Abstract
Salmonella is the most important foodborne pathogen in poultry production systems and can infect humans via consumption of contaminated food. Ducks, an important waterfowl widely raised in China, are also a vehicle that transmits Salmonella through the food supply chain. In this study, 701 samples were collected from each production stage of the duck production chain. Salmonella was isolated and identified, and the isolates were tested for drug sensitivity and molecular typing based on whole genome sequencing (WGS) to explore the prevalence of Salmonella in the duck production chain. Altogether, a total of 180 Salmonella isolates (25.7%) were obtained from the duck production chain, 82 (35.7%) isolates were from hatchery samples, followed by 64 (29.2%) from market samples, 17 (23.6%) from farm samples, and 17 (9.4%) from slaughterhouse samples. All isolates were divided into 9 serotypes, among which S. Typhimurium, S. Anatum, and S. Enteritidis were the dominant serotypes. The S. Typhimurium was distributed in various production stages in the duck production chain. Among the 16 antibiotics, selected 60 isolates were only resistant to NAL, indicating that resistance of Salmonella in the duck production chain was low. WGS phylogenetic relationship results based on core-genome SNPs showed that S. Typhimurium can spread across geographic regions and along between different stages of the duck production chain, eventually reaching the market where it is a potential threat to consumer health. This study explored the prevalence of Salmonella in the duck production chain which will provide data support for proposing some interventions to control Salmonella.
Key words: Salmonella, duck production chain, whole genome sequencing, core-genome SNP
INTRODUCTION
The World Health Organization (WHO) estimated that 715,000 people died from diarrhea each year, and food poisoning accounted for one-third of this number, which Salmonella was one for the important contributor (Besser, 2018). Most human Salmonella infections are associated with the ingestion of contaminated foods, such as poultry, pork, beef, eggs, and milk (Zhao et al., 2008). Salmonella is associated with food and public health safety, and is transmitted by ingestion of contaminated food or water, or by direct contact with infected people or animals (Knodler and Elfenbein, 2019). The sources of Salmonella contamination are relatively diverse, but one of the most important sources is poultry and poultry products (Authority et al., 2019). Poultry products contaminated with Salmonella can seriously impact the life, health, and safety of consumers. Due to human production activities, Salmonella is geographically widely spread, making Salmonella contamination a global burden.
Duck is an important waterfowl widely raised in China. According to the Food and Agriculture Organization (FAO) report, China is the largest producer of duck meat, producing 3 million tons annually, and its consumption continues to increase every year (Wang et al., 2017). Since 1978, China has focused on improving livestock and poultry production, and the livestock and poultry sectors have undergone a scaling-up transformation from traditional household production mainly for self-consumption or local-market distribution, to intensive industrial production (Qian et al., 2018). At present, there are many large duck farms in China that have complete duck production chains, from hatching and breeding to sales. Recent studies have shown that waterfowl, such as ducks, are also important sources of Salmonella (Martelli et al., 2016; Kim et al., 2021). Salmonella infection in ducks is a recessive infection, however, it can also cause serious clinical symptoms with high mortality and threat to food safety (Grigar et al., 2017). Salmonella contamination can occur at any point in the duck production chain, such as production, harvest, processing, storage, transportation, and retail (Park et al., 2013). Therefore, Salmonella contamination is likely to eventually be inadvertently purchased by consumers, thereby causing harm to their health and can potentially be fatal. However, there are few studies on Salmonella contamination in duck production chains.
Many molecular typing techniques are widely used in the field of microbiology and can be used to trace the origins of pathogenic bacteria. Among them, whole genome sequencing (WGS) enables serotyping, antimicrobial resistance, virulence profiling, and subtyping in a single WGS workflow and provides high resolution and precision (Inns et al., 2017). WGS is increasingly used in public health laboratories for typing and characterizing foodborne pathogens. WGS is also frequently implemented in routine surveillance of foodborne pathogens and in foodborne disease outbreak investigations (Leekitcharoenphon et al., 2016). The US Food and Drug Administration (FDA) established a monitoring network using WGS for major foodborne pathogens (Lindsay et al., 2018), and this technique has been successfully used in Salmonella subtyping for monitoring and traceability of Salmonella prevalence (Rounds et al., 2020).
This study aimed to conduct an epidemiological survey of Salmonella in a duck production chain using samples from a hatchery and farm in Gaomi, Shandong Province, and a slaughterhouse and market in Yangzhou, Jiangsu Province. We determined the levels of Salmonella contamination in the duck production chain and used WGS typing technology to assess the clonal relationship between isolates. We also aimed to identify the key stage of Salmonella contamination and provide technical support to prevent and control Salmonella in the duck production chain.
MATERIALS AND METHODS
Sample Collection
In this study, 701 samples were collected from a duck production chain (one hatchery and one farm in Gaomi, Shandong Province; one slaughterhouse and 5 markets in Yangzhou, Jiangsu Province) for Salmonella isolation and identification, respectively. After hatching, the ducks were raised on a farm until adulthood and then transported to slaughterhouses and markets in Yangzhou, 500 km from the farm, for slaughter and sale.
A total of 230 samples were collected from the hatchery, including incubator samples, fertilized egg samples, and weak duckling samples (Table 1). A total of 72 samples were collected from the farm, including stool, feed, and water samples (Table 1). A total of 180 samples were collected from the slaughterhouse and included slaughter table samples, duck organs, duck meat, transport cage samples, and slaughterhouse stool samples from live ducks (Table 1). A total of 219 duck meat product samples were collected from 2 supermarkets and 3 retail markets in Yangzhou (Table 1). Each sample was marked, placed in a sterile plastic sample bag, transported to the laboratory on ice, and processed immediately (within 24 h of sample collection).
Table 1.
Isolation of Salmonella from different duck production stages.
| Location | Stage | Sample | Number of samples | Isolate (%) | Stage separation rate (%) |
|---|---|---|---|---|---|
| Shandong province | Hatchery | Fertilized egg | 171 | 66 (38.6) | 35.7% a |
| Incubator | 50 | 13 (26) | |||
| Weak duckling | 9 | 3 (33.3) | |||
| Farm | Stool | 50 | 17 (34) | 23.6% | |
| Feed | 12 | 0 | |||
| Water | 10 | 0 | |||
| Jiangsu province | Slaughterhouse | Slaughter table | 35 | 1 (2.9) | 9.4% |
| Duck organs | 35 | 5 (14.3) | |||
| Duck meat | 35 | 8 (22.9) | |||
| Transport cage | 35 | 0 | |||
| Slaughterhouse stool | 40 | 3 (7.5) | |||
| Market | Supermarket duck meat | 134 | 57 (42.5) | 29.2% b | |
| Supermarket duck organs | 19 | 0 | |||
| Retail market duck meat | 45 | 6 (13.3) | |||
| Retail market duck organs | 21 | 1 (4.8) | |||
| Total | 701 | 180 (25.7) |
Indicates a significant difference in isolation rate for hatchery (35.7%) to farm (23.6%) or slaughterhouse (9.4%) (P < 0.05).
Indicates a significant difference in isolation rate for market (29.2%) to farm (23.6%) or slaughterhouse (9.4%) (P < 0.05).
Isolation and Identification of Salmonella
Stool Samples
There are many other intestinal bacteria in stool samples that interfere with the isolation of Salmonella. Therefore, Salmonella was isolated from stool samples using a modified semi-solid Rappaport Vassiliadis (MSRV; Difco, BD, Sparks, MD) method (Soria et al., 2012). BPW was added to each sample at 10 times the sample volume. Each sample was incubated for 16 to 18 h at 37°C for preliminary enrichment. BPW (300 μL) was added to MSRV semi-solid medium in 3 aliquots and statically incubated for 24 h at 42°C for selective enrichment. One loopful of each MSRV was then streaked onto xylose lysine tergitol 4 (XLT4; Difco, BD) agar plates, which were incubated at 37°C for 24 h. The suspected colonies (circular, black, with a transparent annulus) were collected from the 4 mL liquid LB medium with a disposable sterile inoculating loop, and cultured for 12 to 14 h at 37°C on a constant temperature shaker for PCR identification (according to the published sequence of enterotoxin STN gene of Salmonella, the sequence of primers Ⅰ was 5′ -CTTTGGTCGTAAAATAAGGCG-3′, and the sequence of primers Ⅱ was 5′ -TGCCCAAAGCAGAGAGATTC-3′; Zhou et al., 2017). The PCR-confirmed colonies were then biochemically identified according to the API-20E Biochemical Reagent Guide. Finally, the confirmed colonies were selected for serotype identification using slide agglutination test with O and H antigens (Tianrun Bio-Pharmaceutical, Ningbo, China) according to the manufacturer's instructions. The serotype results were analyzed and interpreted according to the Kauffmann-White scheme (Wattiau et al., 2011).
Other Samples
For water samples, 100 mL of water were first filtered using 0.22-μm filter (Jinteng Co. Ltd. Tianjin, China), and the filter was collected for the experiment. For swab and water samples, the pre-enrichment step was performed by suspending each sample in 50 mL BPW and incubating the samples at 37°C for 16 to 18 h. For fertilized egg samples and weak duckling samples (liver and yolk), the pre-enrichment step was performed by weighing 10 times the volume of BPW. After weighing, the samples were incubated at 37°C for 16 to 18 h. For the market samples (duck meat and organs), each sample (25 ± 0.5 g) was aseptically weighed and transferred into 225 mL of BPW and incubated at 37°C for 18 h. BPW pre-incubation droplets (1 mL) were added to 10 mL of Rappaport-Vassiliadis R10 broth (RVR10; Difco, BD, Sparks, MD) and statically incubated for 24 h at 42°C for selective enrichment. One loopful of each RVR10 broth culture was then streaked onto XLT4 agar plates, which were incubated at 37°C for 24 h. Salmonella confirmation and identification were performed as for the stool samples.
Antimicrobial Susceptibility Testing
A total of 60 strains of Salmonella isolates were tested for antimicrobial susceptibility using the Kirby–Bauer disk diffusion method (Li et al., 2014). A total of 16 antimicrobial agents were tested: ampicillin (AMP, 10 μg), amoxicillin (AMC 20 μg), meropenem (MEM 10 μg), cefazolin (CFZ, 30 μg), aztreonam (ATM, 30 μg), nalidixic acid (NAL, 30 μg), enrofloxacin (ENR, 5 μg), ciprofloxacin (CIP, 5 μg), tetracycline (TET, 30 μg), chloramphenicol (CHL, 30 μg), kanamycin (KAN, 20 μg), amikacin (AK, 30 μg), gentamicin (GEN, 10 μg), streptomycin (STR, 10 μg), trimethoprim-sulfamethoxazole (SXT, 10 μg), and nitrofurantoin (F, 30 μg). The results were interpreted as sensitive, intermediate, or resistant according to the Clinical and Laboratory Standards Institute guidelines (CLSI, 2013). Escherichia coli ATCC 25922 and Enterococcus faecalis ATCC 29212 were used as quality control strains.
Whole Genome Sequencing and Analysis
A total of 29 S. Typhimurium and 12 S. Enteritidis isolates randomly selected from different production stages were processed for whole genome sequencing. Genomic DNA was extracted from the Salmonella isolates using the TIAN amp Bacteria DNA Kit (Tiangen, Beijing, China) according to the manufacturer's instructions. All the genomes were fragmented with an insertion size of 500 bp to construct the library, and the NEB Next Ultra DNA Library Prey Kit for Illumina (NEB, Beverly, MA) was used to generate sequencing libraries, following the manufacturer's recommendations. The gene libraries were sequenced using the Illumina platform Hiseq 2500 (Illumina, San Diego, CA) by Novogene Co. Ltd. (Beijing, China). All genomes were assembled de novo using SPAdes (version 3.15.3; Bankevich et al., 2012). The serotypes were analyzed using the Salmonella in silico typing resource (SISTR; Yoshida et al., 2016). The Multilocus Sequence Typing (MLST) of isolates was identified in silico with an open-source software, mlst (https://github.com/tseemann/mlst), which incorporates components of the PubMLST database (Jolley and Maiden, 2010). The alleles and STs were assigned according to the MLST scheme at http://mlst.warwick.ac.uk/mlst/dbs/Senterica. The antimicrobial resistance genes of the isolates were analyzed using the ResFinder 4.1 database (https://cge.cbs.dtu.dk/services/ResFinder-4.1/) (Bortolaia et al., 2020). Core-genome SNP analysis of the Salmonella isolates was performed using Snippy (https://github.com/tseemann/snippy). Then, the core-genome SNP-based maximum-likelihood (ML) tree was constructed by Gubbins (Croucher et al., 2015). WGS data of all Salmonella isolates were submitted to the European Nucleotide Archive with the accession number PRJEB51470.
Data Analysis
Data on the prevalence of Salmonella isolates from the study samples were analyzed using the statistical software program SPSS (version 16.0, SPSS, Chicago, IL). The data were compared using the chi-square test, and P < 0.05 was regarded as statistically significant.
RESULTS
Salmonella Isolation Rate in Duck Production Chain
As shown in Table 1, a total of 701 samples were collected in this study, and 180 strains of Salmonella were isolated. The total positive rate of Salmonella isolates was 25.7%. Of all the production stages, the highest rate of Salmonella isolates was 35.7% in the hatchery, followed by 29.2% in the market. The positive rate of Salmonella isolates from the slaughterhouse was 9.4%, and 23.6% from the farm samples. The isolation rate of Salmonella in the hatchery and market samples was significantly higher than that in the farm and slaughterhouse samples (P < 0.05).
Salmonella Serotype Distribution
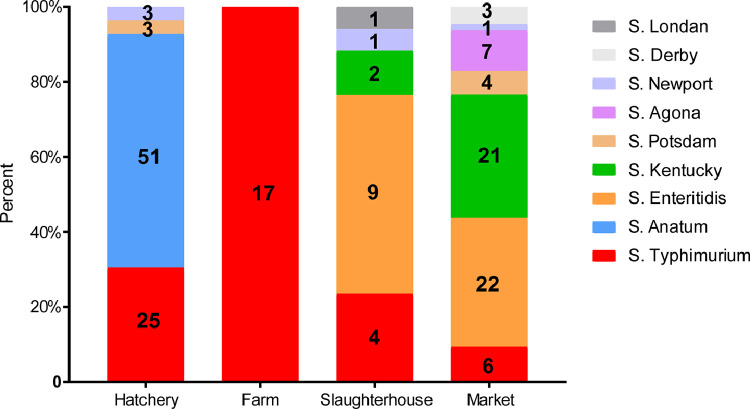
Nine different serovars were identified among the 180 positive Salmonella isolates (Figure 1), of which the isolates from market contained the most serotypes (n = 7). The isolates from hatchery and slaughterhouse contained four serotypes each, and the isolates from farm contained one serotype. In this study, the most prevalent isolated serotype was S. Typhimurium (28.9%, 52/180), followed by S. Anatum (28.3%, 51/180), S. Enteritidis (17.2%, 31/180), S. Kentucky (12.8%, 23/180), S. Potsdam (3.9%, 7/180), S. Agona (3.9%, 7/180), S. Newport (2.8%, 5/180), S. Derby (1.7%, 3/180), and S. Londan (0.6%, 1/180; Figure 1). S. Typhimurium was the most widely distributed and appeared in all stages of the production chain (Figure 1). S. Anatum was mainly concentrated in the hatchery, whereas S. Enteritidis appeared in slaughterhouse and market samples (Figure 1). Other serotypes were isolated in small amounts and concentrated in only one or 2 kinds of samples; for example, S. Derby and S. Newport were only isolated from market samples (Figure 1).
Figure 1.
The prevalence of serotypes of Salmonella isolates in duck production chain. Numbers represent the isolate numbers of different Salmonella serotypes in different stages.
Antimicrobial Resistance Phenotypes
The susceptibility of 60 Salmonella isolates to 16 antibiotics is shown in Table 2. The overall resistance of the Salmonella isolates in this study was low. They were susceptible to β-Lactams, Aminoglycosides, Sulfonamides, Tetracycline, Chloramphenicol, Nitrofuran. However, resistance to NAL was the most commonly observed in the duck production chain isolates. The S.Typhimurium isolates from duck production chain were only resistant to NAL, and the resistance rate was 100% (35/35). The resistance rate of S. Anatum to NAL was 68% (17/25).
Table 2.
Antimicrobial resistance phenotypes of 60 Salmonella isolates.
| Antibiotic | Resistance number of different sample isolates (%) |
||||
|---|---|---|---|---|---|
| Hatchery |
Farm | Slaughterhouse | Market | ||
| Serotype |
S. Typhimurium (n = 15) |
S. Anatum (n = 25) |
S. Typhimurium (n = 10) |
S. Typhimurium (n = 4) |
S. Typhimurium (n = 6) |
| β-Lactams | |||||
| Ampicillin (AMP) | 0 | 0 | 0 | 0 | 0 |
| Amoxicillin (AMC) | 0 | 0 | 0 | 0 | 0 |
| Cefazolin (CFZ) | 0 | 0 | 0 | 0 | 0 |
| Meropenem (MEM) | 0 | 0 | 0 | 0 | 0 |
| Aztreonam (ATM) | 0 | 0 | 0 | 0 | 0 |
| Aminoglycosides | |||||
| Kanamycin (KAN) | 0 | 0 | 0 | 0 | 0 |
| Gentamicin (GEN) | 0 | 0 | 0 | 0 | 0 |
| Streptomycin (STR) | 0 | 0 | 0 | 0 | 0 |
| Amikacin (AK) | 0 | 0 | 0 | 0 | 0 |
| Quinolone | |||||
| Ciprofloxacin (CIP) | 0 | 0 | 0 | 0 | 0 |
| Enrofloxacin (ENR) | 0 | 0 | 0 | 0 | 0 |
| Nalidixic acid (NAL) | 15 (100) | 17 (68) | 10 (100) | 4 (100) | 6 (100) |
| Sulfonamides | |||||
| Trimethoprim-sulfamethoxazole (SXT) | 0 | 0 | 0 | 0 | 0 |
| Tetracycline | |||||
| Tetracycline (TET) | 0 | 0 | 0 | 0 | 0 |
| Chloramphenicol | |||||
| Chloramphenicol (CHL) | 0 | 0 | 0 | 0 | 0 |
| Nitrofuran | |||||
| Nitrofurantoin (F) | 0 | 0 | 0 | 0 | 0 |
Whole Genome Sequences Analysis of S. Typhimurium
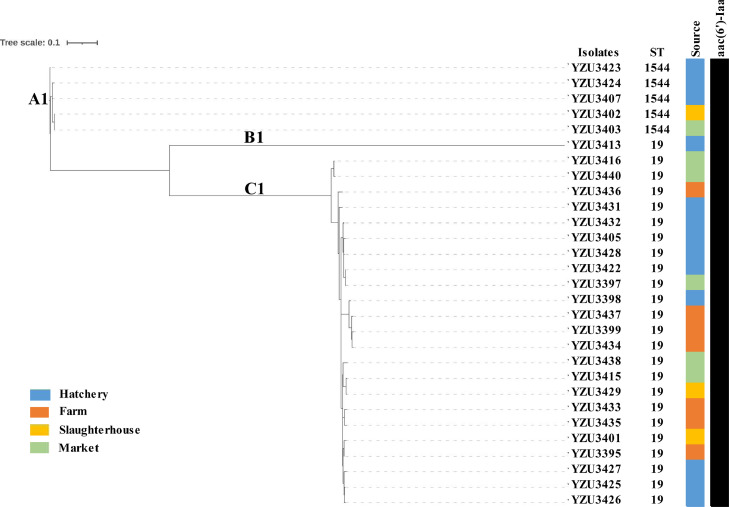
S. Typhimurium was the most predominant serotype isolated in duck production chain. Twenty-nine strains of S. Typhimurium isolates selected randomly from different production stages were used for WGS. The serotypes, MLST, antimicrobial resistance genes, and phylogenetic analysis were performed based on the assembly of whole genome sequences. The WGS data showed that the serotype of 29 Salmonella isolates was S. Typhimurium consistent with the Kauffmann-White scheme result. The MLST results showed that 29 strains of S. Typhimurium isolates were classified into 2 STs, including ST19 (82.8%, 24/29) and ST1544 (17.2%, 5/29; Figure 2). ST19 subtypes were found in all production stages, including hatchery, farm, slaughterhouse, and market. ST1544 subtypes appeared in hatchery, slaughterhouse, and market samples.
Figure 2.
Phylogenetic tree of S. Typhimurium isolates based on core-genome SNPs. The analysis included 29 S. Typhimurium isolates from different duck production stages. The antimicrobial resistant genes were listed according to the WGS data. Abbreviation: WGS, whole genome sequencing.
The antimicrobial resistance genes of 29 strains of S. Typhimurium were analyzed based on WGS. The results showed that only aac(6′)-Iaa (aminoglycoside resistance gene) was identified in the S. Typhimurium isolates (Figure 2), and it was present in all tested isolates (100%, 29/29). Quinolone resistance is usually mediated by mutations in the quinolone resistance-determining regions (QRDRs) of the gyrA, gyrB, parC, and parE genes (Almeida et al., 2018). The mutation of QRDRs in the Salmonella isolates is shown in Table 3. Our results showed that 28 of 29 strains (96.6%) presented mutation points in the gyrA gene, all of which were resistant to nalidixic acid (Table 3). The points of mutation in gyrA included gyrA (D87G) in 5 isolates and gyrA(S83Y) in 23 isolates.
Table 3.
Mutation of the QRDRs in S. Typhimurium and S. Enteritidis.
| Serotypes | gyrA | gyrB | parC | parE |
|---|---|---|---|---|
| S. Typhimurium (n = 29) | D87G (5) S83Y (23) | 0 | 0 | 0 |
| S. Enteritidis (n = 12) | D87Y (6) D87G (1) D83Y (5) | 0 | 0 | 0 |
The phylogenetic relationships between the 29 S. Typhimurium strains were assessed using core-genome SNPs. The resulting phylogenetic tree showed that the S. Typhimurium isolates were grouped into 3 clusters (A1, B1, and C1; Figure 2). The main cluster was C1, which contained 8 closely related isolates from all duck production stages (hatchery [n = 8], farm [n = 4], slaughterhouse [n = 2], and market [n = 5]). Cluster A1 contained Salmonella isolates from the hatchery, slaughterhouse, and market samples, and cluster B1 had only one isolate identified from the hatchery samples.
Whole Genome Sequences Analysis of S. Enteritidis
S. Enteritidis was identified as another prevalent serotype in the slaughterhouse and market in this study. Twelve strains of S. Enteritidis isolates selected randomly for WGS (Figure 3). Two ST patterns were identified in S. Enteritidis isolates. ST11 subtype was found in market. ST8055 which is a new subtype was found in both slaughterhouse and market. By WGS analysis 12 antimicrobial resistance genes were identified in S. Enteritidis isolates. The aminoglycoside resistance gene aac(6′)-Iaa was detected in all the isolates, followed by the beta-lactamase resistance gene blaTEM-1B (75%, 9/12). Out of the 12 S. Enteritidis isolates, 10 (83.3%) carried more than 3 antimicrobial resistance genes. A minimum spanning tree based on the cgSNP analysis showed that the S. Enteritidis isolates are grouped into 3 clusters. Clusters B2 contained ST11 subtypes. The new subtypes ST8055 were belonged to Clusters C2.
Figure 3.
Phylogenetic tree of S. Enteritidis isolates based on core-genome SNPs. The analysis included 12 S. Enteritidis isolates from slaughterhouse and market. The antimicrobial resistant genes were listed according to the WGS data. Abbreviation: WGS, whole genome sequencing.
DISCUSSION
In recent years, the prevalence of Salmonella in ducks has been reported in China (Yang et al., 2019; Chen et al., 2020), but the prevalence in the duck production chain has rarely been studied. We investigated the prevalence of Salmonella in a duck production chain across 2 provinces (Jiangsu and Shandong), and used core-genome SNPs to evaluate the clonal relationship between isolates from different sources. Our results showed that Salmonella could spread between different geographic regions and along the stages of the duck production chain.
Salmonella spp. was recovered from a duck production chain between Yangzhou, Jiangsu Province, and Gaomi, Shandong Province, with an overall prevalence of 25.7%, indicating that Salmonella contamination in the duck production chain is relatively serious. The highest isolation rate of Salmonella (35.7%) was observed at the hatchery stage indicating that a potential threat to the downstream of duck production chain. The isolation rate of Salmonella in farms was 23.6%, similar to the 21.9% found in the study by Martelli et al. (2017). Kim et al. (2021) found that the positive rate of Salmonella contamination increased significantly after the introduction of ducklings to the farm. In contrast, the duck farm in this study had strict biosecurity measures of removing weak ducklings after they hatch, which may lead to a lower rate of Salmonella isolation in the farm than in the hatchery. The isolation rate of Salmonella in the slaughterhouse was 9.4%, which is lower than the 21.7% found by Lee et al. (2016). The isolation rate in the market segment (29.2%) was significantly higher than that in the slaughterhouse (P < 0.05). Among them, the supermarket duck meat samples had the highest isolation rate (42.5%), which may be due to cross-contamination of Salmonella during production, storage, transportation, and sales.
Of the nine serovars identified in this study, the dominant serotypes were S. Typhimurium, S. Anatum, and S. Enteritidis. S. Typhimurium was the most widely distributed in duck production chain. S. Typhimurium is one of the most frequent serotypes causing foodborne diseases and has been proven to exist widely (Roccato et al., 2015). It has disrupted the development of the aquaculture industry and negatively affected human health (Sun et al., 2020). S. Anatum was only isolated in the hatchery and mainly came from fertilized egg samples (70.9%), indicating that it had been present in the hatchery for a long time. In addition, we found that S. Anatum was not isolated from the downstream production stages of the farm, slaughterhouse, and market, indicating that it may lead to dead embryos and weak ducklings, which are probably eliminated before reaching the farm stage. Yang et al. (2019) also isolated a large amount of S. Anatum from a duck farm in Shandong Province. There have been several reports that S. Anatum causes harm to a variety of livestock and poultry (Yang et al., 2019) and salmonellosis infections in humans (Hassan, 2017). In addition, S. Enteritidis and S. Kentucky accounted for a certain proportion of outbreaks, but they only existed in a few sources. S. Enteritidis was only found in the slaughterhouse and the market samples, and S. Kentucky only in the market, which shows that these 2 serotypes are newly introduced serotypes after the farm stage. Therefore, it is necessary to strengthen the disinfection of slaughterhouses and markets. S. Kentucky has been reported to be widespread in poultry such as chickens and turkeys (Vosik et al., 2018), but rarely in ducks. This shows that there is serious Salmonella cross-contamination in the market. S. Kentucky has emerged as a global human pathogen (Xiong et al., 2020) and requires further research attention.
The extensive use of antimicrobial agents has contributed to the development of antimicrobial resistance and multidrug resistance (MDR) in Salmonella (Nguyen et al., 2016). We selected 60 Salmonella isolates to conduct resistance experiments against 16 major antibiotics. 64% of Salmonella isolates from the hatchery and all Salmonella isolates from the farm, slaughterhouse, and market are resistant to NAL. The ducks in the farm are in good health, and NAL is not currently used in the farm to prevent and control bacterial contamination. Therefore, this phenomenon may be due to the large use of quinolone antibiotics in the early stage to prevent and control bacterial contamination. However, with the continuous improvement of relevant policies on the use of antibiotics and the continuous improvement strict biosecurity measures, the sensitivity of Salmonella to antibiotics has increased. The high resistant to NAL deserves further attention because resistance to this antimicrobial agent may lead to the delay or failure of fluoroquinolone therapies and could have serious consequences (Marquez-Ruiz et al., 2008). The aac(6′)-Iaa (aminoglycoside resistance gene) was found in all tested S. Typhimurium, however aac(6′)-Iaa is cryptic gene in Salmonella and cannot confer aminoglycoside resistance (Magnet et al., 1999). Quinolone resistance is typically mediated by mutations in the QRDRs of gyrA, gyrB, parC, and parE, which code for bacterial DNA gyrase, leading to changes in the binding site of the antimicrobial to the enzyme and/or the acquisition of plasmid-mediated quinolone resistance (PMQR) genes (McDermott et al., 2016). Our results showed that most of the nalidixic acid resistant (Quinolone) S. Typhimurium strains presented mutations in gyrA. However, one nalidixic acid-resistant strain did not show any mutation in gyrA, gyrB, parC, and parE, consistent with the results of Almeida et al. (2018) in S. Typhimurium strains isolated from Brazil.
Studies have shown that WGS can be used to perform molecular characterization and phylogenetic analysis of Salmonella spp. (Pearce et al., 2018). Our results showed that Salmonella serotyping based on WGS is consistent with the Kauffmann-White Scheme using antisera. The results indicated that WGS provides an effective alternative tool for genome-based identification serovars. WGS has been frequently used in Salmonella subtyping for outbreak investigation and pathogen source tracking (Hyeon et al., 2021). In our study, S. Typhimurium from the different stages in the duck production chain, clustered together suggesting that S. Typhimurium can spread along the production chain, and eventually flow into the market, becoming a potential threat to human health. Because the hatchery, farm and slaughterhouse, market were located in different geographic regions, S. Typhimurium can spread between disparate regions of the production chain. Similar findings by Karp et al. (2020) showed that Salmonella spread from Asia to the Americas through production pathways. S. Typhimurium which spreads between different regions along the duck production chain, expands the scope of contamination and has a serious impact on the local prevention and control of Salmonella. Therefore, the prevention of Salmonella contamination in food products should not only focus on the downstream market, but also the control strategies at the upstream stages, including hatcheries, farms, and slaughterhouses, to stop the spread of Salmonella along the production chain. The control of Salmonella in the whole duck production chain effectively improves the food safety of final retail products and decreases the risk of transmission to humans.
In conclusion, we examined the epidemiology of Salmonella in a duck production chain in Jiangsu and Shandong provinces. Our results indicate a high prevalence of Salmonella in the duck production chain. The core-genome SNP results showed that Salmonella isolates from different production stages were genetically similar. This shows that Salmonella can spread between different regions along the duck production chain. Therefore, the control of Salmonella in the duck production chain should be strengthened, and relevant regulations should be formulated to prevent the spread of Salmonella from the duck production chain to humans.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China project (31972685, 31902278), the China Postdoctoral Science Foundation (2018M642333), Jiangsu Province Policy Guidance Program (International Science and Technology Cooperation) (BZ2020013), the Research and Development Program of Jiangsu (BE2021354), the Yangzhou University Science and Technology Innovation Team (2018), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The funding bodies had no role in study design, in the collection, analysis, or interpretation of data, in the writing of the report, or in the decision to submit the article for publication.
DISCLOSURES
The authors declare that they have no conflict of interest.
REFERENCES
- Almeida F., Seribelli A.A., Medeiros M.I.C., Rodrigues D.D.P., de MelloVarani A., Luo Y., Allard M.W., Falcao J.P. Phylogenetic and antimicrobial resistance gene analysis of Salmonella Typhimurium strains isolated in Brazil by whole genome sequencing. PLoS One. 2018;13 doi: 10.1371/journal.pone.0201882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Authority, E. F. S., E. F. S. Authority, E. C. D. P. Contro, and ECDC The European Union one health 2018 zoonoses report. EFSA J. 2019;17:e05926. doi: 10.2903/j.efsa.2019.5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser J.M. Salmonella epidemiology: a whirlwind of change. Food Microbiol. 2018;71:55–59. doi: 10.1016/j.fm.2017.08.018. [DOI] [PubMed] [Google Scholar]
- Bortolaia V., Kaas R.S., Ruppe E., Roberts M.C., Schwarz S., Cattoir V., Philippon A., Allesoe R.L., Rebelo A.R., Florensa A.F., Fagelhauer L., Chakraborty T., Neumann B., Werner G., Bender J.K., Stingl K., Nguyen M., Coppens J., Xavier B.B., Malhotra-Kumar S., Westh H., Pinholt M., Anjum M.F., Duggett N.A., Kempf I., Nykasenoja S., Olkkola S., Wieczorek K., Amaro A., Clemente L., Mossong J., Losch S., Ragimbeau C., Lund O., Aarestrup F.M. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020;75:3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.Q., Bai J., Wang S.J., Zhang X.B., Zhan Z.Q., Shen H.Y., Zhang H.X., Wen J.P., Gao Y., Liao M., Zhang J.M. Prevalence, antimicrobial resistance, virulence genes and genetic diversity of Salmonella isolated from retail duck meat in Southern China. Microorganisms. 2020;8:444. doi: 10.3390/microorganisms8030444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical Laboratory Standards Institute (CLSI) Twenty-Third Informational Supplement M100-S23. CLSI; Wayne, PA: 2013. Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- Croucher N.J., Page A.J., Connor T.R., Delaney A.J., Keane J.A., Bentley S.D., Parkhill J., Harris S.R. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigar M.K., Cummings K.J., Rankin S.C. Prevalence of Salmonella among waterfowl along the Texas Gulf coast. Zoonoses Public Health. 2017;64:689–692. doi: 10.1111/zph.12380. [DOI] [PubMed] [Google Scholar]
- Hassan R. Multistate outbreak of Salmonella Anatum infections linked to imported hot peppers - United States, May-July 2016. MMWR Morb. Mortal. Wkly. Rep. 2017;66:838. doi: 10.15585/mmwr.mm6625a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyeon J.Y., Li S.T., Mann D.A., Zhang S.K., Kim K.J., Lee D.H., Deng X.Y., Song C.S. Whole-genome sequencing analysis of Salmonella enterica serotype Enteritidis isolated from poultry sources in South Korea, 2010-2017. Pathogens. 2021;10:45. doi: 10.3390/pathogens10010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inns T., Ashton P.M., Herrera-Leon S., Lighthill J., Foulkes S., Jombart T., Rehman Y., Fox A., Dallman T., De Pinna E., Browning L., Coia J.E., Edeghere O., Vivancos R. Prospective use of whole genome sequencing (WGS) detected a multi-country outbreak of Salmonella Enteritidis. Epidemiol. Infect. 2017;145:289–298. doi: 10.1017/S0950268816001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley K.A., Maiden M.C.J. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp B.E., Leeper M.M., Chen J.C., Tagg K.A., Watkins L.K.F., Friedman C.R. Multidrug-resistant Salmonella serotype Anatum in travelers and seafood from Asia, United States. Emerg. Infect. Dis. 2020;26:1030–1033. doi: 10.3201/eid2605.190992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.S., Kim G.S., Son J.S., Lai V.D., Mo I.P., Jang H. Prevalence, biosecurity factor, and antimicrobial susceptibility analysis of Salmonella species isolated from commercial duck farms in Korea. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler L.A., Elfenbein J.R. Salmonella enterica. Trends Microbiol. 2019;27:964–965. doi: 10.1016/j.tim.2019.05.002. [DOI] [PubMed] [Google Scholar]
- Lee S.K., Choi D., Chon J.W., Seo K.H. Resistance of strains producing extended-spectrum beta-lactamases among Salmonella from duck carcasses at slaughterhouses in three major provinces of South Korea. Foodborne Pathog. Dis. 2016;13:135–141. doi: 10.1089/fpd.2015.2042. [DOI] [PubMed] [Google Scholar]
- Leekitcharoenphon P., Hendriksen R.S., Hello S.Le, Weill F.X., Baggesen D.L., Jun S.R., Ussery D.W., Lund O., Crook D.W., Wilson D.J., Aarestrup F.M. Global genomic epidemiology of Salmonella enterica serovar Typhimurium DT104. Appl. Environ. Microbiol. 2016;82:2516–2526. doi: 10.1128/AEM.03821-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.C., Pan Z.M., Kang X.L., Geng S.Z., Liu Z.Y., Cai Y.Q., Jiao X.A. Prevalence, characteristics, and antimicrobial resistance patterns of Salmonella in retail pork in Jiangsu province, eastern China. J. Food Prot. 2014;77:236–245. doi: 10.4315/0362-028X.JFP-13-269. [DOI] [PubMed] [Google Scholar]
- Lindsay C., Flint J., Lilly K., Hope K., Wang Q., Howard P., Sintchenko V., Durrheim D.N. Retrospective use of whole genome sequencing to better understand an outbreak of Salmonella enterica serovar Mbandaka in New South Wales, Australia. Western Pac. Surveill. Response J. 2018;9:20–25. doi: 10.5365/wpsar.2017.8.4.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnet S., Courvalin P., Lambert T. Activation of the cryptic aac(6′)-Iy aminoglycoside resistance gene of Salmonella by a chromosomal deletion generating a transcriptional fusion. J. Bacteriol. 1999;181:6650–6655. doi: 10.1128/jb.181.21.6650-6655.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez-Ruiz G., Martin-Polvillo M., Velasco J., Dobarganes C. Formation of oxidation compounds in sunflower and olive oils under oxidative stability index conditions. Eur. J. Lipid Sci. Tech. 2008;110:465–471. [Google Scholar]
- Martelli F., Birch C., Davies R.H. Observations on the distribution and control of Salmonella in commercial duck hatcheries in the UK. Avian pathol. 2016;45:261–266. doi: 10.1080/03079457.2016.1146820. [DOI] [PubMed] [Google Scholar]
- Martelli F., Gosling R.J., Callaby R., Davies R. Observations on Salmonella contamination of commercial duck farms before and after cleaning and disinfection. Avian Pathol. 2017;46:131–137. doi: 10.1080/03079457.2016.1223835. [DOI] [PubMed] [Google Scholar]
- McDermott P.F., Tyson G.H., Kabera C., Chen Y.S., Li C., Folster J.P., Ayers S.L., Lam C., Tate H.P., Zhao S.H. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob. Agents Chemother. 2016;60:5515–5520. doi: 10.1128/AAC.01030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D.T.A., Kanki M., Do Nguyen P., Le H.T., Ngo P.T., Tran D.N.M., Le N.H., Dang C.V., Kawai T., Kawahara R., Yonogi S., Hirai Y., Jinnai M., Yamasaki S., Kumeda Y., Yamamoto Y. Prevalence, antibiotic resistance, and extended-spectrum and AmpC beta-lactamase productivity of Salmonella isolates from raw meat and seafood samples in Ho Chi Minh City, Vietnam. Int. J. Food Microbiol. 2016;236:115–122. doi: 10.1016/j.ijfoodmicro.2016.07.017. [DOI] [PubMed] [Google Scholar]
- Park M.K., Park J.W., Wikle H.C., Chin B.A. Evaluation of phage-based magnetoelastic biosensors for direct detection of Salmonella Typhimurium on spinach leaves. Sensor. Actuat. B-Chem. 2013;176:1134–1140. [Google Scholar]
- Pearce M.E., Alikhan N.F., Dallman T.J., Zhou Z.M., Grant K., Maiden M.C.J. Comparative analysis of core genome MLST and SNP typing within a European Salmonella serovar Enteritidis outbreak. Int. J. Food Microbiol. 2018;274:1–11. doi: 10.1016/j.ijfoodmicro.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y., Song K.H., Hu T., Ying T.Y. Environmental status of livestock and poultry sectors in China under current transformation stage. Sci. Total Environ. 2018;622:702–709. doi: 10.1016/j.scitotenv.2017.12.045. [DOI] [PubMed] [Google Scholar]
- Roccato A., Uyttendaele M., Cibin V., Barrucci F., Cappa V., Zavagnin P., Longo A., Ricci A. Survival of Salmonella Typhimurium in poultry-based meat preparations during grilling, frying and baking. Int. J. Food Microbiol. 2015;197:1–8. doi: 10.1016/j.ijfoodmicro.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Rounds J.M., Taylor A.J., Eikmeier D., Nichols M.M., Lappi V., Wirth S.E., Boxrud D.J., Smith K.E., Medus C. Prospective Salmonella Enteritidis surveillance and outbreak detection using whole genome sequencing, Minnesota 2015-2017. Epidemiol. Infect. 2020;148:e254. doi: 10.1017/S0950268820001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria M.C., Soria M.A., Bueno D.J. Comparison of 2 culture methods and PCR assays for Salmonella detection in poultry feces. Poult. Sci. 2012;91:616–626. doi: 10.3382/ps.2011-01831. [DOI] [PubMed] [Google Scholar]
- Sun H.H., Wan Y.P., Du P.C., Bai L. The epidemiology of monophasic Salmonella Typhimurium. Foodborne Pathog. Dis. 2020;17:87–97. doi: 10.1089/fpd.2019.2676. [DOI] [PubMed] [Google Scholar]
- Vosik D., Tewari D., Dettinger L., M'ikanatha N.M., Shariat N.W. CRISPR typing and antibiotic resistance correlates with polyphyletic distribution in human isolates of Salmonella Kentucky. Foodborne Pathog. Dis. 2018;15:101–108. doi: 10.1089/fpd.2017.2298. [DOI] [PubMed] [Google Scholar]
- Wang C., Chen Q., Zhang C., Yang J., Lu Z., Lu F., Bie X. Characterization of a broad host-spectrum virulent Salmonella bacteriophage fmb-p1 and its application on duck meat. Virus Res. 2017;236:14–23. doi: 10.1016/j.virusres.2017.05.001. [DOI] [PubMed] [Google Scholar]
- Wattiau P., Boland C., Bertrand S. Methodologies for Salmonella enterica subsp. enterica subtyping: gold standards and alternatives. Appl. Environ. Microbiol. 2011;77:7877–7885. doi: 10.1128/AEM.05527-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z.Y., Wang S.J., Huang Y.M., Gao Y., Shen H.Y., Chen Z.Q., Bai J., Zhan Z.Q., Wen J.P., Liao M., Zhang J.M. Ciprofloxacin-resistant Salmonella enterica Serovar Kentucky ST198 in broiler chicken supply chain and patients, China, 2010-2016. Microorganisms. 2020;8:140. doi: 10.3390/microorganisms8010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Ju Z.J., Yang Y., Zhao X.N., Jiang Z.Y., Sun S.H. Serotype, antimicrobial susceptibility and genotype profiles of Salmonella isolated from duck farms and a slaughterhouse in Shandong province. China. BMC Microbiol. 2019;19:202. doi: 10.1186/s12866-019-1570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida C.E., Kruczkiewicz P., Laing C.R., Lingohr E.J., Gannon V.P.J., Nash J.H.E., Taboada E.N. The Salmonella in silico typing resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One. 2016;11 doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., White D.G., Friedman S.L., Glenn A., Blickenstaff K., Ayers S.L., Abbott J.W., Hall-Robinson E., McDermott P.F. Antimicrobial resistance in Salmonella enterica serovar Heidelberg isolates from retail meats, including poultry, from 2002 to 2006. Appl. Environ. Microbiol. 2008;74:6656–6662. doi: 10.1128/AEM.01249-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z.H., Li J.W., Zheng H.J., Jin X.C., Shen Y., Lei T.Y., Sun X.Y., Pan Z.M., Jiao X.A. Diversity of Salmonella isolates and their distribution in a pig slaughterhouse in Huaian, China. Food Contr. 2017;78:238–246. [Google Scholar]