Abstract
Vibrio cholerae is an autochthonous inhabitant of riverine and estuarine environments and also is a facultative pathogen for humans. Genotyping can be useful in assessing the risk of contracting cholera, intestinal, or extraintestinal infections via drinking water and/or seafood. In this study, environmental isolates of V. cholerae were examined for the presence of ctxA, hlyA, ompU, stn/sto, tcpA, tcpI, toxR, and zot genes, using multiplex PCR. Based on tcpA and hlyA gene comparisons, the strains could be grouped into Classical and El Tor biotypes. The toxR, hlyA, and ompU genes were present in 100, 98.6, and 87.0% of the V. cholerae isolates, respectively. The CTX genetic element and toxin-coregulated pilus El Tor (tcpA ET) gene were present in all toxigenic V. cholerae O1 and V. cholerae O139 strains examined in this study. Three of four nontoxigenic V. cholerae O1 strains contained tcpA ET. Interestingly, among the isolates of V. cholerae non-O1/non-O139, two had tcpA Classical, nine contained tcpA El Tor, three showed homology with both biotype genes, and four carried the ctxA gene. The stn/sto genes were present in 28.2% of the non-O1/non-O139 strains, in 10.5% of the toxigenic V. cholerae O1, and in 14.3% of the O139 serogroups. Except for stn/sto genes, all of the other genes studied occurred with high frequency in toxigenic V. cholerae O1 and O139 strains. Based on results of this study, surveillance of non-O1/non-O139 V. cholerae in the aquatic environment, combined with genotype monitoring using ctxA, stn/sto, and tcpA ET genes, could be valuable in human health risk assessment.
Toxigenic Vibrio cholerae O1 and V. cholerae O139 are etiological agents of epidemic cholera. However, both V. cholerae O1 strains that do not produce cholera toxin, i.e., that are nontoxigenic (NT), and non-O1/non-O139 strains have also been associated with cholera, gastroenteritis, septicemia, and/or extraintestinal infections (49, 51, 52, 63, 66, 71, 81). Outbreaks of cholera were reported in Brazil during the third (1853 to 1854), fourth (1866 to 1868), and fifth (1893 to 1895) pandemics (3). In the early 1970s, when cholera spread to Africa and Southern Europe, it was forecast to arrive in countries across the Atlantic as well. This prompted the establishment of a surveillance program in S. Paulo State, Brazil, by the WHO (World Health Organization) and CETESB (Companhia de Tecnologia de Saneamento Ambiental, S.P.-Brazil—State Agency for Environmental Control). Sewage samples were monitored for V. cholerae in the community, and 12,867 samples were collected. From these samples, four NT V. cholerae O1 strains were isolated, the first V. cholerae O1 NT strains to be isolated in Brazil. The isolates were from sewage samples collected in 1978, 1980, and 1983 (42). Non-O1 V. cholerae strains were subsequently isolated from sewage (77.3%), seawater (40.4%), and freshwater (33.3%) samples collected in 1982 and 1983, at a time when no cases of cholera or gastroenteritis had been reported in S. Paulo State (41). At the same time, in Rio de Janeiro State, non-O1 V. cholerae was isolated from 12% of seawater and oyster samples (64).
The seventh pandemic reached the Americas on 29 January 1991 in Lima, Peru, and spread rapidly to the Peruvian Northern Andean and Amazon regions (74). Brazil reported its first case of cholera on 8 April 1991 in Tocantins. Cholera cases then occurred in the Amapa, Amazonas, Maranhāo, Mato Grosso, Pará, and Rondonia States. However, in S. Paulo State, a more developed region of the country, the highest incidence of cholera in 1994 was 0.23 per 100,000 inhabitants compared to 33.4, the national average that same year (47).
The pathogenicity of V. cholerae O1 and O139 strains depends on a combination of properties, including enterotoxin (cholera toxin [CT], ctxA) and the ability to adhere to, and colonize, the small intestine (colonization factor, tcpA) (27). The major virulence-associated factors are present in clusters (23), with at least three regions in the V. cholerae chromosome. The first is the CTX genetic element (45), which has now been reported to comprise the genome of a filamentous bacteriophage (CTXΦ) (80). The second region is a large pathogenicity island for V. cholerae (VPI) (35) that encodes a toxin-coregulated pilus (TCP) gene cluster, a type IV pilus that functions as an essential colonization factor (75) and acts as CTXΦ receptor (35). The third gene cluster, the RTX toxin gene cluster, was described by Lin et al. (40) in a V. cholerae O1 El Tor strain and encodes cytotoxic activity for Hep-2 cells in vitro. However, the implication of RTX in pathogenesis has yet to be confirmed (16). Other factors have been associated with enteropathogenicity and include an El Tor-like hemolysin (hlyA) (82), heat-stable enterotoxin (stn/sto) (1, 22, 55), hemagglutinins (14), neuraminidase (nanH) (20), a new CT (33), outer membrane protein (ompU) (72), Shiga-like toxin (stx) (33), a ToxR regulatory protein (46), and a zonula occludens toxin (zot) (18).
V. cholerae can be found in the environment both as a free-living bacterium and in association with zooplankton (30). Therefore, not surprisingly, non-O1/non-O139 V. cholerae is frequently isolated from the aquatic environment and seafood (5, 8, 9, 29, 30, 31, 32, 41, 43, 44, 78). In fact, V. cholerae is a heterogeneous species, with 206 serotypes identified to date (G. B. Nair, personal communication).
The emergence of the new serogroup O139 as a second etiologic agent of cholera epidemics (48), along with the discovery of horizontal and vertical genetic transfer by phages (80) and the elucidation of pathogenicity islands and other mobile genetic elements (36), has revived interest in the non-O1/non-O139 V. cholerae strains that had been previously implicated in cholera-like epidemics (2, 13, 51, 71, 81). In addition, the possible conversion of non-O1 to O1 serotype has provided added interest (10).
Cholera surveillance is now under way in many countries, based primarily on detection of V. cholerae O1 and O139 and determining the presence of cholera toxin using biological and molecular methods. However, virulence-associated factors in V. cholerae isolates from environmental sources are of concern.
The primary objective of this study was to evaluate the presence of virulence-associated factors in V. cholerae populations as potential pathogenic markers suitable for environmental monitoring. Virulence-associated factors studied here included cholera toxin (ctxA), hemolysin (hlyA), non-O1 heat-stable enterotoxin (stn/sto), outer membrane protein (ompU), TCP (tcpA and tcpI), ToxR regulatory protein (toxR), and zonula occludens toxin (zot).
MATERIALS AND METHODS
Bacterial strains.
A total of 69 V. cholerae isolates were included in this study. V. cholerae O1 strains comprised 19 toxigenic clinical and environmental isolates from Brazil (14 isolates), Peru (3 isolates), Chile (1 isolate), and Mexico (1 isolate) and four nontoxigenic V. cholerae O1 isolates from Brazil (1978 to 1980). Thirty-nine environmental isolates of non-O1/non-O139 V. cholerae from Brazil, seven V. cholerae O139 clinical isolates from India, and five V. mimicus environmental isolates from Louisiana (United States) were included in the study (Tables 1 and 2). The Brazilian environmental strains were isolated in the CETESB Laboratory, S. Paulo, Brazil. All of the isolates are part of a culture collection (RRC) at the Center of Marine Biotechnology (Baltimore, Md.). Frozen stock cultures were subcultured on Luria-Bertani (LB) broth (Difco Laboratories, Detroit, Mich.), streaked onto LB agar, and then onto TCBS agar (Oxoid) to verify purity.
TABLE 1.
Genotypic traits of toxigenic and NT V. cholerae O1, V. cholerae O139, and V. mimicus isolates examined in this study
| Strain (no. of strains) | Country | Reference(s) | Yr | Source | Code (n) | Genotype (presence [+] or absence [−] of genes)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ctxA | hlyAET | ompU | stn/sto | tcpAET | tcpI | toxR | zot | ||||||
| Toxigenic V. cholerae O1 (19) | Brazil | 61 | 1991–1994 | Sewage | RC7, RC221, RC226, CT2578, CT8514, CT31644, CT25017, CT24082 (8) | + | + | + | − | + | + | + | + |
| Peru | 61 | 1991, 1993 | Clinical | RC8, IAL 1941 (2) | + | + | + | − | + | + | + | + | |
| Brazil | 61 | 1993 | Sewage | RC243, CT15861 (2) | + | + | + | − | + | − | + | + | |
| Chile | 1991 | Clinical | RC11 | + | + | + | − | + | + | + | + | ||
| Mexico | 1991 | Clinical | RC25 | + | + | + | − | + | + | + | + | ||
| Brazil | 41 | 1994 | River | CT7021 | + | + | + | − | + | + | + | + | |
| Brazil | 61 | 1992 | Clinical | CTMACM14 | + | + | + | − | + | − | + | + | |
| Brazil | 61 | 1993 | Sewage | RC224 | + | + | + | + | + | + | + | + | |
| Peru | 1991 | Clinical | RC24 | + | + | + | + | + | + | + | + | ||
| Brazil | 61 | 1993 | Sewage | CT15989 | + | + | + | ∗b | + | + | + | + | |
| Total positives | 19 | 19 | 19 | 2 | 19 | 16 | 19 | 19 | |||||
| NT V. cholerae O1 (4) | Brazil | 41 | 1978 | Sewage | TM45 | − | + | + | − | + | + | + | − |
| Brazil | 41, 61 | 1978 | Sewage | RC229, TM207831 (2) | − | + | + | − | + | − | + | − | |
| Brazil | 41 | 1980 | Sewage | RC231 | − | + | + | − | − | + | + | − | |
| Total positives | 0 | 4 | 4 | 0 | 3 | 2 | 4 | 0 | |||||
| V. cholerae O139 (7) | India | 61 | 1993 | Clinical | RC4, RC30, RC120, RC138, RC139 (5) | + | + | + | − | + | + | + | + |
| Bangladesh | 1993 | Clinical | RC46 | + | + | + | + | + | − | + | + | ||
| India | 61 | 1993 | Clinical | IGV | + | + | + | − | + | − | + | + | |
| Total positives | 7 | 7 | 7 | 1 | 7 | 5 | 7 | 7 | |||||
| V. mimicus (5) | Louisiana | Environa | RC54 | − | − | + | + | − | − | − | − | ||
| Louisiana | Environa | RC55 | + | + | − | + | − | + | − | + | |||
| Louisiana | Environa | RC56 | + | + | − | − | + | + | − | + | |||
| Louisiana | Environa | RC57 | + | + | − | + | − | − | − | − | |||
| Louisiana | Environa | RC59 | + | − | + | − | − | − | − | + | |||
| Total positives | 4 | 3 | 2 | 3 | 1 | 2 | 0 | 3 | |||||
Environ, environmental.
∗, Amplicon with a different size.
TABLE 2.
Genotypic traits of non-O1/non-O139 V. cholerae Brazilian isolates examined in this study
| Country (no.) | Yr | Reference(s) | Source | Code (n) | Genotype (presence [+] or absence [−] of genes)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ctxA | hlyAET | hlyAClass | ompU | stn/sto | tcpAET | tcpAClass | tcpI | toxR | zot | |||||
| Brazil (9) | 1992 | 43 | Mussels | RC236, RC237, RC238, RC244 (4) | − | + | − | + | − | − | − | + | + | − |
| 1992 | 43, 61 | Mussels | RC245, GM39 (2) | − | + | − | + | − | − | − | − | + | − | |
| 1992 | 43, 61 | Mussels | RC239, GM32 (2) | − | + | − | + | − | + | − | + | + | − | |
| 1992 | 44 | Oysters | RC247 | − | − | − | + | − | − | − | − | + | − | |
| Brazil (9) | 1981, 1982 | — | Seawater | RC63, RC64, RC65 (3) | − | + | − | + | + | − | − | + | + | − |
| 1982 | — | Seawater | RC62 | − | + | − | + | + | − | + | − | + | − | |
| 1983 | 61 | Seawater | TM16589 | − | + | − | + | − | − | − | + | + | − | |
| 1982 | — | Seawater | RC61 | + | + | − | + | + | − | + | + | + | − | |
| 1981 | 41 | Seawater | RC253 | + | + | − | + | − | + | − | − | + | − | |
| 1982 | — | Seawater | TMA52 | − | + | − | + | + | + | + | + | + | − | |
| 1982 | — | Seawater | RC60 | + | − | + | + | + | + | + | + | + | − | |
| Brazil (7) | 1982 | — | Sediment | RC68 | − | + | − | + | ∗a | − | − | − | + | − |
| 1981 | 61 | Sediment | TM50022 | − | + | − | + | − | + | − | − | + | − | |
| 1983 | — | Sediment | RC71 | − | + | − | + | − | + | − | + | + | − | |
| 1983 | — | Sediment | RC70 | − | + | − | + | + | − | − | − | + | − | |
| 1982 | — | Sediment | RC66 | − | + | − | + | + | + | − | − | + | − | |
| 1982 | — | Sediment | RC67 | − | + | − | + | + | − | − | + | + | − | |
| 1983 | — | Sediment | RC69 | − | + | − | + | + | + | + | + | + | − | |
| Brazil (14) | 1977, 1978, 1979 | 41, 61 | Wastewater | RC235, RC248, TM41338, IG4 (4) | − | + | − | − | − | − | − | − | + | − |
| 1977, 1978 | 41 | Wastewater | RC242, RC251 (2) | − | + | − | ∗a | − | − | − | − | + | − | |
| 1979, 1992 | 41 | Wastewater | RC234, RC252 (2) | − | + | − | + | − | − | − | + | + | − | |
| 1992 | 41 | River | RC241 | − | + | − | − | − | − | − | + | + | − | |
| 1983 | 41 | Wastewater | RC240 | − | + | − | − | − | + | − | + | + | − | |
| 1979 | 61 | Wastewater | TM31152 | − | + | − | ∗a | − | − | − | + | + | − | |
| 1992 | 61 | Wastewater | CT3481 | − | + | − | + | − | − | − | − | + | − | |
| 1977 | 41 | Wastewater | RC233 | + | + | − | + | − | + | − | − | + | − | |
| 1982 | 41 | Wastewater | RC246 | − | + | − | + | − | + | − | + | + | − | |
| Total strains (39) | 4 | 37 | 1 | 30 | 11 | 12 | 5 | 22 | 39 | 0 | ||||
∗, Amplicon with different size.
Positive and negative controls.
V. cholerae O1 Classical ATCC 11623, V. cholerae O1 El Tor ATCC 14033 (ctxAB negative), V. cholerae O1 Classical ATCC 14035, V. cholerae non-O1 ATCC 14547, V. cholerae non-O1 ATCC 25872 (ctxAB+), V. cholerae non-O1 ATCC 25874 (ctxAB+), V. mimicus ATCC 33653, V. cholerae O22, and V. cholerae O31 were used as positive controls, and Escherichia coli was used as a negative control.
Chromosomal DNA preparation.
DNA was extracted by the CTAB (cetyltrimethylammonium bromide) method previously described (62). DNA extracts were resuspended in Tris-EDTA (10 mM Tris-HCl, 0.10 mM EDTA [pH 8.0]) buffer and stored at 4°C for further analysis. Dilutions of template DNA were made with sterile distilled water to obtain a concentration of ca. 100 ng/μl.
PCR primers and amplification conditions.
The oligonucleotide primers for each of the selected virulence-associated factors were designed based on available GenBank sequences for V. cholerae O1 Classical and V. cholerae O1 E1 Tor for all genes, except the stn/sto genes, for which V. cholerae non-O1 and V. cholerae O1 sequences were used. The sequence positions and accession numbers of the sequences or sources are listed in Table 3. Oligonucleotide primers were synthesized by Genosys Biotechnologies, Inc.
TABLE 3.
Primers used in this study
| Gene(s), primers, and sequences (5′ to 3′) | Amplicon size (bp) | PCR conditionsa | Source or reference; accession no. |
|---|---|---|---|
| ctxA and ompU (CT subunit A and outer membrane protein) | 60–1 | ||
| 94F, CGG GCA GAT TCT AGA CCT CCT G | 564 (ctxA) | 19 | |
| 614R, CGA TGA TCT TGG AGC ATT CCC AC | |||
| 80F, ACG CTG ACG GAA TCA ACC AAA G | 869 (ompU) | This study; AE004149, AF253529, U73751 | |
| 906R, GCG GAA GTT TGG CTT GAA GTA G | |||
| zot and toxR (zonula occludens toxin and operon ToxR) | 60–1 | ||
| 225F, TCG CTT AAC GAT GGC GCG TTT T | 947 (zot) | This study; AE004224, AF123049, AF175708, M8363 | |
| 1129R, AAC CCC GTT TCA CTT CTA CCC A | |||
| 101F, CCT TCG ATC CCC TAA GCA ATA C | 779 (toxR) | This study; M21249, AE004179 | |
| 837R, AGG GTT AGC AAC GAT GCG TAA G | |||
| tcpA (TCP A [Classical and El Tor]) | 60–1 | ||
| 72F, CAC GAT AAG AAA ACC GGT CAA GAG | 451 (El Tor) | Modified from reference 39; AE004168, UO9807, X64098, X09807 | |
| 477R, CGA AAG CAC CTT CTT TCA CGT TG | |||
| 647R, TTA CCA AAT GCA ACG CCG AAT G | 620 (Classical) | ||
| tcpI (TCP I) | 60–3 | ||
| 132F, TAG CCT TAG TTC TCA GCA GGC A | 862 (tcpI) | This study; AE004168, L25659, X64098 | |
| 951R, GGC AAT AGT GTC GAG CTC GTT A | |||
| hlyA (hemolysin [Classical and El Tor]) | 60–1 | ||
| 489F, GGC AAA CAG CGA AAC AAA TAC C | 481 (El Tor) | Modified from reference 69; AE004362, AF117834, M36855, X51746, Y00557 | |
| 744F, GAG CCG GCA TTC ATC TGA AT | 738/727 (ET/Clas) | ||
| 1184R, CTC AGC GGG CTA ATA CGG TTT A | |||
| stn/sto (non-O1 heat-stable enterotoxin) | 55–1 | ||
| 67F, TCG CAT TTA GCC AAA CAG TAG AAA | 172 (stn/sto) | This study; M85198, X74108 | |
| 194R, GCT GGA TTG CAA CAT ATT TCG C |
Temperature of annealing (°C)–time of extension (min).
Multiplex PCR.
The following reagents were added to each sample PCR mixture: 2.5 μl of 10× amplification buffer A (100 mM Tris-HCl [pH 8.3], 500 mM KCl, 15 mM MgCl2, 0.01% [wt/vol] gelatin) (Promega, Madison, Wis.); 0.5 μl each of 2.5 mM dATP, dCTP, dGTP, and dTTP (Promega); 1.0 μl each of forward and reverse primers (20 μM); 0.125 μl of Taq DNA polymerase at 5 U/μl (Promega); and Milli-Q water (to a final volume of 24 μl). PCR was carried out in 0.2-ml microcentrifuge tubes with 24 μl of the PCR mixture and 1 μl (ca. 0.10 μg) of template DNA. The solution was mixed, centrifuged briefly, and placed in an automated Peltier thermal cycler (PTC-200; M. J. Research).
PCR amplification conditions were as follows: denaturation at 94°C for 2 min, annealing for 1 min at the temperatures shown in Table 3, and extension at 72°C, as given in Table 3; with a final extension step at 72°C for 10 min at the end of 30 cycles, followed by maintenance at 4°C.
PCR products were separated by agarose gel electrophoresis (1.4%) in 1× TAE buffer (0.04 Tris-acetate, 0.001 M EDTA [pH 8.0]), stained with ethidium bromide, and visualized by using UV light.
RESULTS
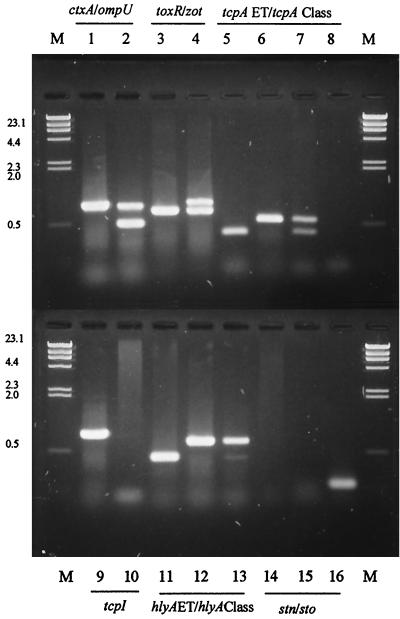
Multiplex PCR was carried out, using positive and negative controls and the primers designed for the genes studied. The size of each amplicon was verified. Optimal PCR conditions were determined for ctxA/ompU-PCR, hlyA (Classical and El Tor), zot/toxR-PCR, and tcpA (Classical and El Tor), using multiplex PCR. For tcpI and stn/sto genes, simple PCR was used. The multiplex and simple PCR products obtained for each gene investigated are shown in Fig. 1, and the corresponding amplicon sizes are given in Table 3.
FIG. 1.
Agarose gel electrophoresis of amplicons obtained using multiplex PCR. Lane M, molecular weight marker (lambda DNA HindIII); lanes 1, 3, 5, 11, and 14, V. cholerae O1 El Tor nontoxigenic ATCC 14033; lanes 2, 6, 9, 12, and 15, V. cholerae O1 Classical ATCC 14035; lane 4, RC224, V. cholerae O1 El Tor (Brazilian isolate from sewage, 1993); lane 7, TMA52, V. cholerae non-O1/non-O139 (Brazilian isolate from seawater, 1982); lane 8, negative control; lane 10, RC46, V. cholerae O139 (tcpI negative); lane 13, RC60, V. cholerae non-O1/non-O139 (Brazilian isolate from seawater, 1982); lane 16, RC66 (Brazilian isolate from sediment, 1982).
The virulence-associated factors for specific V. cholerae serogroups are summarized in Tables 1 and 2.
All of the V. cholerae O1, O139, and non-O1/non-O139 strains, regardless of whether they were toxigenic or NT, were found to possess the toxR regulatory sequence, a gene absent in the five V. mimicus strains tested.
CT and zonula occludens toxin (ZOT) were present in all (100%) of the toxigenic V. cholerae O1 and O139 strains tested and were absent in NT V. cholerae O1. In V. cholerae non-O1/non-O139 strains, we found four strains (RC60, RC61, RC233, and RC253) to be ctxA positive and zot negative. These isolates were positive for ctxA, using multiplex PCR assay, with a 564-bp amplicon identical to that of the O1 strains. The size of ctxA amplicons was confirmed by sequencing (data not shown).
The oligonucleotide primers targeting tcpA exploited sequence differences between the tcpA of the El Tor (ET) and Classical V. cholerae biotypes. All toxigenic V. cholerae O1 and O139 and three of four nontoxigenic V. cholerae O1 isolates examined showed amplicons of the same size as that obtained for the tcpA El Tor gene. Of 39 non-O1/non-O139 V. cholerae strains, 14 yielded amplicons, using tcpA primers. The amplicon size (451 bp) of nine strains (RC66, RC71, RC233, RC239, RC240, RC246, RC253, GM32, and TM50022) was identical to that obtained for V. cholerae El Tor ATCC 14033, while the amplicon size (620 bp) of two strains (RC61 and RC62) was identical to that obtained for V. cholerae O1 Classical ATCC 14035. Interestingly, three strains carried both genes (RC60, RC69, and TMA52) (Table 2).
The tcpI gene was frequently found in toxigenic V. cholerae O1 (84.2%) and O139 (71.4%) serogroups. However, this gene was also present in 50% of the NT V. cholerae O1 and in 56.4% of the non-O1/non-O139 serogroups.
The hlyA El Tor gene was found in all toxigenic and NT V. cholerae O1 and in V. cholerae O139. Among non-O1/non-O139 serogroup strains, 94.9% showed homology to El Tor hemolysin, 2.6% were associated with Classical hemolysin, and 2.5% were negative for both genes. The amplified fragment sizes were 727 bp, specifically for the Classical biotype (ATCC 11623 and ATCC 14035) and both 481 bp and 738 bp for the El Tor biotype. Occasionally, a larger fragment (∼1.4 kb) was observed in the El Tor biotype strains.
The ompU gene was found in all strains of toxigenic and NT V. cholerae O1, V. cholerae O139, and in 76.9% of the environmental non-O1/non-O139 V. cholerae isolates.
Genes homologous to stn/sto were observed in toxigenic V. cholerae O1 (2 of 19), V. cholerae O139 (1 of 7), and non-O1/non-O139 V. cholerae (11 of 39) strains, yielding an amplicon size of 172 bp. However, we observed an amplicon of 800 bp in one isolate from sediment (RC68).
The genotypes found in each V. cholerae serogroup are listed in Table 4. The genotype most frequently observed in clinical toxigenic V. cholerae O1 (68.4%) and O139 (71.4%) isolates was ctxA hlyAET ompU tcpAET tcpI toxR zot. Other genotypes found in toxigenic V. cholerae O1 strains were similar, but the tcpI and/or stn/sto genes were absent. In non-O1/non-O139 V. cholerae serogroups, the genotypes were diverse; however, the most frequent genotypes were hlyAET ompU tcpI toxR (7 of 39) and hlyA toxR (6 of 39).
TABLE 4.
Genotypes of V. cholerae isolates listed by serogroup
| Genotypes | No. of isolates
|
||||
|---|---|---|---|---|---|
| O1 | NT O1 | O139 | Non-O1/non-O139 | Total | |
| ctxA+hlyA+ET ompU+tcpA+ET tcpI+toxR+zot+ | 13 | 0 | 5 | 0 | 18 |
| ctxA+hlyA+ET ompU+tcpA+ET toxR+zot+ | 3 | 0 | 1 | 0 | 4 |
| ctxA+hlyA+ET ompU+stn/sto+tcpA+ET tcpI+toxR+zot+ | 2+1a | 0 | 0 | 0 | 3 |
| ctxA+hlyA+ET ompU+tcpA+ET toxR+ | 0 | 0 | 0 | 2 | 2 |
| ctxA+hlyA+ET ompU+stn/sto+tcpA+ET toxR+zot+ | 0 | 0 | 1 | 0 | 1 |
| ctxA+hlyA+ET ompU+stn/sto+tcpA+Class tcpI+toxR+ | 0 | 0 | 0 | 1 | 1 |
| ctxA+hlyA+Class ompU+stn/sto+tcpA+ET/Class tcpI+toxR+ | 0 | 0 | 0 | 1 | 1 |
| hlyA+ET ompU+tcpI+toxR+ | 0 | 1 | 0 | 7 | 8 |
| hlyA+ET toxR+ | 0 | 0 | 0 | 6 | 6 |
| hlyA+ET ompU+tcpA+ET tcpI+toxR+ | 0 | 1 | 0 | 4 | 5 |
| hlyA+ET ompU+stn/sto+tcpI+toxR+ | 0 | 0 | 0 | 4 | 4 |
| hlyA+ET ompU+toxR+ | 0 | 0 | 0 | 4 | 4 |
| hlyA+ET ompU+tcpA+ET toxR+ | 0 | 2 | 0 | 1 | 3 |
| hlyA+ET ompU+stn/sto+tcpA+ET/Class tcpI+toxR+ | 0 | 0 | 0 | 2 | 2 |
| hlyA+ET tcpI+toxR+ | 0 | 0 | 0 | 2 | 2 |
| hlyA+ET ompU+stn/sto+tcpA+Class toxR+ | 0 | 0 | 0 | 1 | 1 |
| hlyA+ET ompU+stn/sto+tcpA+ET toxR+ | 0 | 0 | 0 | 1 | 1 |
| hlyA+ET tcpA+ET tcpI+toxR+ | 0 | 0 | 0 | 1 | 1 |
1, i.e., an stn/sto amplicon with another size.
The presence of virulence-associated genes in non-O1/non-O139 V. cholerae isolates was analyzed by origin, i.e., water and sediment for marine ecosystems, mussels, and wastewater, and the results are summarized in Table 5.
TABLE 5.
Distribution of virulence-associated genes among non-O1/non-O139 V. cholerae isolates listed by source
| Brazilian source | n | Virulence-associated gene-positive isolatesa
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ctxA |
hlyA
|
stn/sto | ompU |
tcpA
|
tcpI | toxR | zot | |||||
| ET | C | ET | C | ETC | ||||||||
| Mussels | 9 | 0 | 8 | 0 | 0 | 9 | 2 | 0 | 0 | 6 | 9 | 0 |
| Seawater | 9 | 3 | 8 | 1 | 7 | 9 | 1 | 2 | 2 | 7 | 9 | 0 |
| Sediment | 7 | 0 | 7 | 0 | 4 | 7 | 3 | 0 | 1 | 3 | 7 | 0 |
| Wastewater | 14 | 1 | 14 | 0 | 0 | 5 | 3 | 0 | 0 | 6 | 14 | 0 |
| Total | 39 | 4 | 37 | 1 | 11 | 30 | 9 | 2 | 3 | 22 | 39 | 0 |
C, Classical; ETC, both ET and Classical.
DISCUSSION
The majority of V. cholerae strains in the environment are considered to be harmless estuarine microorganisms. However, specific strains appear to have evolved that cause disease in humans by effectively colonizing the small intestine and releasing a potent enterotoxin. Eight chromosomal regions coding for virulence-associated factors in V. cholerae (ctxA, hlyA, ompU, stn/sto, tcpA, tcpI, toxR, and zot) were included in this study.
Distribution of virulence-associated genes in V. cholerae.
The major CTX genetic element has the structure of a compound transposon, with a 4.5-kb central core region (ctxAB, zot, ace, orfU, and cep) that encodes for both CT and for functions required for virion morphogenesis and is flanked by one or more copies of a 2.7-kb repetitive sequence that encodes functions required for regulation, replication, and integration of CTXφ (36, 80). In this region, we tested for the presence of two genes: CT subunit A (ctxA) and ZOT (zot). Interestingly, we found four of the non-O1/non-O139 V. cholerae isolates contained ctxA gene but not the zot gene. The ZOT was described by Fasano et al. (18) as a toxin that increases permeability of the small intestinal mucosa by affecting the structure of the intercellular tight junction, or zonula occludens. However, Waldor and Mekalanos (80) found that zot and orfU correspond to genes involved in CTXΦ morphogenesis and that the biological activity previously designated “zonula occludens toxin” is probably not directly associated with the zot gene product, unless its product possesses a dual function. Additionally, the presence of the zot gene and the absence of the ctx gene in V. cholerae and V. mimicus strains has been reported (7, 21, 38). To explore the CTXΦ genome integrated in the V. cholerae chromosome, sequencing of the CTX genetic element is under way.
The V. cholerae pathogenicity island (VPI) is 39.5 kb and contains genes associated with virulence (TCP-ACF cluster), the regulation of virulence (toxT and tcpP/H), the regulation of chemotaxis (tcpI or acfB), and mobility (int and orfI) (24, 36). Because the genes encoding TCP have been suggested to be part of a larger genetic element consisting of a cluster of genes, we looked for the presence of both tcpA and tcpI genes. The tcpAET gene was present in all toxigenic V. cholerae O1 and O139 isolates, showing that despite different lipopolysaccharide structures, the two share common TCP-associated antigens (60). The tcpAET gene was also found in 3 of 4 strains of NT V. cholerae O1 and in 9 of 39 non-O1/non-O139 Brazilian strains. Interestingly, in the non-O1/non-O139 serogroup, two strains had tcpA genes of the Classical biotype and three had tcpA gene of both biotypes, in agreement with previous reports (6, 54). These results suggest that these strains may have a selective advantage over nonpathogenic strains, with an ability to colonize the human intestine, and become toxigenic (35). However, while the El Tor TcpA pilin is 82% identical to TcpA of the Classical biotype (60), there are at least four major variants of tcpA genes that probably evolved in parallel, though independently, from a common ancestral gene (54). Sequencing of tcpA El Tor amplicons demonstrated the fragment size to be 451 bp, and the amplicon sizes for tcpA Classical was 620 bp in the V. cholerae non-O1/non-O139 strains (data not shown). These variations could indicate other functional significance as well, because TcpA also acts as a coat protein of the bacteriophage VPΦ, produced by vibrios containing VPI (37, 77).
In this study, the presence of strains containing tcpA genes, either identical to those of biotype El Tor or of Classical, is in agreement with results obtained in Australia, where V. cholerae serogroup O6 was shown to contain the tcpAET gene and another strain of serogroup O23 presented as tcpA Classical, both strains having been isolated from water (67).
TcpI, an integral inner membrane protein, involved in environmental sensing and signal transduction (chemosensors), negatively regulates the synthesis of the major pilin subunit of TCP (TcpA) (24, 76). Harkey et al. (24) suggested that regulators such as TcpI, that act downstream of ToxR and ToxT may function to fine-tune the expression of the TCP virulence determinant throughout the pathogenic cycle of V. cholerae. However, our results showing the presence of tcpI gene in 65.5% of V. cholerae strains suggests that the importance of this gene may be physiological and not pathogenesis alone.
Outer membrane protein, OmpU, was reported to be a potential adherence factor for V. cholerae (72). However, later studies suggested that OmpU is not involved in the adhesion of V. cholerae to the intestinal epithelium (53). In this study, the gene was present in 87% of the V. cholerae strains tested, except in nine non-O1/non-O139 V. cholerae strains isolated from wastewater. These findings suggest that this gene may be mainly physiological in its activity.
The hemolysin traditionally has been employed to differentiate between the two biotypes of V. cholerae O1. The sequence of the Classical biotype has an 11-bp deletion within the hlyA structural gene, compared to the El Tor biotype (57). Using information on these genes, we designed primers to differentiate both biotypes and verified the presence of this gene in 98.6% of the V. cholerae strains, in agreement with previous reports (4, 68, 71, 82, 83). The majority belonged to the El Tor biotype, regardless of the hemolytic phenotype (67 of 69). The multiplex PCR for hemolysin effectively distinguished the two biotypes in non-O1/non-O139 V. cholerae strains compared with V. cholerae O1 Classical ATCC 11623 and ATCC 14035 and V. cholerae O1 El Tor ATCC 14033. Interestingly, the hlyA gene is located on chromosome 2 (26).
V. cholerae non-O1/non-O139 strains may also produce a 17-amino-acid heat-stable enterotoxin (NAG-ST) (stn) that is closely related to the heat-stable toxins produced by enterotoxigenic E. coli and other enteric pathogens (1, 55). Also, a heat-stable enterotoxin in V. cholerae O1 strains (sto) has been described (22). We designed primers based on the stn/sto genes associated with heat-stable enterotoxin production and found the genes to be homologous to those of toxigenic V. cholerae O1 (2 of 19), V. cholerae O139 (1 of 7), and non-O1/non-O139 V. cholerae (11 of 39) strains. This is the first report showing higher values than those reported in other studies, using hybridization with a NAG-ST probe, in V. cholerae non-O1/non-O139 population (12, 28, 56, 58). Interestingly, the stn/sto gene occurred more frequently in isolates from seawater (7 of 9) and sediment (4 of 7) and was absent in isolates from sewage (0 of 14) or oysters and mussels (0 of 9). It should be mentioned, however, that Caldini et al. (5) reported the presence of the sto gene in 12.7% (19 of 150) of the V. cholerae non-O1 isolates from freshwater in the river basin of central Italy, but the incidence of NAG-ST in V. cholerae non-O1 was not clearly established. Results of a study involving human volunteers demonstrated that, besides the production of NAG-ST, the virulence of non-O1/non-O139 V. cholerae depends on its ability to colonize the intestine (50).
The regulation and expression of genes for growth and survival depend on the regulon ToxR, coordinately regulated by a cascade mechanism involving three known components: ToxR, ToxS, and ToxT (15). ToxR, a 32-kDa transmembrane protein, is the master regulator, and its expression is dependent upon environmental growth conditions (incubation temperature, pH, osmolarity, bile salts, oxygen tension, hydrostatic pressure, and amino acid composition of the medium) (15, 46). The toxR gene encodes a transcriptional activator controlling CT gene expression (ctxA), TCP biogenesis (tcpA), outer membrane protein expression (ompU), and at least 17 distinct genes in O139 and O1 strains (15, 27, 46). In this study, the presence of the toxR gene was verified in all V. cholerae studied, regardless of serogroup or source of isolation, a finding in agreement with previous reports (21, 71).
The presence of toxR (100%), hlyA (98.6%), ompU (87%), and tcpI (65.5%) genes in the V. cholerae isolates suggest that they are required for functioning of the organism in the environment and are not solely related to pathogenesis.
Five V. mimicus environmental strains were included in our study because of their close relationship to V. cholerae. None carried the toxR gene. The presence of CT in four of the five V. mimicus strains was confirmed, while the zot gene was found in only three of the strains, results similar to those reported earlier by Chowdhury et al. (7). The stn/sto gene was found in three of five environmental strains, as reported elsewhere, using PCR (79). In contrast, Pal et al. (56) reported the presence of NAG-ST gene in 13.7 and 22.6% of V. mimicus isolates from environmental and clinical sources, respectively, and Ramamurthy et al. (59) suggested that V. mimicus may act as a genetic reservoir for these genes.
Multiplex PCR was found to be sensitive and specific for assessing the pathogenicity of clinical and environmental V. cholerae isolates. We tested multiplex PCR for ctxA/tcpA El Tor genes using four primers—94F, 614R, 72F, and 477R—as a primary screening for V. cholerae during epidemiological surveillance with good success (data not shown).
V. cholerae genotypes.
A single factor cannot explain enteropathogenicity. In this study eight virulence-associated genes were detected in V. cholerae strains isolated from both clinical and environment sources. Previous findings, in an earlier study of 172 V. cholerae non-O1/non-O139 environmental isolates from seawater and sediment samples collected in São Paulo, Brazil, showed that 60.4% of the strains were hlyAC+, nanH+, and toxR+ and ctxAB, elt (33), and zot negative as determined by using probes and radioactive hybridization. The frequency of occurrence of the genes among the strains tested was 98.8% for toxR, 97.1% for hlyAC, 66.9% for nanH, 5.8% for slt, 1.2% for zot, and 0.6% for ctxAB (D. E. Alvarado, V. H. Pellizari, T. A. T. Gomes, and I. N. G. Rivera, Abstr. 8th Int. Symp. Microb. Ecol. p. 89, 1998). In this study, we included V. cholerae O1 toxigenic and NT strains and V. cholerae O139 strains, using multiplex PCR, finding that 100% of the V. cholerae O1 and O139 strains carried ctxA, hlyAET, ompU, tcpAET, toxR, zot, and tcpI and sometimes the stn/sto gene.
Non-O1/non-O139 V. cholerae strains tested revealed the genotype hlyAET+ ompU+ tcpI+ toxR+ and negative for ctxA, stn/sto, tcpAET, and zot for isolates from mussels (4 of 9), wastewater (2 of 14), and seawater (1 of 9). Interestingly, we observed the genotype hlyAET+ toxR+ and negative for ctxA, ompU, stn/sto, tcpAET, and zot in 6 of 14 non-O1/non-O139 V. cholerae isolates from wastewater, a finding similar to the virulence pattern obtained for 15 clinical strains associated with an unusual upsurge of cholera-like disease in India (71) and in non-O1/non-O139 V. cholerae strains isolated from volunteers in a vaccine trial in Peru (13).
A low frequency of the combination ctxA+ tcpAClass+ (1 of 39), and ctxA+ tcpAET+ (3 of 39) was observed in environmental non-O1/non-O139 V. cholerae strains, as reported earlier (21). However, we found 23.1% (9 of 39) of CT-negative, but TcpAET-positive strains, potentially convertible to toxigenic strains by CTXΦ, either inside the host intestine or in the environment.
V. cholerae is autochthonous to estuarine and coastal environments (nutrient poor) and also colonize the human intestine (nutrient rich). In the study reported here, we observed a varied incidence of virulence-associated factors in V. cholerae isolates, depending on the source or ecosystem (seawater, seafood, wastewater, and clinical specimens). The response to changing conditions occurs by expressing appropriate sets of genes that promote growth in each niche.
Aquatic and marine ecosystems are subjected to large spatial and temporal nutrient fluxes arising from seasonal and geographic variations in temperature, salinity, nutrient input, pH, oxygen tension, etc. (65). The mechanisms by which environmental conditions affect the coordinated expression of virulence factors by V. cholerae remain poorly understood. Hase and Mekalanos (25) proposed a model where both ToxR and -S and TcpP and -H are involved in sensing various environmental and internal stimuli and are required for the production of TCP in V. cholerae. Furthermore, the V. cholerae genome encodes 43 methyl-accepting chemotaxis proteins that regulate swimming behavior in response to aminoacids, sugars, and oxygen (16).
Non-O1/non-O139 V. cholerae and risk assessment.
Non-O1/non-O139 V. cholerae strains can no longer be ignored. The rationale for continuous monitoring is based first on the emergence of serogroup O139 (Bengal) in Bangladesh (CT positive) and Argentina (CT negative), each of which clearly evolved independently (73). Second, the sixth pandemic, the seventh pandemic, and U.S. Gulf Coast isolates represent three different clones, each independently evolved from environmental non-O1 V. cholerae isolates (34). Third, other epidemic serogroups have emerged, including V. cholerae O31 (50), O37 (11), O53 (52), O10, and O12 (13, 71). Finally, the emergence of a new clone of the V. cholerae O1 El Tor in Calcutta, India (70), and in Bangladesh (17) has been reported. The possibility exists that additional new strains of toxigenic V. cholerae with epidemic potential may emerge in the future.
We reported earlier an ERIC-PCR method for detecting emergent pathogenic V. cholerae strains, whereby specific fingerprints for pathogenic strains (CT and ZOT toxin positive) were different from nonpathogenic strains (61). In this study, we report probable parental toxigenic genotypes present in small numbers in the Brazilian environment. While these strains did not cause epidemics, there are environmental factors that may change, enhance multiplication or dominance, or select for genotypes of V. cholerae, and the strains themselves need to be tested for potential selective advantage under selected environmental conditions.
ACKNOWLEDGMENTS
Irma N. G. Rivera was supported by a Fellowship from UNESCO-ASM during the research period and from CNPQ (National Council for Research Support [Brazil]) during the postdoctoral period.This research was supported by NIH (grant 1R01A139129-01) and EPA (grant R824995-01).
We thank P. S. Sanchez and M. I. Sato from the State Agency for Environmental Control-CETESB, S. Paulo, Brazil, for providing the strains in this study.
REFERENCES
- 1.Arita M T, Takeda T, Honda T, Miwatani T. Purification and characterization of Vibrio cholerae non-O1 heat-stable enterotoxin. Infect Immun. 1986;52:45–49. doi: 10.1128/iai.52.1.45-49.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagchi K, Echevarria P, Arthur J D, Sethabutr O, Serichantalergs O, Hoge C W. Epidemic of diarrhea caused by Vibrio cholerae non-O1 that produced heat-stable toxin among Khmers in a camp in Thailand. J Clin Microbiol. 1993;31:1315–1317. doi: 10.1128/jcm.31.5.1315-1317.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barua D. History of cholera. In: Barua D, Greenough III W B, editors. Cholera. New York, N.Y: Plenum Medical Book Company; 1992. pp. 1–35. [Google Scholar]
- 4.Brown M H, Manning P A. Haemolysin genes of Vibrio cholerae: presence of homologous DNA in non-haemolytic O1 and haemolytic non-O1 strains. FEMS Microbiol Lett. 1985;30:197–201. [Google Scholar]
- 5.Caldini G, Neri A, Cresti S, Boddi V, Rossolini G M, Lanciotti E. High prevalence of Vibrio cholerae non-O1 carrying heat-stable-enterotoxin-climate river basin of central Italy. Appl Environ Microbiol. 1997;63:2934–2939. doi: 10.1128/aem.63.7.2934-2939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakraborty S, Mukhopadhyay A K, Bhadra R K, Ghosh A N, Mitra R, Shimada T, Yamasaki S, Faruque S M, Takeda Y, Colwell R R, Nair G B. Virulence genes in environmental strains of Vibrio cholerae. Appl Environ Microbiol. 2000;66:4022–4028. doi: 10.1128/aem.66.9.4022-4028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chowdhury M A R, Hill R T, Colwell R R. A gene for the enterotoxin zonula occludens toxin is present in V. mimicus and V. cholerae O139. FEMS Microbiol Lett. 1994;119:377–380. doi: 10.1111/j.1574-6968.1994.tb06916.x. [DOI] [PubMed] [Google Scholar]
- 8.Colwell R R, Kaper J B, Joseph S W. Vibrio cholerae and Vibrio parahaemolyticus and other vibrios: occurrence and distribution in Chesapeake Bay. Science. 1977;198:394–396. [PubMed] [Google Scholar]
- 9.Colwell R R, Spira W M. The ecology of Vibrio cholerae. In: Barua D, Greenough III W B, editors. Cholera. New York, N.Y: Plenum Medical Book Company; 1992. pp. 107–127. [Google Scholar]
- 10.Colwell R R, Huq A, Chowdhury M A R, Brayton P R, Xu B. Serogroup conversion of Vibrio cholerae. Can J Microbiol. 1995;41:946–950. doi: 10.1139/m95-131. [DOI] [PubMed] [Google Scholar]
- 11.Dakin S P H, Howel D J, Sutton R G A, O'Keffe M E, Thoms P. Gastroenteritis due to non-agglutinable (non-cholera) vibrios. Med J Aust. 1994;2:490. doi: 10.5694/j.1326-5377.1974.tb70935.x. [DOI] [PubMed] [Google Scholar]
- 12.Dalsgaard A, Serichantalergs O, Shimada T, Sethabutr O, Echevarria P. Prevalence of Vibrio cholerae with heat-stable enterotoxin (NAG-ST) and cholera toxin genes; restriction fragment length polymorphisms of NAG-ST genes among V. cholerae O serogroups from a major shrimp production area in Thailand. J Med Microbiol. 1995;43:216–220. doi: 10.1099/00222615-43-3-216. [DOI] [PubMed] [Google Scholar]
- 13.Dalsgaard A, Albert M J, Taylor D N, Shimada T, Meza R, Serichantalergs O, Echevarria P. Characterization of Vibrio cholerae non-O1 serogroups obtained from an outbreak of diarrhea in Lima, Peru. J Clin Microbiol. 1995;33:2715–2722. doi: 10.1128/jcm.33.10.2715-2722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datta-Roy K, Banerjee K, De S P, Ghose A C. Comparative study of expression of hemagglutinins, hemolysins, and enterotoxins by clinical and environmental isolates of non-O1 of Vibrio cholerae in relation to their enteropathogenicity. Appl Environ Microbiol. 1986;52:875–879. doi: 10.1128/aem.52.4.875-879.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiRita V J. Co-ordinate control of virulence gene expression by ToxR in Vibrio cholerae. Mol Microbiol. 1992;6:451–458. doi: 10.1111/j.1365-2958.1992.tb01489.x. [DOI] [PubMed] [Google Scholar]
- 16.DiRita V J. Genomics happens. Science. 2000;289:1488. doi: 10.1126/science.289.5484.1488. [DOI] [PubMed] [Google Scholar]
- 17.Faruque S M, Albert M J, Mekalanos J J. Epidemiology, genetics and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fasano A, Baudry B, Pumplin D W, Wasserman S S, Tall B D, Ketley J M, Kaper J B. Vibrio cholerae produces a second enterotoxin which affects intestinal tight junctions. Proc Nat Acad Sci USA. 1991;88:5242–5246. doi: 10.1073/pnas.88.12.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fields P I, Popovic T, Wachsmuth K, Olsvik O. Use of polymerase chain reaction for detection ot toxigenic Vibrio cholerae O1 strains from the Latin American cholera epidemics. J Clin Microbiol. 1992;30:2128–2121. doi: 10.1128/jcm.30.8.2118-2121.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galen J E, Ketley J M, Fasano A, Richardson S H, Wasserman S S, Kaper J B. Role of Vibrio cholerae neuraminidase in the function of cholera toxin. Infec Immun. 1992;60:406–415. doi: 10.1128/iai.60.2.406-415.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh C, Nandy R K, Dasgupta S K, Nair G B, Hall R H, Ghose A C. A search for cholera toxin (CT), toxin coregulated pilus (TCP), the regulatory element ToxR and other virulence factors in non-O1/non-O139 Vibrio cholerae. Microb Pathog. 1997;22:199–208. doi: 10.1006/mpat.1996.0105. [DOI] [PubMed] [Google Scholar]
- 22.Guglielmetti P, Bravo L, Zanchi A, Montè R, Lombardi G, Rossolini G M. Detection of the Vibrio cholerae heat-stable enterotoxin gene by polymerase chain reaction. Mol Cell Probes. 1994;8:101–106. doi: 10.1006/mcpr.1994.1005. [DOI] [PubMed] [Google Scholar]
- 23.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 24.Harkey C W, Everiss K D, Peterson K M. The Vibrio cholerae toxin-coregulated-pilus gene tcpI encodes a homolog of methyl-accepting chemotaxis proteins. Infect Immun. 1994;62:2669–2678. doi: 10.1128/iai.62.7.2669-2678.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Häse C C, Mekalanos J J. TcpP protein is a positive regulator of virulence gene expression. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, Gill S R, Nelson K E, Read T D, Tettelin H, Richardson D, Ermolaeva M D, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann R D, Nierman W C, White O, Salzberg S L, Smith H O, Colwell R R, Mekalanos J J, Venter J C, Fraser C M. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–484. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herrington D A, Hall R H, Losonsky G, Mekalanos J J, Taylor R K, Levine M M. Toxin, toxin-coregulated pili and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med. 1988;168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoge C W, Sethabutr O, Bodhidatta L, Echevarria P, Robertson D C, Morris J G., Jr Use of a synthetic oligonucleotide probe to detect strains of non-serovar O1 Vibrio cholerae carrying the gene for heat-stable enterotoxin (NAG-ST) J Clin Microbiol. 1990;28:1473–1476. doi: 10.1128/jcm.28.6.1473-1476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huq A, Small E B, West P A, Huq M I, Rahman R, Colwell R R. Ecology of V. cholerae with special reference to planktonic crustacean copepods. Appl Environ Microbiol. 1983;45:275–283. doi: 10.1128/aem.45.1.275-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huq A, Colwell R R. Vibrios in the marine and estuarine environment: tracking Vibrio cholerae. Ecosystem Health. 1996;2:198–214. [Google Scholar]
- 31.Islam M S, Drasar B S, Sack R B. The aquatic environment as reservoir of Vibrio cholerae: a review. J Diarrhoeal Dis Res. 1994;11:197–206. [PubMed] [Google Scholar]
- 32.Kaper J, Lockman H, Colwell R R, Joseph S W. Ecology, serology and enterotoxin production of Vibrio cholerae in Chesapeake Bay. Appl Environ Microbiol. 1979;37:91–103. doi: 10.1128/aem.37.1.91-103.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaper J B, Fasano A, Trucksis M. Toxins of Vibrio cholerae. In: Wachsmuth I K, Blake P A, Olsvik O, editors. Vibrio cholerae and cholera: molecular to global perspectives. Washington, D.C.: ASM Press; 1994. pp. 145–176. [Google Scholar]
- 34.Karaolis D K R, Lan R, Reeves P R. The sixth and seventh cholera pandemic are due to independent clones separately derived from environmental, non-toxigenic, non-O1 Vibrio cholerae. J Bacteriol. 1995;177:3193–3198. doi: 10.1128/jb.177.11.3191-3198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karaolis D K R, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karaolis D K R, Kaper J B. Pathogenicity islands and other mobile virulence elements of Vibrio cholerae. In: Kaper J B, Hacker J, editors. Pathogenicity islands and other mobile virulence elements. Washington, D.C.: ASM Press; 1999. pp. 167–187. [Google Scholar]
- 37.Karaolis D K R, Somara S, Maneval D R, Jr, Johnson J A, Kaper J B. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature. 1999;399:375–379. doi: 10.1038/20715. [DOI] [PubMed] [Google Scholar]
- 38.Karasawa T, Mihara T, Kurazono H, Nair B G, Garg S, Ramamurthy T, Takeda Y. Distribution of the zot (zonula occludens toxin) gene among strains of Vibrio cholerae O1 and non-O1. FEMS Microbiol Lett. 1993;106:143–146. doi: 10.1111/j.1574-6968.1993.tb05950.x. [DOI] [PubMed] [Google Scholar]
- 39.Keasler S P, Hall R H. Detecting and biotyping Vibrio cholerae O1 with multiplex chain reaction. Lancet. 1993;341:1661. doi: 10.1016/0140-6736(93)90792-f. [DOI] [PubMed] [Google Scholar]
- 40.Lin W, Fullner K J, Clayton R, Sexton J A, Rogers M B, Calia K E, Calderwood S B, Fraser C, Mekalanos J J. Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc Natl Acad Sci USA. 1999;96:1071–1076. doi: 10.1073/pnas.96.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martins M T. Ecologia de Vibrio cholerae no ecossistema aquático. Tese Livre Docente ICB-USP. São Paulo, Brazil: Universidade de São Paulo; 1988. . (In Portuguese.). [Google Scholar]
- 42.Martins M T, Pessõa G V A, Sanchez P S, Sato M I Z, Coimbrão C A, Monteiro C K, Marques E. Occurrence of V. cholerae O1 non-toxigenic in wastewaters from São Paulo, Brazil. Wat Sci Tech. 1991;24:363–366. [Google Scholar]
- 43.Matté G R, Matté M H, Sato M I Z, Sanchez P S, Rivera I G, Martins M T. Potentially pathogenic vibrios associated with mussels from a tropical region on the Atlantic Coast of Brazil. J Appl Bacteriol. 1994;77:281–287. doi: 10.1111/j.1365-2672.1994.tb03075.x. [DOI] [PubMed] [Google Scholar]
- 44.Matte G R, Matté M H, Rivera I G, Martins M T. Distribution of potentially vibrios in oysters from a tropical region. J Food Prot. 1994;57:870–873. doi: 10.4315/0362-028X-57.10.870. [DOI] [PubMed] [Google Scholar]
- 45.Mekalanos J J. Cholera toxin: genetic analysis, regulation, and role in pathogenesis. Curr Top Microbiol Immunol. 1985;118:97–118. doi: 10.1007/978-3-642-70586-1_6. [DOI] [PubMed] [Google Scholar]
- 46.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 47.Ministerio de Saúde-Fundação Nacional de Saúde. MS/FNS, 1999—Boletin epidemiológico CENEPI, 1999. São Paulo, Brazil: Ministerio de Saúde-Fundação Nacional de Saúde, Centro de Vigilância Epidemiologica; 1999. [Google Scholar]
- 48.Mooi F R, Bik E M. The evolution of epidemic Vibrio cholerae strains. Trends Microbiol. 1997;5:161–165. doi: 10.1016/S0966-842X(96)10086-X. [DOI] [PubMed] [Google Scholar]
- 49.Morris J G, Jr, Picardi J L, Lieb S, Lee J V, Roberts A, Hood M, Guun R A, Blake P. Isolation of nontoxigenic Vibrio cholerae O group 1 from a patient with severe gastrointestinal disease. J Clin Microbiol. 1984;19:196–197. doi: 10.1128/jcm.19.2.296-297.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morris J G, Jr, Takeda T, Tall B D, Losonsky G A, Bhattacharya S K, Forrest B D, Kay B A, Nishibuchi M. Experimental non-O1 group Vibrio cholerae gastroenteritis in humans. J Clin Investig. 1990;85:697–705. doi: 10.1172/JCI114494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morris J G. Non-O1 group 1 Vibrio cholerae strains not associated with epidemic disease. In: Wachsmuth I K, Blake P A, Olsvik O, editors. Vibrio cholerae and cholera: molecular to global perspectives. Washington, D.C.: ASM Press; 1994. pp. 103–116. [Google Scholar]
- 52.Mukhopadhyay A K, Saha P K, Garg S, Bhattacharya S K, Shimada T, Takeda T, Takeda Y, Nair G B. Distribution and virulence of Vibrio cholerae belonging to serogroups other than O1 and O139: a nation survey. Epidemiol Infect. 1995;114:65–70. doi: 10.1017/s0950268800051918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakasone N, Iwanaga M. Characterization of outer membrane protein OmpU of Vibrio cholerae O1. Infect Immun. 1998;66:4726–4728. doi: 10.1128/iai.66.10.4726-4728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nandi B, Nandy R K, Vicente A C P, Ghose A C. Molecular characterization of a new variant of toxin-coregulated pilus protein (TcpA) in a toxigenic non-O1/non-O139 strain of Vibrio cholerae. Infect Immun. 2000;68:948–952. doi: 10.1128/iai.68.2.948-952.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogawa A, Kato J, Watanabe H, Nair B G, Takeda T. Cloning and nucleotide sequence of a heat-stable enterotoxin gene from Vibrio cholerae non-O1 isolated from a patient with traveler's diarrhea. Infect Immun. 1990;58:3325–3329. doi: 10.1128/iai.58.10.3325-3329.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pal A, Ramamurthy T, Bhadra R K, Takeda T, Shimada T, Takeda Y, Nair G B, Pal S C, Chakrabarty S. Reassessment of the prevalence of heat-stable enterotoxin (NAG-ST) among environmental Vibrio cholerae non-O1 strains isolated from Calcutta, India, by using a NAG-ST DNA probe. Appl Environ Microbiol. 1992;58:2485–2489. doi: 10.1128/aem.58.8.2485-2489.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rader A E, Murphy J R. Nucleotide sequences and comparison of the hemolysin determinants of Vibrio cholerae El Tor RV79 (Hly+) and RV79 (Hly−) and Classical 569B (Hly−) Infect Immun. 1988;56:1414–1419. doi: 10.1128/iai.56.6.1414-1419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramamurthy T, Bag P K, Pal A, Bhattacharya S K, Shimada T, Takeda T, Karasawa T, Kurasono H, Takeda Y, Nair G B. Virulence patterns of Vibrio cholerae non-O1 strains isolated from hospitalised patients with acute diarrhoea in Calcutta, India. J Med Microbiol. 1993;39:310–317. doi: 10.1099/00222615-39-4-310. [DOI] [PubMed] [Google Scholar]
- 59.Ramamurthy T, Albert M J, Huq A, Colwell R R, Takeda Y, Takeda T, Shimada T, Mandal B K, Nair G B. Vibrio mimicus with multiple toxin types isolated from human and environmental sources. J Med Microbiol. 1994;40:194–196. doi: 10.1099/00222615-40-3-194. [DOI] [PubMed] [Google Scholar]
- 60.Rhine J A, Taylor R K. TcpA pilin sequences and colonization requirements for O1 and O139 Vibrio cholerae. Mol Microbiol. 1994;13:1013–1020. doi: 10.1111/j.1365-2958.1994.tb00492.x. [DOI] [PubMed] [Google Scholar]
- 61.Rivera I G, Chowdhury M A R, Huq A, Jacobs D, Martins M T, Colwell R R. Enterobacterial repetitive intergenic consensus sequences and the PCR to generate fingerprints of genomic DNAs from Vibrio cholerae O1, O139, and non-O1 strains. Appl Environ Microbiol. 1995;61:2898–2904. doi: 10.1128/aem.61.8.2898-2904.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rivera I N G, Chowdhury M A R, Sanchez P S, Sato M I, Huq A, Colwell R R, Martins M T. Detection of cholera (ctx) and zonula occludens (zot) toxin genes in Vibrio cholerae O1, O139, and non-O1 strains. World J Microbiol Biotech. 1995;11:572–577. doi: 10.1007/BF00286376. [DOI] [PubMed] [Google Scholar]
- 63.Rodrigue D C, Popovic T, Wachsmuth I K. Nontoxigenic Vibrio cholerae O1 infections in the United States. In: Wachsmuth I K, Blake P A, Olsvik O, editors. Vibrio cholerae and cholera: molecular to global perspectives. Washington, D.C.: ASM Press; 1994. pp. 69–76. [Google Scholar]
- 64.Rodrigues D P, Hofer E. Vibrio species from the water-oyster ecosystem of Sepetitiba Bay in Rio de Janeiro State, Brazil. Rev Microbiol. 1986;17:332–338. [Google Scholar]
- 65.Roszak D B, Colwell R R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987;51:365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saha P K, Koley H, Mukhopadhyay A K, Bhattacharya S K, Nair G B, Ramakrishnan B S, Krishnan S, Takeda T, Takeda Y. Nontoxigenic Vibrio cholerae O1 serotype Inaba Biotype El Tor associated with a cluster of cases of cholera in Southern India. J Clin Microbiol. 1996;34:1114–1117. doi: 10.1128/jcm.34.5.1114-1117.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Said B, Smith H R, Scotland S M, Rowe B. Detection and differentiation of the gene for toxin co-regulated pili (tcpA) in Vibrio cholerae non-O1 using the polymerase chain reaction. FEMS Microbiol Lett. 1995;125:205–210. doi: 10.1111/j.1574-6968.1995.tb07359.x. [DOI] [PubMed] [Google Scholar]
- 68.Sakazaki R, Gomes C Z, Sebald M. Taxonomical studies of the so-called NAG vibrios. Jpn J Med Sci Biol. 1967;20:265–280. [Google Scholar]
- 69.Shangkuan Y H, Show Y S, Wang T M. Multiplex polymerase chain reaction to detect toxigenic Vibrio cholerae and to biotype Vibrio cholerae O1. J Appl Bacteriol. 1995;79:264–273. doi: 10.1111/j.1365-2672.1995.tb03136.x. [DOI] [PubMed] [Google Scholar]
- 70.Sharma C, Nair G B, Mukhopadhyay A K, Bhattacharya S K, Ghosh R K, Ghosh A. Molecular characterization of Vibrio cholerae O1 biotype El Tor strains isolates between 1992 and 1995 in Calcutta, India: evidence for the emergence of a new clone of the El Tor biotype. J Infect Dis. 1997;175:1134–1141. doi: 10.1086/516453. [DOI] [PubMed] [Google Scholar]
- 71.Sharma C, Thungapathra M, Ghosh A, Mukhopadhyay A K, Basu A, Mitra R, Basu I, Bhattacharya S K, Shimada T, Ramamurthy T, Takeda T, Yamasaki S, Takeda Y, Nair G B. Molecular analysis of non-O1, non-O139 Vibrio cholerae associated with an unusual upsurge in the incidence of cholera-like disease in Calcutta, India. J Clin Microbiol. 1998;36:756–763. doi: 10.1128/jcm.36.3.756-763.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sperandio V, Bailey C, Giron J A, DiRita V J, Silveira W D, Vettore A L, Kaper J B. Cloning and characterization of the gene encoding the OmpU outer membrane protein of Vibrio cholerae. Infect Immun. 1996;64:5406–5409. doi: 10.1128/iai.64.12.5406-5409.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stroeher U H, Jedani K E, Dredge B K, Morona R, Brown M H, Karageorgos L E, Albert M J, Manning P A. Genetic rearrangements in the rfb regions of Vibrio cholerae O1 and O139. Proc Natl Acad Sci USA. 1997;92:10374–10378. doi: 10.1073/pnas.92.22.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tauxe R, Seminario L, Tapia R, Libel M. The Latin American epidemic. In: Wachsmuth I K, Blake P A, Olsvik O, editors. Vibrio cholerae and cholera: molecular to global perspectives. Washington, D.C.: ASM Press; 1994. pp. 321–344. [Google Scholar]
- 75.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taylor R K. Genetic studies of enterotoxin and other potential virulence factors of Vibrio cholerae. In: Hopwood D A, Chater K F, editors. Genetics of bacterial diversity. London, England: Academic Press, Ltd.; 1989. pp. 309–329. [Google Scholar]
- 77.Taylor R K. Virus on virus infects bacterium. Nature. 1999;399:312–313. doi: 10.1038/20565. [DOI] [PubMed] [Google Scholar]
- 78.Twedt R M, Madden J M, Hunt J M, Francis D W, Peeler J T, Duran A P, Hebert W O, McCay S G, Roderick C N, Spite G T, Wazenski T J. Characterization of Vibrio cholerae isolated from oysters. Appl Environ Microbiol. 1981;41:1475–1478. doi: 10.1128/aem.41.6.1475-1478.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vicente A C, Coelho A M, Salles C A. Detection of Vibrio cholerae and V. mimicus heat-stable toxin gene sequence by PCR. J Med Microbiol. 1997;46:398–402. doi: 10.1099/00222615-46-5-398. [DOI] [PubMed] [Google Scholar]
- 80.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 81.World Health Organization. Outbreak of gastroenteritis by nonagglutinable (NAG) vibrio. Wkly Epidemiol Rev. 1969;44:1–28. [Google Scholar]
- 82.Yamamoto K, Al-Omani M, Honda T, Takeda Y, Miwatani T. Non-O1 Vibrio cholerae hemolysin: purification, partial characterization, and immunological relatedness to El Tor hemolysin. Infect Immun. 1984;45:192–196. doi: 10.1128/iai.45.1.192-196.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yamamoto K, Ichinose Y, Nakasone N, Tanabe M, Nagahama M, Sakurai J, Iwanaga M. Identity of hemolysins produced by Vibrio cholerae non-O1 and V. cholerae O1, biotype El Tor. Infect Immun. 1986;51:927–931. doi: 10.1128/iai.51.3.927-931.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]