Abstract
Mounting evidence supports the idea that transcriptional patterns serve as more specific identifiers of active enhancers than histone marks; however, the optimal strategy to identify active enhancers both experimentally and computationally has not been determined. Here, we compared 13 genome-wide RNA sequencing assays in K562 cells and showed that the nuclear run-on followed by cap-selection assay (GRO/PRO-cap) has advantages in eRNA detection and active enhancer identification. We also introduced a tool, Peak Identifier for Nascent Transcript Starts (PINTS), to identify active promoters and enhancers genome-wide and pinpoint the precise location of the 5′ transcription start sites. Finally, we compiled a comprehensive enhancer candidate compendium based on the detected eRNA TSSs available in 120 cell and tissue types that can be accessed at https://pints.yulab.org. With the knowledge of the best available assays and pipelines, this large-scale annotation of candidate enhancers will pave the way for selection and characterization of their functions in a time- and labor-efficient manner in the future.
Introduction
Regulation of transcription is a synergetic process requiring both trans-regulatory factors, like transcription factors, and cis-regulatory elements, like promoters and enhancers. In contrast to promoters, which initiate transcription in their proximal regions to produce stable RNA products, enhancers regulate their target gene(s) distally. Certain epigenomic signatures (enrichment of H3K4me1 and H3K27ac, high chromatin accessibility, and CBP/p300 binding) are considered to be defining features of active enhancer loci1,2. However, studies also revealed that enhancers could themselves produce relatively short-lived divergent transcripts, called enhancer RNAs (eRNAs)3,4. More recent studies further showed that distal divergent transcription events are more reliable marks for active enhancers than epigenomic signatures5,6. Recently we have proposed7,8 and later experimentally verified6 the basic unit of active enhancers that are defined by the transcription start sites (TSSs) of the divergent eRNA transcription, and delimited by the promoter-proximal RNA polymerase II (Pol II) pause sites flanking these TSSs. Therefore, to identify active enhancers genome-wide, it is critical to detect eRNAs and their TSSs with high sensitivity and specificity.
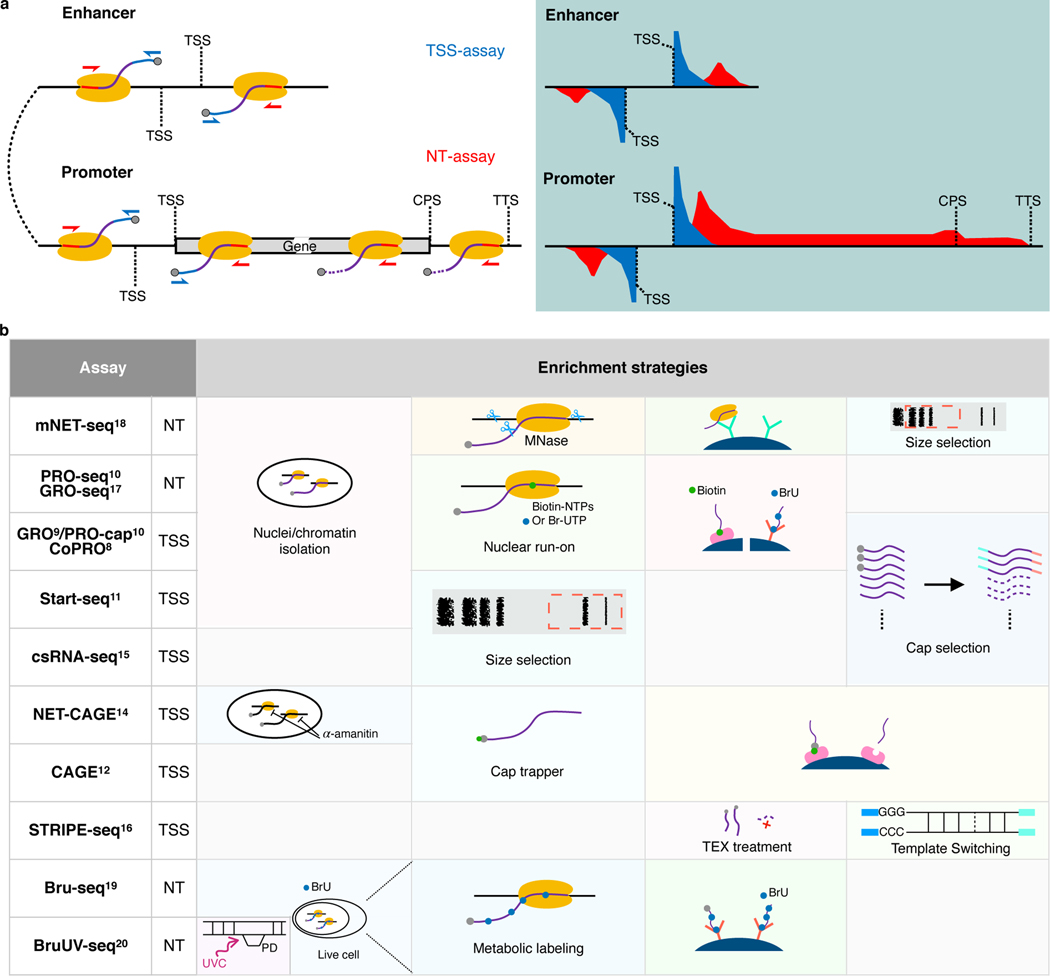
eRNAs are usually in extremely low abundance in cells due to their short half-lives. Therefore, conventional RNA-seq experiments capture eRNAs with very low efficiency3. Recently, two categories of genome-wide RNA sequencing assays have been developed, focusing either on TSSs or on the actively-transcribing polymerase positions (Fig. 1a). We named 8 assays (GRO9/PRO-cap10, CoPRO8, Start-seq11, CAGE12, RAMPAGE13, NET-CAGE14, csRNA-seq15, and STRIPE-seq16) from the former category as TSS-assays, because these assays enrich for active 5′ TSSs of promoters and enhancers (Fig. 1a). We also named 5 assays (GRO-seq17, PRO-seq10, mNET-seq18, Bru-seq19, and BruUV-seq20) from the latter category as Nascent Transcript-assays (NT-assays), because they are designed to trace the elongation or pause status of the polymerases and capture their products (Fig. 1a). To enrich for RNA populations of interests, these assays implement various experimental strategies, including nuclei/chromatin isolation8–11,18, nuclear run-on8–10 or metabolic labeling19,20 with biotin or bromo-tagged nucleotides and affinity purification, Pol II immunoprecipitation18, size selection15,16, and enzymatic elimination of non-capped RNAs8–10,16, or chemical tagging of capped RNAs12–14. We summarized key experimental steps of both TSS- and NT-assays in Fig. 1b. In fact, the list of all assays compared here, plus total RNA-seq21,22, have all been used in some capacity to identify enhancer elements. However, considering that most of these assays are not specifically designed to capture eRNAs, caution should be taken when exploiting these assays and their data to identify active enhancers.
Fig. 1. Comparison of currently available assays for detecting eRNAs.
a, Schematics of enhancer and promoter/gene transcription by RNA Pol II (left panel) and characteristic profiles of TSS- and NT-assays (right panel, light-blue shaded area). Black lines represent genomic DNA; nascent RNAs are purple curved lines with 5′ and 3′ ends colored blue and red, respectively, with gray spheres as caps and yellow ovals indicate RNA Pol IIs. Arrows indicate the direction of sequencing reads, TSS-assay in blue, and NT-assay in red. Representative read density profiles are colored accordingly in blue and red for TSS- and NT-assays, respectively. TSS: transcription start site; CPS: cleavage polyadenylation site; TTS: transcription termination site. b, Enrichment strategies used by different TSS- and NT-assays. TEX: terminator exonuclease. A detailed description is available in Supplementary Notes.
Because these assays were initially designed for different purposes, various computational tools were developed for exploring and interpreting the raw experimental data—for example, Tfit23, dREG24,25, and dREG.HD26 were developed to identify transcriptional regulatory elements (TREs) from some NT-assays, including GRO-seq and PRO-seq; FivePrime27 (based on paraclu28), GROcapTSSHMM7, and HOMER15 were introduced for analyzing data from CAGE, GRO-cap, and csRNA-seq, respectively. While all these tools can potentially be used to identify eRNA transcription and active enhancers, there has not been a systematic evaluation and comparison of their performance with datasets generated by the aforementioned experimental assays.
In this study, we systematically examined 13 experimental assays, including seven TSS-assays, five NT-assays, and total RNA-seq (as the outgroup), in terms of their sensitivity and specificity for capturing eRNAs. We also developed a computational tool, Peak Identifier for Nascent Transcript Starts (PINTS), which is designed to identify enhancer candidates from these assays. Moreover, by comparing PINTS with 8 other widely-used computational tools, we found that PINTS gave the highest overall performance pertaining to robustness, sensitivity, and specificity, especially when analyzing data from TSS-assays. Finally, we constructed a comprehensive enhancer candidate compendium for 120 cells and tissues using the robust and unified definition of active enhancers based on detected eRNA TSSs genome-wide5–7, and developed an online web server (https://pints.yulab.org/) to navigate, prioritize, and analyze enhancers based on a wide range of genomic and epigenomic annotations. We expect our enhancer compendium will be a valuable resource to the research community for the effective selection of candidate enhancers for further functional characterization.
Results
TSS-assays are more sensitive in eRNA detection
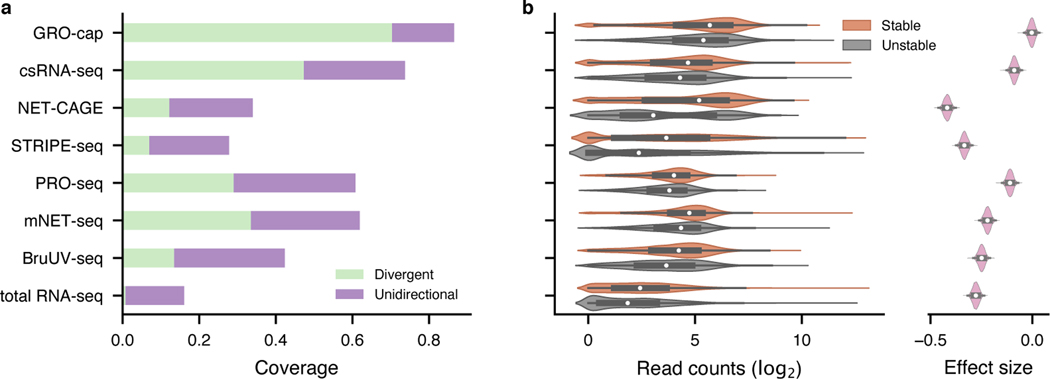
To perform a quantitative comparison of eRNA detection sensitivity, we first normalized all libraries by downsampling them to the same sequencing depth as the library with the lowest depth (18.9 million mappable reads, Supplementary Table 1). We then compared the assays’ sensitivity by examining their coverage in 803 (635 intergenic, 113 intronic, and 55 others) previously identified bona fide enhancers that were validated by CRISPR/Cas9-mediated deletion and CRISPRi in K562 cells29–37 (“CRISPR-identified enhancer set”, Methods, Supplementary Table 2). With the same sequencing depth, GRO-cap ranks first in sensitivity: it covers 86.6% of CRISPR-identified enhancers (70.4% divergent: ≥ 5 reads detected from both strands and 16.2% unidirectional: ≥ 5 reads detected only on one strand; Fig. 2a and top track in Extended Data Fig. 1a). csRNA-seq comes in second place with 73.7% (47.3% divergent and 26.4% unidirectional) coverage of these validated enhancers (Fig. 2a and Extended Data Fig. 1a). We re-evaluated the sensitivity of the 13 assays using another set of reference enhancers (previously validated by Massively Parallel Reporter Assay [MPRA]38–42 and Self-Transcribing Active Regulatory Region Sequencing [STARR-seq]6,43–45), and the results remain the same (Extended Data Fig. 1a, bottom track). Furthermore, to test the robustness of our conclusion, we also evaluated assay sensitivity of 8 assays that have data available in another cell line, GM12878, using STARR-seq-identified enhancers as reference6,46. As with K562 (Extended Data Fig. 1a), GRO-cap is the most sensitive assay to detect active enhancers in GM12878 (Extended Data Fig. 1b).
Fig. 2. Evaluation of assay sensitivity in eRNA detection.
a, The capability of different assays to capture validated enhancers (CRISPR-identified enhancer set). All libraries were downsampled to the same sequencing depth. “Unidirectional” and “divergent” indicate the detection of eRNAs originated from either one or both strands of the enhancer loci, respectively. b, Differences in read coverage among stable () and unstable () transcripts. GRO-cap has the highest coverage of both stable and unstable transcripts and the least preference toward stable transcripts. Preferences (effect sizes) were evaluated as Cohen’s d via bootstrap (). In the box plot, the center dots, box limits, and whiskers denote the median, upper and lower quartiles, and 1.5× interquartile range, respectively.
We further evaluated the sensitivity of these assays by their ability to capture unstable transcripts. eRNAs are usually less stable than mRNAs and this is the case here seen by comparing decay rates of transcripts47 from 514 CRISPR-identified enhancers to mRNAs (p-value from two-sided Mann-Whitney U test < , rank-biserial correlation: 0.442). We then used as the cutoff between stable and unstable transcripts the 95th quantile of decay rates of mRNAs and surveyed the distribution of read counts captured in the two categories among all assays. Consistent with our conclusion above, GRO-cap has the smallest differences in read coverage between stable and unstable transcripts (Cohen’s d: −0.003, 95% CI: [−0.033, 0.023]), indicating assays using nuclear run-on followed by cap-selection have the greatest ability to enrich unstable transcripts, which is of particular importance for detecting eRNAs (Fig. 2b and Extended Data Fig. 1c). We evaluated the effects of technical artifacts, including strand specificity and mis-priming, and our results suggest all libraries have great strand specificity (average: 0.984, SD: 0.019, Extended Data Fig. 2a~c) and low internal priming rates (Supplementary Notes, Extended Data Fig. 2d, and e).
Cap selection or Pol II pausing does not bias eRNA capture
Assays enriching for capped RNAs showed an advantage in detecting eRNAs (Fig. 2a and b). Because not all eRNAs are necessarily capped, we wanted to assess if the fraction of capped eRNAs influences these quantifications of eRNAs. To address this concern, we reanalyzed a previously published dataset6, where libraries were prepared with input RNAs pre-selected for different capping states (capped, uncapped, and unselected), and we assessed the abilities of the different libraries to detect CRISPR-identified enhancers. The difference among libraries prepared with three different inputs is minimal: there is a ~97% overlap between the library prepared with capped RNAs and that with unselected RNAs. Therefore, we consider the bias in enhancer detection as negligible when enriching for capped RNAs (Extended Data Fig. 2f, g, and Supplementary Notes).
Another concern about run-on assays is whether paused polymerases can efficiently resume elongation. A previous study in Drosophila showed that sarkosyl treatment could unleash paused polymerases and allow for efficient elongation of the nascent RNAs that correlated with Pol II levels measured by ChIP-seq48. Here, we calculated the correlation coefficient of promoter reads detected by PRO-seq and POLR2A ChIP-exo49 associated with paused Pol IIs in human K562 cells and found little difference, indicating paused Pol IIs elongate efficiently in run-on assays (Extended Data Fig. 2h).
Gene body reads in NT-assays contribute to lower sensitivity
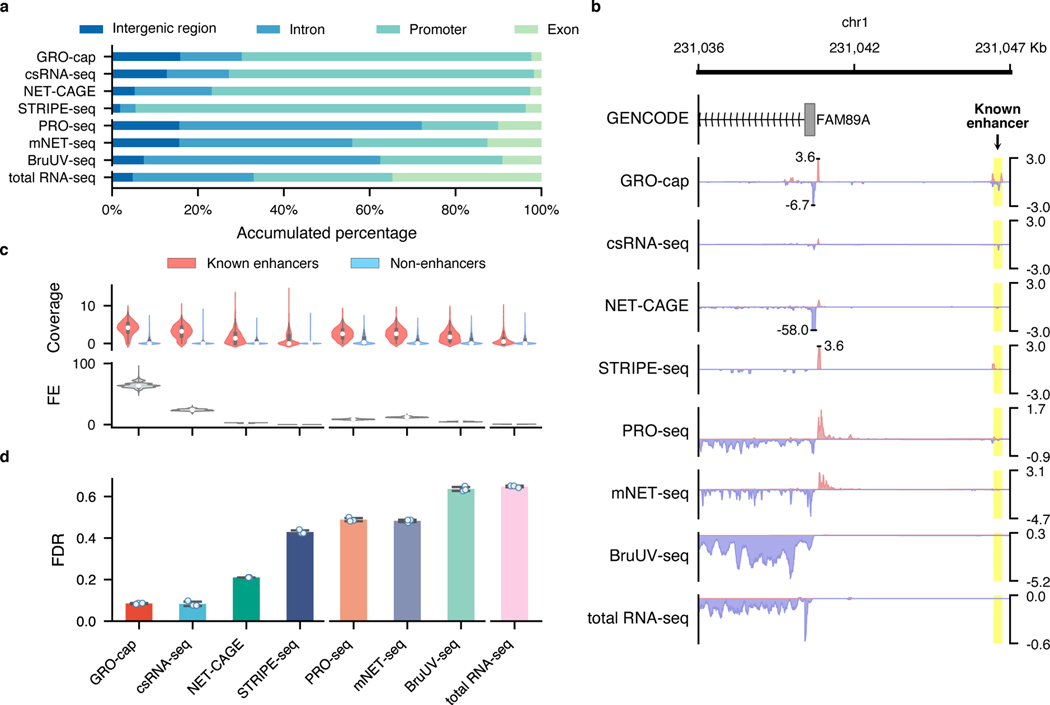
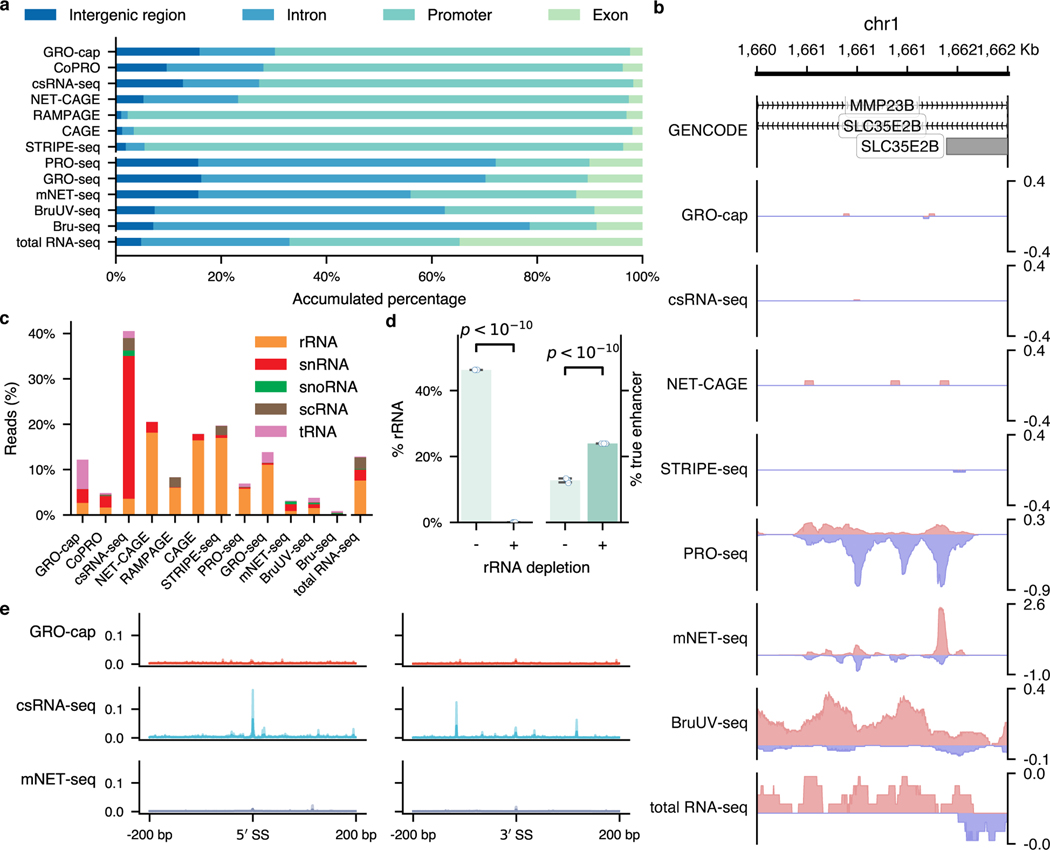
For the two families of assays that we compared in this study, we noticed that TSS-assays are generally more sensitive in detecting eRNAs than NT-assays, even for assays that use very similar enrichment strategies (Fig. 1b). For instance, while both GRO-cap and PRO-seq employ similar nuclear run-on procedures, there is a 41.3% difference between their divergent coverage of the CRISPR-identified enhancer set (Fig. 2a). When inspecting the genome-wide distribution of reads (Fig. 3a and Extended Data Fig. 3a), we noticed that NT-assays have significantly higher proportions of reads coming from gene body regions (mean of NT-assays: 65.6%, mean of TSS-assays: 13.0%, p-value from two-sided Mann-Whitney U test: 0.003), which is not surprising as they are designed to reveal all actively transcribing RNA polymerases, whereas TSS-assays are specifically designed to identify TSSs. Because eRNA transcription is, on average, much lower than that of genes7, such a high portion of gene body reads in NT-assays dilute the signal from eRNAs and significantly lowers their sensitivity in detecting active enhancers. As shown in Fig. 3b, NT-assays detect a substantial number of reads in the FAM89A gene body, whereas TSS-assays only have reads in the promoter regions of the FAM89A gene. As a result, almost all NT-assays (except PRO-seq) have no discernable signal at a distal enhancer locus near the FAM89A gene that was validated by CRISPRi34 (Fig. 3b). Another potential problem for NT-assays is that reads that are mapped to intergenic or intronic regions could be derived from either eRNAs, the unprocessed precursors of RNAs from other categories (e.g., pre-mRNAs), or read-through from an upstream transcription event (Extended Data Fig. 3b), which further reduces their sensitivity for detecting eRNAs.
Fig. 3. Characterization of factors affecting assay sensitivity and evaluation of assay specificity in eRNA detection.
a, Genome-wide distribution of sequencing reads originated from intergenic regions, introns, exons, and promoters detected by different assays. b, A genome browser snapshot of a gene (FAM89A) and its enhancer (highlighted in yellow), demonstrating the different patterns of signals captured by TSS- (enriched in the promoter and enhancer regions) vs. NT- (enriched in the promoter and gene body regions) assays. Signals are normalized by reads per million (RPM). c, Specificity in detecting eRNAs. Top track: differences of read coverage among CRISPR-identified enhancer () and non-enhancer () loci, a pseudo-count (1) was added to each locus, and the coverage was log-transformed. Bottom track: signal-to-noise ratios depicted in forms of fold enrichment (FE), the results are based on bootstrapped samples (), median statistics are used to calculate the fold changes. In the box plot, the center dots, box limits, and whiskers denote the median, upper and lower quartiles, and 1.5× interquartile range, respectively. d, False discovery rates (FDRs) estimated by the overlap between the top 20,000 genomic bins and the reference (803 CRISPR-identified and 6,777 non-enhancer) loci. Downsampled libraries were used (); values and error bars represent the mean and SD.
Most of the RNA transcripts in living cells belong to the families of highly abundant RNAs, e.g., rRNAs and snRNAs. When capturing eRNAs with sequencing-based methods, a successful exclusion of RNAs of these high-abundance families from the sequencing libraries during the preparation process can greatly enhance the efficiency and sensitivity of eRNA identification. We compiled a comprehensive list of high-abundance RNAs in human cells by incorporating annotations from GENCODE50, RefSeq51, and RMSK52. Based on this list, an average of 7.12% of the mappable reads in each assay originated from rRNAs despite most of the assays employed strategies to reduce rRNA inclusion (Extended Data Fig. 3c). By simulating an rRNA-depleted BruUV-seq library, we found that complete depletion of rRNAs could contribute to a 1.76-fold boost in detecting eRNAs (Extended Data Fig. 3d).
When the assay specifically enriches for short transcripts, like csRNA-seq, a relatively large proportion (31.5%) of the mappable reads were found to have originated from snRNAs (Extended Data Fig. 3c). Detection of such a disproportionally large fraction of snRNAs suggests potential contamination in the sequencing library from the splicing intermediates. To test the possibility, we calculated the signal densities at all the splice sites in the human genome according to GENCODE annotation (release 24, comprehensive version)50. As shown in Extended Data Fig. 3e, csRNA-seq detects more signals at the splicing junctions than all the other assays.
TSS-assays, especially GRO-cap, have higher specificity
While genomic regions with detectable transcriptional events account for 75% of the human genome53, many of these events are considered to be spurious transcriptional noise54,55 because of their extremely low transcript yields compared to mRNAs and of the intrinsic promiscuity of RNA Pol II under certain circumstances56. Therefore, it is critical to detect and differentiate the reads that originated from spurious transcription in these assays. To that end, we collected non-enhancer loci from eight MPRA38–42 and STARR-seq6,43,44 studies and further removed elements overlapping with predicted Enhancer-Like Sequence (ELS) or Promoter-Like Sequence (PLS) from candidate cis-regulatory elements (cCRE) annotations57 to generate a set of 7,097 loci (referred to as the “non-enhancer set”, Methods, Extended Data Fig. 4a, Supplementary Table 3). We observed that signal intensities in the CRISPR-identified enhancer set are often higher than that in the non-enhancer set (Fig. 3c and Extended Data Fig. 4b, all of them have p-values from two-sided Mann-Whitney U test < ), with GRO-cap having the highest signal-to-noise ratio (64-fold enrichment, Fig. 3c, Extended Data Fig. 4b for K562 and Extended Data Fig. 4c for GM12878).
We also calculated the false discovery rate (FDR) for each assay based on the overlap of their reads with both the CRISPR-identified enhancer and non-enhancer sets. We found that TSS-assays generally have lower FDRs than NT-assays (TSS-assay mean: 0.232 vs. NT-assay mean: 0.544, p-value from two-sided Mann-Whitney U test: , Fig. 3d and Extended Data Fig. 4d), with assays enriching for capped and short RNAs having the lowest FDR (mean: 0.083, SD: 0.007).
PINTS: Peak Identifier for Nascent Transcript Starts
Two categories of tools are currently available to process data generated by RNA sequencing assays. Tools in the first category predict entire transcription units; they are primarily used for NT-assays and often use a set of cutoffs for fold-changes, e.g., HOMER (GRO-seq)58 or Hidden Markov Models19,59, e.g., groHMM, to determine the start and end positions of transcription units (Extended Data Fig. 5a). Tools in the second category usually identify narrower regions for potential regulatory elements (mainly promoters and enhancers, often referred to as peak callers) and include GROcapTSSHMM7, dREG24, Tfit23, dREG.HD26, TSScall11, HOMER (csRNA-seq)15, and FivePrime27 (based on Paraclu28). Based on previous studies, the peak of divergent TSSs, which is associated with eRNA transcription, is an effective mark for active enhancers5,6. Therefore, in this comparative study, we focused on the second category of computational tools.
To achieve a higher resolution in identifying transcriptional regulatory elements, a common practice is to only look at the ends of the captured transcripts. Two critical issues emerge when evaluating the statistical significance of peaks (i.e., TSSs), especially for the TSS-assays. First, when only considering transcript ends, the fraction of zeros (no mapped reads per base pair) in the local background increases, which deflates read density in the local background and thus inflates the statistical significance of the candidate TSSs and results in false positives. Second, multiple TSSs can localize in close proximity in the genome and therefore inflate the estimation of read density in the local background, resulting in diminished statistical significance for all TSSs in that locus and leading to false negatives in TSS detection. To address these issues, we developed Peak Identifier for Nascent Transcript Starts (PINTS), which uses zero-inflated Poisson models to evaluate local read densities and employs interquartile range (IQR)-based refinement to ameliorate false negatives by conditionally masking candidate TSSs in the local background (Extended Data Fig. 5b). PINTS was inspired by MACS260 with modifications specifically implemented for identifying eRNA TSSs from genome-wide TSS-assays. After evaluating the significance of each TSS, PINTS defines TREs as divergent TSS pairs that are within 300 bp from each other, as suggested by previous studies6,8 (Extended Data Fig. 5b, Methods). We identify candidate enhancers as the distal TREs that are farther than 500 bp away from the known protein-coding gene TSSs6.
Peak callers vary in resolution and computational needs
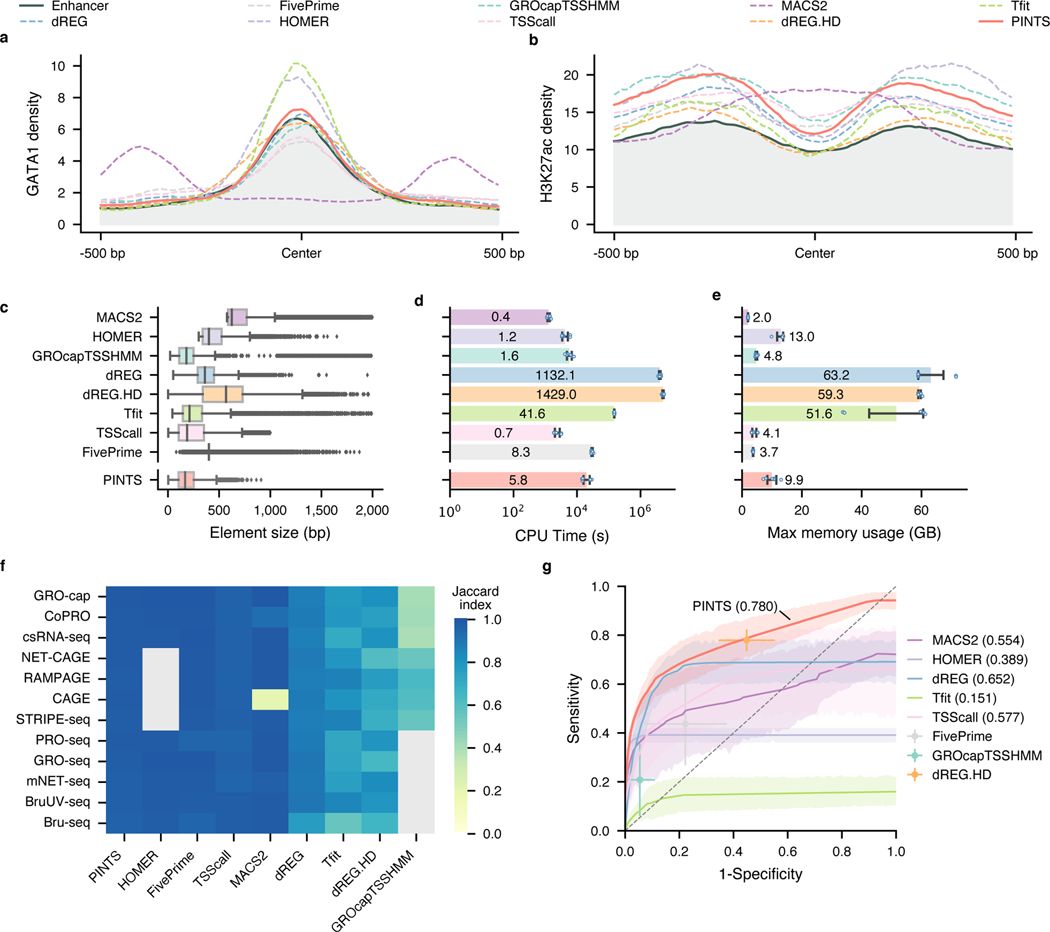
Candidate enhancer loci identified by different algorithms should share the same features as CRISPR-identified enhancers with characteristic epigenomic marks (DHS, H3K27ac, H3K4me3, and H3K4me1) and transcription factor (CBP/p300, GATA1, and CTCF)-binding sites. Indeed, we found that candidate enhancers identified by most tools recapitulate these features of CRISPR-identified enhancers (Fig. 4a, b, and Extended Data Fig. 6a). However, the patterns of epigenomic marks and TF-binding sites of candidate enhancers identified by MACS260, a widely used peak caller for analyzing ChIP-seq data, are distinct from those of the CRISPR-identified enhancers, suggesting the default peak-shifting model of MACS2 may not be suitable for identifying eRNA TSSs of active enhancers (Supplementary Notes and Extended Data Fig. 6b).
Fig. 4. PINTS achieves the best balance among resolution, robustness, sensitivity, specificity, and computational resources required.
a and b, Profiles of GATA1 binding sites and H3K27ac pattern in CRISPR-identified enhancer regions and distal TREs identified by different peak callers. c, Distribution of element sizes identified by different tools. Sample size from the top to the bottom: 38,703, 9,546, 19,886, 51,128, 56,300, 17,152, 54,002, 40,042, 35,998. In the box plot, the center lines, box limits, and whiskers denote the median, upper and lower quartiles, and 1.5× interquartile range, respectively. Points show observations that are not in the 1.5× interquartile range. d, CPU time consumed by peak callers to identify elements from various TSS-assay libraries. The average CPU time labeled inside each bar is in the unit of hours (). The x-axis is in scale. e, Maximum memory usage during peak calling from TSS-assay libraries. The average maximum usage labeled inside or on top of each bar is in the unit of gigabytes (). For d and e, values and error bars represent the mean and SD. f, Robustness of peak calls made by different tools. Libraries were mapped to both hg19 and hg38, and robustness was measured as the Jaccard index between calls from hg19 and hg38 (lifted over). Cells colored in gray indicate either that the tool cannot be applied to the corresponding assays or that one or more required datasets are not available. g, Aggregated ROC curves for each peak caller on all TSS-assay datasets (). The solid lines represent the mean values; the corresponding shaded areas show the 95% confidence interval of the means (via bootstrap). For tools where ROCs cannot be calculated, solid dots represent their performance with default parameters. Values and error bars show the mean and SD.
We further surveyed the size distribution of these elements as an indication of peak-calling resolutions. We found that PINTS, GROcapTSSHMM, Tfit, and TSScall achieve higher resolution (average peak size of 185–300 bp) than other tools when analyzing TSS-assay data (Fig. 4c and Extended Data Fig. 6c). Notably, peaks called by dREG.HD and MACS2 range between 548–751 bp in size, whereas dREG, FivePrime, and HOMER peaks range between 381–460 bp.
The amount of computational resources required by a peak caller greatly affects its general applicability. Here, we compared the total amount of CPU time and peak memory usage that each tool requires for identifying divergent elements from TSS-assays (Fig. 4d and e). Based on our calculation, it is feasible to run PINTS, MACS2, HOMER, GROcapTSSHMM, TSScall, and FivePrime on a typical personal computer.
PINTS achieves high overall performance for TSS-assays
A common task in functional studies of enhancers is to compare active enhancers across different conditions and diseases21,22, which requires the in silico enhancer-predicting tools to be robust against biological and experimental variances. To test the upper bound of robustness for the aforementioned tools, we performed peak calling using these tools on datasets from 12 assays by aligning each dataset to two commonly-used builds of the human reference genome: hg19 and hg38. Because there is no variation in the sequencing datasets themselves and the two reference genome builds are very similar61 (Extended Data Fig. 7a), we expect that the differences in peak calls using these two different genome builds should be minimal across all datasets. We found that by simply changing the reference genome builds, half of them have a Jaccard index smaller than 0.9 for at least one assay (Fig. 4f). Furthermore, we noticed that Jaccard indices were even lower when we tried to evaluate robustness across real technical and biological replicates (average: 0.507, SD: 0.246), especially for TSScall and FivePrime, where their robustness is only 0.401 (SD: 0.104) and 0.384 (SD: 0.203), respectively (Extended Data Fig. 7b). PINTS consistently have great robustness in both cases (average: 0.976, SD: 0.008 between genome builds, and average: 0.728, SD: 0.052 across replicates).
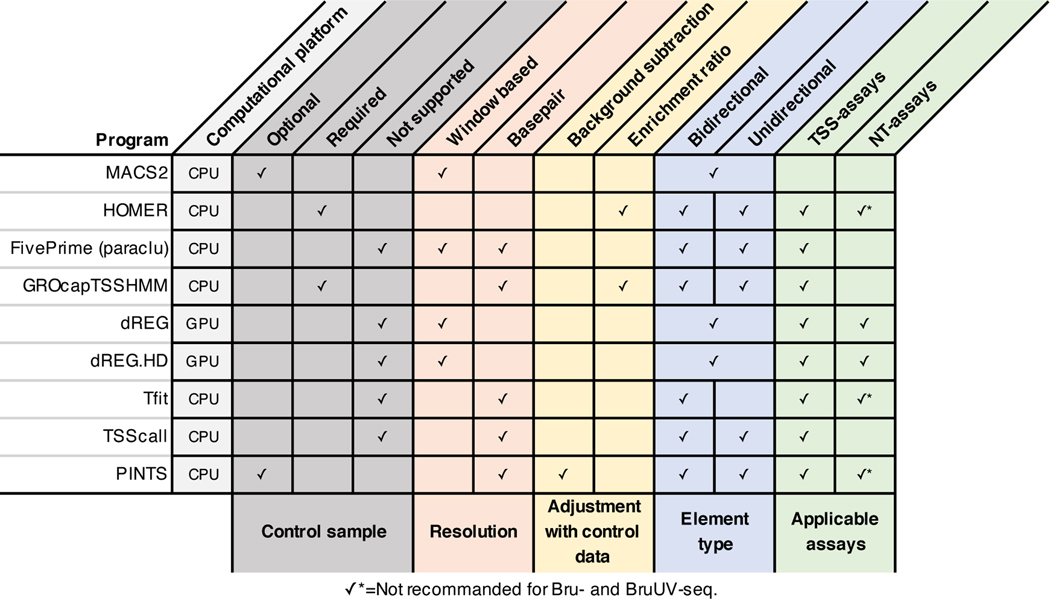
To evaluate sensitivity and specificity, two key performance metrics, we merged the CRISPR-identified enhancer set with the promoter regions from GENCODE v2450 as the positive set, and non-enhancer loci as the negative set (Methods, Extended Data Fig. 8a). We then evaluated each tool’s performance for all TSS-assay datasets (Fig. 4g). The results show that PINTS achieves the best balance between sensitivity and specificity (PINTS mean AUC: 0.780, SD: 0.082; mean AUC for the second-best tool dREG: 0.652, SD: 0.109). Moreover, when we compared the performance of all available tools on sparsely sequenced libraries (with 18.9, 15, and 10 million mappable reads), PINTS still outperforms other tools (Extended Data Fig. 8b~d). When evaluating unique TREs identified by PINTS, we noticed enrichments in epigenomic marks (H3K27ac and H3K4me1, Extended Data Fig. 9a) and motifs for both enhancer-activity-related and cell-type-specific transcription factors (Extended Data Fig. 9b). For all of these computational tools, we summarize their key requirements, main characteristics, and applicability to different RNA sequencing assays in Extended Data Fig. 10.
An enhancer compendium for human cell and tissue types
Previous studies have shown that, compared with histone marks, detecting enhancers by divergent eRNA TSSs has advantages in both resolution and specificity5,6. Introducing an eRNA-centric enhancer compendium, in addition to all the available enhancer datasets based on histone marks57,62, will be an invaluable resource to better understand gene regulation, to functionally annotate the non-coding genome, and to help prioritize non-coding variants across disease cohorts by their potential impact on enhancer activities63. Toward this goal, we applied PINTS to identify candidate enhancers using TSS-assay datasets (i.e., GRO/PRO-cap, CoPRO, csRNA-seq, NET-CAGE, RAMPAGE, CAGE, and STRIPE-seq) across 33 cell lines, 7 in vitro differentiated cells, 35 primary cells, and 45 tissue samples, including all available TSS-assay datasets through the ENCODE portal (Fig. 5a). Such a comprehensive catalog of enhancers across a wide range of human cells and tissues analyzed by the same exact computational pipeline provides an excellent resource to perform meaningful comparative genomic analyses to study the dynamics of enhancers and gene regulation in general, which will help focus on true biological differences while minimizing technical variations. In addition, for 7 human cell lines (K562, GM12878, HepG2, HeLa-S3, MCF-7, H9, and HCT116), we applied all other available tools (FivePrime, HOMER, TSScall, dREG, dREG.HD, and Tfit) to identify candidate enhancers. We believe this unique resource of enhancers in 7 cell lines with multiple TSS-assay datasets analyzed by all available computational tools will greatly help further the studies of enhancers and their key architecture characteristics. All of these candidate enhancer calls are made publicly available through our web server (https://pints.yulab.org) described in detail below. We will regularly update our enhancer compendium as new datasets, especially those in new cell lines or samples, and assays, become available.
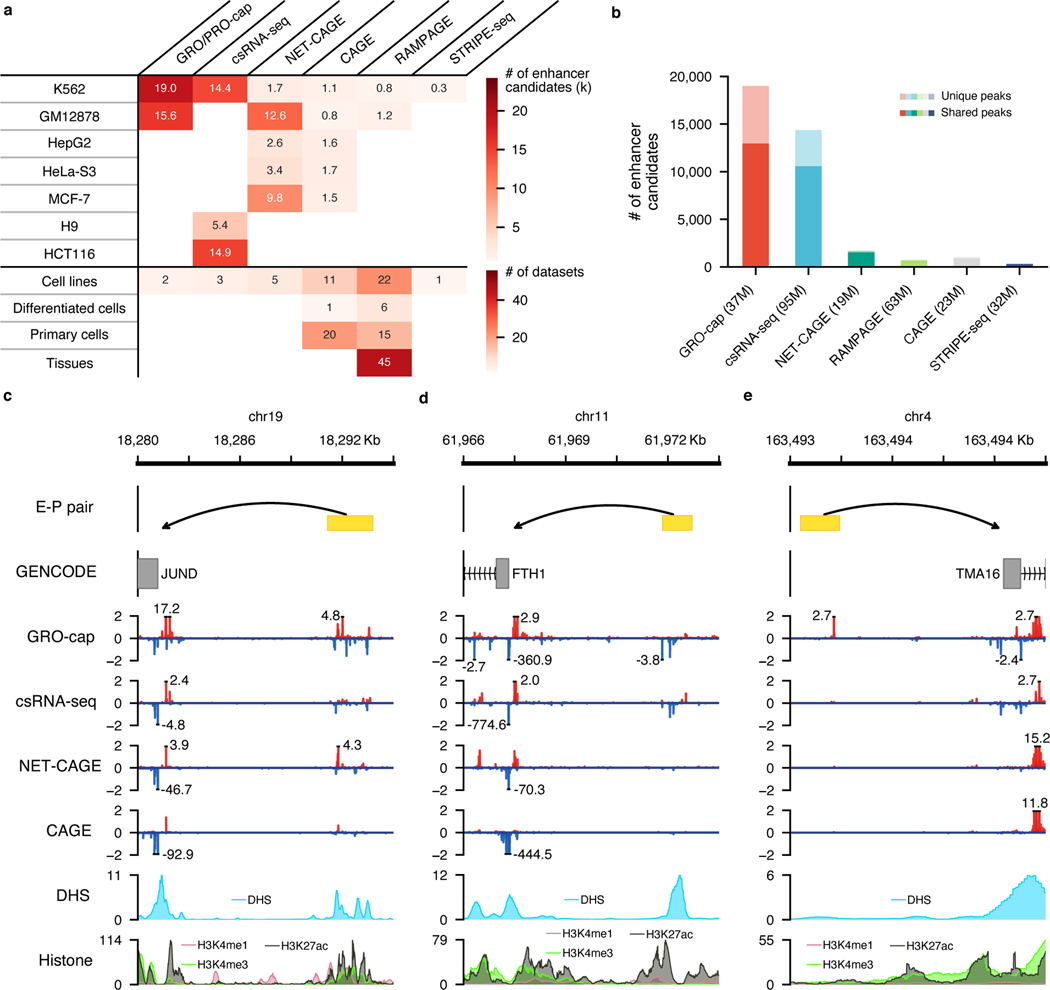
Fig. 5. A comprehensive human enhancer compendium.
a, Summaries of all distal elements identified from different assays with PINTS in 7 cell lines (top) and 131 datasets generated by different assays across 120 biosamples included in our enhancer compendium (bottom). Differentiated cells stand for in vitro differentiated cells. b, The number of distal elements identified by PINTS from different assays in K562. The dark colors indicate the proportion of shared elements identified from at least one other assay; light colors indicate elements unique to the corresponding assay. The number of mappable reads for each assay is listed in the parentheses. c, A known enhancer locus where most of these assays captured signals on both strands. d, A known enhancer locus where only GRO-cap and csRNA-seq captured detectable signals. e, A known enhancer locus where only GRO-cap captures clear signals. Signal tracks in c, d, and e were normalized by their sequencing depths (RPM).
In human K562 cells where datasets are available from all TSS-assays, our results show that GRO-cap has by far the most number of distal TREs (19,006 identified by PINTS with 9,531 unique enhancer calls – not identified by any other assay); the second-best dataset, csRNA-seq, only has 14,375 enhancer calls with 5,048 unique (Fig. 5b). This is not surprising given that GRO-cap has the best sensitivity in detecting eRNA transcription (Fig. 2a and b) and this GRO-cap dataset has the third-highest read depth (Fig. 5b). We selected three CRISPRi-validated enhancer-promoter pairs34 to visualize these differences and showed the variety in signal abundances across different datasets (Fig. 5c~e). For example, the enhancer which regulates the JUND gene (Fig. 5c) has decent accessibility and is supported by epigenomic marks, including H3K27ac and H3K4me1. As expected, all four TSS-assays can identify this enhancer. The expression levels of enhancers are not necessarily proportional to the levels of epigenomic marks, and for eRNAs whose expression levels are lower (e.g., the enhancer that regulates FTH1 in Fig. 5d), assays that are more effective in capturing unstable transcripts are more likely to recover them. Finally, for the enhancer regulating TMA16, signals from histone marks are quite minimal, but GRO-cap still captures clear signals of eRNA transcription at this locus and readily enables the identification of this enhancer (Fig. 5e).
PINTS web server for exploring and analyzing enhancers
To make it easier for biologists to explore the tens of thousands of candidate enhancers in our compendium and to prioritize these enhancers for further studies, we constructed an online web server (https://pints.yulab.org), where users can query any human genomic region of interest in a given biological sample to get a comprehensive list of candidate enhancers detected by any available TSS-assays in that sample using any of the 8 peak callers (PINTS, dREG, dREG.HD, FivePrime, GROcapTSSHMM, HOMER, Tfit, and TSScall). We also included detailed annotations for each candidate enhancer, including epigenetic features57, core promoter elements64, potential transcription factor binding sites65, and presence of population variants and ClinVar mutations66 (Fig. 6). Users can prioritize enhancers by requiring the joint presence of specific annotations. For the transcription factor binding site analysis, our web server will automatically integrate the information from available RNA-seq data to only include factors that are expressed in the selected sample.
Fig. 6. Interactive PINTS web server.
The server is composed of three modules: a catalog database containing our human enhancer compendium across 120 biosamples, an interactive user interface, and analytical engines. When the user provides genomic loci of their interest as search queries, previously annotated information related to the loci will be retrieved from the catalog database. The user can further refine the search results by a series of filters, including the supporting assays, callers, the presence of mutations, transcription factor binding sites, core promoter elements, and epigenomic marks. On the other hand, if the user chooses to upload peak calls generated from their data, the analytical engines will analyze the uploads to provide the same types of annotations as those in the catalog database.
Furthermore, users can upload their own enhancer calls in a human cell line or tissue; our web server will automatically annotate all of their enhancer calls with the same pipeline as described above. We have integrated epigenomic features and RNA-seq data for 197 human-derived biosamples from the ENCODE project in our web server. For user-uploaded enhancer data in these samples, we automatically refine our annotations by only reporting binding sites of expressed transcription factors and associating each enhancer with epigenomic features specific to the corresponding sample.
Users can explore all annotations of their selected enhancers via our integrated genome browser; alternatively, they can easily export all annotations in plain text format for downstream analyses.
Discussion
eRNAs are increasingly being recognized as a critical marker for active enhancers genome-wide5,6; however, the optimal strategy (both experimental assays and their analytical pipelines) to detect eRNAs and thus identify enhancer loci has not been critically evaluated. In this study, we systematically compared 13 in vivo genome-wide RNA sequencing assays in K562 cells and showed that TSS-assays are in general more sensitive than NT-assays to detect eRNAs, because signals will not be diluted by active transcription in gene bodies. One additional and critical advantage of the TSS-assays is that they reveal the precise location of eRNA TSSs, allowing for high-resolution detection and delimitation of enhancer loci genome-wide, as demonstrated in our recent work6. Overall, our results show that GRO/PRO-cap has the best overall performance in detecting active enhancers in terms of both sensitivity and specificity. Thus, for fresh cells and tissue specimens or samples with high RNA quality, we recommend GRO/PRO-cap based on our observations in this study. Because run-on cap assays require an enzymatically-active RNA polymerase and cap structure, other TSS-assays (e.g., FFPEcap-seq67) or some NT-assay18 may perform better in samples where these requirements cannot be met, for example, for paraffin-embedded, formalin-fixed samples.
We noticed that when using current computational tools to identify TREs from various RNA sequencing datasets, minor changes in sample processing could lead to more than 20% of changes in the final results, which brings the robustness of the peak calls into question. To address this issue, we introduced a computational tool, PINTS. Our benchmarks indicate that PINTS achieves the best balance among robustness, applicability, sensitivity, and specificity, especially for TSS-assays capable of detecting the precise location of eRNA TSSs.
In this study, we used CRISPR/Cas9 and CRISPRi validated enhancers29–37 as the positive reference set and MPRA/STARR-seq negative segments6,38–44 as non-enhancers. Although these two sets show quite different epigenomic profiles (Extended Data Fig. 4a), which indicates that our non-enhancer set is depleted of true enhancers, there might still be a few false negatives in the non-enhancer set. This is because, in the published MPRA/STARR-seq datasets, only a very small number of promoters were used to test all candidate elements, but some enhancers might not work with these promoters. Furthermore, some tested elements might be truncated due to synthesis limitations ( bp) or random fragmentation of the genome. However, such cases are not expected to affect our relative ranking of different assays and thus will have minimal impact on our conclusions. We also note that the features defining enhancers and non-enhancers, both at structural and functional levels, are still a work in progress, and while it appears that the vast majority of active enhancers are transcribed, an accurate estimation of the fraction of enhancers that can initiate transcription is still unknown.
Furthermore, we provide a detailed, comprehensive human enhancer compendium with a unified definition6,7 of enhancers based on the detected divergent pairs of eRNA TSSs. Such a robust, unified and comprehensive catalog of enhancers across 120 cell types and tissues is expected to shine a light on the mechanism of gene regulation and architectural details of enhancers in general. The precise definition of enhancer element boundaries afforded by TSS-assays like PRO/GRO-cap would alleviate potential concerns regarding whether full-length enhancer elements were selected and tested in follow-up functional studies, and thus improve coverage of elements by eliminating incomplete or ill-defined candidates. Such a well-defined catalog of enhancers also provides an invaluable resource for follow-up studies to better understand the similarities and key differences in gene regulation across various tissues and conditions, and to identify key enhancers whose malfunctions can lead to specific disorders.
Methods
Data preprocessing
All datasets were managed and analyzed with BioQueue68 (accession numbers for these datasets are available in Supplementary Table 1). Raw reads were preprocessed with fastp69 for adapter trimming. Only reads longer than 14 bp were kept for downstream analyses. All processed reads from RNA assays were aligned using STAR70 to primary assemblies of human reference genome hg38 (GCF_000001305.15) together with rDNA (U13369.1) with parameters as --outSAMattributes All --outSAMmultNmax 1 --outFilterMultimapNmax 50. For studies using Drosophila cells, or other specific samples as spike-ins, Drosophila reference genome (dm6), or the corresponding reference sequences used in the original studies were incorporated into the index (details available in Supplementary Table 1). To measure the robustness of peak prediction, we also mapped reads to primary assemblies of human reference genome hg19 (from UCSC, sequences from alternative loci/haplotypes were removed the same way as for hg38). Reads from DNA assays were aligned and processed using bwa71 and samtools72 with default parameters.
Determining read coverage among reference regions
Sequencing reads of replicates for the same assay were merged together and downsampled to the same sequencing depth (the same number of mappable reads) three times using picard with parameter STRATEGY = Chained. These downsampled data were then converted to bed files to calculate the fraction of overlap between the sequencing reads and the reference regions in a strand-specific manner.
Classifying the transcription units as stable and unstable units with TT-seq
Transcript annotations derived from TT-seq (GSE7579247) were downloaded from the GEO database. Transcription units with NA values were discarded. The 95th quantile of estimated decay rates for mRNAs was used as the cutoff between the unstable (above the cutoff) and stable (below the cutoff) transcription units.
Characterizing the genome-wide distribution of reads
The entire genome was classified into four categories based on the annotations in GENCODE (ver 24)50: exonic and intronic regions were defined as in GENCODE, except that any region with overlapping intronic and exonic annotation was considered as exonic; the 500-bp regions flanking annotated transcription start sites of protein-coding transcripts were annotated as promoters; all other regions were considered as intergenic. Sequencing reads of various assays were assigned to the categories of promoters, introns, exons, or intergenic regions (in the exact order) if they were aligned to the corresponding annotated regions in the genome.
Identifying sequencing reads from splicing intermediates
The exact or approximate positions of transcript termini were inferred from the read ends, and the abundance of their corresponding transcripts was normalized as RPM for this analysis. A list of annotated splice junctions and their 200 bp flanking regions in the human genome was compiled based on GENCODE v2450. For each assay, we iterated through this list and recorded normalized read counts at each position. In Extended Data Fig. 3e, both the average of signals and the 95% confidence interval (estimated by bootstrap) of the averages were reported.
Compiling the reference sets
The experimentally quantified enhancer activity of various DNA elements was collected from previous studies (enhancers: 938 from CRISPR31 or CRISPRi29,30,32–37; non-enhancers: 20,941 from STARR-seq6,43,44 and 17,462 from MPRA38–42). Overlapping elements within the same category were merged until the resulting elements overlap with elements in the other category. Non-enhancer loci were excluded in the final set if they 1) were shorter than 250 bp; 2) overlapped with PLS or ELS predicted by cCRE57, or 3) overlapped with possible promoters (1kb regions flanking TSSs in GENCODE). When selecting MPRA38–42 and STARR-seq6,43–45 identified enhancers in K562, we required the supporting data from at least two independent studies to ameliorate the inclusion of false-positive enhancers resulting from orientation bias (STARR-seq) or promoter activity (MPRA). For GM12878, to ameliorate false positives caused by orientation bias, only orientation-independent enhancer calls from HiDRA were used6,46.
Calculating FDRs for assays
multiBamSummary73 was used to generate tables of read counts across the genome in 500-bp bins. For each assay, the bins were ranked by counts in them, and the top n bins were considered as true signals from the assay (four cutoffs were tested: 5,000, 10,000, 20,000, and 100,000, Extended Data Fig. 4d). If a bin overlapped with a locus in the CRISPR-identified enhancer set, then the bin was considered as a true positive (TP); if a bin overlapped with a locus in the non-enhancer set, it was considered as a false positive (FP). The false discovery rates were calculated as:
PINTS
Briefly, read ends are separated based on their mapping directions on the reference genome (forward or reverse), and the read counts are binned into 100-bp windows. Adjacent windows with reads available are merged to avoid splitting potential TRE elements. Within each window, the algorithm first finds peak seeds using a prominence-based approach. Then with a maximum-scoring pairing strategy28, the nearby seeds will be merged together as peak candidates if the density after merging meets the following condition:
The default value for is 1, and PINTS’ resolution can be further fine-tuned by incorporating reference annotations. For example, when the transcript annotation is available, PINTS will try to avoid the overlap of peak candidates with more than one transcript.
Next, to address the increased sparsity of signals when only the read ends are taken into account, the Expectation-Maximization algorithm is used to fit zero-inflated Poisson (ZIP) models to both peak candidates and their neighborhood regions ( for read density, for the proportion of zeros that are not coming from a Poisson process), the probability mass function of these models has the following form:
Assume an unobservable latent random variable , for a window of observations, the complete log-likelihood is proportional to:
In E-step at the th iteration, is estimated by its conditional expectation:
In M-step, given the estimations of and are updated as follows:
An interquartile range (IQR)-based refinement is applied before fitting ZIP models to the neighborhood regions. In this case, if certain peak candidates in a local environment are considered as outliers by IQR (their densities are above , where ), these candidates will be masked. For libraries with low sequencing depth, instead of simultaneously masking all outlier peak candidates in the local background, PINTS masks one peak candidate at a time and calculates the resulting peak density in the local background. The process will be reiterated until PINTS either identifies the outlier candidates or reports the non-existence of such outliers. The estimated densities are then used to determine the statistically significant peaks, which are further categorized into divergent peak pairs (peaks on opposite strands and within 300 bp) and unidirectional peaks.
For enhancer-like elements that do not pass the statistical cutoff of PINTS but have significant enhancer-related epigenomic marks, PINTS will offer an option to include these elements in the output where they will be labeled as “Marginal” and their corresponding epigenomic marks. To use this feature, the user needs to add --epig-annotation <biosample> when running PINTS.
PINTS depends on open-source Python packages including numpy74, scipy75, pandas, statsmodels76, pybedtools77, pyBigWig, pysam, and biopython78.
Generating peak calls with existing tools
Peak calls for different assays were made using default parameters for other existing peak callers with the following exceptions. For MACS2, --keep-dup all was set so that reads mapped to the same loci would be kept. For FivePrime, parameters , and were optimized according to the sequencing depth of corresponding libraries, and both divergent TSS calls and enhancer calls were combined as the final output. All tools were allowed to create up to 16 threads/subprocesses if they allow multithreading or parallel computing. For peak callers that do not primarily identify divergent peaks, unidirectional peaks were paired as long as they were within 300 bp and on opposite strands. Maximum memory usage and CPU time (sum of all threads) were monitored with the help from BioQueue68. UCSC Genome Browser command line tools (bedToBigBed, bigBedToBed, bigWigToBedGraph, bedGraphToBigWig, and bigWigMerge) and bedtools79 were used for the conversion of file formats. All peak calls were generated on machines with Intel Xeon Gold 6152 CPU @ 2.10 GHz with 88 cores, 1006 GB of RAM running CentOS 7.6.1810.
Evaluating the systematic biases of different peak calling methods
For each assay, divergent elements were identified using all applicable peak callers, including PINTS. To accommodate the size difference in these elements as well as elements in the CRISPR-identified enhancer set, a 1,000-bp region centered around the midpoint of each element was used to evaluate the performance of different methods.
Evaluating the upper bound of peak caller robustness
Sequencing reads were aligned to another popular reference genome sequence, hg19, and divergent elements were identified accordingly with different peak callers. Peak calls generated from both genome releases were cross lifted using UCSC’s liftover and the average between the two Jaccard indices was considered as the upper bound robustness (UBR):
ROC
For each assay, any element from the CRISPR-identified and non-enhancer set was filtered out if there were no sequencing reads aligned to both strands of the element. The positive set was composed of an equal number of randomly sampled promoters (1kb regions flanking TSSs in GENCODE v24) from expressed genes and the filtered enhancers. The negative set was composed of the filtered non-enhancers. ROCs were generated by calculating the number of divergent elements overlapping with the positive and negative sets under different cutoffs of scores: p-values of peaks for PINTS and MACS2, FDRs for TSScall, output SVR scores for dREG, likelihood ratio scores for Tfit, and peak scores for HOMER (findcsRNATSS.pl and findPeak -style TSS). For dREG.HD, GROcapTSSHMM, FivePrime, and HOMER (GRO-seq), since there are either no scores or multiple scores returned in the final output, the sensitivity and specificity were evaluated and reported with their default parameters.
PINTS web server
The PINTS web server is powered by the Django web framework. dbSNP v15380 was used for annotating mutations among TREs; JASPAR 202065 was employed for annotating transcription factor registries, and only TFs expressed in the corresponding biosample (based on RNA-seq data from ENCODE) were included (see Supplementary Table 4 for list of datasets used and accession information). cCRE v25457 was used for epigenomic annotation. Core promoter elements were annotated using the following strategy: for each major TSS (+1), the portal annotated the elements as having an initiator or an initiator-like element when the sequence of −3~+3 matches BBCABW; or the sequence of +1~+2 matches YR, respectively. The TATA box (−32~−21) and DPR elements (+17~+35) were identified using the previously published SVR model64.
Data availability
Processed TRE calls are publicly accessible via our web portal (https://pints.yulab.org). Data that support the findings of this study are available within the paper and its supplementary information files. All sequencing data analyzed in this study were retrieved from public databases (NCBI GEO and ENCODE portal); lists of accessions are available in Supplementary Tables 1 and 4.
Code availability
The source code of PINTS is publicly available at https://github.com/hyulab/PINTS; scripts and pipelines used to generate results that are reported in this study can be retrieved from https://github.com/hyulab/PINTS_analysis.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article
Extended Data
Extended Data Fig. 1. An extended evaluation of eRNA detection sensitivity of different assays.
a and c are the extended versions for Fig. 2a and b, respectively. a and b show the capability of different assays to capture previously identified enhancers. The color of stacked bars indicates the detection of eRNAs originated from either one or both strands of the enhancer loci. The transparency level shows the number of reads for an enhancer locus to be considered as covered. The top track in a is derived from the CRISPR or CRISPRi based reference set (), the bottom track is derived from consensus loci validated by STARR-seq and MPRA (). b, Sensitivity evaluated in the other cell line, GM12878, with orientation-independent enhancers identified from previous studies ()6,46. c, Differences in read coverage among stable () and unstable () transcripts. The error bars in the top track show the extrema of effect sizes (). The center dots, box limits, and whiskers in the bottom track of c denote the median, upper and lower quartiles, and 1.5× interquartile range, respectively.
Extended Data Fig. 2. Effect of technical artifacts on eRNA capture.
a, A new strategy for evaluating strand specificity without the interference from promoter-upstream transcripts (PROMPTs)81. Red and blue colors indicate reads’ mapping direction; the highlighted (yellow) region indicates a previously validated82 PROMPT. Only the first exon in green was used for evaluation. b, Strand specificities of three stranded and unstranded RNA-seq libraries with our strategy. The p-value was estimated by a two-sided t test; c, Strand specificity for all libraries evaluated with our strategy. Values and error bars represent the mean and SD. (GRO-cap, CoPRO, csRNA-seq, PRO-seq, GRO-seq, mNET-seq), (STRIPE-seq), (CAGE and RAMPAGE), (BruUV-seq, total RNA-seq), (Bru-seq). d, Distribution of 3-mers at flush end sites83 for RIP-seq and TGIRT-seq. The dashed red lines stand for the frequency of RT3-mers (sequence identical to the last three nts for the RT primer [for RIP-seq] or the 3′ adapter [for TGIRT-seq]) in the genome. e, Log odds ratios (LORs) of observed RT3-mer at flushing end sites versus in the genome (top) and internal priming rates (bottom) of assays when the internal priming could be detected from the sequencing data. f, The overlap between enhancers in the RppH library (Capped+Uncapped as “C+U”) that are also covered in the Capped library (C). The x-axis shows the minimum number of reads required for an enhancer locus to be considered as covered. g, Difference of log-transformed read counts between the capped (C) and RppH (C+U) libraries. The effect size was measured by Cohen’s d. In the box plot, the center dots, box limits, and whiskers denote the median, upper and lower quartiles, and 1.5× interquartile range, respectively. h, Pearson’s r of log-transformed reads from promoters of expressed transcripts (TPM > 5) was quantified using PRO-seq and POLR2A ChIP-exo. (low), (medium), and (high).
Extended Data Fig. 3. Analyses of factors affecting assays’ sensitivity in detecting eRNAs.
a is the extended version for Fig. 3a. b, An example shows that divergent transcripts detected by NT-assays can originate from two overlapping genes (MMP23B and SLC35E2B) instead of from a regulatory element. Sequencing reads were RPM-normalized. c, Proportion of mappable reads from different assays originated from various abundant RNA families. d, Effects of rRNA depletion in eRNA enrichment. For each category, three downsampled libraries were included. BruUV-seq libraries from a previously published study84 were used for this analysis. The p-value for rRNA percentage was calculated by two proportions z test (two-sided, p-value: 0); the p-value for true enhancer coverage was calculated by McNemar’s test (two-sided, p-value: ). Values and error bars represent the mean and SD. e, The distribution of sequencing reads (in RPM) around GENCODE-annotated splicing junction sites. The shaded area indicates the 95% confidence interval of mean values estimated via bootstrap.
Extended Data Fig. 4. Extended evaluations of assays’ specificity.
a, Epigenomic and transcription factor binding profiles for the enhancer and non-enhancer sets. For H3K27ac and CTCF, the profiles are presented as fold-changes over control; for DHS, the profile is shown as normalized sequencing depth. Solid lines represent mean densities, and shades depict the 95% confidence interval of mean values estimated via bootstrap. KE: known enhancers; NE: non-enhancers. b Signal-to-noise ratios evaluated in K562. for known enhancers, for non-enhancers. c, Signal-to-noise ratios evaluated in GM12878. (Known enhancers), and (Non-enhancers). For b and c, 10,000 bootstrapped samples were used for calculating the fold enrichment (FE). The center dots, box limits, and whiskers in b and c denote the median, upper and lower quartiles, and 1.5× interquartile range, respectively. d, False discovery rates estimated by the overlap between the top 5,000, 10,000, 20,000, and 100,000 genomic bins and the true and non-enhancer sets. Downsampled libraries were used (); values and error bars represent the mean and SD.
Extended Data Fig. 5. Assessments of transcript unit prediction and schematic illustration of PINTS.
a, The consistencies vary greatly between transcription units annotated in GENCODE (Annot.) and those predicted by different tools58,59,85 (Pred.). Lines in the violin plot indicate the 25th, 50th, and 75th quartiles, respectively. b, Schematic plot of PINTS. i, Improvement of TSS identification resolution by focusing only on read ends and using zero-inflated Poisson (ZIP) models to fit local background to address the significantly increased sparsity of signals. The thin grey lines indicate sequencing reads with the 5′ ends highlighted in red. ii, The existence of other potential true peaks (pink) elevates the estimation of read density in the local background. iii, A schematic plot shows how IQR-ZIP works. The blue box shows the read density distribution of the local background; the purple dot shows the density of the peak to be tested; the pink dot shows the density of a potential true peak close to the peak to be tested, whose read density is a clear outlier and thus excluded from local background estimation.
Extended Data Fig. 6. Profiles of peak calls generated by different peak callers for various assays.
a, Aggregated profiles of epigenomic marks, transcription binding sites, and chromatin accessibility in true enhancer regions and distal TREs identified by different peak callers for TSS- and NT-assays. The shaded area indicates the 95% confidence interval of mean values estimated via bootstrap; b, An example demonstrating why MACS2 is not suitable for identifying TREs. c, Distribution of element sizes identified from 12 assays by all applicable peak callers. In the box plot, the center lines, box limits, and whiskers denote the median, upper and lower quartiles, and 1.5× interquartile range, respectively; points show observations that are not in the range of quartiles . A table of sample sizes is available in Supplementary Table 5.
Extended Data Fig. 7. Extended analyses on the robustness of element predictions.
a, A previous study showed that the sequences between hg19 and hg38 are very similar as hg38 has 0.09% more ungapped non-centromeric sequences than hg19, only 0.17% of ungapped hg19 sequences are not in hg3861. Here we show the distribution of sequencing reads in the genome. The read counts of each assay were summarized against their frequency in a log scale with hg19 as blue lines and hg38 as orange lines. The p-values were calculated by two-sided Student’s t tests. b, Robustness (Jaccard index) of different peak callers when applying them to experimental data with technical and biological replicates. Correlations between alignments (Sample cor.) were calculated as Pearson’s r of log-transformed read counts among genomic bins (500 bp).
Extended Data Fig. 8. Performance evaluation of peak callers under different sequencing depths.
a, Epigenomic patterns of the true positive (enhancers, promoters) and true negative (non-enhancers) sets used for ROC calculation for peak calling from GRO-cap. b~d, Sensitivity and specificity of different peak callers when analyzing TSS-libraries (n=7) downsampled to 18.9 (b), 15 (c), and 10 (d) million mappable reads. The corresponding shaded areas show the 95% confidence interval of the means (via bootstrap). For tools where ROCs cannot be calculated, solid dots represent their performance with default parameters. Values and error bars show mean and SD.
Extended Data Fig. 9. Profiles of unique distal elements identified by different tools.
a, Comparison of the epigenomic signals (fold change over control) in elements uniquely identified by PINTS and other tools. b, Enrichment (measured as log odds ratios) of TF-binding motifs in PINTS unique TREs compared to other tools. The circles indicate the corresponding p-values (, two-sided z tests), and the error bars indicate the 90% confidence interval.
Extended Data Fig. 10. A summary of the computational tools compared in this study.
The features of different algorithms are summarized and grouped by their roles in the peak calling procedure (colored blocks). Features utilized by each tool to call peaks from nascent transcript sequencing data are indicated.
Supplementary Material
Acknowledgments
Computation was performed on a cluster administered by the Biotechnology Resource Center at Cornell University. We thank members of the Yu and Lis laboratories and the ENCODE Consortium (specifically Ali Mortazavi, Mats Ljungman, and Jill E. Moore) for helpful discussions and guidance; Hongya Zhu for her suggestions on concept visualization. This work was supported by grants from the National Institutes of Health (UM1HG009393 to J.T.L. and H.Y.; R01DK115398, R01DK127778, and R01HD082568 to H.Y.). L.Y. was supported by the Cornell Presidential Life Sciences Fellowship.
Footnotes
Competing Interests Statement
Authors declare no competing interests.
References
- 1.Heintzman ND et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet 39, 311–318 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Calo E. & Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol. Cell 49, 825–837 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim T-K et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Descostes N. et al. Tyrosine phosphorylation of RNA polymerase II CTD is associated with antisense promoter transcription and active enhancers in mammalian cells. Elife 3, e02105 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersson R. et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tippens ND et al. Transcription imparts architecture, function and logic to enhancer units. Nat. Genet 52, 1067–1075 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Core LJ et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet 46, 1311–1320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tome JM, Tippens ND & Lis JT Single-molecule nascent RNA sequencing identifies regulatory domain architecture at promoters and enhancers. Nat. Genet 50, 1533–1541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kruesi WS, Core LJ, Waters CT, Lis JT & Meyer BJ Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. Elife 2, e00808 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwak H, Fuda NJ, Core LJ & Lis JT Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339, 950–953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henriques T. et al. Widespread transcriptional pausing and elongation control at enhancers. Genes Dev. 32, 26–41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kodzius R. et al. CAGE: cap analysis of gene expression. Nat. Methods 3, 211–222 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Batut P, Dobin A, Plessy C, Carninci P. & Gingeras TR High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res. 23, 169–180 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirabayashi S. et al. NET-CAGE characterizes the dynamics and topology of human transcribed cis-regulatory elements. Nat. Genet 51, 1369–1379 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Duttke SH, Chang MW, Heinz S. & Benner C. Identification and dynamic quantification of regulatory elements using total RNA. Genome Res. 29, 1836–1846 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Policastro RA, Raborn RT, Brendel VP & Zentner GE Simple and efficient profiling of transcription initiation and transcript levels with STRIPE-seq. Genome Res. 30, 910–923 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Core LJ, Waterfall JJ & Lis JT Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nojima T. et al. Mammalian NET-Seq Reveals Genome-wide Nascent Transcription Coupled to RNA Processing. Cell 161, 526–540 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulsen MT et al. Coordinated regulation of synthesis and stability of RNA during the acute TNF-induced proinflammatory response. Proc. Natl. Acad. Sci. U. S. A 110, 2240–2245 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magnuson B. et al. Identifying transcription start sites and active enhancer elements using BruUV-seq. Sci. Rep 5, 17978 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H. et al. A Pan-Cancer Analysis of Enhancer Expression in Nearly 9000 Patient Samples. Cell 173, 386–399.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z. et al. Transcriptional landscape and clinical utility of enhancer RNAs for eRNA-targeted therapy in cancer. Nat. Commun 10, 4562 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azofeifa JG & Dowell RD A generative model for the behavior of RNA polymerase. Bioinformatics 33, 227–234 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danko CG et al. Identification of active transcriptional regulatory elements from GRO-seq data. Nat. Methods 12, 433–438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z, Chu T, Choate LA & Danko CG Identification of regulatory elements from nascent transcription using dREG. Genome Res. 29, 293–303 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu T. et al. Chromatin run-on and sequencing maps the transcriptional regulatory landscape of glioblastoma multiforme. Nat. Genet 50, 1553–1564 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adiconis X. et al. Comprehensive comparative analysis of 5’-end RNA-sequencing methods. Nat. Methods 15, 505–511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frith MC et al. A code for transcription initiation in mammalian genomes. Genome Res. 18, 1–12 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thakore PI et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 12, 1143–1149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fulco CP et al. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science 354, 769–773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wakabayashi A. et al. Insight into GATA1 transcriptional activity through interrogation of cis elements disrupted in human erythroid disorders. Proc. Natl. Acad. Sci. U. S. A 113, 4434–4439 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klann TS et al. CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat. Biotechnol 35, 561–568 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie S, Duan J, Li B, Zhou P. & Hon GC Multiplexed Engineering and Analysis of Combinatorial Enhancer Activity in Single Cells. Mol. Cell 66, 285–299.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Gasperini M. et al. A Genome-wide Framework for Mapping Gene Regulation via Cellular Genetic Screens. Cell 176, 377–390.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fulco CP et al. Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat. Genet 51, 1664–1669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie S, Armendariz D, Zhou P, Duan J. & Hon GC Global Analysis of Enhancer Targets Reveals Convergent Enhancer-Driven Regulatory Modules. Cell Rep. 29, 2570–2578.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schraivogel D. et al. Targeted Perturb-seq enables genome-scale genetic screens in single cells. Nat. Methods 17, 629–635 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kheradpour P. et al. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 23, 800–811 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwasnieski JC, Fiore C, Chaudhari HG & Cohen BA High-throughput functional testing of ENCODE segmentation predictions. Genome Res. 24, 1595–1602 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ulirsch JC et al. Systematic Functional Dissection of Common Genetic Variation Affecting Red Blood Cell Traits. Cell 165, 1530–1545 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ernst J. et al. Genome-scale high-resolution mapping of activating and repressive nucleotides in regulatory regions. Nat. Biotechnol 34, 1180–1190 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maricque BB, Chaudhari HG & Cohen BA A massively parallel reporter assay dissects the influence of chromatin structure on cis-regulatory activity. Nat. Biotechnol (2018) doi: 10.1038/nbt.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rathert P. et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature 525, 543–547 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dao LTM et al. Genome-wide characterization of mammalian promoters with distal enhancer functions. Nat. Genet 49, 1073–1081 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Lee D. et al. STARRPeaker: uniform processing and accurate identification of STARR-seq active regions. Genome Biol. 21, 298 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X. et al. High-resolution genome-wide functional dissection of transcriptional regulatory regions and nucleotides in human. Nat. Commun 9, 5380 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwalb B. et al. TT-seq maps the human transient transcriptome. Science 352, 1225–1228 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Core LJ et al. Defining the status of RNA polymerase at promoters. Cell Rep. 2, 1025–1035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mchaourab ZF, Perreault AA & Venters BJ ChIP-seq and ChIP-exo profiling of Pol II, H2A.Z, and H3K4me3 in human K562 cells. Sci Data 5, 180030 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harrow J. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Leary NA et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 16, 418–420 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Djebali S. et al. Landscape of transcription in human cells. Nature 489, 101–108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Field A. & Adelman K. Evaluating Enhancer Function and Transcription. Annu. Rev. Biochem 89, 213–234 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Andersson R. & Sandelin A. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet 21, 71–87 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Palazzo AF & Koonin EV Functional Long Non-coding RNAs Evolve from Junk Transcripts. Cell 183, 1151–1161 (2020). [DOI] [PubMed] [Google Scholar]
- 57.ENCODE Project Consortium et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang D. et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474, 390–394 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chae M, Danko CG & Kraus WL groHMM: a computational tool for identifying unannotated and cell type-specific transcription units from global run-on sequencing data. BMC Bioinformatics 16, 222 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneider VA et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 27, 849–864 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pennacchio LA, Bickmore W, Dean A, Nobrega MA & Bejerano G. Enhancers: five essential questions. Nat. Rev. Genet 14, 288–295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vo Ngoc L, Huang CY, Cassidy CJ, Medrano C. & Kadonaga JT Identification of the human DPR core promoter element using machine learning. Nature 585, 459–463 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fornes O. et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Landrum MJ et al. ClinVar: improvements to accessing data. Nucleic Acids Res. 48, D835–D844 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vahrenkamp JM et al. FFPEcap-seq: a method for sequencing capped RNAs in formalin-fixed paraffin-embedded samples. Genome Res. 29, 1826–1835 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 68.Yao L, Wang H, Song Y. & Sui G. BioQueue: a novel pipeline framework to accelerate bioinformatics analysis. Bioinformatics 33, 3286–3288 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Chen S, Zhou Y, Chen Y. & Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H. & Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramírez F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harris CR et al. Array programming with NumPy. Nature 585, 357–362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Virtanen P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seabold S. & Perktold J. Statsmodels: Econometric and Statistical Modeling with Python. Proceedings of the 9th Python in Science Conference (2010) doi: 10.25080/majora-92bf1922-011. [DOI] [Google Scholar]
- 77.Dale RK, Pedersen BS & Quinlan AR Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics 27, 3423–3424 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cock PJA et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics vol. 25 1422–1423 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sherry ST et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Preker P. et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322, 1851–1854 (2008). [DOI] [PubMed] [Google Scholar]
- 82.van Arensbergen J. et al. Genome-wide mapping of autonomous promoter activity in human cells. Nat. Biotechnol 35, 145–153 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shivram H. & Iyer VR Identification and removal of sequencing artifacts produced by mispriming during reverse transcription in multiple RNA-seq technologies. RNA 24, 1266–1274 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bedi K, Paulsen MT, Wilson TE & Ljungman M. Characterization of novel primary miRNA transcription units in human cells using Bru-seq nascent RNA sequencing. NAR Genom Bioinform 2, lqz014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zacher B. et al. Accurate Promoter and Enhancer Identification in 127 ENCODE and Roadmap Epigenomics Cell Types and Tissues by GenoSTAN. PLoS One 12, e0169249 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Processed TRE calls are publicly accessible via our web portal (https://pints.yulab.org). Data that support the findings of this study are available within the paper and its supplementary information files. All sequencing data analyzed in this study were retrieved from public databases (NCBI GEO and ENCODE portal); lists of accessions are available in Supplementary Tables 1 and 4.