized, they can be scaled up to achieve yields necessary for structural studies. The same nanodiscs used for structure determination can also be used in biophysical or functional experiments(Alvarez et al., 2010; Nath et al., 2010; Banerjee and Nimigean, 2011; McCoy et al., 2014; Murphy et al., 2019; Evans et al., 2020). While direct transfer of ion channels from nanodiscs into lipid bilayers for single channel recordings has been demonstrated(Banerjee and Nimigean, 2011; Winterstein et al., 2018), we generally prefer liposome-based techniques to study ion channel function since the reconstitution is more versatile and faster.
More recently, styrene maleic acid (SMA) copolymers have been shown to form macroscopic structures similar to nanodiscs (SMA lipid polymer, SMALP)(Knowles et al., 2009; Jamshad et al., 2011; Dörr et al., 2014). These SMALPs can be used to directly extract MPs from membranes thereby conserving the protein-surrounding lipid composition(Dörr et al., 2014). This can be of particular interest for lipidomic studies(Hellwig et al., 2018; Teo et al., 2019), with the downside of losing the controlled conditions necessary for structure-function correlations.
2. Liposome based assays
Classical studies on ion channels are performed on proteins expressed in cells followed by electrophysiological techniques such as two-electrode voltage clamp, whole-cell recordings, or excised-patch recordings(Conti and Neher, 1980). In some cases, functional results obtained from such recordings can be used to correlate structural states to functional states. However, in other cases, the exact environment of the channel protein, such as lipid composition, accessory proteins, or the stoichiometry of modulatory subunits, is important. With purified protein for structural studies in hand it thus makes sense to use the same protein for in-vitro functional assays under defined conditions (Figure 1).
2.1. Radioactive flux assay
In order to get an initial estimation of ion channel activity a radioactive flux assay can be employed(Nimigean et al., 2004; Nimigean, 2006; Mari et al., 2011; Pessoa et al., 2014; Schmidpeter et al., 2018) (Figure 3A,B). This bulk assay relies on a concentration gradient for permeant ions as driving force for extra-vesicular 86Rb+ to enter liposomes and is applicable to K+ as well as non-selective cation channels. The same assay can be used for channels that select for other ions (e.g. Na+, Cl−) provided that a different radioactive species is used, such as 22Na+, 36Cl−(Smith et al., 2001; Yu et al., 2016). Channel protein reconstituted into liposomes at high K+ concentration is subjected to gel filtration to remove K+ from the outside and thus establish the K+ gradient. At this point, 86Rb+ is added to the liposome sample to start the assay. To quantify the amount of 86Rb+ uptake into the liposomes as a function of time, samples are withdrawn at defined time intervals after addition of 86Rb+, and immediately applied to an ion-exchange column to remove extra-liposomal 86Rb+ and thus stop the uptake (Figure 3A). The measured radioactivity of each sample reports the amount of 86Rb+ uptaken into liposomes through the active channels at each time point (Figure 3B).
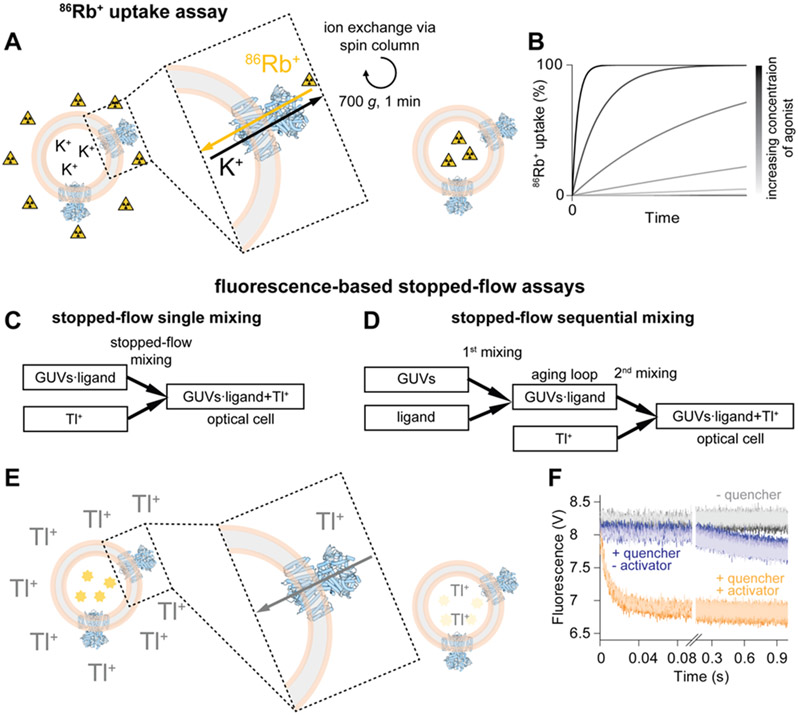
Figure 3: Flux assays.
A Schematic overview of the 86Rb+ uptake assay. The readout reports on the amount of radioactivity inside the liposomes at different times after exposure to 86Rb+ containing uptake solution. B Schematic representation of 86Rb+ uptake over time with increasing concentrations of agonist (from grey to black). C and D Mixing schemes for the single and the sequential mixing stopped-flow assay, respectively. E Principle of the fluorescence quenching. Fluorescent ANTS is shown in bright yellow, faded yellow represents quenched ANTS. F Representative data for an ion channel. The control in the absence of Tl+ (quencher) is shown in grey. In the presence of Tl+ but without agonist (blue) only very slow leakage of the quencher across the liposomal membrane is observed. In the presence of Tl+ and agonist (orange) rapid quenching reflecting channel activity is recorded. For each condition six technical repeats are shown in different shades of grey, blue, or orange, respectively. The ion channel representation (blue) in this figure was prepared from PDB-6CJU, the liposomal membrane is shown in rose and grey.
Since the readout of this assay is radioactivity that accumulates inside liposomes over time, it can even be used for ion channels with very low activity(Nimigean et al., 2004; Mari et al., 2011). The main limitations of this assay are the low time resolution due to manual mixing and necessary chromatography steps as well as its overall qualitative character. Due to the short half-life of 86Rb+ (45 days) the isotope needs to be purchased freshly, while on the other hand only leading to minimal complications in disposing of radioactive material. Other radioactive compounds with different half-lives will require different handling. This assay is useful as an initial test of bulk channel activity and for preliminary screening of conditions that favor channel activity.
2.2. Stopped-flow flux assay
Another, rather new flux assay, is based on stopped-flow fluorescence measurements (Figure 3C-F)(Rusinova et al., 2014; Posson et al., 2018). This assay resembles macroscopic channel recordings and more quantitative (kinetic) information can be extracted compared to the 86Rb+ flux assay. Ion channels are reconstituted into giant unilamellar vesicles (GUVs) that encapsulate the fluorescent dye ANTS (8-Aminonaphthalene-1,3,6-trisulfonic acid). ANTS fluorescence is quenched by Tl+, which is applied to the liposomes by rapid, stopped-flow mixing and can serve as permeant ion to enter the liposomes through open channels and quench the fluorescence (Figure 3E,F)(Ingólfsson and Andersen, 2010). Tl+, as an activity-dependent permeable quencher, can be used for K+ channels, non-selective cation channels, and some less-selective Na+ channels(Ingólfsson and Andersen, 2010; Rusinova et al., 2014; Posson et al., 2015; Menny et al., 2017; Posson et al., 2018; Schmidpeter et al., 2018; Tong et al., 2019). Tl+ can enter the GUVs only through activated channels and accordingly the quenching kinetics are a measure of macroscopic channel activity. When performed with only one mixing step, this assay reads out the steady-state activity of the channel (Figure 3C)(McCoy et al., 2014; Rusinova et al., 2014). Alternatively, in a sequential mixing setup (Figure 3D), the conditions can be changed rapidly in a first mixing step (addition of activator, inhibitor, blocker, etc.) and Tl+ is only added in a second mixing step to probe activity(Posson et al., 2015, 2018; Schmidpeter et al., 2018; Fan et al., 2020; Schmidpeter et al., 2020a). The resulting quenching kinetics provide information about channel activity as function of the incubation time between the two mixing events. In this way, one can obtain information about kinetics of ion channel activation, block, and inactivation with millisecond time resolution after addition of the respective compound(Posson et al., 2015; Schmidpeter et al., 2018; Fan et al., 2020). Recently, we also showed that this sequential mixing assay is suitable to analyze the enzymatic modulation of ion channel activity(Schmidpeter et al., 2020a).
The use of ANTS-Tl+ as fluorophore-quencher pair indicates, that this assay is optimized for K+ channels, but it can also be applied to non-selective cation channels and even some Na+ channels (Ingólfsson and Andersen, 2010; McCoy et al., 2014; Rusinova et al., 2014; Posson et al., 2015; Menny et al., 2017; Tong et al., 2019). A different fluorophore-quencher pair can be used in order to adapt this assay for anion channels(Posson et al., 2018). Since its development, this assay has been used to study a variety of channels(Ingólfsson and Andersen, 2010; McCoy et al., 2014; Rusinova et al., 2014; Posson et al., 2015; Menny et al., 2017; Schmidpeter et al., 2018; Tong et al., 2019; Schmidpeter et al., 2020a). In order to obtain high-quality liposomes it is important to remove all organic solvent from the lipids before the reconstitution. This can be accomplished by drying the lipids into a thin film under a constant N2 stream followed by further drying of the lipids overnight under vacuum. After solubilizing the lipids using CHAPS, ANTS and protein are added, and the mix is incubated for about 20 min before detergent removal is initiated by adding BioBeads. Complete detergent removal is crucial for obtaining tightly formed GUVs and in turn good experimental results. Similar to nanodisc formation, the temperature during detergent removal should be above the phase transition temperature of the lipids. Furthermore, the efficiency of BioBeads is highest between 18 and 24 °C. We usually perform detergent removal for 3 h at 21 °C unless the phase transition temperature of the lipids is higher. Subsequently, GUVs are extruded through a 0.1 μm membrane to obtain a more homogeneous size distribution of GUVs. Extra-vesicular ANTS is removed via a PD-10 desalting column and the final reconstitution is diluted to yield a good signal-to-noise ratio.
It is possible that certain ion channels incorporate into GUVs with an orientational bias. This can be problematic as only the outside of the liposomes is accessible to ligand application after rapid mixing. On the other hand, the fluorescence read-out is very sensitive and the activity of even a fraction of reconstituted channels is frequently sufficient to allow for generating reproducible and interpretable results.
2.3. Bilayer recordings
In contrast to the qualitative nature of the 86Rb+ flux assay (Figure 3A,B) and the macroscopic nature of the Tl+ flux assay (Figure 3E,F), single channel bilayer recordings(Krueger et al., 1983; Miller, 1983; Hanke and Kaupp, 1984; Morera et al., 2007) can provide extraordinary detail about the function of ion channels (Figure 4). For this assay, protein is reconstituted into rather small liposomes (diameter <100 nm) and a range of protein concentrations needs to be tested to obtain single channel incorporations into the bilayer with reasonable frequency. Even under optimized conditions, the single molecule nature of this assay can make detailed analyses rather tedious(Miller, 1983). If the only question to be answered is whether the channel under investigation is active or not, there are more efficient (bulk) assays. On the other hand, analysis of ion channel opening and closing events at the single molecule level can provide a wealth of information into the channel gating mechanism, which few other assays can provide. In addition, the single-channel open probability is instrumental for predicting the fraction of closed and open channel conformations existing in a sample in defined conditions. These state distributions should be qualitatively preserved in a cryo-EM experiment under identical conditions and thus should allow assignment of specific structures to functional ion channel states(Rheinberger et al., 2018; Schmidpeter et al., 2020a; Yu et al., 2021).
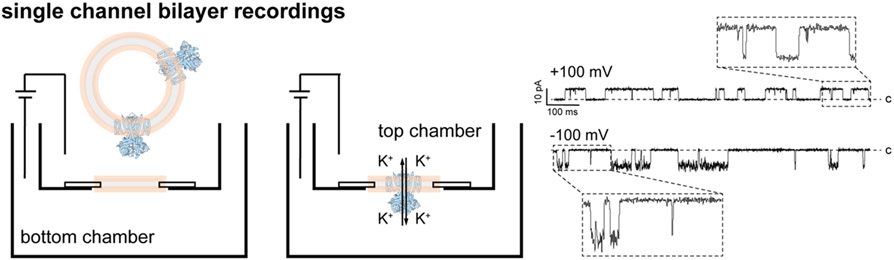
Figure 4: Scheme of a horizontal bilayer setup.
Proteo-liposomes are applied to the top chamber (left). After fusion of the liposomes with the bilayer and channel insertion (middle) the activity of a single ion channel can be recorded (right). Representative recordings at different voltages are shown for an engineered ion channel. The closed level (c) is indicated. PDB-6CJU was used for the channel representations (blue), liposomes or planar bilayers are shown in rose/gray.
Using DPhPC (1,2-diphytanoyl-sn-glycero-3-phosphocholine) as the base lipid to paint bilayers yields reproducibly stable bilayers for the experiment that can be repeatedly perfused with agonist, antagonist, or modulator containing solutions. Unlike channels expressed on a cell membrane, where they are always in the same orientation, incorporation of channels from liposomes into lipid bilayers results in randomly-oriented channels, unless they incorporate with a strong orientational preference into liposomes (Figure 4). To overcome this problem and to silence all channels incorporated in one direction, the bilayer chamber can be easily setup with asymmetric conditions where only one side of the chamber contains either a side-specific stimulant or blocker of channel activity (Figure 4).
In a horizontal setup as shown in Figure 4, the bottom chamber can be repeatedly perfused with different solutions. However, such solution changes need to be performed with great care and at low flow rate in order to preserve the bilayer. As such, single channel bilayer recordings report on steady-state channel activity. The time resolution with which channel openings and closings can be detected is only dependent on the performance of the amplifier and the frequency of the digitizer that is used to read out the signal. Composition of the lipid bilayer can be modified by fusing into a generic planar bilayer proteo-liposomes with different lipid compositions. While this setup can also be used for macroscopic recordings when higher protein concentrations are used, the true power of this technique lies in single channel recordings.
The same proteo-liposomes used for bilayer recordings can be further treated to obtain liposomes suitable for patch-clamp electrophysiology (Delcour et al., 1989; Collins and Gordon, 2013; Matulef and Valiyaveetil, 2018). This can be particularly useful for mechanosensitive (Sotomayor et al., 2007) channels or experiments that need fast solution changes.
3. Examples of studies correlating structure and function of ion channels
In the following, we provide two examples where we used all the techniques described above for detailed studies of two potassium channels. Each channel poses a set of challenges that illustrate the importance of similar experimental conditions between different assays in order to assign structural states to functional states.
3.1. The cyclic nucleotide-gated channel SthK
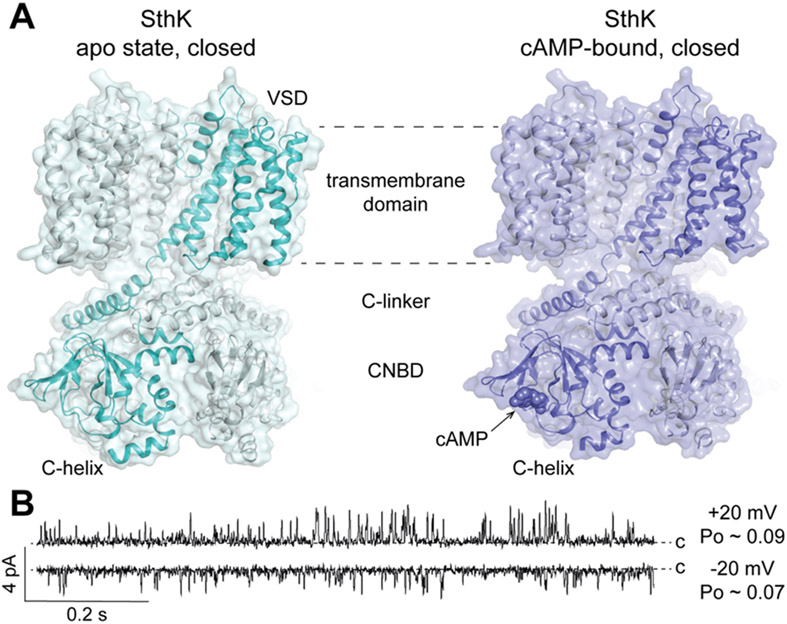
SthK(Brams et al., 2014), from Spirochaeta thermophila, is a bacterial cyclic-nucleotide gated channel that has been used as a model system to decipher channel gating mechanisms of the homologous cyclic nucleotide-gated (CNG) and hyperpolarization-activated and cyclic nucleotide-modulated (HCN) channels in molecular detail (Kesters et al., 2015; Marchesi et al., 2018; Rheinberger et al., 2018; Schmidpeter et al., 2018; Schmidpeter and Nimigean, 2018; Morgan et al., 2019; Evans et al., 2020; Schmidpeter et al., 2020a). Eukaryotic CNG and HCN channels are extremely challenging to express and purify for biophysical studies, making SthK a valuable tool for understanding the function and regulation of these physiologically important proteins. The cAMP dependent activity of SthK was first demonstrated in patch-clamp recordings of SthK channels expressed in Xenopus laevis oocytes(Brams et al., 2014). Optimization of the protein construct, expression, and purification conditions allowed for obtaining pure, homogeneous protein from bacteria for in vitro studies (Schmidpeter et al., 2018; Morgan et al., 2019). Using purified SthK reconstituted into liposomes for the radioactive uptake assay described above (Figure 3A,B) confirmed the cAMP dependent activity, whereas in the presence of cGMP only marginal activity was observed. Furthermore, this assay suggested that negatively charged lipids such as POPG and cardiolipin are crucial for SthK activity. These experiments laid the foundation for single channel bilayer recordings that confirmed the cyclic-nucleotide dependent activity of SthK (Schmidpeter et al., 2018), revealing that SthK is differentially regulated by cAMP and cGMP. The efficacy of cGMP is orders of magnitude lower than that of cAMP even though both ligands display comparable binding affinities for SthK(Schmidpeter et al., 2018; Morgan et al., 2019). Interestingly, while cAMP binding increased channel activity, the maximal open probability in the absence of voltage and in the presence of saturating concentrations of cAMP is only ~0.1. This result means that on average only 10 % of the channels are in the open state under these conditions. Single channel recordings also showed a clear voltage modulation of the cAMP-induced channel activity. The open probability strongly increases with depolarization and reaches ~0.7 at 200 mV. This information was crucial to interpret structural results of SthK reconstituted into lipid nanodiscs obtained from single particle cryo-EM (Figure 5A)(Rheinberger et al., 2018).
Figure 5: Structure and function of SthK.
A Structures of apo SthK and cAMP-bound SthK (PDB: 6CJQ and 6CJU, respectively). B Single-channel recordings of SthK in 5:3:2 DOPC:POPG:Cardiolipin at +/− 20 mV and saturating cAMP concentration demonstrate the low open probability (Po) of SthK at voltages close to zero. Closed levels are indicated.
Structures were solved for apo SthK as well as SthK in complex with cAMP and cGMP, all of which appear to be in closed states(Rheinberger et al., 2018). The liganded structures showed that all subunits had cyclic nucleotide bound to their cyclic nucleotide binding domains (CNBDs). Due to the very low activity of SthK in the presence of cGMP, cGMP-bound SthK can be assigned to a ligand-bound closed state. SthK does not inactivate(Schmidpeter et al., 2018) and accordingly the cAMP-bound structure can also be assigned to a ligand-bound closed state (Figure 5A). In this study, only one structural state could be identified for cAMP-bound SthK, although the open probability of ~0.1 at 0 mV in the presence of saturating cAMP (Figure 5B) predicts that about 10 % of all particles in the cryo-EM sample should be in the open state (with 90 % in the closed state). Several factors can contribute to the absence of this conformation from our final analysis. First, due to the relatively small number of particles (at most 10 % are predicted to be in the open state), it is possible that the cryo-EM analysis could not pick up on this minor class(Rheinberger et al., 2018). This is a general limitation of cryo-EM structure analysis, which could theoretically be overcome by collecting larger datasets with enough particles to resolve even low populated states. Such an approach is possible if storage and processing capacities allow for it. In addition, the temperature change during plunge freezing may shift somewhat the distribution of states and could further decrease the fraction of particles in the open state. Other possibilities to resolve a low populated state (i.e. the open state in the case of SthK) are to screen for mutations that favor such states, for instance using single channel recordings, or to identify conditions that shift the open/closed equilibrium more towards the open state. For SthK, the 86Rb+ flux assay revealed that certain lipids are important for channel function(Schmidpeter et al., 2018). It is possible that these lipids are enriched around SthK in the bacterial membrane. Using SMALPs to extract SthK from the membrane and thereby preserving the lipid composition surrounding the protein might lead to identification of specific lipids which can help towards resolving an open state structure of WT SthK.
In order to obtain information about the activation kinetics of SthK we performed the stopped-flow assay described above. In agreement with the low open probability at 0 mV the fluorescence signal obtained with SthK in this assay is small compared to KcsA or MthK channels who display high open probabilities at 0 mV(McCoy et al., 2014; Rusinova et al., 2014; Posson et al., 2015; Schmidpeter et al., 2018). We observed that SthK activates in two phases with time constants τ of ~20 ms and ~2 s, respectively. Our initial structures of WT SthK could not provide insights into this behavior. However, recently, we were able to identify cis/trans heterogeneity at Pro300 in the siphon region that connects the C-linker to the CNBD to be responsible for the biphasic activation (Schmidpeter et al., 2020a). With trans Pro300, SthK channels display fast-activation and high ligand-binding affinity, whereas with cis Pro300, activation is slow and the apparent affinity for cAMP is lower. Differentiating between cis and trans proline in the structure is only possible at resolutions of ~2.5 Å or better since the backbone trace needs to be unequivocally defined. This resolution currently is not achievable for the siphon, which is inherently flexible, and can explain why the structures of WT SthK only showed one conformational state, presumably with cis Pro300. However, substituting Pro300 with Ala, SthK P300A, can serve as an all-trans mimic and only shows fast-activation and high apparent ligand-binding affinity(Schmidpeter et al., 2020a). The cryo-EM structure of SthK P300A revealed small differences between the CNBDs of this mutant and WT SthK. Furthermore, it provided indications for a different siphon conformation that shows a more homogeneous local resolution than the WT SthK structures. Together, this shows that, despite recent developments in cryo-EM structure analysis, in certain cases mutations are still advantageous over WT proteins in order to populate functionally relevant conformations that otherwise are barely accessible.
3.2. The Calcium-gated potassium channel MthK
MthK(Jiang et al., 2002a), a Ca2+ gated K+ channel from Methanothermobacter thermautotrophicus, is widely used as model system to study the mechanism for Ca2+ gating of the closely related eukaryotic BK channels, which are key players in linking cellular Ca2+ concentration to electrical excitability(Vergara et al., 1998). One difference between MthK and its eukaryotic homologue is the lack of a voltage sensor domain in the bacterial protein. This difference reduces the complexity of the system and allows for studying Ca2+ gating in an isolated manner. To that end, single channel recordings and X-ray crystallography have been used to gain insights into conformational changes and allosteric networks governing the regulation of this channel. However, structures of the MthK channel could only be obtained in the open state using engineered constructs missing half of the regulatory, Ca2+-binding domains(Jiang et al., 2002a, b), and of the isolated pore domain alone(Ye et al., 2010). This left open many important questions about how binding of Ca2+ to the cytosolic domains facilitates channel opening.
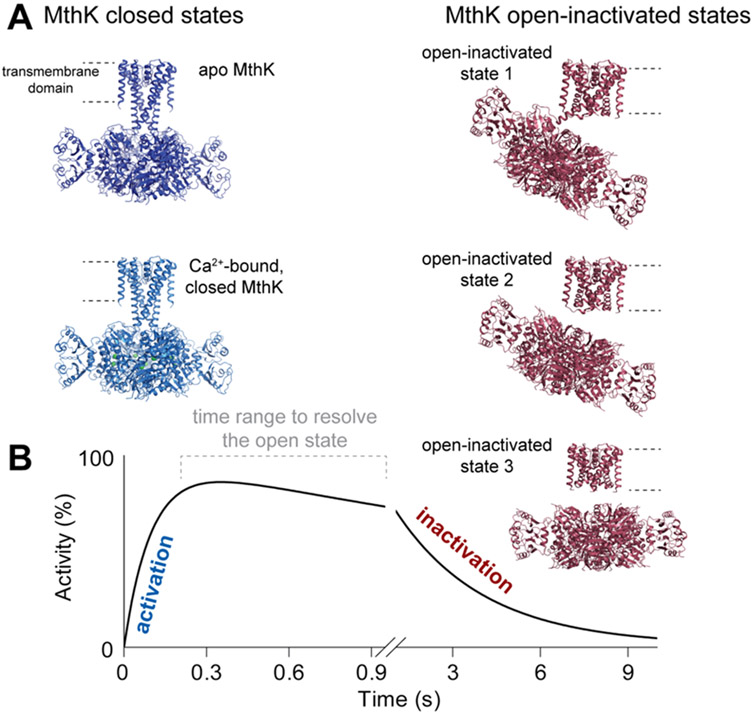
To gain insights into structural rearrangements during gating of MthK, single particle cryo-EM was employed to solve the structure of apo MthK reconstituted in lipid nanodiscs (Figure 6A)(Fan et al., 2020). While the data displayed minor heterogeneity, the functional data from single channel electrophysiology as well as stopped-flow flux assays allowed unambiguous assignment of the observed conformation to a closed channel. Addition of Ca2+ revealed major conformational changes in the cytosolic gating ring as well as in the transmembrane domains (Figure 6A) similar to the open-state crystal structure. Only a minor fraction of particles (~7 %) of MthK in the presence of saturating Ca2+ adopts a structure similar to the closed state (Figure 6A)(Fan et al., 2020). The remaining particles, however, while showing an open bundle crossing gate also showed density for the previously unresolved, cytosolic N-terminus plugging the pore. Stopped-flow data revealed that upon Ca2+ application, the channel inactivates with a time constant τ ~3 s (Figure 6B). Removal of the N-terminus leads to a non-inactivating channel and structures of the open-active MthK channel(Fan et al., 2020). We thus proposed, that this N-terminal peptide is responsible for a ball-and-chain type inactivation in MthK. Only the combination of protein engineering, stopped-flow analysis, and cryo-EM eventually allowed us to decipher the mechanism of Ca2+ gating and inactivation in MthK.
Figure 6: Structure-function correlation for MthK channels.
A Structures of MthK in different states (PDB IDs 6U6D, 6U5R, 6U68, 6U6E, 6U6H) with closed states in blue and open-inactivated states in red. B Schematic representation of the time-dependent activity of MthK as resolved in the sequential-mixing stopped-flow assay. Cryo-EM structures were assigned to functional states based on results as idealized in B.
Obtaining the structures of the open, but non-inactivated, states of the channel from the WT MthK protein will require alternative approaches, since inactivation sets in with a time constant of seconds. Two methods combining cryo-EM with rapid mixing techniques have been described(Chen et al., 2015; Dandey et al., 2020) and both rely on spraying the sample onto the grid while it is plunged into liquid ethane. In the first case(Chen et al., 2015) a microfluidics device has been developed that allows for rapidly mixing two components (such as two interacting proteins or ion channel and agonist). The length of the flow path determines the incubation time of the mixture before it is sprayed onto a grid and plunge frozen. The second approach mixes the two components in the process of spraying them onto the grid. Two piezo dispensing tips are used in sequence to deliver two components onto a self-wicking grid with a time resolution of 10 ms(Dandey et al., 2020).
Such rapid mixing approaches combined with cryo-EM data acquisition and analysis are of great interest to biological processes in general. These methods are highly promising and eventually will allow better correlation between kinetic, functional data and structural data using WT channel proteins in addition to using mutant channel proteins or engineered constructs to structurally resolve transiently populated channel conformations.
4. Protocols
4.1. Materials and equipment
Lipid stock solution in buffer, prepare 100 – 200 mM lipids solubilized in Cholate
Lipid stock solution (25 mg/ml in chloroform)
CHAPS (3-[(3-Cholamidopropyl)-Dimethylammonio]-1-Propane Sulfonate])
ANTS (8-Aminonaphthalene-1,3,6-trisulfonic acid disodium salt), prepare 75 mM stock solution in ddH2O, pH 7.4
NMG (N-methyl-D-glucamine)
Disposable borosilicate glass tubes (VWR)
Desiccator
Water bath sonicator
Spin-X column (Fisher Scientific)
SM-2 BioBeads (Bio-Rad)
Superose6 10/300 column (Cytiva)
Dowex ion exchange resin (Sigma Aldrich)
Sephadex G-50 fine (Cytiva)
General purpose plastic column for gravity, long, 20 ml (Bio-Rad)
Empty spin columns (Bio-Rad)
Mini extruder (Avanti)
PD-10 desalting columns (Cytiva)
Membrane protein purified to homogeneity
Membrane scaffolding protein (MSP), purified as described(Ritchie et al., 2009)
4.2. Nanodisc reconstitution
Detailed protocols for nanodisc reconstitutions have been described before(Ritchie et al., 2009; Kalienkova et al., 2020; Schmidpeter et al., 2020b).
Prepare 1.5 ml tubes for each reconstitution mixture
Add buffer, MSP (produced as previously described(Ritchie et al., 2009)), lipids, and your protein according to the desired MP:MSP:lipid ratio and incubate for 30 – 60 min above the phase transition temperature of the lipids to allow for formation of mixed micelles
Transfer reconstitution mixture to 2 ml tubes containing wet BioBeads equilibrated in reconstitution buffer (25 mg wet BioBeads per 50 μl reconstitution mix) and incubate under gentle agitation at a temperature slightly higher than the phase transition temperature of the lipid mix. Gentle shaking is necessary to prevent BioBeads from settling to the bottom of the tubes and ensure efficient detergent removal.
After 2 h transfer supernatant to new 2 ml tubes with fresh, equilibrated BioBeads and incubate under gentle agitation overnight.
The next day, transfer supernatant to fresh tubes, dilute in sample buffer to reach 500 μl, run sample through Spin-X column (1 min, 10000 g) to remove aggregates and any remaining BioBeads, and subject filtered sample to gel filtration on a Superose 6 10/300 column.
Analyze collected samples on SDS-PAGE to differentiate between the different peaks (Figure 2).
4.3. Reconstitution of ion channels into SUVs
Ion channels can be reconstituted into small unilamellar vesicles for the radioactive 86Rb+ flux assay(Nimigean, 2006) or for single channel bilayer recordings(Morera et al., 2007).
Swell Sephadex G-50 in reconstitution buffer (10 mM Hepes, 400 mM KCl, 5 mM NMG, pH 7.6) overnight.
The next day, degas the swollen Sephadex G-50 under vacuum for at least 2 h.
Dry 5 mg of lipids to a thin film in a disposable glass tube under constant N2 flow.
Wash lipids with 1 ml of n-Pentane and dry again into a thin film under constant N2 flow.
Rehydrate lipids in 500 μl reconstitution buffer (to reach 10 mg/ml concentration).
Add CHAPS powder to reach a final concentration of 33 mM and solubilize lipids by sonication in a water bath sonicator. Perform cycles of 30 s sonication and 5 min incubation at room temperature until the solution is clear.
Add ion channel purified to homogeneity at the desired concentration and incubate for 10 – 30 min to allow for formation of mixed micelles.
Prepare one detergent removal column for each reconstitution by adding Sephadex G-50 in reconstitution buffer to general purpose plastic columns to a final bed volume of 18 ml.
Carefully add reconstitution mix to the top of the column. Add 6 ml (dead volume of the column) of reconstitution buffer and let run through. Add another 3 ml of reconstitution buffer and manually collect 500 μl fractions.
Liposome containing fractions are pooled based on the turbidity of the fractions (as judged by visual inspection).
Prepare 50 μl aliquots in 200 μl tubes, flash freeze in liquid nitrogen and store at −80 °C until use for flux assay or bilayer recordings.
4.4. 86Rb+ flux assay
A detailed protocol for this assay is available(Nimigean, 2006). Make sure to follow all necessary precautions in handling radioactive materials and collect all liquid waste as well as disposables that got in touch with radioactivity in separate containers, suitable for radioactive waste.
Prepare Dowex-NMG before starting the experiment by washing the desired amount of resin with five volumes of 95 % ethanol followed by five volumes of ddH2O. Incubate resin in 1 M NMG for at least 1 h. Wash with ddH2O until the pH of the resin is below 9. Once prepared, Dowex-NMG resin can be stored at 4 °C for a few months.
Thaw 300 μl of proteo-liposomes (obtained as in 4.3) in order to perform the 86Rb+ uptake assay in triplicate. Briefly sonicate liposome solution in a water bath sonicator.
Fill 5 ml disposable spin columns with Sephadex G-50 fine swollen in sorbitol buffer (10 mM Hepes, 400 mM sorbitol, 5 mM NMG, 50 μM KCl, pH 7.6) to reach a bed volume of 1.5 ml and dry by centrifugation (1000 g, 15 s).
Add 100 μl of liposomes to the top of the column bed and spin at 700 g for 1 min to exchange the buffer outside of the liposomes with sorbitol buffer and to establish the K+ gradient across the liposomal membrane. Collect the liposomes.
Immediately start the uptake reaction by adding 2-6 volumes of 86Rb+ uptake buffer (sorbitol buffer supplemented with 0.1 μCi/ml 86Rb+) to the liposomes (e.g. 100 μl liposomes + 600 μl uptake buffer will yield 7 samples, 100 μl each).
At this point prepare 5 ml disposable spin columns filled with 1.5 ml Dowex-NMG resin for ion exchange chromatography, one column for each time point.
Wash columns with 2 ml sorbitol solution (400 mM sorbitol) + 5 mg/ml BSA.
Wash columns with 2 ml sorbitol solution.
At desired time points take 100 μl samples from the uptake reaction mix and immediately pass over a freshly equilibrated ion exchange column, followed by 1.5 ml sorbitol solution. Collect the eluate.
Transfer the eluate into scintillation vials, add 15 ml scintillation fluid and measure the radioactivity.
For the last sample of each series add 1 μg/ml valinomycin to the liposomes, incubate for 2 min and perform ion exchange as described above. This serves as control and provides a readout for the maximal 86Rb+ uptake possible into the liposomes.
4.5. Bilayer recordings
Bilayer recordings are performed in a horizontal setup where the top and bottom chamber are separated by a partition with a 100 μm aperture.
Seal the bottom chamber using vacuum grease and a plastic cover slip.
Separate the bottom and top chambers by placing a partition with a 100 μm aperture in the middle between the two chambers and seal using vacuum grease.
Fill the bottom chamber with solution containing modulators of the desired channel activity. The bottom chamber is easier to perfuse.
Dry a small aliquot of lipids (~70 μl of 10 mg/ml stock) in a disposable glass tube under constant N2 flow, wash with n-Pentane and dry again. Resuspend in ~70 μl decane (for a final 10 mg/ml lipid solution in decane) and briefly sonicate in a water bath sonicator.
Add 1 μl of lipids in decane onto the aperture and let dry. Using 8 mg/ml DPhPC yields reproducibly stable bilayers. Other lipids and lipid mixtures can be used as well, but the concentration of lipids as well as the solvent type need to be optimized to obtain stable bilayers over the aperture with a capacitance between 40 and 60 pF. In addition to n-decane, n-nonane, n-octane, n-dodecane, n-undecane, etc, can also be used(Miller, 1983).
As soon as the lipids are dry, add solution to the top chamber, place Ag/AgCl electrodes into separate wells in the bilayer chamber and connect those wells to the top and bottom chamber by agar bridges made with 100 mM KCl.
Use a lipid-coated 10 μl tip or a fire-polished glass rod to paint bilayers.
Monitor bilayer formation electronically by applying a capacitance step protocol. Useful bilayers should display a capacitance of 40 – 60 pF.
Once good bilayers can be reproducibly formed, thaw one aliquot of liposomes (from 4.3) briefly sonicate the liposomes and apply 1 μl on top of the aperture. Breaking and repainting the bilayer at this step can help to obtain productive fusion of liposomes with the bilayer.
Monitor channel incorporation into the bilayer by applying a constant voltage and monitoring the signal.
Once a channel signal is observed allow for the bilayer to stabilize after the fusion (10 s – 1 min) and start your experiment.
4.6. Reconstitution of channels into GUVs for stopped-flow
The reconstitution of proteins for the stopped-flow assay as well as the assay and data analysis have been described in detail(Rusinova et al., 2014; Posson et al., 2018). We will provide a standard protocol used in our lab that yields 3200 μl of concentrated liposomes before the final desalting step. This amount of liposomes leads to at least 25 separate recordings. The protocol can be linearly scaled to individual needs.
Dry 15 mg of lipids in a disposable glass tube into a thin film under constant N2 flow, further dry lipids overnight under vacuum in a desiccator.
Rehydrate lipids in 1114 μl reconstitution buffer (15 mM Hepes, 150 mM KNO3, pH 7.4), add CHAPS powder to a concentration of 33 mM and solubilize lipids by sonication in a water bath sonicator. Perform cycles of 30 s sonication and 5 min incubation at room temperature until the solution is clear.
Add 557 μl ANTS (75 mM stock solution in ddH2O, pH 7.4) and briefly sonicate.
Add protein to reach the desired concentration, add an additional 500 μl ANTS and incubate for 10 – 30 min.
Add 2 ml of a 50 % BioBead slurry in pre-mix buffer (10 mM Hepes, 140 mM KNO3, pH 7.4) to start detergent removal. At this point, the final reconstitution mixture should have a composition of 10 mM Hepes, 140 mM KNO3, pH 7.4, 25 mM ANTS, 16 mM CHAPS, ~5 mg/ml lipids, and the desired protein concentration.
Incubate reconstitution mix slightly above the phase transition temperature of the lipids for 3 h under constant agitation.
Transfer supernatant to a fresh glass tube and store sample above the phase transition temperature overnight.
The next day, briefly sonicate the solution and extrude through a 0.1 μm membrane using a mini-extruder.
Apply solution to a PD-10 desalting column equilibrated in pre-mix buffer to remove extra-liposomal ANTS.
4.7. Stopped flow assay
Right before starting the assay dilute liposomes 10x for the single-mixing assay or 5x for the double mixing assay, to ascertain a good signal-to-noise ratio. Keep in mind that Tl+ is hazardous to health and environment. Liquid waste from this assay needs to be collected separately, any spillage needs to be removed immediately, and appropriate personal protective equipment should be worn at all times.
Single-mixing assay
For the single-mixing assay (Figure 3C) the channel should be in an equilibrium activity state, which means agonists/antagonists should be present at the desired concentration throughout the entire reconstitution. Mix liposomes with pre-mix buffer (10 mM Hepes, 140 mM KNO3, pH 7.4, supplemented with agonist/antagonist) and record the fluorescence signal for 1 s. Perform at least six technical repeats, no change in fluorescence should be observed.
Next, mix liposomes directly with quenching buffer (10 mM Hepes, 90 mM KNO3, 50 mM TlNO3, pH 7.4, supplemented with the same concentration of agonist/antagonist as used in the pre-mix buffer) and record fluorescence signal for 1 s. Perform at least six technical repeats for each condition.
Perform the same measurements using liposomes without channel protein as control.
Sequential-mixing assay
Prepare sample as described in 4.6. Right before the experiment dilute liposomes 5x with pre-mix buffer and incubate for 10 min. In order to start from an equilibrium activity state supplement pre-mix buffer with agonist/antagonist if necessary.
Mount a 3 ml luer-lock syringe containing liposomes onto the stopped-flow spectrometer.
Mount another 3 ml luer-lock syringe with pre-mix buffer onto the stopped-flow instrument. Pre-mix buffer should be supplemented with channel agonist/antagonist/blocker etc. according to the planned experiment
Mount a 3 ml luer-lock syringe containing recording buffer onto the stopped-flow spectrometer (recording buffer = pre-mix buffer).
The fourth syringe mounted should contain only pre-mix buffer (flush solution to push sample from the aging loop into the sample cell).
If the instrument is equipped with an excitation monochromator set the wavelength to 360 nm, if an LED light source is used make sure that a 360 nm wavelength LED is connected. A 420 nm high-pass filter should be placed between the sample cell and the fluorescence photomultiplier.
Perform 4 technical repeats of a sequential mixing experiment to equilibrate all tubing with the desired solution and adjust the photomultiplier tension to the maximum fluorescence value.
Perform at least another six repeats with adjusted photomultiplier tension as control experiment. No change in fluorescence should be observed.
Refill all syringes and exchange the recording buffer with quenching buffer (10 mM Hepes, 90 mM KNO3, 50 mM TlNO3, pH 7.4). Quenching buffer should contain the same agonist/antagonist/etc concentration as used in the pre-mix buffer.
Set desired sample aging time (i.e. the delay time between the first and the second mixing step) and perform at least six technical repeats for each condition after the tubing has been equilibrated with the new buffer condition.
Either the condition of the pre-mix buffer or the aging time can be varied in this experiment providing extraordinary versatility. For examples see (McCoy et al., 2014; Rusinova et al., 2014; Posson et al., 2015; Menny et al., 2017; Schmidpeter et al., 2018; Tong et al., 2019; Schmidpeter et al., 2020a).
5. Conclusion
In this chapter, we provide broad background about structural and functional assays that can be used together to understand the structure-function relationship of ion channels in detail. The presented protocols are optimized to study K+ and non-selective channels. In principle, however, these assays can be modified for many other ion channels. We further describe two recent examples from our own work where these techniques were used in combination to describe the mechanism of a cyclic nucleotide-gated K+ channel and a Ca2+-gated K+ channel and to correlate structural conformations resolved by single particle cryo-EM with functional states(Rheinberger et al., 2018; Schmidpeter et al., 2018; Fan et al., 2020; Schmidpeter et al., 2020a). Our studies show that steady-state channel activity obtained from single channel recordings can shed light on the distribution of states in cryo-EM samples. However, it also shows that, in order to understand complex biological systems like ion channels, kinetic measurements under defined conditions are crucial to explain structural data(Dandey et al., 2020; Schmidpeter et al., 2020a). While mutations in some cases can be used to selectively populate specific channel conformations, recent developments in cryo-EM sample preparation allow for time resolved, structural studies(Chen et al., 2015; Dandey et al., 2020). This will further improve correlation of kinetic, functional data with structural conformations obtained from cryo-EM data analyses. However, this still leaves the potential discrepancy between liposome-based assays and structural data obtained from protein reconstituted into nanodiscs. Here, the developments in cryogenic electron tomography are promising as this will enable researchers to use the same liposome sample for structural and functional work on ion channels.
Acknowledgements
This work was supported by grants from the National Institutes of Health (R01GM088352 and R01GM124451 to CN) and the American Heart Association (18POST33960309 to PS).
References
- Alvarez FJ, Orelle C, and Davidson AL. 2010. Functional reconstitution of an ABC transporter in nanodiscs for use in electron paramagnetic resonance spectroscopy. J Am Chem Soc. 132:9513–9515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Huber T, and Sakmar TP. 2008. Rapid incorporation of functional rhodopsin into nanoscale apolipoprotein bound bilayer (NABB) particles. J Mol Biol. 377:1067–1081. [DOI] [PubMed] [Google Scholar]
- Banerjee S, and Nimigean CM. 2011. Non-vesicular transfer of membrane proteins from nanoparticles to lipid bilayers. J Gen Physiol. 137:217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayburt TH, Grinkova YV, and Sligar SG. 2002. Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. Nano Letters. 2:853–856. [Google Scholar]
- Bezanilla F 2008. Ion channels: from conductance to structure. Neuron. 60:456–468. [DOI] [PubMed] [Google Scholar]
- Bezanilla F, Rojas E, and Taylor RE. 1970. Sodium and potassium conductance changes during a membrane action potential. J Physiol. 211:729–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brams M, Kusch J, Spurny R, Benndorf K, and Ulens C. 2014. Family of prokaryote cyclic nucleotide-modulated ion channels. Proc Natl Acad Sci U S A. 111:7855–7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Kaledhonkar S, Sun M, Shen B, Lu Z, Barnard D, Lu TM, Gonzalez RL, and Frank J. 2015. Structural dynamics of ribosome subunit association studied by mixing-spraying time-resolved cryogenic electron microscopy. Structure. 23:1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MD, and Gordon SE. 2013. Giant liposome preparation for imaging and patch-clamp electrophysiology. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti F, and Neher E. 1980. Single channel recordings of K+ currents in squid axons. Nature. 285:140–143. [DOI] [PubMed] [Google Scholar]
- Dandey VP, Budell WC, Wei H, Bobe D, Maruthi K, Kopylov M, Eng ET, Kahn PA, Hinshaw JE, Kundu N, Nimigean CM, Fan C, Sukomon N, Darst SA, Saecker RM, Chen J, Malone B, Potter CS, and Carragher B. 2020. Time-resolved cryo-EM using Spotiton. Nat Methods. 17:897–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcour AH, Martinac B, Adler J, and Kung C. 1989. Modified reconstitution method used in patch-clamp studies of Escherichia coli ion channels. Biophys J. 56:631–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov IG, Grinkova YV, Lazarides AA, and Sligar SG. 2004. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J Am Chem Soc. 126:3477–3487. [DOI] [PubMed] [Google Scholar]
- Dittert I, Benedikt J, Vyklický L, Zimmermann K, Reeh PW, and Vlachová V. 2006. Improved superfusion technique for rapid cooling or heating of cultured cells under patch-clamp conditions. J Neurosci Methods. 151:178–185. [DOI] [PubMed] [Google Scholar]
- Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, and MacKinnon R. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77. [DOI] [PubMed] [Google Scholar]
- Dörr JM, Koorengevel MC, Schäfer M, Prokofyev AV, Scheidelaar S, van der Cruijsen EA, Dafforn TR, Baldus M, and Killian JA. 2014. Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: the power of native nanodiscs. Proc Natl Acad Sci U S A. 111:18607–18612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenman G, and Horn R. 1983. Ionic selectivity revisited: the role of kinetic and equilibrium processes in ion permeation through channels. J Membr Biol. 76:197–225. [DOI] [PubMed] [Google Scholar]
- Evans EGB, Morgan JLW, DiMaio F, Zagotta WN, and Stoll S. 2020. Allosteric conformational change of a cyclic nucleotide-gated ion channel revealed by DEER spectroscopy. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Sukomon N, Flood E, Rheinberger J, Allen TW, and Nimigean CM. 2020. Ball-and-chain inactivation in a calcium-gated potassium channel. Nature. 580:288–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Perez V, Xia XM, and Lingle CJ. 2014. Functional regulation of BK potassium channels by γ1 auxiliary subunits. Proc Natl Acad Sci U S A. 111:4868–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagn F, Etzkorn M, Raschle T, and Wagner G. 2013. Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J Am Chem Soc. 135:1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke W, and Kaupp UB. 1984. Incorporation of ion channels from bovine rod outer segments into planar lipid bilayers. Biophys J. 46:587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig N, Peetz O, Ahdash Z, Tascón I, Booth PJ, Mikusevic V, Diskowski M, Politis A, Hellmich Y, Hänelt I, Reading E, and Morgner N. 2018. Native mass spectrometry goes more native: investigation of membrane protein complexes directly from SMALPs. Chem Commun (Camb). 54:13702–13705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin AL, and Huxley AF. 1939. Action Potentials Recorded from Inside a Nerve Fibre. Nature. 144:710–711. [Google Scholar]
- Hodgkin AL, Huxley AF, and Katz B. 1952. Measurement of current-voltage relations in the membrane of the giant axon of Loligo. J Physiol. 116:424–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingólfsson HI, and Andersen OS. 2010. Screening for small molecules' bilayer-modifying potential using a gramicidin-based fluorescence assay. Assay Drug Dev Technol. 8:427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamshad M, Lin YP, Knowles TJ, Parslow RA, Harris C, Wheatley M, Poyner DR, Bill RM, Thomas OR, Overduin M, and Dafforn TR. 2011. Surfactant-free purification of membrane proteins with intact native membrane environment. Biochem Soc Trans. 39:813–818. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Cadene M, Chait BT, and MacKinnon R. 2002a. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 417:515–522. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Cadene M, Chait BT, and MacKinnon R. 2002b. The open pore conformation of potassium channels. Nature. 417:523–526. [DOI] [PubMed] [Google Scholar]
- Kalienkova V, Alvadia C, Clerico Mosina V, and Paulino C. 2020. Single-Particle Cryo-EM of Membrane Proteins in Lipid Nanodiscs. Methods Mol Biol. 2127:245–273. [DOI] [PubMed] [Google Scholar]
- Kesters D, Brams M, Nys M, Wijckmans E, Spurny R, Voets T, Tytgat J, Kusch J, and Ulens C. 2015. Structure of the SthK Carboxy-Terminal Region Reveals a Gating Mechanism for Cyclic Nucleotide-Modulated Ion Channels. PloS one. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles TJ, Finka R, Smith C, Lin YP, Dafforn T, and Overduin M. 2009. Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J Am Chem Soc. 131:7484–7485. [DOI] [PubMed] [Google Scholar]
- Krueger BK, Worley JF, and French RJ. 1983. Single sodium channels from rat brain incorporated into planar lipid bilayer membranes. Nature. 303:172–175. [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W 2014. Cryo-EM enters a new era. Elife. 3:e03678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre R, and Miller C. 1983. Conduction and selectivity in potassium channels. J Membr Biol. 71:11–30. [DOI] [PubMed] [Google Scholar]
- Liao M, Cao E, Julius D, and Cheng Y. 2013. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 504:107–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi A, Gao X, Adaixo R, Rheinberger J, Stahlberg H, Nimigean C, and Scheuring S. 2018. An iris diaphragm mechanism to gate a cyclic nucleotide-gated ion channel. Nat Commun. 9:3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari SA, Pessoa J, Altieri S, Hensen U, Thomas L, Morais-Cabral JH, and Müller DJ. 2011. Gating of the MlotiK1 potassium channel involves large rearrangements of the cyclic nucleotide-binding domains. Proc Natl Acad Sci U S A. 108:20802–20807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matulef K, and Valiyaveetil FI. 2018. Patch-Clamp Recordings of the KcsA K. Methods Mol Biol. 1684:181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy JG, Rusinova R, Kim DM, Kowal J, Banerjee S, Jaramillo Cartagena A, Thompson AN, Kolmakova-Partensky L, Stahlberg H, Andersen OS, and Nimigean CM. 2014. A KcsA/MloK1 chimeric ion channel has lipid-dependent ligand-binding energetics. J Biol Chem. 289:9535–9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menny A, Lefebvre SN, Schmidpeter PA, Drège E, Fourati Z, Delarue M, Edelstein SJ, Nimigean CM, Joseph D, and Corringer PJ. 2017. Identification of a pre-active conformation of a pentameric channel receptor. Elife. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C 1983. Integral membrane channels: studies in model membranes. Physiol Rev. 63:1209–1242. [DOI] [PubMed] [Google Scholar]
- Morera FJ, Vargas G, González C, Rosenmann E, and Latorre R. 2007. Ion-channel reconstitution. Methods Mol Biol. 400:571–585. [DOI] [PubMed] [Google Scholar]
- Morgan JLW, Evans EGB, and Zagotta WN. 2019. Functional characterization and optimization of a bacterial cyclic nucleotide-gated channel. J Biol Chem. 294:7503–7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy RE, Samal AB, Vlach J, Mas V, Prevelige PE, and Saad JS. 2019. Structural and biophysical characterizations of HIV-1 matrix trimer binding to lipid nanodiscs shed light on virus assembly. J Biol Chem. 294:18600–18612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane T, Kotecha A, Sente A, McMullan G, Masiulis S, Brown PMGE, Grigoras IT, Malinauskaite L, Malinauskas T, Miehling J, Uchański T, Yu L, Karia D, Pechnikova EV, de Jong E, Keizer J, Bischoff M, McCormack J, Tiemeijer P, Hardwick SW, Chirgadze DY, Murshudov G, Aricescu AR, and Scheres SHW. 2020. Single-particle cryo-EM at atomic resolution. Nature. 587:152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath A, Trexler AJ, Koo P, Miranker AD, Atkins WM, and Rhoades E. 2010. Single-molecule fluorescence spectroscopy using phospholipid bilayer nanodiscs. Methods Enzymol. 472:89–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher E, and Sakmann B. 1976. Single-channel currents recorded from membrane of denervated frog muscle fibres. Nature. 260:799–802. [DOI] [PubMed] [Google Scholar]
- Nimigean CM 2006. A radioactive uptake assay to measure ion transport across ion channel–containing liposomes. Nature protocols. 1:1207–1212. [DOI] [PubMed] [Google Scholar]
- Nimigean CM, Shane T, and Miller C. 2004. A cyclic nucleotide modulated prokaryotic K+ channel. J Gen Physiol. 124:203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessoa J, Fonseca F, Furini S, and Morais-Cabral JH. 2014. Determinants of ligand selectivity in a cyclic nucleotide-regulated potassium channel. J Gen Physiol. 144:41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov E, Palanivelu D, Constantine M, Rohde PR, Cox CD, Nomura T, Minor DL, and Martinac B. 2013. Patch-clamp characterization of the MscS-like mechanosensitive channel from Silicibacter pomeroyi. Biophys J. 104:1426–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posson DJ, Rusinova R, Andersen OS, and Nimigean CM. 2015. Calcium ions open a selectivity filter gate during activation of the MthK potassium channel. Nat Commun. 6:8342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posson DJ, Rusinova R, Andersen OS, and Nimigean CM. 2018. Stopped-Flow Fluorometric Ion Flux Assay for Ligand-Gated Ion Channel Studies. Methods Mol Biol. 1684:223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinberger J, Gao X, Schmidpeter PA, and Nimigean CM. 2018. Ligand discrimination and gating in cyclic nucleotide-gated ion channels from apo and partial agonist-bound cryo-EM structures. Elife. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, and Sligar SG. 2009. Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464:211–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinova R, Kim DM, Nimigean CM, and Andersen OS. 2014. Regulation of ion channel function by the host lipid bilayer examined by a stopped-flow spectrofluorometric assay. Biophys J. 106:1070–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidpeter PAM, Gao X, Uphadyay V, Rheinberger J, and Nimigean CM. 2018. Ligand binding and activation properties of the purified bacterial cyclic nucleotide-gated channel SthK. J Gen Physiol. 150:821–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidpeter PAM, and Nimigean CM. 2018. Fluorescence Titrations to Determine the Binding Affinity of Cyclic Nucleotides to SthK Ion Channels. Bio Protoc. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidpeter PAM, Rheinberger J, and Nimigean CM. 2020a. Prolyl isomerization controls activation kinetics of a cyclic nucleotide-gated ion channel. Nat Commun. 11:6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidpeter PAM, Sukomon N, and Nimigean CM. 2020b. Reconstitution of Membrane Proteins into Platforms Suitable for Biophysical and Structural Analyses. Methods Mol Biol. 2127:191–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sligar SG, and Denisov IG. 2020. Nanodiscs: A toolkit for membrane protein science. Protein Sci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AJ, Alder L, Silk J, Adkins C, Fletcher AE, Scales T, Kerby J, Marshall G, Wafford KA, McKernan RM, and Atack JR. 2001. Effect of alpha subunit on allosteric modulation of ion channel function in stably expressed human recombinant gamma-aminobutyric acid(A) receptors determined using (36)Cl ion flux. Mol Pharmacol. 59:1108–1118. [DOI] [PubMed] [Google Scholar]
- Sotomayor M, Vásquez V, Perozo E, and Schulten K. 2007. Ion conduction through MscS as determined by electrophysiology and simulation. Biophys J. 92:886–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo ACK, Lee SC, Pollock NL, Stroud Z, Hall S, Thakker A, Pitt AR, Dafforn TR, Spickett CM, and Roper DI. 2019. Analysis of SMALP co-extracted phospholipids shows distinct membrane environments for three classes of bacterial membrane protein. Sci Rep. 9:1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A, Petroff JT, Hsu FF, Schmidpeter PA, Nimigean CM, Sharp L, Brannigan G, and Cheng WW. 2019. Direct binding of phosphatidylglycerol at specific sites modulates desensitization of a ligand-gated ion channel. Elife. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara C, Latorre R, Marrion NV, and Adelman JP. 1998. Calcium-activated potassium channels. Curr Opin Neurobiol. 8:321–329. [DOI] [PubMed] [Google Scholar]
- Winterstein LM, Kukovetz K, Rauh O, Turman DL, Braun C, Moroni A, Schroeder I, and Thiel G. 2018. Reconstitution and functional characterization of ion channels from nanodiscs in lipid bilayers. J Gen Physiol. 150:637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, and Lander GC. 2020. How low can we go? Structure determination of small biological complexes using single-particle cryo-EM. Curr Opin Struct Biol. 64:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Li Y, and Jiang Y. 2010. Novel insights into K+ selectivity from high-resolution structures of an open K+ channel pore. Nat Struct Mol Biol. 17:1019–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellen G 1987. Permeation in potassium channels: implications for channel structure. Annu Rev Biophys Biophys Chem. 16:227–246. [DOI] [PubMed] [Google Scholar]
- Yip KM, Fischer N, Paknia E, Chari A, and Stark H. 2020. Atomic-resolution protein structure determination by cryo-EM. Nature. 587:157–161. [DOI] [PubMed] [Google Scholar]
- Yu HB, Li M, Wang WP, and Wang XL. 2016. High throughput screening technologies for ion channels. Acta Pharmacol Sin. 37:34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zhu H, Lape R, Greiner T, Du J, Lü W, Sivilotti L, and Gouaux E. 2021. Mechanism of gating and partial agonist action in the glycine receptor. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Morais-Cabral JH, Kaufman A, and MacKinnon R. 2001. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 414:43–48. [DOI] [PubMed] [Google Scholar]