Abstract
Patients with estrogen-receptor-positive (ER+) breast cancer, the most common subtype, remain at risk for lethal metastatic disease years after diagnosis. Recurrence arises partly because tumor cells in bone marrow become resistant to estrogen-targeted therapy. Here, we utilized a co-culture model of bone marrow mesenchymal stem cells (MSCs) and ER+ breast cancer cells to recapitulate interactions of cancer cells in bone marrow niches. ER+ breast cancer cells in direct contact with MSCs acquire cancer stem-like (CSC) phenotypes with increased resistance to standard antiestrogenic drugs. We confirmed that co-culture with MSCs increased labile iron in breast cancer cells, a phenotype associated with CSCs and disease progression. Clinically approved iron chelators and in-house lysosomal iron-targeting compounds restored sensitivity to antiestrogenic therapy. These findings establish iron modulation as a mechanism to reverse MSC-induced drug resistance and suggest iron modulation in combination with estrogen-targeted therapy as a promising, translatable strategy to treat ER+ breast cancer.
Introduction
Estrogen receptor-positive (ER+) breast cancer cells in bone marrow typically persist with less differentiated, cancer stem-like cell (CSC) phenotypes that promote resistance to hormone therapy, contributing to bone marrow being the leading site for metastases in ER+ disease (1-3). Overcoming resistance to hormone therapy remains a critical impediment to improving clinical outcomes as over 50% of patients have metastases that tolerate these drugs (4, 5). Multiple cancer cell-intrinsic mechanisms, including estrogen receptor mutations, transcriptional changes, chromatin remodeling, and alterations in microRNAs, contribute to drug resistance (6-10). However, stromal cells in bone marrow and other sites substantially alter drug sensitivities of cancer cells (11). Metastatic cancer cells commonly displace hematopoietic stem cells in bone marrow from mesenchymal stem cells (MSC)-based niches, enabling direct interactions of MSCs with ER+ breast cancer cells (12). Past studies show contradictory effects of MSCs on survival and drug resistance of cancer cells. MSCs may promote resistance of cancer cells through mechanisms including activation of PI3K/Akt/mTOR and/or MAPK pathways and transitions to CSCs (13-17), but others show MSCs may increase sensitivity of cancer cells to some therapies (18). Identifying processes through which MSCs regulate adaptations of ER+ breast cancer cells to bone marrow will advance knowledge of tumor-stromal interactions in this metastatic site and open new paths to increase drug sensitivity of cancer cells.
Past studies identified subsets of CSCs in breast cancer and further subdivided these cells based on molecular markers and cell functions. Mesenchymal-CSCs (M-CSCs), the CD44high/CD24low subpopulation, display migratory, invasive, and drug-resistant phenotypes, while epithelial-CSCs (E-CSCs) display a high aldehyde dehydrogenase (ALDH+), proliferative phenotype (19). Breast cancer cells can switch from E-CSCs to M-CSCs through an epithelial to mesenchymal transition (EMT) process or acquire a hybrid-CSC phenotype (CD44high/CD24low/ALDHhigh) with high plasticity, invasiveness, drug resistance, and metastatic potential (20). Unfortunately, standard antiestrogenic treatments often enrich for CSCs (21). A predominant regulator of CSCs is iron, an essential nutrient involved in cell respiration, DNA synthesis, epigenetic modifications, and oxygen transport (22, 23). Cancers exhibit dysregulated iron homeostasis with increased intracellular iron leading to greater disease progression, increased resistance to standard therapies, and decreased survival (24, 25). Previous work demonstrates that iron depletion by chelation sensitizes breast cancer cells to standard therapy, potentially through epigenetic modulation, and iron supplementation induces CSC phenotypes through SOX9 and other pluripotency factors (22, 23, 26-28). These observations highlight key functions of iron in CSCs and potential therapeutic vulnerabilities arising from iron dependence.
Here we show that direct co-culture of ER+ breast cancer cells with MSCs increases metastatic potential and M-CSCs while reducing sensitivity to standard therapies, due in part due to an increase in intracellular labile iron. Compounds that disrupt iron homeostasis can reverse many of these phenotypes, suggesting an adjuvant treatment strategy to reduce metastatic ER+ breast cancer.
Methods
Cell culture
We purchased HS5 (CRL-11882), HS27a (CRL-2496), MCF7 (HTB-22), and T47D (HTB-133) cells from the American Type Culture Collection (ATCC, Manassas, VA, USA). Dr. James Rae (University of Michigan, Ann Arbor, MI, USA) provided HCC1428 ER+ breast cancer cells. We maintained HS5, MCF7, and T47D cells in supplemented DMEM and HS27a and HCC1428 cells in supplemented RPMI as previously described (29). We performed short tandem repeats analysis to authenticate cell lines and mycoplasma testing at time of initial passaging.
Lentiviral and transposon vectors
We transduced HS5 and HS27a lines with pLVX click beetle red-Flag IRES-blasticidin (CBRed) and MCF7, T47D, and HCC1428 lines with pLVX click beetle green-Flag-IRES-blasticidin pLVX (CBG), respectively, and selected for stable cells with blasticidin as previously described (30). We engineered MCF7, T47D, and HCC1428 lines to stably express mCherry or blue fluorescent protein (BFP) fused to a nuclear localization sequence (NLS) using recombinant lentiviral vectors and sorted for stably expressing cells using flow cytometry. We transduced MCF7 and T47D lines to stably express pLenti.PGK.LifeAct-GFP.W vector (Rusty Lansford, Addgene plasmid # 51010, University of Southern California, Los Angeles, CA) using lentiviral vectors and sorted for stably expressing cells using flow cytometry (31).
In vitro cell studies
We seeded 12,000 total cells per cm2 with 10% cancer cells and 90% MSCs. 24 hours after seeding, we changed medium to low glucose, phenol-red free DMEM (#11054, Gibco, Thermo Fisher, Grand Island, NY, USA) with 1% fetal bovine serum (FBS, HyClone, ThermoScientific, Waltham, MA, USA), 1% GlutaMAX (#35050, Gibco), 1% Penicillin-Streptomycin (#15140, Gibco), and 10 nM β-Estradiol (#E2758, Sigma-Aldrich, Millipore Sigma, Saint Louis, MO, USA). We named this medium LG/LF. For conditioned medium (CM) assays, we seeded MSCs; waited 4 hours for cells to adhere; removed media; and then added LG/LF media. 24 hours later, we collected CM from MSCs; filtered it through a 0.45 μm filter; and added CM at a 1:1 ratio with fresh LG/LF medium to cancer cells.
Animal studies
The University of Michigan Institutional Animal Care and Use Committee approved all procedures involving animals. We housed mice as previously described (32). We separated breast cancer cells in co-culture conditions from MSCs using human CD326 (EpCAM) Microbeads (#130-061-101, Miltenyi, Auburn, California, USA) and LS columns (#130-042-401, Miltenyi) following manufacturer’s recommendations. To generate systemic metastases, we injected 1 × 105 cancer cells into the left ventricle of 10-12-week old female NSG mice (The Jackson Laboratory, Bar Harbor, ME, USA). We randomly assigned animals to each injection group. We performed bioluminescence using an IVIS Spectrum (Perkin Elmer) as previously described (33). An individual blinded to experimental conditions quantified mice with detectable based on signal above background.
Cell-based bioluminescence imaging
For growth and cytotoxicity curves, we performed dual-dolor bioluminescence imaging of CBRed and CBG using an IVIS Lumina Series III (Perkin Elmer, Waltham, MA, USA) as previously described (34). We exported photon flux data from regions of interest defined in Living Image 4.3.1 (Perkin Elmer) and quantified changes in growth or viability with custom MATLAB code.
For cytotoxicity curves, we treated cells with compounds diluted in LG/LF media for three days. We used ferric ammonium citrate (FAC) (#F5879, Sigma Aldrich), fulvestrant (#S1191, Selleckchem), tamoxifen, deferoxamine (DFO), deferasirox (DFX), RSL-3, salinomycin, and erastin (#13258, #14595, #16753, #17754, #19288, and #13579, Cayman Chemical). We manufactured salinomycin analogs AM5 and AM23 as previously described (35).
Microscopy and image analysis
We performed holographic tomography imaging using a 3D Cell explorer-fluo (Nanolive, Switzerland) and epifluorescence imaging for LifeAct-GFP cancer cells using an Olympus IX73 Inverted Microscope. To quantify changes in cellular morphology, we imported epifluorescence images into MATLAB and quantified them using custom code (31).
Flow cytometry
We separated breast cancer cells in co-culture conditions from MSCs using human EpCAM microbeads as described above for CSC and ER staining experiments. We performed CSC staining and analysis as previously described (36).
We stained for ER-α using [E115]-ChIP Grade (#ab32063, Abcam, Waltham, MA, USA) antibody at the manufacturer’s recommended dilution. We then fixed and permeabilized cells using eBioscience Intracellular Fixation and Permeabilization Buffer Set (#88-8824-00, Invitrogen, Waltham, MA, USA). We based flow cytometry gates on secondary only control.
We measured reactive oxygen species (ROS) using CellROX Green Reagent (#C10444, ThermoFisher), labile iron using BioTracker Far-red Labile Fe2+ dye (#SCT037, Sigma Aldrich), intracellular Zn2+ using BioTracker 515 Green Zn2+ DA Dye (#SCT032, Sigma Aldrich), intracellular Cu+ using BioTracker Green Copper Live Cell Dye (#SCT041, Sigma Aldrich), and relative lipid ROS using BODIPY-C11 reagent (#C10445, ThermoFisher) following manufacturer’s recommendations. We separated signals from cancer cells and stromal cells by GFP-tagged cancer cells for iron, mCherry-NLS-tagged cancer cells for ROS, zinc, and copper stains, and BFP-NLS-tagged cancer cells for lipid ROS.
qRT-PCR and RNA sequencing
We used human EpCAM microbeads to isolate breast cancer cells as described above and extracted RNA using an RNeasy Mini Kit following manufacturer’s instructions (#74104, Qiagen, Hilden, Germany). We performed qRT-PCR with SYBR Green detection using a QuantStudio instrument (Applied Biosystems, ThermoFisher) following manufacturer’s recommendations. We normalized RNA levels to β-Actin control and untreated monoculture of that cell line. Primer sequences for target genes are listed below:
Estrogen receptor (ESR1): 5’-AGGGAAAATGTGTAGAGGGC-3’ and 5’-AGGTGGATCAAAGTGTCTGTG-3’
Progesterone receptor: 5’-GGTGTTTGGTCTAGGATGGAG-3’ and 5’-ACTGGGTTTGACTTCGTAGC-3’
EpCAM: 5’-GTATGAGAAGGCTGAGAT-3’ and 5’-CTTCAAAGATGTCTTCGT-3’
Vimentin: 5’-GGAAACTAATCTGGATTCACTC-3’ and 5’-CATCTCTAGTTTCAACCGTC-3’
ZEB1: 5’-AAAGATGATGAATGCGAGTC-3’ and 5’-TCCATTTTCATCATGACCAC-3’
β-Actin: 5-TGTACGTTGCTATCCAGGCTGTGC-3’ and 5-CGGTGAGGATCTTCATGAGGTAGTC-3’
We performed single end, QuantSeq RNA sequencing using the University of Michigan Advanced Genomics core. We deposited data in the Gene Expression Omnibus database (GEO accession number GSE196312). We analyzed RNA using iDEP (37); imported raw count matrices; and applied a filter using only genes with a minimum of 1 CPM in at least 5 samples. We used rlog to transform counts data and treated missing values as zero. For individual genes, we defined significance as an adjusted p-value of less than 0.05 and absolute log2 fold change >2. EMT genes are defined by the Hallmark EMT pathway, metal-associated genes by genes containing a metal-associated term in their GO pathway descriptions, and ROS/redox-associated genes by genes containing a ROS or redox-associated term in their GO pathway. Finally, we performed DEseq2 with batch correction to quantify fold changes and p-values for each gene and GSEA using MSigDB for pathway enrichment analysis (38, 39).
Western blots
To isolate breast cancer cells from MSCs, we used human EpCAM microbeads as described above. We prepared whole cell lysates using RIPA lysis buffer and performed western blotting as previously described (40). We used antibodies LCN2, FTH-1, and β-Actin (#44058, #3998S, #4970 Cell Signaling Technology, Danvers, MA, USA). We normalized protein levels to their β-Actin loading controls and corresponding monoculture condition.
Statistical analysis
Data represent experiments repeated independently three times. We analyzed and plotted data using MATLAB (Mathworks) and GraphPad Prism (San Diego, CA, USA). We used either a one- or two-way two-sided ordinary ANOVA with Tukey correction for multiple comparisons to analyze differences between groups using an alpha value of 0.05. For box and whisker plots, we defined the bottom and top of a box as first and third quartiles and the band inside the box as the median. We defined ends of the whiskers as 5th and 95th percentiles, respectively.
Code availability
All custom MATLAB codes are available upon request through a material transfer agreement.
Results
Direct contact with bone marrow MSCs increases metastasis and growth of ER+ breast cancer cells
To study interactions between ER+ breast cancer cells and bone marrow MSCs, we cultured cancer cells alone (monoculture) or at a 1:9 ratio of cancer to MSCs (co-culture) without changing total cell numbers in each case (Figure 1a). This ratio recapitulates a bone marrow microenvironment of metastases where normal stromal cells greatly outnumber cancer cells while having sufficient cancer cells to analyze. After seeding cells for 24 hours in full medium, we changed to low glucose (5.6 mM), low FBS (1%) medium for an additional 48 or 72 hours to more closely model physiological levels of serum and glucose.
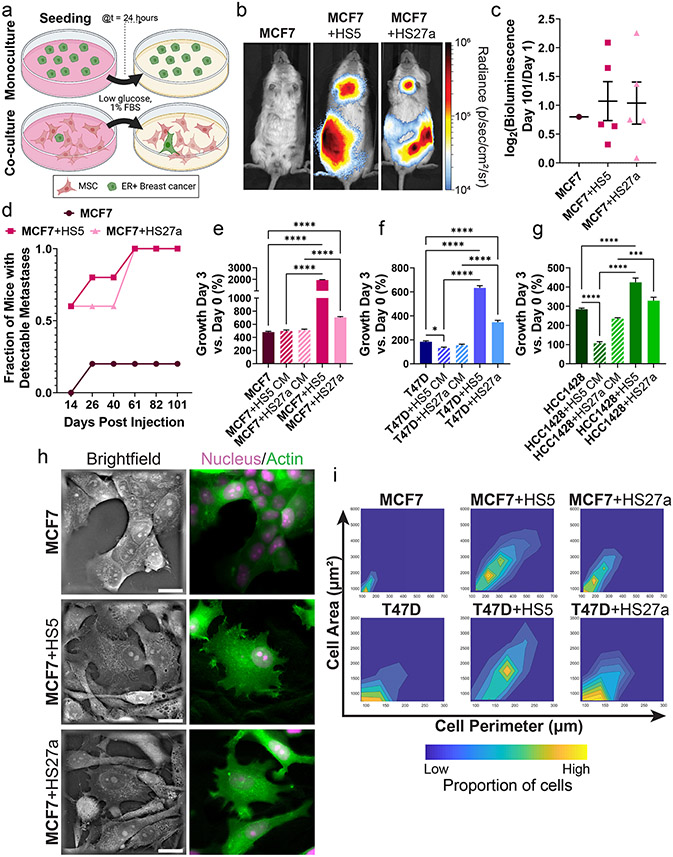
Figure 1. ER+ breast cancer cells co-cultured with MSCs exhibit increased metastases, growth, and mesenchymal morphologies.
For all panels, cell type of interest is bolded in co-culture comparisons. Lines show pair-wise comparisons with *p <0.05,***p<0.001, or ****p<0.0001.
• a: Scheme of the monoculture versus co-culture system for breast cancer cells and MSCs.
• b: Representative whole mouse bioluminescence images of mice 101 days post intracardiac injection. Logarithmic pseudocolor scale depicts range of bioluminescence values with red being highest and blue lowest.
• c: Fraction of mice with metastatic tumor burden detectable by bioluminescence imaging post intracardiac injection over time. N = 5 mice per condition.
• d: Log2 bioluminescence fold change of individual mice on day 101 versus day 1 post intracardiac injection overlayed on the mean value ± standard error of the mean (SEM) for each condition. As MCF7 monoculture only had one mouse with metastases, we did not perform any statistical analyses. N = 5 mice per condition.
• e-g: Mean values ± SEM of percent growth of MCF7 (e), T47D (f), and HCC1428 (g) breast cancer cells grown in monoculture, co-culture, or with conditioned media (CM) from MSCs for three days. N = 8 per condition.
• h: Representative holographic tomography images of MCF7 breast cancer cells marked by LifeAct-GFP and a mCherry nucleus in monoculture versus co-culture with MSCs. Scale bar = 20 μm.
• i: Density plots show morphometric quantification of area and perimeter of cancer cells in monoculture versus co-culture with MSCs. Regions of yellow and blue show higher and lower densities, respectively. N > 200 cells.
We performed intracardiac injection of breast cancer cells into female NSG mice to produce metastases. MCF7 cells previously co-cultured with MSCs all produced metastases after 101 days as analyzed by bioluminescence imaging, while monocultured MCF7 cells formed metastases in only one mouse (n=5, Figure 1b,c). MCF7 cells previously cultured with MSCs displayed increased growth in individual mice versus MCF7 cells grown in monoculture (Figure 1d, S1a).
Next, we quantified growth of cancer cells in vitro in monoculture versus co-culture with MSCs. ER+ cancer cells grown with MSCs displayed increased growth versus monoculture over three days (Figure 1e-g). Cancer cells grown with CM from MSCs failed to recapitulate increases in growth observed in direct co-cultures. Interestingly, CM from HS5 cells decreased growth of T47D and HCC1428 cells relative to monocultures (Figure 1e-g). A prior study reported Inhibitory effects of HS5 CM on growth of ER+ breast cancer cells, mediated in part through interleukin-1 (41).
Cancer cells shifted to more elongated morphology in co-culture with MSCs versus monocultures (Figure 1h, Figure S1b). Holographic tomography images highlighted multiple points of contact between cancer cells and MSCs (Figure 1h). We verified these qualitative observations through morphometric analyses, demonstrating decreased circularity and increased cellular perimeter of ER+ breast cancer cells in co-culture (Figure 1i). These data demonstrate increased metastatic potential and growth in vivo and in vitro for ER+ breast cancer cells grown with bone marrow MSCs. Full effects of MSCs on breast cancer cells require direct cell-to-cell interactions.
MSCs increase resistance of ER+ breast cancer to standard endocrine therapies
We investigated toxicity of two standard antiestrogenic therapies, tamoxifen and fulvestrant, to ER+ breast cancer cells in monocultures or co-cultures with MSCs. We treated cells with increasing concentrations of each drug and quantified relative viability of breast cancer cells and MSCs after three days (Figure 2a-f). Breast cancer cells and MSCs stably expressed CBG or CBRed luciferase, respectively. Since these luciferases require ATP to produce bioluminescence, we can independently quantify cytotoxicity to each cell type in co-cultures by dual-color bioluminescence imaging. Across a range of drug concentrations, cancer cells co-cultured with MSCs showed greater viability than cells grown in monoculture, particularly for MCF7 and HCC1428 cells. Viability of T47D and HCC1428 cells generally increased in co-culture with HS5 versus HS27a MSCs. Interestingly, tamoxifen showed greater toxicity to MSCs than fulvestrant.
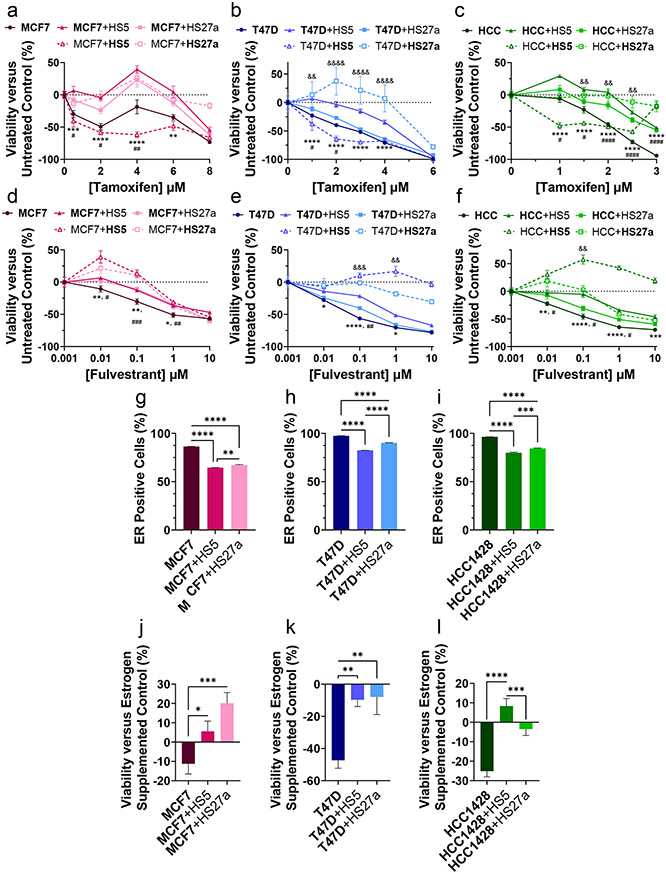
Figure 2. ER+ breast cancer cells co-cultured with MSCs acquire resistance to estrogen targeted therapies.
The cell type of interest is bolded in co-culture comparisons. Lines show pair-wise comparisons with *p <0.05, **p <0.01, ***p<0.001, or ****p<0.0001.
• a-f: Mean responses in cellular viability ± SEM of MCF7 (a, d), T47D (b, e), and HCC1428 (abbreviated as HCC in graph legend; f, c) cells treated for 72 hours with increasing concentrations of estrogen receptor-targeted compounds tamoxifen (a-c) and fulvestrant (d-f). A decrease along the y-axis represents a decrease in cellular viability versus untreated control. Solid lines indicate cancer cells and dashed lines indicate MSCs. Circular symbols indicate monoculture, triangles indicate co-culture with HS5 MSCs, and squares indicate co-culture with HS27a MSCs. Error bars for SEM are smaller than the symbol for some conditions if not evident. * Symbols for p-values indicate the comparison between cancer cells in co-culture with HS5 cells and monoculture, # symbols lines indicate the comparison between cancer cells in co-culture with HS27a cells and monoculture, and & symbols indicate the comparison between cancer cells in co-culture with HS5 cells and HS27a cells. N = 8 per condition.
• g-i: Percentage of ER-presenting MCF7 (g), T47D (h), and HCC1428 (i) cells as measured by flow cytometry. The mean percentage + SEM is plotted for each condition. N = 3.
• j-l: Relative viability of estrogen-depleted MCF7 (j), T47D (k), and HCC1428 (l) cancer cells as a percentage of estrogen supplemented control after 72 hours of estrogen depletion. The mean viability + SEM is plotted for each condition. N = 8.
Both qRT-PCR and flow cytometry revealed reduced ER in cancer cells grown in co-culture versus monoculture (Figure 2g-i, S2a,b,f,g). CM from HS5 cells also has been reported to reduce ER in breast cancer cells (41). Progesterone receptor expression decreased in breast cancer cells in co-cultures with bone marrow MSCs (Figure S2h,i). Furthermore, bone marrow MSCs protected viability of ER+ breast cancer cells cultured without estrogen in charcoal-stripped FBS (Figure 2j-l). Collectively, these data establish that cancer cell interactions with MSCs decrease sensitivity to standard ER+ breast cancer therapies, reduce levels of ER in breast cancer cells, and enable greater survival in estrogen-depleted environments.
Co-culture with bone marrow MSCs shifts ER+ breast cancer cells to hybrid and mesenchymal CSCs
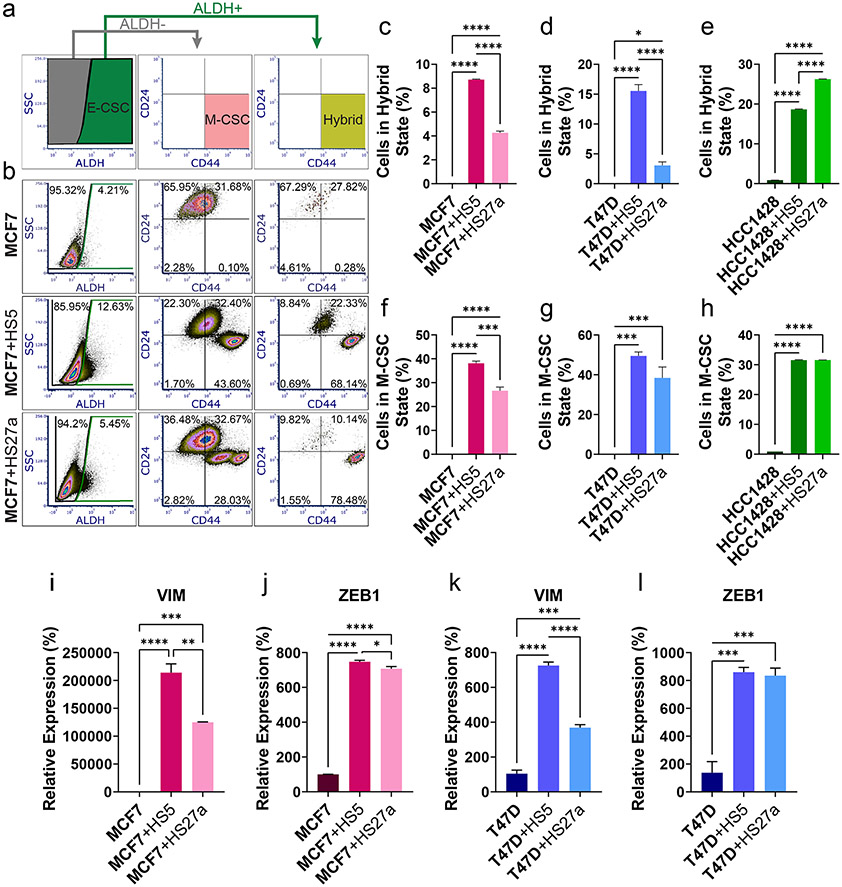
We performed flow cytometry for ALDH activity, CD24, and CD44 to quantify proportions of CSCs (42). ER+ breast cancer cells in monoculture predominantly exhibited non-CSC phenotypes (Figure 3b, S3a-b). In co-culture with bone marrow MSCs, ER+ breast cancer cells overwhelmingly increased hybrid and M-CSC populations (Figure 3c-h). Proportions of E-CSCs did not consistently vary in co-culture versus monoculture except in HCC1428 cells, where co-culture with either MSC line increased E-CSCs (Figure S3c-e). Cancer cells cultured with CM from MSCs did not recapitulate shifts to CSCs seen in co-culture (Figure S3f-n). Furthermore, qRT-PCR showed greater expression of M-CSC markers VIM and ZEB1 in breast cancer cells co-cultured with MSCs (Figure 3i-l). These data indicate that direct interactions with MSCs increase proportions of hybrid and M-CSCs in ER+ breast cancer.
Figure 3. Co-culture with MSCs increases CSC phenotypes in ER+ breast cancer cells.
Cell type of interest is bolded in co-culture comparisons. Lines show pair-wise comparisons with *p <0.05, **p <0.01, ***p<0.001, or ****p<0.0001.
• a: Representative gating scheme for ALDH activity, CD24, and CD44 analyses.
• b: Representative flow cytometry data from MCF7 cells in monoculture versus co-culture with HS5 or HS27a MSCs stained for ALDH activity, CD24, and CD44. N > 10 000 cells.
• c-h: Percentages of hybrid (ALDH+/CD24-/CD44+, c-e) and M-CSC (ALDH-/CD24-/CD44+, f-h) MCF7 (c, f), T47D (d, g), and HCC1428 (e, h) breast cancer cells as measured by flow cytometry. The mean percentage + SEM is plotted for each condition. 3 samples, N > 10 000 cells.
• i-l: Relative expression of vimentin (VIM, i, k) and ZEB1 (j, l) in MCF7 (i, j) and T47D (k, l) ER+ breast cancer cells normalized to monoculture in monoculture or co-culture with MSCs as measured by qRT-PCR. The mean percentage + SEM is plotted for each condition. N=3.
RNA sequencing validates mesenchymal CSC signatures and reveals increases in metal and redox associated genes in co-cultured breast cancer cells
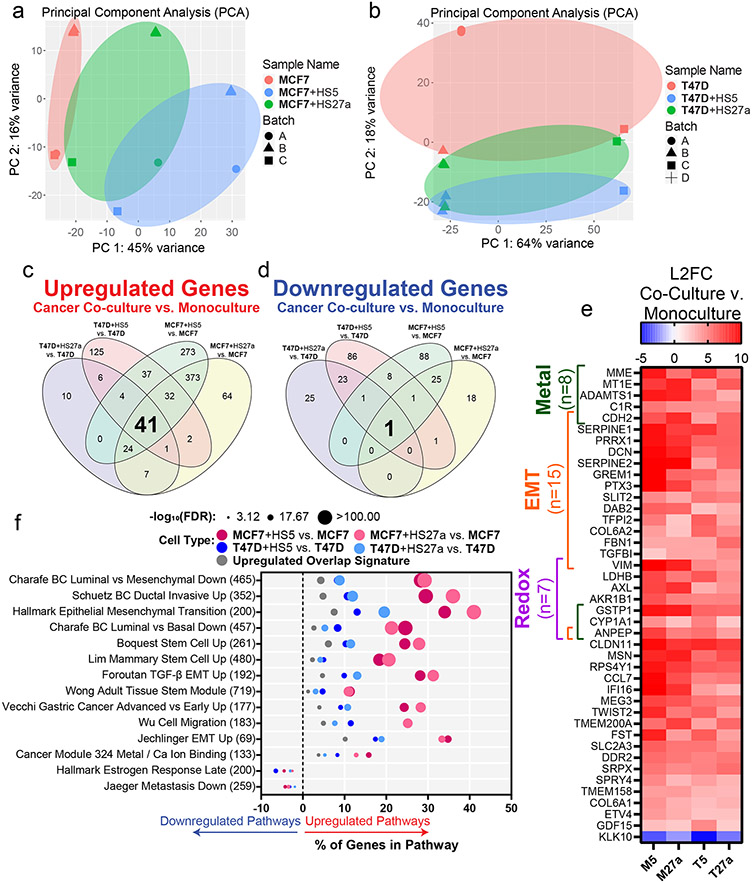
We performed bulk RNA sequencing of MCF7 and T47D ER+ breast cancer cells from monoculture or co-cultures with HS5 or HS27a bone marrow MSCs. PCA analysis of MCF7 and T47D samples showed distinct monoculture and co-culture populations (Figure 4a,b). We defined upregulated and downregulated genes as those with higher or lower expression in co-culture versus monoculture, respectively. We found 41 genes consistently upregulated among all four co-culture versus monoculture comparisons and 1 gene consistently downregulated in all four co-culture versus monoculture conditions (Figure 4c,d). We found upregulation of many metal, EMT, and redox-associated genes in co-culture versus monoculture settings (Figure 4e). Interestingly, the single downregulated gene, kallikrein related peptidase 10 (KLK10), functions as a tumor suppressor in both breast and prostate cancers (43, 44). Relative to HS27a cells, co-culture with HS5 MSCs produced greater transcriptional changes in cancer cells, consistent with previous findings that HS5 MSCs more closely recapitulate primary MSCs and effects on tumor biology (45).
Figure 4. RNA Sequencing of cancer cells in monoculture versus co-culture with MSCs reveals an increase in EMT, metal, and redox genes and pathways.
Cell type of interest is bolded in co-culture comparisons.
• a-b: Principal component analysis of MCF7 (a) and T47D (b) cells in monoculture or co-culture with HS5 or HS27a MSCs before batch correction performed by DESeq2.
• c-d: Venn diagrams show significantly upregulated (c) and downregulated (d) genes in cancer cells in co-culture with MSCs versus monoculture. For individual genes, we defined significance as an adjusted p-value of less than 0.05 and absolute log2 fold change greater than 2.
• e: Heatmap shows the log2 fold change (L2FC) of genes in cancer cells in co-culture versus monoculture: MCF7 co-cultured with HS5 versus MCF7 (M5), MCF7 co-cultured with HS27a versus MCF7 (M27a), T47D co-cultured with HS5 versus T47D (T5), and T47D co-cultured with HS27a versus T47D (T27a).
• f: Significant (false discovery rate (FDR)<0.05) pathways common among all four co-culture versus monoculture conditions with input genes being the upregulated or downregulated genes from each co-culture versus monoculture comparison and the 41 gene upregulated overlap signature from panel e. Larger circles indicate a higher −log10 false discovery rate (FDR) of downregulated genes in that pathway. Pathways upregulated and downregulated in co-culture versus monoculture are right and left of zero on the x-axis, respectively. The number of genes in each pathway are in parentheses to the right of the pathway name.
For each co-culture versus monoculture comparison and the upregulated 41-gene signature, we performed pathway analyses using GSEA/MSigDB of either upregulated or downregulated genes. For upregulated genes, we found pathways associated with increased disease progression, more aggressive breast cancers, stem cells, and EMT (Figure 4f), validating our observations of increased EMT in co-culture (Figure 3). Furthermore, a metal-binding pathway emerged from analyses, consistent with our results showing upregulation of metal genes. Of the downregulated genes, we found pathways associated with less aggressive disease and estrogen responsiveness, reflecting our findings of decreased ER and reliance on estrogen for cancer cells in co-cultures with MSCs (Figure 2).
MSCs in co-culture versus monoculture only had 4 total differentially regulated genes (Figure S4a). Cancer cells supplemented with CM only had 5 consistently differentially regulated genes versus monoculture (Figure S5b), compared to 42 total differentially regulated genes in co-culture versus monoculture, further emphasizing that cell contact is necessary for shifts to EMT. Of the 5 genes, none were significantly higher (p<0.05) in the CM versus monoculture than the co-culture versus monoculture comparison (Figure S5c).
Co-culture with MSCs increases intracellular labile iron in ER+ breast cancer cells
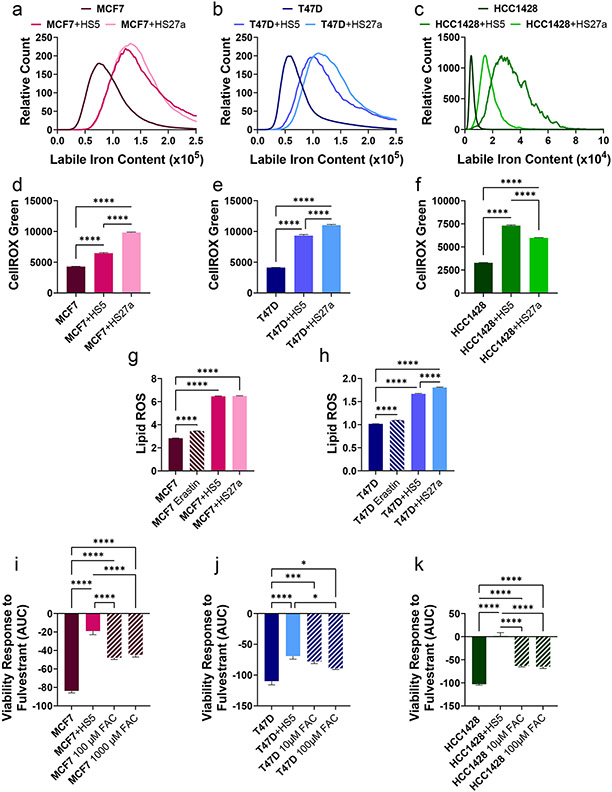
Building upon sequencing data showing an increase in metal-associated genes and pathways, we quantified abundance of selected metals in cancer cells in co-culture versus monoculture. Compared with monocultures, co-culture with bone marrow MSCs increased labile iron in ER+ breast cancer cells in all six co-culture comparisons (Figure 5a-c). We did not observe consistent changes in either intracellular zinc or copper in co-culture versus monoculture (Figure S6a-d). While CM from MSCs modestly increased intracellular labile iron in cancer cells, co-cultures produced a greater effect (Figure S6e-g). HS5 and HS27a MSCs had lower and higher intracellular labile iron relative to ER+ breast cancer cells, respectively. Intracellular labile iron also increased in MSCs co-cultured with ER+ breast cancer cells (Figure S6h-k). Since co-cultured breast cancer cells increased redox-associated genes (Figure 4) and labile intracellular iron can produce ROS (46), we quantified ROS in cancer cells in co-culture. Both ROS and lipid hydroperoxides increased in cancer cells in co-culture relative to monoculture (Figure 5d-h). Next. we cultured cells with increasing amounts of exogenous iron added as FAC. While FAC increased proliferation of T47D and HCC1428 cells in co-culture with MSCs (Figure S7a-c), adding exogenous iron to monocultures decreased sensitivity of all three breast cancer cell lines to fulvestrant, albeit to a lesser extent than co-cultures with MSCs (Figure 5i-k, Figure S7d-f). Overall, co-culture with MSCs increased intracellular labile iron in breast cancer cells and adding exogenous iron reduced sensitivity of ER+ breast cancer cells to antiestrogenic therapy.
Figure 5. Co-culture with MSCs increases labile iron in ER+ breast cancer cells.
Cell type of interest is bolded in co-culture comparisons. Lines show pair-wise comparisons with *p <0.05, **p <0.01, ***p<0.001, or ****p<0.0001.
• a-c: Histograms of labile iron in MCF7 (a), T47D (b), and HCC1428 (c) cancer cells in co-culture with HS5 or HS27a MSCs versus monoculture as measured by flow cytometry. N > 5 000 Cells.
• d-f: Relative levels of intracellular ROS of MCF7 (d), T47D (e), and HCC1428 (f) cancer cells in monoculture or co-culture with MSCs as measured by flow cytometry of CellROX green dye. The mean fluorescence intensity + SEM is plotted for each condition. N> 5 000 cells.
• g-h: Relative levels of lipid ROS as measured by BODIPY-C11 dye in MCF7 (g) and T47D (h) cancer cells in monoculture, in monoculture with Erastin as a positive control, or co-culture with MSCs as measured by flow cytometry. The mean fluorescence ratio + SEM is plotted for each condition. N> 5 000 cells.
• i-k: AUC quantification of cancer cell response in viability to increasing fulvestrant concentrations in monoculture, co-culture, or monoculture with iron supplementation with ferric ammonium citrate (FAC). The mean viability + SEM is plotted for each condition. A decrease along the y-axis indicates an increase in sensitivity to fulvestrant. N=8.
MSCs alter levels of iron-associated proteins in ER+ breast cancer cells
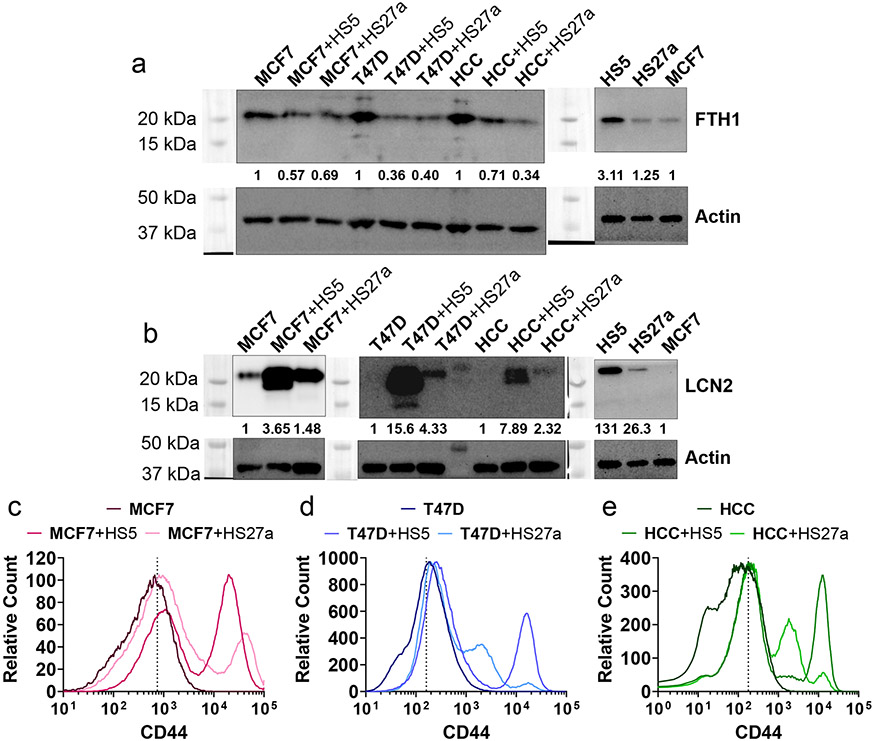
We next investigated molecules that could alter intracellular storage or uptake of iron. Breast cancer cells in co-culture with MSCs exhibited reduced levels of the iron storage protein ferritin heavy chain 1 (FTH1, Figure 6a) and more of the iron-uptake proteins lipocalin 2 (LCN2, Figure 6b) and CD44 (Figure 6c-e). Notably, prior research demonstrated that knockdown of FTH1 produced EMT phenotypes (47), and LCN2 is associated with EMT and other aggressive features of cancer cells (48). CD44 mediates hyaluronate-dependent iron endocytosis and is associated with M-CSCs (42, 49). These data motivate experiments to analyze effects of iron on drug resistance in cancer cells in co-culture with MSCs.
Figure 6. Levels of iron-associated proteins FTH1 decrease and LCN2 and CD44 increase in cancer cells in co-culture with MSCs.
Cell type of interest is bolded in co-culture comparisons.
• a-b: Protein levels quantified by western blot for ferritin heavy chain 1 (FTH1, g) and lipocalin 2 (LCN2, h). Numbers below the protein of interest show relative protein levels.
• c-e: Levels of CD44 measured in single cells by flow cytometry. Dashed line represents gating for positive cells based on CD44-isotype. N>10 000 cells.
Iron modulating compounds sensitize co-cultured ER+ breast cancer cells to antiestrogenic therapy
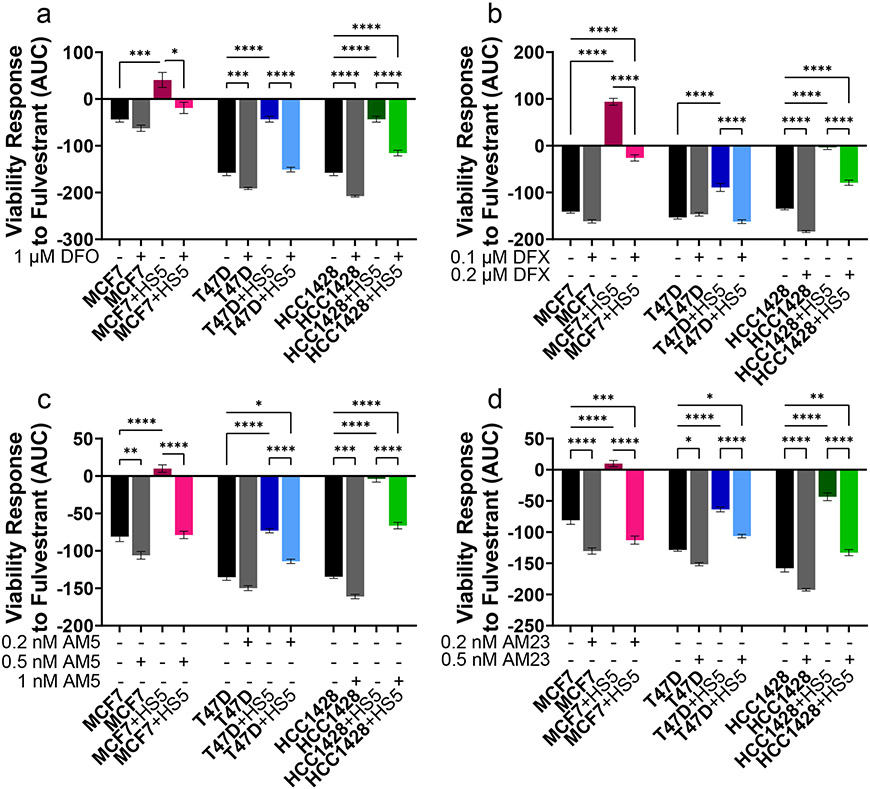
To establish effects of targeting iron on viability of ER+ breast cancer cells in monoculture or co-culture, we treated cells with compounds against various aspects of iron metabolism: ferroptosis activators (erastin or RSL-3), iron chelators DFO or DFX, the CSC inhibition salinomycin, or salinomycin analogs AM5 or AM23 (ironomycin compounds). Ironomycin compounds sequester iron in lysosomes, preferentially causing death of CSCs through a process similar to ferroptosis (35). Neither DFO, DFX, salinomycin, AM5, AM23, nor RSL-3 had consistent effects on viability of ER+ breast cancer cells in co-culture versus monoculture (Figure S8). However, cancer cells in monoculture had increased sensitivity to erastin versus cancer cells co-cultured with MSCs (Figure S8p-r).
We then tested effects of iron-modulating compounds to overcome resistance to fulvestrant of ER+ breast cancer cells in co-culture with MSCs. We chose concentrations of compounds that did not decrease viability of cells without adding fulvestrant. Iron chelators (DFO and DFX) and ironomycin analogs (AM5 and AM23) increased sensitivity of cancer cells in co-culture with MSCs to a range of fulvestrant concentrations (Figure 7, S9). DFO increased sensitivity of cancer cells in monoculture to fulvestrant in T47D and HCC1428 ER+ breast cancer cells. AM23 increased sensitivity of all three ER+ breast cancer cells in monoculture to fulvestrant. These effects were not recapitulated by salinomycin, RSL-3, or erastin, where erastin and RSL-3 further increased drug resistance of cancer cells to fulvestrant in co-culture with MSCs (Figure S10). These data indicate that combination treatment with iron-modulating compounds, specifically iron chelators DFO and DFX and ironomycin analogs AM5 and AM23, sensitize cancer cells in co-culture with MSCs to antiestrogenic therapy.
Figure 7. Iron chelators and ironomycin compounds sensitize ER+ breast cancer cells to fulvestrant.
Cell type of interest is bolded in co-culture comparisons. Lines show pair-wise comparisons with *p <0.05, **p <0.01, ***p<0.001, or ****p<0.0001.
• a-d: AUC quantification of cancer cell response in viability to increasing fulvestrant concentrations in monoculture or co-culture with or without iron chelators Deferoxamine (DFO, a), Deferasirox (DFX, b), AM5 (c), and AM23 (d). AUC responses are calculated from the plots in supplemental figure 12. A decrease along the y-axis indicates an increase in sensitivity to fulvestrant. The mean viability ± SEM is plotted for each condition. N = 6.
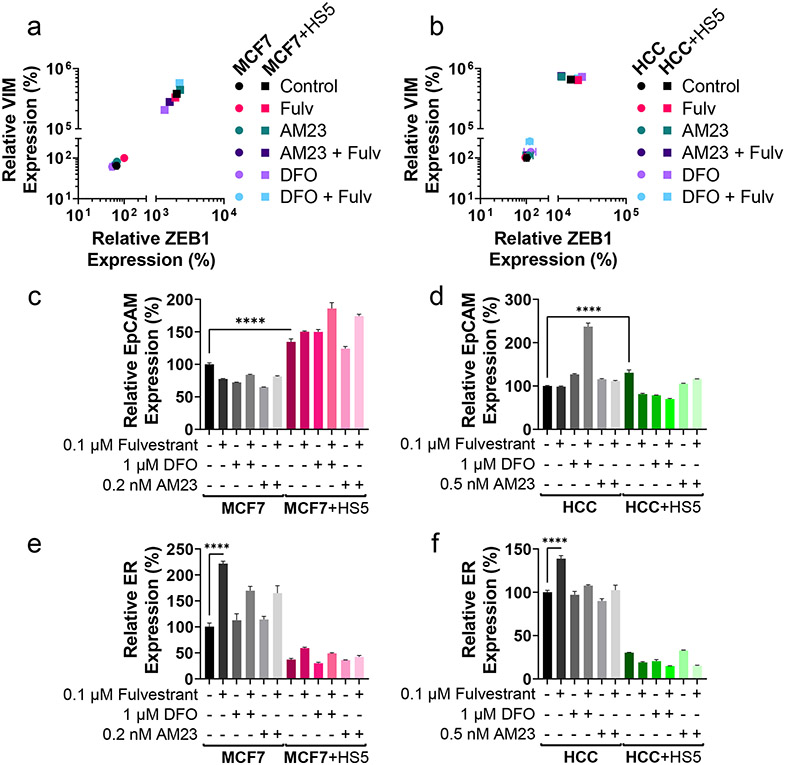
Treatments with iron chelator DFO or ironomycin analog AM23 do not enrich for CSCs in ER+ breast cancer in co-culture or monoculture
As we observed iron chelators DFO and DFX and ironomycin analogs AM5 and AM23 sensitized cancer cells in co-culture with MSCs to fulvestrant, we investigated effects on CSCs because standard hormone therapies often enrich for CSCs (21). Consistent with previous results, co-culture with MSCs elevated VIM and ZEB1 in ER+ breast cancer cells (Figure 8a,b). Co-culture with MSCs increased EpCAM expression and decreased ER expression compared with monoculture (Figure 8c-f). Fulvestrant increased expression of ER in cancer cell monocultures (figure 8e,f) and EpCAM in MCF7 cells co-cultured with HS5 cells (Figure 8c). However, treatments produced only minimal, inconsistent changes in proportions of CSCs and levels of ER in breast cancer cells either in monoculture or in co-culture with MSCs. Unlike many standard cancer therapies, these data indicate that treatment with DFO, AM23, and fulvestrant does not enrich for CSCs in our system.
Figure 8. DFO and AM23 treatment in combination with fulvestrant does not enrich for CSCs.
Cell type of interest is bolded in co-culture comparisons. Lines show pair-wise comparisons with ****p<0.0001.
• a-b: Relative mean expression of EMT markers ZEB1 (x-axis) and VIM (y-axis) in MCF7 (a) and HCC1428 (HCC, b) ER+ breast cancer cells in monoculture (circles) or co-culture (squares) with MSCs in response to treatment with 0.1 μM fulvestrant (FULV), 1 μM DFO, 0.2 nM AM23 (MCF7), 0.5 nM AM23 (HCC1428), or combination. Error bars for SEM are smaller than the symbol for some conditions if not evident. Each monoculture versus co-culture pair is significantly different (p<0.0001) in their ZEB1 and VIM expression. N=3.
• c-f: Relative expression of EpCAM (c,d), and ER (e,f) in MCF7 (c,e) and HCC1428 (HCC,d,f) ER+ breast cancer cells treated with AM23 or DFO in combination with fulvestrant in monoculture or co-culture with MSCs as measured by qRT-PCR. ER expression is significantly decreased (p<0.0001) in each monoculture versus co-culture comparison for the same treatment. Otherwise, pair-wise comparisons are only plotted if the comparison is significantly different and consistent in both MCF7 and HCC1428 comparisons. The mean percentage ± SEM is plotted for each condition. N=3.
Discussion
Our results implicate bone marrow MSCs as regulators of resistance to antiestrogen therapy in ER+ breast cancer and show that direct interactions with MSCs shift ER+ breast cancer cells to drug-resistant, CSCs with an increase in intracellular labile iron. Increases in intracellular iron expose a vulnerability to selected iron-modulating compounds to overcome resistance to ER-targeting drugs.
This work advances knowledge of ER+ breast cancer metastases, building on advantages of our in vitro co-culture model system to recapitulate cancer-stromal interactions in bone marrow and reveal emergent phenotypes. First, ER+ breast cancer cells in direct co-culture with MSCs shift to hybrid and M-CSCs, an effect not recapitulated by CM from MSCs alone. We confirmed this phenotype through flow cytometry for validated markers of CSCs, qRT-PCR for CSC-associated transcripts, and RNA sequencing. Second, ER+ breast cancer cells in direct co-culture with MSCs increased intracellular labile iron, exposing a vulnerability to selected iron chelators and ironomycin analogs to overcome resistance to antiestrogenic therapy. Importantly, combination treatment with iron modulators and fulvestrant did not enrich for CSCs, an adverse effect of many standard therapies.
To our knowledge, the current study shows for the first time that direct contact with MSCs increases labile iron in ER+ breast cancer cells. Previous research demonstrated pivotal functions of iron accumulation in breast cancer and other malignancies, particularly in the context of co-culture and CSCs. Innate immune cells in the tumor microenvironment, such as macrophages and neutrophils, can directly regulate iron in cancer cells, acting as sources of iron and iron-related proteins, including LCN2 and ferritin, that alter iron metabolism in cancer cells (50-52). Additionally, co-culture with carcinoma-associated fibroblasts induces hepcidin, which antagonizes iron export, in epithelial tumor cells (53). Functionally, CSCs more efficiently import iron from the microenvironment than non-CSCs by iron tracing experiments (35). Previous work established cell-intrinsic changes in CSCs of multiple cancer types in various iron uptake, storage, and efflux proteins, including transferrin, transferrin receptor, ferritin, ferroportin, and CD44 (49, 54). While we found an increase in iron-trafficking proteins LCN2 and CD44 and a decrease in the iron storage protein FTH1, some cancers have increased ferritin, suggesting context-dependent ferritin levels (55).
Derivatives of salinomycin have a history of efficacy against drug-resistant cancer cell lines. Salinomycin/ironomycin analogs AM5 and AM23 selectively target CSCs by exploiting lysosomal iron sequestration, eliminating cancer cells before iron accumulation in cellular compartments drives expansion of CSCs (35, 56, 57). Our work showed no consistent changes in CSCs by qRT-PCR after treatment, unlike standard antiestrogenic treatments that often enrich for CSCs (21). Previous work demonstrated that ironomycin analogs reduced CSCs in triple-negative monoculture environments (35), suggesting differences due to molecular subtype and breast cancer cell model system. Iron chelators function by binding iron and thus restricting cellular iron uptake, providing similar benefits to ironomycin analogs, and may prevent cellular iron accumulation before drug resistance arises (58). DFO sensitizes cancer cells to chemotherapeutic agents and affects levels of chromatin remodeling proteins, countering epigenetic modifications caused by intracellular iron (22). Unlike ironomycin analogs and iron chelators, ferroptosis inducers erastin, an inhibitor of cysteine import, and RSL-3, an inactivator of glutathione peroxidase 4, increased resistance of ER+ breast cancer cells to fulvestrant. This difference in effects of compartmental-based iron targeting further demonstrates advantages of targeting lysosomal iron buildup.
Most success of iron chelation therapy in cancer has been restricted to preclinical studies (24), among which DFX has been shown to synergize with multiple chemotherapies to reduce cell proliferation in triple-negative breast cancer (26, 58). Both DFO and DFX have been studied clinically in cancer patients. Treatment with DFO showed partial or complete responses in patients but caused injection site swelling and pain (24). A pilot study with DFX revealed high toxicity and continued tumor progression in advanced hepatocellular carcinoma patients (59). Interestingly, a phase 1 trial of VLX600, an iron chelator, in multiple advanced solid tumors revealed reasonable tolerability (60). While multiple factors may cause anemia in patients with cancer, our data suggest that providing supplemental iron could potentially promote drug resistance of ER+ breast cancer cells in bone marrow. These data indicate that iron modulating compounds hold clinical promise but require further testing to reduce toxicity in patients and define optimal combinations with other cancer therapies.
Our study revealed that direct co-culture with MSCs changed CSC and iron phenotypes in ER+ breast cancer. However, MSCs are just one bone marrow stromal cell type and other cell types, such as macrophages, can affect iron levels in cancer. Further studies should focus on effects of multiple stromal cell types on CSCs and iron content in cancer cells. Here, we studied tumor-stromal interactions in a 2D model system. Interestingly, 3D culture models have shown greater effects than 2D systems on hepcidin and other iron proteins, suggesting we might observe more pronounced phenotypes in a 3D co-culture (53, 61). Further work should explore mechanisms of iron upregulation in co-culture, differential regulation of iron-associated proteins, and increasing cancer cell sensitivity to antiestrogenic therapy with ironomycin analogs and iron chelators.
Supplementary Material
Acknowledgments
We thank Sean Linkes, Ann Marie Deslauriers-Cox, and Michael Pihalja for assistance with Flow cytometry. We acknowledge support from the Advanced Genomics Core, the Flow Cytometry Core, and the Center for Molecular Imaging Core of the University of Michigan Medical School’s Biomedical Research Core Facilities. We acknowledge support to the University of Michigan Rogel Cancer Center through National Institutes of Health grant P30CA046592 for flow cytometry and animal imaging studies.
The authors acknowledge funding from NIH grants R01CA100768, R01CA238042, R01GM138385, R01CA148828, R01CA245546, R01DK095201, R01CA248160, R35CA197585, R01CA2449310, U01CA210152, R01CA238023, R33CA225549, R50CA221807, and R37CA222563 and funding from the Breast Cancer Research Foundation grant BCRF-18-173. RR lab is funded by the European Research Council under the European Union’s Horizon 2020 research and innovation programme grant agreement No 647973 (R.R.), the Foundation Charles Defforey-Institut de France, and Ligue Contre le Cancer Equipe Labellisée. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE 1256260 (J.M.B and A.J.M.). B.A.H., Ph.D., was supported by an American Cancer Society - Michigan Cancer Research Fund Postdoctoral Fellowship, PF-18-236-01-CCG. Z.C.N is supported by the Michigan Postdoctoral Pioneer Program at the University of Michigan Medical School.
Footnotes
Conflict of interest statement
C.A.L. has received consulting fees from Astellas Pharmaceuticals, Odyssey Therapeutics, and T-Knife Therapuetics, and is an inventor on patents pertaining to Kras regulated metabolic pathways, redox control pathways in pancreatic cancer, and targeting the GOT1-pathway as a therapeutic approach (US Patent No: 2015126580-A1, 05/07/2015; US Patent No: 20190136238, 05/09/2019; International Patent No: WO2013177426-A2, 04/23/2015). G.D.L. has received research funding from InterAx Biotech AG and Polyphor (now part of Spexis).
Data availability statement
RNA sequencing data have been deposited in the Gene Expression Omnibus database (GEO accession number GSE196312).
References
- 1.Savci-Heijink CD, Halfwerk H, Hooijer GKJ, Horlings HM, Wesseling J, van de Vijver MJ. Retrospective analysis of metastatic behaviour of breast cancer subtypes. Breast cancer research and treatment. 2015;150(3):547–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel SA, Dave MA, Murthy RG, Helmy KY, Rameshwar P. Metastatic breast cancer cells in the bone marrow microenvironment: novel insights into oncoprotection. Oncology reviews. 2011;5(2):93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pantel K, Alix-Panabieres C. Bone marrow as a reservoir for disseminated tumor cells: a special source for liquid biopsy in cancer patients. Bonekey Rep. 2014;3:584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amanatullah DF, Tamaresis JS, Chu P, Bachmann MH, Hoang NM, Collyar D, et al. Local estrogen axis in the human bone microenvironment regulates estrogen receptor-positive breast cancer cells. Breast Cancer Research. 2017;19(1):121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations—a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015;12(10):573–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katzenellenbogen JA, Mayne CG, Katzenellenbogen BS, Greene GL, Chandarlapaty S. Structural underpinnings of oestrogen receptor mutations in endocrine therapy resistance. Nat Rev Cancer. 2018;18(6):377–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lavinsky RM, Jepsen K, Heinzel T, Torchia J, Mullen TM, Schiff R, et al. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc Natl Acad Sci U S A. 1998;95(6):2920–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnston SR, Lu B, Scott GK, Kushner PJ, Smith IE, Dowsett M, et al. Increased activator protein-1 DNA binding and c-Jun NH2-terminal kinase activity in human breast tumors with acquired tamoxifen resistance. Clin Cancer Res. 1999;5(2):251–6. [PubMed] [Google Scholar]
- 9.Magnani L, Stoeck A, Zhang X, Lánczky A, Mirabella AC, Wang TL, et al. Genome-wide reprogramming of the chromatin landscape underlies endocrine therapy resistance in breast cancer. Proc Natl Acad Sci U S A. 2013;110(16):E1490–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muluhngwi P, Klinge CM. Roles for miRNAs in endocrine resistance in breast cancer. Endocr Relat Cancer. 2015;22(5):R279–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plava J, Cihova M, Burikova M, Matuskova M, Kucerova L, Miklikova S. Recent advances in understanding tumor stroma-mediated chemoresistance in breast cancer. Mol Cancer. 2019;18(1):67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiozawa Y, Pienta KJ, Taichman RS. Hematopoietic stem cell niche is a potential therapeutic target for bone metastatic tumors. Clin Cancer Res. 2011;17(17):5553–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi Z, Yang WM, Chen LP, Yang DH, Zhou Q, Zhu J, et al. Enhanced chemosensitization in multidrug-resistant human breast cancer cells by inhibition of IL-6 and IL-8 production. Breast Cancer Res Treat. 2012;135(3):737–47. [DOI] [PubMed] [Google Scholar]
- 14.Chen DR, Lu DY, Lin HY, Yeh WL. Mesenchymal stem cell-induced doxorubicin resistance in triple negative breast cancer. Biomed Res Int. 2014;2014:532161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeh WL, Tsai CF, Chen DR. Peri-foci adipose-derived stem cells promote chemoresistance in breast cancer. Stem Cell Res Ther. 2017;8(1):177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houthuijzen JM, Daenen LGM, Roodhart JML, Voest EE. The role of mesenchymal stem cells in anti-cancer drug resistance and tumour progression. British journal of cancer. 2012;106(12):1901–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham N, Qian B-Z. Mesenchymal Stromal Cells: Emerging Roles in Bone Metastasis. International journal of molecular sciences. 2018;19(4):1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kucerova L, Skolekova S, Matuskova M, Bohac M, Kozovska Z. Altered features and increased chemosensitivity of human breast cancer cells mediated by adipose tissue-derived mesenchymal stromal cells. BMC Cancer. 2013;13:535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo M, Brooks M, Wicha M. Epithelial-mesenchymal plasticity of breast cancer stem cells: implications for metastasis and therapeutic resistance. Curr Pharm Des. 2015;21(10):1301–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pastushenko I, Brisebarre A, Sifrim A, Fioramonti M, Revenco T, Boumahdi S, et al. Identification of the tumour transition states occurring during EMT. Nature. 2018;556(7702):463–8. [DOI] [PubMed] [Google Scholar]
- 21.Sarmiento-Castro A, Caamaño-Gutiérrez E, Sims AH, Hull NJ, James MI, Santiago-Gómez A, et al. Increased Expression of Interleukin-1 Receptor Characterizes Anti-estrogen-Resistant ALDH+ Breast Cancer Stem Cells. Stem Cell Reports. 2020;15(2):307–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pogribny IP, Tryndyak VP, Pogribna M, Shpyleva S, Surratt G, Gamboa da Costa G, et al. Modulation of intracellular iron metabolism by iron chelation affects chromatin remodeling proteins and corresponding epigenetic modifications in breast cancer cells and increases their sensitivity to chemotherapeutic agents. Int J Oncol. 2013;42(5):1822–32. [DOI] [PubMed] [Google Scholar]
- 23.Recalcati S, Gammella E, Cairo G. Dysregulation of iron metabolism in cancer stem cells. (1873-4596 (Electronic)). [DOI] [PubMed] [Google Scholar]
- 24.Brown RAM, Richardson KL, Kabir TD, Trinder D, Ganss R, Leedman PJ. Altered Iron Metabolism and Impact in Cancer Biology, Metastasis, and Immunology. Frontiers in oncology. 2020;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forciniti S, Greco LA-O, Grizzi FA-OX, Malesci A, Laghi LA-O. Iron Metabolism in Cancer Progression. LID - 10.3390/ijms21062257 [doi] LID - 2257. (1422-0067 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tury SA-OX, Assayag F, Bonin F, Chateau-Joubert S, Servely JL, Vacher S, et al. The iron chelator deferasirox synergises with chemotherapy to treat triple-negative breast cancers. (1096-9896 (Electronic)). [DOI] [PubMed] [Google Scholar]
- 27.Goto W, Kashiwagi S, Asano Y, Takada K, Morisaki T, Takahashi K, et al. Inhibitory effects of iron depletion plus eribulin on the breast cancer microenvironment. BMC Cancer. 2020;20(1):1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chanvorachote P, Luanpitpong SA-O. Iron induces cancer stem cells and aggressive phenotypes in human lung cancer cells. (1522-1563 (Electronic)). [DOI] [PubMed] [Google Scholar]
- 29.Buschhaus JM, Humphries BA, Eckley SS, Robison TH, Cutter AC, Rajendran S, et al. Targeting disseminated estrogen-receptor-positive breast cancer cells in bone marrow. (1476-5594 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buschhaus JM, Luker KE, Luker GD. A Facile, In Vitro 384-Well Plate System to Model Disseminated Tumor Cells in the Bone Marrow Microenvironment. In: Lacorazza HD, editor. Cellular Quiescence: Methods and Protocols. New York, NY: Springer New York; 2018. p. 201–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Humphries BA, Buschhaus JM, Chen YC, Haley HR, Qyli T, Chiang B, et al. Plasminogen Activator Inhibitor 1 (PAI1) Promotes Actin Cytoskeleton Reorganization and Glycolytic Metabolism in Triple-Negative Breast Cancer. Mol Cancer Res. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eckley SS B J, Humphries BA, Robison TH, Luker KE, Luker GD. Short-term environmental conditioning generates cellular memory that enhances tumorigenic potential of triple-negative breast cancer cells. Tomography. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luker G, Pica C, Song J, Luker K, Piwnica-Worms D. Imaging 26S proteasome activity and inhibition in living mice. Nat Med. 2003;9(7):969–73. [DOI] [PubMed] [Google Scholar]
- 34.Cavnar S, Xiao A, Gibbons A, Rickelmann A, Neely T, Luker K, et al. Imaging sensitivity of quiescent cancer cells to metabolic perturbations in bone marrow spheroids. Tomography. 2016;2(2):146–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mai TT, Hamaï A, Hienzsch A, Cañeque T, Müller S, Wicinski J, et al. Salinomycin kills cancer stem cells by sequestering iron in lysosomes. Nature Chemistry. 2017;9(10):1025–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo M, Shang L, Brooks MD, Jiagge E, Zhu Y, Buschhaus JM, et al. Targeting Breast Cancer Stem Cell State Equilibrium through Modulation of Redox Signaling. Cell Metab. 2018;28(1):69–86.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ge SX, Son EW, Yao R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinformatics. 2018;19(1):534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–73. [DOI] [PubMed] [Google Scholar]
- 39.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences. 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ray P, Stacer A, Fenner J, Cavnar S, Mequiar K, Brown M, et al. CXCL12-γ in primary tumors drives breast cancer metastasis. Oncogene. 2015;34(16):2043–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang J, Woods P, Normolle D, Goff JP, Benos PV, Stehle CJ, et al. Downregulation of estrogen receptor and modulation of growth of breast cancer cell lines mediated by paracrine stromal cell signals. Breast Cancer Res Treat. 2017;161(2):229–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colacino JA, Azizi E, Brooks MD, Harouaka R, Fouladdel S, McDermott SP, et al. Heterogeneity of Human Breast Stem and Progenitor Cells as Revealed by Transcriptional Profiling. Stem Cell Reports. 2018;10(5):1596–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu J, Lei H, Fei X, Liang S, Xu H, Qin D, et al. NES1/KLK10 gene represses proliferation, enhances apoptosis and down-regulates glucose metabolism of PC3 prostate cancer cells. Scientific Reports. 2015;5(1):17426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yousef GM, Yacoub GM, Polymeris ME, Popalis C, Soosaipillai A, Diamandis EP. Kallikrein gene downregulation in breast cancer. Br J Cancer. 2004;90(1):167–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adamo A, Delfino P, Gatti A, Bonato A, Takam Kamga P, Bazzoni R, et al. HS-5 and HS-27A Stromal Cell Lines to Study Bone Marrow Mesenchymal Stromal Cell-Mediated Support to Cancer Development. Front Cell Dev Biol. 2020;8:584232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bystrom LM, Guzman Ml Fau - Rivella S, Rivella S. Iron and reactive oxygen species: friends or foes of cancer cells? (1557-7716 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chirillo R, Aversa I, Di Vito A, Salatino A, Battaglia AM, Sacco A, et al. FtH-Mediated ROS Dysregulation Promotes CXCL12/CXCR4 Axis Activation and EMT-Like Trans-Differentiation in Erythroleukemia K562 Cells. Frontiers in oncology. 2020;10:698-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang J, Bielenberg Dr Fau - Rodig SJ, Rodig Sj Fau - Doiron R, Doiron R Fau - Clifton MC, Clifton Mc Fau - Kung AL, Kung Al Fau - Strong RK, et al. Lipocalin 2 promotes breast cancer progression. (1091-6490 (Electronic)). [Google Scholar]
- 49.Müller SA-O, Sindikubwabo F, Cañeque TA-O, Lafon A, Versini A, Lombard B, et al. CD44 regulates epigenetic plasticity by mediating iron endocytosis. (1755-4349 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang W, Ferrara N. Iron Metabolism in the Tumor Microenvironment: Contributions of Innate Immune Cells. Front Immunol. 2020;11:626812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leimberg MJ, Prus E, Konijn AM, Fibach E. Macrophages function as a ferritin iron source for cultured human erythroid precursors. J Cell Biochem. 2008;103(4):1211–8. [DOI] [PubMed] [Google Scholar]
- 52.Ören B, Urosevic J, Mertens C, Mora J, Guiu M, Gomis RR, et al. Tumour stroma-derived lipocalin-2 promotes breast cancer metastasis. J Pathol. 2016;239(3):274–85. [DOI] [PubMed] [Google Scholar]
- 53.Blanchette-Farra N, Kita D, Konstorum A, Tesfay L, Lemler D, Hegde P, et al. Contribution of three-dimensional architecture and tumor-associated fibroblasts to hepcidin regulation in breast cancer. Oncogene. 2018;37(29):4013–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.El Hout M, Dos Santos L, Hamaï A, Mehrpour M. A promising new approach to cancer therapy: Targeting iron metabolism in cancer stem cells. Seminars in Cancer Biology. 2018;53:125–38. [DOI] [PubMed] [Google Scholar]
- 55.Schonberg DL, Miller TE, Wu Q, Flavahan WA, Das NK, Hale JS, et al. Preferential Iron Trafficking Characterizes Glioblastoma Stem-like Cells. Cancer Cell. 2015;28(4):441–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodriguez R, Schreiber SL, Conrad M. Persister cancer cells: Iron addiction and vulnerability to ferroptosis. Molecular cell. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huczyński A, Janczak J, Antoszczak M, Wietrzyk J, Maj E, Brzezinski B. Antiproliferative activity of salinomycin and its derivatives. Bioorg Med Chem Lett. 2012;22(23):7146–50. [DOI] [PubMed] [Google Scholar]
- 58.Katsura Y, Ohara T, Noma K, Ninomiya T, Kashima H, Kato T, et al. A Novel Combination Cancer Therapy with Iron Chelator Targeting Cancer Stem Cells via Suppressing Stemness. Cancers. 2019;11(2):177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saeki I, Yamamoto N, Yamasaki T, Takami T, Maeda M, Fujisawa K, et al. Effects of an oral iron chelator, deferasirox, on advanced hepatocellular carcinoma. World J Gastroenterol. 2016;22(40):8967–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mody K, Mansfield AS, Vemireddy L, Nygren P, Gulbo J, Borad M. A phase I study of the safety and tolerability of VLX600, an Iron Chelator, in patients with refractory advanced solid tumors. Invest New Drugs. 2019;37(4):684–92. [DOI] [PubMed] [Google Scholar]
- 61.Raggi C, Gammella E, Correnti M, Buratti P, Forti E, Andersen JB, et al. Dysregulation of Iron Metabolism in Cholangiocarcinoma Stem-like Cells. Scientific Reports. 2017;7(1):17667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data have been deposited in the Gene Expression Omnibus database (GEO accession number GSE196312).