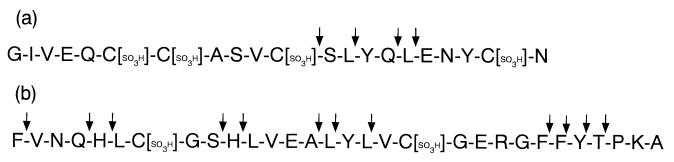Abstract
The gene encoding subtilisin-like protease T. kodakaraensis subtilisin was cloned from a hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. T. kodakaraensis subtilisin is a member of the subtilisin family and composed of 422 amino acid residues with a molecular weight of 43,783. It consists of a putative presequence, prosequence, and catalytic domain. Like bacterial subtilisins, T. kodakaraensis subtilisin was overproduced in Escherichia coli in a form with a putative prosequence in inclusion bodies, solubilized in the presence of 8 M urea, and refolded and converted to an active molecule. However, unlike bacterial subtilisins, in which the prosequence was removed from the catalytic domain by autoprocessing upon refolding, T. kodakaraensis subtilisin was refolded in a form with a putative prosequence. This refolded protein of recombinant T. kodakaraensis subtilisin which is composed of 398 amino acid residues (Gly−82 to Gly316), was purified to give a single band on a sodium dodecyl sulfate (SDS)-polyacrylamide gel and characterized for biochemical and enzymatic properties. The good agreement of the molecular weights estimated by SDS-polyacrylamide gel electrophoresis (44,000) and gel filtration (40,000) suggests that T. kodakaraensis subtilisin exists in a monomeric form. T. kodakaraensis subtilisin hydrolyzed the synthetic substrate N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide only in the presence of the Ca2+ ion with an optimal pH and temperature of pH 9.5 and 80°C. Like bacterial subtilisins, it showed a broad substrate specificity, with a preference for aromatic or large nonpolar P1 substrate residues. However, it was much more stable than bacterial subtilisins against heat inactivation and lost activity with half-lives of >60 min at 80°C, 20 min at 90°C, and 7 min at 100°C.
Hyperthermophilic archaea usually produce highly thermostable proteins. In addition, it has been proposed that these microorganisms retain the nature of the last common ancestor of life most strongly (42). This concept is still widely accepted, although there have been a number recent criticisms (6, 13). Thus, hyperthermophilic archaea are expected to be a valuable source not only to analyze adaptation mechanisms of proteins to extremely high temperatures but probably also to trace the evolution of life.
Thermococcus kodakaraensis KOD1, which had previously been designated Pyrococcus sp. strain KOD1, was isolated from a solfatara at a wharf on Kodakara Island, Kagoshima, Japan (26). The growth temperature of this strain ranges from 65 to 95°C, and the optimal growth temperature is 90°C. The genes encoding various enzymes have been cloned from this strain and overexpressed in Escherichia coli, and the recombinant proteins have been characterized (11). These enzymes are highly stable, much more stable than the mesophilic counterparts, and often show unusual characteristics, such as broad metal ion and nucleoside triphosphate specificities. However, it remains to be determined whether this strain produces serine proteases, although it has been reported that this strain produces at least three proteases, including a hyperthermostable thiol protease (26).
Serine proteases have been well studied from both basic and applied aspects. They have a catalytic triad consisting of Ser, His, and Asp in common. These enzymes are divided into two major groups, subtilisin-like serine proteases (subtilases) and (chymo)trypsin-like serine proteases. The former is distributed in various organisms, including bacteria, archaea, and eucaryotes, more widely than the latter. Based on the difference in the amino acid sequences, subtilases are further classified into six families: subtilisin, thermitase, proteinase K. lantibiotic peptidase, kexin, and pyrolysin (31). Of these families, the subtilisin family, which includes subtilisin E from Bacillus subtilis (33), subtilisin BPN′ from Bacillus amyloliquefaciens (39), and subtilisin Carlsberg from Bacillus licheniformis (20), has been most extensively studied in terms of structure and function. The crystallographic structures of these subtilisins have been determined (2, 21, 43). Because subtilisins are commercially valuable enzymes, there have been extensive attempts to improve their activity and stability with protein engineering technology (34, 40).
Subtilisins are synthesized in the cells as a precursor called preprosubtilisin, in which the presequence and prosequence are attached to the N terminus of the mature protein (20, 33, 39). The presequence acts as a signal peptide that facilitates the secretion of a prosubtilisin across the cytoplasmic membrane. The prosequence acts as an intramolecular chaperone and guides correct folding of the mature protein (7, 19, 28). The prosequence is cleaved from the mature protein through autoproteolysis to produce active mature subtilisin.
In this report, we cloned the gene encoding a subtilisin-like enzyme from T. kodakaraensis KOD1, overexpressed it in E. coli, and purified and characterized the recombinant protein (T. kodakaraensis subtilisin). Subtilases from extreme thermophiles so far identified, except for aerolysin from Pyrobaculum aerophilum (36), belong to the thermitase or pyrolysin family. However, T. kodakaraensis subtilisin showed the highest amino acid sequence identity to members of the subtilisin family, rather than to those of the thermitase or pyrolysin family. Unlike bacterial subtilisins, T. kodakaraensis subtilisin exhibits enzymatic activity in a form with a putative prosequence.
MATERIALS AND METHODS
Cells and plasmids.
T. kodakaraensis KOD1 was isolated in our laboratory (26). E. coli strain HB101 [F− hsdS20(rB− mB−) recA13 ara-13 proA2 lacY1 galK2 rpsL20 (Smr) xyl-5 mtl-1 supE44 λ−] and plasmids pBR322 and pUC18 were from Takara Shuzo Co., Ltd. E. coli BL21-codonPlus(DE3)-RIL [F− ompT hsdS(rB− mB−) dcm+ Tetr galλ(DE3) endA Hte (argU ileY leuW Camr)] was from Stratagene. Plasmid pET25b was from Novagen.
Cloning of the T. kodakaraensis subtilisin gene.
Genomic DNA from T. kodakaraensis KOD1, which was prepared as described previously (18), was digested with HindIII, and the resultant DNA fragments were ligated into the HindIII site of pBR322. The resultant plasmids were used to transform E. coli HB101. Colonies were grown on a plate of LB-casein-agar medium (Luria-Bertani medium supplemented with 1% casein, 50 μg of ampicillin per ml, and 1.5% agar) at 37°C. A replica of this plate was prepared, layered by 1.3% agar containing 1% Tween 20 for lysis of the colonies, and further incubated at 80°C for 2 days for proteolytic degradation of casein. The colonies which gave white halos on a replica plate were judged positive. Plasmid DNAs were isolated from corresponding colonies grown on the original plate and used for further subcloning and sequencing. The DNA sequence was determined by the dideoxy-chain termination method (27) with ABI prism 310 genetic analyzer (Perkin-Elmer). Nucleotide and amino acid sequence analyses, including identification of open reading frames, homology search, and multiple alignment, were performed by using DNASIS software of Hitachi Co., Ltd.
Overproduction and purification of T. kodakaraensis subtilisin.
The gene encoding T. kodakaraensis subtilisin in a form with a putative prosequence was amplified by PCR with a combination of forward (5′-AGTCCCTGCACATATGGGAGAGCAGAATACAATA-3′) and reverse (5′-AGTGGATCCAATCAGCCCAGGGC-3′) primers (the NdeI and BamHI sites are underlined, respectively). Thirty cycles of PCR were performed in a thermal cycler of Perkin-Elmer (GeneAmp PCR System 2400) using Vent polymerase (New England Biolabs). The resultant 1.2-kbp NdeI-BamHI fragment was ligated into the NdeI and BamHI sites of plasmid pET25b to construct plasmid pET25b-Tk-subtilisin. A strain overproducing T. kodakaraensis subtilisin was constructed by transforming E. coli BL21-codonPlus(DE3) with this plasmid. For overproduction, this transformant was grown at 37°C in LB medium containing 50 μg of ampicillin per ml. When the absorbance at 660 nm of the culture reached ca. 0.6, 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture medium and cultivation was continued for an additional 4 h. Cells were then harvested and subjected to the following purification procedures.
All purification procedures were performed at 4°C. Cells were suspended in 20 mM Tris-HCl (pH 9.0), disrupted by sonication, and centrifuged at 15,000 × g for 30 min. The pellet was dissolved in 20 mM Tris-HCl (pH 9.0) containing 8 M urea, dialyzed against 20 mM Tris-HCl (pH 9.0) to remove urea, and centrifuged at 15,000 × g for 30 min. The resultant supernatant was applied to a column (5 ml) of Hi-TrapQ (Pharmacia Biotech) equilibrated with the same buffer. The refolded protein of recombinant T. kodakaraensis subtilisin eluted from the column as a single peak at a NaCl concentration of approximately 0.5 M by linearly increasing the NaCl concentration from 0 to 1.0 M in the same buffer. The purity of the enzyme was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12% polyacrylamide gel (24), followed by staining with Coomassie brilliant blue. The N-terminal amino acid sequence of the protein was determined by a Procise automated sequencer (Perkin-Elmer model 491). The molecular weight of T. kodakaraensis subtilisin was estimated by gel filtration chromatography using a column (1.6 by 60 cm) of Superdex 200 (Pharmacia Biotech) equilibrated with 10 mM Tris-HCl (pH 7.5) containing 150 mM NaCl. Bovine serum albumin (67 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa), and RNase A (13.7 kDa) were used as molecular size standards.
Activity staining of gel.
After conventional SDS–12% PAGE, the gel was washed in 50 mM N-cyclohexyl-3-aminopropane sulfonic acid (CAPS)–NaOH (pH 9.5) containing 2.5% Triton X-100 for 1 h to remove SDS. A replica of this gel was prepared by transferring the proteins in this gel to the 12% polyacrylamide gel containing 0.5% gelatin as described previously (26). The resultant replica of the gel was then incubated at 80°C for 16 h for proteolytic reaction, followed by staining with 0.1% amino black in 100 ml of a solution containing 30% methanol, 10% acetic acid, and 60% water. Protease bands were visualized as clear zones due to the hydrolysis of gelatin.
Enzymatic activity.
The synthetic substrates N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide (AAPF), N-succinyl-Ala-Ala-Pro-Leu-p-nitroanilide (AAPL), and N-succinyl-Ala-Ala-Pro-Asp-p-nitroanilide (AAPD) (Sigma) were used at a concentration of 0.13 mM to determine the enzymatic activity as described previously (17). The buffers used were 50 mM CAPS-NaOH (pH 9.5) containing 5 mM CaCl2 for T. kodakaraensis subtilisin and 50 mM Tris-HCl (pH 8.5) containing 5 mM CaCl2 for subtilisin E, which a kind gift from Takara Shuzo Co., Ltd. The reaction mixture was incubated at 80°C (T. kodakaraensis subtilisin) or 55°C (subtilisin E). The amount of p-nitroaniline released through the reaction was determined from the absorption at 410 nm with the molar absorption coefficient value of 8,900 M−1 cm−1. One unit of enzymatic activity was defined as the amount of the enzyme that produced 1 nmol of p-nitroaniline per min at 80°C for T. kodakaraensis subtilisin and 55°C for subtilisin E. The specific activity was defined as the enzymatic activity per milligram of protein. The protein concentrations of T. kodakaraensis subtilisin and of subtilisin E were determined from the UV absorption at 280 nm with A2800.1% values of 1.24 and 1.25, respectively. These values were calculated by using ɛ values of 1,576 M−1 cm−1 for tyrosine and 5,225 M−1 cm−1 for tryptophan at 280 nm (14).
Thermal stability.
The thermal stability of T. kodakaraensis subtilisin was analyzed by incubating it in 20 mM Tris-HCl (pH 9.0) containing 50 mM CaCl2 at a concentration of 42 μg/ml at 80, 90, and 100°C. At appropriate intervals, an aliquot was withdrawn and the enzymatic activity was determined at 80°C using AAPF as a substrate. The remaining activity was calculated by dividing the activity determined after incubation with that determined before incubation.
Identification of cleavage sites in polypeptides.
Oxidized insulin chains A and B were digested by T. kodakaraensis subtilisin with an enzyme/substrate ratio of 1:10 (by weight) in 20 mM Tris-HCl (pH 9.0) containing 5 mM CaCl2 at 80°C for 30 min. The resultant peptides were separated by reverse-phase high-performance liquid chromatography on a COSMOSIL 5C18-AR column (4.6 by 150 mm) from Nacalai Tesque Co., Ltd. Elution was performed by raising the concentration of acetonitrile linearly from 15 to 50% in 25 min in the presence of 1% acetic acid. The flow rate was 1.0 ml/min, and the peptides were detected by measuring the absorbance at 230 nm. The molecular weights of these peptides were determined with an LCQ Mass Spectrometer System (Finnigan Mat).
CD spectra.
The circular dichroism (CD) spectra were measured on a J-725 automatic spectropolarimeter of Japan Spectroscopic Co., Ltd. The far-UV (200- to 260-nm-wavelength) CD spectrum was obtained at 20°C by using the T. kodakaraensis subtilisin solution (0.11 mg/ml) in 20 mM Tris-HCl (pH 9.0) containing 0.5 M NaCl or the subtilisin E solution (0.11 mg/ml) in 10 mM Tris-HCl (pH 7.5) containing 150 mM NaCl in a cell with an optical path of 2 mm. The mean residue ellipticity, θ, which is measured in degrees square centimeter per decimole, was calculated by using an average amino acid molecular weight of 110.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been deposited in DDBJ with accession number AB056701.
RESULTS
Cloning of the T. kodakaraensis subtilisin gene.
Growing the bacteria on an LB-casein-agar plate is effective in detecting whether E. coli transformants produce highly thermostable proteases, because growth of these transformants on the plate at 37°C, followed by the incubation of the plate at high temperatures, results in the formation of white halos around the colonies, probably due to the precipitation of casein upon proteolytic degradation. Construction of a plasmid library by ligating the HindIII fragments of the T. kodakaraensis KOD1 genome to plasmid pBR322, followed by screening for an E. coli HB101 transformant that forms a halo on an LB-casein-agar plate indicated that a 1.5-kbp HindIII fragment is responsible for the formation of the halo. Determination of the nucleotide sequence indicated that this DNA fragment contains the gene encoding T. kodakaraensis subtilisin with a putative preprosequence (data not shown). T. kodakaraensis subtilisin is composed of 422 amino acid residues with a calculated molecular weight of 43,783 and an isoelectric point of 4.5. A potential Shine-Dalgarno sequence (5′-GGAGGTG-3′), which is complementary to the 3′-terminal sequence (two to eight residues from the 3′ terminus) of the 16S rRNA of T. kodakaraensis KOD1 (26), is located eight bases upstream of the initiation codon for translation. A possible TATA-like promoter site (5′-TTAAAT-3′) and transcription termination site are also located ∼40 bp upstream of the initiation codon for translation and ∼10 bp downstream of the termination codon for translation, respectively.
Amino acid sequence.
Database searches for proteins with amino acid sequences similar to that of T. kodakaraensis subtilisin indicated that this subtilisin is a member of the subtilisin family. Comparison of the amino acid sequence of T. kodakaraensis subtilisin with those of the representative members of the subtilisin family indicates that this subtilisin consists of a putative presequence, a putative prosequence, and a putative catalytic domain, which are composed of 24 (from Met−106 to Ala−83), 82 (from Gly−82 to Pro−1), and 316 (from Ala1 to Gly316) amino acid residues, respectively (Fig. 1). The putative presequence was identified as a secretion signal by the program SignalP version 2.0 world wide server. The putative catalytic domain of T. kodakaraensis subtilisin shows amino acid sequence identities of 45% to aerolysin (36), 44% to subtilisins E (33) and BPN′ (39), and 43% to subtilisin Carlsberg (20). It shows high amino acid sequence identities to the members of other subtilase families as well. It shows amino acid sequence identities of 41% to thermitase (22), 36% to proteinase K, 28% to lactocin leader peptidase (32), 30% to Kex2 protease (12), and 38% to the catalytic core of pyrolysin (37), which represent the thermitase, proteinase K, lantibiotic peptidase, kexin, and pyrolysin families, respectively. Three amino acid residues that form a catalytic triad in subtilases are fully conserved in the T. kodakaraensis subtilisin sequence (Asp33, His71, and Ser242). In addition, the asparagine residue, which is required to form an oxyanion hole, is conserved (Asn182).
FIG. 1.
Alignment of subtilisin sequences. The amino acid sequence of T. kodakaraensis subtilisin (Tk-sub) is compared with those of P. aerophilum aerolysin (Paelys) (accession no. S76079), B. licheniformis subtilisin Carlsberg (BlsCar) (accession no. X03341), B. amyloliquefaciens subtilisin BPN′ (BasBPN) (accession no. X00165), B. subtilis subtilisin E (BssE) (accession no. K01988), and pyrolysin core (Pyroly) (accession no. U55835). Gaps are denoted by dashes. The numbers in parentheses represent the numbers of the amino acid residues inserted or extended at the positions indicated. The conserved amino acid residues are denoted with white letters. The amino acid residues that form a catalytic triad and the asparagine residue that forms an oxyanion hole are denoted by solid and open circles, respectively. The numbers represent the positions of the amino acid residues starting from the N terminus of the mature proteins for bacterial subtilisins and pyrolysin and the positions of putative catalytic domains for T. kodakaraensis subtilisin and aerolysin. The eight α-helices (hA to hH) and nine β-strands (e1 to e9) of subtilisin BPN′ are shown above the sequences.
Overproduction and purification.
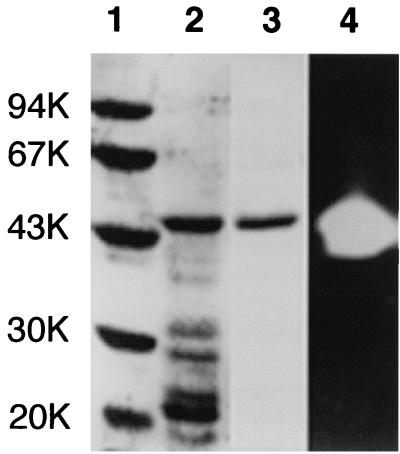
In order to obtain T. kodakaraensis subtilisin in an amount sufficient for biochemical characterizations, we constructed plasmid pET25b-Tk-subtilisin, in which transcription of the gene encoding T. kodakaraensis subtilisin is initiated by the T7 promoter. This T. kodakaraensis subtilisin includes a putative prosequence and is composed of 399 amino acid residues (Met plus Gly−82 to Gly316). We overproduced T. kodakaraensis subtilisin intracellularly in E. coli in this form, because it has previously been shown that prosubtilisin is overproduced in E. coli in an inactive denatured form in inclusion bodies but is effectively converted to the active mature enzyme upon refolding (16). Plasmid pET25b-Tk-subtilisin was used to transform E. coli BL21-codonPlus(DE3)-RIL. Upon induction, recombinant T. kodakaraensis subtilisin accumulated in the cells as inclusion bodies. After lysis of the cells by sonication, the proteins in an insoluble form, which include T. kodakaraensis subtilisin, were collected by centrifugation, solubilized in 20 mM Tris-HCl (pH 9.0) containing 8 M urea, and dialyzed against the same buffer without urea. After this refolding process, several proteins became soluble as revealed by SDS-PAGE (Fig. 2, lane 2). However, gel assay revealed that only the 44-kDa protein exhibits the protease activity (Fig. 2, lane 4). The N-terminal amino acid sequence of this protein was determined to be GEQNTIR, indicating that it represents the refolded protein of T. kodakaraensis subtilisin with the entire putative prosequence. The refolded protein thus obtained will be referred to simply as T. kodakaraensis subtilisin hereafter. Note that the initiation codon for translation is attached to the 5′ terminus of the gene encoding T. kodakaraensis subtilisin. However, the N-terminal methionine residue was posttranslationally removed from the recombinant protein. T. kodakaraensis subtilisin was purified to give a single band on an SDS-polyacrylamide gel (Fig. 2, lane 3) and used for further characterization. The amount of T. kodakaraensis subtilisin purified from 1 liter of culture was roughly 17 mg.
FIG. 2.
Comparison of the purity of T. kodakaraensis subtilisin by SDS-PAGE. Samples were subjected to electrophoresis on a 12% polyacrylamide gel in the presence of SDS. After electrophoresis, the gel was stained with Coomassie brilliant blue (lanes 1 to 3) or stained for protease activity (lane 4). Lane 1, low-molecular-weight, marker kit (Pharmacia Biotech) containing phosphorylase b, bovine serum albumin, ovalbumin, carbonic anhydrase, trypsin inhibitor, and α-lactalbumin; lanes 2 and 4, refolding sample of insoluble fractions obtained from E. coli BL21-codonPlus(DE3) harboring plasmid pET25b-Tk- subtilisin upon lysis by sonication; lane 3, purified T. kodakaraensis subtilisin. The molecular weight (in thousands [K]) of each standard protein is indicated to the left of the gel.
Biochemical properties.
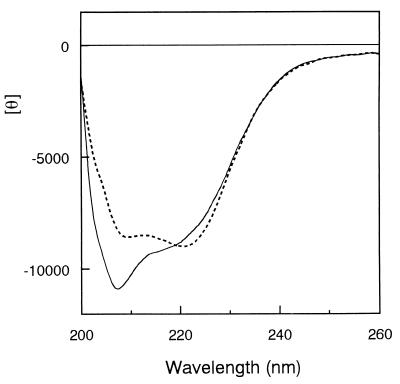
The molecular weight of T. kodakaraensis subtilisin estimated from SDS-PAGE is comparable to that (41,387) calculated from the amino acid sequence. The molecular weight of T. kodakaraensis subtilisin was estimated to be 40,000 by gel filtration column chromatography, which was also comparable to the calculated one (data not shown). These results strongly suggest that T. kodakaraensis subtilisin exists in a monomeric form. The far-UV CD spectrum of T. kodakaraensis subtilisin compared to that of subtilisin E is shown in Fig. 3. These two spectra show a significant difference at around 210 nm. The spectrum of T. kodakaraensis subtilisin gave a trough with the minimum θ value of −11,000 at 208 nm, which was accompanied by a shoulder with a θ value of −9,000 at 220 nm. In contrast, the spectrum of subtilisin E gave a broad trough with the double minimum θ values of −8,500 at 208 nm and −9,000 at 222 nm. These results suggest that the content of the secondary structures varied for these two proteins.
FIG. 3.
CD spectra. The far-UV CD spectrum of T. kodakaraensis subtilisin (solid line) is shown in comparison with that of subtilisin E (broken line). These spectra were measured at 20°C. The mean residue ellipticity θ, which is measured in degrees square centimeter per decimole, was calculated using an average amino acid molecular weight of 110.
Enzymatic activity.
The enzymatic activity of T. kodakaraensis subtilisin was determined by using a synthetic substrate, AAPF. T. kodakaraensis subtilisin required Ca2+ ion for activity and exhibited little enzymatic activity in the absence of the Ca2+ ion, as do other subtilases (31). To examine whether this enzyme exhibits activity in the presence of other metal ions, the enzymatic activity was determined in the presence of various metal ions, such as MgCl2, ZnCl2, CoCl2, FeCl2, CuCl2, MnCl2, NiCl2, SrCl2, and BaCl2. However, T. kodakaraensis subtilisin exhibited little enzymatic activity in the presence of these metal ions. Analysis of the dependence of the T. kodakaraensis subtilisin activity on the CaCl2 concentration indicated that T. kodakaraensis subtilisin gave the highest activity in the presence of 5 mM CaCl2. It exhibited 70% and 80% of the maximal activity in the presence of 1 and 100 mM CaCl2, respectively. Analyses for the pH dependence and temperature dependence of the T. kodakaraensis subtilisin activity indicated that T. kodakaraensis subtilisin gave the highest activity at pH 9.5 and 80°C (data not shown). It exhibited 10 to 20% of the maximal activity at 40 or 90°C and pH 9.5 or at a pH of ∼8.0 or ∼11 and 80°C.
Subtilisins exhibit a broad substrate specificity but prefer large P1 side chains (9). To analyze a substrate specificity of T. kodakaraensis subtilisin briefly, AAPF, AAPL, and AAPD were chosen as representatives of the synthetic P1 substrates, which vary in size and hydrophobicity, and hydrolyzed by T. kodakaraensis subtilisin. The specific activities of T. kodakaraensis subtilisin for the hydrolysis of these substrates are compared with those of subtilisin E in Table 1. Both enzymes hydrolyzed AAPF most effectively, AAPL less effectively, and AAPD very poorly. These results suggest that T. kodakaraensis subtilisin has a substrate specificity similar to those of other subtilisins. The specific activity of T. kodakaraensis subtilisin for the hydrolysis of AAPF under optimal conditions was 30% of that of subtilisin E.
TABLE 1.
Specific activities of T. kodakaraensis subtilisin and subtilisin E toward synthetic substratesa
| Substrate |
T. kodakaraensis
subtilisin
|
Subtilisin E
|
||
|---|---|---|---|---|
| Sp act (U/mg) | Relative activity (%) | Sp act (U/mg) | Relative activity (%) | |
| AAPF | 3,930 | 100 | 13,300 | 100 |
| AAPL | 955 | 24.3 | 2,820 | 21.2 |
| AAPD | <40 | <1.0 | <40 | <0.3 |
The enzymatic activity was determined at pH 9.5 and 80°C for T. kodakaraensis subtilisin or pH 8.5 and 55°C for subtilisin E in the presence of 0.13 mM substrate and 5 mM CaCl2. The experiment was performed in triplicate, and standard errors from the means were within 10% of the values reported.
Cleavage site specificity.
To determine the cleavage site specificity of T. kodakaraensis subtilisin, oxidized insulin chains A and B were digested by T. kodakaraensis subtilisin at 80°C for 30 min. Under these conditions, these insulin chains were not degraded in the absence of T. kodakaraensis subtilisin. Identification of the proteolytic fragments by mass spectrometry indicated that these oxidized insulin chains were digested by T. kodakaraensis subtilisin at the carboxyl termini of the various amino acid residues, such as Tyr, Phe, Leu, Gln, His, Thr, Ser, and Ala, which vary greatly in size and hydrophobicity (Fig. 4). Thus, like other subtilases (25), T. kodakaraensis subtilisin shows a broad substrate specificity with a slight preference to large hydrophobic amino acid residues at the P1 position.
FIG. 4.
Cleavage site specificity of T. kodakaraensis subtilisin. Cleavage sites of oxidized insulin chains A (a) and B (b) by T. kodakaraensissubtilisin are indicated by arrows.
Thermal stability.
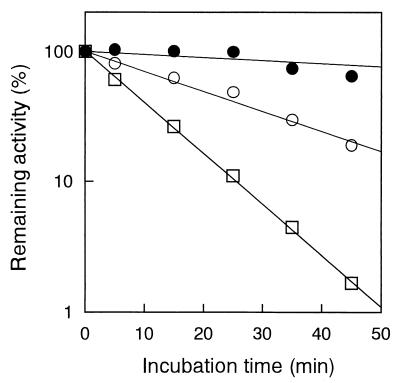
The Ca2+ ion is essential not only for activity but also for stability of subtilases (31). T. kodakaraensis subtilisin lost almost all its enzymatic activity when it was incubated at 90°C for 30 min in the absence of the Ca2+ ion, whereas it retained ∼15, ∼25, and ∼40% of the maximal activity when it was incubated in the presence of 5, 20, and 50 mM CaCl2, respectively. Thus, T. kodakaraensis subtilisin was also stabilized in the presence of the Ca2+ ion. In the presence of 50 mM CaCl2, T. kodakaraensis subtilisin lost enzymatic activity with half-lives of >60 min at 80°C, 20 min at 90°C, and 7 min at 100°C (Fig. 5). In contrast, subtilisin E lost enzymatic activity even at 60°C with a half-life of 18 min (35). Thus, T. kodakaraensis subtilisin is much more stable than subtilisin E.
FIG. 5.
Thermal stability. Semilog plots of the remaining activity versus incubation time are shown. T. kodakaraensis subtilisin was incubated at 80°C (●), 90°C (○), or 100°C (□). The lines were obtained by linear regression of the data.
DISCUSSION
Subtilases from hyperthermophilic archaea.
In this report, we showed that hyperthermophilic archaea produce a second type of subtilases, which are members of the subtilisin family, in addition to the members of the pyrolysin family. The former is represented by T. kodakaraensis subtilisin, and the latter is represented by pyrolysin. Aerolysin from P. aerophilum (36) is a homologue of T. kodakaraensis subtilisin, and stetterlysin from Thermococcus stetteri (38) is a homologue of pyrolysin. The amino acid sequences of the catalytic domains of T. kodakaraensis subtilisin and pyrolysin show relatively high amino acid sequence identities (Fig. 1). However, T. kodakaraensis subtilisin is clearly distinguished from pyrolysin in size. T. kodakaraensis subtilisin is as small as various bacterial subtilisins, whereas pyrolysin is much larger than these subtilisins. Pyrolysin is composed of 1,249 amino acid residues and has large insertions within the catalytic domain, as well as long extensions at the N and C termini of the catalytic domain.
The question of whether these subtilases are universally present in hyperthermophilic archaea then arose. It has been reported that the Pyrococcus furiosus genome contains a gene encoding a small subtilisin-like serine protease, in addition to that encoding pyrolysin (5). This protein may be a member of the subtilisin family. When the genomes of hyperthermophilic archaea Aeropyrum pernix, Archaeoglobus fulgidus, Methanobacterium thermoautotrophicum, Methanococcus jannaschii, Pyrococcus horikoshii, Pyrodictium abyssi, and Thermoplasma acidophilum, whose nucleotide sequences have been completely determined, were examined for the genes encoding subtilases, only the A. pernix genome contains the gene encoding a protein, which shows significant amino acid sequence identity to subtilases. Because it is composed of 440 amino acid residues and its putative catalytic domain shows amino acid sequence identities of 59% to the putative catalytic domain of T. kodakaraensis subtilisin, 43% to subtilisin BPN′, and 36% to the catalytic domain of pyrolysin, there is no doubt that this protease is a member of the subtilisin family. Although the possibility that these genomes contain the genes encoding subtilases with relatively poor sequence similarities cannot be ruled out, these results suggest that neither the T. kodakaraen-sis subtilisin homologue nor the pyrolysin homologue is universally present in hyperthermophilic archaea. Archaea have been shown to consist of three groups, Crenarchaeota, Euryarchaeota, and Korarchaeota (1). Because P. aerophilum and A. pernix are Crenarchaeota, and T. kodakaraensis KOD1 and P. furiosus are Euryarchaeota, and because all of these four archaea produce a member of the subtilisin family, it seems likely that the types of subtilases are not correlated with the archaeal groups.
Role of a putative prosequence.
A prosequence of subtilisins has been shown to function not only as an intramolecular chaperone but also as a template for molecular imprinting that facilitates correct folding of the catalytic domain (29, 30). This prosequence should be removed from the catalytic domain by autoprocessing or by another protease upon completion of the protein folding (31). The removal of this prosequence from the catalytic domain is necessary to generate active subtilisin molecules, because the uncleaved prosequence interacts with the active site of the catalytic domain and thereby inhibits its activity (31), although the inhibitory and chaperone functions of the prosequence are not necessarily linked with each other (10).
In this report, we showed that T. kodakaraensis subtilisin exhibits the enzymatic activity in a form with a putative prosequence. In addition, preliminary studies suggest that T. kodakaraensis subtilisin without a prosequence (Ala1 to Gly316) can be overproduced in E. coli in inclusion bodies and refolded, but this refolded protein does not exhibit the activity at all (data not shown). These results suggest that this putative prosequence does not function as an intramolecular chaperone but is required to keep the conformation of T. kodakaraensissubtilisin functional. Alternatively, it may function as an intramolecular chaperone but is not removed from the catalytic domain upon completion of the protein folding. Comparison of the amino acid sequence of T. kodakaraensis subtilisin with those of bacterial subtilisins indicates that they are rather poorly conserved in the preprosequence region (Fig. 1). The identities of the amino acid sequences between T. kodakaraensis subtilisin and either one of bacterial subtilisins varied from 43 to 45% in the catalytic domain region, whereas they varied from 23 to 29% in the prosequence region. In addition, the T. kodakaraensis subtilisin sequence has a 13-residue insertion between the C terminus of a putative prosequence and the N terminus of a putative catalytic domain. This relatively poor sequence conservation in the prosequence region may be why the putative prosequence of T. kodakaraensis subtilisin is not autoprocessed. Alternatively, the Pro−1 Ala1 bond which connects the putative prosequence and catalytic domain may not be cleaved by T. kodakaraensis subtilisin. Further structural and functional studies will be required to understand the role of the putative prosequence of T. kodakaraensis subtilisin.
Substrate and Ca2+ binding sites.
Subtilases have five substrate binding sites, S4, S3, S2, S1, and S1′, which interact with the substrate amino acid residues, P4, P3, P2, P1, and P1′, respectively (31). The substrate specificities of subtilases are governed mainly by the interactions at the S1 and S4 sites (15). In fact, the members of the subtilisin and thermitase families show a broad substrate specificity, with a preference for aromatic or large nonpolar P4 and P1 substrate residues, because the S4 and S1 sites of these enzymes are large and hydrophobic. Of these substrate binding sites, the S1 site has been well studied because the substrates are hydrolyzed at the C terminus of the P1 residue by subtilases. The S1 site of subtilisin E consists of two side segments (Ser125 to Gly127 and Ala152 to Gly154), and one bottom segment (Val165 to Pro168). Most of these residues are conserved in the T. kodakaraensis subtilisin sequence, suggesting that the S1 site of T. kodakaraensis subtilisin is also large and hydrophobic. In addition, Glu156, which is located near the S1 site and has been shown to be important for substrate binding (41), is conserved in the T. kodakaraensissubtilisin sequence. Because this residue makes contact with the P1 residue of the substrates, subtilisins with Glu at the corresponding position cannot cleave the substrates at the C termini of the acidic residues due to a negative-charge repulsion between the P1 residue and the S1 site. This may be why T. kodakaraensis subtilisin could not hydrolyze AAPD. Thus, the similarity in the substrate specificities between subtilisin E and T. kodakaraensis subtilisin can be explained by the similarity in the size, hydrophobicity, and polarity of their S1 sites.
T. kodakaraensis subtilisin requires Ca2+ ion for activity, as do other subtilases. Crystal structures of subtilisin BPN′ (3) and subtilisin Carlsberg (2) have revealed that these subtilisins have two Ca2+ binding sites, Ca1 and Ca2. The Ca1 site, in which the Ca2+ ion binds with higher affinity, is formed by the side chains of Gln2, Asp41, and several amino acids in a Ca2+-embracing loop (Asn76 to Val81). All of these residues, except for Ser78, are conserved in the T. kodakaraensis subtilisin sequence. Ser78 is replaced by Asp (Asp85) in T. kodakaraensissubtilisin. Likewise, the amino acid residues that form the Ca2 site (Lys170 to Val174 and Glu195 to Asp197), in which the Ca2+ ion binds with lower affinity, are relatively well conserved in the T. kodakaraensis subtilisin sequence. These results suggest that at least two Ca2+ ions bind to T. kodakaraensis subtilisin at the sites, which correspond to the Ca1 and Ca2 sites of bacterial subtilisins.
Thermal stability.
Many subtilases with different optimal temperatures for activity, which varied greatly from 40°C (4, 23) to 115°C (8), have been isolated from various microorganisms. Comparative studies of these enzymes are expected to provide valuable information on the structure-stability-activity relationships of proteins. However, the amino acid sequences of thermostable and thermolabile subtilases often contain a number of insertions and N- or C-terminal extensions compared to those of bacterial subtilisins. The roles of these insertions or extensions on the enzymatic activity and protein stability remain unknown. Without this information, one cannot discuss the stabilization or destabilization mechanism of thermostable or thermolabile subtilases based on the difference in the amino acid sequences in a region which assumes a fold similar to that of the catalytic domain of bacterial subtilisins.
The T. kodakaraensis subtilisin sequence also has two major insertions (Gly124 to Asp143 and Ala258 to Gly270) compared to the bacterial subtilisin sequences (Fig. 1). Assuming that T. kodakaraensis subtilisin shares the three-dimensional structure with bacterial subtilisins, these peptides are inserted into the surface loops between the hD helix and e4 strand and between the hF and hG helices. These loops are located relatively close to each other on the side opposite that of the active site on the surface of the protein molecule. In addition, the peptides from positions 124 to 143 and from positions 258 to 270 are rich in negative and positive charges, respectively. Therefore, it seems likely that these insertions increase the protein stability through electrostatic interactions, without seriously affecting the enzymatic activity.
Note that T. kodakaraensis subtilisin contains two cysteine residues at positions 50 and 65. Cys50 is replaced by Ser (Ser49), and Cys65 is deleted in bacterial subtilisins. However, modeling of the T. kodakaraensis subtilisin structure suggests that these two cysteine residues do not form a disulfide bond (data not shown).
Proteases from T. kodakaraensis KOD1.
We have previously shown that at least three proteases with molecular masses of ∼35, ∼44, and ∼67 kDa are present in the supernatant of the culture of T. kodakaraensis KOD1 (26). Of the three, the 44-kDa protease has been purified to give a single band on a SDS-polyacrylamide gel and identified as a thiol protease (26). This protease has the N-terminal amino acid sequence of VEIXNI and shows optimal temperature and pH for activity at 110°C and pH 7. Therefore, this protease is clearly different from T. kodakaraensis subtilisin. Because T. kodakaraensis subtilisin has a potential secretion signal at the N terminus, it seems likely that this enzyme is secreted to the culture medium. The 44- and 35-kDa proteases are probably natural T. kodakaraensis subtilisin candidates with and without prosequence, respectively. Identification of the 44-kDa protein as a thiol protease does not necessarily indicate that only the 35-kDa protease is a potential candidate, because the possibility that two different proteases with similar sizes are present in the culture supernatant of T. kodakaraensis KOD1 cannot be excluded. Attempts to secrete T. kodakaraensis subtilisin to the periplasmic space of E. coli using pelB signal sequence or its own putative presequence or to culture medium using the secretion system of B. subtilis have so far been unsuccessful. Preparation of antibody against recombinant T. kodakaraensissubtilisin, followed by Western blot analysis, will be necessary to identify natural T. kodakaraensis subtilisin.
ACKNOWLEDGMENTS
We thank S. Fujiwara for helpful discussions and T. Nakamura for technical assistance.
This work was supported in part by grants from the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) and Kato Memorial Bioscience Foundation.
REFERENCES
- 1.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bode W, Papamokos E, Musil D. The high-resolution X-ray crystal structure of the complex formed between subtilisin Carlsberg and eglin c, an elastase inhibitor from the leech Hirudo medicinalis. Structural analysis, subtilisin structure and interface geometry. Eur J Biochem. 1987;166:673–692. doi: 10.1111/j.1432-1033.1987.tb13566.x. [DOI] [PubMed] [Google Scholar]
- 3.Bott R, Ultsch M, Kossiakoff A, Graycar T, Katz B, Power S. The three-dimensional structure of Bacillus amyloliquefaciens subtilisin at 1.8 Åand an analysis of the structural consequences of peroxide inactivation. J Biol Chem. 1988;263:7895–7906. [PubMed] [Google Scholar]
- 4.Davail S, Feller G, Narinx E, Gerday C. Cold adaptation of proteins. J Biol Chem. 1994;269:17448–17453. [PubMed] [Google Scholar]
- 5.de Vos W M, Voorhorst W G B, Dijkgraaf M, Kluskens L D, van der Oost J, Siezen R J. Purification, characterization, and molecular modeling of pyrolysin and other extracellular thermostable serine proteases from hyperthermophilic microorganisms. Methods Enzymol. 2001;330:383–393. doi: 10.1016/s0076-6879(01)30390-7. [DOI] [PubMed] [Google Scholar]
- 6.Doolittle W F. Uprooting the tree of life. Sci Am. 2000;282:90–95. doi: 10.1038/scientificamerican0200-90. [DOI] [PubMed] [Google Scholar]
- 7.Eder J, Rheinnecker M, Fersht A R. Folding of subtilisin BPN′: role of the pro-sequence. J Mol Biol. 1993;233:293–304. doi: 10.1006/jmbi.1993.1507. [DOI] [PubMed] [Google Scholar]
- 8.Eggen R, Geerling A, Watts J, de Vos W D. Characterization of pyrolysin, a hyperthermoactive serine protease from the archaebacterium Pyrococcus furiosus. FEMS Microbiol Lett. 1990;71:17–20. [Google Scholar]
- 9.Estell D A, Graycar T P, Miller J V, Powers D B, Burnier J P, Ng P G, Wells J A. Probing steric and hydrophobic effects on enzyme-substrate interactions by protein engineering. Science. 1986;233:659–663. doi: 10.1126/science.233.4764.659. [DOI] [PubMed] [Google Scholar]
- 10.Fu X, Inouye M, Shinde U. Folding pathway mediated by an intramolecular chaperone. The inhibitory and chaperone functions of the subtilisin propeptide are not obligatorily linked. J Biol Chem. 2000;2:16871–16878. doi: 10.1074/jbc.275.22.16871. [DOI] [PubMed] [Google Scholar]
- 11.Fujiwara S, Takagi M, Imanaka T. Archaeon Pyrococcus kodakaraensisKOD1: application and evolution. Biotechnol Annu Rev. 1998;4:259–284. doi: 10.1016/s1387-2656(08)70073-5. [DOI] [PubMed] [Google Scholar]
- 12.Fuller R S, Brake A J, Thorner J W. Yeast prohormone processing enzyme (KEX2 gene product) is a Ca2+-dependent serine protease. Proc Natl Acad Sci USA. 1989;86:1434–1438. doi: 10.1073/pnas.86.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glansdorff N. About the last common ancestor, the universal life-tree and lateral gene transfer: a reappraisal. Mol Microbiol. 2000;38:177–185. doi: 10.1046/j.1365-2958.2000.02126.x. [DOI] [PubMed] [Google Scholar]
- 14.Good T W, Morton R A. The spectrophotometric determination of tyrosine of tryptophan in proteins. Biochem J. 1946;40:628–632. doi: 10.1042/bj0400628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gron H, Meldal M, Breddam K. Extensive comparison of the substrate preferences of two subtilisins as determined with peptide substrates which are based on the principle of intramolecular quenching. Biochemistry. 1992;31:6011–6018. doi: 10.1021/bi00141a008. [DOI] [PubMed] [Google Scholar]
- 16.Ikemura H, Inouye M. In vitro processing of pro-subtilisin produced in Escherichia coli. J Biol Chem. 1988;263:12959–12963. [PubMed] [Google Scholar]
- 17.Ikemura H, Takagi H, Inouye M. Requirement of pro-sequence for the production of active subtilisin E in Escherichia coli. J Biol Chem. 1987;262:7859–7864. [PubMed] [Google Scholar]
- 18.Imanaka T, Tanaka T, Tsunekawa H, Aiba S. Cloning of the genes for penicillinase, penP and penI, of Bacillus licheniformis in some vector plasmids and their expression in Escherichia coli, Bacillus subtilis, and Bacillus licheniformis. J Bacteriol. 1981;147:776–786. doi: 10.1128/jb.147.3.776-786.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inouye M. Intramolecular chaperone: the role of the pro-peptide in protein folding. Enzyme. 1991;45:314–321. doi: 10.1159/000468904. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs M, Eliasson M, Uhlen M, Flock J I. Cloning, sequencing and expression of subtilisin Carlsberg from Bacillus licheniformis. Nucleic Acids Res. 1985;13:8913–8926. doi: 10.1093/nar/13.24.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain S C, Shinde U, Li Y, Inouye M, Berman H M. The crystal structure of an autoprocessed Ser221Cys-subtilisin E-propeptide complex at 2.0 A resolution. J Mol Biol. 1998;284:137–144. doi: 10.1006/jmbi.1998.2161. [DOI] [PubMed] [Google Scholar]
- 22.Kleine R. Properties of thermitase, a thermostable serine protease from Thermoactinomyces vulgaris. Acta Biol Med Ger. 1982;41:89–102. [PubMed] [Google Scholar]
- 23.Kulakova L, Galkin A, Kurihara T, Yoshimura T, Esaki N. Cold-active serine alkaline protease from the psychrotrophic bacterium Shewanellastrain Ac10: gene cloning and enzyme purification and characterization. Appl Environ Microbiol. 1999;65:611–617. doi: 10.1128/aem.65.2.611-617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Mayr J, Lupas A, Kellermann J, Eckerskorn C, Baumeister W, Peters J. A hyperthermostable protease of the subtilisin family bound to the surface layer of the archaeon Staphylothermus marinus. Curr Biol. 1996;6:739–749. doi: 10.1016/s0960-9822(09)00455-2. [DOI] [PubMed] [Google Scholar]
- 26.Morikawa M, Izawa Y, Rashid N, Hoaki T, Imanaka T. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcussp. Appl Environ Microbiol. 1994;60:4559–4566. doi: 10.1128/aem.60.12.4559-4566.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shinde U, Inouye M. Propeptide-mediated folding in subtilisin: the intramolecular chaperone concept. Adv Exp Med Biol. 1996;379:147–154. doi: 10.1007/978-1-4613-0319-0_16. [DOI] [PubMed] [Google Scholar]
- 29.Shinde U, Fu X, Inouye M. A pathway for conformational diversity in proteins mediated by intramolecular chaperones. J Biol Chem. 1999;274:15615–15621. doi: 10.1074/jbc.274.22.15615. [DOI] [PubMed] [Google Scholar]
- 30.Shinde U P, Liu J J, Inouye M. Protein memory through altered folding mediated by intramolecular chaperones. Nature. 1997;389:520–522. doi: 10.1038/39097. [DOI] [PubMed] [Google Scholar]
- 31.Siezen R J, Leunissen J A M. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 1997;6:501–523. doi: 10.1002/pro.5560060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skaugen M, Abildgaard C I, Nes I F. Organization and expression of a gene cluster involved in the biosynthesis of the lantibiotic lactocin S. Mol Gen Genet. 1997;253:674–686. doi: 10.1007/s004380050371. [DOI] [PubMed] [Google Scholar]
- 33.Stahl M L, Ferrari E. Replacement of the Bacillus subtilissubtilisin structural gene with an in vitro-derived deletion mutation. J Bacteriol. 1984;158:411–418. doi: 10.1128/jb.158.2.411-418.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takagi H. Protein engineering on subtilisin. Int J Biochem. 1993;25:307–312. doi: 10.1016/0020-711x(93)90617-n. [DOI] [PubMed] [Google Scholar]
- 35.Takagi H, Morinaga Y, Ikeura H, Inouye M. Mutant subtilisin E with enhanced protease activity obtained by site-directed mutagenesis. J Biol Chem. 1988;263:19592–19596. [PubMed] [Google Scholar]
- 36.Volkl P, Markiewicz P, Stetter K O, Miller J H. The sequence of a subtilisin-type protease (aerolysin) from the hyperthermophilic archaeum Pyrobaculum aerophilumreveals sites important to thermostability. Protein Sci. 1994;3:1329–1340. doi: 10.1002/pro.5560030819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voorhorst W G B, Eggen R I L, Geerling A C M, Platteeuw C, Siezen R J, de Vos W M. Isolation and characterization of the hyperthermostable serine protease, pyrolysin, and its gene from the hyperthermophilic archaeon Pyrococcus furiosus. J Biol Chem. 1996;271:20426–20431. doi: 10.1074/jbc.271.34.20426. [DOI] [PubMed] [Google Scholar]
- 38.Voorhorst W G B, Warner A, de Vos W M, Siezen R J. Homology modelling of two subtilisin-like proteases from the hyperthermophilic archaea Pyrococcus furiosus and Thermococcus stetteri. Protein Eng. 1997;10:905–914. doi: 10.1093/protein/10.8.905. [DOI] [PubMed] [Google Scholar]
- 39.Wells J A, Ferrari E, Henner D J, Estell D A, Chen E Y. Cloning, sequencing, and secretion of Bacillus amyloliquefaciens subtilisin in Bacillus subtilis. Nucleic Acids Res. 1983;11:7911–7925. doi: 10.1093/nar/11.22.7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wells J A, Estell D A. Subtilisin—an enzyme designed to be engineered. Trends Biochem Sci. 1988;13:291–297. doi: 10.1016/0968-0004(88)90121-1. [DOI] [PubMed] [Google Scholar]
- 41.Wells J A, Cunningham B C, Graycar T P, Estell D A. Recruitment of substrate-specificity properties from one enzyme into a related one by protein engineering. Proc Natl Acad Sci USA. 1987;84:5167–5171. doi: 10.1073/pnas.84.15.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woese C R, Kandler O, Wheelis M L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wright C S, Alden R A, Kraut J. Structure of subtilisin BPN′ at 2.5 angstrom resolution. Nature. 1969;221:235–242. doi: 10.1038/221235a0. [DOI] [PubMed] [Google Scholar]