Abstract
The number of meiotic crossovers is tightly controlled and most depend on pro‐crossover ZMM proteins, such as the E3 ligase HEI10. Despite the importance of HEI10 dosage for crossover formation, how HEI10 transcription is controlled remains unexplored. In a forward genetic screen using a fluorescent crossover reporter in Arabidopsis thaliana, we identify heat shock factor binding protein (HSBP) as a repressor of HEI10 transcription and crossover numbers. Using genome‐wide crossover mapping and cytogenetics, we show that hsbp mutations or meiotic HSBP knockdowns increase ZMM‐dependent crossovers toward the telomeres, mirroring the effects of HEI10 overexpression. Through RNA sequencing, DNA methylome, and chromatin immunoprecipitation analysis, we reveal that HSBP is required to repress HEI10 transcription by binding with heat shock factors (HSFs) at the HEI10 promoter and maintaining DNA methylation over the HEI10 5′ untranslated region. Our findings provide insights into how the temperature response regulator HSBP restricts meiotic HEI10 transcription and crossover number by attenuating HSF activity.
Keywords: Crossover, HEI10, HSBP, HSF, meiosis
Subject Categories: Chromatin, Transcription & Genomics; DNA Replication, Recombination & Repair; Plant Biology
HSBP restrains crossover number in A. thaliana by repressing the transcription of HEI10, encoding an E3 ligase which orchestrates the progression of meiosis.

Introduction
During meiosis, homologous chromosomes (homologs) undergo reciprocal DNA exchanges, called crossovers. Crossovers ensure the correct segregation of homologs during meiosis I and create new combinations of alleles in gametes (Villeneuve & Hillers, 2001; Hunter, 2015). Meiotic recombination is initiated by the formation of DNA double‐strand breaks (DSBs) (Gray & Cohen, 2016). Numerous DSBs are formed, but only about 5% of DSBs are repaired as crossovers, and thus, the number of crossovers is limited to 1–3 per homolog (Mercier et al, 2015). Meiotic DSB ends are resected to generate 3′ single‐stranded DNA ends that are then bound by recombinases DMC1 and RAD51 (Gray & Cohen, 2016). The resulting nucleoprotein complex then invades sister or non‐sister chromatids to produce a displacement loop (D‐loop) structure (Hunter, 2015; Gray & Cohen, 2016). Interhomolog D‐loops are resolved into crossovers by the formation of double Holliday junctions (dHJs). Alternatively, D‐loops are dissolved to produce non‐crossovers (Hunter, 2015; Mercier et al, 2015).
Two crossover pathways are conserved across eukaryotes (Mercier et al, 2015). The class I pathway is responsible for approximately 85–90% of crossovers in Arabidopsis (Arabidopsis thaliana) (Mercier et al, 2015). Class I crossover formation is promoted by a group of ZMM proteins (ZIP4, SHOC1, PTD, MER3, MSH4, MSH5, and HEI10) and MutLγ (MLH1–MLH3) dHJ resolvases (Copenhaver et al, 2002; Higgins et al, 2004; Mercier et al, 2005, 2015; Chelysheva et al, 2012; Duroc et al, 2017; De Muyt et al, 2018). ZMM proteins stabilize interhomolog D‐loops and protect them from anti‐crossover helicases and facilitate the recruitment of MutLγ resolvases at crossover sites (Pyatnitskaya et al, 2019; Cannavo et al, 2020). Class I crossovers are subject to interference, which prevents the formation of another crossover nearby (Berchowitz & Copenhaver, 2010). Conversely, class II crossovers are non‐interfering and formed by MUS81 (Berchowitz et al, 2007). In Arabidopsis, class II crossovers are restricted by anti‐recombination proteins such as FANCM, RECQ4A, and RECQ4B that promote non‐crossovers (Crismani et al, 2012; Girard et al, 2015; Séguéla‐Arnaud et al, 2015).
One of the ZMM proteins required for class I crossover formation is the E3 ubiquitin/SUMO ligase HEI10 (Human enhance of invasion‐10) (Chelysheva et al, 2012; Wang et al, 2012; De Muyt et al, 2014; Qiao et al, 2014). Arabidopsis HEI10 is loaded onto the meiotic chromosome axes as numerous foci during early prophase I, followed by their progressive reduction in numbers during pachytene, with only approximately 10–12 HEI10 foci remaining from late pachytene to diakinesis, marking crossover sites with MLH1 foci (Chelysheva et al, 2012; Morgan et al, 2021). HEI10 interacts with several ZMM proteins in rice and Arabidopsis (Li et al, 2018; Zhang et al, 2019; Nageswaran et al, 2021). The biochemical activity of HEI10 remains elusive in plants, although protein modifications and degradation play critical roles in meiosis (Reynolds et al, 2013; Qiao et al, 2014; Rao et al, 2017; Gao & Colaiácovo, 2018). Studies in Arabidopsis and mice have shown that HEI10 is a dosage‐sensitive pro‐crossover factor (Qiao et al, 2014; Ziolkowski et al, 2017; Serra et al, 2018). HEI10 foci dynamics are also likely associated with crossover interference and the effects of temperature on class I crossover formation (Lloyd et al, 2018; Modliszewski et al, 2018; Morgan et al, 2021). Despite the importance of HEI10 expression in controlling crossover numbers, very little is known about the regulation of HEI10 transcription during meiosis.
In a forward genetic screen using a fluorescent crossover reporter in Arabidopsis, here, we describe the identification of HIGH CROSSOVER RATE2 (HCR2), which encodes HSBP (heat shock factor binding protein), as a repressor of crossover frequency. The hcr2 mutant and meiosis‐specific HSBP knockdown increased HEI10 transcript levels, leading to more crossovers in distal euchromatic regions and lower interference. HSBP is associated with heat shock factors (HSFs) at the HEI10 promoter and maintained DNA methylation over the HEI10 5′ untranslated region. Our work, thus, revealed how the conserved HSBP‐HSF transcriptional module controls HEI10 transcription and restricts class I crossovers during meiosis.
Results
A forward genetic screen identifies hcr2 as a hypomorphic allele (hsbp‐3) of HSBP
To identify new anti‐crossover factors, we performed a forward genetic screen for mutants with an elevated crossover rate using ethyl methanesulfonate (EMS) mutagenesis and the fluorescent recombination reporter 420 in the Arabidopsis Columbia‐0 (hereafter, Col) background (Fig 1A and Appendix Fig S1A−B) (Nageswaran et al, 2021). The 420 reporter system carries two fluorescent reporter transgenes located on the upper arm of chromosome 3 and allows high‐throughput measurements of crossover frequency in individual plants (Melamed‐Bessudo et al, 2005; Ziolkowski et al, 2015, 2017; Nageswaran et al, 2021). We isolated the high crossover rate (hcr) mutants hcr1, hcr2, hcr3, and hcr4 (t‐test, all P < 4.21 × 10−5) (Fig 1A and B and Appendix Table S1) (Nageswaran et al, 2021). We showed previously that HCR1 encodes PROTEIN PHOSPHATASE X‐1 (PPX1), which interacts with ZMM proteins and limits class I crossovers, whereas hcr4 was a fancm mutant allele (Nageswaran et al, 2021). The genetic distance measured between the two 420 fluorescent reporters was 35 cM in hcr2, representing a significantly higher crossover frequency (t‐test, P = 1.32 × 10−10) than the 20 cM in Col, or hcr2/+ heterozygotes (t‐test, HCR2 versus hcr2/+, P = 0.629), indicating that hcr2 is a recessive mutation (Fig 1C and Appendix Table S2). We mapped the causal hcr2 mutation using a BC1F2 population and bulk segregant sequencing (Fig 1C and Appendix Fig S1C and D, and Table S3) (Sun & Schneeberger, 2015; Nageswaran et al, 2021). hcr2 (hereafter hsbp‐3; see below) harbored an EMS‐driven single substitution mutation (C‐to‐T) close to the donor splicing site between the fourth and fifth exons in At4g15802, which encodes heat shock factor binding protein (HSBP) (Fig 1D). HSBP is conserved across eukaryotes and represses transcription by binding to heat shock transcription factors (HSFs) (Appendix Fig S2A−C) (Satyal et al, 1998; Hsu et al, 2010). The fourth intron of Arabidopsis HSBP is of the conserved minor AT‐AC intron splicing class (Fig 1D and Appendix Fig S2D) (Russell et al, 2006). The C‐to‐T substitution in hsbp‐3 resulted in aberrant shorter and longer HSBP splice variants that introduce premature stop codons, compared to Col transcripts (Figs 1E and EV1A and B). We found that HSBP transcripts and HSBP protein levels decreased to approximately 53 and 58%, respectively, of Col levels in hsbp‐3 buds (Figs 1E and F, and EV1B and C). The hsbp‐1 and hsbp‐2, T‐DNA insertion mutants, also showed reduced HSBP transcript levels (hsbp‐1, 70%; hsbp‐2, 9%) and HSBP protein levels (hsbp‐1, 77%; hsbp‐2, 17%) relative to Col, indicating that hsbp alleles are unlikely to be null mutants but instead accumulate HSBP to different levels (Figs 1E and F, and EV1B and C).
Figure 1. The hcr2 mutant is a weak hsbp allele.
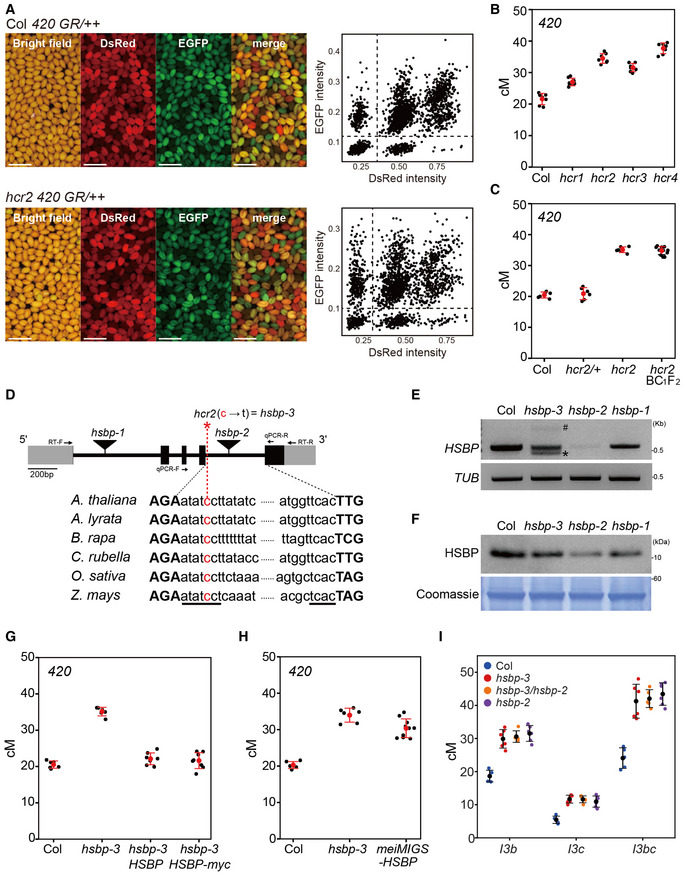
- Representative images of seed fluorescence segregation in 420/++ in wild type (Col) and hcr2. Scatterplots to the right show red (dsRed) and green (eGFP) fluorescence values in 420/++ Col (top) and hcr2 (bottom). Scale bars: 2 mm.
- 420 crossover frequencies (cM) in Col, hcr1, hcr2, hcr3, and hcr4 mutants. n ≥ 7 plants of biological replicates.
- As in (B), 420 crossover frequencies (cM) in Col, hcr2/HCR2, hcr2/hcr2, and individual hcr2 BC1F2 plants. n ≥ 6 plants of biological replicates.
- Schematic diagram of the HSBP locus and position of the hcr2 (hsbp‐3) substitution (red asterisk). Black boxes, exons; gray boxes, UTRs; introns, black lines. The conserved splicing sequence of AT‐AC class introns is underlined. Primer positions for the RT–PCR and RT–qPCR analyses are indicated by arrows.
- End‐point RT–PCR analysis of HSBP in Col, hsbp‐3, hsbp‐2, and hsbp‐1. Hash and asterisk indicate aberrant long and short splicing variants of HSBP in hsbp‐3, respectively. Image J was used to measure relative PCR band intensity for hsbp‐3 (53%), hsbp‐2 (9%), and hsbp‐1 (70%). TUB2 was used as an internal control.
- As in (E), but showing immunoblot analysis of HSBP. hsbp‐3, hsbp‐2 and hsbp‐1 accumulate about 58, 17, and 77% of HSBP levels, respectively. Coomassie‐stained membrane was used as a loading control.
- As in (B), 420 crossover frequencies in Col, hsbp‐3, and hsbp‐3 T1 lines harboring the HSBP or HSBP‐myc transgene. n ≥ 6 plants of biological replicates.
- As in (B), 420 crossover frequencies (cM) in Col, hsbp‐3, and meiMIGS‐HSBP T1 transgenic plants. n ≥ 6 plants of biological replicates.
- As in (B), but showing I3bc crossover frequency (cM) in Col, hsbp‐3, hsbp‐3/hsbp‐2 F1 hybrid, and hspb‐2 plants. n ≥ 5 plants of biological replicates.
Data information: (E, F) Experiments were performed at least three times. (B, G, H) Red dots and horizontal lines indicate mean ± s.d. of cM values from individual plants (one‐sided Welch’s t‐test). Black dots represent cM values of individual plants. (I) Colored dots represent cM values from individual plants.
Source data are available online for this figure.
Figure EV1. Effects of hsbp alleles on HSBP transcripts and HSBP abundance.
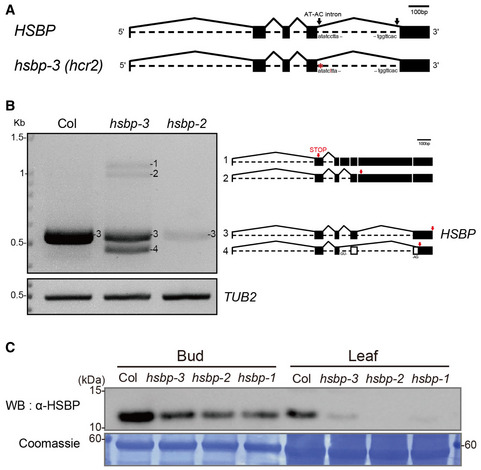
- Schematic representation of HSBP transcripts and position of the hsbp‐3 mutation (red).
- Aberrant long, short splicing variants (1, 2, 4), and normal transcripts (3) of HSBP in hsbp‐3. Arrows (red) indicate the positions of premature stop codons in abnormal splicing variants or the normal stop codon in Col transcripts.
- Immunoblot analysis of HSBP of Col, hsbp‐3, hsbp‐2, and hsbp‐1 in leaves and buds. Coomassie‐stained membrane served as a loading control.
Data information: Experiments were performed at least three times (B, C).
Source data are available online for this figure.
To confirm that HCR2 is HSBP, we generated complementation lines by introducing the entire HSBP genomic region from Col into the 420/++ hsbp‐3 background via transformation (Fig 1G and Appendix Table S4). Primary (T1) transgenic plants harboring genomic HSBP reduced the crossover frequency of the hsbp‐3 mutant to Col levels (t‐test, HSBP P = 0.192, HSBP‐myc P = 0.436) (Fig 1G and Appendix Table S4). We also specifically decreased HSBP transcript levels during meiosis using meiMIGS (meiosis‐specific miRNA induced gene silencing) in the 420/++ background (Figs 1H and EV2A–E) (de Felippes et al, 2012; Nageswaran et al, 2021). These meiMIGSDMC1‐HSBP T1 transgenic plants showed increased 420 crossover frequencies compared with Col plants (t‐test, P = 7.22 × 10−8) (Fig 1H and Appendix Table S5). Importantly, meiMIGSDMC1‐HSBP T2 plants had lower HSBP transcript levels that negatively correlated with 420 crossover frequencies (r = −0.83, P = 1.28 × 10−4) (Fig EV2E).
Figure EV2. Meiosis‐specific miRNA‐induced gene silencing (meiMIGS) of HSBP .
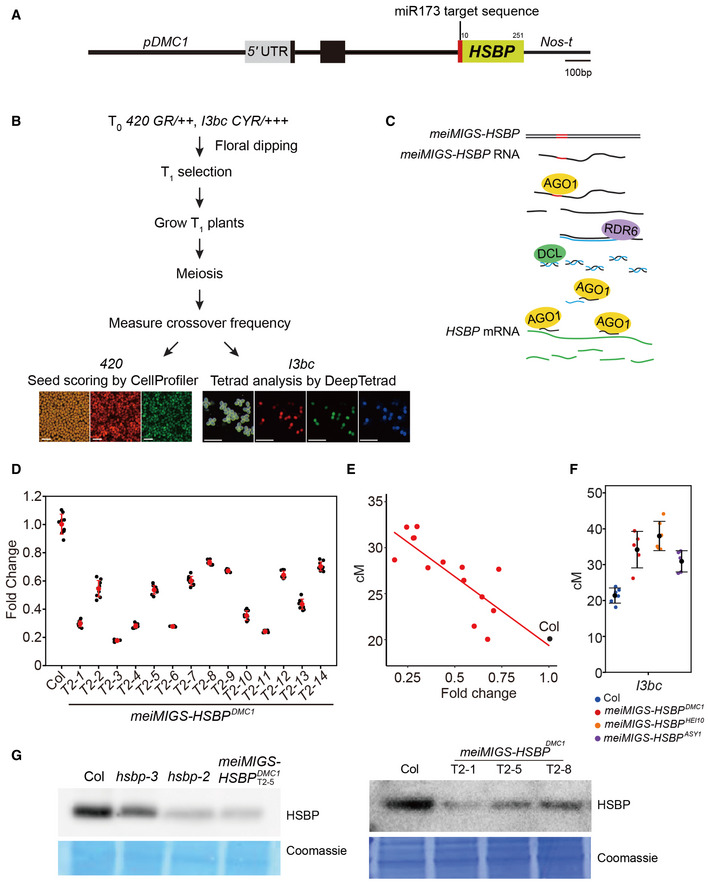
- Schematic diagram of the meiMIGS‐HSBP construct. Scale bar, 100 bp.
- Schematic diagram of the pipeline followed to measure crossover frequency in meiMIGS‐HSBP lines using fluorescent 420 seed or pollen I3bc reporters. Scale bars: 1 mm for seeds, 0.25 mm for pollens.
- Schematic diagram of the meiMIGS‐HSBP mode of action to generate trans‐acting miRNAs during meiosis and silence endogenous HSBP transcripts.
- RT–qPCR analysis of HSBP transcript levels in Col and meiMIGS‐HSBP transgenic lines. TUB2 was used as a reference. Data points (black) represent three biological replicates and three technical repeats per replicate. Red dots and horizontal lines indicate mean ± s.d. of values (one‐sided Welch’s t‐test).
- Correlation between 420 genetic distances (y‐axis, in cM) and HSBP transcript levels in floral buds of Col and meiMIGS‐HSBP lines. The x‐axis indicates fold‐enrichment of HSBP transcript levels compared to those in Col, as determined by RT–qPCR. DMC1 was used as a meiotic gene for normalization. Mean values of triplicate RT–qPCR in Col plants and transgenic lines were used. Col and meiMIGS‐HSBP plants are shown as black and red dots, respectively.
- Crossover frequency (cM) of I3bc in wild‐type Col (blue) and meiMIGS‐HSBP T1 transgenic plants using the meiotic promoters from the genes DMC1 (red), HEI10 (orange), and ASY1 (purple) to drive MIGS‐HSBP expression. Data points (color) indicate cM values from individual plants. Black dots and horizontal lines indicate mean ± s.d. of cM values (one‐sided Welch’s t‐test). n ≥ 5 plants of biological replicates.
- Immunoblot analysis of HSBP abundance in Col, hsbp‐3, hsbp‐2, hsbp‐1, and three meiMIGS‐HSBP T2 transgenic lines in buds. Coomassie‐stained membrane served as a loading control. Experiments were performed at least three times.
Source data are available online for this figure.
We then crossed the 420 hsbp‐3 line with the hsbp‐2 T‐DNA insertion allele to produce F1 hybrid plants (hsbp‐3/hsbp‐2) for an allelism test (Fig 1D). The hsbp‐3/hsbp‐2 F1 hybrid plants exhibited increased 420 crossover frequencies compared to Col plants (t‐test, P = 1.03 × 10−6) (Appendix Fig S3A). However, homozygous hsbp‐2 F2 seeds derived from these F1 plants showed silencing of both fluorescent reporters in the seed coat, which led to altered segregation ratios (Appendix Fig S3B−D), possibly due to the role of HSBP in seed development (Fu et al, 2002; Hsu et al, 2010; Rana et al, 2012). We, therefore, used the three‐color pollen FTL (fluorescence tagged line) I3bc to assess crossover frequencies in hsbp‐2 and hsbp‐3 using DeepTetrad (Lim et al, 2020) (Figs 1I and 2A). We allowed the hsbp‐3/hsbp‐2 F1 plants (I3bc/+++, hsbp‐3/hsbp‐2, qrt1/QRT1) to self‐fertilize and measured crossover frequency in F2 individuals (Fig 1I). Neither hsbp‐2 nor hsbp‐3 mutations led to silencing of the fluorescent reporters in the pollen grains (Appendix Fig S3E). Homozygous plants for hsbp‐3 or, hsbp‐2, as well as hsbp‐3/hsbp‐2 hybrid plants, showed increased crossover frequencies in I3bc compared with Col plants (t‐test, all P < 8.79 × 10−5) but not between them (t‐test, all P > 0.305) (Fig 1I and Appendix Table S6). Together, these results demonstrate that HCR2 encodes HSBP.
Figure 2. hsbp‐3 and meiMIGS‐HSBP increase crossover frequency and reduce interference strength.
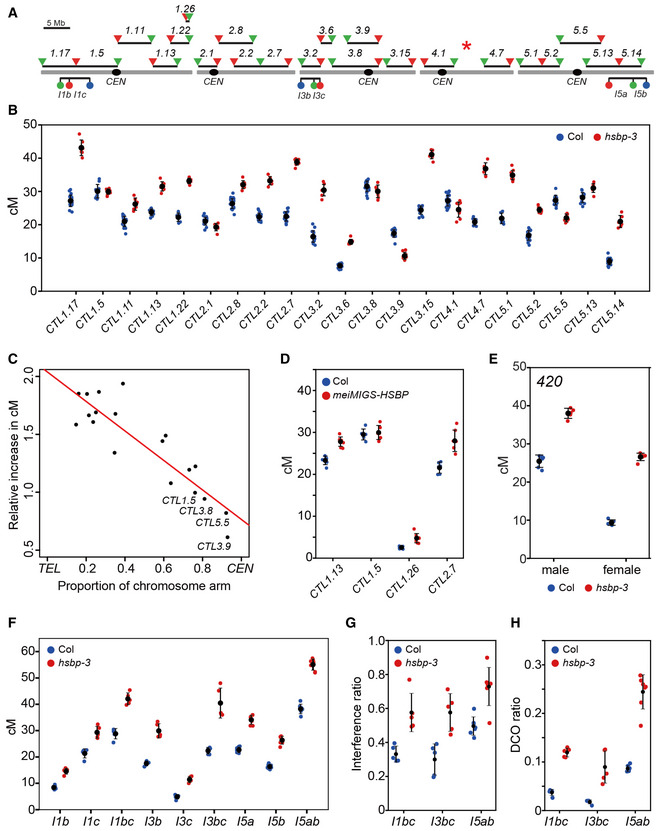
- Seed and pollen FTL T‐DNA intervals throughout the Arabidopsis genome used for crossover frequency measurements. Horizontal lines represent the intervals. Circles and triangles indicate LAT52‐ and NapA‐driven FTL transgenes, respectively. The red asterisk indicates the chromosomal position of hsbp‐3.
- Crossover frequencies of seed FTL/CTL lines in Col (blue) and hsbp‐3 (red). n ≥ 6 plants of biological replicates.
- Correlation between FTL cM changes in hsbp‐3 and the midpoint of the FTL interval analyzed.
- As in (B), crossover frequencies of seed FTL in Col (blue) and meiMIGSDMC1‐HSBP (red). n ≥ 6 plants of biological replicates.
- As in (B), 420 crossover frequencies (cM) in male and female meiosis for Col (blue) and hsbp‐3 (red). n ≥ 5 plants of biological replicates.
- As in (B), crossover frequencies (cM) in pollen FTL I1bc, I3bc, and I5ab in Col and hsbp‐3. n ≥ 5 plants of biological replicates.
- Crossover interference ratios (IFRs) measured using FTL pollen tetrads in Col (blue) and hsbp‐3 (red). n ≥ 5 plants of biological replicates.
- Double crossover (DCO) ratios detected in FTL pollen tetrads in Col (blue) and hsbp‐3 (red). The DCO ratio was calculated as (number of tetrads with more than two crossovers)/(total number of tetrads). n ≥ 5 plants of biological replicates.
Data information: (B, D; E) Mean ± s.d. of cM values are indicated by black dots and horizontal lines (one‐sided Welch’s t‐test). Blue and red dots indicate cM values from individual plants. (G) Mean ± s.d. of IFR values are indicated by black dots and horizontal lines. (H) Mean ± s.d. of DCO ratio values are indicated by black dots and horizontal lines (one‐sided Welch’s t‐test).
hsbp‐3 increases crossover frequency in euchromatic regions
We investigated the effect of hsbp‐3 on crossover frequency in other chromosomal regions. For this, we crossed hsbp‐3 with 22 seed fluorescent recombination reporters, CTLs (Col traffic lines) distributed across the genome, and measured sex‐averaged CTL crossover frequency in individual F2 plants (Fig 2A and Appendix Table S7) (Wu et al, 2015). Homozygous hsbp‐3 plants showed higher crossover frequency than Col plants in CTLs along euchromatic chromosome arms (CTL1.17, CTL1.11, CTL1.13, CTL1.22, CTL2.8, CTL2.2, CTL2.7, CTL3.2, CTL3.6, CTL3.15, CTL4.7, CTL5.1, CTL5.2, and CTL5.14) (t‐test, all P < 3.62 × 10−4), which supports a role for HSBP in repressing crossover frequency outside of the 420 intervals (Fig 2B). However, crossover frequency decreased moderately (CTL2.1, CTL3.9, CTL4.1, CTL5.5) (t‐test, all P < 7.78 × 10−3) or was unchanged (CTL1.5, CTL3.8) (t‐test, CTL1.5 P = 0.847, CTL3.8 P = 0.09) in intervals spanning centromeres (Fig 2B). Indeed, we observed a strong negative correlation between the crossover increase in hsbp‐3 and the proximity of each CTL interval midpoint to the centromere (r = −0.89, R 2 = 0.78, P = 3.145 × 10−7) (Fig 2C). Consistently, the meiMIGS DMC1‐HSBP line also exhibited higher crossover frequencies in the distal intervals CTL1.13, CTL1.26, and CTL2.7 (t‐test, all P < 5.64 × 10−4) but no difference in the centromeric interval CTL1.5 (t‐test, P = 0.598) (Fig 2D and Appendix Table S8).
HSBP limits crossovers in both male and female meiosis
We measured male‐ and female‐specific crossover frequencies by reciprocally crossing 420/++ hsbp‐3 with Col plants. hsbp‐3 significantly elevated 420 crossover frequencies during both male and female meiosis (t‐test, all P < 1.87 × 10−7) (Fig 2E and Appendix Table S9), with a higher crossover frequency increase in female (hsbp‐3, 283%) than male meiosis (hspb‐3, 149%). This result indicated that HSBP restricts crossovers in females more strongly than in males. We further investigated the effects of hsbp‐3 on male crossover frequency using the pollen‐specific FTLs I1bc, I3bc, and I5ab (Fig 2A). The hsbp‐3 mutant showed increased male crossover frequency in all tested FTL intervals (t‐test, all P < 2.35 × 10−4) (Fig 2F and Appendix Table S10). In addition, multiple meiMIGS‐HSBP T1 plants with different meiosis‐specific promoters (DMC1, HEI10, ASY1) displayed elevated I3bc crossover frequency in male meiosis, compared to Col plants (t‐test, all P < 8.38 × 10−3) (Fig EV2F and Appendix Table S11).
hsbp‐3 decreases crossover interference
The crossover interference ratio (IFR) is the ratio between an interval’s map distance (cM) with and without an adjacent crossover, and it can be measured using three‐color pollen FTLs (Francis et al, 2007; Berchowitz & Copenhaver, 2008; Lim et al, 2020). IFR values for FTLs I1bc, I3bc, and I5ab were significantly higher in hsbp compared to their values of Col (t‐test, all P < 5.55 × 10−3), indicating that interference was weaker in hsbp‐3 relative to Col (Fig 2G and Appendix Table S10). Consistently, we detected more double crossovers within FTL intervals in hsbp‐3 compared with those in the Col (t‐test, all P < 8.02 × 10−3) (Fig 2H). However, interference was still evident in hsbp‐3 with IFR values below 1, whereas class II anti‐recombination mutants typically show no interference (IFR = 1) (Crismani et al, 2012; Girard et al, 2015; Séguéla‐Arnaud et al, 2015). These findings indicate that HSBP is required for crossover interference.
Genetic analyses suggest that HSBP restricts class I crossovers
To understand how HSBP limits crossovers, we measured crossover frequency in double or triple mutants between hsbp‐3 and other recombination pathway mutants (Fig 3). We observed an additive increase in crossover frequency in both 420 and CTL1.26 in the fancm hsbp‐3 double mutant compared with either single mutant (t‐test, fancm P = 0.012, hsbp‐3 P = 0.0134) (Fig 3A and B, and Appendix Tables S12 and S13). Similarly, the hcr1 hsbp‐3 double mutant showed a higher crossover frequency in CTL1.26 relative to the single mutants (t‐test, hcr1 P = 4.97 × 10−5, hspb‐3 P = 1.85 × 10−4) (Fig 3B and Appendix Table S13). Using the I3bc FTL, we detected an additive effect of hsbp‐3 on crossover frequency in recq4a recq4b, similar to fancm (t‐test, hsbp‐3 P = 1.30 × 10−4, recq4a recq4b P = 2.49 × 10−3) (Fig 3C and Appendix Table S14). These results indicate that HSBP restricts crossover number independently of FANCM, RECQ4A/4B, and HCR1 (Fig 3A–C). Unlike fancm and recq4a recq4b mutants that restore the low fertility and bivalents of zip4 mutants to Col levels by increasing class II crossovers (Crismani et al, 2012; Séguéla‐Arnaud et al, 2015), hsbp‐3 restored neither zip4 fertility (zip4, ~3.03 seeds/silique; zip4 hsbp‐3, ~2.94 seeds/silique) (Wilcoxon test, P = 0.11) nor bivalents per cell of zip4 (Wilcoxon test, P = 0.17) (Fig 3D–F and Appendix Tables S15 and S16). Furthermore, 420 crossover frequencies in the hsbp‐3 zip4 double mutant did not differ from that of zip4 (t‐test, P = 0.977), indicating that the elevated crossover frequency of hsbp‐3 requires ZIP4 activity (Fig 3A). Together, these genetic analyses indicate that HSBP represses class I crossover formation.
Figure 3. hsbp‐3 leads to an increase in ZMM‐dependent crossovers.
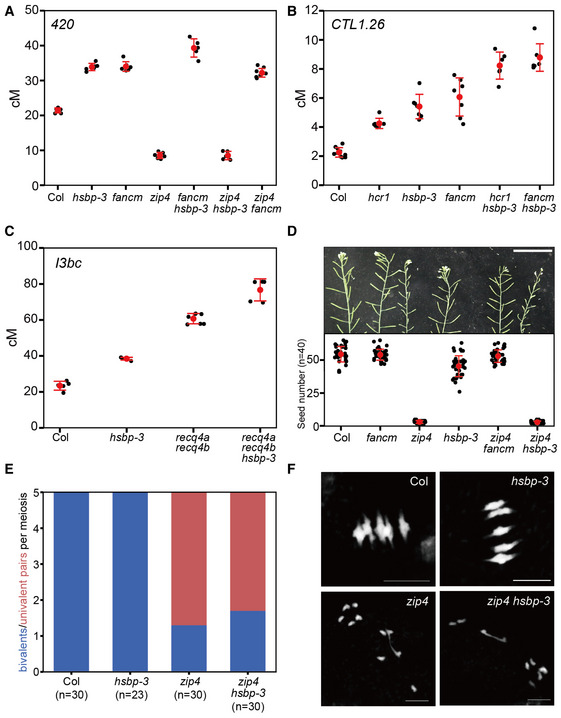
- 420 crossover frequencies (cM) in Col, hsbp‐3, fancm, zip4, fancm hsbp‐3, zip4 hsbp‐3, and zip4 fancm. n ≥ 5 plants of biological replicates.
- As in (A), CTL1.26 crossover frequencies (cM) in Col, hcr1, hsbp‐3, fancm, hcr1 hsbp‐3, and fancm hsbp‐3. n ≥ 6 plants of biological replicates.
- As in (A), I3bc crossover frequencies (cM) in Col, hsbp‐3, recq4a recq4b, and recq4a recq4b hsbp‐3. n ≥ 5 plants of biological replicates.
- Representative silique images and average number of seeds per silique from Col, fancm, zip4, hsbp‐3, zip4 fancm, and zip4 hsbp‐3 plants. Scale bar: 5 cm. Red dots and horizontal lines indicate mean ± s.d. of seed number from siliques. Black dots represent seed number from individual siliques. Significance between genotypes was assessed by Wilcoxon test. n = 40 siliques.
- Average number of bivalents (blue) and pairs of univalent (red) per male meiocyte from Col, hsbp‐3, zip4 and zip4 hsbp‐3. The number of analyzed cells is indicated in parentheses. Significance between genotypes was assessed by Wilcoxon test.
- As in (E), but showing representative metaphase I chromosome spreads stained with DAPI. Scale bar, 10 μm. Images represent three biological replicates.
Data information: (A–C) Red dots and horizontal lines indicate mean ± s.d. of cM values (one‐sided Welch’s t‐test). Black dots represent cM values of individual plants.
Meiotic HSBP knockdown elevates crossovers on chromosome arms in Col/Ler hybrids
Because hsbp‐3 elevated crossover frequency in Col inbred FTL intervals, we investigated the genome‐wide effects of hsbp‐3 on crossover formation in Col/Ler hybrid plants. Accordingly, we mapped genomic crossover sites using genotyping by sequencing (GBS) of F2 individuals derived from a cross between 420 meiMIGS‐HSBP in Col and Ler (Fig 4). We observed increased 420 crossover frequencies in meiMIGS‐HSBP Col/Ler F1 hybrids compared with those in Col/Ler F1 plants (t‐test, P = 8.44 × 10−11) (Fig 4A and B, and Appendix Table S17). We then performed GBS on 288 F2 progeny from one meiMIGS‐HSBP Col/Ler F1 hybrid. Genome‐wide crossover maps of meiMIGS‐HSBP Col/Ler F2 plants revealed more crossovers per individual F2 plant (Wilcoxon test, P = 2.2 × 10−16) and per chromosome, compared with those in Col/Ler F2 plants (n = 240) (Fig 4C and D, and Appendix Table S18). Most of the additional crossovers in meiMIGS‐HSBP occurred within the chromosome arms toward the telomeres (Fig 4E and F), which was consistent with the increased crossover frequency seen in hsbp‐3 FTLs (Fig 2). Collectively, meiotic knockdown of HSBP increased crossovers on chromosome arms in both inbred and hybrid plants. We noticed that the meiMIGS‐HSBP transgenic line might possess a T‐DNA insertion‐mediated chromosomal rearrangement, as evidenced by the suppression of crossovers around the pericentromere and the sharp increase in crossovers at the arms of chromosome 3 (Fig 4F). Therefore, we excluded chromosome 3 in the telomere and centromere analysis (Fig 4E).
Figure 4. Genome‐wide crossover maps in meiMIGS‐HSBP .
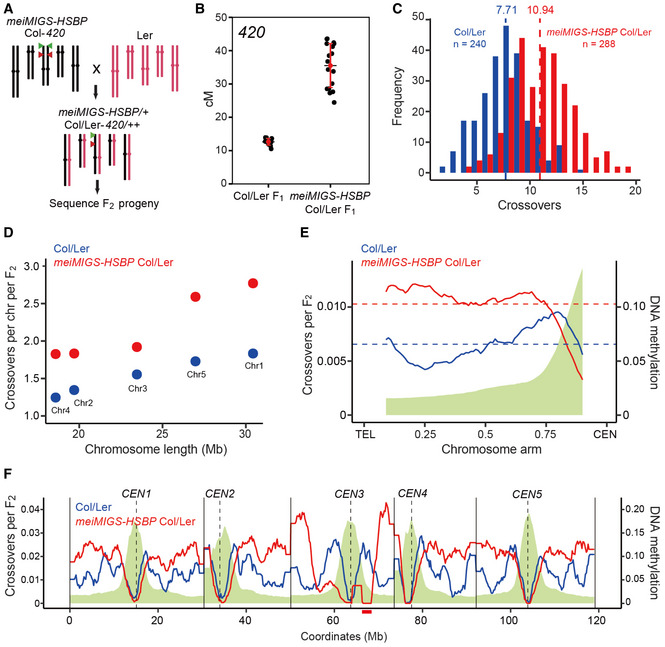
- Schematic diagram of the crossing scheme between meiMIGS‐HSBP Col‐420 (black) and Ler (red) to generate an F2 population for genotyping by sequencing. Green and red triangles indicate the fluorescent reporters in the 420 background on chromosome 3.
- 420 crossover frequencies (in cM) in Col/Ler and meiMIGS‐HSBP Col/Ler F1 hybrids. Red dots and horizontal lines indicate mean ± s.d. of cM values (one‐sided Welch’s t‐test). n ≥ 12 plants of biological replicates.
- Distribution of crossover numbers per F2 individual in Col/Ler (blue) and meiMIGS‐HSBP Col/Ler (red). Vertical dashed lines indicate mean crossover numbers. Significance between genotypes was assessed by one‐sided Welch’s t‐tests.
- Crossover numbers per chromosome in Col/Ler (blue) and meiMIGS‐HSBP Col/Ler (red) F2 populations.
- Normalized crossover frequencies along chromosome arms from the telomere (TEL) to the centromere (CEN) in Col/Ler (blue) and meiMIGS‐HSBP/Ler F2 populations (red). Crossover data for chromosome 3 were excluded due to a possible T‐DNA‐driven chromosome rearrangement. DNA methylation levels are shown in green. Horizontal dashed lines indicate mean values.
- As in (E), without TEL‐CEN scaling. Vertical solid and dashed lines indicate telomeres and centromeres, respectively. The region of T‐DNA‐driven chromosome rearrangement in the pericentromere of chromosome 3 is shown as a solid red underline.
Data information: Significance between genotypes was assessed by one‐sided Wilcoxon tests (D, E).
hsbp and meiMIGS‐HSBP increase HEI10 transcription
Because HSBP interacts with HSF trimers and attenuates their transcriptional activity during the heat shock response (Satyal et al, 1998; Hsu et al, 2010), we performed transcriptome deep sequencing (RNA‐seq) using hsbp‐3 and Col meiocyte‐containing unopened buds (<1 mm) (Fig 5A). Among known meiotic genes, HEI10 and ASY1 transcript levels were significantly higher in hsbp‐3 compared with their levels in Col (Fig 5A). Increased HEI10 transcript levels in hsbp‐3 were consistent with the higher crossover frequencies seen in the mutant (Figs 2 and 3) because HEI10 is a dosage‐dependent pro‐crossover factor in Arabidopsis (Ziolkowski et al, 2017; Serra et al, 2018). We confirmed higher HEI10 and ASY1 transcript levels in hsbp‐3, hsbp‐2, and meiMIGS‐HSBP buds by RT–qPCR (t‐test, all P < 1.39 × 10−2), while DMC1, MLH1, and MUS81 transcript levels were comparable to those of Col (t‐test, P > 0.113) (Appendix Fig S4A and C). To validate the effect of hsbp‐3 on HEI10 transcription during meiosis, we purified male meiocytes and performed RT–qPCR analysis. We again observed elevated HEI10 transcript levels in hsbp‐3 meiocytes compared with those in Col (t‐test, P = 1.15 × 10−9) (Fig 5B). HSBP transcripts were also highly expressed in these purified meiocytes (t‐test, P = 2.93 × 10−14) (Fig 5C) and meiotic buds (t‐test, P = 1.18 × 10−12) (Appendix Fig S4B) compared with their expression in seedlings. Immunoblot analysis of HSBP indicated that HSBP abundance is also higher in buds than in seedlings (Fig EV1C) and reduced in hsbp and meiMIGS‐HSBP buds (Fig EV2G). These results suggest that meiotic HSBP may limit crossover frequency by directly or indirectly repressing HEI10 transcription.
Figure 5. HSBP represses HEI10 transcription via HSF inhibition and DNA methylation.
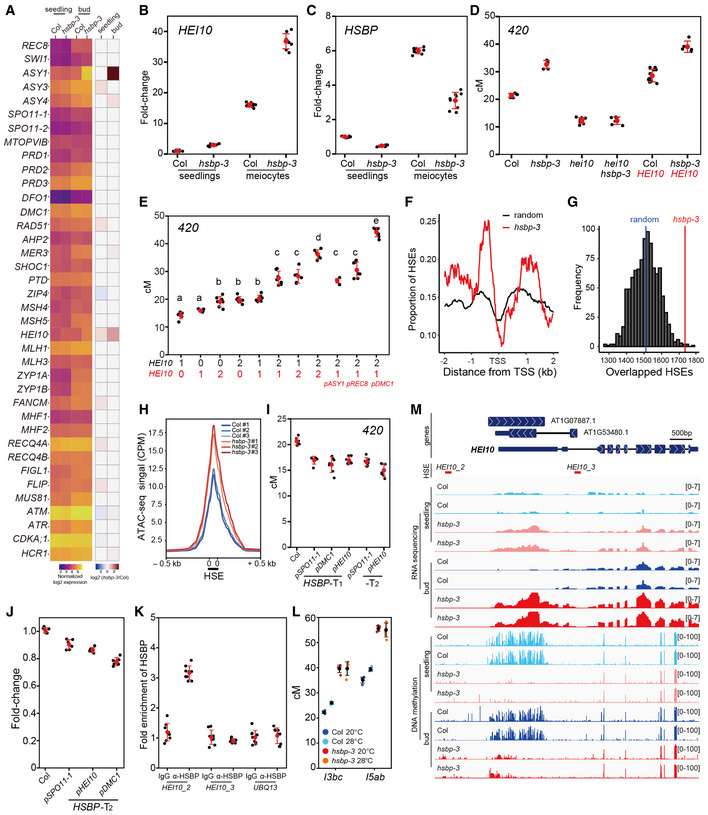
- Heatmap representation of transcript levels for meiotic recombination genes in Col and hsbp‐3 seedlings and buds from RNA‐seq data.
- HEI10 transcript levels in Col and hsbp‐3 meiocytes, compared with seedlings by RT–qPCR. Experiments were performed at least three times. n ≥ 6 two or three technical duplicates of three biological replicates.
- As in (C), HSBP transcript levels. n ≥ 6 two or three technical duplicates of three biological replicates.
- 420 crossover frequencies in Col, hei10, hsbp‐3, hei10 hsbp‐3, HEI10, and HEI10 hsbp‐3. HEI10 (red), HEI10‐myc transgene. n ≥ 6 plants of biological replicates.
- As in (D), plants with different HEI10 dosage and varying meiotic HEI10 expression from the indicated promoters. Black numbers represent HEI10 and the endogenous HEI10 genotype (0, hei10; 1, hei10/HEI10; 2, HEI10/HEI10). Red numbers represent HEI10 and HEI10‐myc transgene copy number using HEI10 or other meiotic gene promoters. One‐way analysis of variance determined significant differences. n ≥ 6 plants of biological replicates.
- Mean coverage of HSE peaks around the transcription start site (TSS) of upregulated genes (n = 983) in hsbp‐3 (red) and 1,000 sets of 983 randomly selected genes (black).
- As in (F), distribution of simulation frequencies (y‐axis) and HSE numbers (x‐axis) in upregulated genes in hsbp‐3 compared with 1,000 simulations of 983 randomly selected genes. Vertical blue line, mean number of the random HSE sets.
- Mean ATAC‐seq signal around HSEs in Col and hsbp‐3 buds. The y‐axis indicates mean CPM (counts per million mapped reads) of ATAC‐seq.
- As in (D), 420 crossover frequencies in Col and transgenic plants expressing HSBP under the indicated promoters. n ≥ 6 plants of biological replicates.
- As in (B), RT–qPCR analysis of HEI10 in Col and meiotic HSBP transgenic plant (T2) buds. n ≥ 6 two or three technical duplicates of three biological replicates.
- HSBP ChIP–qPCR analysis at the HEI10 promoter in buds. The HEI10 primer positions are shown as red lines in (I). UBQ13, negative control. Experiments were performed three times. Data points (black) indicate three technical duplicates of three biological replicates. Red dots and horizontal lines indicate mean ± s.d. values (one‐sided Welch’s t‐test).
- As in (D), but showing crossover frequency of I3bc and I5ab in Col and hsbp‐3 grown under optimal or high temperatures. Black dots and horizontal lines indicate mean ± s.d. of cM values from individual plants. Colored dots represent cM values of individuals. n ≥ 4 plants of biological replicates.
- Integrative genomic viewer window showing the HEI10 region of RNA‐seq and BS‐seq (DNA methylation) data in Col and hsbp‐3.
Data information: (B, C, J) Data points (black) indicate two or three technical duplicates of three biological replicates. Red dots and horizontal lines indicate mean ± s.d. from duplicates (one‐sided Welch’s t‐test). (D, E, I) Red dots and horizontal lines indicate mean ± s.d. of cM values from individual plants (one‐sided Welch’s t‐test). Black dots represent cM values of individual plants.
hsbp‐3 increases HEI10‐dependent crossovers
To test the above hypothesis genetically, we generated a hsbp‐3 hei10 double mutant. 420 crossover frequencies were the same in the hsbp‐3 hei10 double mutant and in hei10 (t‐test, P = 0.985) (Fig 5D and Appendix Table S19), indicating that the increased crossovers in hsbp‐3 depend on HEI10 activity. 420 crossover frequencies also increased additively in Col and hsbp‐3 upon the introduction of a single copy of HEI10‐myc transgene (HEI10, red) (Ziolkowski et al, 2017) (t‐test, hsbp‐3 P = 6.41 × 10−4, HEI10‐myc P = 1.16 × 10−5) (Fig 5D and Appendix Table S19). We confirmed the effect of HEI10 copy number on increasing crossover frequency using a HEI10‐myc transgenic line (one‐way ANOVA test, all P < 1.39 × 10−8) (Fig 5E and Appendix Table S20) as previously reported (Ziolkowski et al, 2017). Varying HEI10 transcript levels using the promoters of other meiotic genes (ASY1, REC8, DMC1) also increased 420 crossover frequencies to variable extents (t‐test, all P < 1.61 × 10−5) (Fig 5E and Appendix Table S20).
HSBP directly represses HEI10 transcription by binding and inhibiting HSFs
Because HSBP inhibits HSF activity by direct binding (Morimoto, 1998; Satyal et al, 1998), we investigated whether HSBP shares target genes with HSFs. We used published genome‐wide HSE (heat stress element) maps from DNA affinity purification sequencing (DAP‐seq) of HSFs (O’Malley et al, 2016). We plotted HSE peaks within 2‐kb windows centered on transcription start sites of genes (n = 983) that are upregulated in hsbp‐3 (Fig 5F and G). We observed a significant enrichment of HSEs in the promoters of these upregulated genes compared with the mean coverage value of HSEs from 1,000 simulations with the same number (n = 983) of randomly selected genes (permutation test, P < 2.2 × 10−16) (Fig 5F and G), which suggests that HSBP and HSFs bind to a common set of genes.
To investigate if HSBP and HSFs control HEI10 transcription in vivo, we performed protoplast transient transfection assays for HSFs, followed by RT–qPCR analysis of HEI10 transcript levels. We selected HSFA1a and HSFA7a among the class A HSF activator family because they are highly expressed in meiotic buds and HSFA7a was induced in hsbp‐3 (Fig EV3A). Transient expression of HSFA1a or HSFA7a increased HEI10 transcription (Fig EV3B and C). Importantly, HSBP inhibited HSF‐mediated HEI10 transcriptional activation when HSBP and HSF were co‐transfected in protoplasts (Fig EV3B and C). To further examine the inhibition of HSF activity by HSBP, we performed ATAC‐seq (assay of transposase accessible chromatin sequencing) in Col and hsbp‐3 buds to analyze DNA accessibility around 42,258 HSEs (Fig 5H) (O’Malley et al, 2016). hsbp‐3 showed elevated DNA accessibility around the HSEs, compared with Col, indicating that HSBP attenuates HSF DNA‐binding and transcriptional activities (Fig 5H). To validate the inhibitory effect of HSBP on crossover frequency in planta, we generated transgenic 420/++ plants that express HSBP additively using the SPO11‐1, DMC1, or HEI10 promoters. These transgenic T1 and T2 plants exhibited lower 420 crossover frequencies (t‐test, all P < 1.03 × 10−5) and lower HEI10 transcript levels (t‐test, all P < 2.78 × 10−4) compared with Col plants (Fig 5I and J, and Appendix Table S21), suggesting that HEI10 transcription is controlled by an HSF–HSBP transcriptional module whereby HSBP inhibits HSF activity during meiosis.
Figure EV3. HSBP represses HEI10 transcription by inhibiting HSFs.
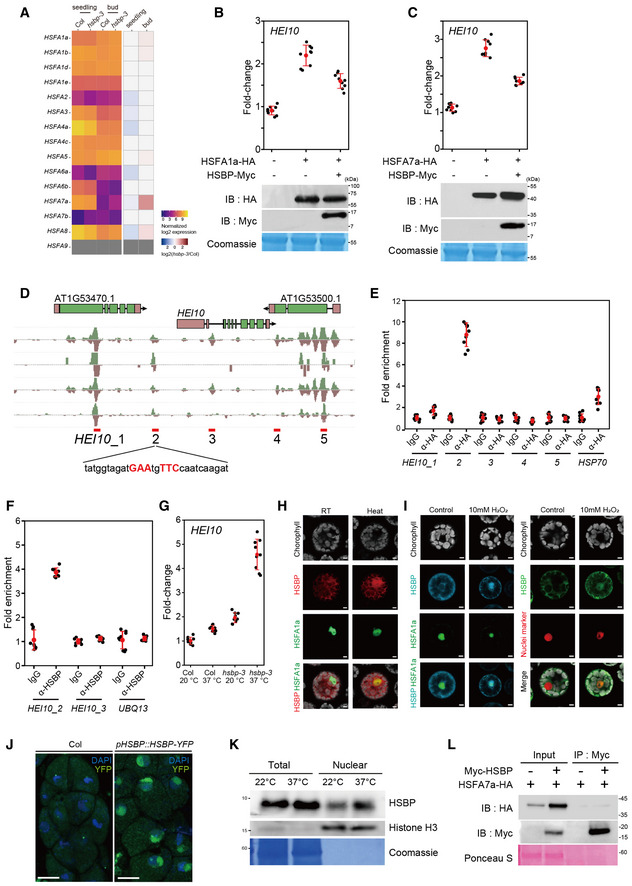
- Heatmap representation of transcript levels in Col‐0 and hsbp‐3 for Arabidopsis class A HSF genes in seedlings and buds from RNA‐seq data.
- RT–qPCR analysis of relative HEI10 transcript levels following transient transfection of HSFA1a and HSBP effector constructs in Arabidopsis protoplasts and after heat treatment (40°C for 1 h). Immunoblots of HSF1a‐HA and HSBP‐Myc in transfected protoplasts. Coomassie‐stained membrane served as a loading control.
- As in B, for HSFA7a and HSBP.
- DAP‐seq data of HSFs at HEI10 regulatory regions. Candidate HSEs (HEI10_1 to 5) around HEI10 are shown.
- As in (B), but showing ChIP–qPCR analysis of HSF7a at HEI10 regulatory regions in protoplasts expressing HSF7a‐HA. IgG was used as a negative control.
- As in (B), but showing HSBP ChIP–qPCR analysis in heat‐treated seedlings using anti‐HSBP antibody. Seedlings (10‐day) were incubated at 37°C for 3 h and used for ChIP analysis.
- RT–qPCR analysis of relative HEI10 transcript levels in Col and hsbp‐3 after heat treatment. Seedlings (10‐day‐old) were incubated at 37°C for 4 h. Data points (color) indicate values of replicates. Black dots and horizontal lines mean ± s.d. values (one‐sided Welch’s t‐test). n ≥ 6 two or three technical duplicates of three biological replicates.
- Co‐localization of the fluorescent fusion proteins HSBP‐RFP and HSFA1a‐GFP in protoplasts. Heat indicates incubation of transfected protoplasts at 40°C for 1 h. Scale bars: 5 μm.
- As in (H), but showing HSBP‐CFP and HSFA1a‐GFP (left), and HSBP‐GFP and nucleus marker (ARR2‐mRFP) (right) after H2O2 treatment. Scale bars: 5 μm.
- Nuclear location of HSBP in male meiocytes of HSBPpro:HSBP‐YFP plants. Nuclei spreads were stained with DAPI. Scale bars: 10 μm.
- Immunoblot analysis of HSBP and histone H3 levels using total proteins and nuclear extracts of Col seedlings. Seedlings (10 days old) were incubated at 37°C for 3 h. The Coomassie‐stained membrane was used as a loading control. Experiments were performed at least three times.
- Co‐immunoprecipitation analysis of Myc‐HSBP with HSF7a‐HA. IB, immunoblot; IP, immunoprecipitation. Ponceau S staining of the membrane, a loading control. Experiments were performed at least three times.
Data information: (B, C, E, F) Experiments were performed three times. Data points (black) indicate three technical duplicates of three biological replicates. Red dots and horizontal lines indicate mean ± s.d. values (one‐sided Welch’s t‐test).
Source data are available online for this figure.
To examine if HEI10 transcription is controlled directly by HSFs and HSBP, we performed chromatin immunoprecipitation, followed by qPCR analysis (ChIP–qPCR) for HSFA7a at the HEI10 locus using a protoplast transient assay and HSF DAP‐seq information (Fig EV3D and E) (O’Malley et al, 2016). We observed a significant enrichment of HSFA7a at one HSE within the HEI10 promoter, thus defining an in vivo binding site of HSFA7a (t‐test, HEI10_2 P = 1.17 × 10−8) (Fig EV3E). Next, we performed ChIP–qPCR analysis for HSBP at the HEI10 promoter in heat‐treated seedlings (37°C, 4 h) and unopened buds (Figs 5K and EV3F). HSBP was enriched at the same HSE in the HEI10 promoter in both buds and seedlings (t‐test, buds, P = 5.29 × 10−12; seedlings, P = 1.30 × 10−9), which demonstrated that HSBP directly represses HEI10 transcription. Exposure to high temperature (37°C) induced HEI10 transcription even in Col seedlings (Fig EV3G), but the hsbp‐3 mutant seedlings displayed a de‐repression of HEI10 transcription under normal growth temperature (20°C), and this was exacerbated at high temperature (Fig EV3G). We also confirmed the high temperature and hydrogen peroxide (H2O2)‐mediated translocation of HSBP from the cytosol to the nucleus, and the co‐localization and co‐immunoprecipitation of HSBP with HSF proteins in protoplasts (EV3H, I, K and L) (Hsu et al, 2010). These results indicate that HSBP represses HEI10 transcription directly by binding and attenuating HSF function at the HEI10 promoter.
HSBP is required for temperature‐sensitive crossover control
High temperature increases class I crossovers compared to the optimal growth temperature of approximately 18°C in Arabidopsis (Lloyd et al, 2018; Modliszewski et al, 2018). We, therefore, examined the effect of temperature (28°C versus 20°C) on crossover frequency in Col and hsbp‐3 using the FTLs I3bc and I5ab. We determined that high temperature increased crossover frequency moderately in I3bc (116.7%, t‐test, P = 3.24 × 10−5) and I5ab (109.7%, t‐test, P = 2.61 × 10−4) as previously reported (Lloyd et al, 2018; Modliszewski et al, 2018), whereas hsbp‐3 showed the same high crossover frequency at both temperatures (I3bc, 98.5%, t‐test, P = 0.95; I5ab,100.8%, t‐test, P = 0.637) (Fig 5L and Appendix Table S22). The effect of high temperature on crossovers was thus compromised in hsbp‐3, indicating that HSBP contributes to the control of crossover formation in response to changes in temperature.
HSBP is required for 5′ UTR DNA methylation and transcriptional repression of HEI10
DNA cytosine methylation was reported to be enriched in the HEI10 5′ untranslated region (5′ UTR) in Col plants (Kawakatsu et al, 2016). To examine if HSBP controls HEI10 transcription via DNA methylation, we performed bisulfite sequencing (BS‐seq) using seedlings and unopened buds of Col and hsbp‐3 (Fig 5M and Appendix Fig S6). Intriguingly, hsbp‐3 led to a loss of DNA methylation at the HEI10 5′ UTR in both seedlings and buds compared with Col (Fig 5M). We also found that the expression of genes associated with DNA demethylation pathways (DEMETER, ROS1, DML2, and IDM1) is induced in hsbp‐3 buds, which may contribute to the loss of methylation in a subset of genes including HEI10 (Appendix Fig S6D−F). In Col tissues, DNA methylation levels at the HEI10 5′ UTR were higher in seedlings than in buds, suggesting that DNA methylation inhibits HEI10 transcription and decreases during early meiosis. Consistent with the BS‐seq results, the RNA‐seq data demonstrated that HEI10 transcript levels are 16‐fold higher in Col meiocytes compared with seedlings and were also higher in hsbp‐3 seedlings and buds relative to Col (Fig 5M). To examine if HSBP is required for maintenance of the DNA methylation at the HEI10 5′ UTR, we performed McrBC–qPCR analysis with the cytosine methylation‐sensitive endonuclease McrBC using seedlings and unopened buds for Col, hsbp‐3, hsbp‐2, and meiMIGS‐HSBP (Appendix Fig S5A). McrBC–qPCR showed that both hsbp‐3 and hsbp‐2 had lower DNA methylation at the HEI10 5′ UTR in seedlings and buds compared with the DNA methylation in Col (Appendix Fig S5A). meiMIGS‐HSBP plants showed a sharp reduction (34.4%) in DNA methylation at the HEI10 5′ UTR in buds but a modest reduction (7.78%) in seedlings (Appendix Figs S4A and S5A). Consistently, meiMIGS‐HSBP did not increase HEI10 transcript levels in seedlings to the same extent as hsbp‐3 or hsbp‐2 (Appendix Fig S4A).
To test the effect of DNA hypomethylation at the HEI10 5′ UTR on HEI10 transcription and crossover frequency, we generated 420/++ plants with epi‐alleles at the HEI10 5′ UTR by crossing 420 to met1 mutant (Appendix Fig S5B). Hypomethylated alleles at the HEI10 5′ UTR exhibited higher 420 crossover frequencies and HEI10 transcription than Col (t‐test, 420 all P < 2.85 × 10−5) (Appendix Fig S5C−E and Table S23). We also identified natural epigenetic variation at the HEI10 5′ UTR in Arabidopsis accession C24 (Kawakatsu et al, 2016), with a loss of DNA methylation that resulted in higher HEI10 transcript levels in C24 seedlings and buds, relative to Col and Cvi (Appendix Fig S5F and G). Together, these results show that HSBP is required to maintain DNA hypermethylation at the HEI10 5′ UTR in both somatic tissue and meiotic buds, and natural variations likely contribute to changes in DNA methylation of HEI10 5′ UTR.
hsbp‐3 shows higher MLH1 and HEI10 foci
We investigated meiosis cytologically using Arabidopsis male chromosome spreads (Fig 6). DAPI (4′,6‐diamidino‐2‐phenylindole) staining of male meiocytes revealed no significant differences between the Col and hsbp‐3, with normal synapsis, bivalent formation, and chromosome segregation (Fig 6A). The hsbp‐3 plants produced shorter siliques and had lower seed fertility (t‐test, silique all P < 1.31 × 10−17, fertility all P < 6.53 × 10−6) (Fig 6B and C, and Appendix Tables S24 and S25), as previously described for other hsbp alleles (Hsu et al, 2010). Pollen viability of hsbp‐3 and hsbp‐2 did not differ from that of Col pollen, as evidenced by Alexander staining (Fig 6D and Appendix Table S26) (t‐test, hsbp‐3 P = 0.465, hsbp‐2 P = 0.334), suggesting that reduced fertility in hsbp‐3 and hsbp‐2 mutants may result from seed abortion during embryogenesis.
Figure 6. Increased MlH1 and HEI10 foci in hsbp‐3 .
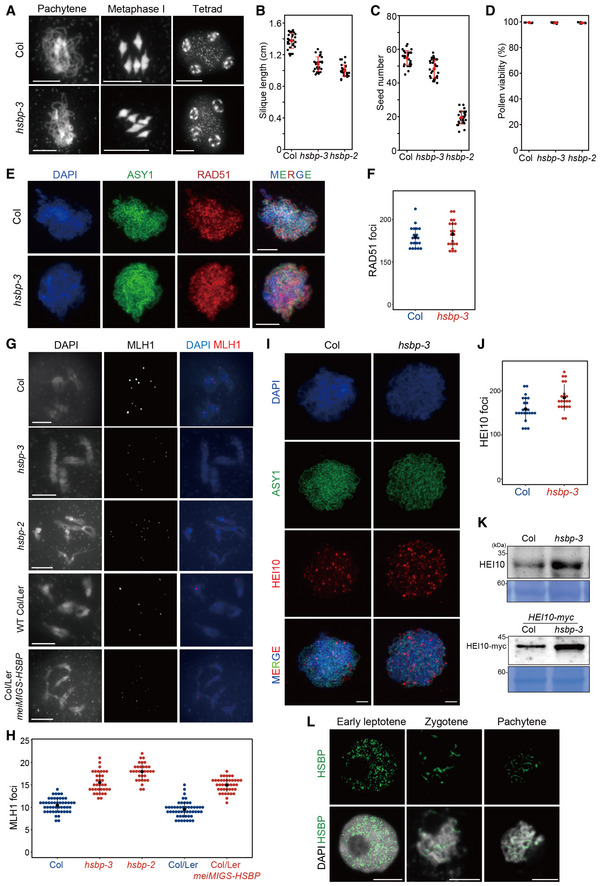
-
ARepresentative images of meiocyte spreads stained with DAPI in Col and hsbp‐3 at the indicated meiotic stages. Scale bars: 10 μm.
-
B–DSilique lengths (B), seed numbers (C), and pollen viability (D) in Col, hsbp‐3, and hsbp‐2. Red dots and horizontal lines indicate mean ± s.d. of cM values (one‐sided Welch’s t‐test). n = 30 siliques of biological replicates (B, C). n = 10 plants of biological replicates (D).
-
ERepresentative images of ASY1 (green) and RAD51 (red) immunostaining in Col and hsbp‐3. Nuclei spreads were stained with DAPI (blue). Scale bars: 10 μm.
-
FQuantification of RAD51 foci numbers per cell in Col (blue) and hsbp‐3 (red). n = 20 cells of biological replicates.
-
GRepresentative images of MLH1 (red) immunostaining in Col, hsbp‐3, hsbp‐2, Col/Ler, and meiMIGS‐HSBP Col/Ler. Nuclear DNA was stained with DAPI. Scale bar: 5 μm.
-
HQuantification of the number of MLH1 foci per cell is shown in (G). n ≥ 32 cells of biological replicates.
-
IRepresentative images of HEI10 (red) and ASY1 (green) immunostaining in Col and hsbp‐3. Scale bar: 2 μm.
-
JQuantification of the number of HEI10 foci per cell is shown in (I). n ≥ 21 cells of biological replicates.
-
KImmunoblot analysis of HEI10 and HEI10‐Myc in Col, hsbp‐3, HEI10‐myc, and HEI10‐myc plants. Experiments were performed at least three times.
-
LRepresentative images of HSBP (green) immunostaining during meiosis. Nuclei spreads were stained with DAPI. Scale bars: 5 μm.
Data information: (F, H, J) Black dots and horizontal lines indicate mean ± s.d. of values (Wilcoxon test).
Source data are available online for this figure.
We counted the number of RAD51 recombinase foci marking meiotic DSB sites along chromosome axes at the leptotene stage using co‐immunostaining with ASY1, a marker of the chromosome axis (Fig 6E). hsbp‐3 and Col had comparable numbers of RAD51 foci (Wilcoxon test, P = 0.588) (Fig 6F and Appendix Table S27). We then investigated the number of MLH1 foci, which mark class I crossover sites (Fig 6G). Significantly more MLH1 foci accumulated in hsbp‐3, hsbp‐2, and meiMIGS‐HSBP Col/Ler than in Col and Col/Ler plants (Wilcoxon test, hsbp‐3, P = 6.81 × 10−14; hsbp‐2, P = 2.95 × 10−14; meiMIGS‐HSBP, P = 4.89 × 10−15) (Fig 6H and Appendix Table S28). We also counted the number of immunostained HEI10 foci per cell from the zygotene to the mid‐pachytene stage (Fig 6I and J). hsbp‐3 showed more HEI10 foci per cell than Col (Wilcoxon test, P = 4.45 × 10−3) (Fig 6J and Appendix Table S29), which correlated with higher HEI10 and HEI10‐myc abundance in the hsbp‐3 background, compared with control plants, as determined by immunoblot analysis (Fig 6K). This observation is consistent with increased HEI10 transcription in hsbp‐3 and the genetic interactions of HSBP with meiotic recombination mutants (Figs 3 and 5). Finally, we determined the localization of HSBP during meiosis using immunostaining with an anti‐HSBP antibody and HSBPpro:HSBP‐YFP plants (Figs 6L and EV3J). We detected abundant HSBP proteins in the nucleus from leptotene to pachytene that overlap with DAPI signals in Col (Fig 6L), whereas HSBP abundance was low in hsbp‐3 and very low in hsbp‐2, as expected from RT–PCR and immunoblot results (Fig 1E and F, and EV3J, and EV4).
Figure EV4. Representative images of HSBP immunostaining in wild‐type Col, hsbp‐3, and hsbp‐2 during meiosis.
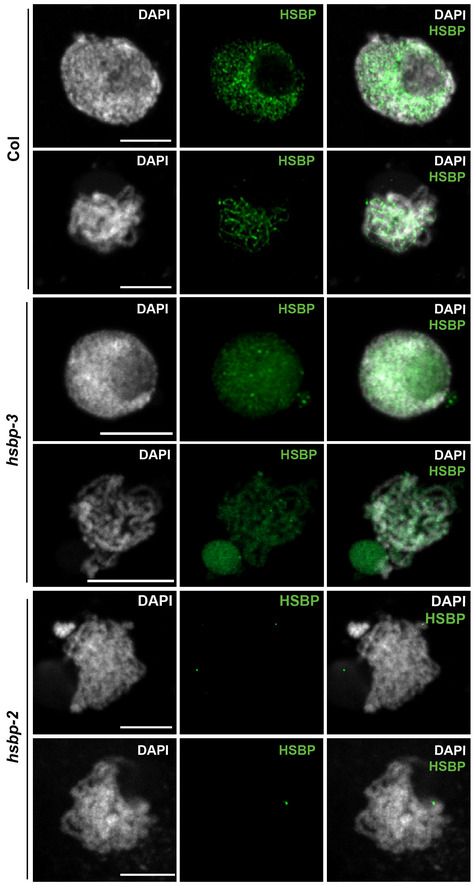
Nuclei spreads were stained with DAPI (white). HSBP signals (green) were reduced in hsbp‐3 and rarely detected in hsbp‐2. Scale bars: 5 μm. Images representative of three biological replicates.
Discussion
We demonstrate that the negative regulator of heat shock response HSBP directly represses HEI10 transcription and restricts crossovers. HSBP forms hexamers that bind to the hydrophobic heptad repeat of HSFs, leading to their dissociation from active HSF trimers to inactive monomers, thereby attenuating HSF transcriptional and DNA‐binding activities (Morimoto, 1998). In plants, transcript levels of heat shock‐responsive genes increase in hsbp mutants and HSBP translocates from the cytosol to the nucleus at high temperatures (Fu et al, 2002; Hsu et al, 2010; Rana et al, 2012). Notably, Arabidopsis HSBP and its rice orthologs are highly expressed in reproductive tissues and are required for embryogenesis (Fu et al, 2002; Hsu et al, 2010; Rana et al, 2012). We determined that Arabidopsis HSBP is abundant in meiocytes and localizes to the nucleus during meiosis (Figs 5 and 6). Our identification of HSBP as a HEI10 transcriptional repressor suggests a possible model for the control of HEI10 expression, whereby HSFs activate HEI10 transcription during early meiosis I, and the activity of HSFs is simultaneously or subsequently attenuated by HSBP, which may determine transcript levels of HEI10 (Fig EV5A–C). We propose that a transcriptional module of HSFs and HSBP contributes to the regulation of meiotic HEI10 transcription, and control of HEI10 protein level during early meiosis I (Chelysheva et al, 2012).
Figure EV5. Proposed model of HSBP for control of HEI10 transcription.
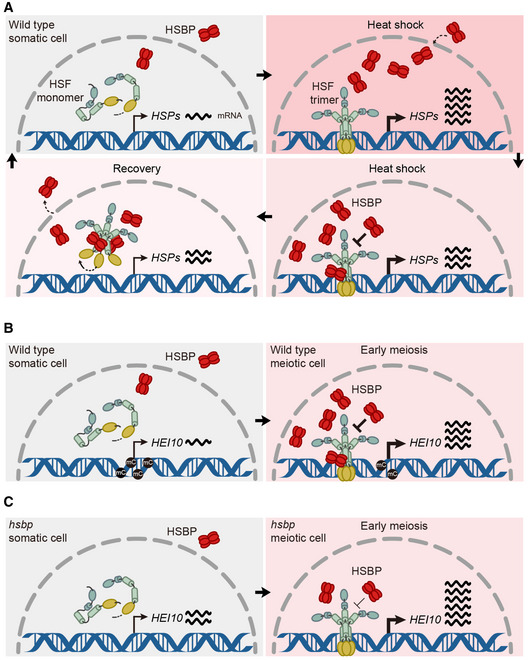
- In somatic cells during heat shock, HSBPs translocate from the cytosol to the nucleus to attenuate the activity of the HSF (heat shock factor) trimer. The activation‐attenuation cycle of HSF‐HSBP controls the transcription of heat shock protein genes (HSPs) and many other genes including HEI10.
- In meiotic cells, during early meiosis, HSFs are highly expressed and activated by unknown developmental factors and signals, potentially including reactive oxygen species (ROS). Plant meiocytes are surrounded by multi‐layered cells and thick callose cell walls, which may induce hypoxia and ROS. Active HSFs contribute to the transcriptional induction of HEI10, in addition to many other genes. Simultaneously or subsequently, HSBP accumulates to high levels and moves to the nucleus during early meiosis. HSBP hexamers bind to HSF trimers and attenuate HSF activity, which decreases HEI10 transcription. The protein levels and activities of HSFs and HSBP are likely important for determining transcript levels of HEI10 during early meiosis.
- In hsbp‐3 meiotic cells, HSBP levels are lower than in wild type during early meiosis, which leads to higher HEI10 transcripts, HEI10 protein levels, and class I crossovers. The lack of DNA methylation at the HEI10 5′ UTR contributes to the initiation of meiosis with higher HEI10 transcript levels in hsbp‐3.
The cycle of HSF activity is dependent on high temperature through trimerization and nuclear translocation, DNA‐binding, and post‐translational modifications (Gomez‐Pastor et al, 2018). In addition to high temperature, HSFs are activated by developmental signals, including reactive oxygen species (ROS) (Ahn & Thiele, 2003; Giesguth et al, 2015; Guo et al, 2016; Gomez‐Pastor et al, 2018). In maize anthers, hypoxia facilitates somatic cells to differentiate as meiocytes (Kelliher & Walbot, 2012). Single‐cell RNA‐seq showed sharp and gradual expression patterns of meiotic recombination genes in maize; however, it remains largely unknown how developmental factors or signals control transcriptional changes of meiotic genes during plant meiosis (Nelms & Walbot, 2019). High HSBP expression levels and nuclear localization of HSBP observed here in meiocytes suggest that HSBP and HSFs may share meiotic signals such as ROS with other transcriptional regulators during transcriptional control of HEI10 and other meiotic genes, including ASY1. Ambient temperatures outside of the optimal range may induce HSF activation and affect the developmental factors that increase class I crossovers in Arabidopsis (Choi et al, 2013; Lloyd et al, 2018; Modliszewski et al, 2018). In barley, a modest temperature shift leads to a higher number of interstitial chiasmata, indicating a conserved temperature effect on crossover formation (Higgins et al, 2012). However, high temperatures only modestly promoted class I crossovers (approximately 10–15%) in Arabidopsis, which is likely due to the inhibitory and buffering roles of HSBP on HSF activity for HEI10 transcriptional control.
DNA methylation at the HEI10 5′ UTR was reduced in hsbp‐3, which correlated with higher HEI10 transcript levels. We found that HSBP is required to maintain DNA hypermethylation at the HEI10 5′ UTR; however, it remains unclear how HSBP maintains DNA hypermethylation specifically at the HEI10 5′ UTR. It is likely that transcription of HEI10 contributes to reduce DNA methylation of the HEI10 5′ UTR during meiosis. hsbp might disrupt the cycle of HSF activity during meiosis, seed development, and responses to diverse environmental stresses. Therefore, hspb may cause the continuous production or accumulation of developmental and environmental stress signals such as protein misfolding and ROS, affecting DNA methylation via misregulation of genes involved in small RNA and DNA methylation pathways (Fig 5M and Appendix Figs S5 and S6F). A modest decrease in crossovers at pericentromeres and centromeres in hsbp‐3 and meiMIGS‐HSBP is likely due to a slight increase in DNA methylation via upregulation of the genes involved in transposon‐associated small RNA production and DNA methylation (Figs 2B and 4E, and Appendix Fig S6A−C and F). It is also worth noting that hsbp mutants may affect a newly identified epigenetic protein complex comprising a J‐domain protein and HSP70 in plants (Ichino et al, 2021) because HSBP associates with HSP70 (Satyal et al, 1998). Determining whether and how HSBP, HSFs, HSPs, and temperature interact to modulate transcription and the epigenetic landscape in Arabidopsis accessions will be instrumental to our understanding of local adaptation and crossover change.
We determined that HSBP represses class I crossovers, adding to the previously described HCR1 and ZYP1 (Capilla‐Pérez et al, 2021; France et al, 2021; Nageswaran et al, 2021). Genetic disruption of HSBP orthologs using genome editing or RNA interference may increase crossovers and accelerate breeding in crop species. Importantly, our findings shed light on how the evolutionarily conserved transcriptional regulators of HSFs and HSBP have been hitchhiked to control transcription during meiosis, epigenetic information, and crossover recombination in plants and other eukaryotes (Abane & Mezger, 2010).
Materials and Methods
Plant materials
The Arabidopsis (Arabidopsis thaliana) accession Col‐0 was used as the wild type and grown in controlled growth rooms (20°C, 50–60% humidity, and 16‐h‐light/8‐h‐dark photoperiod). Seed and pollen FTL lines were used as previously described (Melamed‐Bessudo et al, 2005; Wu et al, 2015). The T‐DNA insertion lines hsbp‐2 (SALK_093051) (Hsu et al, 2010), zip4‐2 (SALK_068052) (Chelysheva et al, 2012), and the fancm‐1 mutant (Crismani et al, 2012) were provided by the Arabidopsis Biological Resource Center (ABRC). Genotyping of hcr2 was performed by PCR using oligonucleotides hcr2‐geno F and R, followed by SspI (NEB, UK) restriction endonuclease digestion. Genotyping of hsbp‐2 was performed by PCR using primers hsbp‐2 geno_F and R for Col and hsbp‐2 geno_R and LBb1.3 for the T‐DNA allele. The oligonucleotides used for genotyping, plasmid constructs, and experiments in this study are listed in Appendix Table S30.
Isolation and mapping of hcr2
The forward genetic screen and mapping of EMS‐derived hcr2 in the 420 GR/++ hemizygous background were performed as described previously (Nageswaran et al, 2021). To map hcr2, the mutant hcr2 in the 420 reporter background (hcr2 420 GR/GR) was backcrossed to Col. The resulting F1 plants (hcr2/HCR2; 420 GR/++) were allowed to self‐fertilize to produce BC1F2 populations (Fig 1 and Appendix Fig S1). F3 seeds from individual BC1F2 plants were harvested and used to measure the 420 crossover frequencies. Fifty F2 plants with high crossover rates, as determined by 420 crossover frequencies in their F3 seeds, were selected and their BC1F3 seeds were pooled. Nuclear genomic DNA (gDNA) of pooled F3 seedlings was isolated and used to construct a DNA sequencing library as described (Nageswaran et al, 2021). The SHOREmap (v.3.0) pipeline was applied to map candidate mutations responsible for the hcr2 (hspb‐3) phenotype, as described (Nageswaran et al, 2021).
Genetic complementation of hcr2 by a genomic copy of HSBP
A 3.8‐kb HSBP gDNA fragment including the promoter (1.0‐kb) and coding regions was PCR amplified using primers HSBP‐genomic F and R (Appendix Table S30). For the HSBP‐myc (6x mycs) transgenic line, the HSBP promoter and coding region without stop codon were PCR amplified using HSBP‐genomic F and HSBP‐myc R primers. The resulting PCR products were cloned into the binary vector pPZP211‐6x myc, which harbors the nopaline synthase (NOS) terminator, as described (Choi et al, 2018). The pPZP211‐HSBP and pPZP211‐HSBP‐myc constructs were electroporated into Agrobacterium (Agrobacterium tumefaciens) strain GV3101‐pSOUP and transformed into Arabidopsis 420/++ F1 Col plants by the floral dip method. T1 plants were selected for kanamycin resistance, grown, and measured for 420 crossover frequencies.
Measurement of crossover frequency using fluorescent seed and pollen FTLs
The CellProfiler image analysis pipeline was used to measure crossover frequency (cM) by analyzing the number of fluorescent and non‐fluorescent seeds from FTL/++ hemizygous plants (Carpenter et al, 2006; Ziolkowski et al, 2015). Crossover frequency (in cM) was calculated by counting green‐alone fluorescent seeds (NGreen), red‐alone fluorescent seeds (NRed), and total seeds (NTotal) using the formula (Melamed‐Bessudo et al, 2005; Ziolkowski et al, 2015). Welch’s t‐test was used to determine the significance of differences in crossover frequency between genotypes. Pollen tetrad FTL‐based measurement of crossover frequency and interference ratio (IFR) were performed using DeepTetrad and pollen FTLs in the qrt1 mutant background, as described (Berchowitz & Copenhaver, 2008; Lim et al, 2020).
Generation of meiMIGS‐HSBP and meiotic HSBP transgenic plants
The vectors for meiosis‐specific microRNA‐mediated gene silencing (meiMIGS) transgenic plants were constructed using Golden Gate cloning, as described (Nageswaran et al, 2021). The HSBP coding sequence (At4g15802) was cloned into the Lv0 vector (pICH41331) following amplification using EJ‐HSBP‐F forward primers, which include the miR173 target sequence and EJ‐HSBP‐R reverse primers (Appendix Table S30). The Lv2 binary vector was electroporated into Agrobacterium strain GV3101‐pSOUP and transformed into Arabidopsis via floral dipping. The promoters of meiotic genes were cloned into Lv0 vectors to drive meiMIGS‐HSBP expression during meiosis. To generate transgenic plants that additively express HSBP, the Lv0 vectors with the DMC1, SPO11‐1, or HEI10 promoters were assembled individually into the Lv1 vector with HSBP Lv0 and pICH41421 terminator vector and subsequently assembled to Lv2 binary vectors.
RT–qPCR analysis
Total RNA was isolated using TRIzol reagent (Invitrogen) and used for reverse‐transcriptase quantitative PCR using a reverse transcription kit (enzynomics, EZ405S). Total RNA of Arabidopsis male meiocytes was isolated from stage 9 floral buds by gently squeezing between a glass slide and coverslip as described (Walker et al, 2018). Quantitative PCR was performed using a CFX real‐time PCR detection system (Bio‐Rad). TUB2 (TUBULIN BETA CHAIN2) was used as a reference for normalization. RT–qPCRs were performed and analyzed for three biological replicates and three technical repeats per replicate.
HSBP protein purification and antibody generation
The coding sequence of HSBP (At4g15802) was amplified by PCR with pET‐HSBP_F and pET‐HSBP_R primers using the cDNA as template. The PCR product was cloned into the NdeI and XhoI restriction sites of pET30a (Novagen) to add a C‐terminal 6x‐his tag using the Gibson assembly cloning system. The resulting construct was transformed into Escherichia coli strain BL21 (DE3) RIL. Bacterial cells harboring the construct were grown in 1 L of LB medium containing kanamycin (50 mg/ml) and chloramphenicol (25 mg/ml) at 37°C. After the addition of 1.0‐mM IPTG (Isopropyl‐β‐D‐thiogalactoside), the culture was maintained at 18°C for 16 h for protein production. Bacterial cells were collected by centrifugation at 11,000 g for 15 min at 4°C and the pellet was resuspended in buffer A (40‐mM Tris–HCl, pH 8.0). The cell pellet was disrupted by sonication and the cell debris was removed by centrifugation at 11,000 g for 30 min at 4°C. The lysate was bound to Ni‐NTA agarose (QIAGEN) and the bound proteins were eluted with 300‐mM imidazole in buffer A. Recombinant HSBP protein was purified by dialysis and used to produce the polyclonal antibody against HSBP by inoculating rabbits (GWVITEK, Korea).
Generation of genome‐wide crossover maps by genotyping by sequencing (GBS)
Col/Ler and meiMIGS‐HSBP Col/Ler F2 individuals were grown on soil for 3 weeks. Genomic DNA (gDNA) from two to three adult leaves per plant was extracted by the CTAB method to prepare sequencing libraries as described (Ziolkowski et al, 2017; Serra et al, 2018; Nageswaran et al, 2021). Then, 150 ng gDNA from each F2 plant was fragmented using dsDNA Shearase (Zymo Research) and used to generate one sequencing library per plant. The 96 barcoded libraries were pooled and subjected to paired‐end 150‐bp sequencing using an Illumina HiSeq X instrument (Macrogen, Korea). The TIGER pipeline was used to analyze the sequencing data and map crossovers as described (Nageswaran et al, 2021).
RNA sequencing
RNA extraction and library construction were performed as described (Choi et al, 2018). Briefly, 5 μg of total RNA was extracted from unopened floral buds (smaller than approximately 1 mm) and 10‐day‐old seedlings using TRIzol reagent (Invitrogen). A Ribo‐Zero magnetic kit (MRZPL116, Epicentre) was used for rRNA depletion from total RNA. Then, 50 ng of rRNA‐depleted RNA was used to construct sequencing libraries using a ScriptSeq v2 RNA‐seq Library Preparation Kit (SSV21124, Epicentre). Twelve PCR cycles were used for amplification of the libraries, which were indexed using ScriptSeq Index PCR Primers (RSBC10948, Epicentre). Sequencing was performed on an Illumina HiSeq instrument (Macrogen, Korea). Adapter sequences were trimmed from the raw reads with Trim Galore (v. 0.6.6) with parameters ‐q 0 ‐‐stringency 3 ‐‐length 20. Trimmed reads were aligned to the TAIR10 reference genome using STAR (v. 2.7.3) (Dobin et al, 2013) with default parameters. The number of reads mapping to exons was calculated using featureCounts (v. 2.0.1) with default parameters (Liao et al, 2014). Differentially expressed genes (DEGs) were identified among meiosis‐related genes (in‐house list) with the R package DESeq2 using a Benjamini–Hochberg adjusted P‐value < 0.01 as cutoff (Love et al, 2014).
ATAC sequencing
The purification of nuclei and ATAC‐seq library construction were performed as described (Maher et al, 2018). Briefly, 1 g of Arabidopsis unopened flower buds was ground in liquid nitrogen. The ground powder was resuspended in nuclei purification buffer (NPB, 20‐mM MOPS, pH 7.0, 40‐mM NaCl, 90‐mM KCl, 2‐mM EDTA, 0.5‐mM spermidine, 0.2‐mM spermine, 0.5‐mM EGTA, 1× Roche Complete protease inhibitor cocktail). Nuclei were isolated by sucrose density gradient centrifugation. Approximately 100,000 nuclei were used for ATAC‐seq library construction by measuring gDNA concentrations using a Qubit™ dsDNA BR Assay Kit (Thermo, Q32850). Tagmentation was performed using the Tagment DNA Enzyme and Buffer kit (Illumina, 20034210). Transposed DNA fragments were purified using AMPure XP beads (Beckman Coulter, A63881). After purification, transposed DNA was PCR amplified with 12 cycles using Next High‐Fidelity 2×PCR Master Mix (NEB, M0541) with Nextera DNA CD Index primers. The indexed libraries were subjected to paired‐end 50‐bp sequencing using an Illumina HiSeq X instrument (Macrogen, Korea).
Genome‐wide bisulfite sequencing (BS‐seq) analysis
For BS‐seq library construction, gDNA was isolated with the DNeasy Plant Mini Kit (Qiagen 69104, USA). The gDNA was fragmented by sonication using a Bioruptor (Diagenode, Belgium) to a mean size of approximately 250 bp, followed by blunt‐ending, 3′‐end dA addition, and adaptor ligation according to the manufacturer's instructions. The ligation products were used for bisulfite conversion using an EZ DNA Methylation‐Gold kit (ZYMO). The different‐sized fragments were separated and collected by electrophoresis on 2% Tris‐acetate EDTA (TAE) agarose gels, followed by fragment purification (QIAquick Gel Extraction kit, Qiagen), PCR amplification, and cyclization. The DNA libraries were sequenced on a DNBseq platform (BGI, Hong Cong). BS‐seq raw reads were aligned to the TAIR10 reference genome allowing one mismatch, and cytosine coverage was calculated using Bismark (v. 0.22.3). Identification of differentially methylated regions (DMRs) was performed as described (Williams & Gehring, 2017). All biological replicates were merged into one, and only the cytosines with a coverage of more than five reads were considered for further analysis. Cytosines with different methylation levels between hsbp‐3 and Col‐0 (CG, 35%; CHG, 20%; CHH, 15%) were identified as differentially methylated cytosines (DMCs). Each 200‐bp bin overlapping by 100 bp was assigned a “DMR score” calculated as:
200‐bp bins with a DMR score higher than 1.5 or lower than −1.5 were defined as hyperDMR or hyperDMR, respectively.
McrBC–qPCR analysis
gDNA was isolated using the DNeasy Plant Mini Kit (Qiagen 69104, USA). Then, 50 ng of gDNA was digested in NEBuffer2 with McrBC (NEB, M0272S) at 37°C for 4 h and inactivated at 65°C for 30 min. Digested DNA was used for quantitative PCR using a CFX real‐time PCR detection system (Bio‐Rad). gDNA in the same digestion reaction without McrBC treatment was used as a control. McrBC–qPCRs were performed and analyzed for three biological replicates and three technical repeats per replicate.
Immunocytological analysis of wild‐type and hsbp‐3 meiocytes
Floral buds containing meiocytes were fixed in 3:1 (v/v) ethanol:acetic acid, and chromosome spreading was performed as described (Ross et al, 1996). The chromatin was stained with DAPI, and immunostaining of MLH1 was performed as described (Lambing et al, 2020). Co‐immunostaining of ASY1, RAD51, and HEI10 was performed on chromosome spreads using Lipsol and fresh anthers, as described (Lambing et al, 2020). Images were captured using a DeltaVision Personal DV microscope (Applied Precision/GE Healthcare) equipped with a CDD CoolSNAP HQ2 camera (Photometrics). Image analyses were performed using softWoRx software version 5.5 (Applied Precision/GE Healthcare) and ImageJ. The following published antibodies were used: α‐ASY1 (rat, 1:200 or 1:500 dilution), α‐MLH1 (rabbit, 1:200 dilution), α‐RAD51 (rabbit, 1:300 dilution), α‐HEI10 (chicken, 1:1,000, a gift from Mathilde Grelon) and α‐HSBP (rabbit, 1:1,000 dilution) (Higgins et al, 2005; Sanchez‐Moran et al, 2007; Chelysheva et al, 2010). Quantification of the number of MLH1 foci per meiotic cell and the number of RAD51 foci per cell associated with the axis protein ASY1 was performed manually. The number of HEI10 foci per cell was automatically counted using CellProfiler. A Wilcoxon test was used to assess significant differences for RAD51, MLH, and HEI10 foci counts between genotypes.
Arabidopsis protoplast transient transfection assays
Vectors for the transient transfection of Arabidopsis protoplasts were constructed using Golden Gate cloning. The full‐length coding sequences of HSBP and HSFs were cloned into the Lv0 universal vector (pICH41331), as described (Nageswaran et al, 2021). Plasmid DNA transfection into protoplasts was performed as described (Nageswaran et al, 2021). To examine the effects of HSF and HSBP transient overexpression on HEI10 transcription, 20 μg of plasmid DNA was transfected into 20 × 103 protoplasts and incubated at room temperature for 12 h, followed by incubation at 40°C for 1 h. Total RNA was isolated using TRIzol reagent (Invitrogen) for RT–qPCR analysis. For translocation of HSBP into the nucleus, colocalization, and co‐immunoprecipitation of HSBP and HSF, 20 μg of total plasmid DNAs (35Spro:HSF‐GFP and 35Spro:RFP‐HSBP) was co‐transfected into protoplasts and incubated at room temperature for 12 h, followed by incubation at 40°C for 1 h. Fluorescence from transfected protoplasts was detected using a confocal microscope (LSM 800, Zeiss). Co‐transfected protoplasts were used for co‐immunoprecipitation and immunoblotting experiments, as described (Nageswaran et al, 2021).
Chromatin immunoprecipitation and quantitative PCR (ChIP–qPCR) analysis
HSF7a ChIP was performed using Arabidopsis protoplasts. Approximately 2 × 107 protoplasts were transfected with 400 μg of plasmid DNA (35Spro:HSF7a‐HA) and incubated at room temperature for 6 h in constant low‐light conditions (50 µmol m−2 s−1), followed by incubation at 40°C for 1 h. Transfected protoplasts were crosslinked in 1% (w/v) formaldehyde for 10 min, then quenched with 0.125 M glycine for 5 min at room temperature. Crosslinked protoplasts were used for nuclei isolation, immunoprecipitation with anti‐HA antibody (ab9110, Abcam), and DNA recovery as described (Choi et al, 2018). HSBP ChIP experiments were performed using 2 g of 10‐day‐old seedlings that were heat treated at 37°C for 4 h and unopened floral buds. Nuclei isolation, chromatin crosslinking, and recovery were performed as described (Choi et al, 2018). Briefly, chromatin was sheared using a Bioruptor pico instrument (Diagenode) for 10 min at high power alternating 30 s on/30 s off. Chromatin immunoprecipitation was performed using an α‐HSBP antibody (10 μg), or normal IgG, and DNA purification was performed as described (Choi et al, 2018). Purified DNA was used for qPCR on a CFX real‐time PCR detection system (Bio‐Rad). All ChIP–qPCRs were performed and analyzed for three biological replicates and three technical repeats per replicate. The oligonucleotides used for the ChIP–qPCR are listed in Appendix Table S30.
Author contributions
Juhyun Kim: Conceptualization; Data curation; Formal analysis; Investigation; Visualization; Methodology; Writing—original draft. Jihye Park: Data curation; Formal analysis; Validation; Investigation; Visualization; Methodology; Writing—original draft. Heejin Kim: Investigation; Visualization; Methodology; Writing—review & editing. Namil Son: Data curation; Software; Formal analysis; Validation; Visualization; Writing—review & editing. Eun‐Jung Kim: Investigation; Methodology; Writing—review & editing. Jaeil Kim: Data curation; Formal analysis; Investigation; Visualization; Methodology; Writing—review & editing. Dohwan Byun: Data curation; Formal analysis; Validation; Investigation; Visualization; Methodology; Writing—review & editing. Youngkyung Lee: Investigation; Methodology; Writing—review & editing. Yeong Mi Park: Validation; Investigation; Methodology; Writing—review & editing. Divyashree C Nageswaran: Investigation; Methodology; Writing—review & editing. Pallas Kuo: Investigation; Methodology; Writing—review & editing. Teresa Rose: Investigation; Methodology. Tuong Vi T Dang: Methodology; Writing—review & editing. Ildoo Hwang: Supervision; Funding acquisition; Writing—review & editing. Christophe Lambing: Supervision; Funding acquisition; Investigation; Visualization; Methodology; Writing—review & editing. Ian R Henderson: Formal analysis; Supervision; Funding acquisition; Writing—review & editing. Kyuha Choi: Conceptualization; Data curation; Formal analysis; Supervision; Funding acquisition; Investigation; Visualization; Methodology; Writing—original draft; Project administration; Writing—review & editing.
In addition to the CRediT author contributions listed above, the contributions in detail are:
JuK, JP, and KC designed experiments. JuK, JP, NS, DB, JaK, CL, IRH, and KC analyzed the data. JuK, JP, HK, NS, CL, EK, JaK, DB, YL, YMP, DCN, PK, TR, TVD, and KC conducted experiments. KC wrote the manuscript with assistance from JuK and JP. All authors reviewed the manuscript.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Source Data for Figure 1
Source Data for Figure 6K
Acknowledgements
We thank Gregory Copenhaver (University of North Carolina), Avraham Levy (The Weizmann Institute), and Scott Poethig (University of Pennsylvania) for providing pollen and seed FTLs. We thank Raphael Mercier (Max Planck Institute, Cologne) for providing fancm and recq4a recq4b seeds. We thank Chris Franklin for providing ASY1 and RAD51 antibodies. We thank the Gurdon Institute for access to their microscopy facilities. We thank Charles Underwood (Max Planck Institute, Cologne), Yumi Kim (Johns Hopkins University), Mathilde Grelon (Institut National de la Recherche Agronomique, France), and Piotr Ziolkowski (Adam Mickiewicz University) for providing helpful comments. This work was funded by the Suh Kyungbae Foundation (SUHF) SUHF‐17020079, Next‐Generation BioGreen 21 Program PJ01337001, New Breeding Technologies Development Program PJ014795, Rural Development Administration, Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education NRF‐2020R1A2C2007763. CL research is funded by a BBSRC grant‐aided support as part of the Institute Strategic Program Designing Future Wheat Grant (BB/P016855/1) and an Institutional Sponsorship Fund as part of the UKRI grant (BB/W510543/1). Work in the IRH group was supported by BBSRC grants BB/S006842/1, BB/S020012/1, and BB/V003984/1, European Research Council Consolidator Award ERC‐2015‐CoG‐681987 ‘SynthHotSpot’ and Marie Curie International Training Network ‘COMREC’.
The EMBO Journal (2022) 41: e109958.
Data availability
Sequencing data of F2 individuals of meiMIGS‐HSBP Col/Ler and Col/Ler have been deposited in the ArrayExpress database at EMBL‐EBI (http://www.ebi.ac.uk/arrayexpress) under accession number E‐MTAB‐10168 (https://www.ebi.ac.uk/arrayexpress/experiments/E‐MTAB‐10168/), E‐MTAB‐10783 (https://www.ebi.ac.uk/arrayexpress/experiments/E‐MTAB‐10783/), and E‐MTAB‐11586 (https://www.ebi.ac.uk/arrayexpress/experiments/E‐MTAB‐11586/). RNA‐seq, BS‐seq, and ATAC‐seq data for the Col and hcr2 (hsbp‐3) have been deposited in the ArrayExpress database at EMBL‐EBI under accessions E‐MTAB‐10791 (https://www.ebi.ac.uk/arrayexpress/experiments/E‐MTAB‐10791/), E‐MTAB‐10657 (https://www.ebi.ac.uk/arrayexpress/experiments/E‐MTAB‐10657/), and E‐MTAB‐10790 (https://www.ebi.ac.uk/arrayexpress/experiments/E‐MTAB‐10790/).
References
- Abane R, Mezger V (2010) Roles of heat shock factors in gametogenesis and development. FEBS J 277: 4150–4172 [DOI] [PubMed] [Google Scholar]
- Ahn SG, Thiele DJ (2003) Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev 17: 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz LE, Copenhaver GP (2008) Fluorescent Arabidopsis tetrads: a visual assay for quickly developing large crossover and crossover interference data sets. Nat Protoc 3: 41–50 [DOI] [PubMed] [Google Scholar]
- Berchowitz LE, Copenhaver GP (2010) Genetic interference: don’t stand so close to me. Curr Genomics 11: 91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz LE, Francis KE, Bey AL, Copenhaver GP (2007) The role of AtMUS81 in interference‐insensitive crossovers in A. thaliana . PLoS Genet 3: e132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E, Sanchez A, Anand R, Ranjha L, Hugener J, Adam C, Acharya A, Weyland N, Aran‐Guiu X, Charbonnier J‐B et al (2020) Regulation of the MLH1–MLH3 endonuclease in meiosis. Nature 586: 618–622 [DOI] [PubMed] [Google Scholar]
- Capilla‐Pérez L, Durand S, Hurel A, Lian Q, Chambon A, Taochy C, Solier V, Grelon M, Mercier R (2021) The synaptonemal complex imposes crossover interference and heterochiasmy in Arabidopsis . Proc Natl Acad Sci U S A 118: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang I, Friman O, Guertin DA, Chang J, Lindquist RA, Moffat J et al (2006) Cell Profiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7: R100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L, Grandont L, Vrielynck N, le Guin S, Mercier R, Grelon M (2010) An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: immunodetection of cohesins, histones and MLH1. Cytogenet Genome Res 129: 143–153 [DOI] [PubMed] [Google Scholar]
- Chelysheva L, Vezon D, Chambon A, Gendrot G, Pereira L, Lemhemdi A, Vrielynck N, Le Guin S, Novatchkova M, Grelon M (2012) The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet 8: e1002799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Zhao X, Kelly KA, Venn O, Higgins JD, Yelina NE, Hardcastle TJ, Ziolkowski PA, Copenhaver GP, Franklin FCH et al (2013) Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat Genet 45: 1327–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Zhao X, Tock AJAJ, Lambing C, Underwood CJCJ, Hardcastle TJTJ, Serra H, Kim JJ, Cho HSHS, Kim JJ et al (2018) Nucleosomes and DNA methylation shape meiotic DSB frequency in Arabidopsis thaliana transposons and gene regulatory regions. Genome Res 28: 532–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver GP, Housworth EA, Stahl FW (2002) Crossover interference in Arabidopsis . Genetics 160: 1631–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crismani W, Girard C, Froger N, Pradillo M, Santos JL, Chelysheva L, Copenhaver GP, Horlow C, Mercier R (2012) FANCM limits meiotic crossovers. Science 336: 1588–1590 [DOI] [PubMed] [Google Scholar]
- De Muyt A, Pyatnitskaya A, Andréani J, Ranjha L, Ramus C, Laureau R, Fernandez‐Vega A, Holoch D, Girard E, Govin J et al (2018) A meiotic XPF–ERCC1‐like complex recognizes joint molecule recombination intermediates to promote crossover formation. Genes Dev 32: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Zhang L, Piolot T, Kleckner N, Espagne E, Zickler D (2014) E3 ligase Hei10: a multifaceted structure‐based signaling molecule with roles within and beyond meiosis. Genes Dev 28: 1111–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duroc Y, Kumar R, Ranjha L, Adam C, Guérois R, Md Muntaz K, Marsolier‐Kergoat M‐C, Dingli F, Laureau R, Loew D et al (2017) Concerted action of the MutLβ heterodimer and Mer3 helicase regulates the global extent of meiotic gene conversion. eLife 6: e21900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felippes FF, Wang J, Weigel D (2012) MIGS: miRNA‐induced gene silencing. Plant J 70: 541–547 [DOI] [PubMed] [Google Scholar]
- France MG, Enderle J, Röhrig S, Puchta H, Franklin FCH, Higgins JD (2021) ZYP1 is required for obligate cross‐over formation and cross‐over interference in Arabidopsis . Proc Natl Acad Sci U S A 118: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis KE, Lam SY, Harrison BD, Bey AL, Berchowitz LE, Copenhaver GP (2007) Pollen tetrad‐based visual assay for meiotic recombination in Arabidopsis . Proc Natl Acad Sci U S A 104: 3913–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S, Meeley R, Scanlon MJ (2002) Empty pericarp2 encodes a negative regulator of the heat shock response and is required for maize embryogenesis. Plant Cell 14: 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Colaiácovo MP (2018) Zipping and unzipping: protein modifications regulating synaptonemal complex dynamics. Trends Genet 34: 232–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesguth M, Sahm A, Simon S, Dietz KJ (2015) Redox‐dependent translocation of the heat shock transcription factor AtHSFA8 from the cytosol to the nucleus in Arabidopsis thaliana . FEBS Lett 589: 718–725 [DOI] [PubMed] [Google Scholar]
- Girard C, Chelysheva L, Choinard S, Froger N, Macaisne N, Lehmemdi A, Mazel J, Crismani W, Mercier R (2015) AAA‐ATPase FIDGETIN‐LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLoS Genet 11: e1005369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez‐Pastor R, Burchfiel ET, Thiele DJ (2018) Regulation of heat shock transcription factors and their roles in physiology and disease. Nat Rev Mol Cell Biol 19: 4–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S, Cohen PE (2016) Control of meiotic crossovers: from double‐strand break formation to designation. Annu Rev Genet 50: 175–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Liu JH, Ma X, Luo DX, Gong ZH, Lu MH (2016) The plant heat stress transcription factors (HSFS): Structure, regulation, and function in response to abiotic stresses. Front Plant Sci 7: 114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Armstrong SJ, Franklin FCH, Jones GH (2004) The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis . Genes Dev 18: 2557–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Perry RM, Barakate A, Ramsay L, Waugh R, Halpin C, Armstrong SJ, Franklin FCH (2012) Spatiotemporal asymmetry of the meiotic program underlies the predominantly distal distribution of meiotic crossovers in barley. Plant Cell 24: 4096–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Sanchez‐Moran E, Armstrong SJ, Jones GH, Franklin FCH (2005) The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev 19: 2488–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SF, Lai HC, Jinn TL (2010) Cytosol‐localized heat shock factor‐binding protein, AtHSBP, functions as a negative regulator of heat shock response by translocation to the nucleus and is required for seed development in Arabidopsis . Plant Physiol 153: 773–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N (2015) Meiotic recombination: the essence of heredity. Cold Spring Harb Perspect Biol 7: 1–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichino L, Boone BA, Strauskulage L, Harris CJ, Kaur G, Gladstone MA, Tan M, Feng S, Jami‐Alahmadi Y, Duttke SH et al (2021) MBD5 and MBD6 couple DNA methylation to gene silencing through the J‐domain protein SILENZIO. Science 372: 1434–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Huang S‐S, Jupe F, Sasaki E, Schmitz RJ, Urich MA, Castanon R, Nery JR, Barragan C, He Y et al (2016) Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166: 492–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher T, Walbot V (2012) Hypoxia triggers meiotic fate acquisition in maize. Science 337: 345–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambing C, Kuo PC, Tock AJ, Topp SD, Henderson IR (2020) ASY1 acts as a dosage‐dependent antagonist of telomere‐led recombination and mediates crossover interference in Arabidopsis . Proc Natl Acad Sci U S A 24: 13647–13658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Qin B, Shen Y, Zhang F, Liu C, You H, Du G, Tang D, Cheng Z (2018) HEIP1 regulates crossover formation during meiosis in rice. Proc Natl Acad Sci U S A 115: 10810–10815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Lim EC, Kim J, Park J, Kim EJ, Kim J, Park YM, Cho HS, Byun D, Henderson IR, Copenhaver GP et al (2020) DeepTetrad: high‐throughput image analysis of meiotic tetrads by deep learning in Arabidopsis thaliana . Plant J 101: 473–483 [DOI] [PubMed] [Google Scholar]
- Lloyd A, Morgan C, Franklin C, Bomblies K (2018) Plasticity of meiotic recombination rates in response to temperature in Arabidopsis . Genetics 208: 1409–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15: 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher KA, Bajic M, Kajala K, Reynoso M, Pauluzzi G, West DA, Zumstein K, Woodhouse M, Bubb K, Dorrity MW et al (2018) Profiling of accessible chromatin regions across multiple plant species and cell types reveals common gene regulatory principles and new control modules. Plant Cell 30: 15–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed‐Bessudo C, Yehuda E, Stuitje AR, Levy AA (2005) A new seed‐based assay for meiotic recombination in Arabidopsis thaliana . Plant J 43: 458–466 [DOI] [PubMed] [Google Scholar]
- Mercier R, Jolivet S, Vezon D, Huppe E, Chelysheva L, Giovanni M, Nogué F, Doutriaux M‐P, Horlow C, Grelon M et al (2005) Two meiotic crossover classes cohabit in Arabidopsis: one is dependent on MER3, whereas the other one is not. Curr Biol 15: 692–701 [DOI] [PubMed] [Google Scholar]
- Mercier R, Mézard C, Jenczewski E, Macaisne N, Grelon M (2015) The molecular biology of meiosis in plants. Annu Rev Plant Biol 66: 297–327 [DOI] [PubMed] [Google Scholar]
- Modliszewski JL, Wang H, Albright AR, Lewis SM, Bennett AR, Huang J, Ma H, Wang Y, Copenhaver GP (2018) Elevated temperature increases meiotic crossover frequency via the interfering (Type I) pathway in Arabidopsis thaliana . PLOS Genet 14: e1007384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan C, Fozard JA, Hartley M, Henderson IR, Bomblies K, Howard M (2021) Diffusion‐mediated HEI10 coarsening can explain meiotic crossover positioning in Arabidopsis . Nat Commun 12: 4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI (1998) Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev 12: 3788–3796 [DOI] [PubMed] [Google Scholar]
- Nageswaran DC, Kim J, Lambing C, Kim J, Park J, Kim E‐J, Cho HS, Kim H, Byun D, Park YM et al (2021) HIGH CROSSOVER RATE1 encodes PROTEIN PHOSPHATASE X1 and restricts meiotic crossovers in Arabidopsis . Nat Plants 7: 452–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelms B, Walbot V (2019) Defining the developmental program leading to meiosis in maize. Science 364: 52–56 [DOI] [PubMed] [Google Scholar]
- O’Malley RC, Huang SSC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR (2016) Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165: 1280–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyatnitskaya A, Borde V, De Muyt A (2019) Crossing and zipping: molecular duties of the ZMM proteins in meiosis. Chromosoma 128: 181–198 [DOI] [PubMed] [Google Scholar]
- Qiao H, Prasada Rao HBD, Yang YE, Fong JH, Cloutier JM, Deacon DC, Nagel KE, Swartz RK, Strong E, Holloway JK et al (2014) Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat Genet 46: 194–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana RM, Dong S, Tang H, Ahmad F, Zhang H (2012) Functional analysis of OsHSBP1 and OsHSBP2 revealed their involvement in the heat shock response in rice (Oryza sativa L.). J Exp Bot 63: 6003–6016 [DOI] [PubMed] [Google Scholar]
- Rao HBDP, Qiao H, Bhatt SK, Bailey LRJ, Tran HD, Bourne SL, Qiu W, Deshpande A, Sharma AN, Beebout CJ et al (2017) A SUMO‐ubiquitin relay recruits proteasomes to chromosome axes to regulate meiotic recombination. Science 355: 306–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A, Qiao H, Yang YE, Chen JK, Jackson N, Biswas K, Holloway JK, Baudat F, de Massy B, Wang J et al (2013) RNF212 is a dosage‐sensitive regulator of crossing‐over during mammalian meiosis. Nat Genet 45: 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross KJ, Fransz P, Jones GH (1996) A light microscopic atlas of meiosis in Arabidopsis thaliana . Chromosom Res 4: 507–516 [DOI] [PubMed] [Google Scholar]
- Russell AG, Charette JM, Spencer DF, Gray MW (2006) An early evolutionary origin for the minor spliceosome. Nature 443: 863–866 [DOI] [PubMed] [Google Scholar]
- Sanchez‐Moran E, Santos J‐L, Jones GH, Franklin FCH (2007) ASY1 mediates AtDMC1‐dependent interhomolog recombination during meiosis in Arabidopsis . Genes Dev 21: 2220–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyal SH, Chen D, Fox SG, Kramer JM, Morimoto RI (1998) Negative regulation of the heat shock transcriptional response by HSBP1. Genes Dev 12: 1962–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séguéla‐Arnaud M, Crismani W, Larchevêque C, Mazel J, Froger N, Choinard S, Lemhemdi A, Macaisne N, Van Leene J, Gevaert K et al (2015) Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc Natl Acad Sci U S A 112: 4713–4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra H, Lambing C, Griffin CH, Topp SD, Nageswaran DC, Underwood CJ, Ziolkowski PA, Séguéla‐Arnaud M, Fernandes JB, Mercier R et al (2018) Massive crossover elevation via combination of HEI10 and recq4a recq4b during Arabidopsis meiosis. Proc Natl Acad Sci U S A 115: 2437–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Schneeberger K (2015) Shoremap v3.0: fast and accurate identification of causal mutations from forward genetic screens. Methods Mol Biol 1284: 381–395 [DOI] [PubMed] [Google Scholar]
- Villeneuve AM, Hillers KJ (2001) Whence meiosis? Cell 106: 647–650 [DOI] [PubMed] [Google Scholar]
- Walker J, Gao H, Zhang J, Aldridge B, Vickers M, Higgins JD, Feng X (2018) Sexual‐lineage‐specific DNA methylation regulates meiosis in Arabidopsis . Nat Genet 50: 130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wang M, Tang D, Shen Y, Miao C, Hu Q, Lu T, Cheng Z (2012) The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet 8: e1002809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BP, Gehring M (2017) Stable transgenerational epigenetic inheritance requires a DNA methylation‐sensing circuit. Nat Commun 8: 2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Rossidivito G, Hu T, Berlyand Y, Poethig RS (2015) Traffic lines: new tools for genetic analysis in Arabidopsis thaliana . Genetics 200: 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wang C, Higgins JD, Kim YJ, Moon S, Jung KH, Qu S, Liang W (2019) A multiprotein complex regulates interference‐sensitive crossover formation in rice. Plant Physiol 181: 221–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziolkowski PA, Berchowitz LE, Lambing C, Yelina NE, Zhao X, Kelly KA, Choi K, Ziolkowska L, June V, Sanchez‐Moran E et al (2015) Juxtaposition of heterozygous and homozygous regions causes reciprocal crossover remodelling via interference during Arabidopsis meiosis. eLife 4: e03708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziolkowski PA, Underwood CJ, Lambing C, Martinez‐Garcia M, Lawrence EJ, Ziolkowska L, Griffin C, Choi K, Franklin FCH, Martienssen RA et al (2017) Natural variation and dosage of the HEI10 meiotic E3 ligase control Arabidopsis crossover recombination. Genes Dev 31: 306–317 [DOI] [PMC free article] [PubMed] [Google Scholar]


