Abstract
The 15 species of small carnivorous marsupials that comprise the genus Antechinus exhibit semelparity, a rare life‐history strategy in mammals where synchronized death occurs after one breeding season. Antechinus males, but not females, age rapidly (demonstrate organismal senescence) during the breeding season and show promise as new animal models of ageing. Some antechinus species are also threatened or endangered. Here, we report a chromosome‐level genome of a male yellow‐footed antechinus Antechinus flavipes. The genome assembly has a total length of 3.2 Gb with a contig N50 of 51.8 Mb and a scaffold N50 of 636.7 Mb. We anchored and oriented 99.7% of the assembly on seven pseudochromosomes and found that repetitive DNA sequences occupy 51.8% of the genome. Draft genome assemblies of three related species in the subfamily Phascogalinae, two additional antechinus species (Antechinus argentus and A. arktos) and the iteroparous sister species Murexia melanurus, were also generated. Preliminary demographic analysis supports the hypothesis that climate change during the Pleistocene isolated species in Phascogalinae and shaped their population size. A transcriptomic profile across the A. flavipes breeding season allowed us to identify genes associated with aspects of the male die‐off. The chromosome‐level A. flavipes genome provides a steppingstone to understanding an enigmatic life‐history strategy and a resource to assist the conservation of antechinuses.
Keywords: Antechinus, chromosome‐level, dasyurid, genome assembly, semelparity, suicidal reproduction
1. INTRODUCTION
For every animal species, the most critical activity is mating and the raising of offspring. Reproduction is a costly process, and animals invest energy into strategies that maximize the number and survival of offspring (Collett et al., 2018). Broadly, these strategies span the so‐called iteroparity–semelparity continuum: from a single (semel) to repeated (itero) reproductive episode(s) before death (reviewed in Hughes, 2017; Krajewski et al., 2000). In mammals, semelparity is observed in some Australian dasyurids (males only) and South American didelphids (both sexes) (Fisher et al., 2013; Leiner et al., 2008). Obligate male semelparity is the most extreme form of semelparity, occurring in the marsupial subfamilies Phascogalinae (15 Antechinus species and three Phascogale species; Bradley, 2003; Naylor et al., 2008) and Dasyurinae (the kaluta, Dasykaluta rosamondae; Hayes et al., 2019). In these dasyurid species, all males die soon after a 1‐ to 3‐week mating period (“die‐off” at 11.5 months of age), and a proportion of females survive to breed in the subsequent year (Baker & Dickman, 2018).
Breeding is a stressful life‐history event, but the stress response enables most animals to cope and to mobilize energy (Romero, 2004). Hormones play a vital role in this process. While in antechinus, both sexes have elevated stress hormone levels (chiefly the corticosteroid cortisol) during breeding, males also exhibit surging plasma testosterone levels (Bradley, 2003; Braithwaite & Lee, 1979; Croft, 2003; McAllan, 2006; Naylor et al., 2008). This interferes with negative feedback mechanisms and reduces levels of corticosteroid binding globulin (CBG), increasing the levels of free (i.e., unbound), active corticosteroids in the blood (Bradley, 2003; Naylor et al., 2008). As the breeding season progresses, corticosteroid levels continue to rise; this ultimately contributes to systemic collapse and death of all males. Corticosteroids are anti‐inflammatory and, at high concentrations, immunosuppressive, triggering a decline of body condition (Manoli et al., 2007; Perogamvros et al., 2012). Stress, combined with malnutrition, results in a significant negative energy balance and is associated with numerous comorbidities (Argiles et al., 2016). Cortisol is also elevated in females throughout the short breeding season, but in the absence of elevated testosterone, free cortisol levels remain relatively constant.
There is mounting evidence that testosterone and cortisol contribute to differences in ageing and related diseases (Austad & Fischer, 2016; Gems, 2014; Lemaitre et al., 2020; Martocchia et al., 2016). Moreover, ageing and obligate semelparity manifest as a decline in immune function—immunosenescence (Childs et al., 2017; Froy et al., 2019). In contrast to current mammal models of ageing, antechinuses are “natural” models of physiological senescence (see Diamond, 1982; McAllan, 2006). For example, the brain of the ageing wild‐type mouse and rat does not accumulate amyloid‐beta (Aβ) (Jucker, 2010). Aβ plaques, which are associated with chronic stress and elevated cortisol levels (e.g., see Lesuis et al., 2018), accumulate at the end of the antechinus lifespan in antechinuses—mirroring Alzheimer's disease neuropathology and potentially making them a suitable disease model (McAllan, 2006; Naylor et al., 2008). A high‐quality genome assembly is critical for future genetic experiments (e.g., CRISPR). Moreover, considering the strong effects of cortisol and testosterone on male obligate semelparity, identifying the expression of genes associated with these hormones and its consequences may help to understand the evolution of this extreme reproductive strategy.
Recent work has uncovered several new antechinus species, bringing the total number of species to 15 (Baker et al., 2013; Baker et al., 2014; Baker et al., 2015; Baker et al., 2012; Mutton et al., 2019). In 2018, Antechinus arktos and A. argentus were listed as Endangered on the Australian Government's threatened species list and were included in the 20 Australian mammals most likely to go extinct in the next two decades (Geyle et al., 2018). Moreover, areas of their habitat were recently ravaged by extensive wildfires and drought. It is estimated that nearly 3 billion animals died in these events in 2020 (Chris Dickman, 2020, University of Sydney, pers. comm.). Genomic resources are urgently required to support research on the population structure, speciation and recovery of antechinus species.
To investigate the phylogeny, demographic history, and genomic diversity of Antechinus flavipes and to pave the way to understanding the evolution of semelparity, high‐quality genomic data are essential. A genome assembly of the brown antechinus (Antechinus stuartii) was recently reported (Brandies et al., 2020a). However, a chromosome‐level genome is not yet available. PacBio long‐read sequencing combined with the chromosome conformation capture method Hi‐C resolves complex regions (such as repeat regions) and can greatly improve downstream analyses (Schloissnig et al., 2021). Here, we employed third‐generation sequencing technologies to generate the first chromosome‐level genome of an antechinus, that of the yellow‐footed antechinus (A. flavipes), as well as draft genomes of three related dasyurids (Figure 1a). We highlight genome features, phylogeny and historical demography, and a gene expression profile of A. flavipes male die‐off.
FIGURE 1.
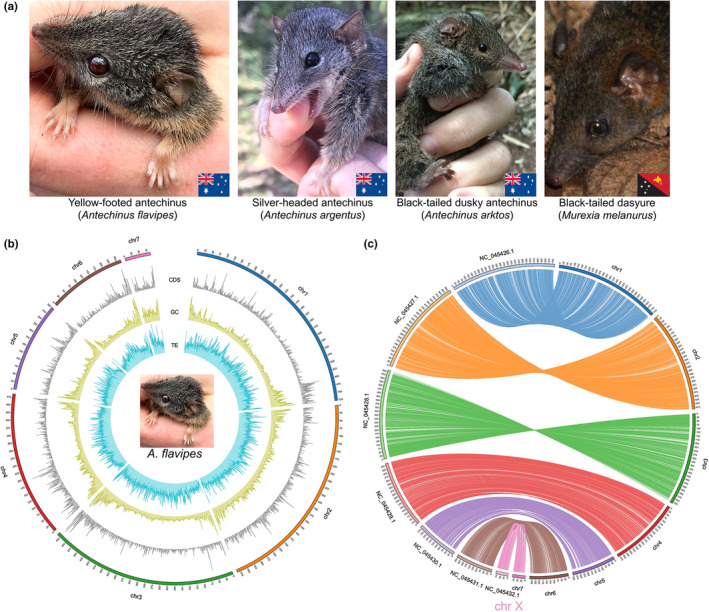
Overview of sequenced species and reference genome assemblies generated in this study. (a) Photographs of the five marsupial species sequenced. A yellow‐footed antechinus (Antechinus flavipes), silver‐headed antechinus (Antechinus argentus), and black‐tailed dusky antechinus (Antechinus arktos) from Australia; and a black‐tailed dasyure (Murexia melanurus) from Papua New Guinea. Antechinus spp. and M. melanurus photographs are courtesy of A.M.B. and John Hornbuckle, respectively. (b) Circos plot of the reference genome assembly of A. flavipes. The outermost segment represents chromosome sequences, with the numbers on the external surface indicating genome size in Mb. The line plots, from outside to inside, represent the distribution of CDS density (from 0 to 0.15), GC content (from 0.30 to 0.65) and TE ratio (from 0.2 to 1.0). Frequencies were calculated in 500‐kb sliding windows. (c) Circos plot showing shared synteny of A. flavipes (chr1‐chr7) and Sarcophilus harrisii (NC_045426.1‐NC_045432.1). Aligned using lastz. The synteny blocks are linked using lines coloured in accordance with the A. flavipes chromosomes. Aligned blocks with length shorter than 20 kb are not shown. Chr7 in A. flavipes corresponds to the X chromosome of Sarcophilus harrisii
2. MATERIALS AND METHODS
2.1. Sample collection
“AdamAnt,” an adult male yellow‐footed antechinus (Antechinus flavipes), was sampled in Samford Valley (greater Brisbane), QLD, Australia (27°22′S, 152°52′E) in September 2018. A second male individual (“Ant2”), sampled at the same location in 2017, was not used for genome assembly but employed in demographic analysis. A total of 106 A. flavipes samples were collected for transcriptome sequencing (RNA‐seq). Ear tissue samples (i.e., ear clippings) for short‐insert whole‐genome sequencing were obtained from the black‐tailed dusky antechinus Antechinus arktos (Lamington National Park, QLD, Australia, 28°22′S, 153°15′E; specimen AA100 sampled by A.M.B. in 2015) and the silver‐headed antechinus Antechinus argentus (from Blackdown Tableland National Park, QLD, Australia; 23°82′S, 149°07′E; specimen BD‐17‐5A sampled by A.M.B. in 2017). Frozen liver tissue from the black‐tailed dasyure (Murexia melanurus; Wau, Morobe, Papua New Guinea, 7°20′S, 146°42′E; voucher specimen ABTC46020 sampled by the South Australian Museum in 1985) was also sequenced. All DNA and RNA were extracted by BGI Hong Kong.
2.2. Reference genome sequencing and assembly
We used a combination of long‐fragment sequencing, short‐insert library sequencing for error correction and gap filling, and chromatin conformation capture (Hi‐C) to generate chromosome‐level semelparous mammal reference genomes. High‐molecular‐weight (HMW) DNA extracted from the testis of the A. flavipes individual “AdamAnt” was used to generate long‐read (PacBio) sequencing data by Annoroad Gene Technology. Briefly, 8 µg of DNA was sheared using a g‐TUBE (Covaris) and concentrated with AMPure PB magnetic beads (PacBio). A total of 323.85 Gb (~100×) A. flavipes PacBio reads were assembled using canu version 1.7 (Koren et al., 2017) with the error correction module. The corrected subreads were used for initial draft assembly using wtdbg2 version 1.2.8 (Ruan & Li, 2020). Paired‐end (2 × 100 bp) BGI‐SEQ500 data were generated from cerebrum, liver, heart and lung tissue from the same individual by BGI‐Qingdao. To reduce base errors, the assembly was “polished” using pilon version 1.23 (Walker et al., 2014) and 151.43 Gb (50×) 100‐bp paired‐end BGISEQ‐500 reads (mapped to the initial PacBio assembly using minimap2 version 2.10: Li, 2018; and samtools version 1.9: Li et al., 2009).
Genome sizes were estimated by k‐mer frequency analysis (Liu et al., 2013). Briefly, 100‐bp paired‐end whole genome sequence (WGS) reads were used as input into gce (Genomic Charactor Estimator) version 1.0.0 (Marcais & Kingsford, 2011) to obtain the k‐mer frequency and the genome size was estimated using the equation “Genome size = k‐mer number/k‐mer depth,” where the “k‐mer number” is the total number of k‐mers and “k‐mer depth” denotes the peak frequency that occurred more than any other frequencies. Genome length was estimated on the basis of the total scaffold length of the assembly. Using the frequency distribution of 17‐mers of short paired‐end reads (Figure S1), the A. flavipes genome was estimated to be 3.2 Gb.
Assembly quality was assessed using busco (Benchmarking Universal Single‐Copy Orthologs) version 5.0.0_cv1 (Seppey et al., 2019), employing the gene predictor augustus version 3.2.1 (Stanke & Waack, 2003) and the 9226‐gene busco mammalian lineage data set (mammalia_odb10). Although, gene centric, the busco Score is a good predictor of genome completeness (Seppey et al., 2019).
2.3. Assignment of scaffolds to chromosomes
A. flavipes Hi‐C libraries from liver tissue were prepared as described previously (Lieberman‐Aiden et al., 2009). Hi‐C libraries were sequenced on the BGISEQ‐500 platform and quality controlled using the hic‐pro (version 2.8.0_devel) pipeline (Servant et al., 2015), resulting in 51.0 Gb uniquely aligned read pairs. Reads validated by hic‐pro were next used to scaffold contigs into seven chromosome clusters (2n‐14 karyotype) using the 3d‐dna version 1.12 pipeline (Dudchenko et al., 2017). The assembly was further improved by interactive correction using juicebox version 1.11.08 (Dudchenko et al., 2018; Durand et al., 2016). A chromatin contact map can be found in Figure S2.
2.4. Draft dasyurid genome assemblies
Specimens of the Australian dasyurids A. arktos and A. argentus and the New Guinean dasyurid M. melanurus were sequenced by short‐insert WGS (see Table S1). Reference‐based (i.e., “pseudo”) nuclear genomes were obtained by mapping to the A. flavipes reference genome (see Feigin et al., 2018 and Supporting Methods).
2.5. RNA‐seq and transcriptome data processing
Eight A. flavipes specimens in 2017 were sequenced by BGI Hong Kong. The RNA of the remainder (seven males and six females collected in 2018) was extracted by BGI Hong Kong and sequenced by BGI Qingdao. Only those samples collected during the 2018 breeding season were used for differential gene expression analysis (the samples collected in 2017 were used for gene annotation only). Raw data were filtered using flexbar version 3.4.0 (Dodt et al., 2012; Roehr et al., 2017) with default settings (removes reads with any uncalled bases). BGISEQ‐500 adapters were obtained from a comparative study of Illumina and BGI sequencing platforms (Mak et al., 2017). Any residual rRNA reads (the majority removed by poly(A) selection prior to sequencing library generation) were removed from the reads using sortmerna version 2.1b (Kopylova et al., 2012) against the SILVA version 119 ribosomal database (Quast et al., 2013). Tissue transcriptomes were de novo assembled using trinity version 2.8.4 (Grabherr et al., 2011; Haas et al., 2013; Henschel et al., 2012) with default parameters. The A. flavipes transcriptome assemblies were assessed using busco (Seppey et al., 2019). Details of transcriptome sequencing data used in study are listed in Tables S2–S4 (also see Supporting Methods).
2.6. Genome annotation
2.6.1. Repeat element annotation
We identified repetitive elements by integrating homology and de novo predictions. Homology‐based transposable elements (TEs) annotations were obtained by interrogating a genome assembly with known repeats in the Repbase database version 16.02 (Bao et al., 2015) using repeatmasker version 4.0.5 (DNA‐level) (Tarailo‐Graovac & Chen, 2009) and repeatproteinmask (protein‐level; implemented in repeatmasker). De novo TE predictions were obtained using repeatmodeler version 1.1.0.4 (Smit & Hubley, 2010) and ltrharvest version 1.5.8 (Ellinghaus et al., 2008) to generate database for a repeatmasker run. tandem repeat finder (version 4.07) (Benson, 1999) was used to find tandem repeats (TRs) in the genome. A nonredundant repeat annotation set was obtained by combining the above data. Genome sequences of other marsupials were also interrogated using the repeat element annotation pipeline. The following were downloaded from DNA Zoo (Virginia opossum [Didelphis virginiana; dv‐2k; Dudchenko et al., 2018]), the NCBI Reference Sequence Database (Release 86) (Pruitt et al., 2007) (gray short‐tail opossum [Monodelphis domestica; MonDom5; GCF_000002295.2], koala [Phascolarctos cinereus; phaCin_unsw_v4.1; GCA_002099425.1], Tasmanian devil [Sarcophilus harrisii; Devil_ref v7.0, also known as sarHar1; GCA_000189315.1], Tammar wallaby [Notamacropus eugenii; Meug_1.1; GCA_000004035.1] and common wombat [Vombatus ursinus; GCF_900497805.2]), and the GigaScience Database (brown antechinus [Antechinus stuartii; Brandies et al., 2020a; Brandies et al., 2020b]).
2.6.2. Protein‐coding gene annotation
Protein‐coding genes of A. flavipes and Didelphis virginiana were annotated using homology‐based prediction, de novo prediction and RNA‐seq‐assisted prediction methods. Sequences of homologous proteins from five mammals (human [Homo sapiens], M. domestica, P. cinereus, S. harrisii and V. ursinus]) were downloaded from NCBI. These protein sequences were aligned to the repeat‐masked genome using blat version 0.36 (Kent, 2002). genewise version 2.4.1 (Birney et al., 2004) was employed to generate gene structures based on the alignments of proteins to the genome assembly. De novo gene prediction was performed using augustus version 3.2.3 (Stanke et al., 2006), genscan version 1.0 (Burge & Karlin, 1997) and glimmerhmm version 3.0.1 (Majoros et al., 2004) with a human training set. Transcriptome data were mapped to the assembled genome using hisat2 version 2.1.0 (Kim et al., 2019) and samtools version 1.9 (Li et al., 2009), and coding regions were predicted using transdecoder version 5.5.0 (Grabherr et al., 2011; Haas et al., 2013). A final nonredundant reference gene set was generated by merging the three annotated gene sets using evidencemodeler version 1.1.1 (EVM) (Haas et al., 2008) and excluding EVM gene models with only ab initio support. The gene models were translated into amino acid sequences and used in local blastp (Camacho et al., 2009) searches against the public databases Kyoto Encyclopedia of Genes and Genomes (KEGG; version 89.1) (Kanehisa & Goto, 2000), Clusters of Orthologous Groups (COG) (Tatusov et al., 2000), NCBI nonredundant protein sequences (NR; version 20170924) (O'Leary et al., 2016), Swiss‐Prot (release‐2018_07) (UniProt Consortium, 2012), TrEMBL (TRanslation of EMBL [nucleotide sequences that are not in Swiss‐Prot]; release‐2018_07) (O'Donovan et al., 2002), and InterPro (version 69.0) (A. L. Mitchell et al., 2019). A total of 18,068 (93.5%) of A. flavipes genes could be functionally annotated. Where specific genes are named in this paper, human nomenclature assignments are used unless otherwise noted.
2.7. Phylogeny and divergence time estimation
Marsupials (metatherians) and eutherians diverged ~160 million years ago (Ma), long before the radiation of extant eutherian clades (~100 Ma) (Luo et al., 2011; Phillips et al., 2009). We employed the platypus (Ornithorhynchus anatinus) as an outgroup as this species is frequently used as an outgroup for both eutherian and marsupial mammals and it has a high‐continuity, well‐annotated genome (contig N50 15.1 Mb; scaffold N50 83.3 Mb). In contrast, the genomes of the most closely related eutherian mammals, the edentates (sloths, armadillos and anteaters) currently have relatively poor‐quality genome assemblies. These include, for example, the armadillo Dasypus novemcinctus (contig N50 0.03 Mb, scaffold N50 1.7 Mb) and the sloth Choloepus hoffmanni (contig N50 0.06 Mb; scaffold N50 0.4 Mb).
We identified 6090 high‐confidence 1:1 orthologues by interrogating the predicted proteins from the gene models of 11 species (ten marsupials and the platypus) using sonicparanoid version 1.3.0 (Cosentino & Iwasaki, 2019). Because the A. arktos, A. argentus and M. murexia pseudogenomes were derived from A. flavipes, their genes were extracted using the A. flavipes gene annotation file. The corresponding coding sequences (CDS) for each species were aligned using prank version 100802 (Loytynoja & Goldman, 2005) and filtered by gblocks version 0.91b (Talavera & Castresana, 2007) to identify conserved blocks (removing gaps, ambiguous sites and excluding alignments less than 300 bp in size), leaving 6090 genes. Maximum‐likelihood (ML) phylogenetic trees were generated using raxml version 7.2.8 (Stamatakis, 2006) and iq‐tree version 2.1.3 (Minh et al., 2020) with three CDS data sets: the whole coding sequence (whole‐CDS), first codon positions and fourfold degenerate (4d) sites. Identical topologies and similar support values were obtained (1000 bootstrap iterations were performed). The divergence time between species was estimated using mcmctree (a Bayesian molecular clock model implemented in paml version 4.7; Yang, 2007) with the GTR+G nucleotide substitution model, and the whole‐CDS ML tree and concatenated whole‐CDS supergenes as inputs. We used 100,000 iterations after a burn‐in of 10,000 iterations. mcmctree calibration points (million years ago; Ma) were obtained from timetree (Kumar et al., 2017): O. anatinus–P. cinereus (~167–192 Ma), V. ursinus–D. virginiana (~72–86 Ma), V. ursinus–M. melanurus (~56–64 Ma), V. ursinus–P. cinereus (~31–39 Ma), M. domestica–D. virginiana (~24–39 Ma) and S. harrisii–M. melanurus (~4–22 Ma). For comparison, phylogenetic trees of marsupials in the marsupial orders Dasyuromorphia and Didelphimorphia were obtained by querying the PHYLACINE (The Phylogenetic Atlas of Mammal Macroecology) resource (Faurby et al., 2018).
2.8. Genomic diversity
Genome‐wide heterozygosity (π) was calculated from (2 × 100 bp) WGS read mapped to the S. harrisii genome. For details, see Supporting Methods.
2.9. Demographic history
We inferred demographic histories by applying the Pairwise sequentially Markovian coalescent (PSMC) method (Li & Durbin, 2011). Short‐insert paired‐end WGS reads of A. flavipes (two individuals), A. arktos, A. argentus, A. stuartii (two individuals; see Brandies et al., 2020a, 2020b) and M. melanurus were mapped to a repeat‐masked S. harrisii genome. Putative X‐linked scaffolds in the S. harrisii assembly were excluded (see Supporting Methods). Geographical maps were generated using the R package “ggmap” (Kahle & Wickham, 2013).
2.10. Transcriptome profiling of Antechinus flavipes breeding
To search for transcripts associated with obligate semelparity, we performed differential gene expression analysis of a single A. flavipes population (Samford Valley, Queensland, Australia) from August to October 2018. Every 2 days, we microscopically examined female urine for cornified, anuclear vaginal epithelial cells, a sign of peak oestrus (Selwood, 1980). Concurrent visual inspections of captured males for behavioural changes (e.g., agitation) revealed evidence of stress beginning in the last week of September, followed by progressive loss of body condition (Figure S3). The practical and ethical challenges of longitudinal sampling from a wild population limited the number of samples that could be obtained. In this study, we present differential expression data of eight male and six female antechinuses (liver, kidney, skeletal muscle, cerebrum and reproductive tissues [testis and ovary]). See Supporting Methods and Figures S4 and S5 for data processing, assessment and differential expression procedures, and Appendix S1 for normalized counts.
3. RESULTS AND DISCUSSION
3.1. Genome assembly and annotation
We used PacBio sequencing and Hi‐C (Burton et al., 2013) to create a 3.2‐Gb (2n = 14) chromosome‐level male Antechinus flavipes nuclear genome assembly with a contig N50 of 51.8 Mb and scaffold N50 of 636.7 Mb (Table 1 and Figure 1). The seven chromosomes ranged in size from 86.6 to 727.3 Mb (Table 2) and covered ~99.7% of the assembly. We also generated A. flavipes reference‐based assemblies of Antechinus arktos (female), Antechinus argentus (female) and Murexia melanurus (male) from short‐insert WGS reads. Most A. flavipes genome regions had a GC‐depth of about 50×, while a small peak at 25× depth revealed heterozygous sex chromosomes (Figure S6). Hi‐C was able to recover an ~86.5‐Mb X chromosome (chromosome 7), while the Y chromosome could not be obtained (see Supporting Methods section S2.0). This is unsurprising, as marsupial Y chromosomes are small (~10–12 Mb; akin to microchromosomes of birds), repeat‐rich and refractory to assembly (Deakin & O'Neill, 2020). The chromosomes of A. flavipes were highly homologous to the related Tasmanian devil (Sarcophilus harrisii) (Figure 1c), supporting the quality of the assembly.
TABLE 1.
Antechinus flavipes genome assembly statistics
| Statistic | Assembly method | |
|---|---|---|
| PacBio (100×) + BGISEQ‐500 (50×) polishing | PacBio + BGISEQ‐500 polishing + Hi‐C | |
| Contig number | 1107 | 1107 |
| Contig length (bp) | 3,192,388,871 | 3,192,388,871 |
| Contig N50 (bp) | 51,843,704 | 51,843,704 |
| Contig max length (bp) | 168,884,005 | 168,884,005 |
| Scaffold number | 1105 | 487 |
| Scaffold N50 (bp) | 51,843,704 | 636,717,922 |
| GC content (%) | 36.21 | 36.21 |
TABLE 2.
Summary of assembled Antechinus flavipes pseudochromosomes
| Chr ID | Sequence number | Sequence length (bp) |
|---|---|---|
| Chr 1 | 88 | 727,308,865 |
| Chr 2 | 83 | 686,337,178 |
| Chr 3 | 146 | 636,717,922 |
| Chr 4 | 50 | 486,195,733 |
| Chr 5 | 104 | 300,479,852 |
| Chr 6 | 63 | 264,590,327 |
| Chr 7 | 91 | 86,570,782 |
The GC content of A. flavipes (~36%) (Table 1) is similar to the brown antechinus (A. stuartii) and S. harrisii. As indicated by 17‐mer frequency analysis (Figure S1), the A. flavipes genome is repeat‐rich. Repetitive elements account for 51.8% (~1.7 Gb) of the assembly, with long interspersed elements (LINEs; ~45%), long terminal retrotransposons (LTRs; 15.4%) and short interspersed elements (SINEs; 6.6%) being the major classes of TEs (Tables S5 and S6). We annotated 24,708 A. flavipes protein‐coding genes (82.2% supported by transcriptome data from 12 tissues). The A. flavipes reference assembly obtained a busco (Benchmarking Universal Single‐Copy Orthologs) (Seppey et al., 2019) genome completeness score of 92.4%—comparable to A. stuartii (92.2% and 92.3% for a female and male assembly, respectively) (Brandies et al., 2020a), the koala (Phascolarctos cinereus; 93.9%) (Johnson et al., 2018) and S. harrisii (91.6%; 2019 assembly mSarHar1.1) (Table S7). The scores of the three dasyurid reference‐based assemblies, here used for phylogenetic and demography analyses, ranged from 74.2% to 90.0% recovered complete BUSCO genes (Table S8).
3.2. Phylogeny and demographic history
To construct a time‐calibrated species tree (Figure 2a), we identified 6090 high‐confidence single‐copy orthologues from 10 marsupial species and a monotreme, the platypus (Ornithorhynchus anatinus). Extant Australasian and American marsupials diverged from a South American ancestor ~80 Ma (Kumar et al., 2017; Nilsson et al., 2010), emphasizing the deep evolutionary history of marsupials. We estimated that the lineages leading to extant Dasyuridae (e.g., Antechinus) and Didelphidae diverged 71.7 Ma (70.1–72.7 Ma 95% credibility interval [CI]). The dasyurid subfamily Phascogalinae comprises three genera: the semelparous Australian Antechinus and Phascogale, which have male die‐off, and the iteroparous New Guinean sister genus Murexia. Small dasyurids from mainland Australia are likely to have dispersed into New Guinea 9–11 Ma (Mitchell et al., 2014). We found the divergence time of Antechinus and Murexia to be about 11.0 Ma (95% CI 8.3–13.3). Our analysis shows that the four antechinus species split from a common ancestor ~6.8 Ma (95% CI 6.1–7.3), while A. flavipes and A. argentus + A. stuartii share an ancestor ~5.9 Ma (95% CI 5.3–6.5). Our phylogeny and divergence times concord with trees generated from the PHYLACINE database (Faurby et al., 2018) (Figure S7) and with recent estimates from a small set of mitochondrial and nuclear genes (Mutton et al., 2019).
FIGURE 2.
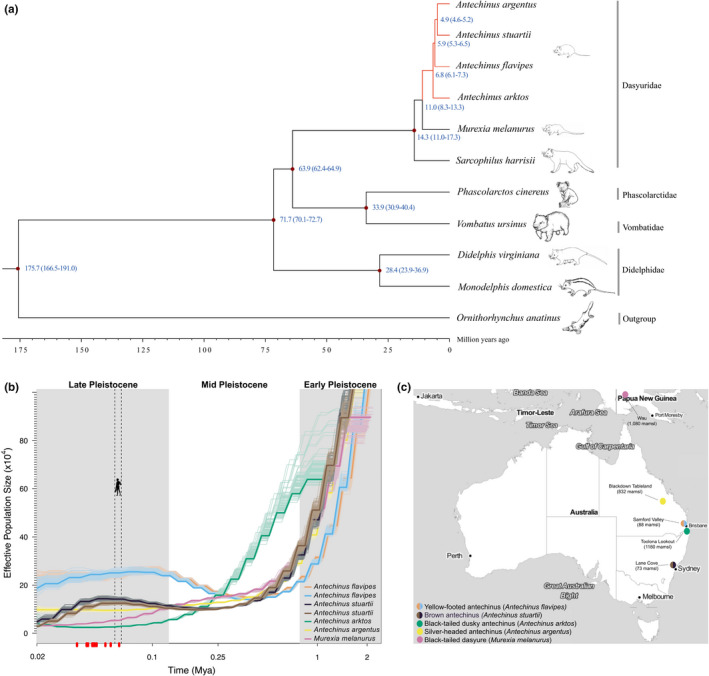
Evolutionary history of semelparous mammals. (a) Inferred phylogeny of ten marsupials and the platypus (outgroup) based on whole‐coding sequences of 6090 1:1 orthologues. Blue numbers at nodes represent the estimated divergence time from present (million years ago; Ma) between lineages. Semelparous lineages are indicated in red. (b) Demography of the dasyurid subfamily Phascogalinae. Historical population sizes (N e) were estimated using the pairwise sequentially Markovian coalescent (PSMC) method (Li & Durbin, 2011) and diploid genome sequences. The x‐axis shows the time before present in years on a log scale; the dashed grey line shows human arrival in Australia 59.3–65 ka (Clarkson et al., 2017); red dots indicate the extinction times of 15 species of Pleistocene Sahul (Saltre et al., 2016). The y‐axis shows N e, with bootstrap estimates indicated by lighter lines. Plots were scaled using a mutation rate (μ) of 1.37 × 10−9 substitutions per nucleotide per generation and species‐specific generation times (g) of 1 year. Two Antechinus flavipes (coloured in orange and blue) and two Antechinus stuartii (in brown and black) individuals were interrogated. (c) Map showing the location (coloured circles) and elevation (m amsl; metres above sea level) of sampled dasyurids (see panel b)
The PSMC method (Li & Durbin, 2011), which can infer changes in the effective population size (N e) over the Pleistocene, was used to evaluate four sequenced antechinus species and M. melanurus (Figure 2b,c). Until the end of the Pleistocene, ~11,700 years ago, mainland Australia, New Guinea and Tasmania formed a single continent, Sahul (Barrows et al., 2002). Glaciation of Sahul was restricted to the Snowy Mountains of southeastern mainland Australia and the Tasmanian highlands, and the continent experienced cycles of aridity and sea‐level fluctuations (Barrows et al., 2002). Evidently, during the Early to Mid‐Pleistocene until about 250,000 years ago, all species had larger population sizes, which is probably a reflection of larger expanses of connected forest habitats (Baker & Van Dyck, 2013). After 250,000 years ago, however, the species occurring in higher‐elevation, wetter habitat, A. arktos, A. argentus and M. melanurus, apparently declined in population size, whereas A. flavipes and A. stuartii populations, which are tolerant of drier conditions, underwent expansion (Figure 2a,b). This pattern persisted into the Late Pleistocene ~11,700–126,000 years ago, a period when temperature fluctuations were large, and many mesic‐adapted species declined, were restricted or became extinct; more arid‐adapted species radiated into burgeoning desert landscapes (Saltre et al., 2016). Our results are generally in accordance with a recent study suggesting that aridity and geographical isolation promoted dasyurid speciation (García‐Navas et al., 2020).
While the PSMC method cannot infer population dynamics after ~20,000 years ago, it is noteworthy that A. flavipes had the largest N e in the most recent PSMC estimate. This species is one of the most common, arid‐adapted and broadly distributed species in the genus Antechinus today (Baker & Van Dyck, 2013; Lada et al., 2008; Mutton et al., 2019). There is an apparent reduction in population size of A. stuartii ~20,000–60,000 years ago. Interestingly, although A. stuartii is generally considered a common antechinus species, evidence is emerging that it should be considered a species complex, representing multiple lineages, and thus possesses a smaller distributional range than presently recognized (Viacava et al., 2021). Moreover, we note that the A. stuartii individuals included here were sampled from Lane Cove National Park, a site with a low current population size (NSW Department of Environment & Conservation, 2004). In conclusion, our analysis suggests that climate change during the Early (~2.5–0.78 Ma) (Lisiecki & Raymo, 2005) and Mid‐Pleistocene (0.78–0.126 Ma) (Elderfield et al., 2012) has shaped the evolution of the dasyurid subfamily Phascogalinae by isolating populations in different habitats, resulting in some species being now greatly restricted in their distribution.
We found that all small dasyurid taxa have much higher (100 orders of magnitude) genome‐wide heterozygosity (π from 0.048 to 0.096) than the endangered S. harrisii (π = 0.00032 (Miller et al., 2011)) (Figure S8). Moreover, the heterozygosity of A. flavipes is twofold higher than that of A. arktos, A. argentus and M. melanurus. This may reflect its larger historical and current population size (Figure 2b,c). Although we are comparing individuals from single populations, combined with factors such as environmental change, loss of habitat and population fragmentation, the lower genetic diversity of A. arktos and A. argentus may reflect their present‐day low population and threatened species status (Mutton et al., 2019; Riordan et al., 2020). Future studies including assessment of geographical population structures and the inclusion of historical and current specimens (Diez‐Del‐Molino et al., 2018) are required to assess the threat level of extant small dasyurids. Our high‐quality A. flavipes assembly serves as an ideal reference genome for population genomic studies of A. flavipes and the 14 other antechinus species.
3.3. A gene expression profile of male die‐off in Antechinus
We profiled gene expression during antechinus reproduction by surveying multiple tissues from an A. flavipes population in Samford Valley in Queensland, Australia, during the 2018 breeding season (Movie S1). We observed reproductive behaviour and peak oestrus on September 15, 2018 (see Supporting Methods for details). Adult male A. flavipes disappear from this site after the first week of October. Antechinus male die‐off is characterized by a dramatic increase in free cortisol (Figure 3a), mediated by a surge in testosterone which decreases the production of corticosteroid‐binding globulin (Boonstra, 2005; Naylor et al., 2008). The potent effects of cortisol mean that the associated gene expression and its consequences (Le Phuc et al., 2005; McDowell et al., 2018) become a suitable backdrop against which comparisons can be made. While corticosteroids regulate a large number of genes via the glucocorticoid receptor (GR) (Le Phuc et al., 2005; McDowell et al., 2018), complicating gene expression analyses, chronic (i.e., breeding male antechinus) exposure to free corticosteroids has distinct outcomes (Manoli et al., 2007). The steroid hormone mediates a stress response, alters metabolism and mobilizes glucose as energy from a series of stores, initially enhancing short‐term survival.
FIGURE 3.
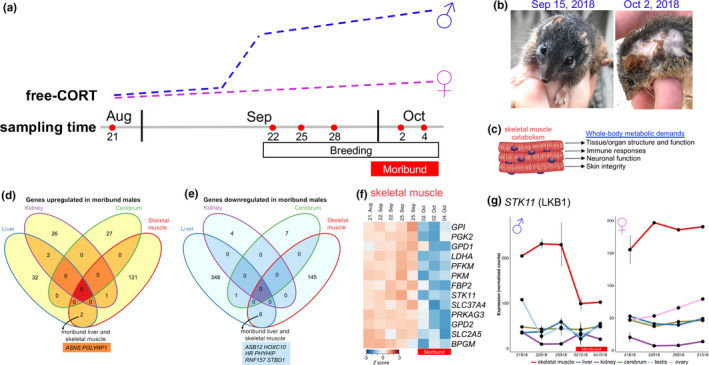
Gene expression profiling of the Antechinus flavipes breeding cycle. (a) Schematic overview of the yellow‐footed antechinus (A. flavipes) breeding season in Samford Valley, Queensland, Australia. Free‐CORT illustrates an increase in the level of free (unbound) stress hormones (chiefly the corticosteroid cortisol) in males during the breeding season of antechinuses (based on three studies illustrated in Naylor et al., 2008). (b) Rapid organismal senescence of male A. flavipes. Left, male (~11 months of age) prior to the breeding season. Right, late breeding‐stage male (~11.5 months of age). Note the loss of fur and deterioration of body condition. (c) In the moribund state, when other energy stores have been depleted, skeletal muscle catabolism (induced by the corticosteroid cortisol) supports whole‐body metabolic demands. (d) Venn diagram for the overlap of significantly up‐regulated (left) or down‐regulated (right) genes in moribund male antechinuses. Called by limma version 3.22.1 (≥1.5‐fold change and p ≤ .01). (e) Heat map of selected energy metabolism genes differentially expressed in the skeletal muscle of moribund male antechinuses. Moribund samples are indicated by a red bar below the map. Scaled TMM‐normalized read counts (denoted as the row Z score) are plotted, with red indicating high expression and blue indicating low expression. BPGM, 2,3‐bisphosphoglycerate mutase; FBP2, fructose‐bisphosphatase 2; GPD1, glycerol‐3‐phosphate dehydrogenase 1; GPD2, glycerol‐3‐phosphate dehydrogenase 2; GPI, glucose‐6‐phosphate isomerase; LDHA, lactate dehydrogenase A; PFKM, phosphofructokinase, muscle; PGK2, phosphoglycerate kinase 2; PKM, pyruvate kinase M1/2; PRKAG3, protein kinase AMP‐activated noncatalytic subunit gamma 3; SLC2A5, solute carrier family 2 member 5; SLC37A4, solute carrier family 37 member 4; STK11, serine/threonine kinase 11. (f) Expression pattern of the AMPK regulator LKB1 (STK11) in male (left) and female (right) tissues throughout the breeding season. Counts per million TMM‐normalized counts, with standard error bars
Because of a limited number of samples in our current data set, we considered genes that were differentially expressed at the end of the breeding season (early October), when all males die (are moribund; right panel in Figure 3b). Each sex was compared separately. There were more differentially expressed genes (limma‐modified Student's t‐test p ≤ .01) in the male liver and skeletal muscle than in the kidney and cerebrum (Figure 3c and Appendix S2), and most of these were downregulated. Genes upregulated in moribund male livers showed enrichment of biological process gene sets attributed to the innate immune response (bootstrap p ≤ .0001) (Table S9). However, several cytokine genes (PDGFRA, PDGFRB, PRNP, CD248, CD79A, CXCR5 and CXCR6) were downregulated in the moribund liver (Appendix S2). Characteristic liver pathology of the moribund male antechinus includes infections by bacteria such as Listeria monocytogenes (Barker et al., 1978; Bradley et al., 1980), viruses (e.g., dasyurid herpesvirus 1; Amery‐Gale et al., 2014) and parasitic infestation (Poskitt et al., 1984). Cortisol‐mediated immunosuppression in the preceding two weeks is likely to lead to poor late‐stage resistance to invading pathogens. We identified eight genes that were downregulated and two genes that were upregulated by both liver and skeletal muscle of moribund males (Figure 3d). This included decreased expression of the glycophagy receptor STBD1 (Johansen & Lamark, 2020) and increased expression of ASNS, a gene induced by amino acid starvation stress (Balasubramanian et al., 2013). Downregulation of SLC2A4 (encodes the glucose transporter GLUT4; Amoasii et al., 2019) and NUR4A2 (NUR11; a regulator of GLUT4 expression; Amoasii et al., 2019) indicates reduced glucose uptake by the skeletal muscle of moribund males. KEGG pathway and biological process enrichment analysis revealed metabolic dysregulation of skeletal muscle in moribund males, with marked downregulation of genes associated with glycolysis, gluconeogenesis and insulin signalling—indicating insulin resistance (bootstrap p ≤ .0001) (Figure 3e; Tables S9 and S10). These genes included PRKAG3 (−4.7‐fold; limma‐modified Student's t‐test, p = 3.7 × 10−4), which encodes a subunit of a key energy‐sensing molecule, AMP‐activated protein kinase, and its regulator LKB1 (STK11; −2.0‐fold, p = 2.4 × 10−3) (Appendix S2). AMPK responds to energetic stress to regulate multiple metabolic pathways. Reduced expression of LKB1 was limited to the skeletal muscle of moribund A. flavipes males (Figure 3f). LKB1 has a tissue‐specific role in energy metabolism. Complete or partial loss of Stk11 expression in mouse skeletal muscle prevents AMPK activation and causes metabolic defects such as acceleration of ageing‐induced myopathy (Bujak et al., 2015; Koh et al., 2006; Sakamoto et al., 2005; Thomson et al., 2007).
Our work is the first gene expression profile of a semelparous mammal. In agreement with observational data from A. flavipes and other antechinuses (Bradley et al., 1980; Naylor et al., 2008; Woollard, 1971), our results indicate near‐complete depletion of fat stores and skeletal muscle atrophy—leading to a catastrophic energy crisis and an inability to maintain cognitive and physical function in moribund males. Further experimental work on the endocrine system of semelparous mammal species is needed to characterize the role of testosterone and the dysregulation of the hypothalamic–pituitary–adrenal axis and negative feedback by cortisol. We were not able to capture gene expression changes in brain regions other than the whole cerebrum of A. flavipes during the breeding season, limiting our insights into the effects on major organs. Nevertheless, the A. flavipes reference genome permits future studies on gene expression and regulation (e.g., glucocorticoid receptor ChIP‐seq [chromatin immunoprecipitation sequencing]).
4. CONCLUSION
Our study and accompanying data sets, including the first chromosome‐level genome of a semelparous mammal, provide a critical resource for future studies on semelparity and its association with the universal ageing process. Complementary genomic data, including the genomes of additional Australasian and South American semelparous and iteroparous marsupials, are forthcoming and promise to shed light on whether semelparity in lineages with a common ancestor ~80 Ma evolved by independent, lineage‐specific molecular changes or by shared molecular convergence. The genomic resources also provide tools for broader studies on the ecology, evolution and conservation of marsupials.
AUTHOR CONTRIBUTIONS
I.S. and G.F. initiated and coordinated the project. C.P., K.M. and A.M.B. collected Antechinus flavipes samples. L.K.C. dissected A. flavipes. A.M.B. provided ear clippings from Antechinus arktos and Antechinus argentus. P.B.T. and L.K.C. provided frozen tissue samples for whole‐genome, Hi‐C and transcriptome sequencing. C.S. led the sequencing and assembly efforts with G.F. R.T., K.H. and C.S. contributed to genome assembly and annotation. R.T., K.H., C.S. and I.S. performed comparative genomics analyses. The de novo and reference‐based assemblies of A. arktos, A. argentus and M. melanurus were generated by I.S. I.S. performed population history analysis. K.H. performed diversity analysis. I.S. assembled transcriptomes and performed gene expression analysis. R.T. performed molecular evolution analyses. L.K.C., D.O.F., N.O.L. and A.M.B. provided direction on marsupial ecology and physiology. R.T., L.K.C., D.O.F., A.M.B. and I.S. wrote the paper with input from all authors.
OPEN RESEARCH BADGES
This article has earned an Open Data Badge for making publicly available the digitally‐shareable data necessary to reproduce the reported results. The data is available at https://github.com/sciseim/semelparity-genomics.
Supporting information
Supplementary Material
Movie S1
Appendix S1
Appendix S2
ACKNOWLEDGEMENTS
This study was supported by the National Natural Science Foundation of China (NSFC) grants 31950410545 (to I.S.) and 31900310 (to R.T.), the Jiangsu Science and Technology Agency (to I.S.), the Queensland University of Technology Faculty of Health/School of Biomedical Sciences Industry Collaboration Preparedness Pilot Research Support Scheme (I.S., L.K.C. and A.M.B.), and a Brazilian Council of Research and Scientific Development (CNPQ/PELD) grant 441225/2016‐0 (to N.O.L.). We thank Dr Penny Jeffery (Queensland University of Technology) for help with sample processing, and Leanne Wheaton (South Australian Museum) for providing the Murexia melanurus tissue sample. We also thank the staff of the Samford Ecology Research Facility (SERF) at Queensland University of Technology for supporting our A. flavipes field collection activities. A. flavipes collection was approved by the Queensland University of Technology University Animal Ethics Committee (permit nos. 1700000661 and 1800000141) and the Queensland Department of Environment and Science (WITK18736718).
Tian, R. , Han, K. , Geng, Y. , Yang, C. , Shi, C. , Thomas, P. B. , Pearce, C. , Moffatt, K. , Ma, S. , Xu, S. , Yang, G. , Zhou, X. , Gladyshev, V. N. , Liu, X. , Fisher, D. O. , Chopin, L. K. , Leiner, N. O. , Baker, A. M. , Fan, G. , & Seim, I. (2022). A chromosome‐level genome of Antechinus flavipes provides a reference for an Australian marsupial genus with male death after mating. Molecular Ecology Resources, 22, 740–754. 10.1111/1755-0998.13501
Ran Tian and Kai Han contributed equally to this work.
Contributor Information
Guangyi Fan, Email: fanguangyi@genomics.cn.
Inge Seim, Email: inge@seimlab.org.
DATA AVAILABILITY STATEMENT
Antechinus flavipes PacBio reads are available at China National GenBank (CNGB) Project ID CNP0001147. Short‐read whole‐genome sequencing and RNA sequencing data (BGISEQ‐500) generated in this project are available at NCBI BioProject PRJNA565840. The A. flavipes genome assembly is available at NCBI Genomes (JADWMD000000000). Pseudo (reference‐based) and de novo genome assemblies of Antechinus arktos, Antechinus argentus and Murexia melanurus (Seim, 2020a, 2020b), a ROPUS Singularity image (Seim, 2020c), and gene annotation files and associated FASTA files for A. flavipes are available at Zenodo (Seim, 2021). Mitochondrial genome assemblies of A. flavipes (MN447797), A. arktos (MK977601) and M. melanurus (MK977600) are available at GenBank. Various scripts used for data processing and analyses are available on GitHub at https://github.com/sciseim/semelparity‐genomics.
REFERENCES
- Amery‐Gale, J. , Vaz, P. K. , Whiteley, P. , Tatarczuch, L. , Taggart, D. A. , Charles, J. A. , Schultz, D. , Ficorilli, N. P. , Devlin, J. M. , & Wilks, C. R. (2014). Detection and identification of a gammaherpesvirus in Antechinus spp. Australia. Journal of Wildlife Diseases, 50(2), 334–339. 10.7589/2013-07-165 [DOI] [PubMed] [Google Scholar]
- Amoasii, L. , Sanchez‐Ortiz, E. , Fujikawa, T. , Elmquist, J. K. , Bassel‐Duby, R. , & Olson, E. N. (2019). NURR1 activation in skeletal muscle controls systemic energy homeostasis. Proceedings of the National Academy of Sciences of the United States of America, 116(23), 11299–11308. 10.1073/pnas.1902490116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argiles, J. M. , Campos, N. , Lopez‐Pedrosa, J. M. , Rueda, R. , & Rodriguez‐Manas, L. (2016). Skeletal muscle regulates metabolism via interorgan crosstalk: Roles in health and disease. Journal of the American Medical Directors Association, 17(9), 789–796. 10.1016/j.jamda.2016.04.019 [DOI] [PubMed] [Google Scholar]
- Austad, S. N. , & Fischer, K. E. (2016). Sex differences in lifespan. Cell Metabolism, 23(6), 1022–1033. 10.1016/j.cmet.2016.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, A. M. , & Dickman, C. (2018). Secret lives of carnivorous marsupials. CSIRO PUBLISHING. [Google Scholar]
- Baker, A. M. , Mutton, T. Y. , & Hines, H. B. (2013). A new dasyurid marsupial from Kroombit Tops, south‐east Queensland, Australia: The Silver‐headed Antechinus, Antechinus argentus sp. nov. (Marsupialia: Dasyuridae). Zootaxa, 3746(2), 201–239. [DOI] [PubMed] [Google Scholar]
- Baker, A. M. , Mutton, T. Y. , Hines, H. B. , & Van Dyck, S. (2014). The Black‐tailed Antechinus, Antechinus arktos sp. nov.: a new species of carnivorous marsupial from montane regions of the Tweed Volcano caldera, eastern Australia. Zootaxa, 3765(2), 101–133. [DOI] [PubMed] [Google Scholar]
- Baker, A. M. , Mutton, T. Y. , Mason, E. , & Gray, E. (2015). A taxonomic assessment of the Australian Dusky Antechinus Complex: A new species, the Tasman Peninsula Dusky Antechinus (Antechinus vandycki sp. nov.) and an elevation to species of the Mainland Dusky Antechinus (Antechinus swainsonii mimetes (Thomas)). Memoirs of the Queensland Museum, 59, 75–126. 10.17082/j.2204-1478.59.2015.2014-10 [DOI] [Google Scholar]
- Baker, A. M. , Mutton, T. Y. , & Van Dyck, S. (2012). A new dasyurid marsupial from eastern Queensland, Australia: The buff‐footed antechinus, Antechinus mysticus sp. nov. (Marsupialia: Dasyuridae). Zootaxa, 3515(1), 1–37. [Google Scholar]
- Baker, A. M. , & Van Dyck, S. (2013). Taxonomy and redescription of the Yellow‐footed Antechinus, Antechinus flavipes (Waterhouse) (Marsupialia: Dasyuridae). Zootaxa, 3649, 1–62. 10.11646/zootaxa.3649.1 [DOI] [PubMed] [Google Scholar]
- Balasubramanian, M. N. , Butterworth, E. A. , & Kilberg, M. S. (2013). Asparagine synthetase: Regulation by cell stress and involvement in tumor biology. American Journal of Physiology. Endocrinology and Metabolism, 304(8), E789–E799. 10.1152/ajpendo.00015.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, W. , Kojima, K. K. , & Kohany, O. (2015). Repbase update, a database of repetitive elements in eukaryotic genomes. Mobile DNA, 6, 11. 10.1186/s13100-015-0041-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker, I. , Beveridge, I. , Bradley, A. , & Lee, A. (1978). Observations on spontaneous stress‐related mortality among males of the dasyurid marsupial Antechinus stuartii Macleay. Australian Journal of Zoology, 26(3), 435–447. 10.1071/ZO9780435 [DOI] [Google Scholar]
- Barrows, T. T. , Stone, J. O. , Fifield, L. K. , & Cresswell, R. G. (2002). The timing of the last glacial maximum in Australia. Quaternary Science Reviews, 21(1–3), 159–173. 10.1016/S0277-3791(01)00109-3 [DOI] [Google Scholar]
- Benson, G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Research, 27(2), 573–580. 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney, E. , Clamp, M. , & Durbin, R. (2004). GeneWise and genomewise. Genome Research, 14(5), 988–995. 10.1101/gr.1865504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonstra, R. (2005). Equipped for life: The adaptive role of the stress axis in male mammals. Journal of Mammalogy, 86(2), 236–247. 10.1644/BHE-001.1 [DOI] [Google Scholar]
- Bradley, A. J. (2003). Stress, hormones and mortality in small carnivorous marsupials. In Jones M., Dickman C., & Archer M. (Eds.), Predators with pouches‐the biology of carnivorous marsupials (pp. 255–267). CSIRO Publishing. [Google Scholar]
- Bradley, A. J. , McDonald, I. R. , & Lee, A. K. (1980). Stress and mortality in a small marsupial (Antechinus stuartii, Macleay). General and Comparative Endocrinology, 40(2), 188–200. 10.1016/0016-6480(80)90122-7 [DOI] [PubMed] [Google Scholar]
- Braithwaite, R. W. , & Lee, A. K. (1979). A mammalian example of semelparity. American Naturalist, 113, 151–155. 10.1086/283372 [DOI] [Google Scholar]
- Brandies, P. A. , Tang, S. , Johnson, R. S. P. , Hogg, C. J. , & Belov, K. (2020a). The first Antechinus reference genome provides a resource for investigating the genetic basis of semelparity and age‐related neuropathologies. Gigabyte, 1, 7. 10.46471/gigabyte.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandies, P. A. , Tang, S. , Johnson, R. S. P. , Hogg, C. J. , & Belov, K. (2020b). Supporting data for "The first Antechinus reference genome provides a resource for investigating the genetic basis of semelparity and age‐related neuropathologies". GigaScience Database, 10.5524/100807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujak, A. L. , Crane, J. D. , Lally, J. S. , Ford, R. J. , Kang, S. J. , Rebalka, I. A. , Green, A. E. , Kemp, B. E. , Hawke, T. J. , Schertzer, J. D. , & Steinberg, G. R. (2015). AMPK activation of muscle autophagy prevents fasting‐induced hypoglycemia and myopathy during aging. Cell Metabolism, 21(6), 883–890. 10.1016/j.cmet.2015.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C. , & Karlin, S. (1997). Prediction of complete gene structures in human genomic DNA. Journal of Molecular Biology, 268(1), 78–94. 10.1006/jmbi.1997.0951 [DOI] [PubMed] [Google Scholar]
- Burton, J. N. , Adey, A. , Patwardhan, R. P. , Qiu, R. , Kitzman, J. O. , & Shendure, J. (2013). Chromosome‐scale scaffolding of de novo genome assemblies based on chromatin interactions. Nature Biotechnology, 31(12), 1119–1125. 10.1038/nbt.2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho, C. , Coulouris, G. , Avagyan, V. , Ma, N. , Papadopoulos, J. , Bealer, K. , & Madden, T. L. (2009). BLAST+: Architecture and applications. BMC Bioinformatics, 10, 421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs, B. G. , Gluscevic, M. , Baker, D. J. , Laberge, R. M. , Marquess, D. , Dananberg, J. , & van Deursen, J. M. (2017). Senescent cells: An emerging target for diseases of ageing. Nature Reviews Drug Discovery, 16(10), 718–735. 10.1038/nrd.2017.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson, C. , Jacobs, Z. , Marwick, B. , Fullagar, R. , Wallis, L. , Smith, M. , Roberts, R. G. , Hayes, E. , Lowe, K. , Carah, X. , Florin, S. A. , McNeil, J. , Cox, D. , Arnold, L. J. , Hua, Q. , Huntley, J. , Brand, H. E. A. , Manne, T. , Fairbairn, A. , … Pardoe, C. (2017). Human occupation of northern Australia by 65,000 years ago. Nature, 547(7663), 306–310. 10.1038/nature22968 [DOI] [PubMed] [Google Scholar]
- Collett, R. A. , Baker, A. M. , & Fisher, D. O. (2018). Prey productivity and predictability drive different axes of life‐history variation in carnivorous marsupials. Proceedings of the Royal Society B, 285(1890), 20181291. 10.1098/rspb.2018.1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosentino, S. , & Iwasaki, W. (2019). SonicParanoid: Fast, accurate and easy orthology inference. Bioinformatics, 35(1), 149–151. 10.1093/bioinformatics/bty631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft, D. B. (2003). Behaviour of carnivorous marsupials. In Jones M., Dickman C., & Archer M. (Eds.), Predators with pouches: The biology of carnivorous marsupials (pp. 332–346). CSIRO Publishing. [Google Scholar]
- Deakin, J. E. , & O'Neill, R. J. (2020). Evolution of marsupial genomes. Annual Review of Animal Biosciences, 8, 25–45. 10.1146/annurev-animal-021419-083555 [DOI] [PubMed] [Google Scholar]
- Diamond, J. M. (1982). Big‐bang reproduction and ageing in male marsupial mice. Nature, 298(5870), 115–116. [DOI] [PubMed] [Google Scholar]
- Diez‐Del‐Molino, D. , Sanchez‐Barreiro, F. , Barnes, I. , Gilbert, M. T. P. , & Dalen, L. (2018). Quantifying temporal genomic erosion in endangered species. Trends in Ecology & Evolution, 33(3), 176–185. 10.1016/j.tree.2017.12.002 [DOI] [PubMed] [Google Scholar]
- Dodt, M. , Roehr, J. T. , Ahmed, R. , & Dieterich, C. (2012). FLEXBAR‐flexible barcode and adapter processing for next‐generation sequencing platforms. Biology (Basel), 1(3), 895–905. 10.3390/biology1030895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko, O. , Batra, S. S. , Omer, A. D. , Nyquist, S. K. , Hoeger, M. , Durand, N. C. , Shamim, M. S. , Machol, I. , Lander, E. S. , Aiden, A. P. , & Aiden, E. L. (2017). De novo assembly of the Aedes aegypti genome using Hi‐C yields chromosome‐length scaffolds. Science, 356(6333), 92–95. 10.1126/science.aal3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko, O. , Shamim, M. S. , Batra, S. S. , Durand, N. C. , Musial, N. T. , Mostofa, R. , Pham, M. , St Hilaire, B. G. , Yao, W. , Stamenova, E. , Hoeger, M. , Nyquist, S. K. , Korchina, V. , Pletch, K. , Flanagan, J. P. , Tomaszewicz, A. , McAloose, D. , Estrada, C. P. , Novak, B. J. , … Aiden, E. L. (2018). The Juicebox Assembly Tools module facilitates de novo assembly of mammalian genomes with chromosome‐length scaffolds for under $1000. bioRxiv, Preprint at http://biorxiv.org/content/ 10.1101/254797v1. [DOI] [Google Scholar]
- Durand, N. C. , Robinson, J. T. , Shamim, M. S. , Machol, I. , Mesirov, J. P. , Lander, E. S. , & Aiden, E. L. (2016). Juicebox provides a visualization system for Hi‐C contact maps with unlimited zoom. Cell Systems, 3(1), 99–101. 10.1016/j.cels.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elderfield, H. , Ferretti, P. , Greaves, M. , Crowhurst, S. , McCave, I. N. , Hodell, D. , & Piotrowski, A. M. (2012). Evolution of ocean temperature and ice volume through the mid‐Pleistocene climate transition. Science, 337(6095), 704–709. 10.1126/science.1221294 [DOI] [PubMed] [Google Scholar]
- Ellinghaus, D. , Kurtz, S. , & Willhoeft, U. (2008). LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics, 9, 18. 10.1186/1471-2105-9-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faurby, S. , Davis, M. , Pedersen, R. O. , Schowanek, S. D. , Antonelli, A. , & Svenning, J. C. (2018). PHYLACINE 1.2: The phylogenetic atlas of mammal macroecology. Ecology, 99(11), 2626. 10.1002/ecy.2443 [DOI] [PubMed] [Google Scholar]
- Feigin, C. Y. , Newton, A. H. , Doronina, L. , Schmitz, J. , Hipsley, C. A. , Mitchell, K. J. , Gower, G. , Llamas, B. , Soubrier, J. , Heider, T. N. , Menzies, B. R. , Cooper, A. , O’Neill, R. J. , & Pask, A. J. (2018). Genome of the Tasmanian tiger provides insights into the evolution and demography of an extinct marsupial carnivore. Nature Ecology & Evolution, 2(1), 182–192. 10.1038/s41559-017-0417-y [DOI] [PubMed] [Google Scholar]
- Fisher, D. O. , Dickman, C. R. , Jones, M. E. , & Blomberg, S. P. (2013). Sperm competition drives the evolution of suicidal reproduction in mammals. Proceedings of the National Academy of Sciences of the United States of America, 110(44), 17910–17914. 10.1073/pnas.1310691110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froy, H. , Sparks, A. M. , Watt, K. , Sinclair, R. , Bach, F. , Pilkington, J. G. , Pemberton, J. M. , McNeilly, T. N. , & Nussey, D. H. (2019). Senescence in immunity against helminth parasites predicts adult mortality in a wild mammal. Science, 365(6459), 1296–1298. 10.1126/science.aaw5822 [DOI] [PubMed] [Google Scholar]
- García‐Navas, V. , Kear, B. P. , & Westerman, M. (2020). The geography of speciation in dasyurid marsupials. Journal of Biogeography, 47, 2042–2053. 10.1111/jbi.13852 [DOI] [Google Scholar]
- Gems, D. (2014). Evolution of sexually dimorphic longevity in humans. Aging (Albany NY), 6(2), 84–91. 10.18632/aging.100640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyle, H. M. , Woinarski, J. C. Z. , Baker, G. B. , Dickman, C. R. , Dutson, G. , Fisher, D. O. , Ford, H. , Holdsworth, M. , Jones, M. E. , Kutt, A. , Legge, S. , Leiper, I. , Loyn, R. , Murphy, B. P. , Menkhorst, P. , Reside, A. E. , Ritchie, E. G. , Roberts, F. E. , Tingley, R. , & Garnett, S. T. (2018). Quantifying extinction risk and forecasting the number of impending Australian bird and mammal extinctions. Pacific Conservation Biology, 24(2), 157–167. 10.1071/PC18006 [DOI] [Google Scholar]
- Grabherr, M. G. , Haas, B. J. , Yassour, M. , Levin, J. Z. , Thompson, D. A. , Amit, I. , Adiconis, X. , Fan, L. , Raychowdhury, R. , Zeng, Q. , Chen, Z. , Mauceli, E. , Hacohen, N. , Gnirke, A. , Rhind, N. , di Palma, F. , Birren, B. W. , Nusbaum, C. , Lindblad‐Toh, K. , … Regev, A. (2011). Full‐length transcriptome assembly from RNA‐Seq data without a reference genome. Nature Biotechnology, 29(7), 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, B. J. , Papanicolaou, A. , Yassour, M. , Grabherr, M. , Blood, P. D. , Bowden, J. , Couger, M. B. , Eccles, D. , Li, B. O. , Lieber, M. , MacManes, M. D. , Ott, M. , Orvis, J. , Pochet, N. , Strozzi, F. , Weeks, N. , Westerman, R. , William, T. , Dewey, C. N. , … Regev, A. (2013). De novo transcript sequence reconstruction from RNA‐seq using the Trinity platform for reference generation and analysis. Nature Protocols, 8(8), 1494–1512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, B. J. , Salzberg, S. L. , Zhu, W. , Pertea, M. , Allen, J. E. , Orvis, J. , White, O. , Buell, C. R. , & Wortman, J. R. (2008). Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biology, 9(1), R7. 10.1186/gb-2008-9-1-r7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, G. , Simmons, L. , Dugand, R. , Mills, H. R. , Roberts, J. , Tomkins, J. , & Fisher, D. (2019). Male semelparity and multiple paternity confirmed in an arid‐zone dasyurid. Journal of Zoology, 308(4), 266–273. 10.1111/jzo.12672 [DOI] [Google Scholar]
- Henschel, R. , Lieber, M. , Wu, L.‐S. , Nista, P. M. , Haas, B. J. , & LeDuc, R. D. (2012). Trinity RNA‐Seq assembler performance optimization. Paper presented at the Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the eXtreme to the campus and beyond, Chicag, IL, USA. [Google Scholar]
- Hughes, P. W. (2017). Between semelparity and iteroparity: Empirical evidence for a continuum of modes of parity. Ecology and Evolution, 7(20), 8232–8261. 10.1002/ece3.3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen, T. , & Lamark, T. (2020). Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. Journal of Molecular Biology, 432(1), 80–103. 10.1016/j.jmb.2019.07.016 [DOI] [PubMed] [Google Scholar]
- Johnson, R. N. , O’Meally, D. , Chen, Z. , Etherington, G. J. , Ho, S. Y. W. , Nash, W. J. , Grueber, C. E. , Cheng, Y. , Whittington, C. M. , Dennison, S. , Peel, E. , Haerty, W. , O’Neill, R. J. , Colgan, D. , Russell, T. L. , Alquezar‐Planas, D. E. , Attenbrow, V. , Bragg, J. G. , Brandies, P. A. , … Belov, K. (2018). Adaptation and conservation insights from the koala genome. Nature Genetics, 50(8), 1102–1111. 10.1038/s41588-018-0153-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jucker, M. (2010). The benefits and limitations of animal models for translational research in neurodegenerative diseases. Nature Medicine, 16(11), 1210–1214. 10.1038/nm.2224 [DOI] [PubMed] [Google Scholar]
- Kahle, D. , & Wickham, H. (2013). ggmap: Spatial visualization with ggplot2. The R Journal, 5(1), 144–161. 10.32614/RJ-2013-014 [DOI] [Google Scholar]
- Kanehisa, M. , & Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research, 28(1), 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W. J. (2002). BLAT–the BLAST‐like alignment tool. Genome Research, 12(4), 656–664. 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Paggi, J. M. , Park, C. , Bennett, C. , & Salzberg, S. L. (2019). Graph‐based genome alignment and genotyping with HISAT2 and HISAT‐genotype. Nature Biotechnology, 37(8), 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, H.‐J. , Arnolds, D. E. , Fujii, N. , Tran, T. T. , Rogers, M. J. , Jessen, N. , Li, Y. , Liew, C. W. , Ho, R. C. , Hirshman, M. F. , Kulkarni, R. N. , Kahn, C. R. , & Goodyear, L. J. (2006). Skeletal muscle‐selective knockout of LKB1 increases insulin sensitivity, improves glucose homeostasis, and decreases TRB3. Molecular and Cellular Biology, 26(22), 8217–8227. 10.1128/MCB.00979-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopylova, E. , Noe, L. , & Touzet, H. (2012). SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics, 28(24), 3211–3217. 10.1093/bioinformatics/bts611 [DOI] [PubMed] [Google Scholar]
- Koren, S. , Walenz, B. P. , Berlin, K. , Miller, J. R. , Bergman, N. H. , & Phillippy, A. M. (2017). Canu: Scalable and accurate long‐read assembly via adaptive k‐mer weighting and repeat separation. Genome Research, 27(5), 722–736. 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewski, C. , Woolley, P. A. , & Westerman, M. (2000). The evolution of reproductive strategies in dasyurid marsupials: Implications of molecular phylogeny. Biological Journal of the Linnean Society, 71(3), 417–435. 10.1111/j.1095-8312.2000.tb01267.x [DOI] [Google Scholar]
- Kumar, S. , Stecher, G. , Suleski, M. , & Hedges, S. B. (2017). TimeTree: A resource for timelines, timetrees, and divergence times. Molecular Biology and Evolution, 34(7), 1812–1819. 10.1093/molbev/msx116 [DOI] [PubMed] [Google Scholar]
- Lada, H. , Thomson, J. R. , Mac Nally, R. , & Taylor, A. C. (2008). Impacts of massive landscape change on a carnivorous marsupial in south‐eastern Australia: Inferences from landscape genetics analysis. Journal of Applied Ecology, 45(6), 1732–1741. 10.1111/j.1365-2664.2008.01563.x [DOI] [Google Scholar]
- Le Phuc, P. , Friedman, J. R. , Schug, J. , Brestelli, J. E. , Parker, J. B. , Bochkis, I. M. , & Kaestner, K. H. (2005). Glucocorticoid receptor‐dependent gene regulatory networks. PLoS Genetics, 1(2), e16. 10.1371/journal.pgen.0010016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiner, N. O. , Setz, E. Z. , & Silva, W. R. (2008). Semelparity and factors affecting the reproductive activity of the Brazilian slender opossum (Marmosops paulensis) in southeastern Brazil. Journal of Mammalogy, 89(1), 153–158. [Google Scholar]
- Lemaître, J.‐F. , Ronget, V. , Tidière, M. , Allainé, D. , Berger, V. , Cohas, A. , Colchero, F. , Conde, D. A. , Garratt, M. , Liker, A. , Marais, G. A. B. , Scheuerlein, A. , Székely, T. , & Gaillard, J.‐M. (2020). Sex differences in adult lifespan and aging rates of mortality across wild mammals. Proceedings of the National Academy of Sciences of the United States of America, 117(15), 8546–8553. 10.1073/pnas.1911999117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesuis, S. L. , Weggen, S. , Baches, S. , Lucassen, P. J. , & Krugers, H. J. (2018). Targeting glucocorticoid receptors prevents the effects of early life stress on amyloid pathology and cognitive performance in APP/PS1 mice. Translational Psychiatry, 8(1), 53. 10.1038/s41398-018-0101-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. (2018). Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics, 34(18), 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , & Durbin, R. (2011). Inference of human population history from individual whole‐genome sequences. Nature, 475(7357), 493–496. 10.1038/nature10231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Handsaker, B. , Wysoker, A. , Fennell, T. , Ruan, J. , Homer, N. , Marth, G. , Abecasis, G. , Durbin, R. , & 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25(16), 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman‐Aiden, E. , van Berkum, N. L. , Williams, L. , Imakaev, M. , Ragoczy, T. , Telling, A. , Amit, I. , Lajoie, B. R. , Sabo, P. J. , Dorschner, M. O. , Sandstrom, R. , Bernstein, B. , Bender, M. A. , Groudine, M. , Gnirke, A. , Stamatoyannopoulos, J. , Mirny, L. A. , Lander, E. S. , & Dekker, J. (2009). Comprehensive mapping of long‐range interactions reveals folding principles of the human genome. Science, 326(5950), 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisiecki, L. E. , & Raymo, M. E. (2005). A Pliocene‐Pleistocene stack of 57 globally distributed benthic δ18O records. Paleoceanography, 20(1). 10.1029/2004PA001071 [DOI] [Google Scholar]
- Liu, B. , Shi, Y. , Yuan, J. , Hu, X. , Zhang, H. , Li, N. , Li, Z. , Chen, Y. , Mu, D. , & Fan, W. (2013). Estimation of genomic characteristics by analyzing k‐mer frequency in de novo genome projects. arXiv preprint arXiv:1308.2012. [Google Scholar]
- Loytynoja, A. , & Goldman, N. (2005). An algorithm for progressive multiple alignment of sequences with insertions. Proceedings of the National Academy of Sciences of the United States of America, 102(30), 10557–10562. 10.1073/pnas.0409137102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Z. X. , Yuan, C. X. , Meng, Q. J. , & Ji, Q. (2011). A Jurassic eutherian mammal and divergence of marsupials and placentals. Nature, 476(7361), 442–445. 10.1038/nature10291 [DOI] [PubMed] [Google Scholar]
- Majoros, W. H. , Pertea, M. , & Salzberg, S. L. (2004). TigrScan and GlimmerHMM: Two open source ab initio eukaryotic gene‐finders. Bioinformatics, 20(16), 2878–2879. 10.1093/bioinformatics/bth315 [DOI] [PubMed] [Google Scholar]
- Mak, S. S. T. , Gopalakrishnan, S. , Carøe, C. , Geng, C. , Liu, S. , Sinding, M.‐H. , Kuderna, L. F. K. , Zhang, W. , Fu, S. , Vieira, F. G. , Germonpré, M. , Bocherens, H. , Fedorov, S. , Petersen, B. , Sicheritz‐Pontén, T. , Marques‐Bonet, T. , Zhang, G. , Jiang, H. , & Gilbert, M. T. P. (2017). Comparative performance of the BGISEQ‐500 vs Illumina HiSeq2500 sequencing platforms for palaeogenomic sequencing. Gigascience, 6(8), 1–13. 10.1093/gigascience/gix049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoli, I. , Alesci, S. , Blackman, M. R. , Su, Y. A. , Rennert, O. M. , & Chrousos, G. P. (2007). Mitochondria as key components of the stress response. Trends in Endocrinology & Metabolism, 18(5), 190–198. 10.1016/j.tem.2007.04.004 [DOI] [PubMed] [Google Scholar]
- Marcais, G. , & Kingsford, C. (2011). A fast, lock‐free approach for efficient parallel counting of occurrences of k‐mers. Bioinformatics, 27(6), 764–770. 10.1093/bioinformatics/btr011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martocchia, A. , Stefanelli, M. , Falaschi, G. M. , Toussan, L. , Ferri, C. , & Falaschi, P. (2016). Recent advances in the role of cortisol and metabolic syndrome in age‐related degenerative diseases. Aging Clinical and Experimental Research, 28(1), 17–23. 10.1007/s40520-015-0353-0 [DOI] [PubMed] [Google Scholar]
- McAllan, B. (2006). Dasyurid marsupials as models for the physiology of ageing in humans. Australian Journal of Zoology, 54(3), 159–172. 10.1071/ZO05073 [DOI] [Google Scholar]
- McDowell, I. C. , Barrera, A. , D'Ippolito, A. M. , Vockley, C. M. , Hong, L. K. , Leichter, S. M. , Bartelt, L. C. , Majoros, W. H. , Song, L. , Safi, A. , Koçak, D. D. , Gersbach, C. A. , Hartemink, A. J. , Crawford, G. E. , Engelhardt, B. E. , & Reddy, T. E. (2018). Glucocorticoid receptor recruits to enhancers and drives activation by motif‐directed binding. Genome Research, 28(9), 1272–1284. 10.1101/gr.233346.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, W. , Hayes, V. M. , Ratan, A. , Petersen, D. C. , Wittekindt, N. E. , Miller, J. , Walenz, B. , Knight, J. , Qi, J. , Zhao, F. , Wang, Q. , Bedoya‐Reina, O. C. , Katiyar, N. , Tomsho, L. P. , Kasson, L. M. , Hardie, R.‐A. , Woodbridge, P. , Tindall, E. A. , Bertelsen, M. F. , … Schuster, S. C. (2011). Genetic diversity and population structure of the endangered marsupial Sarcophilus harrisii (Tasmanian devil). Proceedings of the National Academy of Sciences of the United States of America, 108(30), 12348–12353. 10.1073/pnas.1102838108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh, B. Q. , Schmidt, H. A. , Chernomor, O. , Schrempf, D. , Woodhams, M. D. , von Haeseler, A. , & Lanfear, R. (2020). IQ‐TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution, 37(5), 1530–1534. 10.1093/molbev/msaa015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, A. L. , Attwood, T. K. , Babbitt, P. C. , Blum, M. , Bork, P. , Bridge, A. , Brown, S. D. , Chang, H.‐Y. , El‐Gebali, S. , Fraser, M. I. , Gough, J. , Haft, D. R. , Huang, H. , Letunic, I. , Lopez, R. , Luciani, A. , Madeira, F. , Marchler‐Bauer, A. , Mi, H. , … Finn, R. D. (2019). InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Research, 47(D1), D351–D360. 10.1093/nar/gky1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, K. J. , Pratt, R. C. , Watson, L. N. , Gibb, G. C. , Llamas, B. , Kasper, M. , Edson, J. , Hopwood, B. , Male, D. , Armstrong, K. N. , Meyer, M. , Hofreiter, M. , Austin, J. , Donnellan, S. C. , Lee, M. S. Y. , Phillips, M. J. , & Cooper, A. (2014). Molecular phylogeny, biogeography, and habitat preference evolution of marsupials. Molecular Biology and Evolution, 31(9), 2322–2330. 10.1093/molbev/msu176 [DOI] [PubMed] [Google Scholar]
- Mutton, T. Y. , Phillips, M. J. , Fuller, S. J. , Bryant, L. M. , & Baker, A. M. (2019). Systematics, biogeography and ancestral state of the Australian marsupial genus Antechinus (Dasyuromorphia: Dasyuridae). Zoological Journal of the Linnean Society, 186(2), 553–568. 10.1093/zoolinnean/zly062 [DOI] [Google Scholar]
- Naylor, R. , Richardson, S. J. , & McAllan, B. M. (2008). Boom and bust: A review of the physiology of the marsupial genus Antechinus . Journal of Comparative Physiology B, 178(5), 545–562. 10.1007/s00360-007-0250-8 [DOI] [PubMed] [Google Scholar]
- Nilsson, M. A. , Churakov, G. , Sommer, M. , Tran, N. V. , Zemann, A. , Brosius, J. , & Schmitz, J. (2010). Tracking marsupial evolution using archaic genomic retroposon insertions. PLoS Biology, 8(7), e1000436. 10.1371/journal.pbio.1000436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NSW Department of Environment and Conservation (2004). Systematic survey of vertebrate fauna in Lane Cove National Park. https://www.environment.nsw.gov.au/resources/nature/VertebrateFaunaLaneCoveNP.pdf [Google Scholar]
- O'Donovan, C. , Martin, M. J. , Gattiker, A. , Gasteiger, E. , Bairoch, A. , & Apweiler, R. (2002). High‐quality protein knowledge resource: SWISS‐PROT and TrEMBL. Briefings in Bioinformatics, 3(3), 275–284. 10.1093/bib/3.3.275 [DOI] [PubMed] [Google Scholar]
- O'Leary, N. A. , Wright, M. W. , Brister, J. R. , Ciufo, S. , Haddad, D. , McVeigh, R. , Rajput, B. , Robbertse, B. , Smith‐White, B. , Ako‐Adjei, D. , Astashyn, A. , Badretdin, A. , Bao, Y. , Blinkova, O. , Brover, V. , Chetvernin, V. , Choi, J. , Cox, E. , Ermolaeva, O. , … Pruitt, K. D. (2016). Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Research, 44(D1), D733–745. 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perogamvros, I. , Ray, D. W. , & Trainer, P. J. (2012). Regulation of cortisol bioavailability–effects on hormone measurement and action. Nature Reviews Endocrinology, 8(12), 717–727. 10.1038/nrendo.2012.134 [DOI] [PubMed] [Google Scholar]
- Phillips, M. J. , Bennett, T. H. , & Lee, M. S. (2009). Molecules, morphology, and ecology indicate a recent, amphibious ancestry for echidnas. Proceedings of the National Academy of Sciences of the United States of America, 106(40), 17089–17094. 10.1073/pnas.0904649106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poskitt, D. , Barnett, J. , Duffey, K. , Lee, A. , Kimpton, W. , & Muller, H. (1984). Stress related involution of lymphoid tissues in Australian marsupial mice. Immunobiology, 166(3), 286–295. 10.1016/S0171-2985(84)80046-7 [DOI] [PubMed] [Google Scholar]
- Pruitt, K. D. , Tatusova, T. , & Maglott, D. R. (2007). NCBI reference sequences (RefSeq): A curated non‐redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Research, 35(Database issue), D61–65. 10.1093/nar/gkl842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast, C. , Pruesse, E. , Yilmaz, P. , Gerken, J. , Schweer, T. , Yarza, P. , Peplies, J. , & Glöckner, F. O. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web‐based tools. Nucleic Acids Research, 41(Database issue), D590–596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan, C. E. , Pearce, C. , McDonald, B. J. F. , Gynther, I. , & Baker, A. M. (2020). Vegetation structure and ground cover attributes describe the occurrence of a newly discovered carnivorous marsupial on the Tweed Shield Volcano caldera, the endangered black‐tailed dusky antechinus (Antechinus arktos). Ecology and Evolution, 10(4), 2104–2121. 10.1002/ece3.6045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehr, J. T. , Dieterich, C. , & Reinert, K. (2017). Flexbar 3.0 ‐ SIMD and multicore parallelization. Bioinformatics, 33(18), 2941–2942. 10.1093/bioinformatics/btx330 [DOI] [PubMed] [Google Scholar]
- Romero, L. M. (2004). Physiological stress in ecology: Lessons from biomedical research. Trends in Ecology & Evolution, 19(5), 249–255. 10.1016/j.tree.2004.03.008 [DOI] [PubMed] [Google Scholar]
- Ruan, J. , & Li, H. (2020). Fast and accurate long‐read assembly with wtdbg2. Nature Methods, 17(2), 155–158. 10.1038/s41592-019-0669-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, K. , McCarthy, A. , Smith, D. , Green, K. A. , Grahame Hardie, D. , Ashworth, A. , & Alessi, D. R. (2005). Deficiency of LKB1 in skeletal muscle prevents AMPK activation and glucose uptake during contraction. EMBO Journal, 24(10), 1810–1820. 10.1038/sj.emboj.7600667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltré, F. , Rodríguez‐Rey, M. , Brook, B. W. , Johnson, C. N. , Turney, C. S. M. , Alroy, J. , Cooper, A. , Beeton, N. , Bird, M. I. , Fordham, D. A. , Gillespie, R. , Herrando‐Pérez, S. , Jacobs, Z. , Miller, G. H. , Nogués‐Bravo, D. , Prideaux, G. J. , Roberts, R. G. , & Bradshaw, C. J. A. (2016). Climate change not to blame for late Quaternary megafauna extinctions in Australia. Nature Communications, 7, 10511. 10.1038/ncomms10511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloissnig, S. , Kawaguchi, A. , Nowoshilow, S. , Falcon, F. , Otsuki, L. , Tardivo, P. , Timoshevskaya, N. , Keinath, M. C. , Smith, J. J. , Voss, S. R. , & Tanaka, E. M. (2021). The giant axolotl genome uncovers the evolution, scaling, and transcriptional control of complex gene loci. Proceedings of the National Academy of Sciences of the United States of America, 118(15), e2017176118. 10.1073/pnas.2017176118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seim, I. (2020a). de novo genome assemblies of black‐tailed dusky antechinus (Antechinus arktos), silver‐headed antechinus (Antechinus argentus), and black‐tailed dasyure (Murexia melanurus). 10.5281/zenodo.3735672 [DOI] [Google Scholar]
- Seim, I. (2020b). Gapfilled nuclear genome assemblies of black‐tailed dusky antechinus (Antechinus arktos), silver‐headed antechinus (Antechinus argentus), and black‐tailed dasyure (Murexia melanurus). 10.5281/zenodo.3735686 [DOI] [Google Scholar]
- Seim, I. (2020c). ROPUS ‐ Reference‐guided ortholog pipeline for unannotated species. 10.5281/zenodo.3722900 [DOI] [Google Scholar]
- Seim, I. (2021). Gene annotations and associated FASTA file of Antechinus flavipes (yellow‐footed antechinus). 10.5281/zenodo.5205475 [DOI] [Google Scholar]
- Selwood, L. (1980). A timetable of embryonic development of the dasyurid marsupial Antechinus stuartii (Macleay). Australian Journal of Zoology, 28(4), 645–669. 10.1071/ZO9800649 [DOI] [Google Scholar]
- Seppey, M. , Manni, M. , & Zdobnov, E. M. (2019). BUSCO: Assessing genome assembly and annotation completeness. Methods in Molecular Biology, 1962, 227–245. 10.1007/978-1-4939-9173-0_14 [DOI] [PubMed] [Google Scholar]
- Servant, N. , Varoquaux, N. , Lajoie, B. R. , Viara, E. , Chen, C.‐J. , Vert, J.‐P. , Heard, E. , Dekker, J. , & Barillot, E. (2015). HiC‐Pro: An optimized and flexible pipeline for Hi‐C data processing. Genome Biology, 16, 259. 10.1186/s13059-015-0831-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A. F. , & Hubley, R. (2010). RepeatModeler Open‐1.0. 2008‐2015. http://www.repeatmasker.org [Google Scholar]
- Stamatakis, A. (2006). RAxML‐VI‐HPC: Maximum likelihood‐based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22(21), 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Stanke, M. , Keller, O. , Gunduz, I. , Hayes, A. , Waack, S. , & Morgenstern, B. (2006). AUGUSTUS: Ab initio prediction of alternative transcripts. Nucleic Acids Research, 34(Web Server), W435–W439. 10.1093/nar/gkl200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke, M. , & Waack, S. (2003). Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics, 19(Suppl 2), ii215–ii225. 10.1093/bioinformatics/btg1080 [DOI] [PubMed] [Google Scholar]
- Talavera, G. , & Castresana, J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology, 56(4), 564–577. 10.1080/10635150701472164 [DOI] [PubMed] [Google Scholar]
- Tarailo‐Graovac, M. , & Chen, N. (2009). Using RepeatMasker to identify repetitive elements in genomic sequences. Current Protocols in Bioinformatics, 25(1), 4–10. 10.1002/0471250953.bi0410s25 [DOI] [PubMed] [Google Scholar]
- Tatusov, R. L. , Galperin, M. Y. , Natale, D. A. , & Koonin, E. V. (2000). The COG database: A tool for genome‐scale analysis of protein functions and evolution. Nucleic Acids Research, 28(1), 33–36. 10.1093/nar/28.1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, D. M. , Porter, B. B. , Tall, J. H. , Kim, H. J. , Barrow, J. R. , & Winder, W. W. (2007). Skeletal muscle and heart LKB1 deficiency causes decreased voluntary running and reduced muscle mitochondrial marker enzyme expression in mice. American Journal of Physiology. Endocrinology and Metabolism, 292(1), E196–202. 10.1152/ajpendo.00366.2006 [DOI] [PubMed] [Google Scholar]
- UniProt Consortium (2012). Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Research, 40(Database issue), D71–75. 10.1093/nar/gkr981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viacava, P. , Baker, A. M. , Blomberg, S. P. , Phillips, M. J. , & Weisbecker, V. (2021). Using 3D geometric morphometrics to aid taxonomic and ecological understanding of a recent speciation event within a small Australian marsupial (genus Antechinus). bioRxiv. [Google Scholar]
- Walker, B. J. , Abeel, T. , Shea, T. , Priest, M. , Abouelliel, A. , Sakthikumar, S. , Cuomo, C. A. , Zeng, Q. , Wortman, J. , Young, S. K. , & Earl, A. M. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One, 9(11), e112963. 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woollard, P. (1971). Differential mortality of Antechinus stuartii (Macleay): Nitrogen balance and somatic changes. Australian Journal of Zoology, 19(4), 347–353. [Google Scholar]
- Yang, Z. (2007). PAML 4: Phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution, 24(8), 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Movie S1
Appendix S1
Appendix S2
Data Availability Statement
Antechinus flavipes PacBio reads are available at China National GenBank (CNGB) Project ID CNP0001147. Short‐read whole‐genome sequencing and RNA sequencing data (BGISEQ‐500) generated in this project are available at NCBI BioProject PRJNA565840. The A. flavipes genome assembly is available at NCBI Genomes (JADWMD000000000). Pseudo (reference‐based) and de novo genome assemblies of Antechinus arktos, Antechinus argentus and Murexia melanurus (Seim, 2020a, 2020b), a ROPUS Singularity image (Seim, 2020c), and gene annotation files and associated FASTA files for A. flavipes are available at Zenodo (Seim, 2021). Mitochondrial genome assemblies of A. flavipes (MN447797), A. arktos (MK977601) and M. melanurus (MK977600) are available at GenBank. Various scripts used for data processing and analyses are available on GitHub at https://github.com/sciseim/semelparity‐genomics.


