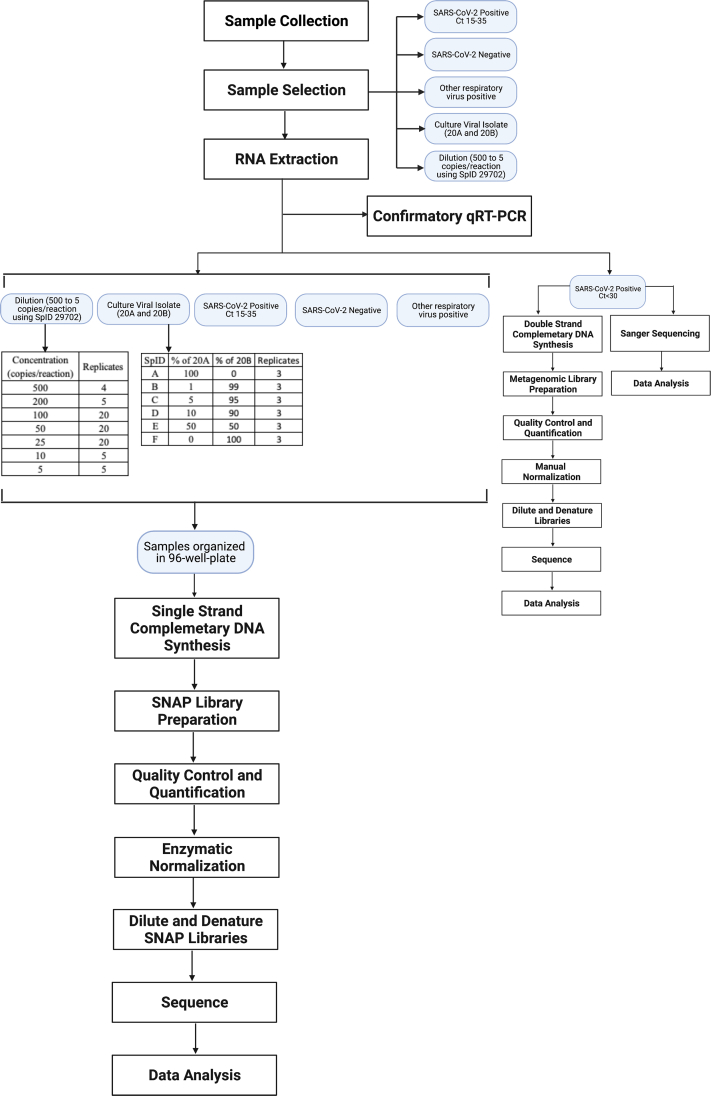
Abstract
Amplicon-based sequencing methods are central in characterizing the diversity, transmission, and evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), but need to be rigorously assessed for clinical utility. Herein, we validated the Swift Biosciences' SARS-CoV-2 Swift Normalase Amplicon Panels using remnant clinical specimens. High-quality genomes meeting our established library and sequence quality criteria were recovered from positive specimens, with 95% limit of detection of 40.08 SARS-CoV-2 copies/PCR. Breadth of genome recovery was evaluated across a range of CT values (11.3 to 36.7; median, 21.6). Of 428 positive samples, 413 (96.5%) generated genomes with <10% unknown bases, with a mean genome coverage of 13,545× ± SD 8382×. No genomes were recovered from PCR-negative specimens (n = 30) or from specimens positive for non–SARS-CoV-2 respiratory viruses (n = 20). Compared with whole-genome shotgun metagenomic sequencing (n = 14) or Sanger sequencing for the spike gene (n = 11), pairwise identity between consensus sequences was 100% in all cases, with highly concordant allele frequencies (R2 = 0.99) between Swift and shotgun libraries. When samples from different clades were mixed at varying ratios, expected variants were detected even in 1:99 mixtures. When deployed as a clinical test, 268 tests were performed in the first 23 weeks, with a median turnaround time of 11 days, ordered primarily for outbreak investigations and infection control.
Since the deposition of the first severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) whole genome sequence (NC_045512.1) in January 2020, >4 million SARS-CoV-2 genomes have been deposited to public data repositories, far exceeding any other human pathogen.1,2 Such a feat has been made possible because of advances in next-generation sequencing (NGS) technologies that enable near real-time genomic surveillance.3, 4, 5, 6, 7, 8, 9 Viral whole-genome sequencing (WGS) of laboratory-confirmed SARS-CoV-2 isolates is frequently used in outbreak investigations, deployment of public health interventions, development of vaccines and therapeutics, and evaluation of vaccine and antiviral effectiveness against emerging variants.4, 5, 6,10, 11, 12, 13, 14, 15 Specific SARS-CoV-2 variants have been associated with higher viral loads, lower vaccine effectiveness, and worse outcomes, such as mortality.16, 17, 18, 19, 20, 21, 22, 23 Notably, B.1.1.7 has been associated with increased disease severity, prolonged hospitalization, and higher mortality risk.16,18,19,21, 22, 23, 24 In addition, recent studies have shown poorer outcomes for patients infected with variants B.1.351 and P.1.23,25 In a clinical setting, identification of specific viral mutations can aid in the selection of monoclonal therapies,26, 27, 28, 29 and viral sequencing can be used to monitor the accumulation of mutations during long-term viral replication in immunocompromised individuals.30 As treatment regimens expand, validated WGS assays are needed not only for genomic surveillance but also for high-quality, clinically actionable data with rapid turnaround times.
Multiplexed amplicon sequencing methods have proven to be faster, more sensitive, and more cost-effective than shotgun and capture-based approaches, enabling genome recovery across a wide range of viral loads.31,32 We previously tested one such panel from Swift Biosciences and demonstrated genome recovery up to a CT of 36 across a broad range of isolates.33 Designed against the SARS-CoV-2 Wuhan–Hu-1 complete genome (https://www.ncbi.nlm.nih.gov/nuccore, accession number NC_045512.2, last accessed July 12, 2022), the Swift Normalase Amplicon Panel (SNAP) primer set amplifies 345 amplicons ranging from 116 to 255 bp (average, 150 bp) in a single tube to cover the approximately 30-kb SARS-CoV-2 genome. This assay can generate libraries in <3 hours using an input concentration of only 10 to 100+ viral copies for single-strand cDNA or double-strand cDNA synthesis. This can be followed by either manual normalization or Swift Biosciences' proprietary enzymatic normalization of multiplexed libraries for equimolar pools.
For clinical use, sequencing assays need to be rigorously validated, documented, and performed in Clinical Laboratory Improvement Amendments–accredited laboratories. However, no specific guidelines currently exist for the development and validation of WGS assays for SARS-CoV-2. We validated the Swift Biosciences' one-tube SARS-CoV-2 SNAP Version 2.0—the first clinical validation of an NGS-based assay for WGS of SARS-CoV-2 to our knowledge—according to US Food and Drug Administration’s “Considerations for Design, Development, and Analytical Validation of Next Generation Sequencing-Based in Vitro Diagnostics Intended to Aid in the Diagnosis of Suspected Germline Diseases” (https://www.fda.gov/media/99208/download, last accessed January 4, 2021) and US Food and Drug Administration's “Submitting Next Generation Sequencing Data to the Division of Antiviral Products Guidance for Industry” Technical Specifications Document (https://www.fda.gov/regulatory-information/search-fda-guidance-documents/submitting-next-generation-sequencing-data-division-antiviral-products-guidance-industry-technical, last accessed August 19, 2021). Using clinical specimens, the analytical sensitivity, analytical specificity, limit of detection, accuracy, and precision (reproducibility and repeatability) of the assay were evaluated, establishing acceptance criteria for sequencing libraries and output genomes. This assay is now available as a clinically orderable test, with results returned to physicians to aid in disease management and treatment. It has been used in multiple vaccine trials, research studies, validation of other NGS-based assays, and sequencing SARS-CoV-2 for public health surveillance and outbreak investigation.4,6,10,34, 35, 36, 37, 38
Materials and Methods
Performance Evaluation
Existing US Food and Drug Administration guidelines were used for designing, developing, and establishing the analytical validity of NGS-based tests for the diagnosis of suspected germline diseases and development of antivirals to clinically validate a multiplex amplicon sequencing method for WGS of SARS-CoV-2 (Supplemental Figure S1). The aim was to determine analytical sensitivity (limit of detection), analytical specificity, accuracy, and precision (reproducibility and repeatability) of the assay using remnant clinical specimens described below.
Clinical Specimens
Use of deidentified remnant clinical specimens from University of Washington Virology Laboratory for SARS-CoV-2 testing was approved by the University of Washington Institutional Review Board. Specimens were tested for SARS-CoV-2 using one of the following PCR assays: CDC-based Laboratory Developed Assays, Abbott m2000 (Abbott Laboratories, Chicago, IL); Roche Cobas (Roche, Basel, Switzerland); or Hologic Panther/Panther Fusion (Hologic, Marlborough, MA).39,40 Samples used for analysis of specificity were tested using University of Washington Virology Laboratory respiratory virus panel to identify non–SARS-CoV-2 respiratory pathogens, including respiratory syncytial virus, influenza virus type A, parainfluenza virus types 1, 2, and 3, and human metapneumovirus.41
Laboratory-confirmed specimens used in this study came from nasal or nasopharyngeal swabs collected in either phosphate-buffered saline or viral transport medium that had >500 μL volume remaining. This included SARS-CoV-2–positive specimens (n = 428), specimens negative for both SARS-CoV-2 and other respiratory viruses (n = 30), and specimens positive only for other respiratory viruses (n = 20). Water was used as a negative control, and previously confirmed and sequenced SARS-CoV-2–positive specimens were used as positive controls.
SARS-CoV-2 Virus Culture Isolates
For detection of within-sample variation, two previously sequenced culture isolates from different clades were used: WA-UW-20236 TM, EPI_ISL_4926371, Nextstrain clade 20A, and WA-UW-19433 TM, EPI_ISL_4926374, 20B. The culture isolates had a viral load of 7 × 104 copies/μL and an approximate CT of 20. Viruses were isolated from original clinical specimens at the University of Vermont Biosafety Level 3 (BSL-3) facility under an approved Institutional Biosafety protocol, as previously.42 Vero E6-TMPRSS2 cells [obtained from the Japanese Collection of Research Bioresources (JCRB) Cell Bank, JCRB number JCRB1819] were inoculated with 100 μL of clinical sample, inoculated for 1 hour at 37°C with rocking, washed with phosphate-buffered saline, and overlaid with 1 mL standard Vero medium containing 2% fetal bovine serum. Wells were monitored daily for the appearance of cytopathic effect.43 Once cytopathic effect was observed, supernatants were clarified to remove cellular debris and prepared for RNA extraction by mixing 1:1 with Qiagen Buffer AVL (Qiagen, Germantown, MD). The high viral loads in supernatants after viral isolation (CT 10 and 9.5, respectively) allowed preparation of multiple dilutions and mixtures. The consensus sequences for the two samples differed at a total of 16 positions, with 6 mutations unique to the 20A specimen (C4633T, C10965T, T14643C, A20268G, C22482T, and C28854T), 10 mutations unique to the 20B specimen [G2144T, G3824A, G13348T, C15933T, G16968T, T19839C, G28083T, and GGG(28881 to 28883)AAC], and 4 mutations common to both samples (C241T, C3037T, C14408T, and A23403G). All positions are relative to the SARS-CoV-2 isolate Wuhan–Hu-1 reference sequence (NC_045512.2).
RNA Extraction and PCR
RNA was extracted from 200 μL of each specimen (nasal or nasopharyngeal swabs in phosphate-buffered saline or viral transport medium) and eluted to 100 μL on the MagNAPure LC (Roche) using the manufacturer's instructions. This was followed by a confirmatory RT-PCR using AgPath-ID One-Step RT-PCR kit (Life Technologies, Carlsbad, CA) with CDC N1/N2 primers and probes for N gene following protocol and concentrations.39,44 Specimens with N1/N2 CT ≤18 were diluted to prevent inhibition in downstream procedures. For measurement of analytical sensitivity, diluted RNA was also quantified by reverse transcription digital droplet PCR (RT-ddPCR) using the One-Step RT-ddPCR Advanced Kit for Probes (Bio-Rad, Hercules, CA) with N2 primer/probe set.
Swift SNAP Library Preparation and Quality Control
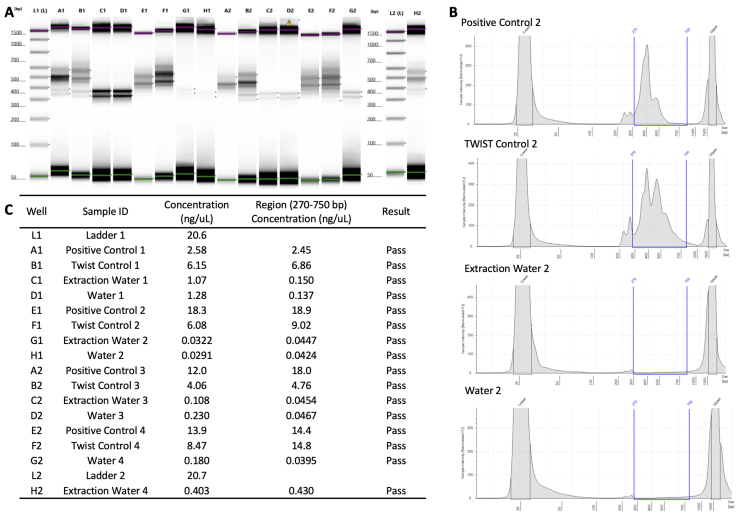
Using SuperScript IV First-Strand Synthesis System (Thermo Fisher, Waltham, MA), 11 μL of extracted RNA was subjected to single-strand cDNA synthesis, and 10 μL of the resulting single-strand cDNA was used for library preparation using the Swift SARS-CoV-2 SNAP Version 2.0 kit (Swift Biosciences, Ann Arbor, MI). First-strand synthesis and library preparation protocols were automated on liquid handlers, Sciclone G3 NGSx iQ Workstation (PerkinElmer, Waltham, MA), following manufacturer's instructions (https://www.protocols.io/view/uw-virology-swift-snapv2-protocol-byw4pxgw, last accessed October 18, 2021). Libraries were quantified on a VICTOR Nivo Multimode Microplate Reader (PerkinElmer) using the Quant-iT dsDNA High Sensitivity kit (Life Technologies). Library quality was inspected on the Agilent 4200 TapeStation (Agilent Technologies, Santa Clara, CA) using TapeStation D1000 DNA ScreenTape. Libraries with concentration ≥1.35 ng/μL on the plate reader and ≥0.5 ng/μL (region concentration from 250 to 750 bp) on the TapeStation were deemed accepted for sequencing. These cutoffs were determined on the basis of multiple runs of negative controls and expected ranges of amplicon sizes (Supplemental Figure S2). Libraries satisfying these criteria were enzymatically normalized using Normalase to generate equimolar pools using the Swift SARS-CoV-2 SNAP Version 2.0 kit. Generated pools were sequenced on the Illumina NextSeq 500 (Illumina, San Diego, CA) using the NextSeq 500/550 High Output Kit version 2.5 (2 × 150 cycles). Positive and negative controls were included on each run, and only sequencing runs with >50% reads passing filter and >60% of bases exceeding Phred quality scores of 30 were accepted.
Instrument Validation
After initial validation on the Illumina NextSeq 500, parallel sets of libraries were run on Illumina NextSeq 2000 and Illumina MiSeq instruments using 2 × 150 kits. Run, library, and genome quality metrics were assessed to ensure they meet passing criteria, and consensus genomes and allele frequencies were compared across the two runs.
Metagenomic Sequencing
Shotgun metagenomic sequencing [metagenomic NGS (mNGS)] was performed as previously described.45,46 Briefly, 18 μL of extracted RNA from selected positive specimens (CT ≤ 25) was subjected to double-stranded cDNA synthesis using the SuperScript IV First-Strand Synthesis System followed by second-strand synthesis using Sequenase version 2.0 (Thermo Fisher) and AMPure XP (Beckman Coulter Life Sciences, Brea, CA) bead-based purification. Libraries were prepared from 6 μL of the cDNA using Nextera Flex (Illumina) following manufacturer's protocols and sequenced on the NextSeq 500/550 (Illumina) using the High Output Kit version 2.5 (75 cycles).
Illumina COVIDseq
A subset of samples was sequenced using an alternative amplicon panel (Illumina COVIDseq) to evaluate the robustness of variant identification among more recent samples. The COVIDseq protocol was implemented on the same liquid handler system (PerkinElmer Sciclone G3 NGSx iQ Workstation) and used to prepare libraries from extracted RNA. COVIDseq libraries were sequenced on an Illumina Novaseq6000 instrument using a 1 × 100 read format with a target of 2 million reads per sample.
Sanger Sequencing (Spike Gene)
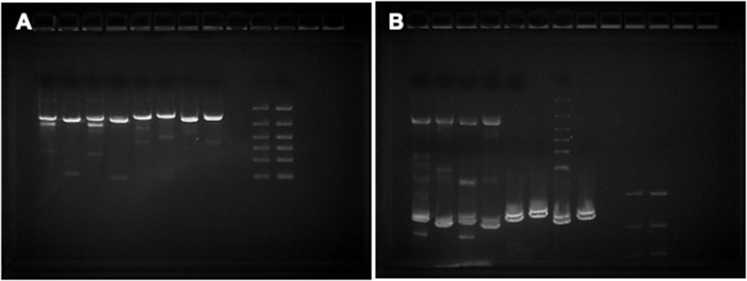
Three amplicons covering the spike coding region (NC_045512.2: 21252 to 22861, 22559 to 24395, and 24072 to 25572) were generated for each sample using SuperScript III One-Step PCR Mix (Thermo Fisher) (Table 1). Amplicons were purified using Qiagen columns and eluted with 50 μL of elution buffer, and 2 μL of each purified sample was run on a FlashGel (Lonza, Basel, Switzerland). Samples with visible bands were Sanger sequenced using four to eight separate sequencing primers (IDT, Coralville, IA) (Table 2) and compared against corresponding NGS samples.
Table 1.
Amplification Primers for Sanger Sequencing
| Region | Primer name | Sequence | Amplicon, bp | ||
|---|---|---|---|---|---|
| 1 | CoV-2 S_Amp-F1a | 5′-TGCGTCATCATCTGAAGCAT-3′ | R1a | R1b | |
| CoV-2 S_Amp-F1b | 5′-CAAACCACGCGAACAAATAG-3′ | F1a | 1743 | 1610 | |
| CoV-2 S_Amp-R1a | 5′-TGCTACCGGCCTGATAGATT-3′ | F1b | 1698 | 1565 | |
| CoV-2 S_Amp-R1b | 5′-AACGCAGCCTGTAAAATCATC-3′ | ||||
| 2 | CoV-2 S_Amp-F2a | 5′-CAAACTTGTGCCCTTTTGGT-3′ | R2a | R2b | |
| CoV-2 S_Amp-F2b | 5′-CCGCATCATTTTCCACTTTT-3′ | F2a | 1840 | 1987 | |
| CoV-2 S_Amp-R2a | 5′-GTGCACTTGCTGTGGAAGAA-3′ | F2b | 1726 | 1873 | |
| CoV-2 S_Amp-R2b | 5′-CAATTTGCACTTCAGCCTCA-3′ | ||||
| 3 | CoV-2 S_Amp-F3a | 5′-CAGATGCTGGCTTCATCAAA-3′ | R3a | R3b | |
| CoV-2 S_Amp-F3b | 5′-TGGTGATTGCCTTGGTGATA-3′ | F3a | 1495 | 1525 | |
| CoV-2 S_Amp-R3a | 5′-AACGCCAACAATAAGCCATC-3′ | F3b | 1470 | 1500 | |
| CoV-2 S_Amp-R3b | 5′-GGAAGCGCTCTGAAAAACAG-3′ | ||||
Two forward and two reverse primers were designed to amplify each of three overlapping 1.5- to 2-kb regions of the spike coding region. Amplicon length for each combination is listed. The primers used for most samples are in bold; the other primers were used if these primers did not generate visible bands on a FlashGel.
Table 2.
Sequencing Primers for Sanger Sequencing
| Region | Part | Primer name | Sequence |
|---|---|---|---|
| 1 | 1 | CoV-2 S_Seq1-F1a | 5′-CAAATCCAATTCAGTTGTCTTCC-3′ |
| CoV-2 S_Seq1-F1b | 5′-TGGAGGAATACAAATCCAATTCA-3′ | ||
| CoV-2 S_Seq1-R1a | 5′-TAAAGCCGAAAAACCCTGAG-3′ | ||
| CoV-2 S_Seq1-R1b | 5′-TGAGGGAGATCACGCACTAA-3′ | ||
| 2 | CoV-2 S_Seq1-F2a | 5′-GGACCTTGAAGGAAAACAGG-3′ | |
| CoV-2 S_Seq1-F2b | 5′-TGCGAATAATTGCACTTTTGA-3′ | ||
| CoV-2 S_Seq1-R2a | 5′-CTTTCCAGTTTGCCCTGGAG-3′ | ||
| CoV-2 S_Amp-R1b∗ | 5′-AACGCAGCCTGTAAAATCATC-3′ | ||
| 2 | 1 | CoV-2 S_Seq2-F1a | 5′-TGCAGATTCATTTGTAATTAGAGG-3′ |
| CoV-2 S_Amp-F2b∗ | 5′-CCGCATCATTTTCCACTTTT-3′ | ||
| CoV-2 S_Seq2-R1a | 5′-TGCACCAATGGGTATGTCAC-3′ | ||
| CoV-2 S_Seq2-R1b | 5′-CGCATATACCTGCACCAATG-3′ | ||
| 2 | CoV-2 S_Seq2-F2a | 5′-CTGCACAGAAGTCCCTGTTG-3′ | |
| CoV-2 S_Seq2-F2b | 5′-CCCTGTTGCTATTCATGCAG-3′ | ||
| CoV-2 S_Seq2-R2a | 5′-TGTACCCGCTAACAGTGCAG-3′ | ||
| CoV-2 S_Seq2-R2b | 5′-GGTTGGCAATCAATTTTTGG-3′ | ||
| 3 | 1 | CoV-2 S_Seq3-F1a | 5′-ACTGTTTTGCCACCTTTGCT-3′ |
| CoV-2 S_Seq3-F1b | 5′-TTAACGGCCTTACTGTTTTGC-3′ | ||
| CoV-2 S_Seq3-R1a | 5′-AACCAGTGTGTGCCATTTGA-3′ | ||
| CoV-2 S_Seq3-R1b | 5′-GACAAATGGCAGGAGCAGTT-3′ | ||
| 2 | CoV-2 S_Seq3-F2a | 5′-CTTCCCTCAGTCAGCACCTC-3′ | |
| CoV-2 S_Seq3-F2b | 5′-GAGGCTGAAGTGCAAATTGA-3′ | ||
| CoV-2 S_Seq3-R2a | 5′-TAGCGCGAACAAAATCTGAA-3′ | ||
| CoV-2 S_Seq3-R2b | 5′-AGGGAGTGAGGCTTGTATCG-3′ |
Each amplicon was designed to be sequenced in two overlapping parts, with two forward and reverse primers for each part. The primers used for most samples are in bold; the other primers were used if these primers did not yield usable Sanger sequences.
Two amplification primers were used as sequencing primers if not used to generate the amplicons.
Genome Assembly and Data Analysis
Genomes from Swift SNAP libraries were assembled using a custom pipeline, TAYLOR (https://github.com/greninger-lab/covid_swift_pipeline, last accessed July 5, 2022), described previously.33,47 Briefly, raw reads were adapter and quality trimmed using BBDuk (https://jgi.doe.gov/data-and-tools/bbtools, last accessed July 5, 2022), aligned to the Wuhan–Hu-1 reference genome (NC_045512.2), and trimmed of PCR primers using Primerclip (https://github.com/swiftbiosciences/primerclip, last accessed July 5, 2022). Consensus genomes and variants were called using BCFtools.48 For a genome to pass acceptability criteria, >1 million raw reads, >750× mean genome coverage, >1000× mean spike gene coverage, 100% of spike gene with at least 200× coverage, and <10% unknown bases (Ns) in the final consensus sequence were required. A minimum of 6× per base coverage was required to call a non-N base in the consensus sequence.
For Sanger sequencing, reads were imported into Geneious version 9.1.8 (Biomatters, Auckland, New Zealand), trimmed with an error probability limit of 0.005, mapped to the reference spike sequence (NC_045512.2: 21563 to 25384), and manually trimmed to the start and stop codons for spike. Trimmed reads were de novo assembled, and the final Sanger consensus sequence for spike was extracted. Samples that passed quality control (QC) for Swift and had a complete spike consensus sequence by Sanger were included in the comparison. Consensus sequences for spike were aligned using MAFFT version 7.45, and the number of pairwise nucleotide differences between sequences was compared.
Shotgun metagenomic sequencing and Illumina COVIDseq libraries were also analyzed using the same bioinformatic pipeline (TAYLOR) but with settings adjusted to omit primer trimming. Consensus sequences were aligned using MAFFT version 7.45 (https://mafft.cbrc.jp/alignment/software) and pairwise identity computed by comparing nucleotide differences between Swift and shotgun sequences. Allele frequencies for all single nucleotide variants called by the TAYLOR pipeline at ≥1% were compared between Swift and shotgun samples.
Probit analysis to determine limit of detection calculations was performed in SPSS version 26 (IBM, Armonk, NY). Other statistical analyses and data visualizations were performed using R (R Foundation for Statistical Computing, Vienna, Austria; https://www.r-project.org).
Clinical Test Result Reporting
For clinical orders (https://testguide.labmed.uw.edu/public/view/SARSEQ and https://testguide.labmed.uw.edu/public/view/SAREPI, last accessed July 5, 2022), downstream analysis is performed on consensus sequences for samples that pass all QC criteria described above. Consensus sequences are analyzed using Pangolin (https://pangolin.cog-uk.io, last accessed July 5, 2022) to obtain PANGO lineage designations and Nextclade (https://clades.nextstrain.org, last accessed July 5, 2022) to generate Nextstrain clade assignments and a list of mutations. The mutation list generated from Nextclade is parsed to extract amino acid changes in spike. For SARSEQ orders, the Phylogenetic Assignment of Named Global Outbreak (PANGO) lineage, Nextstrain clade, and list of amino acid changes in spike are returned along with a free-text interpretation of these results. The interpretation includes information about variants of concern/variants of interest (VOCs/VOIs) and any known phenotypic effects associated with specific mutations, such as increased transmissibility or reduced neutralization by monoclonal antibodies. Example reports are shown in Supplemental Figure S3. For SAREPI orders (requested for outbreak/cluster investigations), consensus sequences passing QC criteria are aligned using MAFFT version 7.45, and the total number of pairwise nucleotide differences between sequences is reported as a table along with SARSEQ results.
Bioinformatic Pipeline Validation
To assess the performance of the bioinformatic pipeline and data processing steps, a set of benchmark data sets from the CDC were used (https://github.com/CDCgov/datasets-sars-cov-2, last accessed October 1, 2021). These data sets contain raw FASTQ files for samples generated using the ARTIC protocol (https://artic.network/ncov-2019, last accessed July 5, 2022), with results indicating PANGO lineages, and designation as VOC/VOI. The TAYLOR pipeline described above was run on the benchmark data sets, with settings to perform primer trimming for ARTIC amplicon primers, and consensus sequences and lineage designations were compared against the results provided. This analysis is performed each time there is a major version change in our bioinformatic pipeline.
Results
Swift SNAP Assay Validation Using Clinical Specimens
Rigorous assay validation was performed using deidentified clinical specimens received at the University of Washington Virology Laboratory between September 21, 2020, and October 19, 2020. Run- and sample-level acceptability criteria were defined (see Materials and Methods), ensuring multiple quality control checks at each stage of library preparation and sequencing. All sequencing runs successfully met run quality metrics with 93.24% average % Phred quality scores of 30 (SD, 4.46%), 92.95% average % passing filter (SD, 5.65%), no amplification of negative controls, and successful recovery of positive controls.
Limit of Detection of Swift SNAP Assay Is 40 Copies per PCR
To determine the limit of detection of the assay, serial dilutions of extracted RNA were prepared from a positive SARS-CoV-2 clinical sample (WA-UW-29702, EPI_ISL_603255). Multiple replicate libraries (n = 4 to 20 replicates) (Table 3) were prepared for each dilution, with target concentrations ranging from 500 to 5 copies/reaction. The concentrations of the prepared dilutions were then verified by quantitative RT-PCR (RT-qPCR) and RT-ddPCR (Table 3). Measured concentrations by RT-qPCR ranged from 698.0 to 7.8 copies/reaction. At concentrations of ≥82 copies/reaction, high-quality genomes were recovered in 100% of attempted replicates (n = 4 to 20) (Table 3 and Supplemental Table S1). On the basis of the RT-qPCR concentrations, the lowest concentration at which acceptable genomes were recovered 95% of the time was defined as the limit of detection and determined by probit analysis to be 40.08 copies/reaction or 1821.81 copies/mL of original sample (Table 3 and Supplemental Table S1).
Table 3.
Limit of Detection Determination Using Serial Dilutions of a Positive Specimen in Replicates
| Concentration, copies/reaction |
Replicates, n | Passing genomes∗ | % Passing† | |
|---|---|---|---|---|
| RT-qPCR | RT-ddPCR | |||
| 698.0 | 1080.7 | 4 | 4 | 100 |
| 309.1 | 599.0 | 20 | 20 | 100 |
| 157.5 | 287.0 | 20 | 20 | 100 |
| 82.0 | ND | 20 | 20 | 100 |
| 51.1 | ND | 20 | 19 | 95 |
| 14.8 | ND | 5 | 4 | 80 |
| 7.9 | ND | 5 | 2 | 40 |
ND, not determined; RT-ddPCR, reverse transcription digital droplet PCR; RT-qPCR, quantitative RT-PCR.
Number of genomes satisfying all acceptability criteria.
Percentage of genomes satisfying all acceptability criteria.
No Cross-Reactive Amplification of Non–SARS-CoV-2 Respiratory Virus Genomes
Assay specificity was evaluated by checking for cross-reactivity to non–SARS-CoV-2 respiratory viruses using clinical specimens positive for seasonal coronaviruses, rhinovirus, human parainfluenza viruses (1, 2, 3, and 4), respiratory syncytial virus, adenovirus, metapneumovirus, influenza A virus, and influenza B virus (n = 20 samples total) (Supplemental Table S2).49 Only 15 of 20 (75%) non–SARS-CoV-2 or other respiratory virus-positive specimens met the library QC criteria to be sequenced. No recoverable genomes were obtained from any of the 15 specimens, and the number of mapped reads ranged between 0 and 3114 reads, which represented <1% of post-trimmed reads in all cases (Supplemental Table S2).
Recovery of High-Quality Genomes from Clinical Samples across a Wide Range of Viral Loads and Lineages
Breadth of genome recovery was evaluated using 428 SARS-CoV-2–positive clinical samples (nasopharyngeal, nasal, or oropharyngeal swabs) across a range of CT values (11.3 to 36.7; median, 21.6) and 30 SARS-CoV-2–negative clinical samples. Of 428 positive samples, 413 (96.5%) generated consensus genomes with <10% Ns, and overall mean genome coverage was 13,545× ± SD 8382× (Table 4 and Supplemental Table S3). The 15 samples that failed to generate acceptable genomes had CT values ranging between 12.2 and 36.7 (median, 18.4). Of these 15 samples, 5 had raw reads <1 million, and all 15 had <100,000 mapped reads, resulting in low mean genome coverage (0.1× to 133×; median, 20×) (Supplemental Table S3).
Table 4.
Genome Recovery from SARS-CoV-2 PCR-Positive and PCR-Negative Clinical Specimens
| CT | Samples, N | Passing genomes, n (%) |
|---|---|---|
| <15 | 60 | 56 (93) |
| 15–20 | 123 | 118 (96) |
| 20–25 | 109 | 106 (97) |
| 25–30 | 97 | 91 (94) |
| >30 | 39 | 35 (90) |
| Negative | 30 | 0 |
To assess genome recovery across lineages, a set of 146 samples that were sequenced by both Swift and an alternate amplicon sequencing approach (Illumina COVIDseq) was used. Of these, 132 samples (144/146 by Swift, and 133/146 by COVIDseq) generated consensus sequences with <10% Ns by both methods and were included in the comparison. These included 53 Alpha, 8 Delta, 16 Epsilon, and 24 Omicron samples, and 19 other VOCs/VOIs, including Beta (1), Gamma (15), Iota (2), and Lambda (1) (Supplemental Table S4). Lineages agreed between COVIDseq and Swift libraries in 129 of 132 pairs (97.7%). Of the three discordant pairs, broader lineages agreed (B.1.1.7 versus Q3, BA.1.1 versus BA.1, and B.1 versus B.1.637) with the Swift library, generating the finer-scale lineage designation.
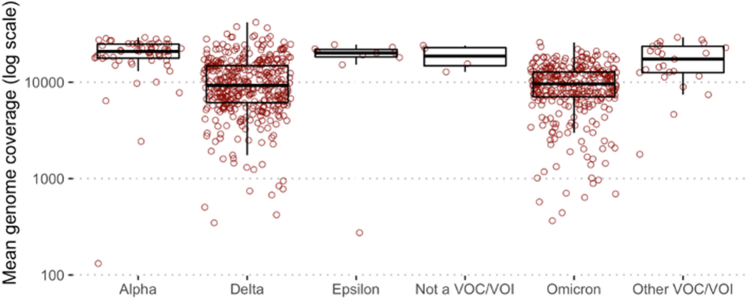
The Swift SNAP panel has also been used to sequence samples for genomic surveillance as part of the CDC's National SARS-CoV-2 Strain Surveillance program. Of 799 samples sequenced by Swift since April 2021, slightly reduced overall genome coverage was observed with Delta (10,987×; n = 387) and Omicron (9815×; n = 314) specimens compared with non-Delta/Omicron lineages (19,063×; n = 98) but we continue to recover high-quality sequences across these lineages (Supplemental Figure S4 and Supplemental Table S5).
Of the 30 samples that were PCR negative for SARS-CoV-2, 17 did not meet presequencing library QC criteria [concentrations, ≥1.35 ng/μL on the plate reader and ≥0.5 ng/μL (region concentration from 250 to 750 bp) on the TapeStation] and were not taken forward for sequencing. Of the remaining 13 sequenced samples, the number of mapped reads ranged between 0 and 5271, representing <0.5% of post-trimmed reads. No SARS-CoV-2 genomes were recovered from any of these samples (Supplemental Table S6).
Accurate Detection of Within-Sample Variation
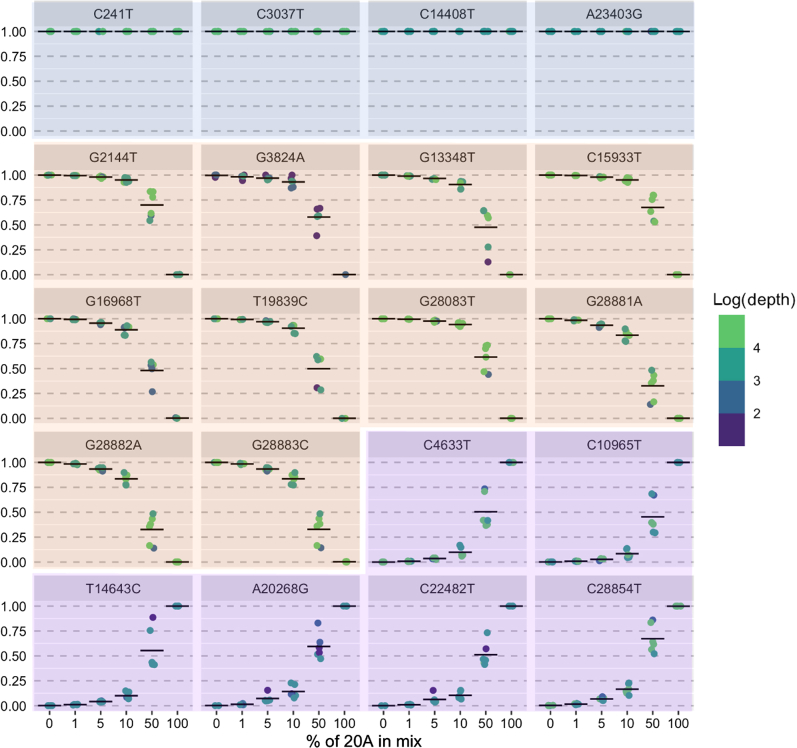
RNA was extracted from two SARS-CoV-2 cultured viral isolates from distinct clades (WA-UW-20236 TM, Nextstrain clade 20A, and WA-UW-19433 TM, 20B) with multiple distinguishing mutations (see Materials and Methods). Mixtures were prepared with ratios of 20A:20B ranging from 0:100 to 50:50 to 100:0. Mutations were called against the Wuhan–Hu-1 reference sequence (NC_045512) and filtered to accept mutations with allele frequency >1%. All expected mutations were identified at mixing ratios of ≥5:95 (Table 5 and Figure 1). At mixing ratios of 1:99, four of six of the expected low-frequency 20A mutations were identified. In the remaining two mixtures, variants were present but at allele frequencies of <1%, as would be expected on the basis of mixing at 1:99 (mean allele frequency and depth: C4633T, 0.78% and 10,025×; C10965T, 0.96% and 10,028×; n = 3 replicates).
Table 5.
Variant Detection in Mixtures Prepared from Two Samples: 20A and 20B
| Mix | % 20A specimen | % 20B specimen | Common variants detected (4 total∗) | 20A variants detected (6 total†) | 20B variants detected (10 total‡) |
|---|---|---|---|---|---|
| 1 | 0 | 100 | 4 | 0 | 10 |
| 2 | 1 | 99 | 4 | 4 | 10 |
| 3 | 5 | 95 | 4 | 6 | 10 |
| 4 | 10 | 90 | 4 | 6 | 10 |
| 5 | 50 | 50 | 4 | 6 | 10 |
| 6 | 100 | 0 | 4 | 6 | 0 |
20A, WA-UW-20236 TM; 20B, WA-UW-19433 TM.
Expected variants common to both samples (n = 4): C241T, C3037T, C14408T, and A23403G.
Variants in 20A sample (n = 6): C4633T, C10965T, T14643C, A20268G, C22482T, and C28854T.
Variants in 20B sample (n = 10): G2144T, G3824A, G13348T, C15933T, G16968T, T19839C, G28083T, and GGG(28881 to 28883) AAC.
Figure 1.
Measured allele frequency (y axis) for all expected mutations in sample mixtures described in Table 5. Blue panels indicate mutations common to WA-UW-20236 TM (20A) and WA-UW-19433 TM (20B) samples; orange panels, mutations expected in 20B sample; and purple panels, mutations expected in 20A samples. n = 20 expected mutations.
High Concordance with Sanger and Shotgun Metagenomic Sequencing
To test whether assay results agreed with alternative sequencing methods, a subset of clinical positive specimens sequenced by Swift was subjected to Sanger sequencing of the spike gene and shotgun mNGS. Full-length spike consensus sequences obtained by Sanger sequencing were compared against their corresponding Swift sequences for 14 positive specimens with CT ≤ 30. All 14 specimens selected for Sanger sequencing passed genome acceptability criteria by Swift, whereas 11 of 14 (78.6%) generated a complete Sanger sequence for spike (Supplemental Figure S5). Pairwise sequence identity between spike consensus sequences for Sanger and Swift was 100% for all 11 samples (Supplemental Table S7).
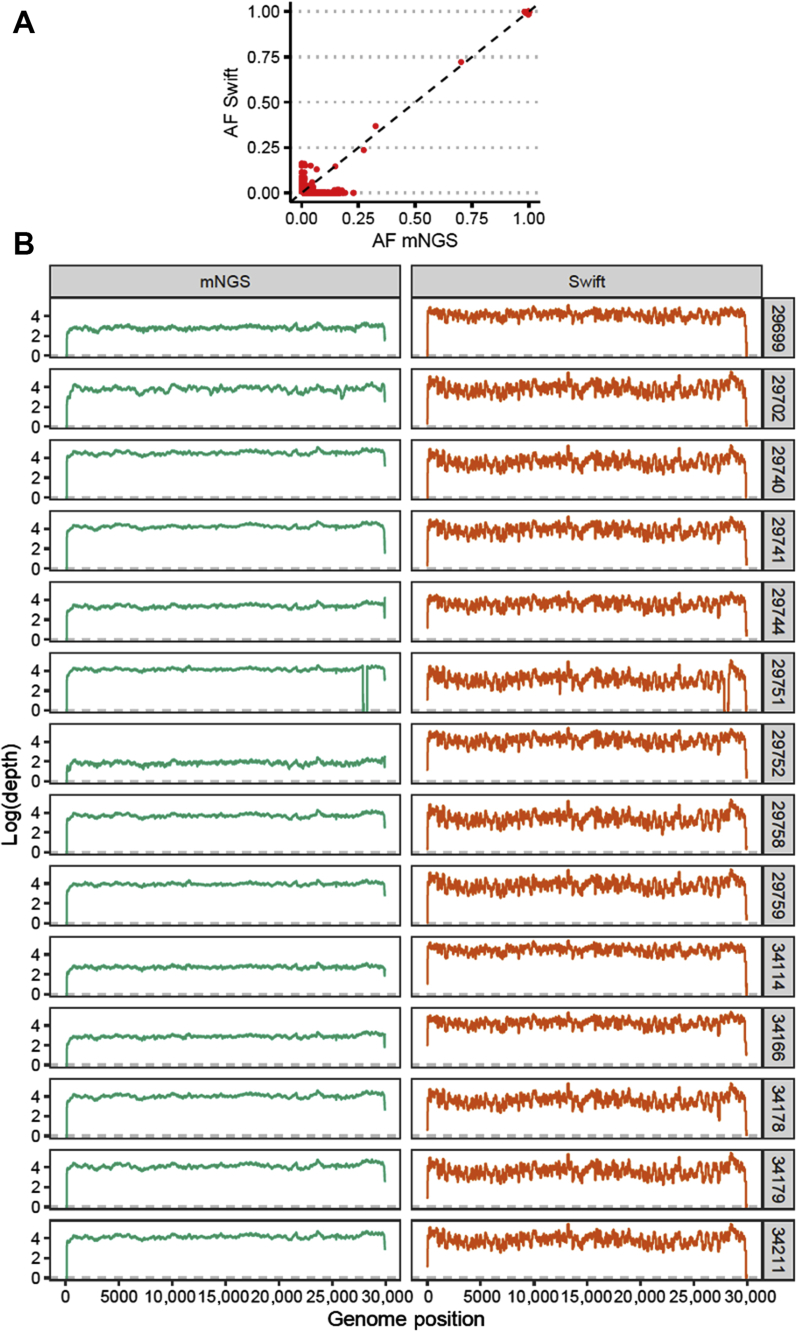
Shotgun mNGS was performed on 14 samples with CT ≤ 25, and consensus sequences were obtained for all 14 samples, with mean genome coverage of 4304× (SD, 9886×) and 0% to 1.3% Ns (Supplemental Table S8). Pairwise sequence identity between shotgun and Swift sequences was 100% for all 14 sample pairs. For all single-nucleotide variants identified at ≥1%, allele frequencies were highly concordant between Swift and mNGS libraries (R2 = 0.99; Pearson correlation coefficient = 0.99; P < 2.2 × 10−16) (Figure 2A). Of these variants, only 2 of 7421 (0.03%) had allele frequencies differing by >20% between library preparation methods, attributable to poor subgenomic RNA mapping in the Swift library or noisy ends of reads in the metagenomic library of one sample (WA-UW-29702). Across the genome, depth of coverage ranged from 0× to 370,627× for Swift and from 0× to 130,281× for mNGS (Figure 2B), with Pielou evenness >0.99 for both mNGS and Swift libraries. The mean whole-genome coverage in the Swift libraries (15,631×) was greater than in the mNGS libraries (9981×). Notably, in one sample (WA-UW-29751), a decrease in coverage at positions 27823 to 28233 in both Swift and mNGS samples, corresponding to a 411-nucleotide deletion in ORF7b/8 that has been found in other samples from Washington state, was observed (data not shown).
Figure 2.
Swift versus shotgun metagenomic next-generation sequencing (mNGS). A: Allele frequencies (AFs) are highly concordant between Swift and mNGS libraries. B: Comparison of depth of coverage (log10 reads) versus genomic position relative to NC_045512 for specimens sequenced with both Swift (right panels,orange) and mNGS (left panels,green). n = 14 samples (A).
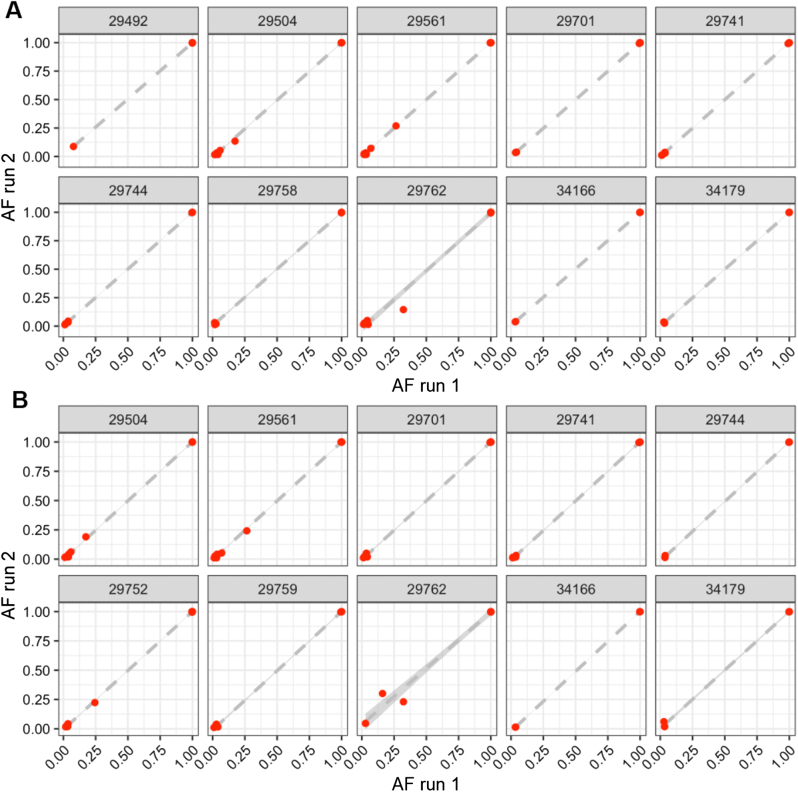
High Assay Precision (Repeatability and Reproducibility)
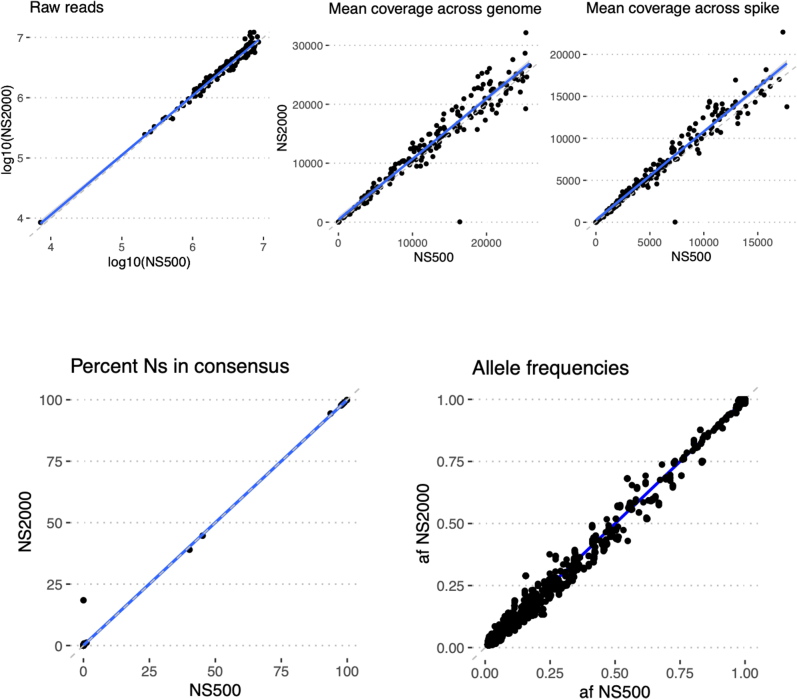
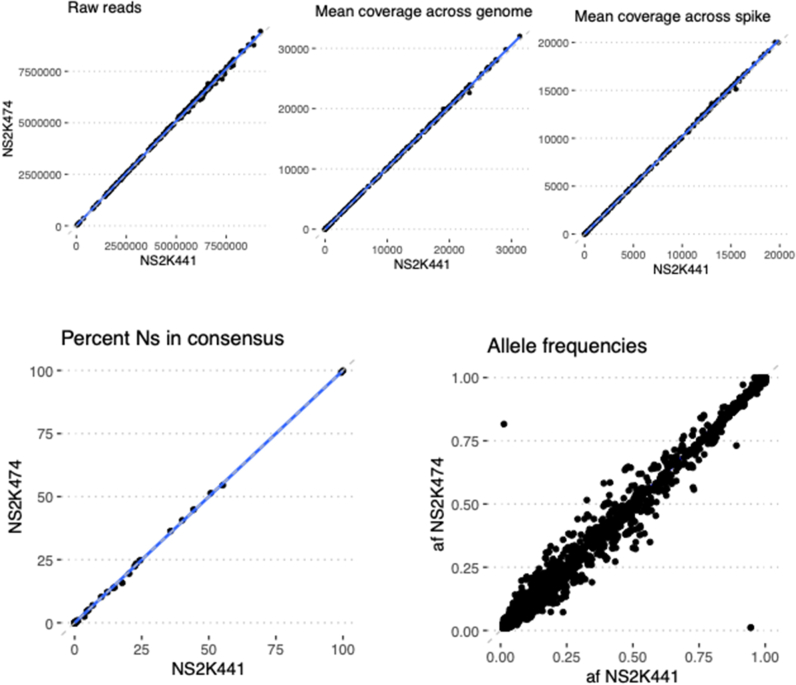
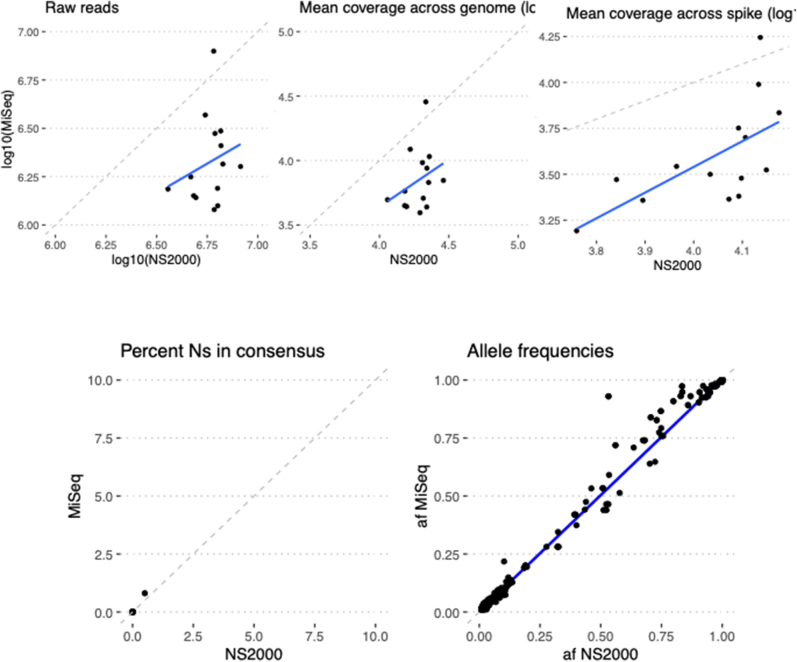
Precision was evaluated by testing the concordance of assay results and quality metrics for specimens tested multiple times on the same run (repeatability) and on different runs (reproducibility). For repeatability, two replicate libraries were prepared for 10 SARS-CoV-2–positive specimens on the same library preparation plate on the liquid handler and the same sequencing run. For reproducibility, 10 SARS-CoV-2–positive specimens had libraries prepared 3 days later and sequenced on a separate run by a different technician. All the replicates of the specimen on both the same run and different runs yielded acceptable genomes with highly correlated allele frequencies (Figure 3). Assay results were also highly reproducible across different instruments, including Illumina NextSeq 500, two separate NextSeq 2000s, and MiSeq (Supplemental Figures S6–S8).
Figure 3.
Highly reproducible allele frequencies (AFs) across replicate libraries of 12 SARS-CoV-2–positive specimens sequenced on the same run (A) and on different runs on a different day by a different technician (B). Dashed gray lines show lines of best fit by linear regression, and shaded gray bands represent 95% CIs for the linear fit.
Robust Pipeline Performance Using Benchmark Data Sets
The performance of our data processing pipeline was evaluated using CDC benchmark data sets of samples confirmed to be VOC/VOI (n = 16) and non-VOC/VOI (n = 39) sequences. The number of raw FASTQ reads ranged from 41,436 to 6,798,138 (median, 602,508) across both data sets (Supplemental Table S9). After processing through our bioinformatic pipeline, consensus genomes with <10% Ns were obtained for all samples, with mean genome coverage between 159× and 28,122× (median, 2598×), and mean spike gene coverage between 138× and 22,099× (median, 1954×). PANGO and Nextclade lineages were compared for consensus sequences generated by our pipeline against benchmark deposited consensus genomes. Lineages agreed in 16 of 16 VOC/VOI samples and 37 of 39 non-VOC/VOI samples. In the two non-VOC samples where the lineages did not agree, the discrepancy was found to be due to mixed populations being reported using International Union of Pure and Applied Chemistry ambiguities in the benchmark set versus the majority consensus base being reported by TAYLOR. After masking ambiguous sites, consensus sequences in these discrepant samples showed 100% pairwise identity.
Deployment of a High-Throughput NGS Assay in a Clinical Setting: The UW Virology Experience
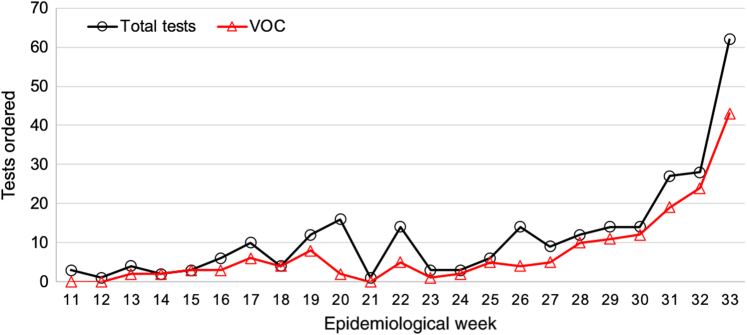
The UW Virology SARS-CoV-2 clinical sequencing assay went live on March 3, 2021, and in the first 23 weeks since launch, a total of 268 cases were processed (Figure 4). The median turnaround time, defined as order to first result, was 11 days. Tests were indicated for infection control (infections among patient-facing staff members), confirmation of patient-staff transmission, and cluster investigations. The diagnostic yield of VOCs identified in this period was 64% (171 samples). Among clinical orders, the first sample of B.1.617.2/Delta was detected in week 22.
Figure 4.
Weekly SARS-CoV-2 clinical whole-genome sequencing test volumes, along with the number of variants of concern (VOCs) detected in each week's batch.
Semi-annual proficiency testing was performed via a sample exchange with a peer Clinical Laboratory Improvement Amendments/College of American Pathologists–certified laboratory in which six blinded specimens were exchanged and sequenced. Unblinding was performed following the completion of the study to compare the strains identified and the spike gene mutations detected. Among proficiency testing samples exchanged, 100% of PANGO lineages and 98% of spike gene mutations were identified correctly.
Discussion
Tiling multiplex PCR amplicon panels are an attractive option for high-throughput sequencing of SARS-CoV-2 as they allow recovery of genomes from a wide range of viral loads with relatively short workflows. Herein, we performed systematic validation of the Swift SNAP protocol using US Food and Drug Administration guidelines for other NGS-based assays, to deploy the assay for routine clinical use in a Clinical Laboratory Improvement Amendments– and College of American Pathologists–approved facility. The Swift SNAP assay demonstrated high sensitivity, with recovery of full-length genomes at viral loads as low as 40.08 copies per reaction, with the ability to faithfully recover allele frequencies in mixed populations. The latter feature is of particular importance in quantifying within-host variation to track mutations with potential clinical significance following vaccination or treatment. Recent studies have suggested that amplicon sequencing provides a more accurate representation of minor allele frequencies than capture-based sequencing.31,50
The Swift SNAP panel produces short amplicons of <255 bp that can be directly sequenced with short-read technologies, like Illumina, without additional fragmentation. In contrast, the popular low-cost ARTIC panel generates amplicons of size approximately 400 bp that require fragmentation before sequencing on Illumina platforms or direct sequencing via Oxford Nanopore Technologies (Oxford, UK).51,52 In addition, the larger number of primers and overlapping amplicons in SNAP results in fewer amplicon dropouts compared with ARTIC-based methods.52
Given the wide range of viral loads among SARS-CoV-2 specimens, it can be challenging to optimize multiplex amplicon panels to simultaneously achieve high sensitivity and evenness of genome coverage. The manufacturer-recommended settings were found to be adequate for genome recovery across the range of viral loads we see in most clinical specimens. At low viral loads, the high background of nonviral RNA can make genome recovery challenging. In addition, the Swift SNAP protocol includes an optional proprietary enzymatic normalization (Normalase) step, which results in equimolar pools with even coverage across samples and amplicons. This allows for batching of samples with minimal sample reorganization. Although additional cycles of amplification may help stretch analytical sensitivity, this can result in an accumulation of primer dimers that can affect overall run quality and may amplify trace contaminants.52 Failure to generate high-quality genomes was generally due to low viral load in the original sample or, less frequently, due to pipetting errors by the liquid handler.
One of the limitations of short-read sequencing using reference-based approaches is the reduced ability to identify large structural variants, long insertions and deletions, and tandem duplicates. Long deletions often manifest as low-coverage regions masked with ambiguous bases (Ns) and require manual review to confirm that they are deletions versus amplicon dropouts. In the current study, we saw one example of this, and we have previously identified similar deletions in ORFs 7a, 7b, and 8 using Swift.53
As SARS-CoV-2 continues to spread globally, accumulating mutations across its genome, it is likely that some of these mutations impact the performance of amplicon panels. Our analysis of VOC/VOI samples shows that the Swift panel continues to perform well against currently circulating variants, albeit with reduced depth of coverage with Delta and Omicron variants. Although the redundancy offered by overlapping amplicons allows the Swift panel to tolerate some divergence, there may be a need to update or spike in primers in the future.
Taken together, our results demonstrate the high sensitivity, specificity, and reproducibility of the Swift SNAP amplicon panel for SARS-CoV-2, which make it ideal for clinical applications. Our protocol is available (https://www.protocols.io/view/uw-virology-swift-snapv2-protocol-byw4pxgw, last accessed October 18, 2021), with options for automation via robotic liquid handling systems. In addition, our study provides a framework for validating amplicon sequencing methods that have proved to be an important tool in our fight against coronavirus disease 2019 (COVID-19) and will be important for other emerging pathogens.
Acknowledgments
We thank Noah Hoffman and Niklas Krumm for guidance on cloud-based storage and analysis; the CDC Technical Outreach and Assistance for States Team for support with the CDC benchmark data sets; and Kanika Sharma (PerkinElmer) for automation help.
Footnotes
Supported by NIHAI027757 (P.R.) and NIHP30GM118228-04 (E.A.B.). Computational analyses were supported by Fred Hutch Scientific Computing (NIH Office of Research Infrastructure Programs grant S10OD028685) and University of Washington Laboratory Medicine Informatics.
A.L.G. and P.R. contributed equally to this work.
Disclosures: A.L.G. reports contract testing from Abbott Laboratories and research support from Gilead and Merck, outside of the described work. All other authors report no other financial or nonfinancial competing interests for this work.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2022.05.007.
Contributor Information
Alexander L. Greninger, Email: agrening@uw.edu.
Pavitra Roychoudhury, Email: proychou@uw.edu.
Supplemental Data
Supplemental Figure S1.
Summary of overall validation steps. 20A, WA-UW-20236 TM; 20B, WA-UW-19433 TM; SNAP, Swift Normalase Amplicon Panel.
Supplemental Figure S2.
A: TapeStation traces for clinical and Twist positive controls and two negative controls (water). Green and pink horizontal bars show lower and upper markers, respectively. B: Examples of electropherogram view of the controls with region view from 270 to 750 bp indicated with blue lines. The upper (right) and lower (left) markers in the High Sensitivity D1000 assay are labeled in each panel. C: Table with the total concentration and the region (270 to 750 bp) concentration of the library fragments for each sample represented in A. ID, identifier.
Supplemental Figure S3.
Sample report for clinical test orders. NCBI, National Center for Biotechnology Information; NGS, next-generation sequencing.
Supplemental Figure S4.
Mean genome coverage for samples sequenced for genomic surveillance shows reduced coverage for Delta and Omicron samples. VOC, variant of concern; VOI, variant of interest.
Supplemental Figure S5.
Gels showing Sanger amplicons for one representative sample (SpID 26865). Lanes from left to right show: amplicon 1 F1a/R1a, F1a/R1b, F1b/R1a, and F1b/R1b; amplicon 2 F2a/R2a, F2a/R2b, F2b/R2a, and F2b/R2b; blank lane; 3 μL of 100- to 3000-bp ladder; and 5 μL ladder (A); and amplicon 3 F3a/R3a, F3a/R3b, F3b/R3a, and F3b/R3b; two blank lanes; and 3 μL ladder (B). Note that the amplicon 1 and amplicon 2 samples from gel 1 are visible at the bottom of gel 2 because samples were run consecutively on the same gel. Most sequencing was performed on amplicons generated using primers F1a/R1b, F2a/R2a, and F3b/R3b.
Supplemental Figure S6.
Instrument validation: identical libraries were sequenced in parallel on the Illumina NextSeq 500 and an Illumina NextSeq 2000 (VH00441). Blue lines show lines of best fit by linear regression, and dashed lines show y = x (perfect concordance). n = 181 identical libraries. Af, allele frequency; Ns, unknown bases.
Supplemental Figure S7.
Instrument validation: identical libraries were sequenced in parallel on two different Illumina NextSeq 2000 instruments (VH00441/NS2K441 versus VH00474/NS2K474). Blue lines show lines of best fit by linear regression, and dashed lines show y = x (perfect concordance). n = 185 identical libraries. Af, allele frequency; Ns, unknown bases.
Supplemental Figure S8.
Instrument validation: identical libraries were sequenced in parallel on an Illumina NextSeq 2000 and an Illumina MiSeq. Blue lines show lines of best fit by linear regression, and dashed lines show y = x (perfect concordance). n = 12 identical libraries. Af, allele frequency; Ns, unknown bases.
References
- 1.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moustafa A.M., Planet P.J. Jumping a moving train: SARS-CoV-2 evolution in real time. J Pediatr Infect Dis Soc. 2021;10:S96–S105. doi: 10.1093/jpids/piab051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paden C.R., Tao Y., Queen K., Zhang J., Li Y., Uehara A., Tong S. Rapid, sensitive, full-genome sequencing of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26:2401–2405. doi: 10.3201/eid2610.201800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fauver J.R., Petrone M.E., Hodcroft E.B., Shioda K., Ehrlich H.Y., Watts A.G., et al. Coast-to-coast spread of SARS-CoV-2 during the early epidemic in the United States. Cell. 2020;181:990–996.e5. doi: 10.1016/j.cell.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng X., Gu W., Federman S., du Plessis L., Pybus O.G., Faria N.R., et al. Genomic surveillance reveals multiple introductions of SARS-CoV-2 into Northern California. Science. 2020;369:582–587. doi: 10.1126/science.abb9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bedford T., Greninger A.L., Roychoudhury P., Starita L.M., Famulare M., Huang M.-L., et al. Cryptic transmission of SARS-CoV-2 in Washington state. Science. 2020;370:571–575. doi: 10.1126/science.abc0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield COVID-19 Genomics Group. Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang J.W., Toovey O.T.R., Harvey K.N., Hui D.D.S. Introduction of the South African SARS-CoV-2 variant 501Y.V2 into the UK. J Infect. 2021;82:e8–e10. doi: 10.1016/j.jinf.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Washington N.L., Gangavarapu K., Zeller M., Bolze A., Cirulli E.T., Barrett K.M.S., et al. Genomic epidemiology identifies emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. medRxiv. 2021 doi: 10.1101/2021.02.06.21251159. [Preprint] doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lieberman N.A.P., Peddu V., Xie H., Shrestha L., Huang M.-L., Mears M.C., Cajimat M.N., Bente D.A., Shi P.-Y., Bovier F., Roychoudhury P., Jerome K.R., Moscona A., Porotto M., Greninger A.L. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLOS Biol. 2020;18 doi: 10.1371/journal.pbio.3000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toyoshima Y., Nemoto K., Matsumoto S., Nakamura Y., Kiyotani K. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J Hum Genet. 2020;65:1075–1082. doi: 10.1038/s10038-020-0808-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. N Engl J Med. 2021;384:1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez Bernal J., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., Groves N., Dabrera G., Myers R., Campbell C.N.J., Amirthalingam G., Edmunds M., Zambon M., Brown K.E., Hopkins S., Chand M., Ramsay M. Effectiveness of Covid-19 vaccines against the B.1.617.2 (Delta) variant. N Engl J Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chemaitelly H., Yassine H.M., Benslimane F.M., al Khatib H.A., Tang P., Hasan M.R., Malek J.A., Coyle P., Ayoub H.H., al Kanaani Z., al Kuwari E., Jeremijenko A., Kaleeckal A.H., Latif A.N., Shaik R.M., Abdul Rahim H.F., Nasrallah G.K., al Kuwari M.G., al Romaihi H.E., Al-Thani M.H., al Khal A., Butt A.A., Bertollini R., Abu-Raddad L.J. mRNA-1273 COVID-19 vaccine effectiveness against the B.1.1.7 and B.1.351 variants and severe COVID-19 disease in Qatar. Nat Med. 2021;27:1614–1621. doi: 10.1038/s41591-021-01446-y. [DOI] [PubMed] [Google Scholar]
- 15.Nasreen S., Chung H., He S., Brown K.A., Gubbay J.B., Buchan S.A., Fell D.B., Austin P.C., Schwartz K.L., Sundaram M.E., Calzavara A., Chen B., Tadrous M., Wilson K., Wilson S.E., Kwong J.C., Investigators on behalf of the CIRN (CIRN) PCN (PCN) Effectiveness of COVID-19 vaccines against variants of concern in Ontario, Canada. medRxiv. 2021 doi: 10.1101/2021.06.28.21259420. [Preprint] doi: [DOI] [Google Scholar]
- 16.Fajnzylber J., Regan J., Coxen K., Corry H., Wong C., Rosenthal A., Worrall D., Giguel F., Piechocka-Trocha A., Atyeo C., Fischinger S., Chan A., Flaherty K.T., Hall K., Dougan M., Ryan E.T., Gillespie E., Chishti R., Li Y., Jilg N., Hanidziar D., Baron R.M., Baden L., Tsibris A.M., Armstrong K.A., Kuritzkes D.R., Alter G., Walker B.D., Yu X., Li J.Z., Massachusetts Consortium for Pathogen Readiness SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun. 2020;11:5493. doi: 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hung I.F.N., Cheng V.C.C., Wu A.K.L., Tang B.S.F., Chan K.H., Chu C.M., Wong M.M.L., Hui W.T., Poon L.L.M., Tse D.M.W., Chan K.S., Woo P.C.Y., Lau S.K.P., Peiris J.S.M., Yuen K.Y. Viral loads in clinical specimens and SARS manifestations. Emerg Infect Dis. 2004;10:1550–1557. doi: 10.3201/eid1009.040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies N.G., Jarvis C.I., CMMID COVID-19 Working Group. Edmunds W.J., Jewell N.P., Diaz-OrdazKarla, Keogh R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593:270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Challen R., Brooks-Pollock E., Read J.M., Dyson L., Tsaneva-Atanasova K., Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ. 2021;372:n579. doi: 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frampton D., Rampling T., Cross A., Bailey H., Heaney J., Byott M., Scott R., Sconza R., Price J., Margaritis M., Bergstrom M., Spyer M.J., Miralhes P.B., Grant P., Kirk S., Valerio C., Mangera Z., Prabhahar T., Moreno-Cuesta J., Arulkumaran N., Singer M., Shin G.Y., Sanchez E., Paraskevopoulou S.M., Pillay D., McKendry R.A., Mirfenderesky M., Houlihan C.F., Nastouli E. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. Lancet Infect Dis. 2021;21:1246–1256. doi: 10.1016/S1473-3099(21)00170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bager P., Wohlfahrt J., Fonager J., Rasmussen M., Albertsen M., Michaelsen T.Y., Møller C.H., Ethelberg S., Legarth R., Button M.S.F., Gubbels S., Voldstedlund M., Mølbak K., Skov R.L., Fomsgaard A., Krause T.G. Risk of hospitalisation associated with infection with SARS-CoV-2 lineage B.1.1.7 in Denmark: an observational cohort study. Lancet Infect Dis. 2021;21:1507–1517. doi: 10.1016/S1473-3099(21)00290-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patone M., Thomas K., Hatch R., Tan P.S., Coupland C., Liao W., Mouncey P., Harrison D., Rowan K., Horby P., Watkinson P., Hippisley-Cox J. Mortality and critical care unit admission associated with the SARS-CoV-2 lineage B.1.1.7 in England: an observational cohort study. Lancet Infect Dis. 2021;21:1518–1528. doi: 10.1016/S1473-3099(21)00318-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paredes M.I., Lunn S.M., Famulare M., Frisbie L.A., Painter I., Burstein R., et al. Associations between severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants and risk of coronavirus disease 2019 (COVID-19) hospitalization among confirmed cases in Washington state: a retrospective cohort study. Clin Infect Dis. 2022 doi: 10.1093/cid/ciac279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golubchik T., Lythgoe K.A., Hall M., Ferretti L., Fryer H.R., MacIntyre-Cockett G., de Cesare M., Trebes A., Piazza P., Buck D., Todd J.A., The COVID-19 Genomics UK (COG-UK) Consortium. Fraser C., Bonsall D. Early analysis of a potential link between viral load and the N501Y mutation in the SARS-COV-2 spike protein. medRxiv. 2021 doi: 10.1101/2021.01.12.20249080. [Preprint] doi: [DOI] [Google Scholar]
- 25.Funk T., Pharris A., Spiteri G., Bundle N., Melidou A., Carr M., et al. Characteristics of SARS-CoV-2 variants of concern B.1.1.7, B.1.351 or P.1: data from seven EU/EEA countries, weeks 38/2020 to 10/2021. Euro Surveill. 2021;26:2100348. doi: 10.2807/1560-7917.ES.2021.26.16.2100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greaney A.J., Starr T.N., Barnes C.O., Weisblum Y., Schmidt F., Caskey M., Gaebler C., Cho A., Agudelo M., Finkin S., Wang Z., Poston D., Muecksch F., Hatziioannou T., Bieniasz P.D., Robbiani D.F., Nussenzweig M.C., Bjorkman P.J., Bloom J.D. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat Commun. 2021;12:4196. doi: 10.1038/s41467-021-24435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S., Mascola J.R., Chang J.Y., Yin M.T., Sobieszczyk M., Kyratsous C.A., Shapiro L., Sheng Z., Huang Y., Ho D.D. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 28.Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., Gilchuk P., Zost S.J., Tahan S., Droit L., Turner J.S., Kim W., Schmitz A.J., Thapa M., Wang D., Boon A.C.M., Presti R.M., O'Halloran J.A., Kim A.H.J., Deepak P., Pinto D., Fremont D.H., Crowe J.E., Corti D., Virgin H.W., Ellebedy A.H., Shi P.-Y., Diamond M.S. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z., VanBlargan L.A., Bloyet L.-M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J., Alford B., Buchser W.J., Ellebedy A.H., Fremont D.H., Diamond M.S., Whelan S.P.J. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488.e4. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., Solomon I.H., Kuo H.-H., Boucau J., Bowman K., Adhikari U.D., Winkler M.L., Mueller A.A., Hsu T.Y.-T., Desjardins M., Baden L.R., Chan B.T., Walker B.D., Lichterfeld M., Brigl M., Kwon D.S., Kanjilal S., Richardson E.T., Jonsson A.H., Alter G., Barczak A.K., Hanage W.P., Yu X.G., Gaiha G.D., Seaman M.S., Cernadas M., Li J.Z. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao M., Liu X., Ji J., Li M., Li J., Yang L., Sun W., Ren P., Yang G., Zhao J., Liang T., Ren H., Chen T., Zhong H., Song W., Wang Y., Deng Z., Zhao Y., Ou Z., Wang D., Cai J., Cheng X., Feng T., Wu H., Gong Y., Yang H., Wang J., Xu X., Zhu S., Chen F., Zhang Y., Chen W., Li Y., Li J. Multiple approaches for massively parallel sequencing of SARS-CoV-2 genomes directly from clinical samples. Genome Med. 2020;12:57. doi: 10.1186/s13073-020-00751-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charre C., Ginevra C., Sabatier M., Regue H., Destras G., Brun S., Burfin G., Scholtes C., Morfin F., Valette M., Lina B., Bal A., Josset L. Evaluation of NGS-based approaches for SARS-CoV-2 whole genome characterisation. Virus Evol. 2020;6 doi: 10.1093/ve/veaa075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Addetia A., Lin M.J., Peddu V., Roychoudhury P., Jerome K.R., Greninger A.L. Sensitive recovery of complete SARS-CoV-2 genomes from clinical samples by use of Swift Biosciences’ SARS-CoV-2 multiplex amplicon sequencing panel. J Clin Microbiol. 2020;59:e02226-20. doi: 10.1128/JCM.02226-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sadoff J., le Gars M., Shukarev G., Heerwegh D., Truyers C., de Groot A.M., Stoop J., Tete S., van Damme W., Leroux-Roels I., Berghmans P.-J., Kimmel M., van Damme P., de Hoon J., Smith W., Stephenson K.E., de Rosa S.C., Cohen K.W., McElrath M.J., Cormier E., Scheper G., Barouch D.H., Hendriks J., Struyf F., Douoguih M., van Hoof J., Schuitemaker H. Interim results of a phase 1–2a trial of Ad26.COV2.S Covid-19 vaccine. N Engl J Med. 2021;384:1824–1835. doi: 10.1056/NEJMoa2034201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sadoff J., Gray G., Vandebosch A., Cárdenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Fennema H., Spiessens B., Offergeld K., Scheper G., Taylor K.L., Robb M.L., Treanor J., Barouch D.H., Stoddard J., Ryser M.F., Marovich M.A., Neuzil K.M., Corey L., Cauwenberghs N., Tanner T., Hardt K., Ruiz-Guiñazú J., le Gars M., Schuitemaker H., van Hoof J., Struyf F., Douoguih M. Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N Engl J Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hilt E.E., Boocock J., Trejo M., Le C.Q., Guo L., Zhang Y., Sathe L., Arboleda V.A., Yin Y., Bloom J.S., Wang P.-C., Elmore J.G., Kruglyak L., Shrestha L., Bakhash S.A.M., Lin M., Xie H., Huang M.-L., Roychoudhury P., Greninger A., Chandrasekaran S., Yang S., Garner O.B. Retrospective detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in symptomatic patients prior to widespread diagnostic testing in southern California. Clin Infect Dis. 2022;74:271–277. doi: 10.1093/cid/ciab360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McEwen A.E., Cohen S., Bryson-Cahn C., Liu C., Pergam S.A., Lynch J., Schippers A., Strand K., Whimbey E., Mani N.S., Zelikoff A.J., Makarewicz V.A., Brown E.R., Bakhash S.A.M., Baker N.R., Castor J., Livingston R.J., Huang M.-L., Jerome K.R., Greninger A.L., Roychoudhury P. Variants of concern are overrepresented among postvaccination breakthrough infections of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in Washington state. Clin Infect Dis. 2022;74:1089–1092. doi: 10.1093/cid/ciab581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin M.J., Rachleff V.M., Xie H., Shrestha L., Lieberman N.A.P., Peddu V., Addetia A., Casto A.M., Breit N., Mathias P.C., Huang M.-L., Jerome K.R., Greninger A.L., Roychoudhury P. Host–pathogen dynamics in longitudinal clinical specimens from patients with COVID-19. Sci Rep. 2022;12:5856. doi: 10.1038/s41598-022-09752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lieberman J.A., Pepper G., Naccache S.N., Huang M.-L., Jerome K.R., Greninger A.L. Comparison of commercially available and laboratory-developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. J Clin Microbiol. 2021;58:e00821-20. doi: 10.1128/JCM.00821-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Degli-Angeli E., Dragavon J., Huang M.-L., Lucic D., Cloherty G., Jerome K.R., Greninger A.L., Coombs R.W. Validation and verification of the Abbott RealTime SARS-CoV-2 assay analytical and clinical performance. J Clin Virol. 2020;129:104474. doi: 10.1016/j.jcv.2020.104474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuypers J., Wright N., Ferrenberg J., Huang M.-L., Cent A., Corey L., Morrow R. Comparison of real-time PCR assays with fluorescent-antibody assays for diagnosis of respiratory virus infections in children. J Clin Microbiol. 2006;44:2382–2388. doi: 10.1128/JCM.00216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bruce E.A., Mills M.G., Sampoleo R., Perchetti G.A., Huang M.-L., Despres H.W., Shirley D.J., Jerome K.R., Greninger A.L., Botten J.W. Predicting infectivity: comparing four PCR-based assays to detect culturable SARS-CoV-2 in clinical samples. medRxiv. 2021 doi: 10.1101/2021.07.14.21260544. [Preprint] doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graham N.R., Whitaker A.N., Strother C.A., Miles A.K., Grier D., McElvany B.D., Bruce E.A., Poynter M.E., Pierce K.K., Kirkpatrick B.D., Stapleton R.D., An G., van den Broek-Altenburg E., Botten J.W., Crothers J.W., Diehl S.A. Kinetics and isotype assessment of antibodies targeting the spike protein receptor-binding domain of severe acute respiratory syndrome-coronavirus-2 in COVID-19 patients as a function of age, biological sex and disease severity. Clin Transl Immunol. 2020;9:e1189. doi: 10.1002/cti2.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nalla A.K., Casto A.M., Huang M.-L.W., Perchetti G.A., Sampoleo R., Shrestha L., Wei Y., Zhu H., Jerome K.R., Greninger A.L. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peddu V., Shean R.C., Xie H., Shrestha L., Perchetti G.A., Minot S.S., Roychoudhury P., Huang M.-L., Nalla A., Reddy S.B., Phung Q., Reinhardt A., Jerome K.R., Greninger A.L. Metagenomic analysis reveals clinical SARS-CoV-2 infection and bacterial or viral superinfection and colonization. Clin Chem. 2020;66:966–972. doi: 10.1093/clinchem/hvaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greninger A.L., Zerr D.M., Qin X., Adler A.L., Sampoleo R., Kuypers J.M., Englund J.A., Jerome K.R. Rapid metagenomic next-generation sequencing during an investigation of hospital-acquired human parainfluenza virus 3 infections. J Clin Microbiol. 2017;55:177–182. doi: 10.1128/JCM.01881-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin M.J., Rachleff V.M., Xie H., Shrestha L., Lieberman N.A.P., Peddu V., Addetia A., Casto A.M., Breit N., Mathias P.C., Huang M.-L., Jerome K.R., Greninger A.L., Roychoudhury P. Host-pathogen dynamics in longitudinal clinical specimens from patients with COVID-19. medRxiv. 2021 doi: 10.1101/2021.04.27.21256149. [Preprint] doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., Li H. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10 doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burd E.M. Validation of laboratory-developed molecular assays for infectious diseases. Clin Microbiol Rev. 2010;23:550–576. doi: 10.1128/CMR.00074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chiara M., D’Erchia A.M., Gissi C., Manzari C., Parisi A., Resta N., Zambelli F., Picardi E., Pavesi G., Horner D.S., Pesole G. Next generation sequencing of SARS-CoV-2 genomes: challenges, applications and opportunities. Brief Bioinform. 2021;22:616–630. doi: 10.1093/bib/bbaa297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tyson J.R., James P., Stoddart D., Sparks N., Wickenhagen A., Hall G., Choi J.H., Lapointe H., Kamelian K., Smith A.D., Prystajecky N., Goodfellow I., Wilson S.J., Harrigan R., Snutch T.P., Loman N.J., Quick J. Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. bioRxiv. 2020 doi: 10.1101/2020.09.04.283077. [Preprint] doi: [DOI] [Google Scholar]
- 52.Itokawa K., Sekizuka T., Hashino M., Tanaka R., Kuroda M. Disentangling primer interactions improves SARS-CoV-2 genome sequencing by multiplex tiling PCR. PLoS One. 2020;15:e0239403. doi: 10.1371/journal.pone.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Addetia A., Xie H., Roychoudhury P., Shrestha L., Loprieno M., Huang M.-L., Jerome K.R., Greninger A.L. Identification of multiple large deletions in ORF7a resulting in in-frame gene fusions in clinical SARS-CoV-2 isolates. J Clin Virol. 2020;129:104523. doi: 10.1016/j.jcv.2020.104523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.