Abstract
A platform to accelerate optimization of proteolysis targeting chimeras (PROTACs) has been developed using a direct-to-biology (D2B) approach with a focus on linker effects. A large number of linker analogs—with varying length, polarity, and rigidity—were rapidly prepared and characterized in four cell-based assays by streamlining time-consuming steps in synthesis and purification. The expansive dataset informs on linker structure–activity relationships (SAR) for in-cell E3 ligase target engagement, degradation, permeability, and cell toxicity. Unexpected aspects of linker SAR was discovered, consistent with literature reports on “linkerology”, and the method dramatically speeds up empirical optimization. Physicochemical property trends emerged, and the platform has the potential to rapidly expand training sets for more complex prediction models. In-depth validation studies were carried out and confirm the D2B platform is a valuable tool to accelerate PROTAC design–make–test cycles.
Keywords: Heterobifunctional degrader, PROTAC, Direct-to-biology, Direct-to-assay, Parallel medicinal chemistry, High-throughput experimentation, Library chemistry
Proteolysis targeting chimeras (PROTACs) are an emerging modality with the potential to modulate protein targets which are challenging to drug with traditional small molecules. By inducing proximity between an intracellular target protein and an E3 ligase complex, these heterobifunctional molecules catalyze formation of a ternary complex, resulting in ubiquitination of the target protein and its proteosome-mediated degradation.1−8 PROTACs consist of a ligand for an E3 ligase, such as cereblon (CRBN) and von Hippel Lindau, and a protein of interest (POI) ligand connected by a covalent linker. Pioneering efforts have resulted in advancement of multiple PROTACs to clinical stage development and sparked high interest in academic and industrial laboratories.9−12
Optimization of PROTACs presents a number of challenges. PROTACs tend to occupy the beyond-rule-of-5 property space associated with low passive permeability, negatively impacting cell penetration, oral bioavailability, and central nervous system exposure.13−16 Structure–activity relationships (SAR) generated in binary complex binding assays—that is, measuring ligand binding individually to either target protein or E3 ligase—are valuable but omit cooperativity effects in the ternary complex. Assessment of functional ternary complex formation is often performed in cell-based degradation assays, where SAR can be confounded by permeability effects. While landmark structural studies have characterized degrader ternary complexes,17,18 multiple potential complexes may exist in solution, and structure-guided optimization may not be possible in every case. Degrader optimization continues to be primarily an empirical process driven by many cycles of assay data collected from synthesized compounds.
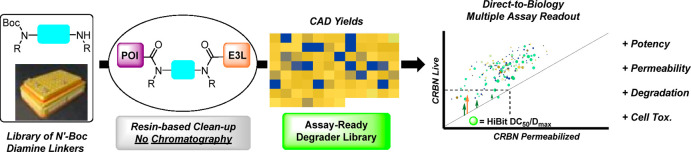
To accelerate PROTAC optimization, we developed a novel platform combining high-throughput chemistry with high-throughput cell-based assays. This platform substantially increases the datasets in each cycle of PROTAC optimization, speeding up empirical optimization and expanding training datasets for predictive modeling. We applied a “direct-to-biology” (D2B) approach.19−22 D2B involves synthesizing large libraries on a very small scale (<10 μmol reactions in plate-based formats) and assaying them as unchromatographed mixtures. Extensive controls and validation enable generation of quantitative SAR across 102–103 analogs, despite the presence of impurities.
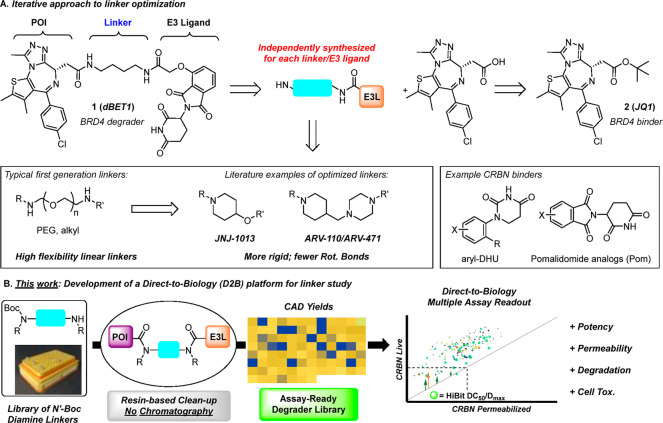
PROTAC linkers are hypothesized to play an important role in degradation,23,24 ternary complex formation,25 and ADME properties;26−28 therefore, we designed D2B PROTAC linker libraries. First-generation PROTACs, such as dBET1 (1), contain flexible linkers, and more recent PROTACs, such as ARV-110 and JNJ-1013, introduce rigid linkers (Scheme 1A). In our design, a highly diverse set of linkers was explored. Test compounds were evaluated in four cell-based assays,29,30 enabling assessment of in-cell E3 ligase target engagement, degradation, permeability, and cell toxicity. Physicochemical property trends emerged from this dataset, and unexpected aspects of linker SAR were discovered as well. These data, along with in-depth validation studies, confirm the D2B platform is a valuable tool to accelerate PROTAC design–make–test cycles.
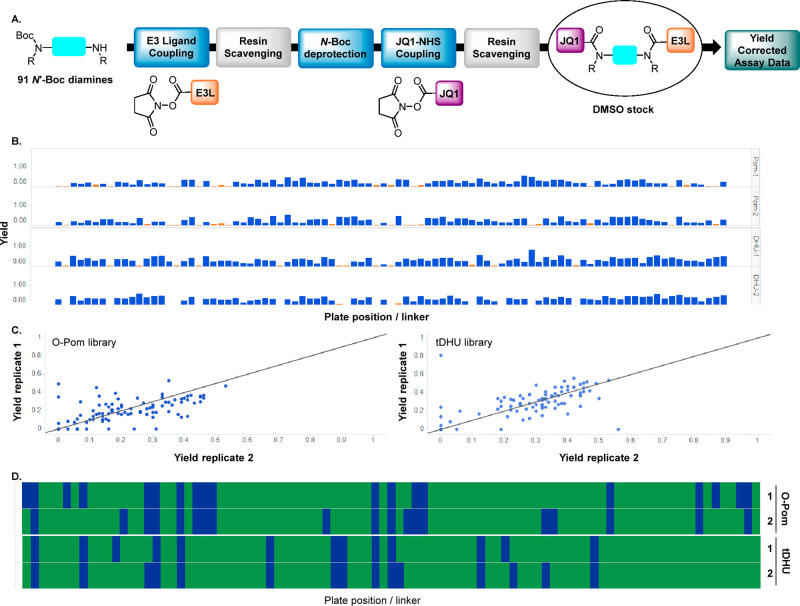
Scheme 1. (A) PROTAC Structure and Typical Synthesis, Including Time-Intensive Chromatographic Purification, and (B) Overview of the Direct-to-Biology (D2B) Strategy, Which Streamlines Plate-Based Synthesis and Evaluation of Unchromatographed PROTACs.
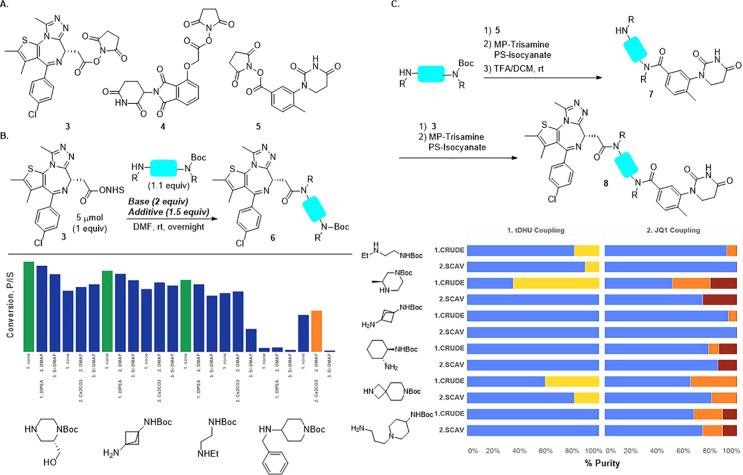
The D2B synthesis library was designed using dBET1 (1, Scheme 1) as a model system to investigate linker SAR for degraders. Using JQ1 (2) as the POI ligand linked to CRBN E3 ligase ligands, such as O-linked pomalidomide (O-Pom) or tolyl-dihydrouracil (tDHU),31 linker diversity was incorporated using mono-N-Boc diamines as the key building blocks, and the synthesis consisted of amide bond formation, TFA-mediated Boc deprotection, and a second amide bond formation. Pilot studies using a set of representative mono-N-Boc diamines demonstrated robust scope for amide formation under a single set of conditions using N-hydroxysuccinimide (NHS) esters (Scheme 2B). JQ1 (2), activated as its NHS ester (3), was treated with a panel of mono-N-Boc diamines representing varying steric and electronic contexts. Primary, secondary, and cyclic amines gave high conversion to amide using DIPEA as base (see the Supporting Information (SI), section III, for additional details). Highly hindered acyclic secondary amines were nearly unreactive, and this relatively small class was excluded from the D2B library.
Scheme 2. (A) NHS Esters Used in D2B Synthesis, (B) Optimization of JQ1 Amide Formation via HTE Base and Additive Screening Using Four Representative Mono-N-Boc Diamines, and (C) Three-Step D2B PROTAC Synthesis.
Final conditions shown as green bars.
Bar graphs represent purity of unchromatographed PROTACs and demonstrate purity enrichment though scavenging resin treatment. Blue: desired PROTAC; yellow, orange, red: reaction byproducts.
Development of the telescoped three-step sequence was accomplished using the cereblon E3 ligase ligand based on tDHU and six model mono-N-Boc-diamines (Scheme 2C).32,33 Following initial amide formation, removal of unreacted starting materials was accomplished using resin-bound scavengers,34 which maximized conversion and purity. The resulting solutions, after concentration, were subjected to TFA-mediated N-Boc deprotection, concentrated, and carried into a second amide formation using JQ1 NHS ester. A second treatment with resin-bound scavengers provided PROTAC product solutions which, after concentration, were dissolved in DMSO and carried directly into assays.
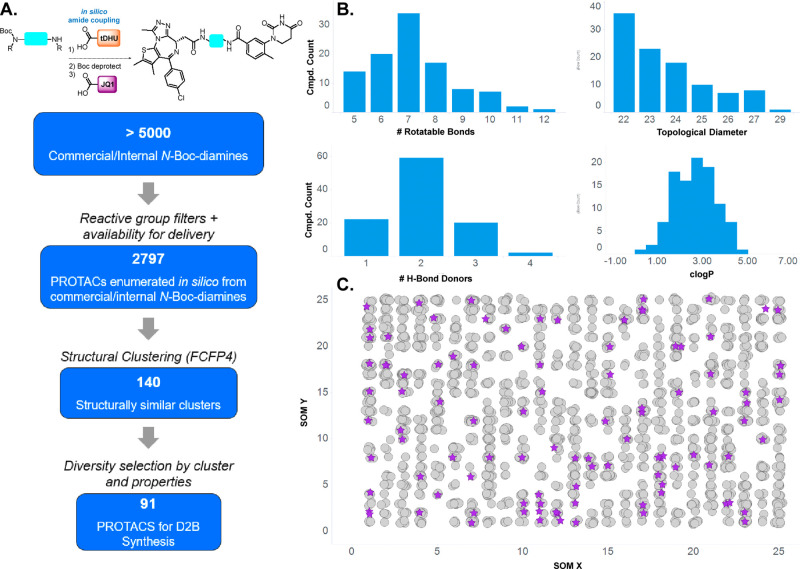
Linker selection (Figure 1) was performed from a pre-filtered collection of virtual N-Boc diamines (>5000) pooled from commercial and internal sources. Additional filtering for incomptable reactive groups and availability yielded nearly 2800 linkers which were enumerated into full-length PROTACs, followed by structure-based clustering (FCFP4 fingerprints) and calculation of multiple properties (H-bond donor/acceptor count, molecular weight, topological diameter, rotatable bonds) (Figure 1A). From this virtual library, 91 PROTACs were selected for synthesis to represent the distribution of calculated property space (Figure 1B) and chemical space (Figure 1C), as represented by a self-organizing map (SOM). Having validated the D2B synthesis and identified a valuable linker set, we prepared two PROTAC D2B libraries, both containing nearly identical sets35 of 91 linkers: JQ1/variable linker/tDHU and JQ1/variable linker/O-Pom.
Figure 1.
(A) Linker selection funnel from commercial and internal pools of potential linkers to the diversity set for D2B synthesis. (B) Distribution of calculated properties within the selected D2B linker set. (C) Self-organizing map depicting structural diversity of selected targets (purple stars) versus the larger set.
Results of the D2B synthesis are summarized in Scheme 3. DMSO solutions of unchromatographed PROTACs were analyzed using a charged aerosol detector (CAD), and CAD peak area for peaks exhibiting the desired m/z were determined. Concentrations were calculated on the basis of a CAD calibration curve using noscapine as a standard, and yields for the three-step D2B sequence were derived. To further validate the D2B methods, each library was prepared and evaluated in duplicate. Yields varied across the 91 diamine linkers, and the O-Pom and tDHU libraries exhibited similar average yields—32% and 27%, respectively (Scheme 3B and SI). Duplicate D2B synthesis yields were generally consistent (Scheme 3C), with a small number showing significant variation; this observation indicates that replicate D2B syntheses reduce false negatives. Figure 3D represents the synthesis success rate across the 91 linkers using a cutoff of 10% yield and shows the similarity between the O-Pom and tDHU libraries (80% success)—this is notable considering the O-Pom glutarimide is chemically sensitive versus the tDHU.36 This finding demonstrates that the D2B PROTAC synthesis and resin scavenging conditions are mild and enables side-by-side assessment of O-Pom and tDHU libraries in cellular assays.
Scheme 3. (A) General Sequence for D2B Synthesis Using NHS Esters and Resin Scavengers, (B) Bar Graph of CAD Yields for O-Pom and tDHU Libraries Aligned by Plate Position, (C) Scatter Plots Comparing PROTAC Yield across Duplicates, for O-Pom (left) and DHU (right), and (D) Synthesis Success Rate Aligned by Plate Position.
Green indicates CAD yield >10%, blue indicates CAD yield <10%. Top two plots are duplicates of O-Pom library; bottom two plots are duplicates of tDHU library.
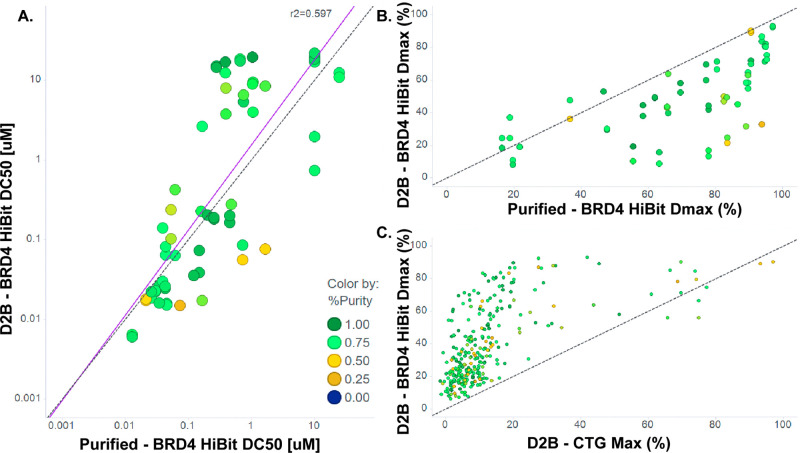
Figure 3.
Purified versus D2B data for BRD4 HiBit and CTG assays. (A) HiBit BRD4 DC50. (B) HiBit BRD4 Dmax. (C) Dmax is not correlated with CTG maximum effect. Colors indicate purity. Dotted line is unity; solid line is best fit.
The two D2B PROTAC linker libraries, JQ-1/variable linker/tDHU and JQ1/variable linker/O-Pom, were profiled in four cell-based assays in 11-point dose–response curves. CRBN target engagement was measured using a nanoBRET assay in HEK293 cells under live-cell and permeabilized-cell conditions. Degradation of BRD4 was assessed using a HEK293 HiBit-tagged BRD4 line, reading out BRD4 concentration to quantitate DC50 and Dmax. To deconvolute on-target BRD4 degradation from general cell toxicity, Cell Titer Glo (CTG) was performed. Individual D2B synthetic samples were sufficient for all four assays, which were tested in parallel.
Control compounds exhibited expected results across all four assays and were included in each D2B assay plate (Table 1). JQ1 carboxylic acid and its NHS ester were inactive across all assays, as was NHS itself. O-Pom and tDHU acids are inactive in the live-cell CRBN target engagement assay and show weak activity in permeabilized cells; neither exhibits BRD4 degradation activity or cell toxicity. NHS esters of O-Pom and tDHU engage CRBN in intact cells at high concentrations and are left-shifted in permeabilized cells, without degradation activity or cell toxicity. An authentic sample of dBET1 exhibits a profile consistent with published results,29 and a D2B sample is within 3-fold across assays with successful degradation of BRD4.
Table 1. D2B Control Compounds and Resultsa.
| CRBN live cell | CRBN perm cell | BRD4 degradation | CTG cell viability | |
|---|---|---|---|---|
| control | nanoBRET, IC50 (μM) | nanoBRET, IC50 (μM) | DC50 (μM)/Dmax (%) | EC50 (μM)/Emax (%) |
| JQ1-CO2H | >50 | >50 | >50/14.2 | >50/14.23 |
| JQ1-NHS ester (3) | >50 | >50 | >50/11.85 | >50/5.26 |
| tDHU-CO2H | >50 | 1.94 | >21.65/20.37 | >50/7.84 |
| tDHU-NHS ester (5) | 22.16 | 6.04 | >50/15.21 | >50/9.24 |
| O-Pom-CO2H | >50 | 0.83 | >50/8.1 | >50/7.58 |
| O-Pom-NHS (4) | 7.42 | 1.05 | >50/11.97 | >50/11.93 |
| N-hydroxysuccinimide | >50 | >50 | >50/15.34 | >50/10.78 |
| dBET1 | 2.2 | 0.09 | 0.48/92.26 | ND/25.16 (10 μM)b |
| dBET1 (D2B synthesis) | 0.93 | 0.056 | 0.28/49.48 | ND/21.72 (12 μM)b |
Target engagement for CRBN was assessed by nanoBRET CRBN-tracer assay in live and permeabilized (digitonin) HEK293 cells. Degradation and cell viability were assessed by HiBit-BRD4 by Cell-Titer-Glo assays, respectively, in HEK293 cells. See Supporting Information for additional details.
Concentration at Emax.
To gauge robustness and intrinsic variability of the four D2B assays, parameters including the Z′ factor, signal-to-background (S/B) ratio, and control compound potency and standard deviation were calculated. The RZ′ values in a 384-well plate across all four assays was >0.6, indicating robust performance (SI, Table S5).
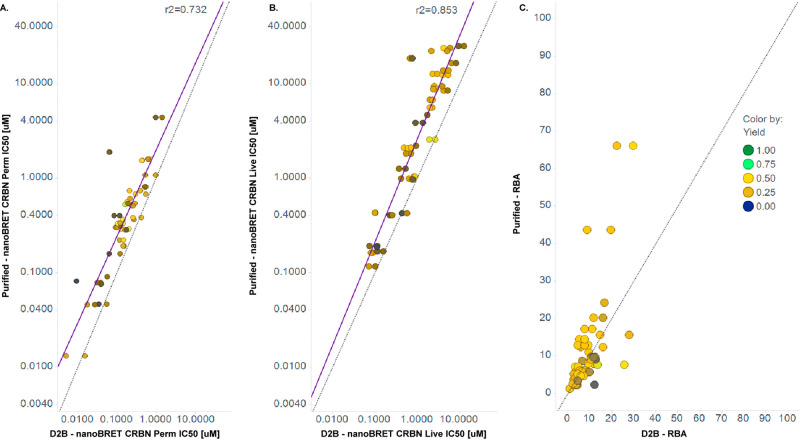
To validate the D2B method for quantitative PROTAC SAR, we compared assay data for D2B samples with those for independently synthesized and purified samples. CRBN nanoBRET target engagement data exhibit an excellent correlation between D2B and purified samples across 3 orders of magnitude in both permeabilized and live cells (Figure 2A,B). The strong correlation between D2B and purified samples was not dependent on product yield and suggests that quantification by CAD provides accurate PROTAC concentrations and dose–response curves.
Figure 2.
Purified versus D2B data for CRBN nanoBRET assays. (A) Permeabilized cells. (B) Live cells. (C) Relative binding affinity (RBA). Colors indicate CAD yield. Solid purple line is regression, and dashed line is unity.
Measurement of CRBN target engagement in live and permeabilized cells enabled calculation of relative binding affinities (RBAs) for test compounds in D2B.37 The RBA, defined as the ratio of CRBN target engagement in live versus permeabilized cells (IC50-live/IC50-permeabilized), estimates cellular permeability differences among test compounds, with larger values indicating lower permeability. As expected, D2B PROTACs exhibit a range of RBAs, and RBA values calculated from D2B and purified samples were generally in good agreement (Figure 2C).
D2B and purified sample data were well correlated for the HiBit BRD4 degradation assay as well, although greater variability in DC50 was observed compared to that with CRBN nanoBRET (Figure 3A). Inspection of dose–response curves showed that D2B Dmax values were reduced versus those of purified samples (Figure 3B) and should be accounted for in their interpretation. Importantly, BRD4 HiBit Dmax is not correlated with CTG, indicating that degradation activity is not generally an artifact of cell toxicity (Figure 3C).
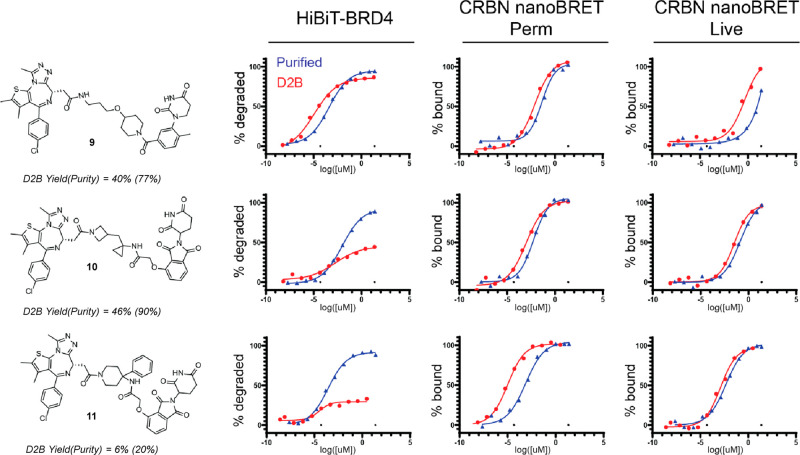
Representative dose–response curves for D2B and purified samples (Figure 4) are instructive for how to prioritize PROTACs emerging from D2B libraries. The most straightforward case, when CRBN target engagement and HiBit degradation are potent and reach high Dmax, clearly warrants follow-up purification and characterization (9). When CRBN target engagement is potent, HiBit DC50 or inflection point is in a potent range, and Dmax is marginal, follow-up should be strongly considered, especially when yield or purity is lower (10 and 11). Reduced Dmax values in D2B can lead to high Dmax values upon purification and recharacterization.
Figure 4.
Representative dose–response curves comparing D2B and purified samples.
The reduced Dmax in D2B versus purified samples is presumably due to the presence of reaction byproducts containing POI or E3 ligand partial PROTACs in D2B samples. While CRBN nanoBRET assays, where D2B versus purified sample variability is low, reflect target engagement, degradation is event-driven and catalytic. In a D2B experiment, binding competition between desired PROTACs and reaction byproducts may decrease the ternary complex concentration; this effect is amplified over many catalytic cycles, leading to reduced Dmax. This observation supports the use of multiple assays—CRBN target engagement, BRD4 degradation, and CTG—to prioritize D2B PROTACs for follow-up.
Validated, high-quality D2B data enabled comparison of assay results and calculated physicochemical properties, and multiple trends emerged for CRBN nanoBRET RBAs. For example, reduced RBA—indicating higher permeability—was associated with fewer hydrogen-bond donors and acceptors (see SI, Figure S7A,B). In addition, lower RBA correlated with increasing clogD, reaching a consistent value for clogD > 4.0 (see SI, Figure S7C). These trends are consistent with expectations and represent the starting point for analyzing D2B datasets. Indeed, recent reports identify conformational effects such as intramolecular hydrogen-bonding as contributors to PROTAC cell permeability,38 and the large datasets enabled by the PROTAC D2B approach will accelerate exploration of more complex prediction models.
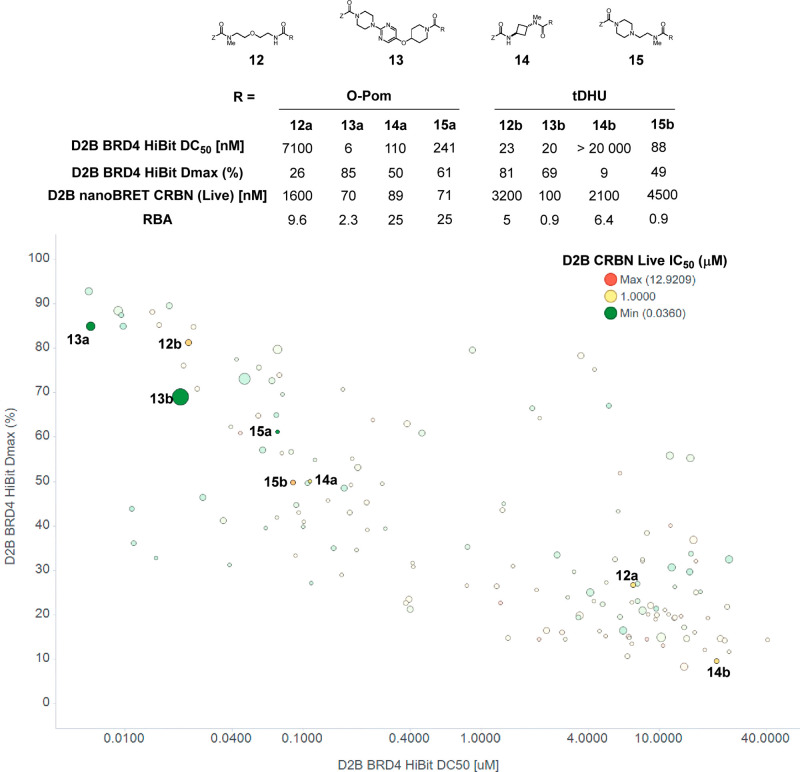
Additional SAR emerged from the D2B dataset, and specific examples are illustrated graphically in Figure 5. BRD4 HiBit Dmax is plotted as a function of DC50, which positions the most attractive compounds toward the upper left quadrant; markers are sized by RBA (larger corresponds to higher permeability) and colored by CRBN live cell target engagement. A range of profiles are observed, and matched-pair analysis reveals significant and unexpected differences between structurally similar compounds. For example, the O-Pom PROTAC containing a flexible ether linker (12a) is a significantly more potent BRD4 degrader versus the corresponding tDHU PROTAC (12b), DC50 = 23 vs 7100 nM. Notably, potent degradation with 12a is observed despite weak CRBN target engagement in the live cell nanoBRET (1600 nM); this underscores the value of evaluating PROTACs in target engagement and degradation assays. The divergent profiles of 12a and 12b are not recapitulated across O-Pom/tDHU matched pairs; for example, 13a and 13b, which contain the same larger, more rigid linker, exhibit nearly identical profiles across degradation, CRBN target engagement potency, and RBA. The size and diversity of the D2B library enabled observation of additional SAR trends highlighting the complex relationship between linker and E3 ligase ligand components. For example, a shorter, more rigid linker with O-Pom (14a) was found to improve both CRBN engagement and degradation compared to the more flexible 12a; however, when tDHU was instead employed the opposite was true, with 14b being essentially inactive in BRD4 degradation with only micromolar CRBN target engagement. Furthermore, the library data revealed that linkers bearing basic amines (15) appear to retain degradation for both O-Pom and tDHU. The O-Pom analog 15a was significantly more potent in the live CRBN nanoBRET; however, the degradation potency was comparable to that of the tDHU (15b) analog, further illustrating the complex relationship between target engagement and degradation efficiency.
Figure 5.
Representative D2B SAR. Table: matched-pair analysis of O-Pom and tDHU PROTACs. Scatter plot: BRD4 HiBit Dmax versus DC50, colored by nanoBRET CRBN live cell IC50, sized by inverse RBA (larger indicates higher permeability).
Evaluation of PROTACs in a telescoped D2B synthesis provides a platform to accelerate SAR of linkers in E3 ligase target engagement, protein degradation potency, permeability, and cell toxicity in cell assays. The synthetic approach leverages the high diversity of diamine building blocks and provides degraders with sufficient quantity and purity for these assays from a single synthesis sample. The throughput of the D2B methodology enables exploration of a large number of linkers, informing the relationship between linker structure and E3 ligase ligand. Extensions of this method, to broaden its utility for PROTAC optimization, include using linking chemistry beyond amide coupling as well as incorporation of other assay types, such as ternary complex readouts. Scalability to higher density plate formats can be explored, as well as adaptation to automation. Lastly, D2B offers the opportunity to accelerate empirical optimization while also building toward predictive modeling through creation of large training datasets. Developments on these enhancements to the methods will be reported in due course.
Acknowledgments
The authors would like to acknowledge Cristina Grosanu and Alex De Vera for assistance in purification of isolated products. We would like to thank Kuanchang Chen for assistance with Charged Aerosol Detector quantification of D2B plate yields. We would like to thank Larry Szewczuk for helpful discussions.
Glossary
Abbreviations
- HTE
high-throughput experimentation
- D2B
direct-to-biology
- PROTACs
proteolysis targeting chimeras
- DMPK
drug metabolism and pharmacokinetics
- RBA
relative binding affinity
- CRBN
cereblon
- O-Pom
oxygen-linked pomalidomide
- tDHU
tolyl dihydrouracil
- BRD4
bromodomain-4
- NHS
N-hydroxysuccinimide
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.2c00124.
Protocols for direct-to-biology experiments, a full list of linkers and products, CAD data, control compound synthesis, synthesis optimization data, assay protocols, and supporting NMR spectra (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Chamberlain P. P.; Hamann L. G. Development of targeted protein degradation therapeutics. Nat. Chem. Biol. 2019, 15, 937–944. 10.1038/s41589-019-0362-y. [DOI] [PubMed] [Google Scholar]
- Hu Z.; Crews C. M. Recent Developments in PROTAC-Mediated Protein Degradation: From Bench to Clinic. ChemBioChem 2022, 23, e202100270 10.1002/cbic.202100270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust T. B.; Donovan K. A.; Yue H.; Chamberlain P. P.; Fischer E. S. Small-Molecule Approaches to Targeted Protein Degradation. Annu. Rev. Can. Bio. 2021, 5, 181–201. 10.1146/annurev-cancerbio-051420-114114. [DOI] [Google Scholar]
- Burslem G. M.; Crews C. M. Small-Molecule Modulation of Protein Homeostasis. Chem. Rev. 2017, 117, 11269–11301. 10.1021/acs.chemrev.7b00077. [DOI] [PubMed] [Google Scholar]
- Dale B.; Cheng M.; Park K.-S.; Kaniskan H. Ü.; Xiong Y.; Jin J. Advancing targeted protein degradation for cancer therapy. Nat. Rev. Cancer 2021, 21, 638–654. 10.1038/s41568-021-00365-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A. C.; Crews C. M. Induced protein degradation: an emerging drug discovery paradigm. Nat. Rev. Drug Discovery 2017, 16, 101–114. 10.1038/nrd.2016.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira M.; Calabrese M. F.; Bullock A. N.; Crews C. M. Targeted protein degradation: expanding the toolbox. Nat. Rev. Drug Discovery 2019, 18, 949–963. 10.1038/s41573-019-0047-y. [DOI] [PubMed] [Google Scholar]
- Schneider M.; Radoux C. J.; Hercules A.; Ochoa D.; Dunham I.; Zalmas L.-P.; Hessler G.; Ruf S.; Shanmugasundaram V.; Hann M. M.; Thomas P. J.; Queisser M. A.; Benowitz A. B.; Brown K.; Leach A. R. The PROTACtable genome. Nat. Rev. Drug Discovery 2021, 20, 789–797. 10.1038/s41573-021-00245-x. [DOI] [PubMed] [Google Scholar]
- Garber K. The PROTAC gold rush. Nat. Biotechnol. 2022, 40, 12–16. 10.1038/s41587-021-01173-2. [DOI] [PubMed] [Google Scholar]
- Mullard A. Targeted protein degraders crowd into the clinic. Nat. Rev. Drug Discovery 2021, 20, 247–250. 10.1038/d41573-021-00052-4. [DOI] [PubMed] [Google Scholar]
- Sakamoto K. M.; Kim K. B.; Kumagai A.; Mercurio F.; Crews C. M.; Deshaies R. J. Protacs: Chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. P. Natl. Acd. Sci. USA 2001, 98, 8554–8559. 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G. E.; Buckley D. L.; Paulk J.; Roberts J. M.; Souza A.; Dhe-Paganon S.; Bradner J. E. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 2015, 348, 1376–1381. 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva S.-L.; Crews C. M. Targeted protein degradation: elements of PROTAC design. Curr. Opin. Chem. Bio. 2019, 50, 111–119. 10.1016/j.cbpa.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson S. D.; Yang B.; Fallan C. Proteolysis targeting chimeras (PROTACs) in ’beyond rule-of-five’ chemical space: Recent progress and future challenges. Bioorg. Med. Chem. Lett. 2019, 29, 1555–1564. 10.1016/j.bmcl.2019.04.030. [DOI] [PubMed] [Google Scholar]
- Cantrill C.; Chaturvedi P.; Rynn C.; Petrig Schaffland J.; Walter I.; Wittwer M. B. Fundamental aspects of DMPK optimization of targeted protein degraders. Drug Discovery Today 2020, 25, 969–982. 10.1016/j.drudis.2020.03.012. [DOI] [PubMed] [Google Scholar]
- Pike A.; Williamson B.; Harlfinger S.; Martin S.; McGinnity D. F. Optimising proteolysis-targeting chimeras (PROTACs) for oral drug delivery: a drug metabolism and pharmacokinetics perspective. Drug Discovery Today 2020, 25, 1793–1800. 10.1016/j.drudis.2020.07.013. [DOI] [PubMed] [Google Scholar]
- Schiemer J.; Horst R.; Meng Y.; Montgomery J. I.; Xu Y.; Feng X.; Borzilleri K.; Uccello D. P.; Leverett C.; Brown S.; Che Y.; Brown M. F.; Hayward M. M.; Gilbert A. M.; Noe M. C.; Calabrese M. F. Snapshots and ensembles of BTK and cIAP1 protein degrader ternary complexes. Nat. Chem. Bio. 2021, 17, 152–160. 10.1038/s41589-020-00686-2. [DOI] [PubMed] [Google Scholar]
- Gadd M. S.; Testa A.; Lucas X.; Chan K.-H.; Chen W.; Lamont D. J.; Zengerle M.; Ciulli A. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 2017, 13, 514–521. 10.1038/nchembio.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesmundo N. J.; Sauvagnat B.; Curran P. J.; Richards M. P.; Andrews C. L.; Dandliker P. J.; Cernak T. Nanoscale synthesis and affinity ranking. Nature 2018, 557, 228–232. 10.1038/s41586-018-0056-8. [DOI] [PubMed] [Google Scholar]
- Kitamura S.; Zheng Q.; Woehl J. L.; Solania A.; Chen E.; Dillon N.; Hull M. V.; Kotaniguchi M.; Cappiello J. R.; Kitamura S.; Nizet V.; Sharpless K. B.; Wolan D. W. Sulfur(VI) Fluoride Exchange (SuFEx)-Enabled High-Throughput Medicinal Chemistry. J. Am. Chem. Soc. 2020, 142, 10899–10904. 10.1021/jacs.9b13652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J. B.; Roughley S. D.; Matassova N.; Brough P. A. Off-Rate Screening (ORS) By Surface Plasmon Resonance. An Efficient Method to Kinetically Sample Hit to Lead Chemical Space from Unpurified Reaction Products. J. Med. Chem. 2014, 57, 2845–2850. 10.1021/jm401848a. [DOI] [PubMed] [Google Scholar]
- Tang W.; Luo T.; Greenberg E. F.; Bradner J. E.; Schreiber S. L. Discovery of histone deacetylase 8 selective inhibitors. Bioorg. Med. Chem. Lett. 2011, 21, 2601–2605. 10.1016/j.bmcl.2011.01.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva S.-L.; Crews C. M Targeted protein degradation: elements of PROTAC design. Curr. Opin. Chem. Biol. 2019, 50, 111–119. 10.1016/j.cbpa.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maple H. J.; Clayden N.; Baron A.; Stacey C.; Felix R. Developing degraders: principles and perspectives on design and chemical space. Med. Chem. Comm. 2019, 10, 1755–1764. 10.1039/C9MD00272C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak R. P.; DeAngelo S. L.; Buckley D.; He Z.; Donovan K. A.; An J.; Safaee N.; Jedrychowski M. P.; Ponthier C. M.; Ishoey M.; Zhang T.; Mancias J. D.; Gray N. S.; Bradner J. E.; Fischer E. S. Plasticity in binding confers selectivity in ligand-induced protein degradation. Nat. Chem. Biol. 2018, 14, 706–714. 10.1038/s41589-018-0055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis T. A.; La Clair J. J.; Burkart M. D. Unraveling the Role of Linker Design in Proteolysis Targeting Chimeras. J. Med. Chem. 2021, 64, 8042–8052. 10.1021/acs.jmedchem.1c00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue C.; Cubillos-Rojas M.; Gutierrez-Prat N.; Sanchez-Zarzalejo C.; Verdaguer X.; Riera A.; Nebreda A. R. Optimal linker length for small molecule PROTACs that selectively target p38α and p38β for degradation. Eur. J. Med. Chem. 2020, 201, 112451. 10.1016/j.ejmech.2020.112451. [DOI] [PubMed] [Google Scholar]
- Troup R. I.; Fallan C.; Baud M. G. J. Current strategies for the design of PROTAC linkers: a critical review. Explor. Target Antitumor. Ther. 2020, 1, 273–312. 10.37349/etat.2020.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riching K. M.; Mahan S.; Corona C. R.; McDougall M.; Vasta J. D.; Robers M. B.; Urh M.; Daniels D. L. Quantitative Live-Cell Kinetic Degradation and Mechanistic Profiling of PROTAC Mode of Action. ACS Chem. Bio. 2018, 13, 2758–2770. 10.1021/acschembio.8b00692. [DOI] [PubMed] [Google Scholar]
- Daniels D. L.; Riching K. M.; Urh M. Monitoring and deciphering protein degradation pathways inside cells. Drug Discovery Today. Technol. 2019, 31, 61–68. 10.1016/j.ddtec.2018.12.001. [DOI] [PubMed] [Google Scholar]
- Arista L.; Hebech C.; Hollingworth G. J.; Holzer P.; Imbach-Weese P.; Lorber J.; Machauer R.; Schmiedeberg N.; Vulpetti A.; Zoller T.. Preparation of N-[3-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)phenyl]benzamide derivatives as BTK inhibitors. WO2019186343A1, Oct 3, 2019.
- Boichenko I.; Bär K.; Deiss S.; Heim C.; Albrecht R.; Lupas A. N.; Hernandez Alvarez B.; Hartmann M. D. Chemical Ligand Space of Cereblon. ACS Omega 2018, 3, 11163–11171. 10.1021/acsomega.8b00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev A.; Krasavin M. Ligands for cereblon: 2017–2021 patent overview. Expert Opin. Ther. Pat. 2022, 32, 171–190. 10.1080/13543776.2022.1999415. [DOI] [PubMed] [Google Scholar]
- See the Supporting Information for details on scavengers used for D2B cleanup.
- Some linkers were different between the OPom and tDHU libraries due to limited availability. See the Supporting Information for a full description of the linker set for each plate.
- Bricelj A.; Dora Ng Y. L.; Ferber D.; Kuchta R.; Müller S.; Monschke M.; Wagner K. G.; Krönke J.; Sosič I.; Gütschow M.; Steinebach C. Influence of Linker Attachment Points on the Stability and Neosubstrate Degradation of Cereblon Ligands. ACS Med. Chem. Lett. 2021, 12, 1733–1738. 10.1021/acsmedchemlett.1c00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasta J. D.; Corona C. R.; Robers M. B. A High-Throughput Method to Prioritize PROTAC Intracellular Target Engagement and Cell Permeability Using NanoBRET. Methods Mol. Biol. 2021, 2365, 265–282. 10.1007/978-1-0716-1665-9_14. [DOI] [PubMed] [Google Scholar]
- Atilaw Y.; Poongavanam V.; Svensson Nilsson C.; Nguyen D.; Giese A.; Meibom D.; Erdelyi M.; Kihlberg J. Solution Conformations Shed Light on PROTAC Cell Permeability. ACS Med. Chem. Lett. 2021, 12, 107–114. 10.1021/acsmedchemlett.0c00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.