Abstract
The application of polymer models to chromosome structure and dynamics is a powerful approach for dissecting functional properties of the chromosome. The models are based on well-established bead-spring models of polymers and are distinct from molecular dynamics studies used in structural biology. In this work, we outline a polymer dynamics model that simulates budding yeast chromatin fibers in a viscous environment inside the nucleus using DataTank as a user interface for the C++ simulation. We highlight features for creating the nucleolus, a dynamic region of chromatin with protein-mediated, transient chromosomal cross-links, providing a predictive, stochastic polymer-physics model for versatile analyses of chromosome spatiotemporal organization. DataTank provides real-time visualization and data analytics methods during simulation. The simulation pipeline provides insights into the entangled chromosome milieu in the nucleus and creates simulated chromosome data, both structural and dynamic, that can be directly compared to experimental observations of live cells in interphase and mitosis.
Keywords: Polymer physics model, Chromatin, Nucleolus, Chromosomal cross-links, All chromosome simulation, Data visualization, DataTank
1. Introduction
Chromosome segregation is performed largely at the intersection of chromosomes and microtubule plus-ends, known as the kinetochore, through the interaction of the kinetochore with the mitotic spindle. Much focus has been paid to the centromere and understanding its role in dictating kinetochore assembly. However, the bulk of the chromosome arm is not typically considered beyond mechanisms required for its condensation. One of the major hurdles in studying bulk chromosomes is the sheer mass of DNA. From the biochemical perspective, there is a dearth of information beyond the nucleosome. Likewise, structural biological approaches toward understanding higher-order chromosome structure are limited due to computational cost of resolved simulations at the required spatial and temporal scales for detailed structure across the entire genome. The application of stochastic bead-spring models of entropic polymers, derived from statistical physics, has revolutionized the field of chromosome structure and dynamics. These models provide a remarkably accurate approximation for chromosome motion under a variety of conditions (such as DNA damage), organization of the nucleolus and other sub-compartments, and higher-order structures such as the bottle brush organization in centromeres and the condensed mitotic chromosome.
The spatial structure of chromosomes furthermore dictates control of genome expression beyond the linear arrangement of nucleotides that encodes the gene map [1-3]. Yet direct observation of chromosomes fails to bring detailed structural information, due to compactness and the intense dynamics of chromosomes inside the nucleus. Numerical simulations extend the possibility for clear and concise observation of chromosome spatial structure and dynamics. These simulations can be validated through direct comparison with experimental results in terms of statistical and stochastic analyses, including but not limited to signal size and intensity, spatial distribution of chromosome territories, and the underlying intra- and inter-chain interactions that reproduce observed genome self-organization.
Modeling DNA as bead-spring polymer chains is widely adopted in the field and has made a tremendous impact on studying genome organization [4-6].
In this work, we introduce the model developed for simulating dynamics of budding yeast chromosomes inside the nucleus in the G1 stage of interphase [7, 8]. Each chromatin fiber is modeled as a flexible polymer consisting of beads connected by nonlinear springs in a viscous environment. 32 polymer chains, representing the arms of the 16 yeast chromosomes with biologically accurate lengths, are generated to directly compare to experimental observations. The dynamics of each bead, representing 5000 bp of DNA, is joined to adjacent beads with nonlinear springs that obey a worm-like chain force regime [9, 10]. Moreover, the polymers are confined within a 1 μm sphere representing the nucleus, and both ends of the 32 polymer chains are tethered on the nuclear membrane to generate a Rabl conformation [4]. Both confinements resemble configurations of the nucleus inspired by experimental results. This stochastic simulation of coupled, entropy-driven, tethered, geometrically confined, bead-spring polymer chains shows many consistent features in terms of dynamic properties of chromatin fibers inside the nucleus.
Additionally, this model simulates the nucleolus, a region inside the nucleus where ribosomal RNA is synthesized [11]. The nucleolus is represented as a specific region on chromosome XII with an enhanced activity specified by intra- and inter-chain interactions. These interactions are modeled by the formation of transient cross-links between beads on the same chain or across separate chains [7]. This approach simulates high-level chromatin interactions with structural proteins such as structural maintenance of chromosome (SMC) proteins in the nucleolus.
The simulation pipeline is entirely accomplished using DataTank. In the .tank file, DataTank offers a graphical interface for setting all inputs. The input parameters are passed to the simulation program attached to the .tank file. After compiling, the program runs the simulation, and DataTank captures the streaming results and simultaneously outputs the nuclear configuration. When the simulation is finished, the results are stored in DataTank. Users have the option to either continue analyzing the data using DataTank or export the data to other file formats for additional analysis. We provide the script for an interphase simulation, Yeast_Chromosome_Dynamics_Simulator.tank, and the simulation program attached to it. Moreover, we provide a number of tools we developed using DataTank for visualization and analysis of the 32-chromosome arm simulation. Lastly, we provide a quick guide for researchers to use DataTank to read their own simulation results and conduct downstream analyses utilizing DataTank’s user-friendly interface and visualization features.
2. Materials
The simulation pipeline uses DataTank, a numerical working environment, as the user interface. DataTank is developed by David Adalsteinsson; researchers can go to the following website for a user license and to download the software https/www.visualdatatools.com/DataTank/. DataTank provides powerful visualization and analysis tools that are compatible with various types of data. ImageTank is an additional data visualization tool that further streamlines data analysis (see Note 3). DataTank employs embedded data-oriented functions and is itself a visual programming environment. Programs created by standard utilities or the built-in C++ framework can be launched in DataTank for numerical purposes. The simulation results are parsed by DataTank in real time for downstream data visualization and analyses.
Yeast_Chromosome_Dynamics_Simulator.tank – The main file for the simulation pipeline contains: (1) The interface to enter input parameters and settings. (2) The port connecting to the simulation program. (3) The visualization module. (4) Modules for stochastic analyses.
Microscope_Simulator.tank—The microscope simulator pipeline functions to convert the numerical results to experimental microscope images. This enables direct statistical comparison of geometries between simulated chromosomes and experimental observations.
Community_Detection_Analysis.tank—The community detection pipeline applies a multilayer modularity optimization process to detect community behavior during chromosomal interactions in simulation results.
All DataTank scripts and attachments are available at https://github.com/heyunyan19930924/Yeast-Chromosome-Dynamic-Simulaion. See Note 1 for online help.
3. Methods
We now show how to run the simulation program using DataTank. After the simulation data is generated, we show ways to store the results in different file formats. We show how to visualize the results as the simulation is progressing. Lastly, we introduce scripts for data analytics of the simulated, all chromosome, time series, consisting of a Microscope Simulator script that passes the simulated data through a microscope imaging pipeline, and a Community Detection script that detects dynamic gene clusters (communities) at the scale they exist.
3.1. Set Inputs and Launch the Simulation
Download and install DataTank. Acquire the license by following instructions on the website. Download the simulation script Yeast_Chromosome_Dynamics_Simulator.tank.
Run Yeast_Chromosome_Dynamics_Simulator.tank through DataTank. Set inputs before running the simulation. Default setting simulates DNA dynamics inside the nucleus of a budding yeast cell. First set the initial geometry by inputting the number of arms, lengths of each arm, and connections at the nuclear wall in the “lengths” and “connections” modules. By default, there are 32 polymer arms simulating 16 chromosomes. “Coefficients” dictionary contains parameters regarding the dynamics, including simulation scale, persistence length of polymers, environmental parameters, and so on. “Evolution” dictionary controls the simulation time scale, timestep, and which timesteps to save (the save time “stride”). The fundamental model introduced so far already builds the basis for polymer dynamics simulation, and there are additional features specifically for simulating chromosomes. Beads are subject to a repulsive potential and the corresponding parameters are in the “Excluded volume” dictionary. The nuclear membrane is modeled as a spherical surface and all arms are restricted within. The membrane location and volume are set in “Void formation” dictionary. By default, both ends of all polymer arms are attached to fixed sites on the membrane. Moreover, this work simulates chromatin interactions as transient cross-links in the nucleolus, and the nucleolus is modeled as a specified region (the default is on chromosome XII) specified in the “Terms” dictionary. Users can toggle all modules listed above to modify the simulation parameters.
- The simulation can be run in two different ways. The user can either run the simulation locally or submit it to a remote computing cluster.
- Local. This approach utilizes the local machine’s CPUs and therefore potentially prevents users from multitasking, especially for long simulations, but it allows users to view the results simultaneously. One can create a “Task” module and add dependencies to all relevant dictionaries simply by dragging the dictionaries in the module. By linking the C++ simulation script to the module as the kernel, the simulation is ready to run.
- Remote. Submit the simulation and run it on a server with legal access to avoid local occupancy. This approach forbids simultaneous observation; however, one can download the results and visualize them locally at any time, even when the job is still running. To create a job, DataTank offers the “Parameter run” module that does the submission and online manipulation. Likewise, once the “Task” module is created, one can link all dependent parameters and simulation script to the task. One can conduct multiple runs in each submission by setting variables as “Time sequences.” DataTank automatically recognizes the format and creates multiple runs for all time values.
- DataTank is only compatible with Xcode in MacOS as its C++ program compiler. It also provides built-in DTSource library as data structures and ports specifically for DataTank modules. The simulation C++ script is developed based on the DTSource library and is attached to Yeast_Chromosome_Dynamics_Simulator.tank.
- Set the application to the attached script in “Parameter run” or “Task” module by clicking the “Set” button; find the script under Yeast_Chromosome_Dynamics_Simulator.tank file.
- Compile or profile the C++ script in Xcode, and one will find that in the scroll down list “Support,” either the “Debug version” or the “Release version” has been lit up. See Note 2 for troubleshooting in compiling.
- Select either the “Debug version” or the “Release version” as the project.
- If the simulation is being run locally, click the “Run” button and the simulation starts to progress.
- If the simulation is being submitted to a remote computing cluster, click the “Create” button to create a Parameter_run.dtask file that stores all required scripts and commands for submitting the job. Navigate to “Task > Run remotely > Add machine” and enter server information and access. Now click the “submit” button, and the job is successfully submitted to the server. The output data is also stored in the same Parameter_run.dtask file. While the file is open, DataTank periodically checks with the server for progress and downloads temporary results.
- To get the simulation results, the approach depends whether the simulation is run locally or online.
- For local simulations, the output is saved by DataTank into a local cache. By accessing the “Task” module itself, we parse the results. If one intends to store the output locally, make a switch in the “Directory” line from “Temp” to a custom directory in the “Task” module before running the program. The output will be stored in a custom .mat file as the simulation progresses.
- For online simulations, the output data is saved inside Parameter_run.dtask file.
3.2. Access Simulation Results
Simulation data stored as .mat file contains the coordinates of each bead at each timestep in a table and stores all tables in a corresponding .mat file. DataTank can parse the .mat file as input and convert to a time series of “3D Path,” a built-in data structure in the DTSource library. By creating a “Data File” module in DataTank, the .mat data could be opened in the module which contains the main output. Three variables are contained in .mat files, and the position information is saved as variable “Var.”
Simulation results saved in .dtask files can be read by “File Name” module in DataTank. Choose “From Parameter Run” from the scroll down bar in the module and specify the file to be read. Unlike “Data File” module, “File Name” module cannot detect the underlying data structure automatically. Manually create a “Group” structure, link the group to the “File Name” module, and one can extract the three variables within the file including the “3D Path” by pressing “Suggest” button.
3.3. Visualize Simulation Data
DataTank possesses powerful data visualization modules that are compatible with various data formats and real–time updates (Fig. 1a-c).
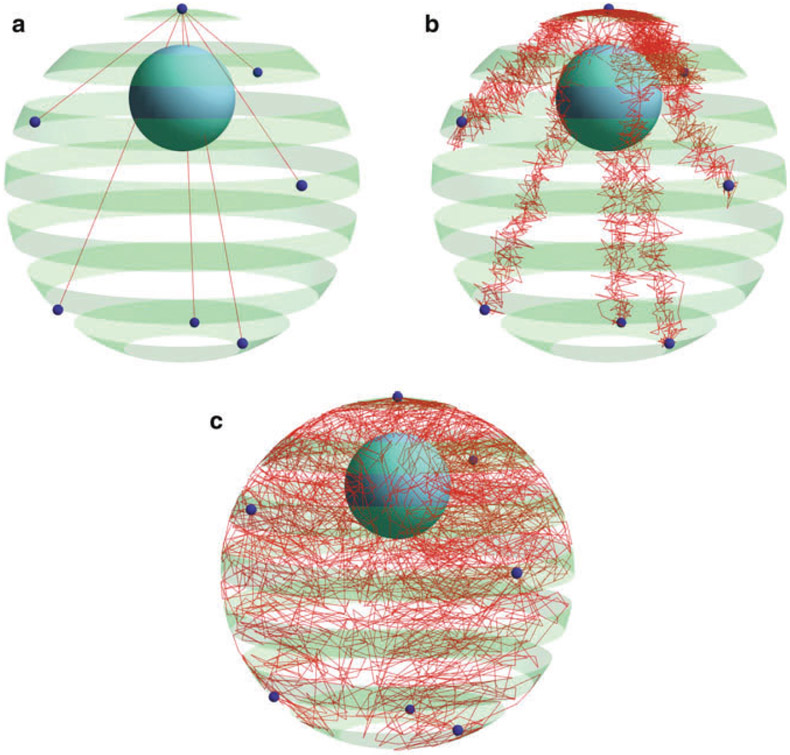
Fig. 1.
Simulation result configuration in DataTank. Red chains correspond to chromosome arms. The green spherical shell represents the nuclear membrane. Dark small spheres on the membrane are tethering spots of all 16 chromosomes—the common centromere and six distal telomeres. The nucleolus region appears in the simulation as the solid blue sphere inside the nucleus. (a) Initial configuration. Each straight line is an initial superposition of the multiple chromosome arms sharing the same tethering spots. (b) Midterm configuration. (c) Equilibrium configuration. Chromosomes spread across the whole nucleus after undergoing entropic dynamics in G1 of interphase
To observe numerical data, drag the “3D Path” variable to the “Variable Monitor” panel on the right side of DataTank. The positions of all beads at a particular timestep will be displayed in the panel. The duration time for a time series can be toggled in the “Time” sliding bar at the bottom.
To create figures, navigate to “Drawing” and create a “3D Space - Rendered” imaging module. A new figure window will be created. Drag the “3D Path” variable into the window, and the chromosome beads and springs will be displayed in the image window. Likewise, one can toggle the “Time” sliding bar to observe chromosome structure at a specific timestep. One can also tweak parameters in the image window to modify visualization features.
By default, a “3D Surface” variable has been created in order to represent the nucleus membrane in the figure. It attains the exact parameter settings from the simulation. Drag this variable into the image window to create a sphere around the chromatin chains representing the nucleus membrane. Set the sphere to be semitransparent.
3.4. Analytics Tools
3.4.1. Microscope Simulator
To compare statistical and stochastic properties of simulated data with experimental observations, a DataTank script, Microscope_-Simulator.tank, applies a three-dimensional point-spread function to the simulation data to simulate fluorescent microscopy images.
Download the script Microscope_Simulator.tank. Compile C++ script in “Microscope Simulator Code” module inside Microscope_Simulator.tank. Add the module as global via clicking the “support” dropdown box and choose “Create as Module…”. Now Microscope Simulator works as a global module and can be executed in all DataTank files.
To apply the simulator, create a “3D Mesh” variable by navigating to “Variable > 3D > 3D Mesh.” Select “Microscope Simulator” in the dropdown box. Three input arguments are required. “Points” takes the list of points to simulate. “Bbox” indicates the bounding box. “Element” reads the mesh basis.
The pipeline outputs the simulated experimental 3D signal from simulation data. One can apply further analysis and compare with experimental observations. Since experimental observations are single image planes in most circumstances, use of the Slicing 3D signal is required (Fig. 2).
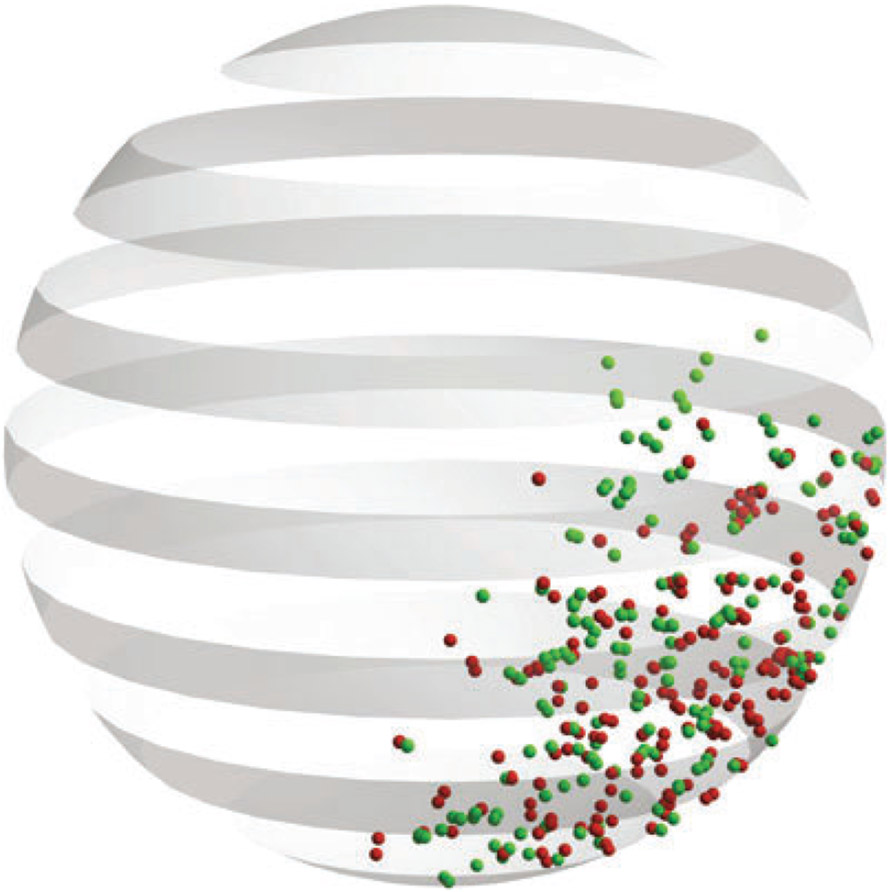
Fig. 2.
Simulated chromosomal signal under the microscope by Microscope_Simulator.tank. This feature uses simulation data, labels specific regions to simulate the experimental observation approach using green fluorescent proteins for labeling, and then converts the simulation signal from the labeling region to a transformed signal. Vertical slices of the transformed 3D signal shown in the figure resemble what the eye or camera observes under the microscope
3.4.2. Community Detection
The transient cross-linking interactions of chromosome segments in the nucleolus have been shown to lead to a range of bead clustering behavior, tuned by the cross-linking affinity [12]. Whatever clustering in the nucleolus exists, the cluster membership of each bead over time can be computed using the clustering script. If there is no discernable clustering, each bead is its own cluster. In all cases, each bead is assigned to one cluster at each timestep, and the clustering algorithm accounts for both spatial proximity and persistence over time. The script applies the GenLouvain algorithm for multilayer modularity optimization ([13, 14], ). See [12] for more details on the application of this module to the simulation model.
Download Community_Detection_Analysis.tank and run the script. To configure, set the “Configuration > Matlab setup > matlabpath” variable to the path to a valid Matlab executable. Set “Configuration > Input” to a simulation data file (see Subheading 3.2). Ensure that all external executables required by the script have been compiled. Clustering parameters may be set in “Configuration > Parameters.” “Start time” and “End time” define the time interval over which clustering is computed. A distinct partition is computed on each of the course-grained time windows, whose widths are set in “Window size.” The variables “Gamma” and “Omega” define the resolution and interlayer coupling parameters. They may be left at the default values or altered to affect the spatiotemporal scale favored by the algorithm (see [12] for further details on these parameters). Specify the portion of beads to be analyzed by community detection pipeline by setting the “Input Processing > Portion of Points on Var.”
The result of the clustering, with points labeled by cluster membership, can be accessed in the variable “Final output - points labeled by cluster.” Note that the cluster identities only change at a temporal frequency defined by the window size. The results can be visualized in the “Beads colored by cluster” drawing window.
4. Notes
For further help regarding DataTank, refer to documentations via “Help.” Online help is also available via “Help > Online Help.”
When compiling attached code through Xcode, issue may occur failing to compile due to outdated default C++ standard library. To solve this issue, click on the project icon, navigate to “Build Settings > Apple Clang – Language – C++ > C++ Standard Library,” and choose “libc++” with C++11 supported. Remote simulation may trigger similar issue because of outdated compiler on server.
ImageTank is another data visualization tool distributed by Visual Data Tool, Inc., also created by David Adalsteinsson, to further streamline data analysis https://visualdatatools.com/ImageTank/ [15]. In addition to all features of DataTank, ImageTank enhances the user interface, image processing, and computing.
Contributor Information
Yunyan He, Department of Mathematics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
David Adalsteinsson, Department of Mathematics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Benjamin Walker, Department of Mathematics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Josh Lawrimore, Department of Biology, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
M. Gregory Forest, Department of Mathematics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA; Department of Applied Physical Sciences, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Kerry Bloom, Department of Biology, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA; Curriculum in Genetics and Molecular Biology, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
References
- 1.Cubenas-Potts C, Corces VG (2015) Topologically associating domains: an invariant framework or a dynamic scaffold? Nucleus 6(6):430–434. 10.1080/19491034.2015.1096467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dekker J, Misteli T (2015) Long-range chromatin interactions. Cold Spring Harb Perspect Biol 7(10):a019356. 10.1101/cshperspect.a019356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verdaasdonk JS, Bloom K (2011) Centromeres: unique chromatin structures that drive chromosome segregation. Nat Rev Mol Cell Biol 12(5):320–332. 10.1038/nrm3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tjong H, Gong K, Chen L, Alber F (2012) Physical tethering and volume exclusion determine higher-order genome organization in budding yeast. Genome Res(6):1295–1305. 10.1101/gr.129437.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vasquez PA, Bloom K (2014) Polymer models of interphase chromosomes. Nucleus 5(5):376–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verdaasdonk JS, Vasquez PA, Barry RM, Barry T, Goodwin S, Forest MG, Bloom K (2013) Centromere tethering confines chromosome domains. Mol Cell 52(6):819–831. 10.1016/j.molcel.2013.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hult C, Adalsteinsson D, Vasquez PA, Lawrimore J, Bennett M, York A, Cook D, Yeh E, Forest MG, Bloom K (2017) Enrichment of dynamic chromosomal crosslinks drive phase separation of the nucleolus. Nucleic Acids Res 45(19):11159–11173. 10.1093/nar/gkx741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasquez PA, Hult C, Adalsteinsson D, Lawrimore J, Forest MG, Bloom K (2016) Entropy gives rise to topologically associating domains. Nucleic Acids Res 44(12):5540–5549. 10.1093/nar/gkw510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marko JF, Siggia ED (1994) Fluctuations and supercoiling of DNA. Science 265 (5171):506–508 [DOI] [PubMed] [Google Scholar]
- 10.Marko JF, Siggia ED (1995) Statistical mechanics of supercoiled DNA. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics 52(3):2912–2938 [DOI] [PubMed] [Google Scholar]
- 11.Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI (2007) The multi-functional nucleolus. Nat Rev Mol Cell Biol 8 (7):574–585. 10.1038/nrm2184 [DOI] [PubMed] [Google Scholar]
- 12.Walker B, Taylor D, Lawrimore J, Hult C, Adalsteinsson D, Bloom K, Forest MG (2019) Transient crosslinking kinetics optimize gene cluster interactions. PLoS Comput Biol 15(8):e1007124. 10.1371/journal.pcbi.1007124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. http://netwiki.amath.unc.edu/GenLouvain/GenLouvain .
- 14.Mucha PJ, Richardson T, Macon K, Porter MA, Onnela JP (2010) Community structure in time-dependent, multiscale, and multiplex networks. Science 328(5980):876–878. 10.1126/science.1184819 [DOI] [PubMed] [Google Scholar]
- 15.O’Shaughnessy EC, Stone OJ, LaFosse PK, Azoitei ML, Tsygankov D, Heddleston JM, Legant WR, Wittchen ES, Burridge K, Elston TC, Betzig E, Chew TL, Adalsteinsson D, Hahn KM (2019) Software for lattice light-sheet imaging of FRET biosensors, illustrated with a new Rap1 biosensor. J Cell Biol 218(9):3153–3160. 10.1083/jcb.201903019 [DOI] [PMC free article] [PubMed] [Google Scholar]