Abstract
Denitrifying Betaproteobacteria play a key role in the anaerobic degradation of monoaromatic hydrocarbons. We performed a multi‐omics study to better understand the metabolism of the representative organism Georgfuchsia toluolica strain G5G6 known as a strict anaerobe coupling toluene oxidation with dissimilatory nitrate and Fe(III) reduction. Despite the genomic potential for degradation of different carbon sources, we did not find sugar or organic acid transporters, in line with the inability of strain G5G6 to use these substrates. Using a proteomics analysis, we detected proteins of fumarate‐dependent toluene activation, membrane‐bound nitrate reductase, and key components of the metal‐reducing (Mtr) pathway under both nitrate‐ and Fe(III)‐reducing conditions. High abundance of the multiheme cytochrome MtrC implied that a porin–cytochrome complex was used for respiratory Fe(III) reduction. Remarkably, strain G5G6 contains a full set of genes for aerobic toluene degradation, and we detected enzymes of aerobic toluene degradation under both nitrate‐ and Fe(III)‐reducing conditions. We further detected an ATP‐dependent benzoyl‐CoA reductase, reactive oxygen species detoxification proteins, and cytochrome c oxidase indicating a facultative anaerobic lifestyle of strain G5G6. Correspondingly, we found diffusion through the septa a substantial source of oxygen in the cultures enabling concurrent aerobic and anaerobic toluene degradation by strain G5G6.
Introduction
Mono‐ and multi‐aromatic compounds form a severe threat to environmental and human health. In situ microbial conversion is a sustainable and feasible option to remove these compounds from contaminated sites (Lueders, 2017); however, understanding of the metabolism of the responsible microorganisms is needed to implement and optimize removal. Denitrifying Betaproteobacteria are among the key players in anaerobic degradation of monoaromatic hydrocarbons (Lueders, 2017). Georgfuchsia toluolica strain G5G6T was isolated from the Banisveld aquifer (the Netherlands) polluted with low concentrations of benzene, toluene, ethylbenzene and xylene isomers (BTEX) (Weelink et al., 2009). Analysis of functional genes of anaerobic BTEX degradation and 16S rRNA genes has revealed widespread environmental occurrence of Georgfuchsia. For instance, 89% of the sequences encoding the key enzyme benzylsuccinate synthase (bssA) retrieved from the Banisveld aquifer were closely affiliated with G. toluolica indicating its important role in the anaerobic degradation of toluene and xylene (Staats et al., 2011). Moreover, Georgfuchsia 16S rRNA sequences have been detected in a German coal tar contaminated aquifer (Sperfeld et al., 2018), a Chinese drinking water treatment plant (Dong et al., 2019), a pilot‐scale bioreactor mediating sulfide‐driven denitrification in Brazil (Saia et al., 2019), and from toluene‐degrading and Fe(III)‐reducing microcosms prepared from contaminated sites in Germany (Pilloni et al., 2011) and United States (Sun et al., 2014).
Strain G5G6 uses nitrate, Fe(III), and Mn(IV) as electron acceptors and monoaromatic compounds such as toluene, ethylbenzene, phenol, cresol, benzaldehyde and hydroxybenzoate as electron donors. Strain G5G6 is the only isolate of the genus Georgfuchsia and has a number of unique physiological features making it distinct among microbial isolates capable of anaerobic degradation of aromatic hydrocarbons: (i) it is the only known betaproteobacterial isolate capable of coupling toluene oxidation to reduction of nitrate, Fe(III), or Mn(IV), whereas known alpha‐ and betaproteobacterial anaerobic toluene degraders within the genera Azoarcus, Thauera, Aromatoleum, and Magnetospirillum cannot couple toluene oxidation to Fe(III) reduction (Evans et al., 1991; Biegert et al., 1996; Shinoda et al., 2004; Rabus et al., 2005; Shinoda et al., 2005; Blázquez et al., 2018; Meyer‐Cifuentes et al., 2020), (ii) strain G5G6 is a toluene‐oxidizing, Fe(III)‐reducing bacterium that does not belong to the Geobacteraceae, (iii) strain G5G6 is dedicated to the degradation of monoaromatic compounds and does not use more general environmentally available carbon sources such as acetate, lactate and glucose, (iv) strain G5G6 does not seem to grow aerobically, but its draft genome sequence showed presence of genes involved in aerobic monoaromatic degradation (Oosterkamp, 2013), and (v) compared to other toluene‐degrading isolates and consortia, strain G5G6 displays significantly different values in dual isotope (carbon and hydrogen) analyses during toluene degradation coupled to nitrate, Fe(III), or Mn(IV) reduction suggesting variations in the enzymatic transition state depending on the available terminal electron acceptor (Dorer et al., 2016). These traits make strain G5G6 an appealing model bacterium to study toluene degradation using different electron acceptors. Here, we investigated the metabolism of toluene under nitrate‐ and Fe(III)‐reducing conditions using a multi‐omics approach and found an unexpected redundancy in peripheral metabolic pathways for toluene degradation. Most surprisingly, strain G5G6 expresses a full set of genes for aerobic toluene degradation indicating a facultative lifestyle.
Results and discussion
To obtain insight into the metabolism of G. toluolica strain G5G6T (JCM 14632, DSM 19032), we sequenced its genome and performed comparative proteomics of cells grown on toluene with either nitrate or Fe(III) as electron acceptor. Strain G5G6 was grown in anoxic medium using toluene (0.4 mM) and nitrate (5 mM) as described (Weelink et al., 2009). Resazurin (0.005 g/l) and l‐cysteine (1 mM) were added as redox indicator and reducing reagent, respectively. Genomic DNA was extracted using MasterPure™ Gram Positive DNA Purification Kit (Epicentre, WI, USA). The genome of strain G5G6 was sequenced using Single Molecule, Real‐Time (SMRT) sequencing technology with the PacBio RSII platform (Novogene, Hong Kong), assembled using PacBio's dedicated SMRT analysis software suite (SMRT Link, v5.9), and annotated using MicroScope (Microbial Genome Annotation & Analysis Platform) (Vallenet et al., 2020). The resulting genome of strain G5G6 consisted of two contigs: a long contig of 3 465 762 bp and a short contig of 36 233 bp (accession number ERZ1938153). Proteomic analysis was done from triplicate cultures grown on toluene (0.4 mM) and nitrate (5 mM) or toluene (0.4 mM) and Fe(III) citrate (20 mM) using procedures described previously (Peng et al., 2020). All cultures had been adapted to the respective growth condition by at least three previous transfers (5% v/v) to fresh media, and three spikes of toluene (0.4 mM at each spike) during each transfer.
Growth restriction to monoaromatic compounds
Strain G5G6 was reported to use a restricted range of carbon sources including toluene, ethylbenzene, phenol, p‐cresol, m‐cresol, benzaldehyde and p‐hydroxybenzoate (Weelink et al., 2009). Strain G5G6 was not able to grow on carboxylic acids (e.g. pyruvate, acetate), amino acids (cysteine, glutamate, aspartate and alanine), sugars (glucose, fructose, xylose and mannitol), alcohols (ethanol and methanol), cholesterol, hydrogen and yeast extract (Weelink et al., 2009). Our analysis of the genome of strain G5G6 revealed that most of the genes for glycolysis/gluconeogenesis and the tricarboxylic acid (TCA) cycle are present. We also found proteins of these pathways in our proteomic analysis. However, no sugar transporters were encoded in the genome of strain G5G6 indicating that these enzymes are most likely used for biosynthetic purposes. The lack of sugar transporters is also in line with the inability of strain G5G6 to grow on external sugars. While transporter annotations are often ambiguous, most microorganisms contain a set of various imprecisely annotated major facilitator superfamily (MFS) sugar transporters or ABC‐type transport systems. The genome of strain G5G6 encodes only few substrate transporters, most of which are not related to sugar or dicarboxylate transport. Moreover, phosphotransferase systems (PTS) were absent, with only two IIA components encoded in the genome. Despite the presence of fumarate reductase (GTOL_10413–GTOL_10416), no growth with fumarate as substrate or electron acceptor was reported (Weelink et al., 2009). This implies that fumarate reductase is likely used for succinate conversion to fumarate, which is a vital component of anaerobic toluene degradation. Lactate dehydrogenase is not encoded in the genome, in line with the observation that strain G5G6 cannot grow using lactate as the sole carbon and energy source (Weelink et al., 2009). These metabolic features make strain G5G6 a niche specialist dedicated to degradation of monoaromatic compounds, an appealing trait for bioremediation and restoration of hydrocarbon‐contaminated sites. As such, the growth of strain G5G6 in the environment can indicate the presence of monoaromatic compounds. Such cause‐and‐effect relationships between dedicated degraders and corresponding contaminants have been shown for certain members of Dehalococcoidia restricted to anaerobic respiration with organohalogens as terminal electron acceptors (Atashgahi et al., 2016). Therefore, tracking the abundance of the 16S rRNA gene of strain G5G6 could aid in monitoring the fate of monoaromatic compounds, especially at sites with elevated nitrate or Fe(III) availability, which would allow strain G5G6 to outcompete the sulfate‐dependent degradation processes.
Toluene degradation
Toluene degradation was coupled to nitrate reduction to nitrite (Fig. 1A and C), or Fe(III) reduction to Fe(II) (Fig. 1B and D). Despite stoichiometric conversion of nitrate to nitrite (nitrate:nitrite ratio of 1:1.08), the stoichiometry of toluene oxidation relative to nitrite formation was 1:4.3 which is similar to the previously reported 1:5.5 ratio (Weelink et al., 2009), but quite different from the expected ratio of 1:18. Similarly, the toluene oxidation relative to Fe(II) formation (1:23.7) was sub‐stoichiometric compared to the expected ratio of 1:36, as was noted previously (1:13.6) (Weelink et al., 2009). The same phenomenon was reported in microcosms prepared from marine sediments (1:20) (Kim et al., 2014). Such deviations were attributed to the precipitation of Fe(II) in the medium (Weelink et al., 2009; Kim et al., 2014). However, our analysis showed an Fe(III):Fe(II) ratio of 1:0.94, indicating marginal Fe(II) precipitation. Although a part of the carbon and electrons is incorporated into biomass, such a sub‐stoichiometric conversion was not expected. Uninoculated control bottles sealed with Teflon‐lined butyl rubber septa and sampled overtime did not show toluene loss during incubation, thus excluding abiotic toluene loss (data not shown). It is currently not known how the remaining electrons derived from toluene oxidation are dissipated. Potential explanations could be incomplete toluene degradation and/or production of storage compounds such as polyhydroxybutyrate (PHB). Future experiments using 13C‐toluene and 15N‐nitrate and tracking metabolites may allow to further resolve the observed electron imbalance.
Fig. 1.
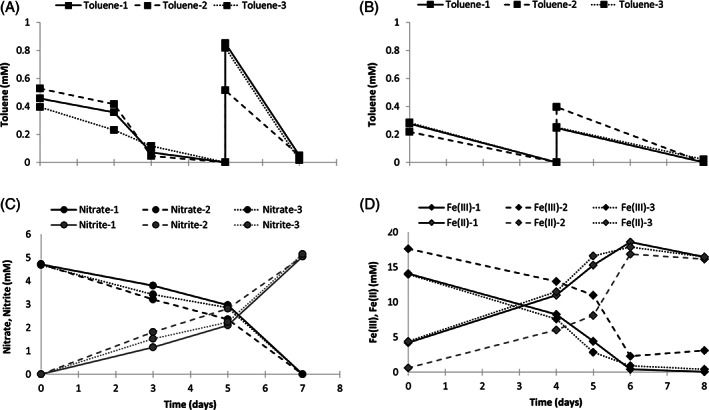
Toluene degradation by strain G5G6 coupled to nitrate reduction (A, C) or Fe(III)citrate reduction (B, D). Triplicate cultures are shown individually.
Anaerobic toluene degradation pathway
The first step in the anaerobic catabolism of toluene is the addition of fumarate to the methyl group of toluene. This reaction is mediated by benzylsuccinate synthase (BSS, encoded by bssABCDEFG), an oxygen‐sensitive glycyl radical enzyme complex (Boll et al., 2014; Boll et al., 2020). The product of the reaction, benzylsuccinate, is converted to succinyl‐CoA and benzoyl‐CoA through a series of β‐oxidation‐like reactions involving enzymes encoded by the bbs genes (Biegert et al., 1996; Carmona et al., 2009). The bss and bbs genes are present in the genome of G. toluolica in the same order and orientation as for other known anaerobic toluene degraders (Fig. 2) indicating their involvement in anaerobic toluene degradation. Interestingly, the genes of anaerobic toluene degradation are also present on the short contig, indicating gene duplication. Compared to the long contig, this contig has a lower GC content (55.2 vs. 58.6%), shows markedly increased coverage and encodes two transposases. This indicates that the small contig is likely a plasmid or part of a large repetitive region in the genome and implies potential distribution of xenobiotic degradation genes by horizontal gene transfer (Top and Springael, 2003). We detected the BssABCDEFG and BbsABDEFG proteins in the proteome, which were among the most prevalent proteins detected under both nitrate‐ and Fe(III)‐reducing conditions (Supporting Information Table S1). Benzoyl‐CoA is the central metabolite of the anaerobic degradation of aromatic compounds and is reductively dearomatized by benzoyl‐CoA reductase (BCR) prior to ring cleavage. BCR is a highly oxygen‐sensitive enzyme within the benzoyl‐CoA pathway (Carmona et al., 2009). High abundance of BcrABCD under the tested conditions indicates that ATP‐dependent class I BCR enzymes are employed by strain G5G6, unlike other strict anaerobes that use ATP‐independent class II BCR enzymes (Boll et al., 2014; Boll et al., 2020). All downstream enzymes of anaerobic benzoyl‐CoA degradation were found in our proteomic analysis under both nitrate‐ and Fe(III)‐reducing conditions (Supporting Information Table S1).
Fig. 2.
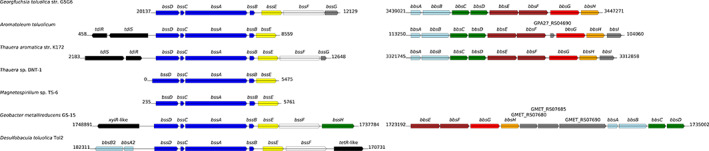
Genomic organization of the bss (left hand) and bbs gene (right hand) regions in strain G5G6 and selected bacteria capable of anaerobic toluene degradation. The flanking numbers indicate the relative positions of genes identified within the genome of these bacteria. The size and orientation of the genes indicate their relative lengths and directions in which they are transcribed. The names of the genes and the corresponding encoded enzymes are listed in the Supporting Information Table S1. The colour codes of the arrows are adapted from Blázquez et al. (2018).
Aerobic toluene degradation pathways
Interestingly, gene clusters encoding enzymes for aerobic degradation of toluene including the initial activation of toluene to m‐cresol by toluene‐4‐monooxygenase (tmoADE), m‐cresol hydroxylation to 3‐methylcatechol by phenol hydroxylase (dmpABCDEF), and subsequent ring opening by catechol 2,3‐dioxygenase (xylE) are located directly next to the genes of anaerobic toluene degradation. Remarkably, proteins of aerobic toluene degradation were also detected under both nitrate‐ and Fe(III)‐reducing conditions. Except for toluene‐4‐monooxygenase (TmoAE), all other proteins of aerobic toluene degradation were significantly (P < 0.05) more abundant under nitrate‐reducing conditions (Supporting Information Table S2). Moreover, we found proteins of the box pathway, the unusually aerobic hybrid benzoate oxidation pathway characterized in Azoarcus evansii (Gescher et al., 2002), including benzoate‐CoA ligase (GTOL_11785), benzoyl‐CoA oxygenase component B (GTOL_11792), ring‐opening benzoyl‐CoA‐dihydrodiol lyase (GTOL_11793), and benzaldehyde dehydrogenase (GTOL_11795) (Supporting Information Table S2). These proteins were also significantly (P < 0.05) more abundant under nitrate‐reducing conditions. Our findings indicate redundancy in peripheral metabolic pathways of toluene degradation in strain G5G6 (Fig. 3). Other oxygenases found under both conditions were phenylpropionate ring‐hydroxylating dioxygenase (GTOL_11036), protocatechuate 4,5‐dioxygenase beta chain (GTOL_11170), and 2‐nitropropane dioxygenase (GTOL_11446).
Fig. 3.
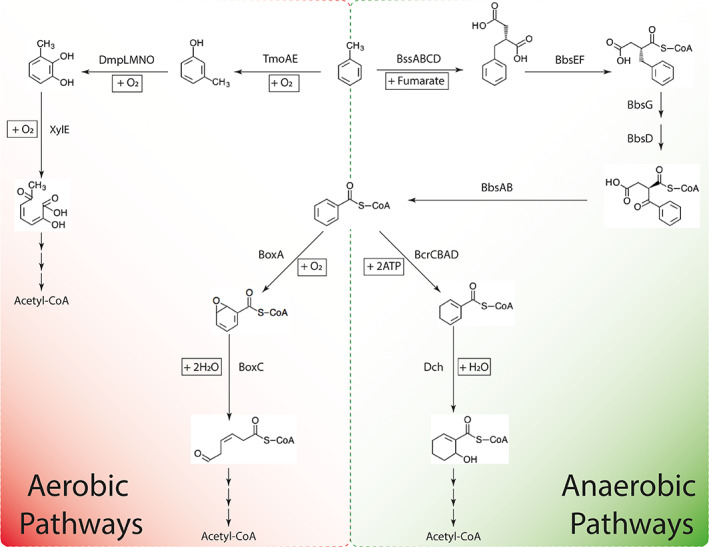
Proposed peripheral toluene degradation by strain G5G6 according to genomic and proteomic analyses.
These observations are remarkable, especially since strain G5G6 was considered as a strict anaerobe and no toluene degradation was observed when 5 or 10% oxygen was used in the headspace (Weelink et al., 2009). Consequently, it is likely that strain G5G6 is capable of toluene degradation under hypoxic conditions, as, for instance, encountered in groundwater systems. Strain G5G6 shows some traits typical of facultative anaerobic toluene degraders: (i) the box pathways are preferentially active under reduced oxygen tension (Denef et al., 2006) and fit into the lifestyle of facultative anaerobes. This pathway needs only one molecule of oxygen for ring activation and employs CoA thioesters that are also intermediates in anaerobic toluene degradation (Fuchs et al., 2011). Therefore, once the substrate‐CoA thioester is formed under hypoxic conditions, it can be metabolized by either the aerobic or the anaerobic degradation strategy; (ii) strain G5G6 has ATP‐dependent class I BCR enzymes, although using ATP for ring reduction is not common for strict anaerobes (Fuchs et al., 2011); (iii) the genomes of strain G5G6 harbours a suite of genes encoding enzymes for oxygen reduction and reactive oxygen species (ROS) detoxification, including high‐affinity cbb3‐type cytochrome c oxidase (GTOL_10453 ‐ GTOL_10457) and Cytochrome bd ubiquinol oxidase (GTOL_10941 ‐ GTOL_10942). The presence of high‐affinity terminal oxidases in the genome may enable oxygen respiration at nanomolar concentrations (Baughn and Malamy, 2004; Stolper et al., 2010) facilitating adaption of strain G5G6 to hypoxic aquifers, and protecting oxygen‐sensitive enzymes from oxidative damage. Some of these proteins were also found in our proteomic analysis, including cytochrome c oxidase (GTOL_10454, GTOL_10455), superoxide dismutase (GTOL_11750), and catalase‐related peroxidase (GTOL_12777).
Identifying the source of oxygen: intracellular oxygen production or oxygen contamination?
Intracellular oxygen production using nitric oxide dismutase (Nod) during nitrate reduction has been observed during anaerobic hydrocarbon degradation coupled to denitrification (Ettwig et al., 2010; Zedelius et al., 2011). However, the genome of strain G5G6 lacks any identifiable nod genes. Another possibility is oxygen contamination. Although we applied anaerobic cultivation procedures, oxygen intrusion cannot fully be excluded for example during repeated substrate addition and sampling for headspace and liquid measurements. Whereas Viton stoppers were used for the isolation of the strain G5G6 (Weelink et al., 2009), we applied Teflon‐lined butyl rubber septa (VWR, the Netherlands) to seal our microcosms, which might be more prone to oxygen leakage than thick Viton stoppers. To test the possibility of oxygen contamination, we used miniaturized luminescence‐based optical sensors (LUMOS, Luminescence Measuring Oxygen Sensor) capable of measuring dissolved oxygen in the nanomolar range (Lehner et al., 2015). The miniaturized sensors were applied within the serum bottles, and the measurement was performed through the bottles as optical windows as outlined previously (Lehner et al., 2015). Triplicate bottles were prepared containing all medium components except inoculum. The bottles were stirred at 300 rpm using magnet stirrers, and samples were taken at regular time intervals. Oxygen concentration remained below 0.5 μM in bottles containing cysteine (Fig. 4, top panels). However, when cysteine was excluded, oxygen concentration increased drastically exceeding the upper detection limit of the applied sensors (10 μM) within 10–13 days (Fig. 4, bottom panels) indicating severe oxygen contamination. Considering the slow reaction of cysteine with oxygen, toluene activation by oxygen and hence concurrent anaerobic and aerobic toluene degradation by strain G5G6 adapted to hypoxic aquifers is likely. However, even if 10 μM oxygen was used for toluene degradation, it can degrade only ~1.3 μM toluene and hence cannot explain the sub‐stoichiometric toluene degradation coupled to dissimilatory Fe(III) and nitrate reduction (Fig. 1). Future experiments using 13C‐toluene in absence of amended electron acceptors and in presence and absence of cysteine may indicate if oxygen can be used as the electron acceptor. Application of oxygen for toluene activation may corroborate the observed redundancy in peripheral metabolic pathways of toluene degradation (Fig. 3) and explain widespread detection of strain G5G6 in natural and built environments around the globe (Pilloni et al., 2011; Staats et al., 2011; Sun et al., 2014; Sperfeld et al., 2018; Dong et al., 2019; Saia et al., 2019). Simultaneous toluene activation by benzylsuccinate synthase and toluene‐4‐monooxygenase (based on our proteomic data) may also explain the significantly different toluene isotopic values observed for strain G5G6 compared to other toluene degraders (Dorer et al., 2016), since methyl group hydroxylation by monooxygenases is known to be associated with strong carbon and especially hydrogen isotope fractionation (Vogt et al., 2016). Dorer et al. (2016) also used Teflon‐coated butyl septa in their isotope fractionation experiments and sampled individual culture bottles several times; hence, a significant penetration of oxygen into the bottles during these experiments cannot be excluded. The facultative anaerobic lifestyle is also intriguing considering that strain G5G6 was first described as a strict anaerobe and named after Georg Fuchs (Weelink et al., 2009), for his scientific contribution to understanding of anaerobic aromatic hydrocarbon degradation (Evans and Fuchs, 1988; Biegert et al., 1996; Fuchs et al., 2011). Oxygen contamination of microcosm experiments designed to investigate ‘anaerobic oxidation’ is a well‐known phenomenon for chlorinated ethene degradation experiments, despite the addition of reducing agents to the medium and lack of colour change of the redox indicator resazurin (Gossett, 2010; Fullerton et al., 2013), which may lead to misinterpretation of the results. For instance, Dechloromonas aromatica strain RCB was introduced as a benzene‐degrading denitrifying bacterium; however, the genome of strain RCB contains no identifiable genes for anaerobic benzene degradation but encodes several aerobic pathways for aromatic degradation (Salinero et al., 2009).
Fig. 4.
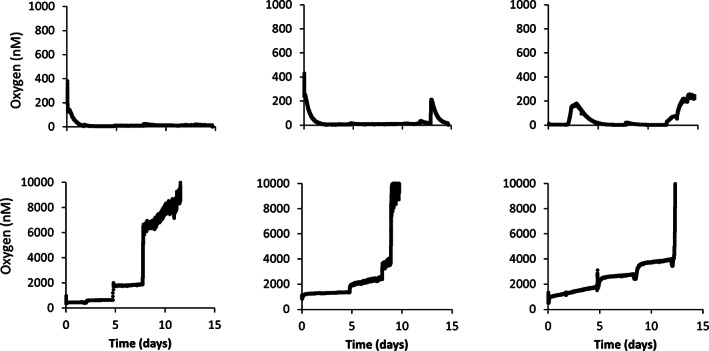
Measurement of oxygen intrusion into 120 ml serum bottles containing growth medium with (top panels) and without cysteine (bottom panels). Liquid samples (1.5 ml) were taken at day 1, 2, 5, 8, 12 and 15 to simulate actual sampling. All syringes were flushed thoroughly using bottles containing sterile N2 and sealed with butyl rubber septa. Each panel represents results from a single bottle. Note that for the Y axes of top and bottom panels different concentration scales are used.
How aerobic and anaerobic metabolism could be reconciled
It is not clear how strain G5G6 would make use of the oxygenases at the same time as oxygen‐sensitive BSS and BCR enzymes in media reduced with l‐cysteine. BSS is a glycyl radical enzyme that is cleaved at the glycyl radical site in the presence of oxygen (Heider et al., 2016). For instance, despite transcription of the bssA gene, peptides of BSS were never detected in Magnetospirillum sp. strain 15–1 grown with toluene and oxygen (Meyer‐Cifuentes et al., 2020) nor in the rhizosphere of toluene‐degrading constructed wetlands from which strain 15–1 was isolated (Lünsmann et al., 2016). Similarly, in toluene‐degrading cells of Thauera sp. strain DNT‐1 (Shinoda et al., 2004) and Magnetospirillum sp. strain TS‐6 (Shinoda et al., 2005) grown under oxic conditions, transcription of bbsA and bcr genes was reported. On the other hand, the bbsA gene was not transcribed under these conditions in cells of Azoarcus sp. strain CIB (Blázquez et al., 2018). One option to protect the oxygen‐sensitive glycyl radical enzyme is the presence of bacterial microcompartments (BMCs) in which the enzymatic core is encapsulated by a selectively permeable protein shell (Kerfeld et al., 2018). However, the genome of strain G5G6 does not encode any known BMC proteins. Alternatively, the identified ROS detoxification enzymes and the hybrid box pathway of strain G5G6 may protect oxygen‐sensitive enzymes. Accordingly, the box pathway was proposed to constitute an alternative oxygen‐scavenging mechanism that may assist in a rapid shift to aerobic degradation if oxygen levels become higher (Valderrama et al., 2012; Díaz et al., 2013). We recently found transcription of box genes in a continuous benzene‐degrading denitrifying culture that displayed concurrent anaerobic and aerobic benzene degradation pathways (Atashgahi et al., 2018). Further studies will be necessary to resolve how aerobic and anaerobic metabolism could be reconciled.
Nitrate reduction
Our genome analysis shows that besides the nitrate reductase complex, strain G5G6 also encoded a nitric oxide reductase complex (Supporting Information Table S3). However, genes for nitrite and nitrous‐oxide reductases were absent. Consequently, nitrite should be the end product of nitrate reduction (Fig. 1), although complete denitrification was previously proposed based on mass balance analysis (Weelink et al., 2009). We detected NarGHI and NorC under both tested conditions, even though no nitrate was present under Fe(III)‐reducing conditions, thus questioning their role during growth with Fe(III) as electron acceptor.
Fe(III) reduction
Unlike nitrate and oxygen, Fe(III) minerals cannot cross the bacterial outer membrane owing to their insolubility. Therefore, rather than using inner membrane terminal reductases, Gram‐negative bacteria have evolved strategies to transfer electrons from the inner membrane to the iron minerals at the extracellular face of the outer membrane (Xie et al., 2021). The multiprotein porin–cytochrome c complex MtrCAB of Shewanella strains is the best studied electron‐transfer model for the respiratory reduction of mineral oxides (Pitts et al., 2003; Edwards et al., 2020). MtrA and mtrC encode two decahaem c‐type cytochromes, and mtrB a transmembrane β‐barrel that forms a pore in the membrane. The complex of these three proteins allows electrons from the cytoplasmic respiratory chain to be transferred via the periplasmic MtrA through MtrB to the outer‐membrane decahaem MtrC. Homologues of mtrAB (GTOL_11386 and GTOL_11387) are present in the genome of strain G5G6. Further analysis using FeGenie (a tool for identification of iron‐related genes) (Garber et al., 2020) indicated that GTOL_11385 is a homologue of mtrC, which encodes for a putative decahaem protein with 10 CxxCH motifs. As such, the mtrCAB genes in strain G5G6 are organized in the same sequential order as mtrCAB operons known from other Fe(III) oxide‐reducing bacteria. MtrCB were found under both Fe(III)‐ and nitrate‐reducing conditions indicating their constitutive expression (Supporting Information Table S4). In the vicinity of mtrCAB, three additional MtrC homologues were encoded with 10 cytochrome c binding sites and signal peptides for export into the periplasm (GTOL_11402, GTOL_11403 and GTOL_11405) that were among the most abundant proteins detected under Fe(III)‐reducing conditions (Supporting Information Table S4). These proteins are homologous to the outer‐membrane multiheme cytochrome MtrC/OmcA of Rhodoferax ferrireducens and Shewanella baltica. Whereas the role of MtrC in Fe(III) reduction is well‐known, three additional co‐located mtrC genes in a single operon and their strong expression under Fe(III)‐reduction conditions have not been reported in dissimilatory Fe(III)‐reducing prokaryotes (Fig. 5), further signifying the unique genomic and physiological features of strain G5G6.
Fig. 5.
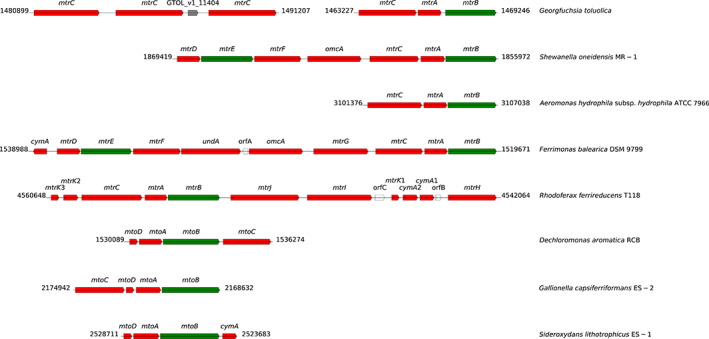
Genomic organization of the mtr clusters identified in strain G5G6 and selected bacteria capable of dissimilatory Fe(III)‐reduction. Three additional co‐localized mtrC genes in a genomic region were only found in the genome of strain G5G6. The flanking numbers indicate the relative positions of genes identified within the genome of these bacteria. The size and orientation of the genes indicate their relative lengths and directions in which they are presumed to be transcribed. The genes predicted to encode c‐type cytochromes are labelled in red and those predicted to encode β‐barrel outer membrane proteins are labelled in green. The figure is modified after Shi et al. (2012).
Conclusions
Unlike metabolically versatile monoaromatic‐degrading microbes, strain G5G6 is restricted to degradation of monoaromatic hydrocarbons. Strain G5G6 shows, however, more versatility in exploiting electron acceptors, and couples toluene oxidation to the reduction of nitrate, Fe(III), Mn(IV), and likely oxygen at low concentrations. These traits signify the importance of strain G5G6 as a niche specialist dedicated to degradation of monoaromatic compounds in hypoxic environments. Strain G5G6 displays redundancy in peripheral metabolic pathways of toluene degradation that seems to be important in environmental isolates (Denef et al., 2006). The box pathway seems to be an adaptation to low or fluctuating oxygen concentrations. Benzoyl‐CoA can be diverted to aerobic or anaerobic metabolic routes, depending on the oxygen supply. Detection of oxygen in glass vials sealed with butyl rubber septa indicate that diffusion through the septa may be a substantial source of contamination. Hence, care should be taken in interpreting the results of microcosm experiments designed to investigate anaerobic oxidation of, for example, hydrocarbons. Substantial oxygen contamination can slow down or inhibit anaerobic microbes/pathways, and/or induce aerobic ones. This study casts new light on the physiology of an understudied yet environmentally relevant microorganism. Further studies will be necessary to resolve how aerobic and anaerobic metabolism could be reconciled, and to elucidate strategies employed to protect oxygen‐sensitive enzymes of the anaerobic toluene degradation pathway under constant oxygen intrusion.
Data availability
The genome sequence of strains G5G6 was deposited in the European Bioinformatics Institute (accession number ERZ1938153). A list of proteins detected from strain G5G6 under nitrate‐ and Fe(III)‐reducing conditions is available in Supporting Information Table S5.
Supporting information
Table S1 Abundance of proteins of anaerobic toluene degradation detected from G5G6 cultures grown on toluene and Fe(III) (TF) or toluene and nitrate (TN).
Table S2. Abundance of proteins of aerobic toluene degradation detected from G5G6 cultures grown on toluene and Fe(III) (TF) or toluene and nitrate (TN).
Table S3. Abundance of proteins involved in denitrification detected from G5G6 cultures grown on toluene and Fe(III) (TF) or toluene and nitrate (TN).
Table S4. Abundance of proteins involved in Fe(III)citrate reduction detected from G5G6 cultures grown on toluene and Fe(III) (TF) or toluene and nitrate (TN).
Table S5 List of proteins detected from strain G5G6 under F(III)‐reducing (TF) and nitrate‐reducing (TN) conditions.
Acknowledgements
The authors thank Arkadiy Garber for his advice on using FeGenie tool. This work was supported by Wageningen University & Research through its investment theme Resilience, the Technology Foundation (STW), the Applied Science Division of the Dutch Research Council (NWO; project 08053), NWO grant 016.Vidi.189.050, and a Gravitation grant of the Netherlands Ministry of Education, Culture and Science and NWO (project 024.002.002 SIAM). B.K. was supported by the Villum foundation, Denmark (VYI Grant 25491).
References
- Atashgahi, S. , Lu, Y. , and Smidt, H. (2016) Overview of known organohalide‐respiring bacteria—phylogenetic diversity and environmental distribution. In Organohalide‐Respiring Bacteria. Adrian, L. , and Loffler, F. (eds.). Berlin, Heidelberg: Springer‐Verlag, pp. 63–105. [Google Scholar]
- Atashgahi, S. , Hornung, B. , Van Der Waals, M.J. , Da Rocha, U.N. , Hugenholtz, F. , Nijsse, B. , et al. (2018) A benzene‐degrading nitrate‐reducing microbial consortium displays aerobic and anaerobic benzene degradation pathways. Sci Rep 8: 4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughn, A.D. , and Malamy, M.H. (2004) The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature 427: 441–444. [DOI] [PubMed] [Google Scholar]
- Biegert, T. , Fuchs, G. , and Heider, J. (1996) Evidence that anaerobic oxidation of toluene in the denitrifying bacterium Thauera aromatica is initiated by formation of benzylsuccinate from toluene and fumarate. Eur J Biochem 238: 661–668. [DOI] [PubMed] [Google Scholar]
- Blázquez, B. , Carmona, M. , and Díaz, E. (2018) Transcriptional regulation of the peripheral pathway for the anaerobic catabolism of toluene and m‐xylene in Azoarcus sp CIB. Front Microbiol 9: 506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll, M. , Estelmann, S. , and Heider, J. (2020) Catabolic pathways and enzymes involved in the anaerobic degradation of monocyclic aromatic compounds. In Anaerobic Utilization of Hydrocarbons, Oils, and Lipids. Handbook of Hydrocarbon and Lipid Microbiology. Boll, M. (ed.). Cham: Springer, pp. 85–133. [Google Scholar]
- Boll, M. , Löffler, C. , Morris, B.E. , and Kung, J.W. (2014) Anaerobic degradation of homocyclic aromatic compounds via arylcarboxyl‐coenzyme a esters: organisms, strategies and key enzymes. Environ Microbiol 16: 612–627. [DOI] [PubMed] [Google Scholar]
- Carmona, M. , Zamarro, M.T. , Biázquez, B. , Durante‐Rodríguez, G. , Juárez, J.F. , Valderrama, J.A. , et al. (2009) Anaerobic catabolism of aromatic compounds: a genetic and genomic view. Microbiol Mol Biol Rev 73: 71–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denef, V. , Klappenbach, J. , Patrauchan, M. , Florizone, C. , Rodrigues, J. , Tsoi, T. , et al. (2006) Genetic and genomic insights into the role of benzoate‐catabolic pathway redundancy in Burkholderia xenovorans LB400. Appl Environ Microbiol 72: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz, E. , Jiménez, J.I. , and Nogales, J. (2013) Aerobic degradation of aromatic compounds. Curr Opin Biotechnol 24: 431–442. [DOI] [PubMed] [Google Scholar]
- Dong, S. , Liu, L. , Zhang, Y. , and Jiang, F. (2019) Occurrence and succession of bacterial community in O(3)/BAC process of drinking water treatment. Int J Env Res Pub Health 16: 3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer, C. , Vogt, C. , Neu, T.R. , Stryhanyuk, H. , and Richnow, H.‐H. (2016) Characterization of toluene and ethylbenzene biodegradation under nitrate‐, iron(III)‐ and manganese(IV)‐reducing conditions by compound‐specific isotope analysis. Environ Pollut 211: 271–281. [DOI] [PubMed] [Google Scholar]
- Edwards, M.J. , White, G.F. , Butt, J.N. , Richardson, D.J. , and Clarke, T.A. (2020) The crystal structure of a biological insulated transmembrane molecular wire. Cell 181: 665–673. e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwig, K.F. , Butler, M.K. , Le Paslier, D. , Pelletier, E. , Mangenot, S. , Kuypers, M.M. , et al. (2010) Nitrite‐driven anaerobic methane oxidation by oxygenic bacteria. Nature 464: 543–548. [DOI] [PubMed] [Google Scholar]
- Evans, P. , Mang, D. , Kim, K.S. , and Young, L. (1991) Anaerobic degradation of toluene by a denitrifying bacterium. Appl Environ Microbiol 57: 1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, W.C. , and Fuchs, G. (1988) Anaerobic degradation of aromatic compounds. Annu Rev Microbiol 42: 289–317. [DOI] [PubMed] [Google Scholar]
- Fuchs, G. , Boll, M. , and Heider, J. (2011) Microbial degradation of aromatic compounds—from one strategy to four. Nat Rev Microbiol 9: 803–816. [DOI] [PubMed] [Google Scholar]
- Fullerton, H. , Crawford, M. , Bakenne, A. , Freedman, D.L. , and Zinder, S.H. (2013) Anaerobic oxidation of ethene coupled to sulfate reduction in microcosms and enrichment cultures. Environ Sci Technol 47: 12374–12381. [DOI] [PubMed] [Google Scholar]
- Garber, A.I. , Nealson, K.H. , Okamoto, A. , McAllister, S.M. , Chan, C.S. , Barco, R.A. , and Merino, N. (2020) FeGenie: a comprehensive tool for the identification of iron genes and iron gene neighborhoods in genome and metagenome assemblies. Front Microbiol 11: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gescher, J. , Zaar, A. , Mohamed, M. , Schägger, H. , and Fuchs, G. (2002) Genes coding for a new pathway of aerobic benzoate metabolism in Azoarcus evansii . J Bacteriol 184: 6301–6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossett, J.M. (2010) Sustained aerobic oxidation of vinyl chloride at low oxygen concentrations. Environ Sci Technol 44: 1405–1411. [DOI] [PubMed] [Google Scholar]
- Heider, J. , Szaleniec, M. , Martins, B.M. , Seyhan, D. , Buckel, W. , and Golding, B.T. (2016) Structure and function of benzylsuccinate synthase and related fumarate‐adding glycyl radical enzymes. J Mol Microbiol Biotechnol 26: 29–44. [DOI] [PubMed] [Google Scholar]
- Kerfeld, C.A. , Aussignargues, C. , Zarzycki, J. , Cai, F. , and Sutter, M. (2018) Bacterial microcompartments. Nat Rev Microbiol 16: 277–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.J. , Park, S.J. , Cha, I.T. , Min, D. , Kim, J.S. , Chung, W.H. , et al. (2014) Metabolic versatility of toluene‐degrading, iron‐reducing bacteria in tidal flat sediment, characterized by stable isotope probing‐based metagenomic analysis. Environ Microbiol 16: 189–204. [DOI] [PubMed] [Google Scholar]
- Lehner, P. , Larndorfer, C. , Garcia‐Robledo, E. , Larsen, M. , Borisov, S.M. , Revsbech, N.‐P. , et al. (2015) LUMOS‐A sensitive and reliable optode system for measuring dissolved oxygen in the nanomolar range. PLoS One 10: e0128125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lueders, T. (2017) The ecology of anaerobic degraders of BTEX hydrocarbons in aquifers. FEMS Microbiol Ecol 93: fiw220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lünsmann, V. , Kappelmeyer, U. , Taubert, A. , Nijenhuis, I. , Von Bergen, M. , Heipieper, H.J. , et al. (2016) Aerobic toluene degraders in the rhizosphere of a constructed wetland model show diurnal polyhydroxyalkanoate metabolism. Appl Environ Microbiol 82: 4126–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer‐Cifuentes, I. , Gruhl, S. , Haange, S.‐B. , Lünsmann, V. , Jehmlich, N. , Bergen, M.V. , et al. (2020) Benzylsuccinate synthase is post‐transcriptionally regulated in the toluene‐degrading denitrifier Magnetospirillum sp. strain 15–1. Microorganisms 8: 681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterkamp, M.J. (2013) Physiology and biochemistry of aromatic hydrocarbon‐degrading bacteria that use chlorate and/or nitrate as electron acceptor. Wageningen University & Research, Wageningen, The Netherlands.
- Peng, P. , Goris, T. , Lu, Y. , Nijsse, B. , Burrichter, A. , Schleheck, D. , et al. (2020) Organohalide‐respiring Desulfoluna species isolated from marine environments. ISME J 14: 815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilloni, G. , von Netzer, F. , Engel, M. , and Lueders, T. (2011) Electron acceptor‐dependent identification of key anaerobic toluene degraders at a tar‐oil‐contaminated aquifer by Pyro‐SIP. FEMS Microbiol Ecol 78: 165–175. [DOI] [PubMed] [Google Scholar]
- Pitts, K.E. , Dobbin, P.S. , Reyes‐Ramirez, F. , Thomson, A.J. , Richardson, D.J. , and Seward, H.E. (2003) Characterization of the Shewanella oneidensis MR‐1 decaheme cytochrome MtrA expression in Escherichia coli confers the ability to reduce soluble Fe (III) chelates. J Biol Chem 278: 27758–27765. [DOI] [PubMed] [Google Scholar]
- Rabus, R. , Kube, M. , Heider, J. , Beck, A. , Heitmann, K. , Widdel, F. , and Reinhardt, R. (2005) The genome sequence of an anaerobic aromatic‐degrading denitrifying bacterium, strain EbN1. Arch Microbiol 183: 27–36. [DOI] [PubMed] [Google Scholar]
- Saia, F.T. , de Souza, T.S.O. , Pozzi, E. , Duarte, R.T.D. , and Foresti, E. (2019) Sulfide‐driven denitrification: detecting active microorganisms in fed‐batch enrichment cultures by DNA stable isotope probing. Mol Biol Rep 46: 5309–5321. [DOI] [PubMed] [Google Scholar]
- Salinero, K.K. , Keller, K. , Feil, W.S. , Feil, H. , Trong, S. , Di Bartolo, G. , and Lapidus, A. (2009) Metabolic analysis of the soil microbe Dechloromonas aromatica str. RCB: indications of a surprisingly complex life‐style and cryptic anaerobic pathways for aromatic degradation. BMC Genomics 10: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, L. , Rosso, K.M. , Zachara, J.M. , and Fredrickson, J.K. (2012) Mtr extracellular electron‐transfer pathways in Fe (III)‐reducing or Fe (II)‐oxidizing bacteria: a genomic perspective. Biochem Soc Trans 40: 1261–1267. [DOI] [PubMed] [Google Scholar]
- Shinoda, Y. , Sakai, Y. , Uenishi, H. , Uchihashi, Y. , Hiraishi, A. , Yukawa, H. , et al. (2004) Aerobic and anaerobic toluene degradation by a newly isolated denitrifying bacterium, Thauera sp. strain DNT‐1. Appl Environ Microbiol 70: 1385–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda, Y. , Akagi, J. , Uchihashi, Y. , Hiraishi, A. , Yukawa, H. , Yurimoto, H. , et al. (2005) Anaerobic degradation of aromatic compounds by Magnetospirillum strains: isolation and degradation genes. Biosci Biotechnol Biochem 69: 1483–1491. [DOI] [PubMed] [Google Scholar]
- Sperfeld, M. , Rauschenbach, C. , Diekert, G. , and Studenik, S. (2018) Microbial community of a gasworks aquifer and identification of nitrate‐reducing Azoarcus and Georgfuchsia as key players in BTEX degradation. Water Res 132: 146–157. [DOI] [PubMed] [Google Scholar]
- Staats, M. , Braster, M. , and Röling, W.F.M. (2011) Molecular diversity and distribution of aromatic hydrocarbon‐degrading anaerobes across a landfill leachate plume. Environ Microbiol 13: 1216–1227. [DOI] [PubMed] [Google Scholar]
- Stolper, D.A. , Revsbech, N.P. , and Canfield, D.E. (2010) Aerobic growth at nanomolar oxygen concentrations. Proc Natl Acad Sci 107: 18755–18760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, W. , Sun, X. , and Cupples, A.M. (2014) Presence, diversity and enumeration of functional genes (bssA and bamA) relating to toluene degradation across a range of redox conditions and inoculum sources. Biodegradation 25: 189–203. [DOI] [PubMed] [Google Scholar]
- Top, E.M. , and Springael, D. (2003) The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr Opin Biotechnol 14: 262–269. [DOI] [PubMed] [Google Scholar]
- Valderrama, J.A. , Durante‐Rodríguez, G. , Blázquez, B. , García, J.L. , Carmona, M. , and Díaz, E. (2012) Bacterial degradation of benzoate cross‐regulation between aerobic and anaerobic pathways. J Biol Chem 287: 10494–10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallenet, D. , Calteau, A. , Dubois, M. , Amours, P. , Bazin, A. , Beuvin, M. , et al. (2020) MicroScope: an integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res 48: D579–D589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt, C. , Dorer, C. , Musat, F. , and Richnow, H.‐H. (2016) Multi‐element isotope fractionation concepts to characterize the biodegradation of hydrocarbons—from enzymes to the environment. Curr Opin Biotechnol 41: 90–98. [DOI] [PubMed] [Google Scholar]
- Weelink, S.A.B. , Van Doesburg, W. , Saia, F.T. , Rijpstra, W.I.C. , Röling, W.F.M. , Smidt, H. , and Stams, A.J.M. (2009) A strictly anaerobic betaproteobacterium Georgfuchsia toluolica gen. Nov., sp. nov. degrades aromatic compounds with Fe(III), Mn(IV) or nitrate as an electron acceptor. FEMS Microbiol Ecol 70: 575–585. [DOI] [PubMed] [Google Scholar]
- Xie, Q. , Lu, Y. , Tang, L. , Zeng, G. , Yang, Z. , Fan, C. , et al. (2021) The mechanism and application of bidirectional extracellular electron transport in the field of energy and environment. Crit Rev Environ Sci Technol 51: 1924–1969. [Google Scholar]
- Zedelius, J. , Rabus, R. , Grundmann, O. , Werner, I. , Brodkorb, D. , Schreiber, F. , et al. (2011) Alkane degradation under anoxic conditions by a nitrate‐reducing bacterium with possible involvement of the electron acceptor in substrate activation. Environ Microbiol Rep 3: 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Abundance of proteins of anaerobic toluene degradation detected from G5G6 cultures grown on toluene and Fe(III) (TF) or toluene and nitrate (TN).
Table S2. Abundance of proteins of aerobic toluene degradation detected from G5G6 cultures grown on toluene and Fe(III) (TF) or toluene and nitrate (TN).
Table S3. Abundance of proteins involved in denitrification detected from G5G6 cultures grown on toluene and Fe(III) (TF) or toluene and nitrate (TN).
Table S4. Abundance of proteins involved in Fe(III)citrate reduction detected from G5G6 cultures grown on toluene and Fe(III) (TF) or toluene and nitrate (TN).
Table S5 List of proteins detected from strain G5G6 under F(III)‐reducing (TF) and nitrate‐reducing (TN) conditions.
Data Availability Statement
The genome sequence of strains G5G6 was deposited in the European Bioinformatics Institute (accession number ERZ1938153). A list of proteins detected from strain G5G6 under nitrate‐ and Fe(III)‐reducing conditions is available in Supporting Information Table S5.


