Abstract
A random transposon-based mutagenesis system was optimized for the purple sulfur phototrophic bacterium Thiocapsa roseopersicina BBS. Screening for hydrogenase-deficient phenotypes resulted in the isolation of six independent mutants in a mini-Tn5 library. One of the mutations was in a gene showing high amino acid sequence similarity to HypF proteins in other organisms. Inactivation of hydrogen uptake activity in the hypF-deficient mutant resulted in a dramatic increase in the hydrogen evolution capacity of T. roseopersicina under nitrogen-fixing conditions. This mutant is therefore a promising candidate for use in practical biohydrogen-producing systems. The reconstructed hypF gene was able to complement the hypF-deficient mutant of T. roseopersicina BBS. Heterologous complementation experiments, using hypF mutant strains of T. roseopersicina, Escherichia coli, and Ralstonia eutropha and various hypF genes, were performed. They were successful in all of the cases tested, although for E. coli, the regulatory region of the foreign gene had to be replaced in order to achieve partial complementation. RT-PCR data suggested that HypF has no effect on the transcriptional regulation of the structural genes of hydrogenases in this organism.
Hydrogenases catalyze the reversible oxidation of molecular H2. Two major classes are distinguished: Fe-only hydrogenases and [NiFe] hydrogenases (6). The latter is typically composed of an electron transfer small subunit and a catalytic large subunit. The formation of an active [NiFe] hydrogenase involves a complex maturation process. The major steps include biosynthesis of the unprocessed, inactive subunits, incorporation of Fe-S clusters into the small subunit, assembly of Ni, Fe, and the diatomic ligands (CN and CO) into the active center in the large subunit, and C-terminal cleavage of the large subunit by a specific protease (22). Accessory proteins are involved in the assembly of the active center, while other gene products participate in the regulation of biosynthesis and electron transport (8, 19, 20, 22). Several microorganisms harbor multiple, distinct hydrogenases (4, 28, 31, 32). In these cases each hydrogenase has a set of specific maturation proteins (e.g., protease). Beside the specific proteins, pleiotropic (Hyp) maturation proteins are also involved in hydrogenase biosynthesis. Without the action of these hyp gene products (primarily HypB, HypC, HypD, HypE, and HypF), the Ni, Fe, and diatomic ligands are not inserted into the active center and maturation stops. A notable exception is the chaperon-like HypC protein of Escherichia coli, which is mainly involved in the maturation of hydrogenase 3 (17, 21), and a homologous protein, HybG, is involved in the maturation of hydrogenases 1 and 2. The homologous protein is pleiotropic in Ralstonia eutropha (formerly Alcaligenes eutrophus) (12). It has been shown that the maturation of the H2 sensor regulatory hydrogenase, involved in the transcriptional regulation of some hydrogenases, is dependent on at least one of the pleiotropic proteins (HypF). In these cases, a mutation in hypF apparently represses the transcription of the structural genes via the blocked maturation of the regulatory hydrogenase (10, 33).
The purple sulfur photosynthetic bacterium Thiocapsa roseopersicina BBS has two sets of hydrogenase genes (hydS isp1 isp2 hydL and hupSLCDHIR), coding for structural, anchoring/electron transport, and some specific accessory and regulatory proteins (9, 18, 28). HupD, -H, and -I show homology to specific maturation proteins, while HupR has a sequence related to the response regulator of the two-component regulatory system (13). Other specific and/or pleiotropic accessory genes and components of the regulatory machinery have not been found in the vicinity of these structural gene clusters (9, 28). The Hyd hydrogenase is remarkably stable and it is expressed under both nitrogenase-repressed and derepressed conditions. The molecular architecture responsible for the stable hydrogenase function helps the rational design of biocatalysts for fuel cell applications. Under nitrogen-fixing conditions, an unstable hydrogenase activity can also be detected; the hup genes are believed to code for this inducible, unstable enzyme (28). It is reasonable to assume that in T. roseopersicina the transcription of hupSL is H2 regulated via a regulatory mechanism similar to that of Rhodobacter capsulatus (10, 13) and R. eutropha. (19, 33). The remarkable feature of T. roseopersicina is that it contains two hydrogenases having very similar structural and localization features but very dissimilar stabilities, providing a particularly attractive model system for comparative molecular investigations.
Accessory genes participating in the formation of active [NiFe] hydrogenase play important roles in the complex process of hydrogenase assembly, and some of them are needed for the eventual controlled production of this class of biocatalyst for practical use. The goal of our study was the identification of the minimal number of accessory genes needed for expression of the active hydrogenases in this organism. In order to achieve this goal, transposon mutagenesis and a screening procedure for T. roseopersicina have been developed. This is the first report of transposon mutagenesis of a purple sulfur photosynthetic bacterium. Several hydrogenase-deficient mutants have been isolated; one of them is a hypF mutant (M539). A number of hypF genes and hypF-deficient strains from various organisms have been used in heterologous complementation experiments. The aim has been to investigate the ability of HypF to cooperate with the maturation machinery from various backgrounds. It is demonstrated that HypF of T. roseopersicina and HypF proteins from other organisms can function heterologously. The level of heterologous complementation does not usually reach the level measured in the wild-type strain. No significant effect on the transcriptional level of hup or hyd can be detected by RT-PCR when hypF has been inactivated.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains and plasmids are listed in Table 1. T. roseopersicina strains were maintained in Pfenning's mineral medium, with or without 0.1% NH4Cl, and grown photoautotrophically and anaerobically in liquid cultures for 3 to 4 days (27). Plates were solidified with 7 g of Phytagel (Sigma) per liter, supplemented with acetate (2 g/liter) during selection for transconjugants or for screening, and incubated for 2 weeks in anaerobic jars using the GasPak (BBL) or AnaeroCult (Merck) system. Cultures were illuminated with continuous light at 27 to 30°C. E. coli strains were maintained on Luria-Bertani agar plates. TGYEP medium supplemented with 5 μM NiCl2, 1 μM Na2SeO3, and 1 μM Na2MoO4 was used for in vivo H2 gas production (23). For in vitro uptake measurements, the medium described by Sawers and Boxer (30) was slightly modified (0.5% fumarate, 0.4% glucose, 0.5% peptone, 0.1% tryptone, 1 mM MgCl2, 1 μM NiCl2, 1 μM Na2SeO3, 1 μM Na2MoO4; pH 6.5), and cells were grown under a 5% H2–95% N2 atmosphere at 30 or 37°C. R. eutropha was maintained in FN medium; for hydrogenase derepression, FGN medium was used (12). Mineral medium without a carbon source was used for chemolithoautotrophic growth under an atmosphere of H2-O2-CO2 in an 8:1:1 volumetric ratio (12). Antibiotics were used in the following concentrations (micrograms per milliliter): for E. coli, streptomycin (50), ampicillin (100), chloramphenicol (25), and gentamicin (25); for T. roseopersicina, kanamycin (5), gentamicin (5), and ampicillin (200); for R. eutropha, gentamicin (400).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| T. roseopersicina | ||
| BBS | Wild type | 5 |
| M539 | BBS; hypF::Tn5 (Kmr) | This work |
| E. coli | ||
| XL1-Blue MRF′ | Δ(mcrA) 183 Δ(mcrCB-hsdSMR-mrr) 173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)]c | Stratagene |
| S17-1(λpir) | 294 (recA pro res mod) Tpr Smr (pRP4-2-Tc::Mu-Km::Tn7) λpir | 16 |
| MC4100 | F−araD193 Δ(argf-lac)U169 ptsF25 relA1 flB5301 rpsl150, λ− | 17 |
| DHP-F | MC4100; ΔhypF | 23 |
| R. eutropha | ||
| H16 | Wild type | 34 |
| HF441 | ΔhypF1 ΔhypF2 | 34 |
| Plasmids | ||
| pUTKm | Ampr; Tn5-based minitransposon delivery plasmid with Kmr | 16 |
| RP4 | Ampr Kmr Tcr IncP tra+mob+ | 11 |
| pAC145 | Tcr 6.7-kb HindIII insert with hupT, hupU, hupV, and hypF of R. capsulatus | 14 |
| pBluescript SK(+) | Ampr; cloning vector | Stratagene |
| pGEX-3X | AmprlacIq; expression vector | Amersham Pharmacia Biotech |
| pTETR | pBluescribe19(+) (PstI−) carrying the tetR promoter and RBS | Mária Takács (unpublished data) |
| pHRP309 | IncQ Gmrmob+ | 24 |
| pAF1 | Cmr; carrying the E. coli hypF gene | 23 |
| pM539 | 7-kb ApaI fragment harboring the transposon from M539 in pBluescript SK(+) | This work |
| pTRF | pBluescript SK(+) carrying the T. roseopersicina hypF gene | This work |
| pTRFM | pHRP309 carrying the T. roseopersicina hypF gene | This work |
| pRHF10/1 | pHRP309 carrying the R. capsulatus hupTUV and hypF genes | This work |
| pRHF11/7 | pHRP309 carrying the R. capsulatus hypF gene | This work |
| pEHF7 | pGEX-3X carrying the T. roseopersicina hypF gene | This work |
| pTeHF | pTETR carrying the T. roseopersicina hypF gene | This work |
| pHF1-2 | TRHFO1 and TRHFO2 PCR fragment in SmaI of pBluescript SK(+) | This work |
Conjugation.
The conjugation method of Pattaragulwanit and Dahl (26) was modified as follows. T. roseopersicina was grown in Pfenning's mineral medium for 3 to 4 days to reach late logarithmic or early stationary phase (∼108 to 109 CFU/ml). E. coli was grown to mid-logarithmic phase (optical density at 600 nm = 0.7) in Luria-Bertani medium. Three milliliters of the E. coli donor was filtered on a nitrocellulose membrane and washed three times with 5 ml of Pfenning's mineral medium without Na2S. Then 10 ml of the recipient T. roseopersicina was filtered onto the same membrane. Controls contained only donor or recipient and were handled in the same way. Filters were incubated overnight in a light room aerobically at 27 to 30°C on PNA plates (Pfenning's mineral medium without Na2S, supplemented with 0.2% acetate and 0.2% nutrient broth [BBL] solidified with 1.5% agar [Gibco BRL]). Selection was done on Pfenning's mineral medium supplemented with 0.2% acetate and the appropriate antibiotics. Plates were solidified with Phytagel and incubated in anaerobic jars for 2 weeks. Conjugation into R. eutropha was done as described previously (12).
Transposon mutagenesis.
The minitransposon delivery plasmid pUTKm was mobilized from E. coli S17-1(λpir) by the method described above. One hundred colonies were randomly selected from each mating and screened for the hydrogenase-deficient phenotype.
Screening for hydrogenase mutants.
Fifty colonies were grown on each plate. Replica plates were made and used in the screening procedure. The colonies were transferred onto filter paper (Whatman) and put on top of a stack of three filter papers soaked in 100 mM potassium phosphate buffer (pH 9.4) supplemented with 20 mM oxidized methyl viologen in a petri dish. The petri dishes were incubated under an atmosphere of 100% H2 (traces of O2 were eliminated with palladium catalyst) optionally following a 75°C heat treatment for 1 to 2 h. Hydrogenase-positive colonies started to turn blue after 0.5 to 5 h, while hydrogenase-negative colonies stayed purple overnight.
DNA manipulations, PCR, sequencing, Southern blotting, and sequence analysis.
Preparation of genomic DNA, plasmids, cloning, and Southern blots were done according to general practice (1, 29), or the manufacturers' instructions. PCR was done in a PTC-150 MiniCycler (MJ Research). The primers used for the amplification from hypF were the following: TRHFO1 [5′ GCGGCCCATCTCGGCCATCC 3′]; TRHFO2 [5′ CACCGCCCTGGAGTCGCTGG 3′]; TRHFO6N [5′ ATGACCGCCGAGTCGATTCG 3′] (Fig. 1).
FIG. 1.
Restriction map of the chromosomal region of M539 and subclones (the transposon is represented by a stem loop). cya and hypF represent the ORFs that were identified. The transposon is present only in pM539. TRHF06N, TRHF02, and TRHF01 primers were used in PCR. Restriction enzymes: A, ApaI; E, Eco147I; EV, EcoRV; N, NcoI.
Sequencing was done on both strands by automatic DNA sequencing with an Applied Biosystems 373 Stretch DNA sequencer. The searches in the NBRF, SwissProt, combined EMBL-GenBank, and Prosite databases were carried out with the programs FASTA, TFASTA, and BLAST (University of Wisconsin Genetics Computer Group, version 8.0). The multiple alignments were done with the programs CLUSTALW, MOTIFS, PRETTYBOX, and PRETTYPLOT (University of Wisconsin Genetics Computer Group).
Constructs used in this study.
For relevant features of the constructs, see Fig. 1 and Table 1.
(i) Isolation, subcloning, and sequencing of the chromosomal region harboring the hypF gene.
M539 was obtained by cloning a 7-kb ApaI fragment (containing Tn5) from a partial genomic library into a pBluescript SK(+) ApaI site. The region was mapped, subcloned, and sequenced on both strands. The region surrounding the transposon insertion site was amplified from the genome of the wild-type strain with primers TRHFO1 and TRHFO2 (Fig. 1) at a 65°C annealing temperature in order to confirm the wild-type sequence. The product was cloned into the SmaI site of pBluescript SK(+), yielding pHF1-2 (Fig. 1), and the insert was sequenced.
(ii) Reconstruction of the T. roseopersicina hypF gene with its regulatory region for homologous and heterologous complementation.
The 3,346-bp NcoI fragment of pM5/2 was ligated into the NcoI site of pHF1-2, resulting in pTRF; thus, this plasmid carried the reconstructed hypF gene. The whole insert was recloned into a pHRP309 mobilizable vector using enzymes cutting only in the polylinker of the pTRF: the pTRF was digested with NotI, blunted with T4 polymerase, and cut with KpnI to yield a 4,021-bp fragment. The plasmid pHRP309 was digested with HindIII, blunted, and digested with KpnI. The 4,021-bp pTRF fragment was cloned into pHRP309, giving pTRFM.
(iii) Replacement of the T. roseopersicina hypF regulatory region with E. coli regulatory signals. (a) Simple promoter change.
In pEHF7, the 3.3-kb EcoRV-Eco147I fragment of pTRF (which contains the 3′ end of cya and the full hypF gene [Fig. 1]) was cloned in the SmaI site of pGEX-3X. In this construct the cya gene fragment was fused to the gst gene in frame.
(b) Replacement of the transcriptional and translational signals.
The pTeHF construct was made as follows. The hypF gene was amplified with primers TRHF06N (beginning at the start codon of the hypF) and TRHF01 using Pfu DNA polymerase (Fig. 1). The PCR product was treated with polynucleotide kinase and cloned into the PstI-digested, blunted, CIAP-treated pTETR vector. The construct was confirmed by sequencing.
(iv) Recloning of the R. capsulatus hypF gene into mobilizable vector usable in T. roseopersicina.
The pRHF10/1 construct was made by cloning the 6.7-kb HindIII fragment of pAC145 into pHRP309. Plasmid pRHF11/7 was constructed by inserting the 4.2-kb HindIII-StuI fragment of pAC145 into HindIII-SmaI-digested pHRP309.
RNA isolation, RT, and PCR.
RNA was isolated from cells using the TRIzol reagent (Gibco BRL), following the manufacturer's recommendation. RNA was treated with RNase-free DNase I at 37°C for 60 min in a total volume of 40 μl (40 mM Tris-HCl [pH 7.5], 20 mM MgCl2, 20 mM CaCl2, 4 U of RNase-free DNase) prior to reverse transcription (RT)-PCR. After phenol-chloroform extraction and ethanol precipitation, the RNA was dissolved in 20 μl of H2O. RT-PCR experiments were done as described previously (2), except that 100 U of Moloney murine leukemia virus reverse transcriptase (Promega) was used at 37°C in the presence of 40 U of recombinant RNasin RNase inhibitor (Promega) for 60 min. The following primers were used: for hydS, TRHYD01 (5′CCATGGCTGCCCGTAACCCCACTGAT3′ ) and TPE1(5′GGTGCATTCCTGGAACGACAGCCAGATGACCGA3′ ); for hupS, HUPORT01 (5′GCACCTGCTGCTCCGAGTCC3′) and HUPORT2 (5′AGGATGTAGGTCAGGACCCCG3′).
In vivo H2 gas production.
T. roseopersicina and E. coli cultures were grown anaerobically in gas-tight vials (Hypo-Vial; Pierce) in media described above. the H2 content of the headspace was determined using a Hitachi 263-50 gas chromatograph (3). The values obtained were normalized to the cell densities of the cultures.
Hydrogenase activity measurements.
E. coli was anaerobically grown to an optical density at 600 nm of 0.5 to 0.6 on the medium described by Sawers and Boxer (30). T. roseopersicina and R. eutropha were grown on Pfenning's mineral medium and FGN medium, respectively, to the late logarithmic phase. Crude extracts from E. coli and T. roseopersicina were prepared by treating the cells with lysozyme and three freeze-thaw cycles. R. eutropha cells were sonicated for 1 min. Separation of the soluble hydrogenase (SH) and membrane-bound hydrogenase (MBH) of R. eutropha and solubilization of the MBH were done as previously described (31, 32). An in vitro hydrogen evolution assay was carried out in Hypo-Vials for T. roseopersicina (3). H2 uptake coupled to benzyl viologen reduction was assayed spectrophotometrically (UV2 Unicam) at 37°C for E. coli and at 55°C for T. roseopersicina. The same assay with methylene blue redox dye at 52°C was used for the solubilized MBH of R. eutropha. For the SH of the same organism, H2 uptake coupled to NAD+ reduction was assayed spectrophotometrically at 33°C (32). The anaerobically sealed (SubaSeal and Parafilm) cuvettes, containing the samples with the appropriate electron acceptor and buffer, were flushed with N2 followed by H2, each for 10 min.
Nucleotide sequence accession number. The 5,307-nt long sequence has been deposited in GenBank under the accession number AF292554.
RESULTS
Transposon mutagenesis.
The Tn5-based minitransposon delivery vector, pUTKm, was transferred into T. roseopersicina from E. coli S17-1(λpir) via conjugation. Anaerobic incubation of the matings, as described for Allochromatium vinosum (26), yielded operational frequencies of 10−8, practically giving rise to 0 to 10 colonies per selective plate. However, an increase in efficiency of 2 to 3 orders of magnitude was observed when the T. roseopersicina matings on PNA plates were incubated aerobically. Thus, 1,000 to 2,500 mutant colonies per selective plate could be generated. To confirm that kanamycin-resistant colonies were the result of transposition and the vector was not present, 250 colonies were replicated onto ampicillin (200 μg/ml) plates. The wild-type T. roseopersicina carrying the Ampr RP4 was used as a positive control. The random insertion of the transposon was confirmed by Southern blotting of digested genomic DNA from randomly chosen mutant colonies and hybridization using the labeled mobile element of pUTKm as a probe (data not shown).
Identification of hydrogenase-negative mutants.
One hundred colonies were picked from every mating, and a library of 1,600 kanamycin-resistant colonies was screened as described in Materials and Methods. One in every ∼260 colonies showed a hydrogenase-deficient phenotype, which further substantiated the random insertion of the transposon. Six independent mutants were isolated, five of which appeared pleiotropic, lacking all hydrogenase activity under every condition tested. In one, the remaining hydrogenase activity could be inactivated by heat treatment (75°C for 1 to 2 h) under air; the same treatment did not abolish the HydSL-linked hydrogenase activity of the wild-type T. roseopersicina.
Identification and sequence of hypF.
The ∼7-kb ApaI genomic fragment of one of the pleiotropic mutants (M539) carrying the transposon was cloned and subcloned (Fig. 1). More than 5 kb was sequenced on both strands. The codon usage was similar to that in other genes studied in T. roseopersicina. One open reading frame (ORF), from bp 1777 to 4197, was identified as hypF on the basis of sequence homology. The translated polypeptide, consisting of 806 amino acids, was homologous to HypF proteins of other organisms: 50, 44, and 42% identity and 60, 56, and 52% similarity to R. capsulatus, E. coli, and R. eutropha HypF2, respectively. Sequence elements typical of HypF proteins could be identified: two zinc-finger-like motifs (CXXCX18CXXC) were separated by 24 amino acids (from Cys117 to Cys192) at the N terminus (22), and a motif located between residues 10 and 36 (GXVQGVX2RX13GX3N) indicating acylphosphatases could also be identified in this region. An HHXAH motif typical of HypF proteins was also present from 516 to 520 (25). The transposon insertion took place at 162 bp (54 amino acids) upstream from the C-terminus-encoding region of hypF. No other ORF resembling sequences of genes of proteins involved in hydrogenase biosynthesis could be recognized on this genomic DNA fragment. Upstream of hypF, a putative adenylate cyclase (cya) gene from bp 42 to 1775 was found (Fig. 1).
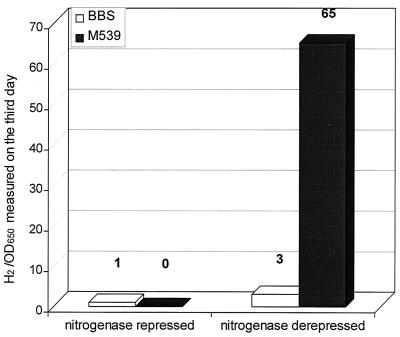
It should be noted that the hypF-deficient strain was devoid of all [NiFe] hydrogenase activities. An important consequence of the mutation was the remarkably pronounced H2-evolving activity under nitrogen fixing conditions (Fig. 2), although no [NiFe] hydrogenase activity could be detected in vitro using the standard assays. Hence, this high hydrogen evolution activity is most likely due to the nitrogenase complex.
FIG. 2.
Relative hydrogen production of the wild-type and the hypF-defective (M539) T. roseopersicina strains in vivo under nitrogenase-repressed and derepressed conditions (see Materials and Methods). Samples were measured after cultivation for 3 days. The amount of H2 evolved by the wild-type strain under non-nitrogen-fixing conditions was set at 1. OD650, optical density at 650 nm.
Complementations using the original regulatory region of hypF genes.
The vectors pTRFM, harboring hypF of T. roseopersicina with its original promoter and ribosome binding site (RBS) and pHRP309 (negative control) were transferred to the transposon-mediated hydrogenase-deficient mutants of T. roseopersicina. The pTRFM construct complemented hydrogenase activity in strain M539 in all in vitro assays (for uptake activity, see Table 2.). None of the other five mutants could be complemented with pTRFM, which supported the idea that independent mutants had been isolated. The level of hydrogenase activity of the complemented M539 mutant did not reach the level of the wild-type strain. Next we tested whether HypF of R. capsulatus was able to complement the M539 mutant, since it showed the highest homology to the HypF of T. roseopersicina. Construct pRHF10/1 harbors the hupTUV and hypF genes of R. capsulatus. The hypF gene can be transcribed from its own promoter and/or from the hupTUV promoter. In construct pRHF11/7, only hypF of R. capsulatus is present with its own regulatory region. In both cases, M539 could be complemented just as well as with pTRFM (Table 2). To further investigate the possibilities of heterologous complementation, these constructs were used to complement the HF441 strain of R. eutropha. All three constructs restored hydrogenase activity, although 100% activity was never measured. The heterologously complemented R. eutropha HF441 strains were able to grow and form colonies under chemolithoautotrophic conditions, although they did so two to four times slower than the wild-type strain, while the negative controls did not have this ability.
TABLE 2.
H2 uptake activities in heterologous complementation experiments
| Gene | Construct | H2 uptake activitya in hypF mutant strain
|
|||
|---|---|---|---|---|---|
| E. coli DHP-F |
R. eutropha HF441
|
T. roseopersicina M539 | |||
| SH | MBH | ||||
| E. coli hypF | pAF1 | 44 ± 4.1 | NR | NR | NR |
| T. roseopersicina hypF | pTRF | 0 | NR | NR | NR |
| pEHF7 | 1.4 ± 0.8 | NR | NR | NR | |
| pTeHF | 4 ± 1.0 | NR | NR | NR | |
| pTRFM | 0 | 58 ± 5.8 | 49 ± 7.8 | 56 ± 11.5 | |
| R. eutropha hypF | pRHF10/1 | 0 | 27 ± 4.3 | 17 ± 3.8 | 70 ± 10.5 |
| pRHF11/7 | 0 | 35 ± 6.1 | 29 ± 5.3 | 58 ± 9.2 | |
Results are percentages of the values for the wild-type strains. Positive controls were the wild-type strains and the HypF-deficient mutant strain carrying the original wild-type gene on a plasmid when the appropriate construct was available. Negative controls were hypF mutant strains and the same strains carrying the vector without any hypF gene. Each experiment was repeated at least three (six for E. coli) times with independent samples prepared from single colonies from different streaks. Values are means ± standard deviations. NR, the plasmid is non replicating in that organism.
The three constructs mentioned above and pTRF (hypF gene of T. roseopersicina in the opposite orientation with respect to the lacZ promoter of pBluescript) were also tested in E. coli. Hydrogenase activity was never restored with these constructs.
Complementations using expression cassette-hypF hybrids.
To determine if the unsuccessful heterologous complementations in E. coli were caused by the inefficient expression of the genes, additional constructs containing various combinations of regulatory elements and the hypF gene of T. roseopersicina were tested. In pEHF7, hypF expression was under the control of the inducible Ptac, but the original hypFRBS of T. roseopersicina was used. In pTeHF the Plac and PtetR promoters and the tetRRBS directed the expression of the hypF gene. To test the level of transcription from different constructs, RT-PCR was performed on variants of strain DHP-F each carrying pTRF, pEHF7, or pTeHF. The level of transcription was lower in the case of pTRF than with the other two (data not shown), supporting the assumption that the low expression from the original T. roseopersicina promoter was not sufficient. The pTeHF and pEHF7 constructs were used in the following experiments. The hydrogenase 2 activity was measured in uptake direction in the cells grown under the conditions where the expression of the E. coli hydrogenase 3 was repressed and hydrogenase 1 was underrepresented (30). Compared to the results with the wild type, significant but low activity could be detected with both pEHF7 and pTeHF (1.4 and 4%, respectively) (Table 2). The formation of active hydrogenase 3 was monitored via in vivo H2 gas production by cells grown in TGYEP medium. The complementation levels (data not shown) corresponded to those obtained by uptake measurements (Table 2), indicating that HypF participated in the maturation of both hydrogenases 2 and 3 to similar extent. In the case of the pEHF7 vector, expression could be induced by various amounts of IPTG (isopropyl-β-d-thiogalactopyranoside; 0, 0.04, 0.4, and 4 mg/ml, respectively). By increasing the level of HypF expression, hydrogenase activity decreased (3.5, 2.9, 1.3, and 0.7%, respectively [100% is the wild-type activity]).
Effect of HypF on the transcription of hydSL and hupSL
The stable hydrogenase (HydSL) is present in T. roseopersicina under both nitrogenase-repressed and derepressed conditions. A considerable unstable hydrogen uptake activity (assumed to be HupSL) could be detected under nitrogen-fixing conditions (28). Inducible hydrogenases are often transcriptionally regulated via a regulatory hydrogenase-linked signal transduction pathway (10). HypF was shown to participate in the maturation of regulatory hydrogenases (10, 33) and to have an effect through this mechanism on the regulation of transcription of the structural genes. RT-PCR was performed on both the wild-type and M539 mutant strains to see if the transcription of hydSL and hupSL was affected by HypF under nitrogenase-repressed or derepressed conditions (Fig. 3). No significant difference in the transcription of hydSL and hupSL between the wild-type and the M539 mutant strains was observed.
FIG. 3.
(A) RT-PCR performed on the wild-type and M539 mutant strains of T. roseopersicina. Primers TRHYD01 and TPE1 were used to detect hydSL, and HUPORT01 and HUPORT02 were used to detect hupSL. The transcript level was monitored as a function of PCR cycle number (shown below the lanes). (B) Control PCRs made on RNA preparations without reverse transcription after 31 PCR cycles. Wt, wild type; gC, control PCR made on genomic DNA; M, marker.
DISCUSSION
A gene transfer system for the purple sulfur photosynthetic bacterium T. roseopersicina BBS was developed. Using an RP4 plasmid-based conjugation system, a Tn5 minitransposon delivery suicide vector (pUTKm) was introduced into this strain from E. coli. An important finding was that the operational frequency increased 100 to 1,000 times in the presence of O2 (compared to anaerobic conditions) during incubation of the matings. Environmental factors (e.g., oxygen) can affect the activity of mobile genetic elements (7, 15), which may explain our observations. It was proven that the transposon insertions took place randomly, which was corroborated by the appearance of a pigment mutant in every ∼3,000 mutant colonies. This is the first report on transposon mutagenesis in purple sulfur bacteria.
To isolate mutants deficient in H2 uptake activity, a reliable screening procedure was developed based on color change of the viologen dyes upon redox transition and on the distinct heat stability of hydrogenases in T. roseopersicina. One of every ∼260 colonies was deficient in hydrogenase activity, and six such mutants were isolated.
In this study the M539 mutant was characterized in detail. No [NiFe] hydrogenase activity was detectable in strain M539 under nitrogen-fixing or nonfixing conditions. The chromosomal region, surrounding the transposon, revealed an ORF coding for a protein with high sequence homology to HypF in other organisms. HypF has been shown to be essential in [NiFe] hydrogenase maturation in other microbes (8, 12, 23, 34) and assumed to participate in the synthesis of the CO and/or CN ligands in E. coli (25).
Remarkably, the hypF mutant strains showed a very intense H2 evolution under nitrogen-fixing conditions (Fig. 2). Firstly, this observation indicated that a substantial amount—probably all—of hydrogen uptake activity has been inactivated in the hypF mutant cells. Secondly, hypF mutation clearly offers an important avenue for the development of efficient biological hydrogen production systems using this organism. Growing under phototrophic and nitrogen-fixing conditions, T. roseopersicina can evolve hydrogen at a practically significant level. Thirdly, the results suggest the existence of a single hypF gene in T. roseopersicina, indicating that at least some elements needed for [NiFe] hydrogenase maturation are shared and that the biosynthesis of the unstable and stable hydrogenases has common elements in this organism. This information is relevant for the understanding of the formation of the two enzymes having similar functions and primary structures but dissimilar stability properties in a single organism.
In order to confirm that the mutation in hypF caused the loss of hydrogenase activity, homologous complementation was carried out with the reconstructed T. roseopersicina hypF (pTRFM) in the M539 mutant. The plasmid-borne hypF restored H2 uptake hydrogenase activity, although the level of complementation did not reach the level measured in the wild-type strain. This might be caused by the alteration of the ratio of the components playing role in the maturation process, unless the process kinetics was affected. Similar observations were reported for homologous complementation experiments with other hyp genes in E. coli (17). In related studies (12, 17), the loss of the pleiotropic hyp function was associated with the accumulation of the unprocessed hydrogenase large subunit protein, which could be detected by Western blotting. This approach was not applicable in our case, because the HupL and HydL proteins were similar in size, 64 and 65 kDa, respectively. The processed and unprocessed forms therefore cannot be distinguished within the resolution limits of gel electrophoresis.
Heterologous complementation experiments were performed to examine if HypF proteins were functionally conserved enough to cooperate with the rest of the maturation machinery in various organisms. The data presented in this study provided enough information to conclude that heterologous complementation with HypF was possible. Using the original T. roseopersicina hypF gene, promoter, and Shine-Dalgarno sequences, heterologous complementation was observed in an R. eutropha recipient but not in E. coli recipients. Upon replacement of the T. roseopersicina promoter alone or the promoter and RBS with those of E. coli, the hypF gene complementation was detectable in E. coli, but only at a low level. The promoter of the pEHF7 construct was inducible with IPTG. Induction with increasing amounts of IPTG had an inverse effect on the complementation level, which supported the idea that in the maturation process the ratio of the accessory proteins was a substantial determinant. In the case of the R. eutropha recipient, the T. roseopersicina hypF gene restored the biosynthesis of active hydrogenase from its own promoter and RBS. Similarly, hypF of R. capsulatus complemented the T. roseopersicina and R. eutropha HypF-deficient recipients without the requirement for the replacement of the R. capsulatus promoter. Heterologous complementation levels were close to activities measured in the homologous complementation systems, but remarkably, both were significantly lower than the activities of the wild-type strains. This study is the first report providing direct experimental proof that interactions involving heterologous Hyp proteins may result in functionally active hydrogenases. The findings also suggest that E. coli may not be the ideal host for large-scale heterologous production of [NiFe] hydrogenases.
A direct effect of HypF on the maturation of the H2-sensing regulatory hydrogenase in R. capsulatus and R. eutropha has been shown (10, 33). As a consequence of this function, HypF affects the transcription of the hydrogenase structural genes. It is important to note that in our case a complete loss of [NiFe] hydrogenase activity is accompanied by unchanged hup and hyd transcript levels in the hypF-deficient mutant (M539) compared to the wild type, T. roseopersicina BBS.
The genetic methods presented here will be useful in the identification of other maturation elements participating in the hydrogenase biosynthesis in T. roseopersicina. Understanding the strain-specific and interchangeable elements participating in hydrogenase biosynthesis will also be important in heterologous production of stable [NiFe] hydrogenase enzymes for practical purposes.
ACKNOWLEDGMENTS
This work was funded by grants from the EU 5th FP QLRT-1999-01267, OMFB-00017/99, and PHARE HU9606-02-01-62.
We thank Ronald E. Hurlbert (Washington State University, Pullman) for S17-1(λpir) and pUTKm, Bärbel Friedrich (Humboldt University, Berlin, Germany) for R. eutropha H16 and HF441, Annette Colbeau (DBMS, CEA-CENG, Grenoble, France) for pEC4 and pAC145, and August Böck and Axel Magalon (Ludwig-Maximilian University, Münich, Germany) for MC4100, DHP-F, and pAF1. B.F. and A.T.K. are grateful to Peter Lindblad for their visit to his lab in Uppsala, which was sponsored by the Swedish Royal Academy of Sciences and Hungarian Academy of Sciences.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Wiley; 1996. [Google Scholar]
- 2.Axelson R, Oxelfelt F, Lindblad P. Transcriptional regulation of Nostoc uptake hydrogenase. FEMS Microbiol Lett. 1999;170:77–81. doi: 10.1111/j.1574-6968.1999.tb13357.x. [DOI] [PubMed] [Google Scholar]
- 3.Bagyinka C, Zorin N A, Kovács K L. Unconsidered factors affecting hydrogenase activity measurement. Anal Biochem. 1984;142:7–15. doi: 10.1016/0003-2697(84)90509-8. [DOI] [PubMed] [Google Scholar]
- 4.Ballantine S P, Boxer D H. Nickel-containing hydrogenase isoenzymes from anaerobically grown Escherichia coli K-12. J Bacteriol. 1985;163:454–459. doi: 10.1128/jb.163.2.454-459.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogorov L V. About the properties of Thiocapsa roseopersicina BBS, isolated from estuaria of White Sea. Mikrobiologija. 1974;43:326–332. [PubMed] [Google Scholar]
- 6.Cammack R, Fernandez V M, Hatchikian E C. Nickel-iron hydrogenases. Methods Enzymol. 1994;243:43–68. [Google Scholar]
- 7.Chow K C, Tung W L. Magnetic field exposure stimulates transposition through the induction of DnaK/J synthesis. Biochem Biophys Res Commun. 2000;270:745–748. doi: 10.1006/bbrc.2000.2496. [DOI] [PubMed] [Google Scholar]
- 8.Colbeau A, Richaud P, Toussaint B, Caballero F J, Elster C, Delphin C, Smith R L, Chabert J, Vignais P M. Organization of genes necessary for hydrogenase expression in Rhodobacter capsulatus. Sequence analysis and identification of two hyp regulatory mutants. Mol Microbiol. 1993;8:15–29. doi: 10.1111/j.1365-2958.1993.tb01199.x. [DOI] [PubMed] [Google Scholar]
- 9.Colbeau A, Kovács K L, Chabert J, Vignais P M. Cloning and sequences of the structural (hupSLC) and accessory (hupDHI) genes for hydrogenase biosynthesis in Thiocapsa roseopersicina. Gene. 1994;140:25–31. doi: 10.1016/0378-1119(94)90726-9. [DOI] [PubMed] [Google Scholar]
- 10.Colbeau A, Elsen S, Tomiyama M, Zorin N A, Dimon B, Vignais P M. Rhodobacter capsulatus HypF is involved in regulation of hydrogenase synthesis through the HupUV proteins. Eur J Biochem. 1998;251:65–71. doi: 10.1046/j.1432-1327.1998.2510065.x. [DOI] [PubMed] [Google Scholar]
- 11.Datta N, Hedges R W, Shaw E S, Sykes R B, Richmond M H. Properties of an R-factor from Pseudomonas aeruginosa. J Bacteriol. 1971;108:1244–1249. doi: 10.1128/jb.108.3.1244-1249.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dernedde J, Eitinger T, Patange N, Friedrich B. hyp gene products in Alcaligenes eutrophus are part of a hydrogenase-maturation system. Eur J Biochem. 1996;235:351–358. doi: 10.1111/j.1432-1033.1996.00351.x. [DOI] [PubMed] [Google Scholar]
- 13.Dischert W, Vignais P M, Colbeau A. The synthesis of Rhodobacter capsulatus HupSL hydrogenase is regulated by the two-component HupT/HupR system. Mol Microbiol. 1999;4:995–1006. doi: 10.1046/j.1365-2958.1999.01660.x. [DOI] [PubMed] [Google Scholar]
- 14.Elsen S, Richaud P, Colbeau A, Vignais P M. Sequence analysis and interposon mutagenesis of the hupT gene, which encodes a sensor protein involved in repression of hydrogenase synthesis in Rhodobacter capsulatus. J Bacteriol. 1993;175:7404–7412. doi: 10.1128/jb.175.22.7404-7412.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghanekar K, McBride A, Dellagostin O, Thorne S, Mooney R, McFadden J. Stimulation of transposition of the Mycobacterium tuberculosis insertion sequence IS6110 by exposure to a microaerobic environment. Mol Microbiol. 1999;33:982–993. doi: 10.1046/j.1365-2958.1999.01539.x. [DOI] [PubMed] [Google Scholar]
- 16.Herrero M, Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobi A, Rossmann R, Böck A. The hyp operon gene products are required for the maturation of catalytically active hydrogenase isoenzymes in Escherichia coli. Arch Microbiol. 1992;158:444–451. doi: 10.1007/BF00276307. [DOI] [PubMed] [Google Scholar]
- 18.Kovács K L, Rákhely G, Pott A S, Takács M, Tóth A, Bratu H, Gyõrfi K, Fodor B, Tusz J, Dahl C. Genes involved in hydrogen and sulfur metabolism in photosynthetic bacteria. Pflueg Arch Eur J Physiol. 2000;439:R81–R83. [Google Scholar]
- 19.Lenz O, Strack A, Tran-Becke A, Friedrich B. A hydrogen-sensing system in transcriptional regulation of hydrogenase gene expression in Alcaligenes species. J Bacteriol. 1997;179:1655–1663. doi: 10.1128/jb.179.5.1655-1663.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lutz S, Jacobi A, Schlensog V, Böhm R, Sawers G, Böck A. Molecular characterization of an operon (hyp) necessary for the activity of the three hydrogenase isoenzymes in Escherichia coli. Mol Microbiol. 1991;5:123–135. doi: 10.1111/j.1365-2958.1991.tb01833.x. [DOI] [PubMed] [Google Scholar]
- 21.Magalon A, Böck A. Analysis of the HypC-HycE complex, a key intermediate in the assembly of the metal center of the Escherichia coli hydrogenase 3. J Biol Chem. 2000;275:21114–21120. doi: 10.1074/jbc.M000987200. [DOI] [PubMed] [Google Scholar]
- 22.Maier T, Böck A. Nickel incorporation into hydrogenases. In: Hausinger R, Eichlorn G L, Marzilli L G, editors. Advances in inorganic biochemistry. New York, N.Y: VHC Publishers Inc.; 1996. pp. 173–192. [Google Scholar]
- 23.Maier T, Binder U, Böck A. Analysis of the hydA locus of Escherichia coli: two genes (hydN and hypF) involved in formate and hydrogen metabolism. Arch Microbiol. 1996;165:333–341. doi: 10.1007/s002030050335. [DOI] [PubMed] [Google Scholar]
- 24.Parales R E, Harwood C S. Construction and use of a new broad-range lacZ transcriptional fusion vector, pHRP309, for Gram− bacteria. Gene. 1993;133:23–30. doi: 10.1016/0378-1119(93)90220-w. [DOI] [PubMed] [Google Scholar]
- 25.Paschos A, Glass R S, Böck A. Carbamoylphosphate requirement for synthesis of the active center of [NiFe]-hydrogenases. FEBS Lett. 2001;488:9–12. doi: 10.1016/s0014-5793(00)02408-x. [DOI] [PubMed] [Google Scholar]
- 26.Pattaragulwanit K, Dahl C. Development of genetic system for a purple sulfur bacterium: conjugative plasmid transfer in Chromatium vinosum. Arch Microbiol. 1995;164:217–222. [Google Scholar]
- 27.Pfenning N, Trüper H G. The family Chromatiaceae. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. Berlin, Germany: Springer; 1991. pp. 3200–3221. [Google Scholar]
- 28.Rákhely G, Colbeau A, Garin J, Vignais P M, Kovács K L. Unusual organization of the genes coding for HydSL, the stable (NiFe)hydrogenase in the photosynthetic bacterium Thiocapsa roseopersicina BBS. J Bacteriol. 1998;180:1460–1465. doi: 10.1128/jb.180.6.1460-1465.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Maniatis T, Fritsch E F. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Sawers G R, Boxer D H. Purification and properties of membrane-bound hydrogenase isoenzyme 1 from anaerobically grown Escherichia coli K12. Eur J Biochem. 1986;156:265–275. doi: 10.1111/j.1432-1033.1986.tb09577.x. [DOI] [PubMed] [Google Scholar]
- 31.Schink B, Schlegel H G. The membrane-bound hydrogenase of Alcaligenes eutrophus. I. Solubilization, purification, and biochemical properties. Biochim Biophys Acta. 1978;569:315–324. doi: 10.1016/0005-2744(79)90117-7. [DOI] [PubMed] [Google Scholar]
- 32.Schneider K, Schlegel H G. Purification and properties of soluble hydrogenase from Alcaligenes eutrophus H16. Biochim Biophys Acta. 1976;452:66–80. doi: 10.1016/0005-2744(76)90058-9. [DOI] [PubMed] [Google Scholar]
- 33.Schwartz E, Buhrke T, Gerischer U, Friedrich B. Positive transcriptional feedback controls hydrogenase expression in Alcaligenes eutrophus H16. J Bacteriol. 1999;181:5684–5692. doi: 10.1128/jb.181.18.5684-5692.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolf I, Buhrke T, Dernedde J, Pohlmann A, Friedrich B. Duplication of hyp genes involved in maturation of [NiFe] hydrogenases in Alcaligenes eutrophus H16. Arch Microbiol. 1998;170:415–419. doi: 10.1007/s002030050666. [DOI] [PubMed] [Google Scholar]