Abstract
A detailed analysis of the molecular epidemiology of non-O157:H7 Shiga toxin-producing Escherichia coli (STEC) was performed by using isolates from sporadic cases of hemolytic-uremic syndrome (HUS), animal reservoirs, and food products. The isolates belonged to the O91 and OX3 serogroups and were collected in the same geographical area over a short period of time. Five typing methods were used; some of these were used to explore potentially mobile elements like the stx genes or the plasmids (stx2-restriction fragment length polymorphism [RFLP], stx2 gene variant, and plasmid analyses), and others were used to study the whole genome (ribotyping and pulsed-field gel electrophoresis [PFGE]). The techniques revealed that there was great diversity among the O91 and OX3 STEC strains isolated in central France. A close relationship between strains of the same serotype having the same virulence factor pattern was first suggested by ribotyping. However, stx2-RFLP and stx2 variant analyses differentiated all but 5 of 21 isolates, and plasmid analysis revealed further heterogeneity; a unique combination of characteristics was obtained for all strains except two O91:H21 isolates from beef. The latter strains were shown by PFGE to be the most closely related isolates, with >96% homology, and hence may be subtypes of the same strain. Overall, our results indicate that the combination of stx2-RFLP, stx2 variant, and plasmid profile analyses is as powerful as PFGE for molecular investigation of STEC diversity. Finally, the non-O157:H7 STEC strains isolated from HUS patients were related to but not identical to those isolated from cattle and food samples in the same geographical area. The possibility that there are distinct lineages of non-O157:H7 STEC, some of which are more virulent for humans, should be investigated further.
Shiga toxin-producing Escherichia coli (STEC) strains have been associated with human diseases ranging from uncomplicated diarrhea to hemorrhagic colitis and hemolytic-uremic syndrome (HUS). They have been implicated both in outbreaks and in sporadic cases of infection. STEC infections are mainly food borne, and bovine feces are the main source of food contamination. The ability of STEC strains to cause serious disease in humans is related to their ability to produce one or more Shiga toxins (Stx1, Stx2, and variants of Stx2) (21; M.S. Jacewicz, H. Trachtman, D. S. Newburg, and D. W. K. Acheson, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., abstr. B-102, p.64, 2000). The variants of Stx2, some of which are thought to be less pathogenic for humans, include the Stx2c variants Stx2vh-a and Stx2vh-b, the Stx2d variants Stx2d-Ount and Stx2d-OX3a, Stx2e, and Stx2f (19, 29, 37, 39). The Shiga toxin-encoding genes (stx) are present in the genomes of temperate, lambdoid bacteriophages, which appear to regulate Shiga toxin expression as part of their lytic switch (36).
There seem to be additional factors involved in STEC virulence. Among these is the locus of enterocyte effacement, which contains genes encoding proteins responsible for the attaching and effacing lesions in epithelial cells (41). This locus has been identified as a pathogenicity island, a region of the bacterial chromosome transmitted by horizontal transfer (27). Large plasmids of STEC encode determinants that are thought to be additional virulence factors. These include enterohemolysin, which is encoded by the ehxA gene and acts as a pore-forming cytolysin on eukaryotic cells (34), and secreted serine protease (EspP), which can cleave human coaggulation factor V (13). All the virulence factors currently described in STEC are thus encoded by accessory genetic elements, which have probably been acquired by horizontal DNA transfer.
Among the STEC strains, E. coli serotype O157:H7 is the main causative agent of large-scale outbreaks. Recent studies using molecular fingerprinting methods have revealed significant genomic diversity among O157:H7 isolates possessing the same known virulence determinants (1, 2, 24, 31, 32). A comparison of human and bovine isolates demonstrated that there are two distinct lineages of E. coli O157:H7 and suggested that one of the lineages was either less virulent for humans or inefficiently transmitted to humans from bovine sources (24). Because of the increasing prevalence of non-O157:H7 serotypes in human diseases, comprehensive data on the molecular epidemiology and virulence properties of STEC strains are needed.
During a 1-year survey performed in the Auvergne region of central France, 220 STEC strains were isolated from bovine feces, food samples, and asymptomatic children (30). Twenty-two isolates were selected and compared with three HUS-associated strains isolated from the stools of adult patients in the same geographical area (10). All 25 STEC isolates were stx2 positive and belonged to serotypes O91:H10, O91:H21, OX3:H−, and OX3:H21, which have been shown to be associated with severe human disease (23; http://www.who.int/emc-documents/zoonoses/whocsraph988c.html). Identical virulence gene patterns and identical serotypes suggested that the strains had a common clonal origin. In this study, we performed a detailed analysis of the molecular epidemiology of non-O157:H7 STEC strains by using ribotyping, pulsed-field gel electrophoresis (PFGE) typing, stx2-restriction fragment length polymorphism (RFLP) and plasmid analyses. Our aims were (i) to analyze the clonal relatedness among STEC isolates obtained from sporadic cases of HUS, cattle, and food products in the same geographical area over a short period of time and (ii) to compare the stx2-RFLP and plasmid profiles with the results of classical subtyping techniques, ribotyping, and PFGE.
MATERIALS AND METHODS
STEC isolates.
A total of 25 STEC isolates collected in the same geographical area (central France) were examined in this study. Three human STEC isolates belonging to serotypes O91:H21, O91:H10, and OX3:H− were isolated from sporadic cases of HUS between March 1997 and June 1997 in the teaching hospital in Clermont-Ferrand, France (10). Nineteen isolates (six O91:H10, three O91:H21, one OX3:H−, and nine OX3:H21 isolates) were obtained from the feces of healthy cattle at the city slaughterhouse from November 1997 to September 1998 (30). Three O91:H21 isolates, obtained from food products (two beef samples and one cheese) between December 1997 and June 1998, were included (30). E. coli EDL933 (= ATCC 43895) (serotype O157:H7) was used as a reference strain, and for PFGE experiments NV95, a bovine O157:H7 strain isolated in France, was also used (30).
Ribotyping.
Total genomic DNA of STEC isolates were prepared from 10-ml overnight cultures in Muller-Hinton broth (Biokar Diagnostics, Beauvais, France) by the method described by Picard-Pasquier et al. (28). Approximately 3 μg of genomic DNA was digested independently with two restriction endonucleases, EcoRI and HindIII (Boehringer Mannheim, Meylan, France) according to the manufacturer's instructions. The digested DNA was separated by electrophoresis in a 0.8% agarose gel at 100 V for 6 h in TAE buffer (40 mM Tris-acetate [pH 8], 5 mM sodium acetate, 2 mM EDTA). DNA restriction fragments were then transferred to Hybond N+ nylon membranes (Amersham Pharmacia Biotech, Orsay, France) by standard methods. The rrnB7 probe was prepared from a 7.0-kb restriction fragment carrying the entire rrnB operon of E. coli K-12 and flanking sequences (3, 11). The 7.0-kb restriction fragment was purified with a 0.22-μm-pore-size filter (SPIN-X; Costar, Cambridge, Mass.) and radiolabeled with [α-32P]dCTP (Amersham Pharmacia Biotech) by using a random primed DNA labeling kit (Boehringer Mannheim) according to the manufacturer's specifications. The probe was separated from unincorporated nucleotides with a Sephadex G-50 Fine (Amersham Pharmacia Biotech) column. Southern hybridization was performed by using a rapid hybridization buffer (Amersham Pharmacia Biotech) as indicated by the manufacturer. The hybridized membranes were washed once at 65°C for 20 min with 0.1% sodium dodecyl sulfate–300 mM NaCl–30 mM sodium citrate and twice at 65°C (20 min each) with 0.1% sodium dodecyl sulfate–75 mM NaCl–7.5 mM sodium citrate, and then they were exposed to Hyperfilm MP film (Amersham Pharmacia Biotech) and processed with an automated film developer (Hyperprocessor; Amersham Pharmacia Biotech). On the basis of its restriction fragment profile, each strain was assigned a ribotype pattern. The groups were each identified by a letter (each group differed from the other groups by more than two profile bands), and a number was used to identify minor differences (in one or two profile bands) within a group. The RFLP Extension Ladder System (Life Technologies, Cergy Pontoise, France) was used as the molecular weight marker.
PFGE analysis.
Genomic DNA was prepared by using the protocol described by Bohm and Karch (9). Restriction endonuclease digestion was performed with 50 U of XbaI (Life Technologies) at 37°C for 18 h. PFGE was performed in 1.2% agarose gels by using a contour-clamped homogeneous electric field PFGE Gene Navigator apparatus (Pharmacia, Uppsala, Sweden) in 0.5× Tris-borate-EDTA buffer at 14°C and 200 V. The pulse time was increased from 10 to 40 s over a 24-h period. Lambda ladder (Bio-Rad, Ivry, France) was used as the size marker. The Dice similarity coefficient and GelCompar software (Applied Maths, Kortrijk, Belgium) were used to compare PFGE profiles. Cluster analysis was performed by using the hierarchic unweighted pair group arithmetic average algorithm.
stx2-RFLP analysis.
Genomic DNA restriction fragments digested by EcoRI or HindIII were Southern blotted as described above and hybridized with an stx2-specific DNA probe. The probe was prepared from a 587-bp PCR product of the strain EDL933 stx2 gene obtained by using the LP43-LP44 primer pair (Table 1) (15). Probe labeling, hybridization, washing, and the procedure used for autoradiography of the membranes were performed as described above. We assigned different profiles to the strains tested on the basis of the size and number of bands obtained. The RFLP Extension Ladder System (Life Technologies) was used as the molecular weight marker.
TABLE 1.
Primer sequences used in PCR to amplify specific fragments from the stx2, ehxA, and espP genes
| Target(s) | Primer | Oligonucleotide sequence (5′-3′) | Location in the gene | Size of amplified product (bp) | Reference |
|---|---|---|---|---|---|
| stx2 | LP43 | ATC CTA TTC CCG GGA GTT TAC G | bp 57 to 643 of B subunit | 587 | 15 |
| LP44 | GCG TCA TCG TAT ACA CAG GAG C | ||||
| stx2, stx2vh-a, stx2vh-b | VT2-c | AAG AAG ATG TTT ATG GCG GT | bp 4 of B subunit to bp 21 upstream | 285 | 39 |
| VT2-d | CAC GAA TCA GGT TAT GCC TC | ||||
| stx2d-Ount, stx2d-OX3a | VT2-cm | AAG AAG ATA TTT GTA GCG G | bp 4 to 259 of B subunit | 256 | 29 |
| VT2-f | TAA ACT GCA CTT CAG CAA AT | ||||
| stx2e | VTe-a | CCT TAA CTA AAA GGA ATA TA | bp -25 downstream to bp 205 of A subunit | 230 | 19 |
| VTe-b | CTG GTG GTG TAT GAT TAA TA | ||||
| ehxA | RH35 | CAC ACG GAG CTT ATA ATA TTC TGT CA | bp 1645 to 1965 | 321 | 18 |
| RH37 | AAT GTT ATC CCA TTG ACA TCA TTT GAC T | ||||
| espP | esp-A | AAA CAG CAG GCA CTT GAA CG | bp 544 to 2373 | 1,830 | 13 |
| esp-B | GGA GTC GTC AGT CAG TAG AT |
Detection of stx2 variants by PCR.
The primers used for stx2 variant analysis are shown in Table 1. The VT2-c–VT2-d primer pair was used in a PCR protocol to detect the stx2, and stx2vh-a, and stx2vh-b genes. RFLP analysis of the amplicons (PCR-RFLP analysis) identifies the Stx2 and Stx2c variants (Stx2vh-a and Stx2vh-b) (39). The VT2-cm–VT2-f primer pair was used to specifically detect the genes coding for the Stx2d variants (Stx2d-Ount and Stx2d-OX3a) (29). The Stx2e variant was detected with the VT2e-a–VT2e-b primer pair (19). The DNA to be amplified was released from whole organisms by boiling. The amplification reactions were performed with a Perkin-Elmer Cetus DNA thermal cycler 2400. DNA from E. coli EDL933 (O157:H7; Stx2), B2F1 (O91:H21; Stx2vh-a and Stx2vh-b), Fac9 (Stx2e), OX3:H11 (Stx2d), and HB101 (negative control) were included in each PCR analysis.
Preparation of plasmid DNA and detection of ehxA.
Plasmid DNA of STEC isolates were prepared from 1.5-ml overnight cultures of bacteria grown in Luria broth (Difco Laboratories, Detroit, Mich.) by the alkaline lysis method described by Kado and Liu (20). Plasmid DNA patterns were obtained by electrophoresing the DNA on 0.7% agarose gels in TAE buffer, and the gels were stained with ethidium bromide and observed under UV light. The molecular sizes of the plasmids were estimated by comparing their mobilities with those of plasmids of known molecular weights extracted from the E. coli EDL933 reference strain. An alphabetical code was used to describe the plasmid profile established for each isolate. The probe used for detection of ehxA was a 321-bp fragment obtained by PCR from strain EDL933 by using primers RH35 and RH37 (Table 1) (18).
Detection of espP by colony blot hybridization.
The espP gene was detected by colony blot hybridization by using the classic procedure of Maas (25). The probe used for detection of espP was a 1,830-bp fragment that was obtained by PCR from strain EDL933 by using primers shown in Table 1 and was labeled as described above.
RESULTS
Ribotyping profiles of the STEC isolates.
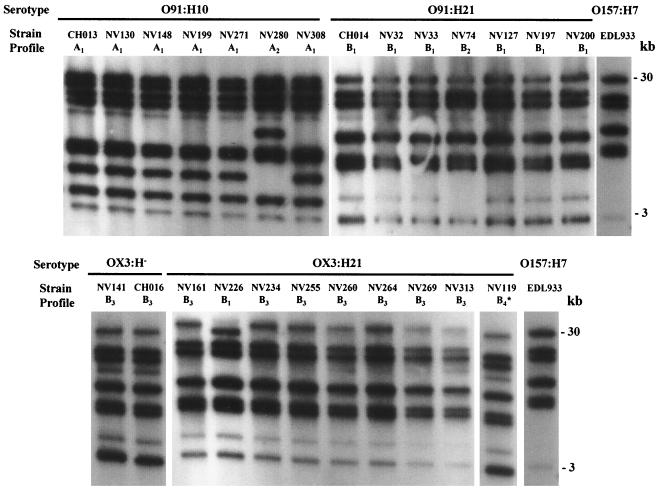
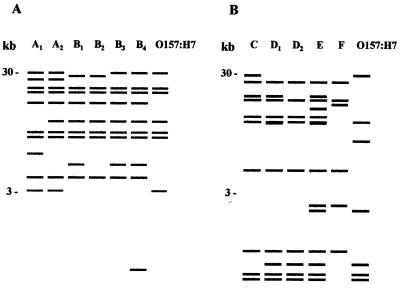
Twenty-two STEC isolates, obtained from cattle and food samples between November 1997 and September 1998 in central France, were analyzed by ribotyping with the rrnB7 probe and were compared with three isolates obtained from HUS patients in the same area. All of the isolates belonged to serotype O91:H10, O91:H21, OX3:H−, or OX3:H21. Between 8 and 10 fragments were obtained with HindIII-digested genomic DNA, and 10 to 18 fragments were obtained with EcoRI; the fragments ranged from 1 to 30 kb long. Figure 1 shows fragment profiles generated from the isolates with restriction enzyme HindIII. Each strain was assigned a ribotype pattern; a letter indicated the group, and a subscript number indicated minor differences (one or two bands) within a group. When HindIII was used, the isolates were classified into two major groups (groups A and B) comprising six different ribotypes designated ribotypes A1 to A2 and B1 to B4. When the EcoRI enzyme was used, four major groups (groups C to F) comprising five ribotypes were identified for the 25 STEC isolates. Figure 2 shows representative patterns generated with the HindIII and EcoRI enzymes. Six and four bands were common to all of the strains for the HindIII and EcoRI profiles, respectively. The profiles obtained for O157:H7 strain EDL933 differed from the non-O157 STEC profiles by at least three bands.
FIG. 1.
Ribotype patterns generated with HindIII-digested genomic DNA. The positions of molecular size standards are indicated on the right. The strains and patterns are shown in Table 2. One additional 1.5-kb fragment could not be shown in pattern B4.
FIG. 2.
Representative ribotype patterns obtained for the STEC strains. For each pattern the letter indicates the group and the subscript number indicates minor differences within a group. The positions of molecular size standards are indicated on the left. (A) Six ribotype patterns obtained with HindIII (patterns A1 to A2 and B1 to B4) for the 25 STEC analyzed in this study compared to the pattern of O157:H7. (B) Five ribotype patterns obtained with EcoRI (patterns C, D1 to D2, E, and F) compared to the pattern of O157:H7. The O157:H7 profile was obtained with the EDL933 reference strain.
When the HindIII and EcoRI ribotypes were combined (Table 2), eight distinct patterns were identified. Two similar patterns were distinguished for the O91:H10 STEC isolates. The NV280 isolate, which was recovered from a bovine sample, produced the A2-C pattern, and the six other O91:H10 isolates (one, CH013, recovered from a patient suffering from HUS and five from bovine samples) produced identical A1-C patterns. A major B1-D1 pattern was identified for six of the seven O91:H21 isolates (one from a patient with HUS, two from beef samples, and three from bovine feces). The 091:H21 isolate obtained from cheese, which had a specific stx1-stx2-ehxA virulence gene profile, had a distinct B2-E pattern. The predominant B3-D2 pattern of five of the nine OX3:H21 isolates was the same as that of the OX3:H− isolates, which suggests that the two serotypes are closely related. In contrast, 13 OX3:H2 strains had ribotype profiles that were completely distinct from those of the OX3:H− strains (data not shown), which indicates a more distant relationship. Because they were clearly different from the HUS-associated OX3:H− isolate, the four remaining OX3:H21 and the OX3:H2 strains were not used for further analysis. Taken together, these data indicate that HindIII and EcoRI ribotyping could reveal only minor differences in non-O157:H7 strains belonging to the same serotype.
TABLE 2.
Origins, virulence gene profiles, and ribotype patterns of the 25 STEC strains, sorted according to serotype
| Serotype | Strain | Origina | Virulence gene profile | Ribotype patternb
|
|
|---|---|---|---|---|---|
| HindIII | EcoRI | ||||
| O91:H10 | CH013 | HUS | stx2 | A1 | C |
| NV130 | Cattle | stx2 | A1 | C | |
| NV148 | Cattle | stx2 | A1 | C | |
| NV199 | Cattle | stx2 | A1 | C | |
| NV271 | Cattle | stx2 | A1 | C | |
| NV308 | Cattle | stx2 | A1 | C | |
| NV280 | Cattle | stx2 | A2 | C | |
| O91:H21 | CH014 | HUS | stx2, ehxA | B1 | D1 |
| NV127 | Cattle | stx2, ehxA | B1 | D1 | |
| NV197 | Cattle | stx2, ehxA | B1 | D1 | |
| NV200 | Cattle | stx2, ehxA | B1 | D1 | |
| NV32 | Beef | stx2, ehxA | B1 | D1 | |
| NV33 | Beef | stx2, ehxA | B1 | D1 | |
| NV74 | Cheese | stx1, stx2, ehxA | B2 | E | |
| OX3:H− | CH016 | HUS | stx2 | B3 | D2 |
| NV141 | Cattle | stx2 | B3 | D2 | |
| OX3:H21 | NV234 | Cattle | stx2 | B3 | D2 |
| NV255 | Cattle | stx2 | B3 | D2 | |
| NV260 | Cattle | stx2 | B3 | D2 | |
| NV269 | Cattle | stx2 | B3 | D2 | |
| NV313 | Cattle | stx2 | B3 | D2 | |
| NV161 | Cattle | stx2 | B3 | F | |
| NV264 | Cattle | stx2 | B3 | F | |
| NV226 | Cattle | stx2 | B1 | F | |
| NV119 | Cattle | stx2 | B4 | F | |
HUS, isolate obtained from HUS patient; cattle, isolate obtained from bovine feces; beef, isolate obtained from beef sample; cheese, isolate obtained from cheese sample.
The letters differentiate groups of isolates and the subscript numbers indicate minor differences between isolates belonging to the same group.
Toxin gene RFLP patterns of STEC isolates.
To establish more discriminative clonal relatedness among STEC isolates, the stx2-RFLP method was used. This method has been used for molecular epidemiological investigations of STEC and has been shown to be sufficiently sensitive to allow interstrain differentiation of STEC belonging to the same serotype (33). For the RFLP procedure we used the EcoRI and HindIII restriction enzymes, which do not have a restriction site in the stx2 toxin gene and allow detection of restriction site polymorphism near the toxin genes. Hybridization of digested genomic DNA with the stx2 probe was performed for 21 isolates, including 16 isolates belonging to serotypes O91:H10, O91:H21, and OX3:H− and the 5 OX3:H21 isolates which produced the B3-D2 ribotype pattern (Table 3). Ten and 11 DNA band size profiles were obtained with HindIII- and EcoRI-digested genomic DNA, respectively. Between one and three fragments of different sizes (ranging from approximately 3.5 to 30 kb for HindIII and from 4.5 to 15 kb for EcoRI) were obtained for STEC isolates with each of the restriction enzymes. For the 21 E. coli isolates, the combined data resulted in 15 distinct RFLP patterns.
TABLE 3.
Comparison of ribotype patterns stx2-RFLP profiles, stx2 types and plasmid profiles of the STEC strains
| Serotype | Strain | Origina | Ribotype patternb |
stx2-RFLP profilec
|
stx2 typed | Plasmid profilee | |
|---|---|---|---|---|---|---|---|
| HindIII | EcoRI | ||||||
| O91:H10 | CH013 | HUS | A1-C | 26, 7 | 10, 7.5 | 2, 2vh-a, 2vh-b | A |
| NV148 | Cattle | A1-C | 26 | 12 | 2vh-a | C | |
| NV271 | Cattle | A1-C | 26 | 12 | 2vh-a | E | |
| NV308 | Cattle | A1-C | 26 | 15 | 2vh-b | D | |
| NV130 | Cattle | A1-C | 26, 20 | 15, 10 | 2vh-a, 2vh-b | B | |
| NV199 | Cattle | A1-C | 26, 20 | 15, 12 | 2vh-a, 2vh-b | D | |
| NV280 | Cattle | A2-C | 26, 11 | 12 | 2vh-a | F | |
| O91:H21 | CH014 | HUS | B1-D1 | 26, 20 | 10, 7.5 | 2, 2vh-a, 2vh-b | G |
| NV127 | Cattle | B1-D1 | 26, 20 | 10, 7.5 | 2vh-a, 2vh-b | H | |
| NV32 | Beef | B1-D1 | 5.5 | 4.5 | 2vh-b | H | |
| NV33 | Beef | B1-D1 | 5.5 | 4.5 | 2vh-b | H | |
| NV200 | Cattle | B1-D1 | 5.5 | 4.5 | 2, 2vh-b | G | |
| NV197 | Cattle | B1-D1 | 20, 9, 5.5 | 12, 5, 4.5 | 2, 2vh-b | J | |
| NV74 | Cheese | B2-E | 5.5 | 4.5 | 2vh-b | I | |
| OX3:H− | CH016 | HUS | B3-D2 | 12 | 15 | 2vh-a | G |
| NV141 | Cattle | B3-D2 | 30, 26 | 12, 4.5 | 2vh-b | G | |
| OX3:H21 | NV234 | Cattle | B3-D2 | 3.5 | 13 | 2vh-b | G |
| NV260 | Cattle | B3-D2 | 3.5 | 13 | 2vh-a | G | |
| NV255 | Cattle | B3-D2 | 3.5 | 12 | 2vh-b | G | |
| NV313 | Cattle | B3-D2 | 4, 3.5 | 12, 4.5 | 2vh-b | G | |
| NV269 | Cattle | B3-D2 | 5.5 | 13 | 2vh-a | G | |
HUS, isolate obtained from HUS patient; cattle, isolate obtained from bovine feces; beef, isolate obtained from beef sample; cheese, isolate obtained from cheese sample.
The letters differentiate groups of isolates, and the subscript numbers indicate minor differences between isolates of the same group.
The numbers indicate the sizes (in kilobases) of the different stx2 fragments obtained.
2, stx2; 2vh-a, stx2vh-a; 2vh-b, stx2vh-b
The letters differentiate isolates on the basis of the number and size of the plasmids.
Most of the O91:H10 strains produced stx2 fragments of different sizes; the only exceptions were two cattle isolates (NV148 and NV271), which produced the same pattern (two bands at approximately 26 and 12 kb with HindIII and EcoRI, respectively). The seven O91:H21 strains produced three distinct RFLP patterns; three isolates from food (NV32, NV33, and NV74) and one isolate from cattle (NV200) produced the same pattern (one band at 5.5 kb with HindIII and one band at 4.5-kb with EcoRI), one bovine isolate (NV197) produced a distinct pattern (three bands at 20, 9, and 5.5 kb and three bands at 12, 5, and 4.5 kb with HindIII and EcoRI, respectively), and the pattern produced by isolate NV127 was indistinguishable from that produced by O91:H21 HUS isolate CH014 (26- and 20-kb HindIII bands and 10- and 7.5-kb EcoRI bands). In contrast, the RFLP patterns of the two OX3:H− isolates, one from a HUS patient and one from cattle feces, differed from each other and from those of the OX3:H21 isolates.
Analysis of the stx2 gene variants.
To further assess the diversity of non-O157:H7 STEC, the stx2 gene types present in each isolate were identified by specific PCR methods. The results of this analysis are summarized in Table 3. The 21 isolates tested were found to contain stx2, stx2vh-a, or stx2vh-b, but none was found to contain stx2d or stx2e. The strains contained either one stx2 gene type or a combination of two or three. The serotypes were not linked to a specific stx genotype.
Three of the seven O91:H10 isolates contained only the stx2vh-a target sequence, one contained only the stx2vh-b gene, and two contained both stx2vh-a and stx2vh-b genes. Interestingly, the isolate from a HUS patient, CH013, contained a combination of the three sequences. Among the O91:H21 STEC, the three isolates obtained from food samples contained only the stx2vh-b gene. Two isolates from cattle contained the stx2 and stx2vh-b sequences, while the other bovine isolate contained stx2vh-a and stx2vh-b. The O91:H21 isolate obtained from a HUS patient contained the three variants. When the OX3:H− and OX3:H21 serotypes were examined, only one stx2 gene type was found for each isolate. One OX3:H isolate from a HUS patient and two OX3:H21 isolates from cattle contained only stx2vh-a, while the other OX3:H− isolate and three OX3:H21 isolates, all obtained from cattle, contained stx2vh-b. No correlation was found between stx2 variant type and stx2-RFLP band size, serotype, or origin of the STEC strains.
Plasmid studies.
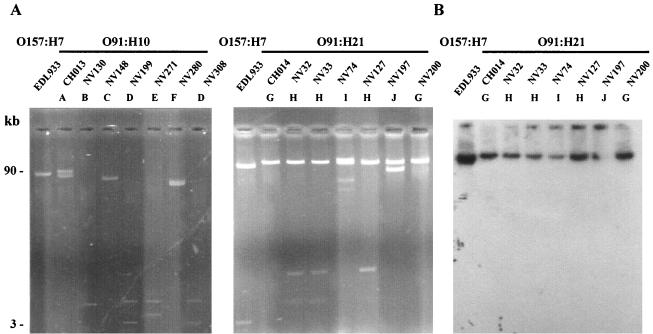
Since analysis of plasmids was found to be helpful for studying the variability of non-O157:H7 STEC (42), we investigated the presence of plasmids in the 21 test strains. Plasmid pO157, the 90-kb virulence plasmid of strain EDL933, an isolate from an outbreak in Michigan that occurred in 1982, was used as a control. The plasmid profiles obtained for the O91:H10 and O91:H21 isolates are shown in Fig. 3A. All isolates carried one to four plasmids whose sizes ranged from approximately 3 to more than 90 kb. Ten different plasmid profiles, profiles A to J, were obtained for the 21 isolates tested (Table 3). Endonuclease restriction analysis of the plasmid content with EcoRI showed that strains with the same plasmid profile had the same cleavage pattern (data not shown).
FIG. 3.
Representative plasmid profiles of the STEC strains. (A) Plasmid DNA of the O91:H10 and O91:H21 STEC strains. The profiles of OX3 isolates are not shown; these isolates produced plasmid profile G similar to the profile of O91:H21 strain CH014. The positions of molecular size standards are indicated on the left. (B) Southern blot hybridization of plasmid DNA of the O91:H21 strains with a specific ehxA probe. The strains and profiles are described in Table 3.
Six plasmid profiles, profiles A to F, were obtained for the seven O91:H10 E. coli strains. Only two isolates from cattle (NV199 and NV308) produced the same profile, profile D. For the O91:H21 strains, we identified four distinct plasmid patterns. All of the strains contained at least one large plasmid (more than 50 kb long), and sometimes the strains contained additional small plasmids (Fig. 3A). Profile G was obtained for the CH014 HUS strain and one bovine isolate (NV200), profile H was obtained for one bovine isolate (NV127) and two beef isolates (NV32 and NV33), profile I was characteristic of the NV74 cheese isolate, and profile J was characteristic of O91:H21 isolate NV197. In contrast, the two OX3:H− and five OX3:H21 E. coli strains produced the same plasmid profile, profile G, which was further evidence of the close relationship of these strains.
Analysis of plasmid-encoded determinants.
Of all the strains analyzed in this study, only the serotype O91:H21 strains contained the ehxA gene (Table 2) (30), which was originally found on large plasmid pO157 of O157:H7 E. coli strains (7). All seven O91:H21 strains contained at least one such large plasmid (Fig. 3A), which was hybridized with a probe complementary to the ehxA sequence. The ehxA gene was present on one of the large plasmids in all of the O91:H21 STEC strains (Fig. 3B). Since the espP gene is also known to be located on pO157, we checked for its presence by performing colony blot hybridization with an espP-specific probe. The probe did not hybridize with any of the O91:H21 STEC strains. These results underline the heterogeneity of the STEC plasmids. Most strains had a unique combination of characteristics; the only exceptions were two O91:H21 beef isolates (NV32 and NV33), which were identical (Table 3).
PFGE analysis.
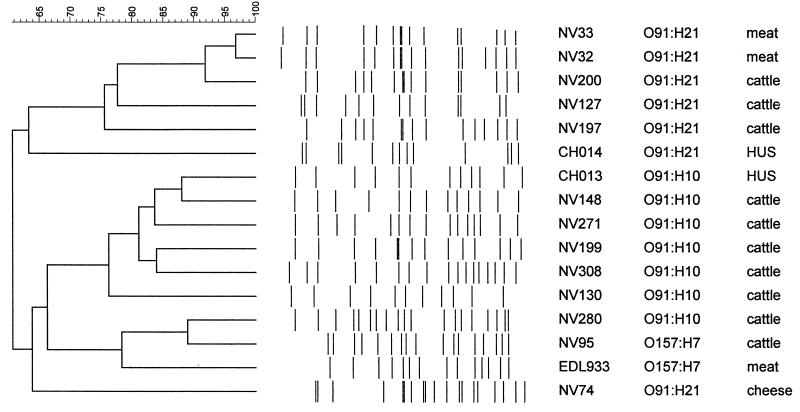
Numerous reports have shown that PFGE typing is a highly discriminatory and reproducible method (2, 6, 26, 32, 38), and we decided to compare this method with ribotyping and the molecular techniques used in this study. To determine clonal relatedness among human, bovine, and food isolates belonging to the same serotypes (O91:H10, O91:H21, OX3:H−, and OX3:H21), we analyzed their XbaI patterns by PFGE. However, several efforts to obtain intact DNA from the OX3 isolates were unsuccessful, and DNA degradation was always observed, which prevented pattern analysis. Thus, 14 O91 isolates (7 O91:H10 and 7 O91:H21 isolates) and two O157:H7 strains, used as controls, were analyzed (Fig. 4). PFGE produced 11 to 19 fragments ranging in size from approximately 50 to 580 kb. A dendrogram analysis of PFGE patterns (Fig. 4) revealed that the isolates were distributed among two main branches of the phylogenetic tree. One cluster included all the O91:H21 isolates except the NV74 strain from cheese. In this cluster, the PFGE patterns of strains from cattle and beef were more than 75% similar whereas the HUS isolate was more distantly related (63% similarity). Only two O91:H21 beef isolates (NV32 and NV33) produced closely related restriction patterns, and there was just one band difference (38). O91:H21 cattle isolate NV200 may be related to isolates NV32 and NV33 since there were only five and four band differences, respectively, which is consistent with two independent genetic events (38). The other O91:H21 isolates were not genetically related according to the criteria of Tenover et al. (38), since their PFGE patterns had more than five band differences.
FIG. 4.
Dendrogram showing the clonal relationships of bovine, food, and HUS non-O157:H7 isolates, as determined by macrorestriction fragment analysis by PFGE, after digestion of genomic DNA by XbaI. The tree was constructed by using the unweighted pair group arithmetic average method. Strain characteristics are described in Table 3.
Two main subgroups were identified in the second cluster. The first subgroup included O91:H10 isolates from cattle and patients with HUS, which were more than 76% similar. Interestingly, HUS isolate CH013 and cattle isolates NV148 and NV271 may be related since there were only three and five band differences, respectively. The second subgroup contained the two O157:H7 isolates included in this study (cattle isolate NV95 and reference strain EDL933) and one O91:H10 isolate from cattle, which were more than 78% similar. The O91:H21 isolate from cheese (NV74) was distantly related to the second cluster, with only 63.7% similarity.
DISCUSSION
In this study, we used a collection of 25 STEC isolates from sporadic cases of HUS, cattle, and food products, obtained in the same geographical area over a short period of time, to perform a detailed analysis of the molecular epidemiology of non-O157:H7 STEC. We compared different typing methods; some of these methods were used to explore potentially mobile elements like the stx genes or plasmids (stx2-RFLP analysis, stx2 gene variant analysis, and plasmid analysis), and others were used to study the whole genome (ribotyping and PFGE).
Identical or very similar ribotype profiles were obtained for members of each serotype. Using two enzymes, we identified eight different profiles for 25 isolates belonging to four serotypes. A total of 19 of 25 isolates could not be differentiated, including six of seven O91:H10 isolates, six of seven O91:H21 isolates, and 7 of 11 OX3:H21/H− isolates. This indicates that ribotyping, which can be a very useful tool for epidemiological investigation, was not able to discriminate between STEC isolates belonging to the same serotype. We therefore used stx2-RFLP analysis, a sensitive and specific method for differentiation of STEC strains, which has also been used in epidemiological investigations of outbreaks and sporadic cases of infection (16, 33). With this technique, 10 of 21 isolates could not be differentiated; two O91:H10 isolates had identical profiles, as did two O91:H21 isolates, four other O91:H21 isolates and two OX3:H21 isolates (Table 3). When this technique was combined with stx2 variant analysis, only 5 of 21 isolates could not be differentiated (O91:H10 isolates NV148 and NV271 and O91:H21 isolates NV32, NV33, and NV74 [NV74 produced a distinct ribotype pattern]). Plasmid analysis revealed further heterogeneity of the STEC isolates, except for those belonging to the OX3 serogroup (all of which harbored a >90-kb plasmid). Taken together, the data obtained with the stx2-RFLP, stx2 variant, and plasmid profile analyses were very discriminatory, since only 2 of 21 isolates (the two O91:H21 beef isolates, NV32 and NV33) could not be differentiated. We then compared the results with those obtained by PFGE. This technique is probably the most powerful tool available for strain differentiation and has been used for a broad range of bacterial pathogens (4, 17, 38). It has also been used in investigations of several outbreaks of E. coli O157 infection and has been shown to discriminate between strains belonging to the same ribotype (2, 6, 26, 31, 32). In our study, PFGE had the greatest discriminatory power for non-O157:H7 STEC isolates, since each isolate produced a unique PFGE pattern. Interestingly, NV32 and NV33, the two O91:H21 beef isolates that could not be distinguished by the stx2 and plasmid analyses, were shown to be the most closely related isolates by PFGE, with >96% homology, and one of these isolates may have been derived from the other by a single genetic event (38). They were isolated from two beef samples purchased from the same butcher's shop 2 days apart and are probably subtypes of the same strain, which confirms the discriminatory power of PFGE. Overall, our results indicate that the combination of stx2-RFLP, stx2 variant, and plasmid profile analyses was as powerful as PFGE for molecular investigation of STEC diversity.
Sequences that are homologous to the stx2 probe are assumed to possess neither EcoRI nor HindIII restriction sites; hence, the smallest number of stx2 genes in the genome was estimated for each strain. Since the HUS-associated strains were more cytotoxic than their bovine counterparts that belonged to the same serotype (30), we tried to establish the relationship between the number of stx2 genes and stx2 variants and cytotoxicity. We were unable to establish any correlation between the number of stx2 genes and cytotoxicity for Vero cells, which was determined in a previous study, except for the two HUS-associated isolates, CH013 and CH014, both of which contained a combination of three variants and were the most cytotoxic isolates among the 25 isolates tested. Several factors may contribute to the degree of cytotoxicity of STEC strains, including not only the number of stx2 genes present in the genome but also the regulation of expression of these genes, which are carried by the genomes of different phages (40). Further studies at the RNA and DNA levels involving sequence analysis of the regulatory stx2-flanking regions will be necessary to compare the STEC isolates. Most serotypes were not linked to a specific stx genotype, and different combinations of stx genes were found in strains belonging to the same serotype. No correlation could be established between the stx2 variant and stx2-RFLP band size or any other characteristic of the strains (serotype, origin, or level of cytotoxicity for Vero cells). Finally, the number of EcoRI and HindIII fragments that hybridized with the stx2 gene probe was greater than the number of stx2 variants in three strains (O91:H21 isolate NV197 and OX3 isolates NV141 and NV313 [Table 3]), which suggests that several copies of the stx2 gene may be present.
As STEC isolates belonging to different serotypes often harbor one or more small or large plasmids, plasmid profile analysis was used in this study as a typing method. The plasmid profiles of the O91:H10 and O91:H21 serotypes varied widely (nine patterns were identified for 14 isolates). The differences were not correlated with the origin, with the serotype, or with other molecular characteristics of the strains. The presence of an approximately 90-kb plasmid carrying several genes encoding functions associated with virulence is a characteristic attribute of many STEC strains (8). All of our strains carried similarly large plasmids, but the plasmids were heterogeneous in terms of their virulence gene contents. The variations were serotype related. All of the O91:H21 strains contained large plasmids encoding the ehxA gene but not the espP gene. None of the O91:H10 or OX3:H− large plasmids contained the ehxA or espP gene. In contrast, 90-kb plasmid pO157 of strain EDL933 (O157:H7) contained both genes (14). Other studies have shown that STEC isolates have different plasmid contents based on serotype (12, 35), and our data confirmed the high level of plasmid diversity among STEC isolates, even within a single serotype.
One of the aims of our study was to determine whether the non-O157:H7 STEC strains isolated from HUS patients were related to those isolated from cattle and food samples in the same geographical area. A close relationship among strains of human, animal, and food origins was suggested by previous serotyping and virulence factor pattern analyses. Most of the strains belonging to the same serotype produced identical ribotype patterns, but both PFGE and the combination of stx2-RFLP, stx2 variant, plasmid profile analyses showed that the strains were clearly different. HUS-associated isolate CH014 was distantly related to the O91:H21 isolates from cattle and food, with only 63.3% similarity as determined by PFGE, although it differed from bovine isolate NV127 only by the stx2 variants and by the absence of a small plasmid (Table 3 and Fig. 3). The other HUS-associated isolate, CH013, was closely related to O91:H10 cattle isolate NV148 as determined by PFGE (88% similarity), indicating that cattle may be a reservoir for pathogenic STEC in France. However, CH013 differed from NV148 by additional stx2 and stx2vh-b genes, stx2-RFLP pattern, and plasmids, suggesting that such pathogenic strains may have evolved from bovine strains by acquisition of additional virulence factors. Finally, the data obtained by examining mobile elements (stx2-RFLP, stx2 variant, and plasmid analyses) were found to be complementary to those provided by PFGE.
With regard to clonal relatedness, the two OX3:H− strains (one from a HUS patient and one from cattle) and five of the nine OX3:H21 isolates had similar ribotypes and identical plasmid profiles. In contrast, 13 OX3:H2 STEC strains obtained from bovine and food sources during the same survey (30) had different virulence profiles and different ribotype patterns (data not shown). We presumed that the OX3:H− strain obtained from a HUS patient was a nonmotile variant of an OX3:H21 strain. OX3:H21 strains have been associated previously with severe human diseases (22). The HUS OX3:H− isolate differed from its OX3:H− and OX3:H21 environmental counterparts only by the stx2 type and stx2-RFLP profile. However, we were not able to further explore the clonal relatedness between these OX3 isolates by PFGE since DNA degradation made typing impossible. The high nuclease activity observed in the OX3 isolates, by limiting the spread of exogenous plasmids, could account for the fact that only one plasmid profile was obtained.
Several studies have shown that there is significant genomic diversity and great variability in virulence among O157:H7 isolates that have the same known virulence determinants (5, 24). A comparison of human and bovine isolates demonstrated that there are two distinct lineages of E. coli O157:H7 and suggested that one of these lineages may be less virulent for humans or may not be efficiently transmitted to humans from bovine sources (24). We postulated that such pathogenic lineages might exist for non-O157:H7 STEC. However, we could not identify any characteristic that was specific to HUS-associated isolates. Further studies are needed to identify the specific properties of the pathogenic clones that distinguish them from their nonpathogenic counterparts.
In conclusion, several powerful molecular techniques showed that there was great diversity among non-O157:H7 STEC strains isolated in central France. The close relationship among strains of human, animal, or food origin belonging to serotypes O91:H10, O91:H21, OX3:H−, and OX3:H21 was initially suggested by serotyping and virulence factor pattern analysis. Most of the strains belonging to the same serotype produced identical ribotype patterns but unique typing patterns when they were subjected to PFGE and analysis of mobile elements. The non-O157:H7 STEC strains isolated from HUS patients were related to, but not identical to, those isolated from cattle and food samples in the same geographical area. Overall, our findings indicate that the combination of stx2-RFLP, stx2 variant, and plasmid profile analyses is as powerful as PFGE for molecular investigation of STEC diversity.
ACKNOWLEDGMENTS
We thank C. P. Vivares from the Laboratoire Parasitologie Moléculaire et Cellulaire, UMR CNRS 6023, Université Blaise Pascal, for providing the Gene Navigator apparatus and advice concerning PFGE experiments. We also thank Liliane Millet from the Laboratoire UR545 Recherche Fromagères INRA Aurillac, for her help with the profile comparison in which the GelCompar software was used and Katy Durieux for technical assistance with ribotyping.
This study was supported in part by the Ministère de l'Enseignement supérieur et de la Recherche (grant EA2148) and by the Ministère de l'Aménagement du Territoire et de l'Environnement (Programme Environnement Santé EN 98-17).
REFERENCES
- 1.Allison L, Stirrat A, Thomson-Carter F M. Genetic heterogeneity of Escherichia coli O157:H7 in Scotland and its utility in strain subtyping. Eur J Clin Microbiol Infect Dis. 1998;17:844–848. doi: 10.1007/s100960050204. [DOI] [PubMed] [Google Scholar]
- 2.Allison L J, Carter P E, Thomson-Carter F M. Characterization of a recurrent clonal type of Escherichia coli O157:H7 causing major outbreaks of infection in Scotland. J Clin Microbiol. 2000;38:1632–1635. doi: 10.1128/jcm.38.4.1632-1635.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alos J-I, Lambert T, Courvalin P. Comparison of two molecular methods for tracing nosocomial transmission of Escherichia coli K1 in a neonatal unit. J Clin Microbiol. 1993;31:1704–1709. doi: 10.1128/jcm.31.7.1704-1709.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arbeit R D, Arthur M, Dunn R, Kim C, Selander R K, Goldstein R. Resolution of recent evolutionary divergence among Escherichia coli from related lineages: the application of pulsed field electrophoresis to molecular epidemiology. J Infect Dis. 1990;161:230–235. doi: 10.1093/infdis/161.2.230. [DOI] [PubMed] [Google Scholar]
- 5.Baker D R, Moxley R A, Francis D H. Variation in virulence in the gnotobiotic pig model of O157:H7 Escherichia coli strains of bovine and human origin. Adv Exp Med Biol. 1997;412:53–58. doi: 10.1007/978-1-4899-1828-4_6. [DOI] [PubMed] [Google Scholar]
- 6.Barrett T J, Lior H, Green J H, Khakhira R, Wells J G, Bell B P, Greene K D, Lewis J, Griffin P M. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J Clin Microbiol. 1994;32:3013–3017. doi: 10.1128/jcm.32.12.3013-3017.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauer M, Welch R A. Characterization of an RTX toxin from enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 1996;64:167–175. doi: 10.1128/iai.64.1.167-175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beutin L, Geier D, Zimmermann S, Karch H. Virulence markers of Shiga-like toxin-producing Escherichia coli strains originating from healthy domestic animals of different species. J Clin Microbiol. 1995;33:631–635. doi: 10.1128/jcm.33.3.631-635.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bohm H, Karch H. DNA fingerprinting of Escherichia coli O157:H7 strains by pulsed-field gel electrophoresis. J Clin Microbiol. 1992;30:2169–2172. doi: 10.1128/jcm.30.8.2169-2172.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonnet R, Souweine B, Gauthier G, Rich C, Livrelli V, Sirot J, Joly B, Forestier C. Non-O157 Stx2-producing Escherichia coli strains associated with sporadic cases of hemolytic-uremic syndrome in adults. J Clin Microbiol. 1998;36:1777–1780. doi: 10.1128/jcm.36.6.1777-1780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brosius J, Ullrich A, Raker M A, Gray A, Dull T J, Gutell R R, Noller H F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981;6:112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- 12.Brunder W, Schmidt H, Frosch M, Karch H. The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology. 1999;145:1005–1014. doi: 10.1099/13500872-145-5-1005. [DOI] [PubMed] [Google Scholar]
- 13.Brunder W, Schmidt H, Karch H. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7, cleaves human coagulation factor V. Mol Microbiol. 1997;24:767–778. doi: 10.1046/j.1365-2958.1997.3871751.x. [DOI] [PubMed] [Google Scholar]
- 14.Burland V, Shao Y, Perna N T, Plunkett G, Sofia H J, Blattner F R. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 1998;26:4196–4204. doi: 10.1093/nar/26.18.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cebula T A, Payne W L, Feng P. Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J Clin Microbiol. 1995;33:248–250. doi: 10.1128/jcm.33.1.248-250.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grimm L M, Goldoft M, Kobayashi J, Lewis J H, Alfi D, Perdichizzi A M, Tarr P I, Ongerth J E, Moseley S L, Samadpour M. Molecular epidemiology of a fast-food restaurant-associated outbreak of Escherichia coli O157:H7 in Washington State. J Clin Microbiol. 1995;33:2155–2158. doi: 10.1128/jcm.33.8.2155-2158.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grundmann H, Schneider C, Hartung D, Daschner F D, Pitt T L. Discriminatory power of three DNA-based typing techniques for Pseudomonas aeruginosa. J Clin Microbiol. 1995;33:528–534. doi: 10.1128/jcm.33.3.528-534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall, R. H., and J. G. Xu. December 1995. A new and distinctive DNA sequence of E. coli O157:H7 and its use for the rapid, sensitive and specific detection of O157:H7 and other enterohemorrhagic E. coli. International patent WO 95/34682.
- 19.Johnson W M, Pollard D R, Lior H, Tyler S D, Rozee K R. Differentiation of genes coding for Escherichia coli verotoxin 2 and the verotoxin associated with porcine edema disease (VTe) by the polymerase chain reaction. J Clin Microbiol. 1990;28:2351–2353. doi: 10.1128/jcm.28.10.2351-2353.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karmali M A, Petric M, Lim C, Fleming P C, Arbus G S, Lior H. The association between idiopathic hemolytic-uremic syndrome and infection by verotoxin-producing Escherichia coli. J Infect Dis. 1985;151:775–782. doi: 10.1093/infdis/151.5.775. [DOI] [PubMed] [Google Scholar]
- 22.Keskimäki M, Ikäheimo R, Kärkkäinen P, Scheutz F, Ratiner Y, Puohiniemi R, Siitonen A. Shiga toxin-producing Escherichia coli serotype OX3:H21 as a cause of hemolytic-uremic syndrome. Clin Infect Dis. 1997;24:1278–1279. doi: 10.1086/513668. [DOI] [PubMed] [Google Scholar]
- 23.Keskimäki M, Saari M, Heiskanen T, Siitonen A. Shiga toxin-producing Escherichia coli in Finland from 1990 through 1997: prevalence and characteristics of isolates. J Clin Microbiol. 1998;36:3641–3646. doi: 10.1128/jcm.36.12.3641-3646.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, Nietfeldt J, Benson A K. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc Natl Acad Sci USA. 1999;96:13288–13293. doi: 10.1073/pnas.96.23.13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maas R. An improved colony hybridization method with significantly increased sensitivity for detection of single genes. Plasmid. 1983;10:296–298. doi: 10.1016/0147-619x(83)90045-8. [DOI] [PubMed] [Google Scholar]
- 26.Martin I E, Tyler S D, Tyler K D, Khakhira R, Johnson W M. Evaluation of ribotyping as epidemiologic tool for typing Escherichia coli serogroup O157 isolates. J Clin Microbiol. 1996;34:720–723. doi: 10.1128/jcm.34.3.720-723.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oelschlaeger T A, Hacker J. Verocytotoxigenic E. coli in Europe. Liège, Belgium: The National Food Centre; 1999. Pathogenicity islands and their role in virulence of bacteria; pp. 73–85. [Google Scholar]
- 28.Picard-Pasquier N, Ouagued M, Picard B, Goullet P, Krishnamoorthy R. A simple, sensitive method of analyzing bacterial ribosomal DNA polymorphism. Electrophoresis. 1989;10:186–189. doi: 10.1002/elps.1150100306. [DOI] [PubMed] [Google Scholar]
- 29.Pierard D, Muyldermans G, Moriau L, Stevens D, Lauwers S. Identification of new verocytotxin type 2 variant B-subunit genes in human and animal Escherichia coli isolates. J Clin Microbiol. 1998;36:3317–3322. doi: 10.1128/jcm.36.11.3317-3322.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pradel N, Livrelli V, Champs C D, Palcoux J-B, Reynaud A, Scheutz F, Sirot J, Joly B, Forestier C. Prevalence and characterization of Shiga toxin-producing Escherichia coli isolated from cattle, food, and children during a one-year prospective study in France. J Clin Microbiol. 2000;38:1023–1031. doi: 10.1128/jcm.38.3.1023-1031.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Preston M A, Johnson W, Khakhria R, Borczyk A. Epidemiologic subtyping of Escherichia coli serogroup O157 strains isolated in Ontario by phage typing and pulsed-field gel electrophoresis. J Clin Microbiol. 2000;38:2366–2368. doi: 10.1128/jcm.38.6.2366-2368.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rios M, Prado V, Trucksis M, Arellano C, Borie C. Clonal diversity of Chilean isolates of enterohemorrhagic Escherichia coli from patients with hemolytic-uremic syndrome, asymptomatic subjects, animal reservoirs, and food products. J Clin Microbiol. 1999;37:778–781. doi: 10.1128/jcm.37.3.778-781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samadpour M. Molecular epidemiology of Escherichia coli O157:H7 by restriction fragment length polymorphism using Shiga-like toxin genes. J Clin Microbiol. 1995;33:2150–2154. doi: 10.1128/jcm.33.8.2150-2154.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt H, Beutin L, Karch H. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect Immun. 1995;63:1055–1061. doi: 10.1128/iai.63.3.1055-1061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt H, Geitz C, Tarr P I, Frosch M, Karch H. Non-O157:H7 pathogenic Shiga toxin-producing Escherichia coli: phenotypic and genetic profiling of virulence traits and evidence for clonality. J Infect Dis. 1999;179:115–123. doi: 10.1086/314537. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt H, Köhler B, Unkmeir A, Bielaszewska M, Karch H. Verocytotoxigenic E. coli in Europe. Liège, Belgium: The National Food Centre; 1999. The role of Stx-encoding bacteriophages in pathogenicity and virulence of Shiga toxin-producing Escherichia coli (STEC) pp. 11–17. [Google Scholar]
- 37.Schmidt H, Scheef J, Morabito S, Caprioli A, Wieler L H, Karch H. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Appl Environ Microbiol. 2000;66:1205–1208. doi: 10.1128/aem.66.3.1205-1208.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tyler S D, Johnson W M, Lior H, Wang G, Rozee K R. Identification of verotoxin type 2 variant subunit B genes in Escherichia coli by the polymerase chain reaction and restriction fragment length polymorphism analysis. J Clin Microbiol. 1991;29:1339–1343. doi: 10.1128/jcm.29.7.1339-1343.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Unkmeir A, Schmidt H. Structural analysis of phage-borne stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect Immun. 2000;68:4856–4864. doi: 10.1128/iai.68.9.4856-4864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu J, Kaper J B. Cloning and characterization of the eae gene of enterohemorrhagic Escherichia coli O157:H7. Mol Microbiol. 1992;6:411–417. doi: 10.1111/j.1365-2958.1992.tb01484.x. [DOI] [PubMed] [Google Scholar]
- 42.Zhang W-L, Bielaszewska M, Liesegang A, Tschäpe H, Schmidt H, Bitzan M, Karch H. Molecular characteristics and epidemiological significance of Shiga toxin-producing Escherichia coli O26 strains. J Clin Microbiol. 2000;38:2134–2140. doi: 10.1128/jcm.38.6.2134-2140.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]