Abstract
Plant-bacterial combinations can increase contaminant degradation in the rhizosphere, but the role played by indigenous root-associated bacteria during plant growth in contaminated soils is unclear. The purpose of this study was to determine if plants had the ability to selectively enhance the prevalence of endophytes containing pollutant catabolic genes in unrelated environments contaminated with different pollutants. At petroleum hydrocarbon contaminated sites, two genes encoding hydrocarbon degradation, alkane monooxygenase (alkB) and naphthalene dioxygenase (ndoB), were two and four times more prevalent in bacteria extracted from the root interior (endophytic) than from the bulk soil and sediment, respectively. In field sites contaminated with nitroaromatics, two genes encoding nitrotoluene degradation, 2-nitrotoluene reductase (ntdAa) and nitrotoluene monooxygenase (ntnM), were 7 to 14 times more prevalent in endophytic bacteria. The addition of petroleum to sediment doubled the prevalence of ndoB-positive endophytes in Scirpus pungens, indicating that the numbers of endophytes containing catabolic genotypes were dependent on the presence and concentration of contaminants. Similarly, the numbers of alkB- or ndoB-positive endophytes in Festuca arundinacea were correlated with the concentration of creosote in the soil but not with the numbers of alkB- or ndoB-positive bacteria in the bulk soil. Our results indicate that the enrichment of catabolic genotypes in the root interior is both plant and contaminant dependent.
The potential to use plants to remediate polluted soils has recently attracted considerable interest (19, 28, 29, 34). Plants can stimulate contaminant disappearance by accumulation and transformation (30), by extracellular transformation (13, 37) and by stimulating microbial degradative activity in the rhizosphere (3, 35). Several authors have investigated the role of microorganisms in phytoremediation, and they have found that certain plant-bacterial associations can increase degradation (1, 8, 11, 33). This suggests that, under certain circumstances, such as in microbially inoculated plants, microorganisms play an important role in phytoremediation systems, but it is not clear what role they play in phytoremediation systems having only indigenous microbial populations.
Plants routinely encounter allelopathic compounds, many of which are analogous to organic contaminants (9), and thus there may be a plant response that stimulates microbial defenses against a soil toxicant or toxin (48). If this is true, plants could recruit bacteria that contain genotypes specific for toxicant degradation into the rhizosphere and root interior, and this selection should be contaminant specific. These bacteria would presumably protect the plant from the phytotoxic effects of the contaminants. In a similar manner, plants growing in herbicide-contaminated soils have high herbicide mineralization activity in their rhizospheres (2). Inoculating herbicide-degrading bacteria into the rhizosphere also protects plants from the phytotoxic effects of the herbicides (17, 32).
The selective pressure of plants on bacterial populations has its maximum effect near the root surface or in the root interior (16, 21, 23). For example, the maximum impact of altering plant genotypes was observed in the root interior of canola with substantially less impact observed on the rhizosphere microbial community (36, 40). Endophytic bacteria enhance the ability of plants to resist pathogens (49), herbivores (10), and other plants (44). Thus, if bacteria do play a role in the plant's ability to tolerate contaminants, we hypothesized that this effect would occur in the endophytic zone.
In this study, gene probes for alkane monooxygenase (alkB) (47), naphthalene dioxygenase (ndoB) (41), and catechol-2,3-dioxygenase (xylE) (24) were used to assess the prevalence of bacteria involved in petroleum hydrocarbon degradation. Gene probes developed for the ntdAa and the ntnM genes (38) were used to test for the prevalence of bacteria involved in nitrotoluene metabolism. The ntdAa gene of Pseudomonas sp. strain JS42 encodes the reductase component of the three-component dioxygenase system that converts 2-nitrotoluene to 3-methylcatechol (27). The ntnM gene from Pseudomonas putida encodes the hydroxylase component of nitrotoluene monooxygenase (18).
The purpose of this study was to assess the influence of plants on contaminant-degrading bacteria in upland terrestrial or freshwater intertidal environments. The effect of different contaminants, petroleum hydrocarbons, or nitroaromatics was also assessed. A wide range of environments and contaminants were purposely selected to investigate if plant selection of catabolic genotypes occurred as a general phenomenon or rather as a special case of a particular plant-soil-contaminant combination.
MATERIALS AND METHODS
Terrestrial petroleum site.
The terrestrial petroleum contaminated study site was located at the Port Hueneme, Calif., Department of Defense National Test Site. Each plot was approximately 30 by 50 m and was contaminated intermittently over a 20-year period with 1.5 g of total petroleum hydrocarbons per kg of soil originating from diesel fuel and heavy oil. Heavy metal concentrations were not significantly above background levels. There were three treatments with four replicate plots each of mixture 1, consisting of Bromus hordeaceous, Festuca arundinacea, Trifolium fragiferum, T. hirtum, and Vulpia microstachys; mixture 2, consisting of B. carinatus, Elymus glaucus, F. ruba, Hordeum californicum, Leymus triticoides, and Nassella pulchra; and an unvegetated plot. These plant mixtures were selected for their suitability to the northern California climate. Mixture 1 contained two clovers, which may increase the nitrogen supply to the soil organisms, whereas mixture 2 only contained native grasses. Plants were seeded in August 1997. Both plant mixtures were sampled at the petroleum site in September, January, and April 1998 and in August 1999 for a total of 16 replicates. The culturable and nonculturable community was analyzed for the F. arundinacea and T. fragiferum treatments and for a composite sample of mixture 2. The soil had a pH of 7.2 and an organic matter content of 2.1% and was composed of 59% sand, 26% silt, and 15% clay. The site was irrigated as needed and fertilized with 11.3 kg of urea and 11.3 kg of diammonium phosphate every other month.
Terrestrial nitroaromatic site.
The nitroaromatic site is a former 2,4,6-trinitrotoluene (TNT) manufacturing facility near Montreal, Quebec, Canada. Soil from which plants were sampled was contaminated with approximately 390 mg of TNT, 12 mg of 2,4-dinitrotoluene, 12 mg of 2,6-dinitrotoluene, 0.38 mg of 2-nitrotoluene, and 1.39 mg of 1,3,5-trinitrobenzene per kg. Production on site was terminated in 1972 and has not been in use since this time. The site was naturally vegetated by a variety of indigenous grasses since production stopped. The soil was a clay-loam with a pH ranging between 6.8 and 7.8. In June, July, and August 1998, the nitroaromatic site was sampled at three replicate locations, with three replicates per site for a total of 27 replicates. This site was not irrigated or fertilized during this study.
Freshwater intertidal petroleum site.
Eight replicate plots (5 by 4 m) were constructed in an intertidal region on the St. Lawrence River, Quebec, Canada. Four plots were contaminated at 0.6 liters/m2 or 12 g kg of sediment−1 with weathered light crude oil in June 1999. Oil was raked into the top 5 cm of the sediment during low tide. Nitrogen and phosphorus as nutrients [NH4NO3 + Ca(H2PO4)2 · H2O] were added initially at a rate of 1,000 g of N and 300 g of P per plot. After 4 weeks, all fertilized plots received weekly applications of fertilizers. Bulk soil and endophytic samples were collected from the on-site natural vegetation (Scirpus pungens) 6 and 8 weeks after contamination for a total of eight replicates per treatment.
Growth chamber study.
Control garden soil was a sandy loam consisting of 70% sand, 7% clay, and 23% silt with a pH of 7.9, water-holding capacity of 76%, total organic C of 11.2%, and Kjeldahl N of 0.38% (14). The garden soil was mixed with soil obtained from an active wood treatment facility contaminated with 43 g of C10 to C50 aliphatic hydrocarbons kg of soil−1 and 11 g of polycyclic aromatic hydrocarbons kg of soil−1 at five different concentrations (0, 5, 10, 20, and 40% [wt/wt]). The soil from the wood treatment facility had aged, and there was intermittent fresh contamination as a result of spills that had occurred during treatment operations. Soil from the nitroaromatic site was mixed with uncontaminated garden soil for a final concentration of nitroaromatics of 340 mg of TNT, 0.4 mg of 2,4-dinitrotoluene, and 0.3 mg of 1,3,5-trinitrobenzene per kg. The contaminated soil was placed in 5-cm pots, F. arundinacea was planted, and pots were placed in a greenhouse with an average daytime temperature of 18°C, a night-time temperature of 5°C, a relative humidity of 40%, and a light intensity of 400 μmol s−1 m−2. Plants were thinned to five per pot 14 days after planting and harvested 6 weeks after planting.
Catabolic gene probe analysis of the cultured and noncultured communities.
Bacteria were extracted from the root interior, rhizosphere, and bulk soil as previously described (36) with the exception that bacteria were resuspended in 0.1% (wt/vol) tetrasodium pyrophosphate (pH 7.0). Aliquots (0.1-ml) of serial dilutions in tetrasodium pyrophosphate (10−2, 10−3, and 10−4) were spread plated onto triplicate plates containing 250 mg of yeast extract (Becton Dickinson, Cockeysville, Md.), 250 mg of tryptone (Difco Laboratories, Detroit, Mich.), 250 mg of soluble starch (Anachemia, Montreal, Quebec, Canada), and 15 g of granulated agar (Becton Dickinson) per liter of tap water. Bacterial colonies were counted and lifted onto nylon membranes after incubation at room temperature (21 to 24°C) for 2 weeks. Cells adhering to nylon membranes were lysed, and the DNA was denatured, fixed, and cross-linked to the membrane and hybridized to the alkB (17), ndoB (41), ntdAa (27), ntnM (18), or xylE (24) gene probes as previously described (12).
The total microbial community DNA was extracted from the soil and rhizosphere by chemical lysis (12), and purified on polyvinylpolypyrrolidone spin columns, (5), and 100 ng was dot blotted in triplicate on Zeta-Probe membranes (Bio-Rad Laboratories, Hercules, Calif.) (39). The concentration of total microbial community DNA applied to membranes was determined by agarose gel electrophoresis as follows. Total DNA (5 μl) was run on a 0.7% agarose gel with a 0.5, 1, and 2 μl of a 100-bp DNA ladder (MBI Fermentas, Inc., Burlington, Ontario, Canada) and quantified by ethidium bromide staining and spot densitometry using a ChemiImager (Alpha Innotech Corp., San Leandro, Calif.). Scintillation counting of cut dot blot membranes after overnight hybridization (65°C) with 32P-labeled oligonucleotide probes was performed with a Tri-Carb scintillation counter model 2100TR (Packard Instruments Co., Meriden, Conn.). Standard curves constructed with total genomic DNA of P. oleovorans for alkB, P. putida for ndoB, and Pseudomonas sp. JS42 for ntdAa were used to estimate the amount of bound probe. The response values, termed genome equivalents, for alkB, ndoB, and ntdA were expressed as nanograms of genomic DNA/100 ng of total community DNA.
Statistics and data transformation.
Typically, the average bacterial population density in the root interior is between 103 and 105 CFU g of root−1 (16) compared to population densities in soil of 107 and 108 CFU g of soil−1. Therefore, in order to compare populations between habitats, genotype prevalence is commonly expressed as the percent composition of the community present in the habitat of interest (21), which in this case is the root interior, rhizosphere, and bulk soil. The percent composition data were arcsine transformed to approximate the normal distribution, and a Skewness and Kurtosis analysis of the data indicated that the data were normally distributed (43). Canonical correlation and regression analysis were performed using Systat 10.
RESULTS
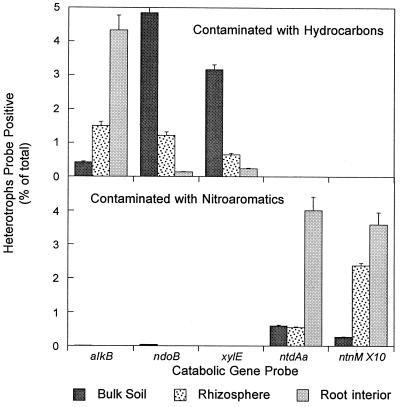
At the terrestrial petroleum hydrocarbon contaminated site, the alkB genotype was 10 times more prevalent in the culturable endophytic microbial community (4.3% of 21,182 colonies probed from 288 plates) compared to the bulk soil microbial community (0.42% of 25,700 colonies probed from 96 plates) (Fig. 1). Similarly, alkB was more prevalent (P < 0.05; Student's t test) in the total community DNA extracted from the rhizosphere, 5.6%, compared to that from the bulk soil, 3.9% (data not shown). In contrast, there were fewer ndoB-positive bacteria present in the root interior (0.13%) than in the bulk soil (4.8%), and no significant difference was seen in ndoB prevalence in the total community DNA of the rhizosphere (5.1%) compared to the bulk soil (4.7%). The xylE results from the culturable community were similar to ndoB, with more xylE-positive bacteria detected in the bulk soil (3.2%) than in the root interior (0.23%). The gene associated with mononitroaromatic metabolism, ntdAa, was not detected at the petroleum-contaminated site.
FIG. 1.
Relative prevalence of catabolic genotypes in the bulk soil, rhizosphere, and root interior at two contaminated field sites. Values for the nitroaromatic site are the average of 27 replicates taken over the course of the summer of 1998. The values for the petroleum-contaminated site are the average of 32 replicates taken in 1998 and 1999. The error bars show the standard error of the mean.
At the nitroaromatic site, ntdAa-positive bacteria were more prevalent in the root interior (4% of 14,000 colonies probed or 6.2 × 104 positive CFU g of root−1) than in the bulk soil (0.6% of 21,000 colonies probed or 1.7 × 106 positive CFU g of soil−1) (Fig. 1). Similarly, the ntdAa gene was six times more prevalent in the total community DNA extracted from the rhizosphere, 0.24%, compared to the bulk soil, 0.04% (data not shown). The ntnM genotype was also more prevalent inside the root, with ntnM-positive bacteria 14-fold more prevalent in the root interior (0.35%) than in the bulk soil (0.03%). There were no endophytic bacteria positive for alkB or ndoB, and very few alkB (0.01%)- or ndoB (0.03%)-positive bacteria in the bulk soil microbial communities of the nitroaromatic site.
The increase in catabolic genotypes in the rhizosphere and root interior was dependent on the catabolic genotype being investigated as well as the plant treatment (Table 1). All plant treatments had an increased prevalence of alkB-positive bacteria in their root interior with little difference seen between plant treatments. However, in the rhizosphere of Rose Clover or mixture 2, alkB bacteria were more prevalent compared to the bulk soil and levels were greater than observed in the rhizosphere of the Tall Fescue. In contrast, ndoB prevalence displayed an opposite trend. The prevalence of ndoB-positive endophytes was lower compared to the rhizosphere in all three plant treatments, and ndoB-positive bacteria in the root interior of Rose Clover and mixture 2 were significantly less prevalent compared to Tall Fescue. Only Tall Fescue increased the prevalence of ndoB-positive bacteria in its rhizosphere compared to bulk soil. Bacteria positive for xylE were not enhanced in the rhizosphere or root interior by any plant treatments.
TABLE 1.
Percent total heterotrophs isolated from the root interior or the rhizosphere that were positive for selected catabolic gene probes in three different treatments of a petroleum hydrocarbon phytoremediation study
| Plant species | % Total heterotrophs (SD)a
|
|||||
|---|---|---|---|---|---|---|
|
alkB
|
ndoB
|
xylE
|
||||
| Root interior | Rhizosphere | Root interior | Rhizosphere | Root interior | Rhizosphere | |
| Tall Fescue | 2.69 (0.1674) | 1.38 (0.1349) | 0.285 (0.05515) | 7.93 (0.3125) | 0.233 (0.04988) | 0.140 (0.04324) |
| Rose clover | 3.54 (0.1912) | 2.52 (0.1813) | 0.101 (0.03286) | 1.69 (0.1491) | 0.242 (0.05083) | 0.440 (0.07655) |
| Composite of mixture 2 | 3.64 (0.1937) | 2.21 (0.1700) | 0.038 (0.02016) | 2.17 (0.1685) | 0.086 (0.0303) | 0.000 |
The standard deviation of the binomial distribution is presented in parentheses. The numbers of colonies assessed for gene presence in the root interior and rhizosphere were 9,345 and 7,476, respectively.
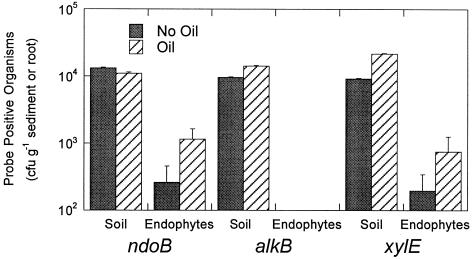
The enrichment of catabolic gene-containing bacteria in the root interior of S. pungens at the freshwater intertidal site was dependent on the presence of contaminants. When plants were exposed to crude oil, the numbers of ndoB- or xylE-positive bacterial endophytes increased (P < 0.05) by an order of magnitude (Fig. 2). There was not a corresponding increase in ndoB-positive organisms in the bulk sediment. Total endophytic or soil heterotrophs were not significantly affected by hydrocarbon contamination with little difference observed between treatments. The numbers of xylE-positive bacteria in the bulk sediment increased significantly, which may explain the corresponding increase in xylE probe-positive organisms in the root interior.
FIG. 2.
Numbers of probe-positive bulk sediment bacteria or endophytes present in field-grown S. pungens. Eight 5-by-4-m plots containing S. pungens were established in the intertidal zone, and oil was applied to four of these plots in June 1999. At 6 and 8 weeks after the oil application, the genotype prevalence in the root interior and bulk sediment was assessed. Each bar shows the average of eight replicates. The error bars show the standard error of the mean.
Interestingly, no alkB-positive endophytes were found, and the presence of oil did not increase the prevalence of alkB-positive bacteria in the bulk sediment. This may be the result of using an alkB DNA probe derived from the terrestrial bacterium P. oleovorans (47) on bacteria extracted from a freshwater sediment environment. Smits and colleagues (42) demonstrated a wide divergence in DNA sequences of alkane monooxygenase genes among microorganisms.
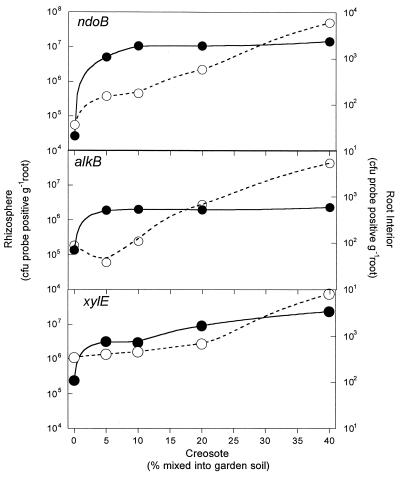
In the growth chamber experiment, the enrichment of alkB- or ndoB-positive endophytic bacteria in F. arundinacea was correlated to the concentration of contaminant in the soil. The numbers of endophytic bacteria that were positive for alkB or ndoB were proportional (r2 = 0.988, P < 0.001 for alkB; r2 = 0.967, P < 0.001 for ndoB) to the soil creosote concentration and not to the prevalence of alkB- or ndoB-positive bacteria in the rhizosphere (Fig. 3). In contrast, the prevalence of rhizosphere bacteria positive for alkB or ndoB was not correlated (r2 = 0.111, P = 0.308 for alkB; r2 = 0.189, P = 0.259 for ndoB) with increasing soil creosote concentrations. The numbers of xylE-positive bacteria increased in the rhizosphere (r2 = 0.673, P = 0.052), as well as in the root interior (r2 = 0.891, P < 0.001). The numbers of endophytic (ca. 4 × 106 CFU g of root−1) or rhizosphere (ca. 3 × 109 CFU g of soil−1) heterotrophs did not differ with increasing creosote concentrations (data not shown). The prevalence of the ndoB genes in the culturable community of the root interior increased 20 times from 0.01% in plants grown with no creosote to 0.2% of the total heterotrophic community.
FIG. 3.
Numbers of probe-positive rhizosphere bacteria (●) or endophytes (○) present in F. arundinacea in response to increasing creosote concentrations in soil. F. arundinacea was planted in garden soil mixed with five concentrations (0, 5, 10, 20, and 40%) of soil heavily contaminated with creosote (43 g C10 to C50 aliphatic hydrocarbons and 11 g of polycyclic aromatic hydrocarbons kg of soil−1). Plants were grown in replicate pots (n = 5) for 7 weeks in a greenhouse, and the genotype prevalence in the bulk soil and root interior was determined. Each point is the average of five replicates. The error bars are obscured by the symbols.
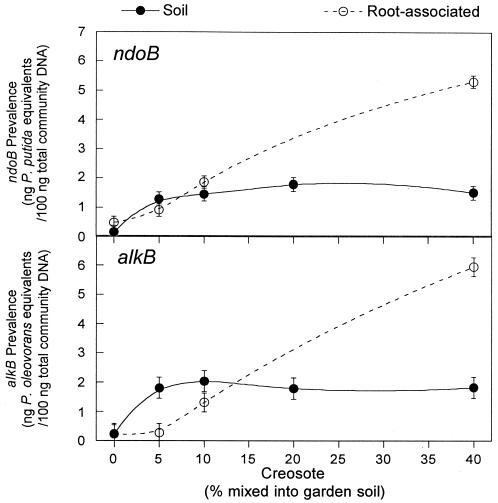
Similar trends were seen in the total community DNA extracted from the root system (Fig. 4), with alkB or ndoB increasing in prevalence as the creosote concentrations in soil increased. For example, the concentration of alkB or ndoB genomic equivalents in the total community DNA extracted from the roots of plants grown in the 40% creosote soil increased by 25 or 10 times, respectively, over plants grown in control soil in a concentration-dependent manner (r2 = 0.994, P < 0.001 for alkB; r2 = 0.998, P < 0.001 for ndoB). No bacteria positive for the ntdAa genotype were found in the creosote-contaminated soils (data not shown).
FIG. 4.
Prevalence of ndoB or alkB genotypes in total community DNA extracted from soil or from the roots of F. arundinacea. Each point is the average of five replicates. The results for root-associated DNA at 20% creosote were lost and not reported. The error bars show the standard error of the mean.
When F. arundinacea was grown in the presence of nitroaromatics, there was no enrichment for ntdAa-positive endophytes, with 0.3% (1,660 colonies probed or 1.3 × 105 positive CFU g of soil−1) of the bulk soil population positive for ntdAa and only 0.05% (1,224 colonies probed or 8.7 × 101 CFU g of root−1) of the endophyte population (data not shown). The numbers of alkB- or ndoB-positive endophytes did not increase in the presence of nitroaromatics in bulk soil. In contrast, the numbers of xylE-positive endophytic bacteria increased from 339 CFU g of root−1 in control plants to 1,040 CFU g of root−1 in plants grown in the presence of nitroaromatics.
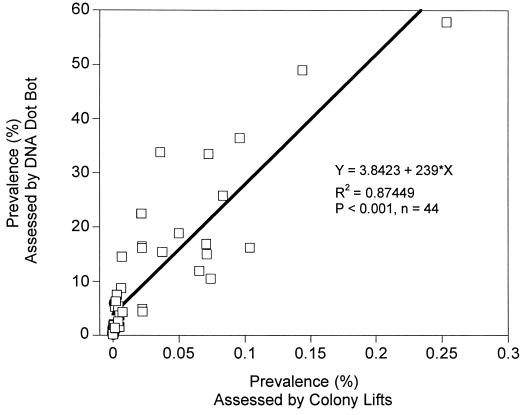
The results of colony lifts obtained using the ndoB, alkB, and ntdAa gene probes for both soil and root interior samples were compared with values obtained by dot blot analysis of the total community DNA. Catabolic gene probe results obtained with colony lifts were well correlated (r2 = 0.875, P < 0.001, n = 44) with results obtained from dot blots (Fig. 5), and a strong linear relationship was observed between the two parameters.
FIG. 5.
Correlation between assessments (n = 44) of the prevalence of ndoB, alkB, and ntdAa by culture-dependent and culture-independent methods in soil and root interior environments. Each point represents a sample analyzed by culture-independent and -dependent processes for the same catabolic gene probe.
DISCUSSION
Our results indicate that certain bacterial catabolic genotypes (alkB, ndoB, ntdAa, and ntnM) were enriched in the interior of plant roots in response to pollution in a contaminant-dependent manner. In contrast, xylE increased in response to all classes of contaminants. Enrichment of specific catabolic genotypes was also dependent on plant species in combination with environmental parameters. For example, S. pungens exposed to oil enriched ndoB but not alkB, whereas the phytoremediation mixture enriched alkB but not ndoB. Similarly, F. arundinacea exposed to nitroaromatics did not increase the prevalence of ntdAa in the root interior, but the grass community at a nitroaromatic-contaminated site had increased prevalence of ntdAa in the root interior. An increase in contaminant-degrading bacteria in the rhizosphere of plants exposed to petroleum hydrocarbons was observed previously (26). Our results extend this observation to other contaminants and indicate that the enrichment is dependent on the type and amount of contaminant present in soil. At terrestrial upland sites, endophytic bacteria only contained the petroleum hydrocarbon-degrading genes, alkB or ndoB, when petroleum hydrocarbons were present in the surrounding soil and/or sediment. Similarly, genes encoding nitrotoluene degradation were only identified when nitrotoluenes were present in the environment. Furthermore, the numbers of endophytic catabolic genotypes were not related to the presence of bacteria containing catabolic genotypes in the surrounding soil but rather with contaminant concentrations in soil.
The increase in endophytic catabolic genotypes may be a plant-dependent or -independent process. The plant may be selectively enriching catabolic genotypes in its root interior via some as-yet-unknown plant process. Roots are known to exert a selective influence on the root-associated bacterial communities (15, 20, 25, 50), and the influence of roots is at least partly plant specific (36). Alternatively, the numbers of endophytic catabolic genotypes may increase due to contaminant flux through the root system. Both explanations account for the contaminant-dependent increase in endophytic catabolic genotypes. It is likely that the enrichment is plant mediated because this explanation accounts for the differences observed between plant species and the dependence of catabolic gene prevalence in the root interior on contaminant levels. For example, alkB-positive endophytes of F. arundinacea increased in proportion to the amount of creosote present in soil, but when this grass was exposed to nitroaromatics no enrichment was detected. In contrast, grasses revegetating a nitroaromatic contaminated terrestrial site contained significant levels of nitroaromatics in their root interior. Thus, if F. arundinacea did not influence the enrichment process, its roots would need to be permeable to alkane hydrocarbons but not nitrotoluenes. Typically, nitrotoluenes are readily absorbed by grasses, so this is probably not the case (30, 31, 45).
The enrichment of contaminant-degrading bacteria in the root interior is at least partially plant species specific. F. arundinacea did not promote an increase in ntdAa-positive endophytic bacteria despite being exposed to nitroaromatic concentrations comparable to that seen at a contaminated field site. For example, there were 6.2 × 104 ntdAa-positive endophytes g of root−1 in plants growing at the contaminated field site but only 8.7 × 101 ntdAa endophyte CFU g of root−1 in F. arundinacea grown in the growth chamber experiment. This plant species is not exceptionally tolerant of nitroaromatics and is used for the phytoremediation of petroleum hydrocarbon-contaminated sites rather than nitroaromatic contaminated sites (38). Similarly, at the petroleum-contaminated site, F. arundinacea had a significantly greater prevalence of ndoB in its root interior compared to T. fragiferum, whereas T. fragiferum had more alkB endophytes than F. arundinacea. In addition to the effects of specific plants, environment plays an important role in altering endophytic community composition. S. pungens, grown inside an intertidal zone, promoted ndoB prevalence inside the root, whereas grass mixtures grown in a terrestrial upland site promoted alkB. It is difficult to distinguish plant effects from those of the environment. The composition of the endophytic microflora is known to be dependent on both plant and soil type (15, 20, 25, 50). There is likely a similar interaction between contaminant, soil, and plant species that determines the level and identity of catabolic genes enriched.
Phytoremediation systems that use microbial inoculants are typically more effective than noninoculated plant systems (2, 8, 11, 33). This work examined one of the mechanisms, i.e., root interior colonization, that may be occurring to explain the increased success of plant-bacterial associations in degrading contaminants. In a similar manner, plant growth-promoting activity has been linked to colonization of the root interior (4, 46). Previous research has correlated the success of a plant-bacterial association to remediate nitroaromatics, in which the ntdAa genotype prevalence in the rhizosphere was promoted (38), but gene prevalence in endophytes was not investigated.
Our results indicated that culture-independent methods of detecting catabolic gene prevalence were more sensitive compared to culture-dependent methods. Gene probes have consistently been shown to be an effective method of monitoring gene prevalence in the environment (6) but do not estimate the in situ activity of these genes. Further, gene probes may underestimate actual prevalence of contaminant-degrading bacteria. Gene probes such as ndoB detect less than 50% of bacteria capable of polycyclic aromatic hydrocarbon metabolism (22). Similarly, sequence divergence between petroleum hydrocarbon genes (7, 42) implies that catabolic gene probes will underestimate the prevalence of bacteria capable of contaminant degradation. More work is required to assess gene probe efficacy in different environmental compartments.
The results from this study indicate that the enrichment of catabolic genotypes in the root interior occurs in different plants in a variety of environments and in response to different contaminants. The enrichment is dependent on plant species and the contaminant in which the plant is growing. The process by which this occurs is not yet known and is the subject of current investigations. The flexible and specific nature of the selective pressure observed here suggests that the mechanism by which plants control the composition of the endophytic microbial community is much more dynamic and responsive than previously believed. The impact of this dynamic response on plant survival at contaminated sites has yet to be determined.
REFERENCES
- 1.Alvey S, Crowley D E. Survival and activity of an atrazine-mineralizing bacterial consortium in rhizosphere soil. Environ Sci Technol. 1996;30:1596–1603. [Google Scholar]
- 2.Anderson T A, Kruger E L, Coats J R. Enhanced degradation of a mixture of three herbicides in the rhizosphere of a herbicide-tolerant plant. Chemosphere. 1994;28:1551–1557. [Google Scholar]
- 3.Banks M K, Lee E, Schwab A P. Evaluation of dissipation mechanisms for benzo [a]pyrene in the rhizosphere of tall fescue. J Environ Qual. 1999;28:294–298. [Google Scholar]
- 4.Bent E, Chanway C P. The growth-promoting effects of a bacterial endophyte on lodgepole pine are partially inhibited by the presence of other rhizobacteria. Can J Microbiol. 1998;44:980–988. [Google Scholar]
- 5.Berthelet M, Whyte L G, Greer C W. Rapid, direct extraction of DNA from soils for PCR analysis using polyvinylpolypyrrolidone spin columns. FEMS Microbiol Lett. 1996;138:17–22. doi: 10.1111/j.1574-6968.1996.tb08128.x. [DOI] [PubMed] [Google Scholar]
- 6.Brockman F J. Nucleic-acid-based methods for monitoring the performance of in situ bioremediation. Mol Ecol. 1995;4:567–578. [Google Scholar]
- 7.Churchill S A, Harper J P, Churchill P F. Isolation and characterization of a Mycobacterium species capable of degrading three- and four-ring aromatic and aliphatic hydrocarbons. Appl Environ Microbiol. 1999;65:549–552. doi: 10.1128/aem.65.2.549-552.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crowley D E, Brennerova M V, Irwin C, Brenner V, Focht D D. Rhizosphere effects on biodegradation of 2,5-dichlorobenzoate by a bioluminescent strain of root-colonizing Pseudomonas fluorescens. FEMS Microbiol Ecol. 1996;20:79–89. [Google Scholar]
- 9.Donnelly P K, Hegde R S, Fletcher J S. Growth of PCB-degrading bacteria on compounds from photosynthetic plants. Chemosphere. 1994;28:981–988. [Google Scholar]
- 10.Elliot S L, Savelis M W, Janssen A, van der Geest L P S, Beerling E A M, Fransen J. Can plants use entomopathogens as bodyguards. Ecol Lett. 2000;3:228–235. [Google Scholar]
- 11.Ellis R J, Thompson I P, Bailey M J. Metabolic profiling as a means of characterizing plant-associated microbial communities. FEMS Microbiol Ecol. 1995;16:9–18. [Google Scholar]
- 12.Fortin N, Fulthorpe R R, Allen D G, Greer C W. Molecular analysis of bacterial isolates and total community DNA from kraft pulp mill effluent treatment systems. Can J Microbiol. 1998;44:537–546. [PubMed] [Google Scholar]
- 13.Garcia C, Roldan A, Hernandez T. Changes in microbial activity after abandonment of cultivation in a semi-arid Mediterranean environment. J Environ Qual. 1997;26:285–291. [Google Scholar]
- 14.Gong P, Siciliano S D, Greer C W, Paquet L, Hawari J, Sunahara G I. Effects and bioavailability of 2,4,6-trinitrotoluene in spiked and field-contaminated soils to indigenous microorganisms. Environ Toxicol Chem. 1999;18:2681–2688. [Google Scholar]
- 15.Grayston S J, Wang S, Campbell C D, Edwards A C. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol Biochem. 1998;30:369–378. [Google Scholar]
- 16.Hallmann J, Quadt-Hallmann A, Mahaffee W F, Kloepper J W. Bacterial endophytes in agricultural crops. Can J Microbiol. 1997;43:895–914. [Google Scholar]
- 17.Hsu T-S, Bartha R. Accelerated mineralization of two organophosphate insecticides in the rhizosphere. Appl Environ Microbiol. 1979;37:36–41. doi: 10.1128/aem.37.1.36-41.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James K D, Williams P A. Ntn genes determining the early steps in the divergent catabolism of 4-nitrotoluene and toluene in Pseudomonas sp. strain TW3. J Bacteriol. 1998;180:2043–2049. doi: 10.1128/jb.180.8.2043-2049.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kreslavski V D, Vasilyeva G K, Comfort S D, Drijber R A, Shea P J. Accelerated transformation and binding of 2,4,6-trinitrotoluene in rhizosphere soil. Bioremediation. 1999;3:59–67. [Google Scholar]
- 20.Lemanceau P, Corberand T, Gardan L, Latour X, Laguerre G, Boeufgras J-M, Alabouvette C. Effect of two plant species Flax (Linum usitatissinum L.) and tomato (Lycopersicon esculentum Mill.), on the diversity of soilborne populations of fluorescent pseudomonads. Appl Environ Microbiol. 1995;61:1004–1012. doi: 10.1128/aem.61.3.1004-1012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lilley A K, Fry J C, Bailey M J, Day M J. Comparison of aerobic heterotrophic taxa isolated from four root domains of mature sugar beet (Beta vulgaris) FEMS Microbiol Ecol. 1996;21:231–242. [Google Scholar]
- 22.Lloyd-Jones G, Laurie A D, Hunter D W F, Fraser R. Analysis of catabolic genes for naphthalene and phenanthrene degradation in contaminated New Zealand soils. FEMS Microbiol Ecol. 1999;29:69–79. [Google Scholar]
- 23.Marilley L, Aragno M. Phylogenetic diversity of bacterial communities differing in degree of proximity of Lolium perenne and Trifolium repens roots. Appl Soil Ecol. 1999;13:127–136. [Google Scholar]
- 24.Nakai C, Kagamiyama H, Nozaki M, Nakazawa T, Inouye S, Ebina Y, Nakazawa A. Complete nucleotide sequence of the metapyrocatechase gene on the TOI plasmid of Pseudomonas putida mt-2. J Biol Chem. 1983;258:2923–2928. [PubMed] [Google Scholar]
- 25.Nehl D B, Allen S J, Brown J F. Deleterious rhizosphere bacteria: an integrating perspective. Appl Soil Ecol. 1997;5:1–20. [Google Scholar]
- 26.Nichols T D, Wolf D C, Rogers H B, Beyrouty C A, Reynolds C M. Rhizosphere microbial populations in contaminated soils. Water Air Soil Pollut. 1997;95:165–178. [Google Scholar]
- 27.Parales J W, Kumar A, Parales R E, Gibson D T. Cloning and sequencing of the genes encoding 2-nitrotoluence dioxygenase from Pseudomonas sp. JS42. Gene. 1996;181:57–61. doi: 10.1016/s0378-1119(96)00462-3. [DOI] [PubMed] [Google Scholar]
- 28.Pradhan S P, Conrad J R, Paterek J R, Srivastava V J. Potential of phytoremediation for treatment of PAHs in soil at MGP sites. J Soil Cont. 1998;7:467–480. [Google Scholar]
- 29.Salt D E, Smith R D, Raskin I. Phytoremediation. Annul Rev Plant Physiol Plant Mol Biol. 1999;49:643–668. doi: 10.1146/annurev.arplant.49.1.643. [DOI] [PubMed] [Google Scholar]
- 30.Schneider K, Oltmanns J, Radenberg T, Schneider T, Pauly-Mundegar D. Uptake of nitroaromatic compounds in plants. Environ Sci Pollut Res. 1996;3:135–138. doi: 10.1007/BF02985519. [DOI] [PubMed] [Google Scholar]
- 31.Sens C, Scheidemann P, Werner D. The distribution of 14C-TNT in different biochemical compartments of the monocotyledonous Triticum aestivum. Environ Pollut. 1999;104:113–119. [Google Scholar]
- 32.Shann J R. The role of plants and plant/microbial systems in the reduction of exposure. Environ Health Persp Suppl. 1995;103:13–15. doi: 10.1289/ehp.95103s413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siciliano S D, Germida J J. Degradation of chlorinated benzoic acid mixtures by plant-bacteria associations. Environ Toxicol Chem. 1998;17:728–733. [Google Scholar]
- 34.Siciliano S D, Germida J J. Mechanisms of phytoremediation: biochemical and ecological interactions between plants and bacteria. Environ Rev. 1998;6:65–79. [Google Scholar]
- 35.Siciliano S D, Germida J J. Enhanced phytoremediation of chlorobenzoates in rhizosphere soil. Soil Biol Biochem. 1999;31:299–305. [Google Scholar]
- 36.Siciliano S D, Germida J J. Taxonomic diversity of bacteria associated with the roots of field grown transgenic Brassica napus cv. Quest, compared to the non-transgenic B. napus cv. Excel and B. rapa cv. Parkland FEMS Microbiol Ecol. 1999;29:263–272. [Google Scholar]
- 37.Siciliano S D, Goldie H, Germida J J. Enzymatic activity in root exudates of Dahurian wild rye (Elymus dauricus) that degrades 2-chlorobenzoic acid. J Agri Food Chem. 1998;46:5–7. doi: 10.1021/jf9708195. [DOI] [PubMed] [Google Scholar]
- 38.Siciliano S D, Greer C W. Plant-bacterial combinations to phytoremediate soil contaminated with high concentrations of 2,4,6-trinitrotoluene. J Environ Qual. 2000;29:311–316. [Google Scholar]
- 39.Siciliano S D, Roy R, Greer C W. Reduction in denitrification activity in field soils exposed to long term contamination by 2,4,6-trinitrotoluene (TNT) FEMS Microbiol Ecol. 2000;32:61–68. doi: 10.1111/j.1574-6941.2000.tb00699.x. [DOI] [PubMed] [Google Scholar]
- 40.Siciliano S D, Theoret C M, de Freitas J R, Huel P J, Germida J J. Differences in the microbial communities associated with the roots of different cultivars of canola and wheat. Can J Microbiol. 1998;44:844–851. [Google Scholar]
- 41.Simon M J, Osslund T D, Saunders R, Ensley B D, Suggs S, Harcourt A, Suen W, Cruden D L, Givson D T, Zylstra G L. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB 9816–4. Gene. 1993;127:31–37. doi: 10.1016/0378-1119(93)90613-8. [DOI] [PubMed] [Google Scholar]
- 42.Smits T H M, Röthlisberger M, Witholt B, van Bellen J B. Molecular screening for alkane hydroxylase genes in Gram-negative and Gram-positive strains. Environ Microbiol. 1999;1:307–317. doi: 10.1046/j.1462-2920.1999.00037.x. [DOI] [PubMed] [Google Scholar]
- 43.Sokal R R, Rohlf F J. Biometry: the principles and practice of statistics in biological research. W. H. New York, N.Y: Freeman and Co.; 1995. [Google Scholar]
- 44.Sturz A V, Christie B R, Matheson B G. Associations of bacterial endophyte populations from red clover and potato crops with potential for beneficial allelopathy. Can J Microbiol. 1998;44:162–167. [Google Scholar]
- 45.Sun W-H, Horst G L, Drijber R A, Elthon T E. Fate of 2,4,6-trinitrotoleune in axenic sand culture systems containing smooth bromegrass. Environ Toxicol Chem. 2000;19:2038–2046. [Google Scholar]
- 46.Toxler J, Zala M, Natsch A, Moene-Loccoz Y, Défago G. Autecology of the biocontrol strain Pseudomonas fluorescens CHA0 in the rhizosphere and inside roots at later stages of plant development. FEMS Microbiol Ecol. 1997;23:119–130. [Google Scholar]
- 47.van Beilen J B, Wubbolts M G, Witholt B. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation. 1994;5:161–174. doi: 10.1007/BF00696457. [DOI] [PubMed] [Google Scholar]
- 48.Walton B T, Hoylman A M, Perez M M, Anderson T A, Johnson T R, Guthrie E A, Christman R F. Rhizosphere microbial communities as a plant defense against toxic substances in soils. In: Anderson T A, Coats J R, editors. Bioremediation through rhizosphere technology. ACS Symposium Series 563. Washington, D.C.: American Chemical Society; 1994. pp. 82–92. [Google Scholar]
- 49.Wei G, Kloepper J W, Tuzun S. Induced systemic resistance to cucumber diseases and increased plant growth by plant growth-promoting rhizobacteria under field conditions. Phytopathology. 1996;86:221–224. [Google Scholar]
- 50.Westover K M, Kennedy A C, Kelleys S E. Patterns of rhizosphere microbial community structure associated with co-occurring plant species. J Ecol. 1997;85:863–873. [Google Scholar]