Abstract
Combining single-cell transcriptomics with mitochondrial DNA (mtDNA) variant detection can be used to establish lineage relationships in primary human cells, but current methods are not scalable to interrogate complex tissues. Here, we combine common 3' single-cell RNA-sequencing protocols with mitochondrial transcriptome enrichment to increase coverage by more than 50-fold, enabling high-confidence mutation detection. The method successfully identifies skewed immune-cell expansions in primary human clonal hematopoiesis.
Single-cell RNA-sequencing (scRNA-seq) enables the unbiased assessment of cell states in health and disease1,2. Combined acquisition of cell state and genetic information can provide additional insight, such as targeted enrichment of cancer driver mutations from single-cell transcriptomes3,4. Separately, combining scRNA-seq with genetic cell barcodes can reveal clonal relationships and the evolutionary dynamics of cells within organisms5,6. However, this has largely been limited to experimental model systems that can be genetically manipulated to insert cell barcodes. To infer clonal dynamics in primary human cells, recent methods have detected and utilized mitochondrial DNA (mtDNA) mutations as naturally occurring genetic cell barcodes7-9. The combination of scRNA-seq with mtDNA mutation detection can inform clonal relationships with high confidence, but is currently restricted to expensive, low-throughput, full-length transcript sequencing technologies like SmartSeq27,10. To enable the reconstruction of clonal relationships in complex human tissues, we developed a method that captures genetic variants from high-throughput scRNA-seq platforms: MAESTER, or Mitochondrial Alteration Enrichment from Single-cell Transcriptomes to Establish Relatedness (Fig. 1a). MAESTER is compatible with the most common high-throughput scRNA-seq platforms, including 10x Genomics 3′ protocols, Seq-Well S3, and Drop-seq (Supplementary Fig. 1-3)11,12. An intermediate step in each of these platforms yields full-length cDNA transcripts, from which we enrich all 15 mitochondrial transcripts using pools of primers, while maintaining cell-identifying barcodes (Fig. 1b, Supplementary Fig. 4). Standard next-generation sequencing with 250 bp reads is then used to obtain the sequence of the amplified mitochondrial transcripts (Fig. 1a). We developed a computational toolkit to call mtDNA variants from MAESTER data, the Mitochondrial Alteration Enrichment and Genome Analysis Toolkit (maegatk; Supplementary Fig. 5; Methods). Building on previous tools that we developed8 for mtDNA variant detection from single-cell ATAC (Assay for Transposase-Accessible Chromatin) or SmartSeq2, maegatk specifically handles technical biases implicit in high-throughput transcriptomic libraries. Maegatk uses unique molecular identifiers (UMIs) to collapse multiple sequencing reads of the same starting transcript, creating a consensus call for every nucleotide based on the most common call and base quality. This approach mitigates sequence errors introduced during PCR and sequencing and is essential to obtain high-confidence variant calls from high-throughput scRNA-seq protocols. We also incorporate indel calling and provide a resource to evaluate the potential functional impact of variants (Supplementary Fig. 6, Supplementary Table 1). Alterations in mtDNA are then used to infer relatedness between cells.
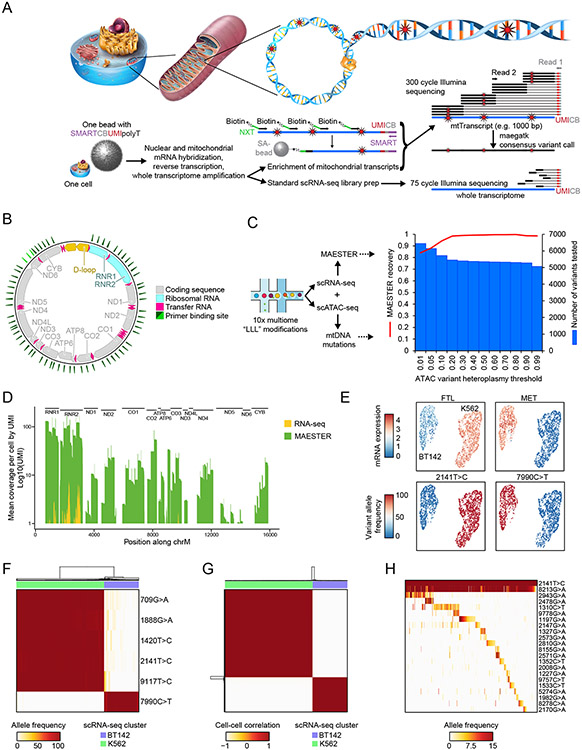
Figure 1. Targeted enrichment of mitochondrial transcripts enables discrimination between genetic clones.
A. Schematic shows the procedures for lineage inference from single-cell transcriptomes using MAESTER. Following mRNA capture and whole transcriptome amplification, part of the cDNA is used for standard scRNA-seq, and another part is used for PCR-based enrichment of mitochondrial transcripts. 300 bp sequencing reads maximize mitochondrial genome coverage to call variants. B. Diagram depicts the circular mitochondrial genome with annotated features. The green triangles indicate where MAESTER primers bind. C. Barplot shows the number of mtDNA variants that were detected by ATAC-seq (blue bars) and their recovery by MAESTER (red line). DNA (ATAC) and RNA (MAESTER) were acquired from the same K562 cells using the 10x multiome workflow. D. Barplot shows coverage of the mitochondrial genome with and without amplification from Seq-Well libraries using MAESTER. Mean coverage of 2,482 K562 and BT142 cells is shown. Each of the transcripts (UMIs) was sequenced ≥3 times. E. UMAPs show detection of cell type-specific (top) gene expression from scRNA-seq and (bottom) homoplasmic mtDNA variants from MAESTER. F. Heatmap depicts separation of 1,523 K562 and BT142 cells (columns) based on six mtDNA variants (rows). Cell type annotation from scRNA-seq is shown on top. G. Correlation matrix shows cell similarity based on the allele frequencies of six homoplasmic variants (rows and columns depict 1,523 cells). Unsupervised clustering identified two clusters that correlate with cell annotations from scRNA-seq. H. Heatmap shows VAF of 21 mtDNA variants detected by MAESTER (rows) for 588 K562 cells (columns, 44.4% of all K562 cells) with informative variants. Homoplasmic K562 variant 2141T>C is shown for comparison. Heatmap is organized by clonal structure (Methods). For E-H, only cells with >3-fold coverage of the indicated variants are shown. SMART and NXT are specific primer binding sequences, SA: streptavidin, CB: cell barcode, UMI: unique molecular identifier.
To verify the recovery of variants in our approach, we sought to measure mitochondrial DNA and RNA from the same individual cells. To achieve this, we performed MAESTER on DOGMA-seq libraries, which enable the concomitant detection of accessible DNA (including mtDNA) and transcriptome-wide RNA via the 10x Genomics multiome kit13 (Fig. 1c). After identifying variants on mtDNA8, we examined the proportion of variants recovered by MAESTER. Our analyses revealed that MAESTER recovered 94.1% of variants at a single-cell heteroplasmy of >10% (Fig. 1c). These results confirm that MAESTER recovers true mitochondrial DNA variants from transcriptomic data.
We established the feasibility and efficiency of MAESTER in standard high-throughput scRNA-seq methods using human cell mixing experiments. Chronic myelogenous leukemia cells (K562) were mixed with brain tumor cells (BT142) and analyzed using Seq-Well S3 and 10x Genomics 3′ v3 protocols. MAESTER dramatically increased the coverage of mitochondrial transcripts compared to scRNA-seq data alone (mean coverage per cell 0.2–0.7-fold for RNA-seq, 52–217-fold for MAESTER, Fig. 1d, Supplementary Fig. 7a-d), enabling reliable mtDNA variant calling with high confidence across many of the transcripts. Using Uniform Manifold Approximation and Projection (UMAP) for dimensionality reduction, the two cell populations cluster based on mRNA expression data (Fig. 1e). MAESTER enabled the identification of six homoplasmic mtDNA variants that distinguished between cell types (Fig. 1e-g, Supplementary Fig. 7d-f). Combining data from all six informative variants cleanly separates cell types and demonstrates 100% concordance with mRNA clusters (Supplementary Fig. 7g,h). Of note, MAESTER identified the same six variants in the Seq-Well and 10x libraries (Supplementary Fig. 8a-d).
To benchmark MAESTER’s ability to identify clonal structure at a more granular resolution, we performed a clonal expansion experiment. One hundred K562 cells were plated and allowed to expand for 14 days (doubling time ~24h), followed by scRNA-seq with MAESTER. We identified 21 informative mtDNA variants that revealed clonally related populations of K562 cells (Fig. 1h, Supplementary Fig. 8e), which were validated by orthogonal bulk ATAC-seq (Supplementary Fig. 8f). These data demonstrate the faithfulness of mtDNA variants enriched from mtRNA and the capacity of this method to resolve subpopulations within closely related cells.
We next applied MAESTER to derive clonal structure within primary human patient specimens. We first utilized a bone marrow aspirate from a patient with clonal hematopoiesis. The clonal hematopoiesis had evolved into blastic plasmacytoid dendritic cell neoplasm (BPDCN), as the patient had skin tumors at the time of collection. However, the concurrent bone marrow aspirate we utilized showed no tumor involvement (Methods). We performed 10x single-cell sequencing with MAESTER on this bone marrow aspirate and identified 9,346 high-quality cells, including all expected cell types, with an abundance of cytotoxic T cells (CTLs), likely due to hemodilution of the bone marrow sample with peripheral blood and possibly related to his evolving malignancy (Fig. 2a, Supplementary Fig. 9). We found that MAESTER coverage largely depends on the mtRNA content per cell (Supplementary Fig. 10) and tested different thresholds to select informative variants (Supplementary Fig. 11). We plotted the largest and most distinct 23 clones using 26 informative mtDNA variants (14.9% of cells were assigned to these clones, Fig. 2b), indicating MAESTER can resolve clonal populations in primary human specimens.
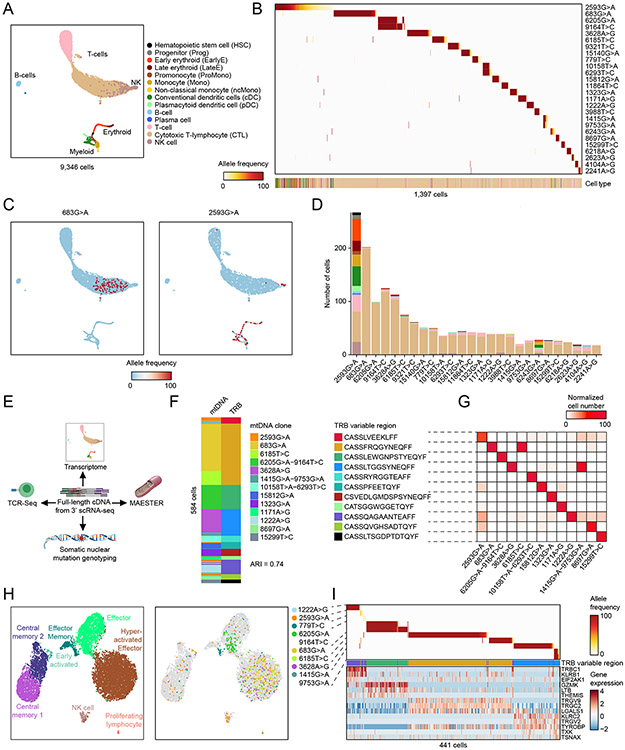
Figure 2. Genetic clones exhibit lineage bias in clonal hematopoiesis.
A. UMAP of all 9,346 cells profiled by 10x scRNA-seq from a bone marrow aspirate from an individual with clonal hematopoiesis. B. Heatmap shows VAF of 26 informative mtDNA variants detected by MAESTER and maegatk (rows) for 1,397 cells (columns) with at least 1% VAF for one of the 26 variants. Heatmap is organized by clones and sorted by clone size. Cell type is listed on the bottom by color according to legend in A. C. UMAPs display VAF in each cell for mtDNA variants 683G>A (left) and 2593G>A (right). D. Stacked bar graph of the number of cells in each clone, with cell type denoted by color according to legend in A. E. Schematic depicts multimodal analysis we performed on the same single cells. F. Plot shows cells (rows) in which both mtDNA and TRB clonal markers were detected. Clones are indicated by colors and defined by mtDNA variants and the TRB variable region, respectively. G. Confusion matrix shows concordance between mtDNA clones and TRB clonotypes. H. UMAPs of 8,382 T-cells in the clonal hematopoiesis sample, with state annotated by transcriptional signatures (left), and selected mtDNA clones (right). I. Heatmap of T-cells (columns) in the selected mtDNA clones with mtDNA VAF (top), TRB sequence (middle), and state-defining transcript expression (bottom). ARI: Adjusted Rand Index.
Many of the mtDNA clones clustered together in the RNA-based UMAP (Fig. 2c, Supplementary Fig. 12). Indeed, we found that many mtDNA clones were lineage biased with 9/23 clones skewed towards CTLs and 2 clones with myeloid lineage bias (Fig. 2d, Supplementary Fig. 13).
The abundance of T cells in the sample provided an opportunity to validate the mtDNA clones with an orthogonal assessment of clonality using the T-cell receptor (TCR) variable region. Building on a TCR enrichment method for Seq-Well14, we developed a protocol for TCR sequencing from 10x 3′ scRNA-seq cDNA. We termed the protocol T-cell Receptor Enrichment to linK clonotypes by sequencing (TREK-seq, Supplementary Fig. 14) and applied this to the bone marrow sample, adding an additional modality to the same single cells (Fig. 2e). TRA and TRB variable regions were detected in T-cells but not other lineages and were highly concordant, confirming reliable TCR enrichment (Supplementary Fig. 15). When comparing TRB variable regions to mtDNA variants, we noted high overlap of the orthogonal clonal markers (ARI = 0.74, Fig. 2f,g). The mtDNA clones that were skewed towards CTLs (e.g. 6205G>A-9164T>C), suggesting the mtDNA mutation occurred after TCR rearrangement, were largely restricted to a single T-cell state (Fig. 2h). In contrast, mtDNA clones with all hematopoietic cell types (e.g. 2593G>A), indicating the mtDNA mutation occurred within a multipotent HSC, contributed to multiple T-cell states and clonotypes (Fig. 2h). Combining MAESTER, TCR-sequencing, and transcriptional states (Fig. 2i) can provide independent validation of clonal relationships and new opportunities to study T-cell biology.
We also identified two clones with myeloid lineage bias, identified by mtDNA alterations 2593G>A and 6243G>A (Supplementary Fig. 13,16a). Given the patient’s clonal hematopoiesis, we sought to understand if these represented expanded clones. We utilized Genotyping of Transcriptomes (GoT)3 to identify the patient’s known ASXL1 and TET2 loss-of-function mutations in single cells. We found that cells within the two myeloid-biased clones contained a high fraction of mutated transcripts (Supplementary Fig. 16b). Of cells in the 2593G>A clone, 44% and 40% had TET2.S792X and TET2.Q1034X mutations, respectively. This is consistent with bi-allelic TET2 inactivation, a recurrent feature in myeloid malignancies15. No mutated transcripts were identified in cells from the other 21 clones we identified, providing evidence that the myeloid-biased clones identified by MAESTER represent cells derived from the patient’s clonal hematopoiesis.
GoT only captured wild-type or mutant transcripts in 3.5% of all 9,346 cells for TET2, and 0.4% for ASXL1. This relative lack of genotyping efficiency is related to gene expression, variant position, and amplicon size, and is similar to other protocols that genotype somatic mutations from high-throughput scRNA-seq libraries4. In contrast, MAESTER captured the mtDNA genotype at the 2593 position in 21,767 transcripts in 1,396 cells (99.9% of cells, an average of 16 transcripts per cell). Combining driver mutation with mtDNA variant detection facilitates phylogeny reconstruction (Supplementary Fig. 16c,d). This allowed us to explore cells marked by 2593G>A as clonally expanded cells with loss of TET2.
To interrogate this population further, we compared the myeloid differentiation trajectory of 2593G>A cells to other cells in the bone marrow using pseudotime analysis (Supplementary Fig. 16e)16. We found the clonal population was skewed towards less mature cell types, consistent with HSC expansion observed in Tet2 knockout mice17. While analysis of more cells and biological replicates is required to generalize these results, our data suggest the utility of MAESTER to identify and investigate pre-malignant cell expansions.
Finally, we applied MAESTER to a primary solid tumor tissue to demonstrate compatibility with complex tissues requiring cell dissociation. In tumor and peripheral blood samples from a glioblastoma patient, we identified a mtDNA deletion in malignant cells that was absent in the blood and we found region-specific clonal populations of malignant cells (Supplementary Fig. 17a-e). We also discovered tumor-associated myeloid cells that were derived from peripheral blood cells in the patient (Supplementary Fig. 17f).
In conclusion, MAESTER enables mtDNA variant detection in high-throughput 3′-biased scRNA-seq data, which was previously limited to ATAC-seq or full-length scRNA-seq. As with all methods that utilize mtDNA mutations to infer clonal relationships, there are limitations inherent to mitochondrial biology. It is currently not possible to track each cell division as single-molecule mutations typically cannot be detected above background due to their low mtDNA heteroplasmy. For MAESTER, the VAF needs to reach >1% for confident detection. The mtDNA copy number per cell and rate of cell proliferation impact the time it takes to reach 1% VAF. Further improvements in mutation detection efficiency (mitochondrial or nuclear) will enable increasingly granular studies of clonal dynamics. Studies that require short-term, per division tracking still require tunable and engineered lineage tracing methods. In contrast, mtDNA variants are suitable to determine clonal relationships between subsets of cells that are more divergent, providing a tool to study in vivo cellular dynamics and human biology. In addition, due to the widespread use of 3′-biased scRNA-seq, the development of MAESTER makes mtDNA variant detection accessible to more research laboratories and a wider range of experimental contexts. The accompanying maegatk software uses UMIs to increase confidence in mtDNA variant calls, an advance over previous methods. MAESTER can be implemented on new or prior scRNA-seq datasets by using the amplified cDNA that is stored as a standard practice. The high-throughput nature of 3′-biased scRNA-seq and MAESTER enables the study of clonal relationships and evolutionary dynamics of cells within complex primary human tissues. The combination of MAESTER with other modalities such as TCR-sequencing, somatic variant detection, and RNA-seq creates synergies that enable analyses and discoveries that are not possible with each method alone. By developing MAESTER, we democratize and expand the use of naturally occurring barcodes created by mtDNA alterations to enable discoveries in human biology.
Methods
Cell lines and culturing.
Human chronic myelogenous leukemia K562 cells (ATCC CCL-243) were cultured in RPMI 1640 Medium with GlutaMAX (Gibco 61870127), supplemented with 10% fetal calf serum (FCS) and penicillin-streptomycin. The BT142 gliomasphere line18 (ATCC ACS-1018) was maintained in Neurobasal media supplemented with 20 ng/mL recombinant EGF (R and D Systems), 20ng/mL FGF2 (R and D Systems), 1X B27 supplement (Invitrogen), 0.5X N2 supplement (Invitrogen), 3 mM L-glutamine, and penicillin/streptomycin19. 25% conditioned media was carried over each passage. Cultures were confirmed to be mycoplasma-free and their identity was verified by STR analysis. K562 and BT142 cells from the same passage were used for Seq-Well scRNA-seq + MAESTER, 10x 3′ v3 scRNA-seq + MAESTER, and bulk ATAC-seq.
Primary human samples
The patients in this study consented to all study procedures under Dana-Farber Cancer Institute IRB-approved research protocols. The patient with clonal hematopoiesis had a history of leukopenia and thrombocytopenia. The bone marrow sample we analyzed was an aspiration in the context of evaluation for skin-only BPDCN. Histologic evaluation of the concurrent bone marrow core biopsy was normal and did not show involvement of malignant BPDCN cells. Targeted sequencing of the bone marrow aspirate identified alterations in ASXL1 and TET2 indicating clonal hematopoiesis (Supplemental Figure 9B). Mononuclear cells were isolated from a bone marrow aspirate by density centrifugation and cryopreserved with 10% DMSO in liquid nitrogen. Cells were thawed using standard procedures; since viability (independently assessed by Trypan and propidium iodide staining) exceeded 90%, unsorted cells were used for scRNA-seq using the 10x 3' v3 protocol. A high proportion of cytotoxic T-cells was recovered, consistent with an expansion of large granular lymphocytes in the peripheral blood of this patient as demonstrated by routine clinical evaluation and confirmatory flow cytometry (Supplemental Figure 9A). This specimen is likely hemodiluted with a contribution from the peripheral blood as the bone marrow core biopsy did not contain this high fraction of T-cells. The scRNA-seq data from this sample is also being utilized in an independent manuscript not involving the MAESTER technique and is under consideration elsewhere (Patient 10, Griffin et al., manuscript under review).
Single-cell RNA-sequencing
For Seq-Well S3 experiments, cells were processed as described previously11. A complete, updated protocol for Seq-Well S3 is hosted on the Shalek Lab website (www.shaleklab.com). Briefly, an array with ~90,000 nanowells was first loaded with barcoded mRNA capture beads, then 10-15,000 cells were added dropwise onto the surface of the array. After cells were allowed to settle into the wells, the array was sealed with a semi-permeable polycarbonate membrane. Cells were lysed and mRNA transcripts were hybridized to the bead contained within the same well at the polyT sequence of the barcoded oligonucleotides. The beads were then used to generate cDNA via reverse transcription. A second strand synthesis step using a random octamer was performed to recover transcripts in which template switching during reverse transcription was not successful. Whole transcriptome amplification (WTA) PCR was performed and the product underwent a combination of tagmentation and PCR to generate dual indexed sequencing libraries. Libraries were sequenced using a 75 cycle kit on the Illumina NextSeq500 with custom read 1 (CR1P) and custom i5 primers (SW-Ci5P, Supplementary Table 2), 20 cycles for Read 1 (cell barcode or CB + UMI), 56 cycles for Read 2 (transcript sequence), and 2 x 8 bp library barcodes.
For 10x Genomics experiments, we used 3’ Single Cell Gene Expression v3 reagents, following all manufacturer’s recommendations. Briefly, 5,000 cells were loaded per well and captured in gel bead-in emulsions. Captured mRNAs were reverse transcribed into cDNAs and amplified to generate WTAs. Library construction involves fragmentation, adapter ligation, and a sample index PCR. Libraries were sequenced using a NovaSeq SP 100 cycle kit with 28 cycles for Read 1 (CB + UMI), 91 cycles for Read 2 (transcript sequence) and an 8 bp library barcode.
For the cell line mixing experiments, we analyzed cells from the same passage using two Seq-Well S3 arrays and two 10x 3′ v3 wells, yielding a similar number of cells and data quality. For the clonal hematopoiesis sample, we used four 10x 3’ v3 wells.
Mitochondrial alteration enrichment for MAESTER from Seq-well or Drop-seq
Similar to a method we initially developed for the detection of somatic mutations4, the starting material for targeted amplification of mtDNA transcripts is the product of the Seq-Well WTA reaction (only a fraction of which is used for scRNA-seq). The general method consists of two PCR reactions with a streptavidin bead enrichment in between (Supplementary Figures 1-2). The first PCR reaction serves to add a biotin tag and Nextera adapter (NEXT) to mitochondrial transcripts while retaining the UMI and CB of the transcripts. The second PCR is used to append Illumina adapters (P5, P7), dual index barcodes to identify the sample, and sequencing primer binding sites.
PCR1
We designed biotinylated primers to tile across the entire mitochondrial transcriptome. Twelve primer mixes were created using 2-11 of these primers at a concentration of 1 μM each (10-fold relative to the final concentration). The SMART-AC primer, which is common to all PCR1 reactions, was included in each primer mix at 10 μM (Figure 1B, Supplementary Figure 4, Supplementary Table 2).
As a template, WTA products from an individual sample were pooled and diluted to be used at 20 ng in a total volume of 10 μl per reaction. Next, 2.5 μl of the primer mix and 12.5 μl of KAPA HiFi Hotstart ReadyMix (Fisher Scientific KK2602) were added to the template, and PCR was performed using the following conditions: initial denaturation at 95°C for 3 minutes, followed by 6 cycles of 98°C for 20 seconds, 65°C for 15 seconds, and 72°C for 3 minutes, ending with a final extension at 72°C for 5 minutes. There were 12 reactions in total for each sample, as each primer mix is used in a single reaction.
Following amplification, the PCR product is pooled and purified with 0.8x AMPure XP beads (Beckman Coulter A63881). Pooling ratios of PCR1 products were empirically determined to obtain a more equal distribution of reads across the mitochondrial transcriptome (Supplementary Figure 4D). Using Streptavidin-coupled Dynabeads, only biotinylated fragments containing the amplicons of interest are captured (following manufacturer’s instructions, ThermoFisher 60101). Dynabeads/DNA-complex is eluted in 23 μl H2O and used as a template for the second PCR.
PCR2
To add Illumina adapters (P7, P5), index barcodes to identify the library (i7, i5), and sequencing primer binding sites to the fragments, a second PCR is performed using 23 μl of streptavidin-bound template, with 2 μl of a 5 μM primer mix (N70D_P7_BCXX and N70_P5_BCXX; Supplementary Table 2) and 25 μl PFU Ultra II HS 2xMasterMix (ThermoFisher Q32854). The parameters used for PCR2 are an initial denaturation at 95°C for 2 minutes, then 6 cycles of 95°C for 20 seconds, 65°C for 20 seconds, and 72°C for 2 minutes, and then a final extension at 72°C for 5 minutes. After the second PCR, the streptavidin beads are magnetized to collect the supernatant, from which DNA is purified with 0.7x AMPure XP beads. After elution in 22 μl TE, the supernatant is transferred to a new tube and saved for sequencing.
The resulting libraries are similar to Seq-Well scRNA-seq libraries but with targeted integration of the sequencing primer binding site at the regions of interest. The libraries were generally 2-10 ng/μl with sizes ranging from 250-1000 bp. Libraries were sequenced on the Illumina NovaSeq SP 300 cycle kit with the forward strand workflow and the CR1P primer, using 20 cycles for Read 1, 264 cycles for Read 2, and 2 x 8 bp index barcodes.
Mitochondrial alteration enrichment for MAESTER from 10x Genomics
Enrichment of mitochondrial transcripts from 10x Genomics 3’ v3 cDNA was very similar to the protocol for Seq-Well or Drop-Seq described above. The main differences are the use of primer sequences specific to 10x and the omission of the biotin enrichment step (Supplementary Figures 1, 3).
PCR1
We designed primers to tile across the entire mitochondrial transcriptome. Twelve primer mixes were created using 2-11 of these primers at a concentration of 1 μM each (10-fold relative to the final concentration). A barcoded GoT-P5-i5-BCXX primer was included in each primer mix at 10 μM for sample indexing (Supplementary Figure 4, Supplementary Table 3).
As a template, cDNA products from an individual sample were pooled and diluted to be used at 20 ng in a total volume of 16 μl per reaction. Next, 4 μl of the primer mix and 20 μl of KAPA HiFi Hotstart ReadyMix (Fisher Scientific KK2602) were added to the template, and PCR was performed using the following conditions: initial denaturation at 95°C for 3 minutes, followed by 6 cycles of 98°C for 20 seconds, 65°C for 15 seconds, and 72°C for 3 minutes, ending with a final extension at 72°C for 5 minutes. There were 12 reactions in total for each sample, as each primer mix is used in a single reaction.
Following amplification, the PCR product is pooled and purified with 1x AMPure XP beads to remove primers (Beckman Coulter A63881). Pooling ratios of PCR1 products were empirically determined to obtain a more equal distribution of reads across the mitochondrial transcriptome (Supplementary Figure 4D, all volumes multiplied by 1.6). After AMPure XP purification, the pooled PCR1 product was eluted in 20 ul H2O.
PCR2
To add Illumina adapters (P7, P5), index barcodes to identify the library (i7, i5), and sequencing primer binding sites to the fragments, a second PCR is performed using 18 μl of the eluate, with 2 μl of a 5 μM primer mix (P5-generic and XV-P7-i7-BCXX; Supplementary Table 3) and 20 μl KAPA HiFi Hotstart ReadyMix (Fisher Scientific KK2602). The parameters used for PCR2 are an initial denaturation at 95°C for 3 minutes, then 6 cycles of 98°C for 20 seconds, 60°C for 30 seconds, and 72°C for 3 minutes, and then a final extension at 72°C for 5 minutes. After the second PCR, the DNA is purified with 0.8x AMPure XP beads. The DNA is eluted in 20 μl TE, the supernatant is transferred to a new tube and saved for sequencing.
The resulting libraries are similar to 10x scRNA-seq libraries but with targeted integration at the regions of interest. The libraries were generally 2-100 ng/μl with sizes ranging from 300-1500 bp. Libraries were sequenced on the Illumina NovaSeq SP 300 cycle kit with 28 cycles for Read 1, 256 cycles for Read 2, and 2 x 8 bp index barcodes. For the NovaSeq Forward Strand Workflow, no custom sequencing primers are required. For the NovaSeq Reverse Complement Workflow, custom index primers should be used instead of the Illumina standards (10x-Ci7P and 10x-Ci5P, Supplementary Table 3).
10x Multiome sequencing
To assess the recall of MAESTER in identifying mitochondrial variants, we conducted an experiment to genotype both mtDNA and mtRNA in the same individual cells. We performed DOGMA-seq13 with LLL lysis on peripheral blood mononuclear cells from a consented healthy donor. As DOGMA-seq utilizes the 10x Genomics Multiome ATAC + Gene Expression kit to capture both DNA (via ATAC) and RNA from the same individual cells, our experimental framework provided the means to verify the detection of mutations on mtDNA via RNA. Though this lysis buffer yielded a low mtRNA copy number as previously described13, we amplified mitochondrial transcripts from the full-length cDNA using MAESTER. We sequenced the corresponding ATAC (containing mtDNA), gene expression, and amplified mtRNA libraries separately.
Bulk ATAC-sequencing
K562 cells from the same passage used for the cell line mixing experiments were analyzed by bulk ATAC-seq for orthogonal validation of mtDNA variants. Cells were washed in PBS, pelleted by centrifugation and ~12,000 cells were lysed and tagmented in 1x TD buffer, 2.5 μl Tn5 (Illumina), 0.1% NP40 and 0.3x PBS in a 50 μl reaction volume as described20. Samples were incubated at 37°C for 30 min at 300 rpm. Tagmented DNA was purified using the MinElute PCR kit (Qiagen). The complete eluate underwent PCR with initial extension and 5 cycles of pre-amplification using indexed primers and NEBNext High-Fidelity 2X PCR Master Mix (NEB). Then, the number of additional cycles was assessed by quantitative PCR using SYBR Green. Seven additional cycles were run. The final library was purified using a MinElute PCR kit (Qiagen). Libraries were sequenced on a NextSeq 500 instrument with paired-end 38 bp reads and dual library indices of 8 bp each.
T-cell Receptor Enrichment to linK clonotypes by sequencing (TREK-seq)
We adapted a previously described TCR sequencing protocol14, developed for Seq-Well, for use with 10x Genomics 3’ v3 cDNA libraries (Supplementary Figure 14). The modifications to the original protocol are as follows: in the TCR enrichment master mix, we added PartialRead1 and PartialTSO primers at a final concentration of 1.25 μM each (Supplementary Table 3). For amplification of TCR transcripts following enrichment, we used the same primers at a final concentration of 0.4 μM each. For the final PCR, to add the Illumina P5 and P7 sequences we used UPS2-N70x and 10X_SI-PCR_P5 primers at a final concentration of 0.2 μM each. The libraries were sequenced using a 150 cycle kit on the Illumina MiSeq loaded at a final DNA concentration of 10 pM, aiming for a cluster density of roughly 450k/mm2. 28 cycles were used for Read 1, which reads the cell barcode and UMI. 150 cycles were used for Index 1, which reads the TCR region. TCRα and TCRβ-specific custom sequencing primers were used for Index 1 at a final concentration of 2.5 μM (aTCR-Seq and bTCR-Seq, Supplementary Table 3).
Single-cell RNA-seq read processing
For Seq-Well, sequencing data was demultiplexed using bcl2fastq2. Read 1 yielded 20 bp reads (12 bp CB and 8 bp UMI), Read 2 yielded 56 bp reads (transcript sequence) and i7 and i5 indices to identify the library were 8 bp each. Reads associated with CBs occurring less than 100 times were removed, and the list of remaining CBs was used to generate Read 2 fastq files in which the library barcode, the CB, and the UMI were appended to the read identifier. For 10x, scRNA-seq data was processed using cellranger mkfastq to demultiplex into fastq files and cellranger count to quantify gene expression.
To generate the reference genome, we used hg38 sequences and annotations (v99) from Ensembl with the addition of RNA18S and RNA28S annotations from UCSC. Annotations were filtered using cellranger mkgtf with recommended attributes as well as the gene biotypes gene_biotype:Mt_rRNA and gene_biotype:rRNA. The reference genome was then generated with cellranger mkref which includes STAR indexing. To align Seq-Well scRNA-seq data to this reference, we used STAR with the options --outSAMtype BAM SortedByCoordinate and --quantMode TranscriptomeSAM. To align 10x data scRNA-seq data to this reference, we used cellranger count which implements STAR.
MAESTER read processing
MAESTER fastqs include Read 1 encompassing the CB and UMI (20 bp for Seq-Well, 28 bp for 10x), Read 2 covering mitochondrial transcript sequences (264 bp for Seq-Well, 256 bp for 10x), and 2 x 8 bp for dual-indexed library barcodes. We used Illumina bcl2fastq for demultiplexing with both indices. Reads associated with CBs occurring less than 100 times were removed, and the list of remaining CBs was used to generate Read 2 fastq files in which the library barcode, the CB, and the UMI were appended to the read identifier. We trimmed the first 24 bp from these fastqs using homerTools to avoid using primer binding sequences for variant calling. Next, we aligned the fastq files with STAR (--outSAMtype BAM SortedByCoordinate) to the same hg38 reference genome we used for scRNA-seq alignment above. More than 90% of MAESTER reads aligned to chrM. See Supplementary Figure 5 for an overview of these procedures.
maegatk - mitochondrial genome variant calling
To facilitate the analysis of MAESTER data, we developed maegatk, a Python package, as an extension of our previously described mgatk pipeline8. Maegatk specifically handles technical biases implicit in high-throughput scRNA-seq to facilitate the identification of mtDNA variants. First, maegatk takes inputs of a single-cell bam file following the 10x Genomics SAM tag conventions, a valid list of CBs, and more than 20 customizable command-line arguments. Next, the software collapses duplicate reads based on UMI, start position, and CB. Unlike most existing variant calling pipelines, including GATK and mgatk that select a representative read based on highest mean base quality score, maegatk identifies the most likely consensus nucleotide across sequencing read replicates via the CallMolecularConsensusReads (v1.1) tool from fgbio. Further, maegatk provides a --min-reads command-line argument that specifies the minimum number of sequence reads needed for a UMI to be considered for variant calling. This workflow minimizes artifacts due to PCR amplification and sequencing error compared to the standard Picard MarkDuplicates. By calling maegatk-indel, our software enables indel calling by implementing FreeBayes on a per-cell basis, which we validated using simulated mtRNA read data (Supplementary Figure 6). After consensus read deduplication, per-cell, per-position nucleotide counts are enumerated and used for downstream analysis.
For use in our analysis with maegatk, we ensured that all bam files contained the CB and UMI SAM tags according to 10x conventions (CB:Z and UB:Z). We selected reads aligning to chrM and merged bam files from scRNA-seq and MAESTER (Aligned.sortedByCoord.out.bam from STAR or possorted_genome_bam.bam from cellranger). We generated a list of CBs by intersecting CBs with ≥100 alignments to chrM and high-quality CBs from scRNA-seq. Maegatk was then executed with the options --input merged.bam --mito-genome chrM.fa --barcodes CBs.txt --min-reads 3. The last option specifies only UMIs with three reads are used to increase confidence in variant calls. Upon completion, maegatk saves mutation calls as a maegatk.rds file in the SummarizedExperiment format21 for convenient intersection with other modalities and downstream analysis in R.
Multiome analysis
Multiome libraries (ATAC and gene expression) were aligned using CellRanger-Arc to a modified hg38 reference genome with regions of mitochondrial genome homology hard-masked in the nuclear genome8. Mitochondrial DNA from the ATAC library was processed using the mgatk workflow8. In parallel, we applied MAESTER and maegatk to enrich mitochondrial variants from the mRNA library using an unmodified hg38 reference genome. Next, we identified a “gold-standard” dataset of variants from the mgatk ATAC library and examined the concordance of variants that were covered at a minimum of 5x in both DNA (ATAC) and RNA (MAESTER) within individual cells. For a heteroplasmy threshold value X (as shown on the x-axis of Figure 1C), we quantified the recovery of variants by MAESTER as the number of variants that exceeded X in the MAESTER library over the number of variants that exceeded X in the ATAC library.
Single-cell RNA-seq clustering and cell type annotation
For Seq-Well and 10x scRNA-seq data alike, we filtered for cells with ≥2,000 UMIs, ≥1,000 genes, ≤20% alignment to rRNA genes, and ≤20% alignments to genes on chrM. Genes from chrX and chrY were removed from the count matrix. Next, we used Seurat for standard scRNA-seq processing steps including the functions NormalizeData, FindVariableFeatures, ScaleData, and RunPCA (similar to https://satijalab.org/seurat/archive/v3.2/pbmc3k_tutorial.html)22. We implemented graph-based clustering with FindNeighbors with 6 PCA dimensions and FindClusters with a resolution of 0.05-0.1, as we only aimed to distinguish K562 and BT142 cells. We determined the top 10 cell type-specific genes by fold change using the FindAllMarkers function with only.pos = TRUE, min.pct = 0.25 and logfc.threshold = 0.25.
For cell line mixing experiments, we used decontX from the R package celda to remove cells with high ambient RNA23. We supplied the decontX function with the count matrix to calculate per-cell contamination scores. We also scored each cell for both cell type-specific gene signatures using the Seurat function AddModuleScore. Finally, we excluded cells that exceeded a contamination score of 0.05 and had a high module score for both cell type-specific signatures. For Seq-Well, this removed 218/2525 (8.6%) of cells, with 1387 K562 and 920 BT142 cells remaining. For 10x, this removed 112/2778 (4.0%) of cells, with 1310 K562 and 1356 BT142 cells remaining.
For the clonal hematopoiesis sample, we used the cell type annotations that were established using a Random Forest Classifier based on healthy donor populations (Griffin et al., manuscript under review). Further annotation of T and NK cell subsets was done by evaluating cluster-specific gene expression using the FindAllMarkers function with logfc.threshold = 0.25, min.pct = 0.1, test.use = "roc", return.thresh = 0.4, only.pos = TRUE (cluster-identifying marker genes are on https://github.com/petervangalen/MAESTER-2021/tree/main/5_TCR-Seq). Filtering for high-quality cells, dimensionality reduction, and removal of cells with ambient RNA were performed using similar procedures as we used for the cell line mixing experiments except for decontX.
Identification of informative mtDNA variants
To establish clonal relationships between cells, we first select informative variants, and then select cells that are positive for any of these informative variants (i.e. have a VAF of >1%). For informative variant selection, we first calculated an allele frequency matrix of all possible variants (rows) and cells (columns) from the output of the maegatk software. The total number of possible variants is 3x16,569+1=49,708 because the mitochondrial genome (NC_012920) is 16,569 bp with three possible variants each, except base 3,107, which has four possible variants (A, C, T, G) because the reference is N. Next, we generated a table with features for every variant: the mean allele frequency, mean coverage, mean quality score, and the VAF in percentiles of rank sorted cells or the number of cells exceeding a chosen VAF. This allowed us to select informative variants by applying filters such as a mean coverage of >20, mean quality of >30, VAF of <10% in at least 10% of cells, and a VAF of >90% in at least 10% of cells to distinguish cell lines. For the clonal hematopoiesis sample, we selected variants with a mean coverage of >5 per cell, mean quality of >30, VAF of 0% in at least 90% of cells, and a VAF of >50% in at least 10 cells (Supplementary Figure 11). This allowed us to use 26 variants to identify the 23 largest clones, which together made up 14.9% of cells. Selected informative variants were highly enriched for transitions vs. transversions, as expected (>89%). Additional filters to remove artifacts were included for subclone identification, for example, variants informing K562 subclones should be absent in BT142 cells. The classification of K562 and BT142 cells by mtDNA variants was determined by the sum of calls at all homoplasmic variants for either cell line (Supplementary Figure 7G-H).
Having identified informative variants, we assessed their VAFs in single cells by UMAP visualization (Figures 1E, 2C, Supplementary Figures 8B, 12). We used a VAF threshold of 1% to consider a cell positive for an mtDNA variant. We combined highly correlated variants into groups or clones (e.g. 10158T>A and 6293T>C, Figure 2B). To visualize clonal structures in VAF heatmaps, we sorted clones (rows) by their size, and sorted cells (columns) within each clone from high to low VAF or clustered the cells by Pearson correlation (Figure 1H, 2B, 2I, Supplementary Figure 8E, 11C).
Bulk ATAC-seq analysis
For the analysis of ATAC-seq as an orthogonal validation of the presence of mtDNA variants in K562 cells, we aligned demultiplexed fastq files to hg38 using STAR. We subsetted the bam file for alignments to the mitochondrial genome and used Picard-Tools MarkDuplicates to remove duplicates. Next, we ran bam-readcount with the option -w 5 to generate a table of read metrics for every position along the mitochondrial genome. From this table, we extracted informative K562 variants that were identified by MAESTER. We then compared the VAF determined from mRNA transcripts captured by MAESTER to the VAF determined from mtDNA fragments captured by bulk ATAC-seq, the latter being calculated as the number of reads supporting the variant allele over the total sequencing depth at that position.
TREK-seq analysis
Following sequencing of enriched TCR regions, the MiSeq run was demultiplexed using bcl2fastq v2.20.0.422 and a SampleSheet with 150xN as the index sequence. CDR3 sequences were aligned as outlined previously14. Briefly, Hamming errors in cell barcodes were repaired using a whitelist of cell barcodes generated from whole transcriptome sequencing data and a single-base error tolerance. UMIs for each cell barcode were then collapsed with a single-base error tolerance. The repaired reads were then aggregated by CB and UMI, and UMIs with fewer than ten total reads were discarded. The remaining reads were mapped against TCRV and TCRJ IMGT (http://imgt.org/) reference sequences with IgBlast. CDR3 sequences were called by identifying the 104-cysteine and 118-phenylalanine according to IMGT references and translating the amino acid sequences in between. UMIs with a V-gene consensus frequency of less than 0.9 were discarded. Processed TCR sequences were paired with the single-cell transcriptome data by matching of cell barcodes. We recovered TRB in 5,288 (63.1%) and TRA in 4,175 (49.8%) of the total 8,382 T-cells. If multiple TRA or TRB sequences were detected for a single cell barcode, then the corresponding sequence with the highest number of UMIs and raw reads was retained. Next, we stringently selected TRB calls present in at least 10 cells and subsequently selected TRA calls present in at least 10 cells. We visualized the overlap of cells with both TRB and TRA calls and calculated the Adjusted Rand Index using the R package mclust (Supplementary Figure 15). To test overlap with mtDNA variant calls, we selected TRB clones with an mtDNA variant in at least 5 cells and vice versa. These filtered TRB and mtDNA clonal determinations are visualized in Figure 2F-G and intersected with cell type classification and gene expression in Figure 2H-I.
Genotyping of Transcriptomes analysis
To analyze reads from the GoT analysis for the clonal hematopoiesis sample, we utilized IronThrone-GoT (https://github.com/landau-lab/IronThrone-GoT)3. We provided the IronThrone-GoT function with fastq files and a whitelist of CBs for 10x 3′ v3 scRNA-seq. From the summTable, we selected high-confidence transcript calls by filtering for UMIs that were sequenced ≥3x with ≥3x more wild-type than mutant calls or vice versa. We then generated a table with CBs, the number of wild-type transcripts, and the number of mutant transcripts per cell. These calls were intersected by CBs with cells of different subclones, as shown in Supplementary Figure 16B-D.
Pseudotime analysis
We assigned pseudotime values to cells using the R slingshot library16. We selected relevant cell types for four differentiation trajectories from the clonal hematopoiesis sample and provided the slingshot function with the UMAP coordinates. The predicted trajectory (black curve) and assigned pseudotime values (colors) are shown in Supplementary Figure 16E, which is an enlargement of Figure 2A. The same pseudotime values are used for the horizontal axis of adjacent density plots.
Functional annotation of mitochondrial variants
Mitochondrial genes encode factors involved in the respiratory chain. Variants that impact function are thus expected to alter energy metabolism. Since genes required for mitochondrial biogenesis and replication reside in the nuclear genome, mtDNA variants are less likely to impact the intrinsic replication rate of the mitochondrion in which the mutation occurs and hence the heteroplasmy level. However, beneficial/detrimental mutations could affect the fitness of the host cell and the potential functional impact of variants that are used to establish lineage relationships should be assessed in this context. We generated a table of mitochondrial variants, their effects on the resultant proteins, and potential functional consequences by merging the Ensembl Variant Effect Predictor (VEP) (https://useast.ensembl.org/info/docs/tools/vep/index.html) and MitoMap (https://www.mitomap.org/MITOMAP). First, a five-column table of variants was generated using the reference mitochondrial genome (https://www.genome.jp/dbget-bin/www_bget?-f+refseq+NC_012920), with columns as follows: Chromosome number (MT), starting nucleotide, ending nucleotide, substitution (e.g., A/T), and strand (+), as described in https://useast.ensembl.org/info/website/upload/var.html. This format is used as the default input for VEP. Second, VEP was run using the variant table as input via the web interface to predict the effect of variants on genes, proteins, and regulatory regions. Additional configurations included ‘Protein’ and ‘UniProt’ (identifiers), ‘Identify canonical transcripts’ (transcript annotation), and ‘Protein domains’ (protein annotation). The same results can be acquired by running VEP on the command line with the following code:
./vep --af --appris --biotype --buffer_size 500 --canonical --check_existing --distance 0 -- domains --mane --polyphen b --protein --pubmed --regulatory --sift b --species homo_sapiens -- symbol --transcript_version --tsl --uniprot --cache --input_file [input_data] --output_file [output_file]
Third, the VEP output was further processed to retain only the following columns prior to integration with MitoMap data: Uploaded_variation, Location, Allele, Consequence, SYMBOL, Gene, Feature_type, Feature, BIOTYPE, Amino_acids, Codons, SIFT, PolyPhen. Finally, Disease and Polymorphism data were downloaded from the API resources in MitoMap in VCF format (https://www.mitomap.org/foswiki/bin/view/MITOMAP/Resources). These data were merged with the VEF data using the dplyr package in R.
Software used for analysis and figures
Geneious Prime version 2019.1.3 with Primer3 was used for primer design. Read processing was performed using command-line tools including bcl2fastq v2.20.0, Samtools version 1.8, cellranger 3.1.0, homerTools 4.10, STAR version 2.6.0c, and IronThrone-GoT version 1.0. Quality controls and downstream analyses were performed with R for Statistical Computing version 3.6.1 with RStudio version 1.2.5042. We used the tidyverse version 1.3.0 collection of packages including ggplot2 version 3.3.2, Seurat version 3.2.2, SummarizedExperiment 1.24.0 for maegatk output, celda version 1.5.6 for decontX, and slingshot version 2.2.0 for trajectory analyses as described in the provided Github scripts. Mclust version 5.4.9 was used for ARI analysis. Maegatk is a python package and therefore python was utilized as part of maegatk. FlowJo was used for FACS analysis. We also used GraphPad Prism, Microsoft Excel version 16.56, and Adobe Illustrator 2021 for additional statistical analyses and visualization.
Supplementary Material
Supplementary Table 1 This table includes all variants that were used to inform clonal structures in this manuscript, along with functional annotations. This is an excerpt from a table with annotations for every possible mitochondrial variant (see Methods).
Supplementary Table 2 This table includes all oligo sequences used for MAESTER on Seq-Well scRNA-seq cDNA.
Supplementary Table 3 This table includes all oligo sequences used for MAESTER and TREK-seq on 10x Genomics 3’ v3 scRNA-seq cDNA.
Acknowledgements
We thank patients for donating cells, Charles Couturier and Martin Villanueva from the Alex Shalek lab for sequencing, Antonia Kreso, Volker Hovestadt, and Ang Andy Tu for helpful discussions, and Patricia Rogers for technical support. P.V.G., A.A.L., and B.E.B. are supported by the Ludwig Center at Harvard. P.V.G. and V.G.S. are supported by the Harvard Medical School Epigenetics & Gene Dynamics Initiative. P.V.G. is supported by the National Institutes of Health (NIH) R00 Award (CA218832), Gilead Sciences, and the Bertarelli Rare Cancers Fund, and is a Glenn Foundation for Medical Research and AFAR Grant for Junior Faculty awardee. T.E.M. is supported by the American Brain Tumor Association Basic Research Fellowship in honor of Joel A. Gingras, Jr. T.E.M. and J.A.V. are supported by the UK Brain Tumour Charities Future Leaders Award, GN-000701. C.A.L. is supported by a Stanford Science Fellowship and Parker Scholar award. A.T.S. is supported by the National Institutes of Health grant U01CA260852, the Cancer Research Institute Technology Impact Award, and a Pew-Stewart Scholars for Cancer Research Award. LSL is supported by an Emmy Noether fellowship by the German Research Foundation (DFG, LU 2336/2-1). J.C.L and D.M.M were supported in part by the Koch Institute Support (core) NIH Grant P30-CA14051 from the National Cancer Institute, as well as the Koch Institute - Dana-Farber/Harvard Cancer Center Bridge Project and the Food Allergy Science Initiative at the Broad Institute. V.G.S. is supported by the New York Stem Cell Foundation (NYSCF), a gift from the Lodish family to Boston Children’s Hospital, and NIH grant R01 DK103794. V.G.S. is a NYSCF-Robertson Investigator.
Footnotes
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Code availability
maegatk is available at https://github.com/caleblareau/maegatk and a table with functional annotation of all possible mtDNA variants is available at https://github.com/EDePasquale/Mitochondrial_variants. Computational analyses are described on https://github.com/petervangalen/MAESTER-2021.
Competing interests
B.E.B. discloses financial interests in Fulcrum Therapeutics, HiFiBio, Arsenal Biosciences, and Cell Signaling Technologies. V.G.S. serves as an advisor to and/or has equity in Novartis, Forma, Cellarity, Ensoma, and Branch Biosciences. T.E.M. discloses financial interest in Telomere Diagnostics and Reify Health. A.T.S. discloses financial interests in Immunai and Cartography Biosciences. J.C.L. has interests in Honeycomb Biotechnologies. J.C.L.’s interests are reviewed and managed under the Massachusetts Institute of Technology’s policies for potential conflicts of interest. J.C.L. and the Massachusetts Institute of Technology have filed patents related to the single-cell sequencing methods used in this work. A patent application covering MAESTER has been filed by the Broad Institute of MIT and Harvard. The remaining authors declare no competing interests.
Data availability
Raw and processed data are deposited in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE182685). Single-cell gene expression matrices, mtDNA variant calls and GoT results are available at https://vangalenlab.bwh.harvard.edu/resources/maester-2021/.
References
- 1.Giladi A & Amit I Single-Cell Genomics: A Stepping Stone for Future Immunology Discoveries. Cell vol. 172 14–21 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Acosta J, Ssozi D & Van Galen P Single-Cell RNA Sequencing to Disentangle the Blood System. Arterioscler. Thromb. Vasc. Biol 41, 1012–1018 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nam AS et al. Somatic mutations and cell identity linked by Genotyping of Transcriptomes. Nature 571, 355–360 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Galen P et al. Single-Cell RNA-Seq Reveals AML Hierarchies Relevant to Disease Progression and Immunity. Cell 176, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner DE & Klein AM Lineage tracing meets single-cell omics: opportunities and challenges. Nat. Rev. Genet 21, 410–427 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liggett LA & Sankaran VG Unraveling Hematopoiesis through the Lens of Genomics. Cell 182, 1384–1400 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ludwig LS et al. Lineage Tracing in Humans Enabled by Mitochondrial Mutations and Single-Cell Genomics. Cell (2019) doi: 10.1016/j.cell.2019.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lareau CA et al. Massively parallel single-cell mitochondrial DNA genotyping and chromatin profiling. Nat. Biotechnol 39, 451–461 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu J et al. Single-cell lineage tracing by endogenous mutations enriched in transposase accessible mitochondrial DNA. Elife 8, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Velten L et al. Identification of leukemic and pre-leukemic stem cells by clonal tracking from single-cell transcriptomics. Nat. Commun 12, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes TK et al. Second-Strand Synthesis-Based Massively Parallel scRNA-Seq Reveals Cellular States and Molecular Features of Human Inflammatory Skin Pathologies. Immunity 53, 878–894.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macosko EZ et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mimitou EP et al. Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells. Nat. Biotechnol (2021) doi: 10.1038/s41587-021-00927-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tu AA et al. TCR sequencing paired with massively parallel 3′ RNA-seq reveals clonotypic T cell signatures. Nat. Immunol 20, 1692–1699 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdel-Wahab O et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood 114, 144–147 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Street K et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moran-Crusio K et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20, 11–24 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luchman HA et al. An in vivo patient-derived model of endogenous IDH1-mutant glioma. Neuro. Oncol 14, 184–191 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flavahan WA et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins, and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgan M, Obenchain V, Hester J & Pagès H SummarizedExperiment: SummarizedExperiment container. (2019). [Google Scholar]
- 22.Stuart T et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang S et al. Decontamination of ambient RNA in single-cell RNA-seq with DecontX. Genome Biol. 21, 57 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 This table includes all variants that were used to inform clonal structures in this manuscript, along with functional annotations. This is an excerpt from a table with annotations for every possible mitochondrial variant (see Methods).
Supplementary Table 2 This table includes all oligo sequences used for MAESTER on Seq-Well scRNA-seq cDNA.
Supplementary Table 3 This table includes all oligo sequences used for MAESTER and TREK-seq on 10x Genomics 3’ v3 scRNA-seq cDNA.
Data Availability Statement
Raw and processed data are deposited in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE182685). Single-cell gene expression matrices, mtDNA variant calls and GoT results are available at https://vangalenlab.bwh.harvard.edu/resources/maester-2021/.