Abstract
Surface plasmon resonance (SPR) is a widely used method to study ligand–protein interactions. The throughput and sensitivity of SPR has made it an important technology for measuring low-affinity, ultralow weight fragments (<200 Da) in the early stages of drug discovery. However, the biochemistry of membrane proteins, such as G-protein-coupled receptors (GPCRs), makes their SPR fragment screening particularly challenging, especially for native/wild-type, nonthermostabilized mutant receptors. In this study, we demonstrate the use of SPR-based biosensors to study the entire human family of adenosine receptors and present biologically active novel binders with a range of selectivity to human adenosine 2a receptor (hA2AR) from an ultralow weight fragment library and the public GlaxoSmithKline (GSK) kinase library. Thus, we demonstrate the ability of SPR to screen ultra-low-affinity fragments and identify biologically meaningful chemical equity and that SPR campaigns are highly effective “chemical filters” for screening small building block fragments that can be used to enable drug discovery programs.
Keywords: surface plasmon resonance, GPCRs, fragments, screening
The adenosine 2a receptor (A2AR) belongs to a clade of nucleoside sensing G-protein-coupled receptors (GPCRs) that are stimulated by the ATP metabolite adenosine.1 Adenosine levels fluctuate, accumulating over time during the wake cycle2,3 and with increasing utilization of cellular energy. Adenosine receptors allow cells to sense these changes and respond by adapting metabolic processes at the cellular and organismal level.4,5 High concentrations of adenosine in muscle tissue increase blood flow by promoting local vasodilation following activation of adenosine receptors.6 Lipolysis7 and insulin secretion8 are also influenced by adenosine receptors responding to dynamic adenosine levels. A well-known effect of adenosine accumulation in the brain is a sense of fatigue, a sensation that can be counteracted by human adenosine 2a receptor (hA2AR) antagonists such as caffeine.9 High adenosine levels also suppress inflammation during periods of high-energy use.10 In this way, adenosine receptors function as a rheostat for energy homeostasis related processes, providing healthy responses to increased energy utilization. Adenosine signaling can also support pathological states. Tumor microenvironments (TMEs) frequently contain high levels of adenosine which, via hA2AR activity, suppresses antitumor immune responses5,11 such as neutrophil invasion,12 natural killer cell maturation,13,14 and inhibiting cytotoxic CD8+ T cell tumor cell killing.15 Thus, there is significant interest in understanding adenosine receptor pharmacology and physiology.
Targeting adenosine receptor subtypes in drug discovery has been challenging. There is a high level of endogenous expression in commonly used cellular systems for drug screening,16 which may contribute a significant background signal, thus complicating the determination of selectivity. Biochemical methods using purified receptors are also challenging due to the difficulty in purifying large quantities of functional membrane proteins and limitations of guanosine triphosphate (GTP)-loading assays. Moreover, both methods lack the sensitivity required to detect low-affinity interactions. Conversely, biophysical methods such as surface plasmon resonance (SPR) utilizing exceptionally small quantities of protein can be used in a high-throughput manner to screen for even ultra-low-affinity chemical fragments.17−19 Utilizing drug-like chemical fragments allows the efficient assaying of large quantities of accessible chemical space. This results in the identification of starting points to seed structure–activity relationship (SAR)-driven medicinal chemistry campaigns to increase the affinity of generated molecules for use in downstream cellular efficacy and translational assays. Because of the efficiency in material requirements and throughput, SPR can be used to determine selectivity of molecules during hit-finding and in all stages of development, including acting as a chemical filter to eliminate nonbinding molecules preceding informative biological and translational assays. Purifying wild-type functional GPCRs for this purpose is especially challenging, and many groups have utilized thermostable mutants20 which may impart non-native binding and functional limitations.21,22 In this study, we sought to identify a diverse set of chemical scaffolds that could support a selectivity-focused A2AR drug discovery program using purified wild-type/native GPCRs. We conducted a screen of 656 fragments (supplied by Drug Discovery Unit, University of Dundee) and the GlaxoSmithKline (GSK) kinase library set (367 compounds, supplied by GSK) against human wild-type A2AR (A2AR), established their selectivity profile against the entire human adenosine receptor family (wild-type A1R, A2BR, A3R), and characterized their functional activity in controlled classical live-cell signaling assays designed to allow for the detection of effects of low-affinity interacting fragment-like molecules. We identified 16 fragments, with the exception of one we could validate bona fide biological activity in live-cell assays. These data demonstrate conclusively the effectiveness of SPR-driven hit-finding campaigns in accurately filtering large swathes of chemical space, identifying scaffolds that could be used in developing hA2AR-targeting therapeutics.
Assay Development for the Adenosine A2A Receptor
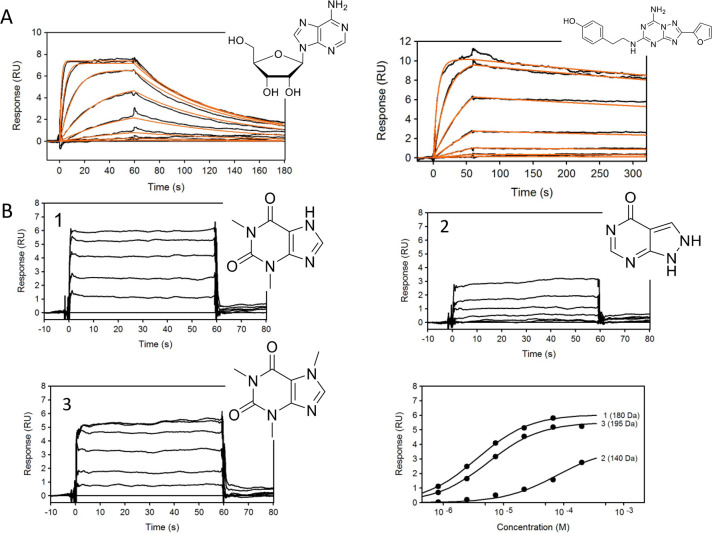
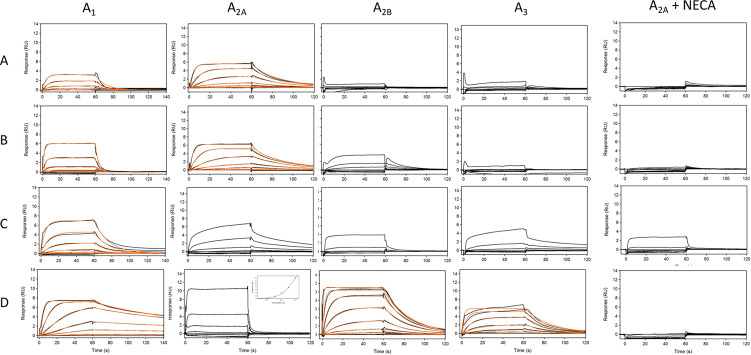
The SPR A2A assay was validated using known activity probes such as the agonist adenosine, the antagonist ZM 241385, and three fragment-like molecules theophylline (mol wt = 180 Da), allopurinol (mol wt = 140 Da), and caffeine (mol wt = 195 Da). The binding sensorgrams obtained are shown in Figure 1A,B. Binding affinities and kinetic parameters are summarized in Table 1 and correspond well to the literature results.17,23 Although the response levels for fragments were low, it was possible to detect fragments as small as 140 Da with affinities ranging from 3.6 to 77 μM.
Figure 1.
(A) Binding sensorgrams of control compounds adenosine (left) and ZM 241385 (right). Adenosine was injected from 7.8 nM to 1 μM, and ZM 241385 was injected in concentration series from 91.4 pM to 200 nM. Black lines represent binding sensorgrams, and red lines represent 1:1 kinetic fit. (B) Binding sensorgrams of fragment-like compounds theophylline (1), allopurinol (2), and caffeine (3) injected at a concentration series from 0.823 to 200 μM. Affinity was calculated using equilibrium 1:1 binding model.
Table 1. Control Compound Affinities/Kinetic Parameters Binding to Adenosine Receptors A1, A2A, A2B, and A3a.
| receptor | compound | ka (M–1 s–1) | kd (s–1) | KD | Ki (literature) | figure |
|---|---|---|---|---|---|---|
| A2A | adenosine | 9.53(±0.05) × 105 | 0.016(±0.008) | 17.3(±0.1) nM | 150 nM | 1A |
| A2A | ZM 241385 | 2.42(±0.08) × 106 | 6.92(±0.03) × 10–4 | 286(±1) pM | 395 pM | |
| A2A | theophylline | N/A | N/A | 3.63(±0.05) μM | N/A | 1B |
| A2A | allopurinol | N/A | N/A | 77(±3) μM | N/A | |
| A2A | caffeine | N/A | N/A | 5.51(±0.08) μM | N/A | |
| A1 | SLV320 | 6.27(±0.01) × 105 | 0.0034(±0.0004) | 5.46(±0.01) nM | 1 nM | 2C–F |
| A2A | CV1808 | 2.93(±0.03) × 106 | 0.027(±0.002) | 9.1(±0.1) nM | 76 nM | |
| A2B | LUF5834 | 1.10(±0.06) × 105 | 0.0086(±0.0004) | 78.2(±0.03) nM | 12 nM | |
| A3 | adenosine | 2.49(±0.03) × 104 | 0.077(±0.007) | 3.07(±0.01) μM | 290 nM |
ka, on-rate; kd, off-rate; KD, affinity; Ki, inhibition constant from literature.
A2A Fragment Screening
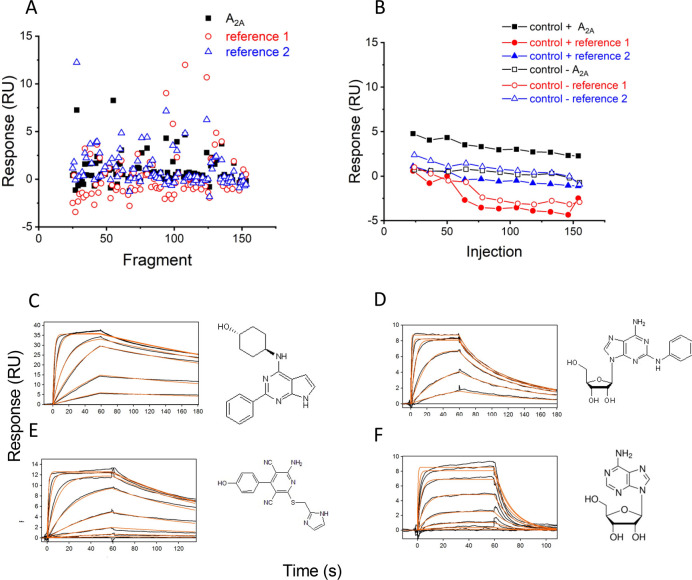
Fragments were screened against the A2A receptor and two additional reference receptors at a single concentration of 50 μM. During each screen, the positive control theophylline and negative control sulpiride were also injected over all receptors. The fragment library was split into small subsets in order to screen each subset within 12 h on a Biacore S200 to avoid significant loss of the receptor from the surface. Figure 2A shows an example of single concentration responses for each fragment from one subset binding to the A2A receptor and the reference receptors. Control binding responses against target and reference surfaces are shown in Figure 2B. A subset of fragments was selected based on the binding response to a target versus reference surfaces and screened in a concentration series to confirm binding and affinity against the A2A receptor.
Figure 2.
(A) Responses of fragments binding to the A2A receptor (black squares), reference 1 (red empty circle) and reference 2 (blue empty triangle) read immediately prior to the end of injection. (B) Binding responses of positive control, theophylline (solid symbols), and negative control, sulpiride (empty symbols), binding to the A2A receptor (square) and reference proteins (circle, triangle). (C) Binding sensorgrams of SLV320 binding to the A1 receptor at a 3-fold concentration series ranging from 4.57 nM to 10 μM. (D) Binding sensorgrams of CV1808 binding to the A2A receptor at a 3-fold concentration series ranging from 4.57 nM to 10 μM. (E) Binding sensorgrams of LUF5834 binding to the A2B receptor at a 3-fold concentration series ranging from 4.57 nM to 10 μM. (F) Binding sensorgrams of adenosine binding to the A3 receptor at a 3-fold concentration series ranging from 0.0457 to 100 μM. Black lines represent binding sensorgrams for each concentration, and red lines are a 1:1 kinetic fit.
Adenosine Receptor Selectivity Assay
To determine whether the fragment hits showed selectivity to the A2A receptor relative to the remaining adenosine receptor family, we developed SPR-based assays for adenosine receptors A1, A2B, and A3. To assess the conformational activity of the receptors, we screened classically selective (Table 1) ligands for each receptor and compared the results to literature data. Binding sensorgrams for SLV320, CV1808, LUF5834, and adenosine binding to A1, A2A, A2B, and A3, respectively, are shown in Figure 2C–F. Binding affinities are summarized in Table 1 and correspond well to literature data.17,24−26
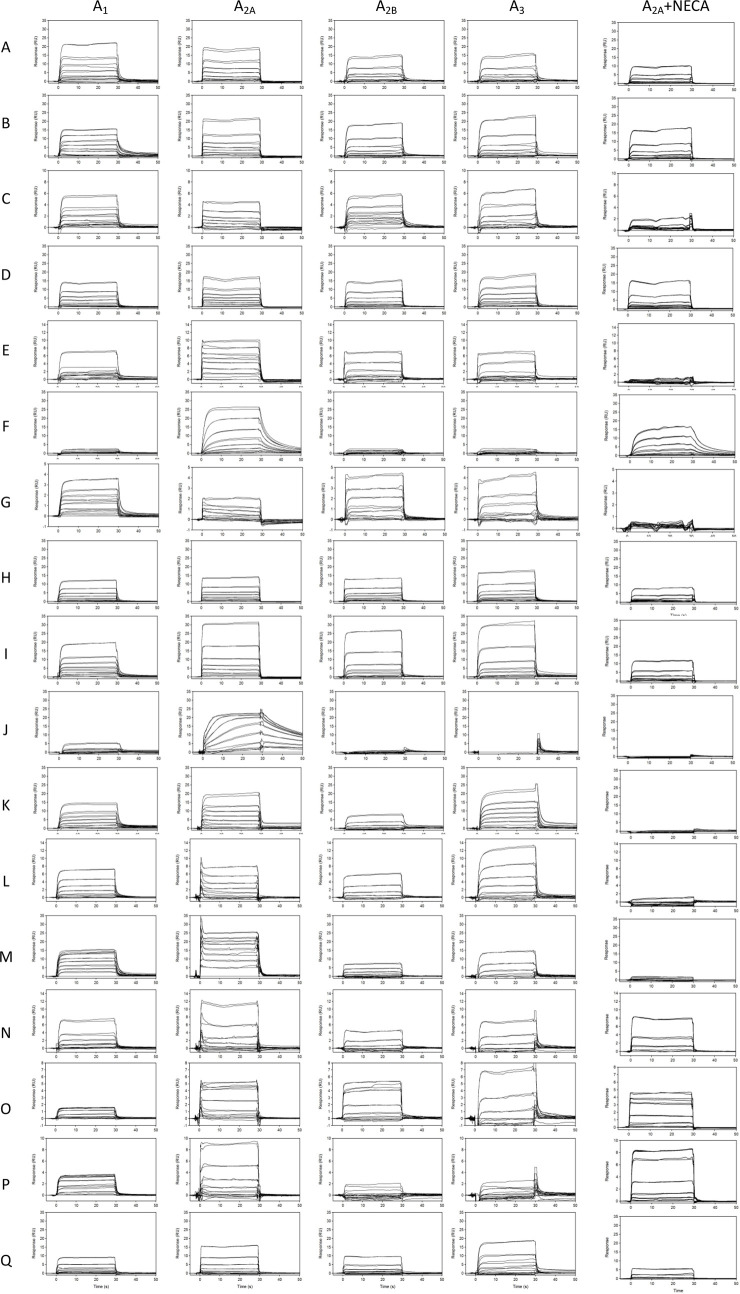
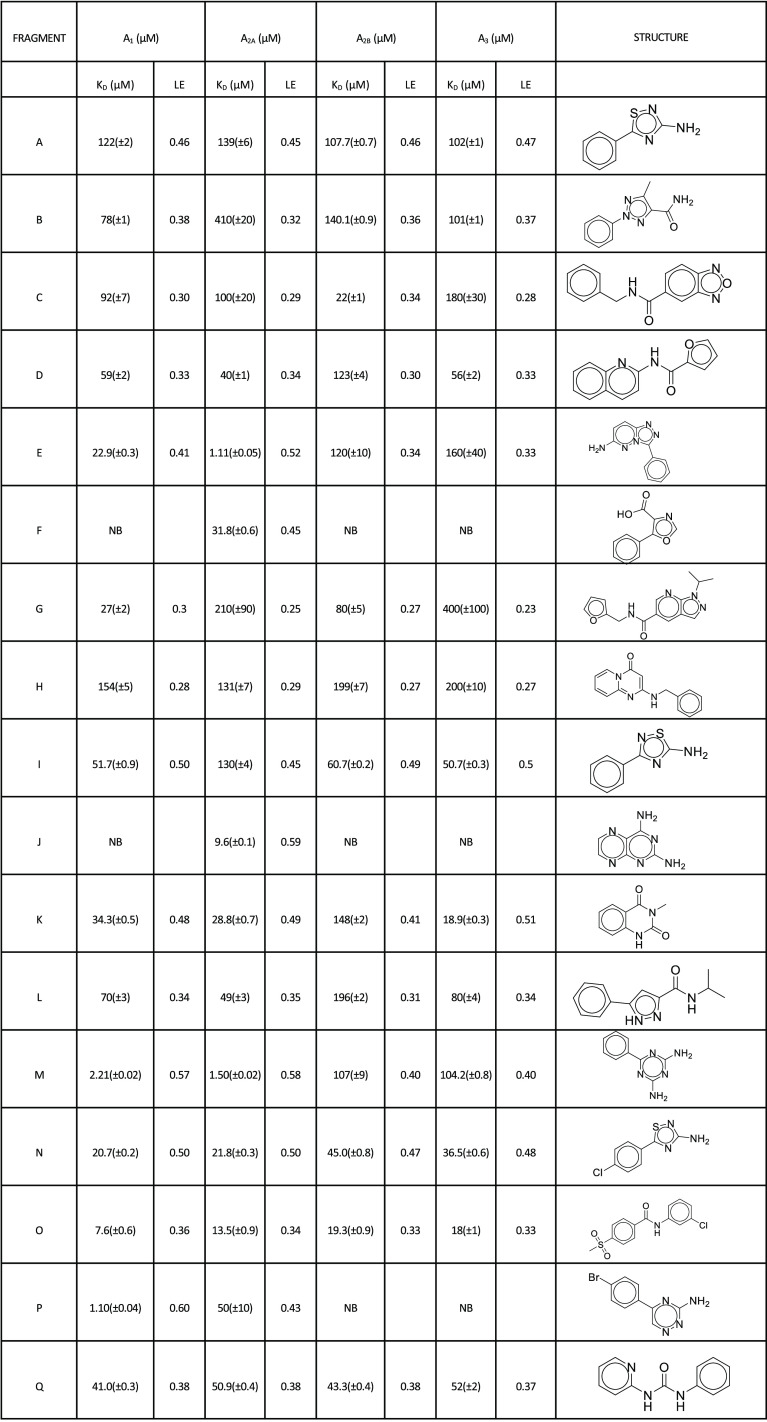
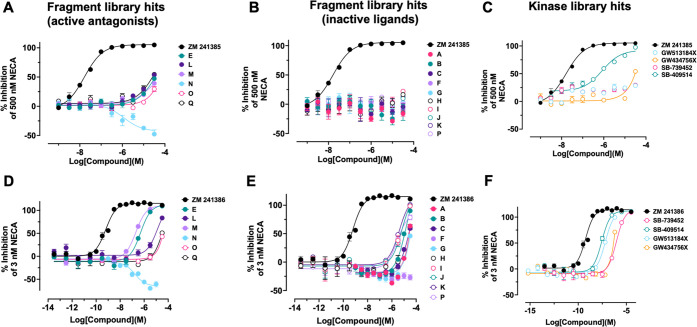
The 17 confirmed fragment hits (fragments A–Q) for the A2A receptor were screened at a concentration series against all adenosine receptors. Affinities were measured at a steady state for fragments that did not show curvature in the association/dissociation phase (with the exception of fragments F and J). The accuracy of curve fits in these instances is lower toward weaker binders as, at testing concentrations, fragments do not reach saturation. Increasing concentration beyond these levels can result in substantial nonspecific interactions with the target, limited solubility, or aggregation. In these cases, fixing Rmax based on the normalized saturating response of other fragments or those that show kinetic binding allows for effective estimates of affinity. The binding sensorgrams are shown in Figure 3, and structures and affinity fits are summarized in Table 2 and Supporting Information Figure S2. Interestingly, several fragments showed high similarity score (>0.5) that could be used for SAR optimization (fragments A, B, F, I, M, N, O, and Q). All fragments, with the exception of fragments F and J, showed binding responses to all adenosine receptors. Unlike the rest of the hits, fragments F and J show not only selectivity to the A2A receptor but also slower off-rates. Fragment M showed high affinity to the A2A receptor at 1.5 μM but also to the A1 receptor at 2.2 μM. Some fragments, such as O, showed higher affinity to the A1 receptor (1.1 μM) compared to that with the A2A receptor (50 μM); however, the fragment is binding at lower Rmax to A1 compared to A2A. All fragments were also screened against the A2A receptor in the presence of 1 μM 5′-N-ethylcarboxamide adenosine (NECA), which is well above NECA’s binding affinity of 34 nM (Figure S4) needed to saturate all available “orthosteric” binding sites for this compound (Figure 3). Interestingly, only fragments A, C, E, G, I, J, K, L, M, and Q showed significant competition with NECA in the solution. Fragments B, D, F, H, M, N, O, and P showed partial or no competition suggesting these fragments bound either to the unoccupied binding pocket on the A2A receptor or to a different (“allosteric”) binding site. To contextualize these findings, a similarity analysis of the novel structures of fragments A–Q was performed to identify the nearest neighbors in the EBI’s ChEMBL database with reported adenosine binding compounds (Table S1). To further the analysis, we also compared fragment hit structures to nearest neighbors within the fragment library that did not show activity (Table S2).
Figure 3.
Binding sensorgrams of fragments A–Q binding to A1, A2A, A2B, and A3. Each compound is injected at concentrations specific to each receptor ranging from 250 nM to 300 μM. Right panel: binding of fragments A–Q to A2A receptor in the presence of 1 μM NECA.
Table 2. Chemical Structures and Binding Affinities of Each Fragment Binding to Adenosine Receptors A1, A2A, A2B, and A3a.
NB = no binding.
Kinase Library Screening against A2A Receptor
To determine the suitability of the SPR assay for larger molecule screening, we measured binding of 367 small molecule compounds from the GSK kinase library set against the adenosine A2A receptor. Each compound was screened at three concentrations to determine the specificity of binding. We selected 19 compounds as possible binders to A2A and screened these against four adenosine receptors at eight concentrations to determine binding affinity and kinetic parameters. We confirmed four compounds as binders: SB-739452, SB-409514, GW513184X, and GW434756X (Figure S1). The data are shown in Figure 4, and kinetic parameters are reported in Table 3. We found that compound SB-739452 is selective to the A2A receptor with only weak binding to the A1 receptor. Compound SB-409514 bound with nanomolar affinity to A2A, and only a weak response was detected to A1 and A2B receptors. Compound GW513184X showed a complex aggregation binding profile to A2A, A2B, and A3 receptors but 1.2 μM binding to the A1 receptor. Interestingly, compound GW434756X was found to be a weak binder to A2A but showed very high affinity binding to the A1 receptor (6 nM) and also nanomolar binding affinities to A2B and A3 receptors (283 and 742 nM). Compounds that predominantly bound to the A1 and A2A receptors (SB-739452, SB-409514, and GW513184X) were previously shown to be potent GSK-3 inhibitors with IC50 values of 7, 94, and 100 nM, respectively.27−29 Compound GW434756X bound weakly to the A2A receptor but with high affinities to A1, A2B, and A3 receptors identified as ∼120 nM inhibitor of p38 kinase.30 All compounds were competitive with 1 μM NECA to the A2A receptor (Figure 4).
Figure 4.
Binding sensorgrams of selected hits from GSK kinase library binding to adenosine receptors A1, A2A, A2B, and A3. Compound A = SB-739452, compound B = SB-409514, compound C = GW513184X, and compound D = GW434756X. Right panels: binding of compounds A–D to A2A receptor in the presence of 1 μM NECA. Orange traces represent a 1:1 kinetic fit.
Table 3. Kinetic Parameters and Binding Affinity of Four Hits from GSK Kinase Library against Adenosine Receptors A1, A2A, A2B, and A3.
| receptor | A1 |
A2A |
||||
|---|---|---|---|---|---|---|
| compound | ka (M–1 s–1) | kd (s–1) | KD | ka (M–1 s–1) | kd (s–1) | KD |
| SB-739452 | 4.25(±0.08) × 104 | 0.09(±0.01) | 2.12(±0.04) μM | 7.62(±0.01) × 104 | 0.04(±0.01) | 0.56(±0.03) μM |
| SB-409514 | 1.21(±0.01) × 105 | 0.27(±0.03) | 2.21(±0.02) μM | 7.23(±0.01) × 105 | 0.079(±0.01) | 0.11(±0.01) μM |
| GW513184X | 5.33(±0.04) × 104 | 0.06(±0.04) | 1.21(±0.01) μM | complex binding | ||
| GW434756X | 1.63(±0.02) × 106 | 0.01(±0.001) | 6.05(±0.04) × 10–3 μM | N/A | N/A | 8.9(±0.4) μM |
| A2B | A3 | |||||
|---|---|---|---|---|---|---|
| SB-739452 | no binding | no binding | ||||
| SB-409514 | weak binding | no binding | ||||
| GW513184X | weak binding | complex binding | ||||
| GW434756X | 1.52(±0.02) × 105 | 0.043(±0.005) | 0.28(±0.02) μM | 5.60(±0.01) × 104 | 0.041(±0.001) | 0.74(±0.01) μM |
Functional Characterization of Fragment and Kinase Library Hits at the A2A Receptor in Cyclic AMP (cAMP) Accumulation and Gs Bioluminescence Resonance Energy Transfer (BRET) TRUPATH Assays
Fragment hits A–Q and the four kinase targeting compounds were screened in agonist and antagonist modes at the A2A receptor. The A2A receptor agonist, NECA, potently stimulated the accumulation of cAMP (pEC50 = 7.64 ± 0.071; n = 12; Figure S4A) at the A2A receptor, corresponding to previous reports.31 Fragments A–Q and the kinase library hits were unable to stimulate cAMP accumulation in HEK293T cells transiently expressing the hA2A receptor, indicating that these compounds lack agonist activity (Figure S4A,B).
To determine if the fragment and kinase library hits exhibit A2A receptor antagonist activity, concentration–response curves of the test compounds were competed with an EC80 of NECA (500 nM) in cAMP accumulation assays (Figure 5A–C). For comparison, the potency of the known A2A receptor antagonist, ZM 241385, was also determined. The reference antagonist, ZM 241385, potently and fully inhibited the cAMP accumulation stimulated by 500 nM of NECA (pIC50 = 8.39 ± 0.80) at the A2A receptor (Figure 5A–C and Table S3). Five fragments (fragments E, L, M, O, Q) were able to partially inhibit the cAMP accumulated by 500 nM of NECA, but their effects in this assay did not reach a saturable limit (Figure 5A), likely a consequence of their low binding affinity for the receptor as seen in the SPR assays (Figure 3 and Table 2). In contrast, fragment N potentiated the cAMP signal stimulated by NECA-stimulated A2A receptor (Table S3). These data suggest that fragment N may be acting as an allosteric modulator, as it did not stimulate cAMP accumulation in the absence of NECA (Figure S4A) and was not competitive with NECA in the SPR binding assay. Under these assay conditions, all other fragments appeared to be inactive as antagonists. SB-409514 fully inhibited NECA-induced cAMP accumulation, whereas GW434756X only partially inhibited the cAMP response at its highest concentration tested (Figure 5C). Both GW513184X and SB-739452 were unable to antagonize the NECA-stimulated cAMP response under these conditions.
Figure 5.
Antagonism of NECA-induced A2A receptor-mediated cAMP accumulation as measured by a GloSensor assay. Ability of the A2A receptor antagonist, ZM 241385, fragment library hits (A,B,D,E) or kinase library hits (C,F) to inhibit the accumulation of cAMP as induced by an effective 80% concentration (EC80) of NECA (500 nM) or effective 20% concentration (EC20) of NECA (3 nM). Data are expressed as a percentage of the maximum NECA response and represent the mean ± SEM of at least three independent experiments performed in technical duplicate.
Since the binding-hit compounds had been identified to bind to the A2A receptor with a range of affinities ranging from low to high micromolar (Table 2), it was hypothesized that the lack of apparent antagonist activity for some compounds may be a consequence of their affinity being too weak to overcome and block cAMP accumulation stimulated by an EC80 of NECA. Antagonist experiments were repeated, but test compounds were instead competed with a stimulation EC20 of NECA (3 nM; Figure 5D–F). Fragments E and M now behaved as full antagonists with potency values in the sub-micromolar range. Fragments L, O, and P had greater activity at their highest concentration (Figure 5D and Table S3). Fragment N, which demonstrated putative positive allosteric modulation, was approximately 7-fold more potent when interacted with an EC20 of NECA (Figure 5D and Table S3). Fragments that were inactive as antagonists under the previous conditions showed some inhibitory activity when interacted with an EC20 of NECA (Figure 5E and Table S3). Similarly, SB-409514 was 76-fold more potent as a full antagonist (Figure 5F and Table S3). Both SB-739452 and GWS513184X now demonstrated full antagonist activity. Due to compound availability, GW513184X and GW434756X were tested at 0.63 mM as the maximum concentration, so it is possible that GW434756X would demonstrate full antagonist activity if the assay was repeated at a higher maximum concentration of the compound.
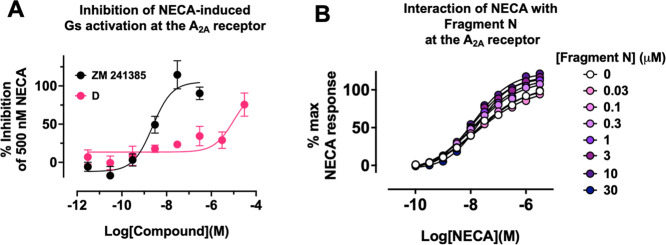
All fragments were inactive in the GloSensor assay in cells lacking transfected A2A (Figure S4C). We did notice that the baseline luciferase signal in this assay was inhibited by fragment D though not the other fragments. We surmised that this may be due to inhibition of the firefly luciferase in the GloSensor protein and so excluded fragment D from these specific analyses. Interestingly, analysis by the similarity ensemble approach32 predicted that fragment D could interact with a luciferase related to that used in the GloSensor assay (luciferin 4-monoxygenase) based on similarity to known inhibitors. Fragment D, for instance, is identical to ZINC152092 (a potent firefly luciferase inhibitor33) with the exception that the furan of fragment D is replaced by a phenyl. Because the GloSensor relies on an ATP-dependent firefly luciferase, it is reasonable to assume that fragments that bind to adenosine nucleotide binding pockets may yet still bind and alter the pharmacology of nucleotide binding receptors such as A2A. To determine whether fragment D was still biologically active, we performed an orthologous TRUPATH assay which did not rely on the firefly luciferase enzyme or any other ATP-dependent enzymes.34−36 The reference antagonist ZM 241385 completely inhibited the activation of the Gs short protein induced by 500 nM of NECA at the A2A receptor (pIC50 = 8.82 ± 0.25; Figure 6A). Fragment D also demonstrated antagonist activity in this assay but did not saturate (estimated pIC50 = 6.78 ± 2.44; Figure 6A) but, unlike the GloSensor assay, did not exhibit a comparable effect in pcDNA transfected control cells (Figure S4D).
Figure 6.
(A) Ability of ZM 241385 or fragment D to inhibit NECA-induced activation of Gs proteins at the A2A receptor in a Gs TRUPATH BRET assay performed in HEK293T cells transiently transfected with the A2A receptor and G proteins. Data are expressed as either a percentage of the maximum NECA response or as a percentage inhibition of the response generated by 500 nM NECA. (B) Effect of the putative allosteric modulator, fragment N, on NECA-stimulated cAMP accumulation at the A2A receptor in HEK293T cells. Data represent the mean ± SEM of three to four experiments performed in at least duplicate.
Since fragment N demonstrated no agonist activity in the GloSensor cAMP accumulation assay but potentiated the ability of an EC80 and EC20 of NECA to stimulate cAMP accumulation (Figure 5A,D and Table S3), we hypothesized that the fragment may be acting via an allosteric mode of action. To further understand the mechanism underlying the activity of fragment N, a cAMP accumulation assay was performed in cells transiently expressing the A2A receptor to determine the degree at which fragment N can modulate NECA agonism (Figure 6B). Interaction of increasing concentrations of fragment N with a concentration–response curve of NECA increased both the potency and Emax of the NECA curve as determined by F-test when the data were fitted to a standard four-parameter logistic function (pEC50p = 0.003; Emaxp = 0.0003). Application of the operational model of allosterism to the data set did not yield a reliable fit of the model,37−39 likely due to the low affinity and potency of the fragment for the A2A receptor. Notably the concentrations tested did not appear to saturate even at 30 μM.
SPR is a highly sensitive assay and with specific data evaluation procedures can be used to detect even extremely weak interactions between chemical entities and target proteins. The approach has been historically beset by limitations that restricted its use to soluble targets. Expanding purification techniques for membrane proteins via the use of novel detergents and lipid formulations, polymers for the production of nano- and lipodiscs, will increase the viability of this approach to wider groups of therapeutic targets. While SPR can screen fragment-like molecules that allow for extrapolation of extremely large and diverse chemical spaces, it cannot predict the biological activity of these binders. Here, we have presented an example of an integrated pharmacology pipeline that takes advantage of the high sensitivity of SPR to interrogate an extremely difficult target class (wild-type GPCRs) and downstream pharmacological activity. Thus, SPR can be used to sample and filter pharmacological space to eliminate the need for high-throughput biological screens that often require higher-affinity compounds and may exclude perfectly viable starting material for drug design. Additionally, by winnowing down the hit material, it becomes far more practical to design experimental assays that are sensitive to the requirements of the screening material. Unlike crystallography, SPR requires lower amounts of proteins, and assays can be optimized to cover proteins with poor thermostability. Due to its amenity for screening diverse libraries of molecules from small fragments to drug-like compounds, SPR can be used during SAR optimization of fragments to larger, more potent molecules. SPR for screening of membrane receptors thus continues to expand the capability to identify novel and selective matter for drug design and development, especially when combined with careful biological assays to establish a binding mode and biological functionality. We also presented SPR assays developed for the family of adenosine receptors which could point to selectivity properties of fragments at early stages of hit discovery. We found that, at the fragment stage, selectivity was limited to higher-affinity fragments such as J and F, suggesting that we could obtain more selectivity for compounds once the fragments are optimized as larger molecules.
Acknowledgments
The work was partially supported by BBSRC CASE PhD Studentship BB/J011770/1, IMI-K4DD, Kinetic Discovery and Exscientia. The GSK compound library published kinase inhibitor set (PKIS) was kindly provided by GSK. We would like to thank Cytiva (formerly GE Healthcare Life Sciences) for access to Biacore S200, and Jeremy Besnard (Exscientia) for running compound similarity evaluations.
Glossary
Abbreviations
- A2AR
adenosine 2a receptor
- BRET
bioluminescence resonance energy transfer
- cAMP
cyclic AMP
- Da
dalton
- GPCR
G-protein-coupled receptor
- GSK
GlaxoSmithKline
- GTP
guanosine triphosphate
- hA2AR
human adenosine 2a receptor
- NECA
5′-N-ethylcarboxamide adenosine
- SAR
structure–activity relationship
- SPR
surface plasmon resonance
- TME
tumor microenvironment
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.2c00099.
All materials and methods; supplementary figures include the chemical structures of kinase compounds, affinity and kinetic fits for fragments against the adenosine receptots, NECA binding to the A2A receptor and A2A receptor-mediated cAMP accumulation measured by a GloSensor assay; supplementary tables include similarity analysis of both fragment structures to known adenosine receptor binders from ChEMBL and nearest inactive neighbors from the fragment library, and inhibitory and potency efficacy estimates of fragment and kinase hits in a cAMP GloSensor assay (PDF)
Author Contributions
⊥ C.S., S.R., and A.B. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Borea P. A.; Gessi S.; Merighi S.; Vincenzi F.; Varani K. Pharmacology of Adenosine Receptors: The State of the Art.. Physiol Rev. 2018, 98 (3), 1591–1625. 10.1152/physrev.00049.2017. [DOI] [PubMed] [Google Scholar]
- Basheer R.; Strecker R. E.; Thakkar M. M.; McCarley R. W. Adenosine and Sleep–Wake Regulation.. Prog. Neurobiol 2004, 73 (6), 379–396. 10.1016/j.pneurobio.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Dworak M.; McCarley R. W.; Kim T.; Kalinchuk A. V.; Basheer R. Sleep and Brain Energy Levels: ATP Changes during Sleep.. J. Neurosci. 2010, 30 (26), 9007–9016. 10.1523/JNEUROSCI.1423-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredholm B. B.; Johansson S.; Wang Y.-Q. Chapter 3 Adenosine and the Regulation of Metabolism and Body Temperature.. Adv. Pharmacol. 2011, 61, 77–94. 10.1016/B978-0-12-385526-8.00003-5. [DOI] [PubMed] [Google Scholar]
- Jiang Y.; Li Y.; Zhu B. T-Cell Exhaustion in the Tumor Microenvironment.. Cell Death Dis 2015, 6 (6), e1792–e1792. 10.1038/cddis.2015.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layland J.; Carrick D.; Lee M.; Oldroyd K.; Berry C. Adenosine Physiology, Pharmacology, and Clinical Applications.. Jacc Cardiovasc Interventions 2014, 7 (6), 581–591. 10.1016/j.jcin.2014.02.009. [DOI] [PubMed] [Google Scholar]
- Koupenova M.; Ravid K. Adenosine, Adenosine Receptors and Their Role in Glucose Homeostasis and Lipid Metabolism.. J. Cell Physiol 2013, 228 (8), 1703–1712. 10.1002/jcp.24352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merighi S.; Borea P. A.; Gessi S. Adenosine Receptors and Diabetes: Focus on the A2B Adenosine Receptor Subtype.. Pharmacol. Res. 2015, 99, 229–236. 10.1016/j.phrs.2015.06.015. [DOI] [PubMed] [Google Scholar]
- Davis J. M.; Zhao Z.; Stock H. S.; Mehl K. A.; Buggy J.; Hand G. A. Central Nervous System Effects of Caffeine and Adenosine on Fatigue.. Am. J. Physiology-regulatory Integr Comp Physiology 2003, 284 (2), R399–R404. 10.1152/ajpregu.00386.2002. [DOI] [PubMed] [Google Scholar]
- Haskó G.; Cronstein B. Regulation of Inflammation by Adenosine.. Front Immunol 2013, 4, 85. 10.3389/fimmu.2013.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavis P. A.; Divisekera U.; Paget C.; Chow M. T.; John L. B.; Devaud C.; Dwyer K.; Stagg J.; Smyth M. J.; Darcy P. K. Blockade of A2A Receptors Potently Suppresses the Metastasis of CD73+ Tumors.. Proc. National Acad. Sci. 2013, 110 (36), 14711–14716. 10.1073/pnas.1308209110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barletta K. E.; Ley K.; Mehrad B. Regulation of Neutrophil Function by Adenosine.. Arteriosclerosis Thrombosis Vasc Biology 2012, 32 (4), 856–864. 10.1161/ATVBAHA.111.226845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A.; Ngiow S. F.; Gao Y.; Patch A.-M.; Barkauskas D. S.; Messaoudene M.; Lin G.; Coudert J. D.; Stannard K. A.; Zitvogel L.; Degli-Esposti M. A.; Vivier E.; Waddell N.; Linden J.; Huntington N. D.; Guimaraes F. S.-F.; Smyth M. J. A2AR Adenosine Signaling Suppresses Natural Killer Cell Maturation in the Tumor Microenvironment.. Cancer Res. 2017, 78 (4), 1003–1016. 10.1158/0008-5472.can-17-2826. [DOI] [PubMed] [Google Scholar]
- Chambers A. M.; Lupo K. B.; Matosevic S. Tumor Microenvironment-Induced Immunometabolic Reprogramming of Natural Killer Cells.. Front Immunol 2018, 9, 2517. 10.3389/fimmu.2018.02517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskovalova T.; Lokshin A.; Huang X.; Su Y.; Mandic M.; Zarour H. M.; Jackson E. K.; Gorelik E. Inhibition of Cytokine Production and Cytotoxic Activity of Human Antimelanoma Specific CD8+ and CD4+ T Lymphocytes by Adenosine-Protein Kinase A Type I Signaling.. Cancer Res. 2007, 67 (12), 5949–5956. 10.1158/0008-5472.CAN-06-4249. [DOI] [PubMed] [Google Scholar]
- Atwood B. K.; Lopez J.; Wager-Miller J.; Mackie K.; Straiker A. Expression of G Protein-Coupled Receptors and Related Proteins in HEK293, AtT20, BV2, and N18 Cell Lines as Revealed by Microarray Analysis.. Bmc Genomics 2011, 12 (1), 14–14. 10.1186/1471-2164-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aristotelous T.; Ahn S.; Shukla A. K.; Gawron S.; Sassano M. F.; Kahsai A. W.; Wingler L. M.; Zhu X.; Tripathi-Shukla P.; Huang X.-P.; Riley J.; Besnard J.; Read K. D.; Roth B. L.; Gilbert I. H.; Hopkins A. L.; Lefkowitz R. J.; Navratilova I. Discovery of Β2 Adrenergic Receptor Ligands Using Biosensor Fragment Screening of Tagged Wild-Type Receptor.. ACS Med. Chem. Lett. 2013, 4 (10), 1005–1010. 10.1021/ml400312j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber S.; Casagrande F.; Hug M. N.; Wang L.; Heine P.; Kummer L.; Plückthun A.; Hennig M. SPR-Based Fragment Screening with Neurotensin Receptor 1 Generates Novel Small Molecule Ligands.. PLoS One 2017, 12 (5), e0175842. 10.1371/journal.pone.0175842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D.; Errey J. C.; Heitman L. H.; Marshall F. H.; Ijzerman A. P.; Siegal G. Fragment Screening of GPCRs Using Biophysical Methods: Identification of Ligands of the Adenosine A(2A) Receptor with Novel Biological Activity.. ACS Chem. Biol. 2012, 7 (12), 2064–2073. 10.1021/cb300436c. [DOI] [PubMed] [Google Scholar]
- Robertson N.; Jazayeri A.; Errey J.; Baig A.; Hurrell E.; Zhukov A.; Langmead C. J.; Weir M.; Marshall F. H. The Properties of Thermostabilised G Protein-Coupled Receptors (StaRs) and Their Use in Drug Discovery.. Neuropharmacology 2011, 60 (1), 36–44. 10.1016/j.neuropharm.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Serrano-Vega M. J.; Magnani F.; Shibata Y.; Tate C. G. Conformational Thermostabilization of the Β1-Adrenergic Receptor in a Detergent-Resistant Form.. Proc. National Acad. Sci. 2008, 105 (3), 877–882. 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidehi N.; Grisshammer R.; Tate C. G. How Can. Mutations Thermostabilize G-Protein-Coupled ReceptorsŒ. Trends Pharmacol. Sci. 2016, 37 (1), 37–46. 10.1016/j.tips.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congreve M.; Rich R. L.; Myszka D. G.; Figaroa F.; Siegal G.; Marshall F. H. Chapter Five Fragment Screening of Stabilized G-Protein-Coupled Receptors Using Biophysical Methods.. Methods Enzymol 2011, 493, 115–136. 10.1016/B978-0-12-381274-2.00005-4. [DOI] [PubMed] [Google Scholar]
- Navratilova I.; Besnard J.; Hopkins A. L. Screening for GPCR Ligands Using Surface Plasmon Resonance.. ACS Med. Chem. Lett. 2011, 2 (7), 549–554. 10.1021/ml2000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navratilova I.; Dioszegi M.; Myszka D. G. Analyzing Ligand and Small Molecule Binding Activity of Solubilized GPCRs Using Biosensor Technology.. Anal. Biochem. 2006, 355 (1), 132–139. 10.1016/j.ab.2006.04.021. [DOI] [PubMed] [Google Scholar]
- Navratilova I. H.; Aristotelous T.; Bird L. E.; Hopkins A. L. Surveying GPCR Solubilisation Conditions Using Surface Plasmon Resonance.. Anal. Biochem. 2018, 556, 23–34. 10.1016/j.ab.2018.06.012. [DOI] [PubMed] [Google Scholar]
- Witherington J.; Bordas V.; Gaiba A.; Naylor A.; Rawlings A. D.; Slingsby B. P.; Smith D. G.; Takle A. K.; Ward R. W. 6-Heteroaryl-Pyrazolo[3,4-b]Pyridines: Potent and Selective Inhibitors of Glycogen Synthase Kinase-3 (GSK-3).. Bioorg. Med. Chem. Lett. 2003, 13 (18), 3059–3062. 10.1016/S0960-894X(03)00646-2. [DOI] [PubMed] [Google Scholar]
- Smith D. G.; Buffet M.; Fenwick A. E.; Haigh D.; Ife R. J.; Saunders M.; Slingsby B. P.; Stacey R.; Ward R. W. 3-Anilino-4-Arylmaleimides: Potent and Selective Inhibitors of Glycogen Synthase Kinase-3 (GSK-3).. Bioorg. Med. Chem. Lett. 2001, 11 (5), 635–639. 10.1016/S0960-894X(00)00721-6. [DOI] [PubMed] [Google Scholar]
- Peat A. J.; Boucheron J. A.; Dickerson S. H.; Garrido D.; Mills W.; Peckham J.; Preugschat F.; Smalley T.; Schweiker S. L.; Wilson J. R.; Wang T. Y.; Zhou H. Q.; Thomson S. A. Novel Pyrazolopyrimidine Derivatives as GSK-3 Inhibitors.. Bioorg. Med. Chem. Lett. 2004, 14 (9), 2121–2125. 10.1016/j.bmcl.2004.02.036. [DOI] [PubMed] [Google Scholar]
- Cheung M.; Harris P. A.; Badiang J. G.; Peckham G. E.; Chamberlain S. D.; Alberti M. J.; Jung D. K.; Harris S. S.; Bramson N. H.; Epperly A. H.; Stimpson S. A.; Peel M. R. The Identification of Pyrazolo[1,5-a]Pyridines as Potent P38 Kinase Inhibitors.. Bioorg. Med. Chem. Lett. 2008, 18 (20), 5428–5430. 10.1016/j.bmcl.2008.09.040. [DOI] [PubMed] [Google Scholar]
- Dionisotti S.; Ongini E.; Zocchi C.; Kull B.; Arslan G.; Fredholm B. B. Characterization of Human A2A Adenosine Receptors with the Antagonist Radioligand [3H]-SCH 58261.. Br. J. Pharmacol. 1997, 121 (3), 353–360. 10.1038/sj.bjp.0701119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser M. J.; Roth B. L.; Armbruster B. N.; Ernsberger P.; Irwin J. J.; Shoichet B. K. Relating Protein Pharmacology by Ligand Chemistry.. Nat. Biotechnol. 2007, 25 (2), 197–206. 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- Heitman L. H.; van Veldhoven J. P. D.; Zweemer A. M.; Ye K.; Brussee J.; IJzerman A. P. False Positives in a Reporter Gene Assay: Identification and Synthesis of Substituted N-Pyridin-2-Ylbenzamides as Competitive Inhibitors of Firefly Luciferase.. J. Med. Chem. 2008, 51 (15), 4724–4729. 10.1021/jm8004509. [DOI] [PubMed] [Google Scholar]
- DiBerto J. F.; Smart K.; Olsen R. H. J.; Roth B. L. Agonist and Antagonist TRUPATH Assays for G Protein-Coupled Receptors.. Star Protoc 2022, 3 (2), 101259. 10.1016/j.xpro.2022.101259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H. J.; DiBerto J. F.; English J. G.; Glaudin A. M.; Krumm B. E.; Slocum S. T.; Che T.; Gavin A. C.; McCorvy J. D.; Roth B. L.; Strachan R. T. TRUPATH, an Open-Source Biosensor Platform for Interrogating the GPCR Transducerome.. Nat. Chem. Biol. 2020, 16, 841–849. 10.1038/s41589-020-0535-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBerto J.; Olsen R. H.; Roth B. L.. TRUPATH, an Open-Source Biosensor Platform for Interrogating the GPCR Transducerome. In Bioluminescence: Methods and Protocols, 4th ed.; Kim S.-B., Ed.; Methods in Molecular Biology; Humana Press, 2022, in press. [DOI] [PubMed] [Google Scholar]
- Leach K.; Loiacono R. E.; Felder C. C.; McKinzie D. L.; Mogg A.; Shaw D. B.; Sexton P. M.; Christopoulos A. Molecular Mechanisms of Action and In Vivo Validation of an M4Muscarinic Acetylcholine Receptor Allosteric Modulator with Potential Antipsychotic Properties.. Neuropsychopharmacol 2010, 35 (4), 855–869. 10.1038/npp.2009.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berizzi A. E.; Gentry P. R.; Rueda P.; Den Hoedt S.; Sexton P. M.; Langmead C. J.; Christopoulos A. Molecular Mechanisms of Action of M5Muscarinic Acetylcholine Receptor Allosteric Modulators.. Mol. Pharmacol. 2016, 90 (4), 427–436. 10.1124/mol.116.104182. [DOI] [PubMed] [Google Scholar]
- Berizzi A. E.; Bender A. M.; Lindsley C. W.; Conn P. J.; Sexton P. M.; Langmead C. J.; Christopoulos A. Structure–Activity Relationships of Pan-Gαq/11 Coupled Muscarinic Acetylcholine Receptor Positive Allosteric Modulators.. ACS Chem. Neurosci. 2018, 9 (7), 1818–1828. 10.1021/acschemneuro.8b00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.