Abstract
Patients with COVID-19 have high prevalence of albuminuria which is used as a marker of progression of renal disease and is associated with severe COVID-19. We hypothesized that SARS-CoV-2 spike protein (S protein) could modulate albumin handling in proximal tubule epithelial cells (PTECs) and, consequently contribute to the albuminuria observed in patients with COVID-19. In this context, the possible effect of S protein on albumin endocytosis in PTECs was investigated. Two PTEC lines were used: HEK-293A and LLC-PK1. Incubation of both cell types with S protein for 16 h inhibited albumin uptake at the same magnitude. This effect was associated with canonical megalin-mediated albumin endocytosis because: (1) DQ-albumin uptake, a marker of the lysosomal degradation pathway, was reduced at a similar level compared with fluorescein isothiocyanate (FITC)-albumin uptake; (2) dextran-FITC uptake, a marker of fluid-phase endocytosis, was not changed; (3) cell viability and proliferation were not changed. The inhibitory effect of S protein on albumin uptake was only observed when it was added at the luminal membrane, and it did not involve the ACE2/Ang II/AT1R axis. Although both cells uptake S protein, it does not seem to be required for modulation of albumin endocytosis. The mechanism underlying the inhibition of albumin uptake by S protein encompasses a decrease in megalin expression without changes in megalin trafficking and stability. These results reveal a possible mechanism to explain the albuminuria observed in patients with COVID-19.
Keywords: Proximal tubule, Albumin endocytosis, Spike protein, SARS-CoV-2, Megalin, COVID-19
1. Introduction
Severe COVID-19 involves multiple organ damage (MOD), including kidney damage [1], [2]. Some studies have reported a correlation between acute kidney injury (AKI) and worse outcome in patients with COVID-19 [3], [4], [5], with increased morbidity and mortality [6]. However, this scenario could be worse because some patients with COVID-19 present silent renal injury (subclinical acute kidney injury [subAKI]) characterized by tubular damage without changes in the estimated glomerular flow rate (eGFR) [3], [4]. Accordingly, a higher prevalence of albuminuria in patients with COVID-19 has been observed, even in those who do not develop AKI [7]. Albuminuria is a well-known marker of progression of renal disease and is associated with severe COVID-19 [3]. To date, the mechanism underlying albuminuria in patients with COVID-19 requires elucidation. In this context, its identification could open new perspectives to understand the pathogenesis of kidney damage in patients with COVID-19.
One important clue is provided by the observation that some patients with COVID-19 develop Fanconi-like syndrome characterized by proximal tubule (PT) injury associated with low-molecular-weight proteinuria and albuminuria. This syndrome encompasses changes in PT protein endocytic machinery [8], [9]. Protein reabsorption in proximal tubule epithelial cells (PTECs) occurs mainly via a canonical receptor-mediated endocytosis where the receptor is formed by the assembly of three proteins: megalin, cubilin and amnionless (AMN) [10]. Megalin (LRP2) is a scavenger receptor that belongs to the LDL-like receptor family of proteins [11]. It works in association with the complex cubilin and amnionless (CUBAM) to promote protein reabsorption in PTECs [10], [11]. Megalin plays a crucial role in the internalization of this complex with protein targeted to lysosome degradation. Kidney biopsy samples from patients with COVID-19 revealed a decrease in megalin expression in PTECs without changes in aquaporin 1 (AQ1) [8]. These observations suggest that megalin-mediated albumin reabsorption in PTECs could be involved in the albuminuria observed in COVID-19.
SARS-CoV-2 infection initially involves engagement of the SARS-CoV-2 spike protein (S protein) to angiotensin converting enzyme type 2 (ACE2) present at the cell surface of diverse cell types [12]. However, the mechanism of COVID-19 pathogenesis could be more complex than has been imagined so far. One attractive hypothesis is the possible role of different proteins of SARS-CoV-2 in this process. Some authors have proposed that S protein, a structural glycoprotein, could also play an important role in the pathogenesis of COVID-19 [13], [14], [15], [16]. It was observed that isolated S protein modulates cellular function in different cell types such as monocytes and endothelial and heart pericytes [14], [15], [16]. In addition, S protein was found in the urine of patients with COVID-19, indicating its presence in the luminal side of PTECs and suggesting a possible effect on modulation of the function of PTECs [17].
Based on these observations, we hypothesized that isolated S protein could modulate albumin handling in PTECs and, consequently contribute to the albuminuria observed in patients with COVID-19. In the present study, we investigated the possible effect of S protein on albumin endocytosis in PTECs. LLC-PK1, a porcine PTEC cell line, and HEK-293, a human embryonic kidney cell line, were used. S protein inhibited megalin-mediated albumin endocytosis in both cell types. This inhibitory effect was associated with a decrease in megalin expression.
2. Methods
2.1. Materials and reagents
LLC-PK1 and HEK-293A cells were obtained from ATCC (Manassas, VA, USA). Thincert cell culture inserts for 12-well plates (0.4-μm diameter) were obtained from GreinerBio-One GmbH (Kremsmünster, Upper Austria, Austria). Bovine serum albumin (BSA), fluorescein isothiocyanate (FITC)-conjugated BSA (BSA-FITC), FITC-dextran and cycloheximide were purchased from Sigma-Aldrich (St Louis, MO, USA). Low- or high-glucose Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), antibiotic-antimycotic (#15240062), DQ green BSA, polyclonal Alexa-488 conjugated anti-rabbit IgG antibody (A32731), polyclonal Alexa Fluor 594-conjugated anti-rabbit IgG antibody (A32754), polyclonal Alexa-488 conjugated anti-mouse IgG antibody (A32723), polyclonal Alexa Fluor 594-conjugated anti-mouse IgG antibody A32744) were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Monoclonal mouse anti-HA antibody (HA.11) was purchased from Covance (Princeton, NJ, USA). Polyclonal rabbit anti-megalin (ab76969), monoclonal rabbit anti-ACE2 (ab108252) and monoclonal mouse anti-aquaporin 1 (AQP1) (ab9566) antibodies were purchased from Abcam (Boston, MA, USA). Monoclonal mouse anti-SARS-CoV/SARS-CoV-2 (COVID-19) spike antibody (GTX632604) was purchased from Genetex (Irvine, CA, USA). Monoclonal rabbit anti-LAMP1 (9091), polyclonal rabbit anti-EEA1 (#2411), monoclonal rabbit anti-β-actin (4970), HRP-conjugated anti-mouse IgG (7076) and HRP-conjugated anti-rabbit IgG (7074) antibodies were obtained from Cell Signaling Tech. (Danvers, MA, USA). VECTASHIELD antifade mounting medium was obtained from Vector Laboratories (Burlingame, CA, USA). Polyvinylidene difluoride (PVDF) membranes were obtained from Merck KGaA (Darmstadt, Germany). All other reagents were purchased from Sigma-Aldrich.
2.2. Isolation of SARS-COV-2 spike protein
Recombinant Wuhan S protein with the same sequence as previously published by Wrapp et al. [18] was kindly provided by Dr. Leda Castillo of Cell Culture and Engineering Laboratory at Federal University of Rio de Janeiro. Briefly, it was produced by heterologous expression in HEK-293-3F6 cells and purified using affinity chromatography as described by Cunha et al. [19].
2.3. Culture of kidney cell lines
LLC-PK1 cells, a well-characterized porcine PT cell line, were cultured in low-glucose DMEM supplemented with 10 % heat-inactivated FBS and 1 % antibiotic-antimycotic at 37 °C in 5 % CO2 [20]. HEK-293A cells, a parental human embryonic kidney cell line, were cultured in high-glucose DMEM supplemented with 10 % heat-inactivated FBS and 1 % antibiotic-antimycotic at 37 °C in 5 % CO2 [21]. When indicated, cells were grown in Thincert cell culture inserts (0.4-μm diameter), 13-mm-diameter round glass coverslips or culture dishes. After reaching the indicated confluence, cells were incubated with 5 μg/mL S protein and/or different compounds as indicated in the figure legends. After treatment, the cells were used in different experimental assays.
2.4. Plasmid and siRNA transfection
LLC-PK1 cells were stably transfected with MegT0-HA plasmid, which contains a truncated form of megalin that lacks the extracellular domain and is tagged with hemagglutinin (HA), as previously described [20]. Porcine Megalin siRNA (BIAGB-000005) and siGENOME control pool non-targeting (D-001206-13-05) were purchased from Dharmacon (Lafayette, CO, USA). LLC-PK1 cells were transfected with lipofectamine 2000 reagent (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer's instructions. For stable transfection with MegT0-HA, transfected cells were selected using 1.0 mg/mL G418 (Sigma-Aldrich, St. Louis, MO, USA). After 9 days, cells were maintained at 0.5 mg/mL G418 and used for the subsequent experiments. For siRNA experiments, cells were used after 72 h of transfection.
2.5. Measurement of receptor-mediated albumin endocytosis, fluid-phase albumin endocytosis and albumin lysosomal degradation
BSA-FITC, dextran-FITC or DQ-albumin were used as markers to assess receptor-mediated albumin endocytosis, fluid-phase albumin endocytosis and lysosomal albumin degradation, respectively [20]. Cells were incubated with 100 μg/mL of these compounds for 30 min (BSA-FITC, dextran-FITC) or 60 min (DQ-albumin) at 37 °C. Blank conditions were obtained in cells co-incubated with 100 mg/mL non-labeled BSA or in the absence of dextran-FITC. Non-internalized tracers were removed by washing the cells using ice-cold Ringer's solution (20 mM HEPES-Tris [pH 7.4] containing 140 mM NaCl, 2.7 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2 and 5 mM D(+)-glucose) at least 11 times. Cells were incubated with ice-cold lysis buffer (0.1 % Triton X-100 diluted in 20 mM MOPS-Tris buffer [pH 7.4]). The samples were harvested and the cell-associated fluorescence intensity was determined by fluorimetry. Relative fluorescence units (excitation = 480 nm, emission = 520 nm) were obtained using a microplate spectrofluorimeter (SpectraMax M5, Molecular Devices, San Jose, CA, USA). Protein concentration was evaluated by the Folin phenol method [22]. The specific measurements were calculated as arbitrary units: (relative fluorescence units of the sample − relative fluorescence units of the blank) and normalized by the total protein concentration.
LLC-PK1 and HEK-293A cells were cultured in glass coverslips for measurements using fluorescence microscopy. Nuclei were stained with DAPI and the coverslips were mounted using VECTASHIELD antifade mounting medium. The cell images were acquired using a Nikon80i epifluorescence microscope (Nikon Corporation, Tokyo, Japan) and cellular fluorescence intensity was determined using Fiji software. Specific BSA-FITC uptake was calculated as arbitrary units: (fluorescence units of the sample − fluorescence units of the blank) / cell number (counted by DAPI+ nuclei). Data were expressed as relative units (compared with control conditions).
2.6. Immunofluorescence and confocal analysis
Immunofluorescence and confocal analysis were carried out as reported previously [18]. Cells were fixed with 4 % paraformaldehyde for 15 min, permeabilized with 0.05 % saponin in PBS for 15 min, and non-specific binding sites blocked with 5 % BSA in PBS for 1 h at room temperature. Primary antibodies were incubated for 1 h at room temperature following the manufacturer's instructions. The cells were then washed with PBS and the secondary antibodies were incubated for 1 h at room temperature following the manufacturer's instructions. When indicated, the nuclei were stained with DAPI. Samples were mounted with VECTASHIELD antifade mounting medium and cell images were acquired and analyzed in a confocal microscope (Leica TCS SP8; Leica, Wetzlar, Germany). Image processing and analysis was performed with Fiji software. The colocalization rate was determined by the Pearson coefficient using the Coloc2 plugin (Fiji software).
2.7. Megalin expression determined by immunofluorescence
Total megalin expression was measured by immunofluorescence as described previously [23]. LLC-PK1 and HEK-293A cells were grown in coverslips until 60 %–70 % confluence was reached. Cells were incubated with 1:100 polyclonal rabbit anti-megalin antibody following the manufacturer's instructions. After washing out, the cells were incubated with 1:200 polyclonal Alexa-488 conjugated anti-rabbit IgG antibody. The levels of megalin expression were determined by the fluorescence intensity per cell (counted by DAPI+ nuclei).
2.8. Assessment of LLC-PK1 cell polarity
Cells were grown in Thincert cell culture inserts and allowed to grow until 90 % confluence was reached. Then, the inserts were recovered and mounted to assess luminal megalin expression, used to determine the cell polarity, through confocal microscopy. Intensity plots (gray values) show the distribution of megalin expression across the cell height. Data are presented as relative units.
2.9. Colocalization between S protein and ACE2
LLC-PK1 and HEK-293A cells were grown in coverslips until reach 60 %–70 % confluence was reached. Cells were incubated with 1:100 monoclonal mouse anti-S protein and 1:100 monoclonal rabbit anti-ACE2 antibodies. After washing out, the cells were incubated with 1:200 polyclonal Alexa Fluor 488-conjugated anti-mouse IgG and 1:200 polyclonal Alexa Fluor 594-conjugated anti-rabbit IgG antibodies. Coverslips were mounted, and the cell images were analyzed by confocal microscopy. Data are presented as the Pearson coefficient obtained per cell.
2.10. Assessment of cellular megalin trafficking
The assessment of cellular megalin trafficking was performed as published previously [23]. Initially, LLC-PK1 cells, stably transfected with megT0-HA, were seeded and grown in coverslips until reach 60 %–70 % confluence was reached. Colocalization between megT0-HA with EEA1 (a marker of early endosomes) or LAMP1 (a marker of lysosomes) was assessed to assess megalin trafficking to early endosomes or lysosomes, respectively. Cells were incubated for 1 h with 1:100 monoclonal mouse anti-HA and 1:50 polyclonal rabbit anti-EEA1 or monoclonal rabbit anti-LAMP1 antibodies simultaneously. Polyclonal Alexa Fluor 594-conjugated anti-mouse IgG (dilution 1:200) and polyclonal Alexa Fluor 488-conjugated anti-rabbit IgG (dilution 1:100) antibodies were used as secondary antibodies. Coverslips were mounted and the cell images were analyzed by confocal microscopy. Data are presented as the Pearson coefficient obtained per cell.
2.11. Protein expression measurement by flow cytometry
Flow cytometry was performed as published previously [23]. After incubation of LLC-PK1 and HEK-293A cells with S protein, the cells were washed out twice with PBS, harvested with trypsin/EDTA for 3 min and centrifuge at 2500 ×g for 5 min. Pellets were resuspended in PBS containing 5 % FBS, fixed with 4 % paraformaldehyde and permeabilized with 0.1 % Triton X-100. Primary antibodies for monoclonal mouse anti-S protein, monoclonal rabbit anti-ACE2, polyclonal rabbit anti-megalin or monoclonal rabbit anti-AQP1 (dilutions 1:100) were used according to the manufacturer's instructions. After washing the unbound antibodies using the permeabilization solution, the samples were incubated with appropriate secondary antibodies (dilutions 1:200) as indicated in the manufacturer's instructions. Specificity control was carried out by incubation of the secondary antibody alone. The flow cytometry analysis was carried out using a BD FACSCalibur cytometer (BD Biosciences, San Jose, CA) and the data were analyzed with FlowJo software (version 10.8; BD Biosciences).
2.12. Cell viability
Cell viability was determined by Annexin V and propidium iodide (PI) staining as recommended by the manufacturer (catalog no. 556547; BD Biosciences). Briefly, after the incubation with S protein, cells were washed out twice with PBS and harvested with trypsin-EDTA solution for 2 min. Then, DMEM supplemented with 10 % FBS was added and the cells were centrifuged at 2500 ×g for 5 min; 10−6 cells were resuspended in buffer solution (catalog no. 556547; BD Biosciences). Annexin V and PI were added and the cells were incubated in the dark for 15 min. Cells were acquired in a FACSCalibur cytometer (BD Biosciences) and analyzed using the FlowJo software (version 10.8; BD Biosciences).
2.13. Cell proliferation
Cell proliferation was assessed using CellTrace CFSE as described by the manufacturer (catalog no. C34554; Invitrogen, Thermo Fisher Scientific). Briefly, the cell suspension was centrifuged at 2500 ×g for 5 min and the cells were resuspended in CellTrace CFSE staining solution for 20 min at 37 °C. The cells were washed out, centrifuged for 2500 ×g for 5 min, resuspended in DMEM with 10 % FBA and grown in six-well plates for 3 days. At this point, an aliquot was taken to analyze the maximum CFSE staining. S protein was added in the last 16 h. At the end, CFSE staining was analyzed to assess cell proliferation. Cells were acquired in a FACSCalibur cytometer (BD Biosciences) and analyzed using the FlowJo software (version 10.8; BD Biosciences).
2.14. Assessment of megalin stability
Megalin stability was assessed using the cycloheximide pulse-chase method [24]. Briefly, LLC-PK1 cells transfected with MegT0-HA were incubated or not with 5 μg/mL S protein overnight. Then, 10 μg/mL cycloheximide (a protein synthesis inhibitor) was added at different periods as indicated in the figure legends. Cells were washed out with PBS and incubated with lysis buffer (20 mM HEPES [pH 7.4], 2 mM EGTA, 1 % Triton X-100, 0.01 % SDS, 50 mM sodium fluoride, 5 mM sodium pyrophosphate, 5 mM sodium orthovanadate, 10 mM sodium β-glycerophosphate, 1 mM PMSF, 1× complete protease inhibitor [Sigma]) for 40 min. Then, the samples were cleared by centrifugation (1000 ×g for 10 min at 4 °C) and the supernatant was recovered and solubilized in Laemmli sample buffer. Megalin expression was determined by immunoblotting. Proteins (50–60 μg) were resolved in 9 % SDS-PAGE and transferred to PVDF membranes. Detection of megT0-HA was performed using monoclonal mouse anti-HA (dilution 1:1000, overnight, 4 °C) and HRP-conjugated anti-mouse IgG (dilution 1:4000, 1 h, room temperature). Detection of β-actin was performed using rabbit anti-β-actin (dilution 1:1000, overnight, 4 °C) and HRP-conjugated anti-rabbit IgG (dilution 1:5000, 1 h, room temperature) antibodies. After antibody labeling, detection was performed using ECL prime as substrate. Images were obtained using the Image Quant LAS4000 Image processing system (GE Healthcare Life Sciences, Pittsburgh, PA, USA) and analyzed using NIH ImageJ software (version 1.6.0). The level of megalin expression was calculated by the ratio of the optical densities of megT0-HA and β-actin. Data are presented as arbitrary units.
2.15. Measurement of angiotensin II levels
Angiotensin II (Ang II) concentrations were determined by ELISA as previously published [25], [26]. After treatment, the cell supernatants were harvested and clarified by centrifugation at 1000 ×g for 10 min to remove any cell debris. Then, 1× complete protease inhibitor (Sigma-Aldrich) was added to avoid peptide degradation. The concentration of Ang II was determined using an Angiotensin II EIA Kit (#RAB0010) purchased from Sigma-Aldrich, following the manufacturer's instructions.
2.16. Statistical analysis
The results are presented as median (interquartile range). Each point (n) represents the mean of 3 different triplicates used in independent experiments. The statistical analysis was performed using GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA; www.graphpad.com). The normality distribution was assessed by the Shapiro-Wilk test. An unpaired Student's t-test or Mann-Whitney test was used to compare two groups as indicated in the figure legends. For comparison among three or more groups, one-way ANOVA followed by Tukey's post hoc test or Kruskal-Wallis test followed by Dunn's post hoc test was used as indicated in the figure legends. Significance was determined as P < 0.05.
3. Results
3.1. SARS-CoV-2 spike protein modulates albumin endocytosis in proximal tubule epithelial cells
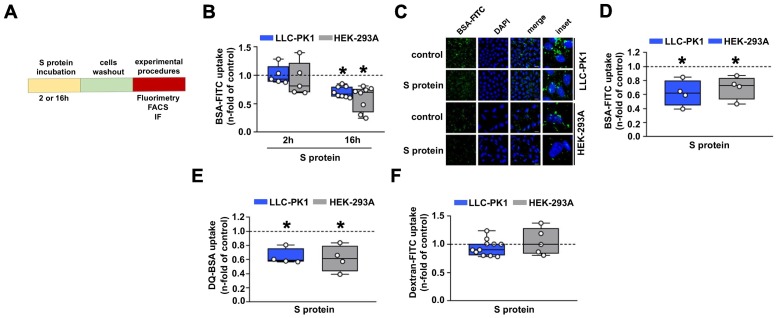
Initially, the effect of S protein on the albumin uptake in PTECs was assessed. LLC-PK1 and HEK-293A cells were incubated for 2 or 16 h with 5 μg/mL of S protein and then albumin endocytosis was measured by different methods (Fig. 1A). Using cell-associated fluorescence, it was observed that overnight incubation of these cells with S protein decreased the uptake of BSA-FITC (Fig. 1B). This effect was not observed when cells were incubated for 2 h. Similar results were obtained when BSA-FITC uptake was assessed by fluorescence microscopy, as observed from BSA-FITC+ puncta inside cells (Fig. 1C, D). It is worth to mention that the capacity of BSA-FITC uptake is higher in LLC-PK1 cells than in HEK-293A cells (2.5 fold). DQ-BSA was used as a tracer to identify if internalized albumin was degraded in lysosomes [27]. S protein also inhibited lysosomal albumin degradation, indicating that albumin uptake follows the canonical pathway (Fig. 1E). On the other hand, fluid-phase albumin endocytosis measured by dextran-FITC uptake was not changed (Fig. 1F). To date, cell death, measured by Annexin V and PI staining, and cell growth, measured by FACS using a CFSE dilution assay, were not changed by S protein in both LLC-PK1 and HEK-293A cells. These results indicate that the effect of S protein is not associated with cell viability (Supplementary Fig. 1A, B).
Fig. 1.
SARS-CoV-2 inhibits canonical megalin-mediated albumin endocytosis in proximal tubule cells. (A) Scheme showing the experimental design used. LLC-PK1 (blue) and HEK-293A (gray) cells were treated with 5 μg/mL S protein for either 2 or 16 h. Cells were washed out, and different experimental procedures were carried out. (B) BSA-FITC uptake was carried out in LLC-PK1 and HEK-293A cells by cell-associated fluorescence (n = 5–8 as indicated at the top of the bars) or (C, D) fluorescence confocal microscopy (n = 4). (C) Representative image and (D) quantification of the fluorescence intensity. (E) DQ-BSA uptake (n = 4) or (F) dextran-FITC uptake (n = 10 in LLC-PK1 cells and n = 5 in HEK-293A) were analyzed by cell-associated fluorescence. LLC-PK1 and HEK-293A cells were treated with S protein for 16 h in (C)–(F). Dashed line represents the control, which was taken as 1.0. Data are presented as median (interquartile range). *P < 0.05 versus control.
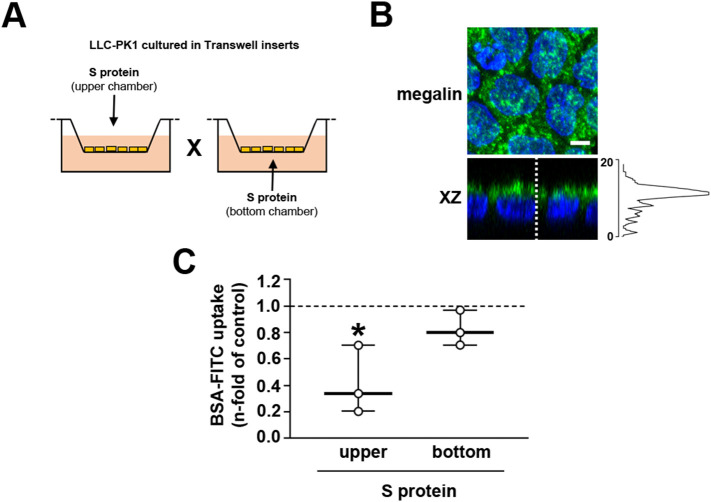
We then investigated the laterality of the effect of S protein on albumin endocytosis. Because HEK-293A cells do not polarize [28], we used only LLC-PK1 cells to address this issue. To address this question, LLC-PK1 cells were grown in Transwell inserts and, after confluence was reached, S protein was added to the upper (luminal side) or lower (basolateral side) chambers (Fig. 2A). Cell polarization was assessed by the expression of megalin at the luminal side (Fig. 2B). The effect of S protein on albumin endocytosis was observed only when LLC-PK1 cells were incubated at the upper chamber (Fig. 2C).
Fig. 2.
The S protein effect is associated with the luminal membrane. (A) Scheme showing the experimental design used. LLC-PK1 cells were seeded on Transwell inserts as described in the Materials and Methods section. S protein was added in the upper chamber or bottom chamber to assess the effect on the luminal and basolateral membrane, respectively. (B) Representative immunofluorescence images of LLC-PK1 cells grown in Transwell inserts (n = 3). Megalin (green) and DAPI (blue) are shown. The XZ axis shows polarized megalin expression in the apical membrane with the intensity plot (gray value) shown at the right. Scale bar, 5 μm. (C) BSA-FITC uptake was measured by cell-associated fluorescence in cells treated in the upper chamber or the bottom chamber with 5 μg/mL S protein for 16 h (n = 3). Dashed line represents the control, which was taken as 1.0. Data are presented as median (interquartile range). *P < 0.05 versus control.
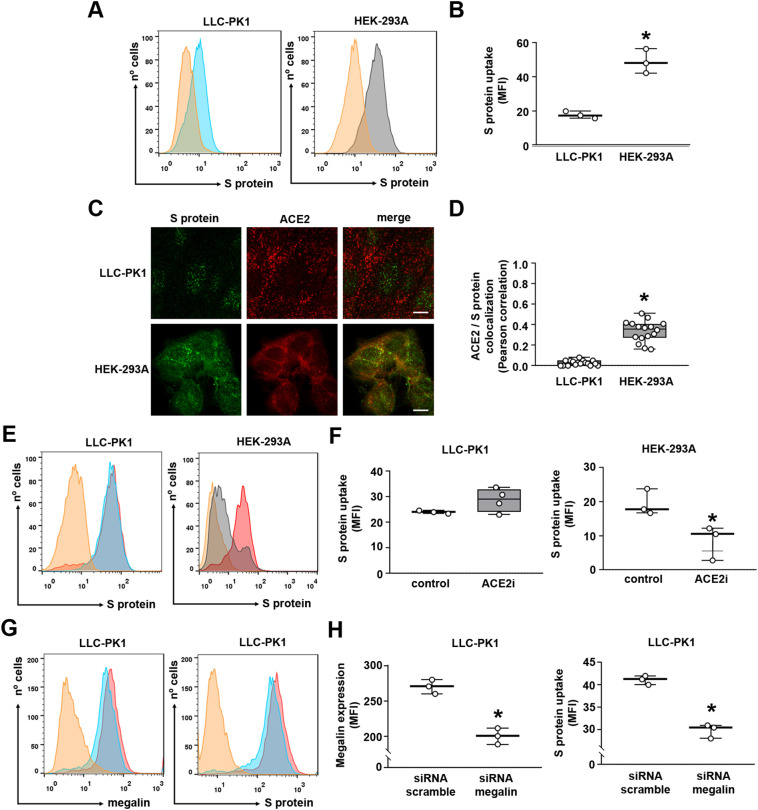
Because megalin is a scavenger receptor involved in the reabsorption of different proteins and peptides, and ACE2 is reported to bind to S protein, we decided to investigate the possible involvement of these proteins in the uptake of S protein in PTECs. Initially, it was observed that the uptake of S protein was higher in HEK-293A cells than in LLC-PK1 cells (Fig. 3A, B). Colocalization of S protein uptake (green color) and ACE2 expression (red color) were assessed using the indirect immunofluorescence technique. Although LLC-PK1 and HEK-293A cells uptake S protein and express ACE2, colocalization was only observed in HEK-293A cells (Fig. 3C, D). In agreement, the catalytical ACE2 inhibitor MLN-4760 (ACE2i) almost completely abolished S protein uptake in HEK-293A cells but not in LLC-PK1 cells (Fig. 3E, F). To test the involvement of megalin in S protein uptake in LLC-PK1 cells, the cells were transfected with siRNA for megalin (Fig. 3G, H). Megalin expression was decreased in 26 % in the cells transfected with siRNA compared with those transfected with scramble siRNA (scrRNA). This decrease was followed by a proportional decrease in S protein uptake.
Fig. 3.
Differential role of megalin and ACE2 in S protein internalization in proximal tubule epithelial cells. (A) S protein uptake in LLC-PK1 cells (blue) and HEK-293A cells (gray) (n = 3). (B) Quantification of S protein uptake in each cell line. (C) Representative confocal images of immunofluorescence for S protein (green) and ACE2 (red) in LLC-PK1 and HEK-293A cells (n = 3). Scale bar, 10 μm. (D) Colocalization of S protein and ACE2 in LLC-PK1 and HEK-293 cells, measured by Pearson coefficient (each point represents single cells of three different experiments). (E, F) Flow cytometry analysis of S protein uptake in LLC-PK1 and HEK-293 cells pre-treated or not with 100 nM MLN-4760, ACE2 inhibitor (ACE2i), for 2 h (n = 3 or 4 as indicated at the top of the bars). (G, H) Flow cytometry analysis of megalin expression (left) and S protein uptake (right) in LLC-PK1 cells transfected with siRNA scramble or siRNA megalin, as described in the Materials and Methods section (n = 3). MFI was calculated as the difference between control (scrRNA, red curve) and siRNA conditions (blue curve) after subtracting the negative control (orange curve). Data are presented as median (interquartile range). *P < 0.05 versus control (F, H) or versus LLC-PK1 cells (B, D).
So far, our results show that S protein has a specific inhibitory effect on canonical megalin-mediated albumin endocytosis in both LLC-PK1 and HEK-293A cells. On the other hand, S protein uptake is mediated preferentially by different pathways in these cells.
3.2. ACE2 and the inhibitory effect of S protein on albumin uptake
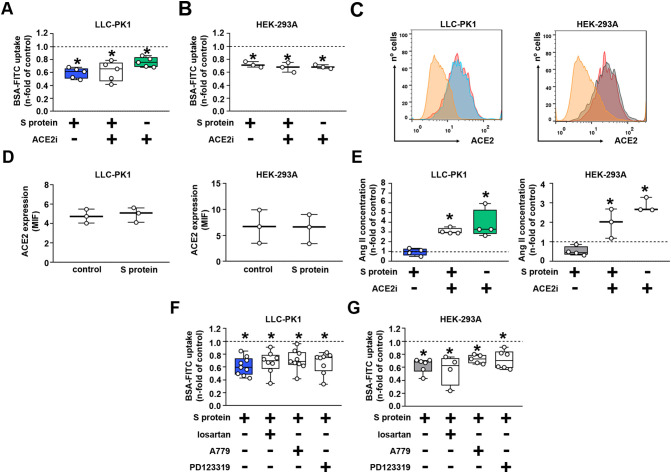
We then decided to investigate the possible role of ACE2 on the inhibitory effect of S protein on albumin uptake. The incubation of LLC-PK1 or HEK-293A cells with ACE2i decreased BSA-FITC uptake in a similar and non-additive manner to S protein (Fig. 4A, B). In addition, we observed that S protein did not change ACE2 expression in LLC-PK1 and HEK-293A cells (Fig. 4C, D). Because ACE2 breaks down octapeptide angiotensin II (Ang II) into heptapeptide angiotensin-(1-7) [Ang(1-7)] [29], the Ang II level was assessed in the luminal medium of both cells. The Ang II level was not changed by S protein (Fig. 4E). On the other hand, the luminal level of Ang II was increased when both cell types were incubated with ACE2i alone or simultaneously with S protein. Moreover, the inhibitory effect of S protein on albumin uptake was not modified by addition of losartan, A779, and PD123319, antagonists of AT1, MAS, and AT2 receptors, respectively (Fig. 4F, G). Together, these data show that ACE2 is not involved in the inhibitory effect of S protein on albumin endocytosis.
Fig. 4.
The S protein effect does not involve the ACE2/Ang II/AT1R axis. (A,B) BSA-FITC uptake in (A) LLC-PK1 and (B) HEK-293 cells treated with 5 μg/mL S protein, 100 nM ACE2i or both components for 16 h (n = 3 or 5 as indicated at the top of the bars). (C,D) Quantification of ACE2 expression by flow cytometry in LLC-PK1 (left) and HEK-293 cells (right) treated with 5 μg/mL S protein for 16 h (n = 3). (E) Quantification of angiotensin II (Ang II) levels through ELISA in the supernatant of LLC-PK1 (left) and HEK-293 cells (right) treated with 5 μg/mL S protein, 100 nM ACE2i inhibitor, or both components for 16 h (n = 3). (F,G) BSA-FITC uptake in (F) LLC-PK1 and (G) HEK-293. Cells were treated with 5 μg/mL S protein incubated simultaneously with 100 nM losartan (an AT1R antagonist), 100 nM A779 (a MASR antagonist), or 100 nM PD123319 (an AT2R antagonist) (n = 4–9 as indicated in the bars). Dashed lines represent the control, which was taken as 1.0. Data are presented as median (interquartile range). *P < 0.05 versus control.
3.3. Megalin expression is changed by S protein
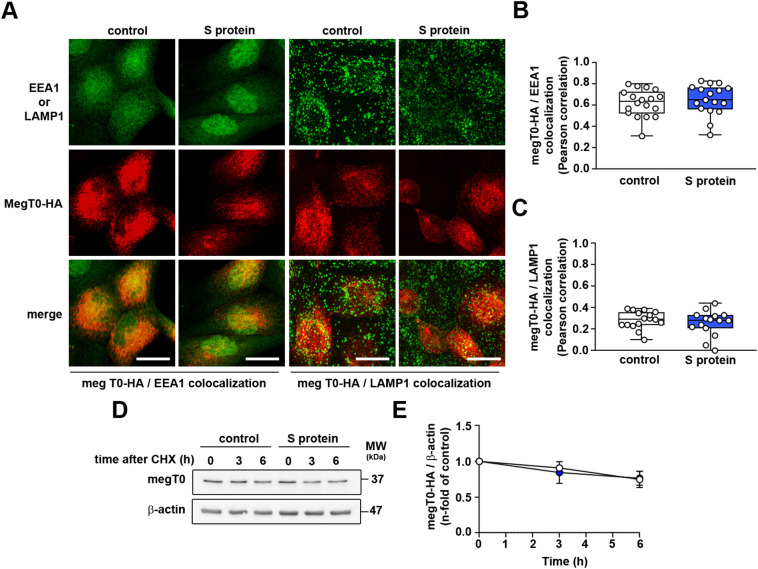
In this step, we investigated the possible effect of S protein on cellular megalin trafficking, stability and/or megalin expression in LLC-PK1 cells. LLC-PK1 cells were transfected with megalin construct lacking the whole extracellular domain tagged with HA epitope (MegT0-HA) [7]. This approach is well known to study the traffic and half-life of megalin [20], [24]. EEA1 and LAMP1 were used as markers of early endosomes and lysosomes, respectively. S protein did not change the colocalization between MegT0-HA with EEA1 (Fig. 5A, B) or LAMP1 (Fig. 5A, C). In addition, megalin stability was not changed by incubation of LLC-PK1 cells with S protein, assessed using the cycloheximide (CHX) pulse-chase approach (Fig. 5D, E).
Fig. 5.
S protein does not change megalin trafficking or stability. (A–C) Representative confocal images of LLC-PK1 cells stably expressing MegT0-HA, a truncated form of megalin. (A) Immunofluorescence for EEA1 (green) and MegT0-HA (red) or LAMP1 (green) and MegT0-HA (red) was carried out. Graphic representations of the colocalization between EEA1 and MegT0-HA (B) or LAMP1 and MegT0-HA (C) are shown, quantified by the Pearson coefficient. Each point represents single cells of three different experiments. Scale bar, 20 μm. (D, E) Kinetics of megalin degradation was accessed by a cycloheximide pulse-chase assay. LLC-PK1 cells stably expressing MegT0HA were treated with 100 μM cycloheximide for 0 h, 3 h, and 6 h, in the presence or absence of treatment with 5 μg/mL S protein for 16 h. Cells were harvested, and immunoblotting was performed for detection of MegT0-HA and β-actin. (E) Quantification of MegT0-HA expression is shown (n = 4). Data are presented as median (interquartile range). NS, P > 0.05.
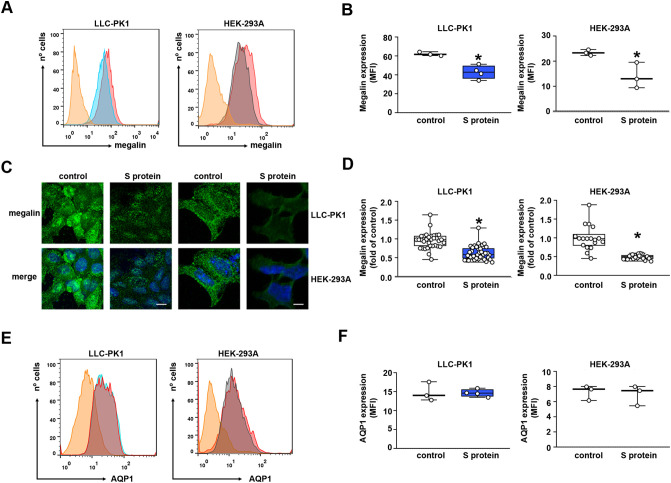
On the other hand, megalin expression was reduced in LLC-PK1 and HEK-293A cells measured by FACS analysis (Fig. 6A–B). Similar inhibition of megalin expression was also observed when immunofluorescence visualized by confocal microscopy was used (Fig. 6C, D). These results agree with the observed inhibitory effect of S protein on albumin endocytosis (Fig. 1B–H). Overnight incubation of the cells with S protein had no significant effect on aquaporin-1 (AQ1) expression in LLC-PK1 and HEK-293A cells (Fig. 6E, F), indicating its specific effect on the albumin endocytosis machinery.
Fig. 6.
S protein inhibits megalin expression in proximal tubule epithelial cells. (A, B) Flow cytometry analysis for megalin expression in LLC-PK1 (left) and HEK-293A cells (right) was performed. (B) Graphic showing expression of megalin, obtained by mean fluorescence intensity (MFI) (n = 3). (C, D) Representative confocal images of megalin expression (green) in LLC-PK1 cells (left) and HEK-293A cells (right). Nuclei are stained with DAPI (blue). Scale bar, 10 μm. (D) Quantification of megalin expression. Each point represents a single cell of four different experiments. (E, F) Flow cytometry analysis for aquaporin-1 expression, AQP1, in LLC-PK1 cells (left) and HEK-293A cells (right) was performed. (F) Graphic showing respective expression of AQP1, obtained by mean fluorescence intensity (MFI) (n = 3–4, as indicated at the top of the bars). Data are presented as median (interquartile range). *P < 0.05 versus control.
4. Discussion
Our study provides a new insight regarding the mechanism underlying albuminuria in patients with COVID-19. This process encompasses direct modulation of canonical megalin-mediated albumin endocytosis in PTECs by S protein due to the reduction in megalin expression. In the present study, we used two renal tubular cell lines: LLC-PK1 and HEK-293A. Despite being embryonic cells, HEK-293A cells express all components of the protein endocytic machinery similar to LLC-PK1 cells [30], [31]. LLC-PK1 cells form a polarized monolayer and present the main characteristic of PTECs [30]. Transcriptomic analysis indicates that LLC-PK1 cells have high similarity to mice PTECs [32].
The classic mechanism proposed to underly the pathogenesis of COVID-19 involves binding of the virus to a specific receptor, usually ACE2, which allows infection of the cells, triggering injury and organ damage [1], [2], [12]. Some evidence indicates that S protein, a structural protein of SARS-CoV-2, could trigger a cellular response and modulate cellular function in different cell types participating in the pathogenesis of COVID-19. It was observed that S protein induces a non-infective cellular dysfunction of heart pericytes, which could be associated with microvascular injury [16]. In addition, Buzhdygan et al. [33] showed that S protein induces rupture of endothelial barrier integrity. An S protein effect was also observed in C57BL/6J male mice infected with spike protein-pseudotyped lentivirus [34]. The presence of spike protein in different organs associated with acute pneumonia and the pro-inflammatory profile was detected [34]. In the present work, we used S protein at 5 μg/mL, a concentration that has been previously used [35]. Serum S protein concentration was shown to range from 0.005 to 0.3 μg/mL [16]. So far, there are no studies regarding the concentration of S protein in the lumen of PTs. Massive water reabsorption occurs in PTs [36], therefore it is plausible to imagine that the S protein concentration could reach a higher value in PTs than in serum. In a study with urine collected from 132 participants 25 % presented a mean concentration of S protein in the urine of 0.033 μg/μg creatinine [17]. Therefore, the S protein concentration at 5 μg/mL could be higher than that detected in human PTs of COVID-19 patients what could represent a limitation to our study.
S protein cannot be freely filtrated in the glomerulus due to its size (>150 kDa) [37]. Here, we showed that S protein modulates albumin endocytosis in LLC-PK1 cells only when added at the luminal side. Then, what is the site of origin of the S protein observed in the tubular lumen? One possibility could be secretion of S protein to the luminal side of SARS-CoV-2-infected tubular and glomerular cells [38]. Once in the lumen of PTECs, S protein could modulate the function of PTECs. Strengthening this view, George et al. [17] showed that 25 % of SARS-CoV-2-positive patients presented urinary S protein, indicating its presence in the lumen of tubular segments. They also found a statistical correlation between urinary S protein and albuminuria in these patients.
One important concern is regarding the species of the cells used in the present work. Hoffman et al. [39] showed a lower rate of infection of LLC-PK1 cells with pseudovirus containing S protein of SARS-CoV-2 compared with human cell lines. This was associated with a specimen-specific interaction between the receptor binding domain (RBD), located in the S1 domain of S protein, and ACE2 receptor. Accordingly, we observed that HEK-293A cells have more S protein uptake than LLC-PK1 cells. Despite the use of cells from different species, S protein showed a similar capacity to inhibit albumin uptake and megalin expression. These results indicate that the inhibitory effect of S protein on albumin endocytosis is dissociated from S protein uptake. Accordingly, Avolio et al. [16] showed that activation of extracellular signal-regulated kinase 1/2 (ERK1/2) by S protein, in heart pericytes, is mediated by a plasma membrane CD147 receptor.
It is well known that ACE2 is highly expressed at the luminal membrane of PTECs and responsible for the breakdown of Ang II into Ang(1-7) [1], [12], [39]. The interaction between S protein and ACE2 could modulate its activity and, consequently, the balance between these peptides [29]. Furthermore, activation of the Ang II/AT1R axis was shown to inhibit albumin endocytosis in PTECs [40], [41]. Therefore, one attractive hypothesis could be involvement of the ACE2/Ang II/AT1R axis in the effect of S protein on albumin endocytosis. In agreement, Lei et al. [15] showed that S protein induces vascular endothelial cell injury due to downregulation of ACE2. Despite all this evidence, the following results indicate that the inhibitory effect of S protein canonical megalin-mediated albumin endocytosis in LLC-PK1 and HEK-293A cells does not involve ACE2 protein: (1) S protein did not change the expression of ACE2 and the Ang II level; (2) AT1R, AT2R, and MASR antagonists do not modify the inhibitory S protein; (3) ACE2 inhibitor increased the Ang II level contrary to S protein. Further experiments are necessary to identify the receptor that mediates the effect of S protein on albumin uptake.
Megalin has been shown to be a target of different compounds involved in the modulation of albumin reabsorption in PTECs [23], [40]. Furthermore, some syndromes linked to changes in the function of PTECs such as Donnai-Barrow syndrome and Dent disease, have decreased surface megalin expression associated with an albuminuria phenotype [42], [43]. In the present work, we showed that the reduction in albumin uptake induced by S protein involves a decrease in total megalin expression without changes in cellular megalin recycling and stability. In agreement, Werion et al. [8] showed in post-mortem patients stricken by COVID-19 a specific reduction in megalin expression in PTECs associated with low-molecular-weight proteinuria, neutral aminoaciduria, and urinary β2-microglobulin, markers of PT injury. Accordingly, Korman et al. [9], using a cohort of 42 patients with COVID-19, observed a high frequency of acute Fanconi-like syndrome associated with a proteinuria phenotype that could precede initiation of AKI. Interestingly, changes in megalin-mediated albumin endocytosis have been associated with a pro-inflammatory phenotype and tubule-interstitial injury observed in a subAKI animal model [44]. This phenomenon has been postulated to precede the development of AKI in patients with COVID-19 [4].
Our results are in line with an increasing number of reports demonstrating that SARS-CoV-2 S protein per se modulates cellular function as observed previously in other cell types [14], [15], [16], [45]. Thus, in addition to viral infection, viral proteins could be a new source of cellular response during the onset and progression of COVID-19, playing a role in its pathogenesis. Our results suggest that viral S protein could participate in the genesis of tubular proteinuria observed with high prevalence in patients with COVID-19.
The following is the supplementary data related to this article.
Supplementary Fig. 1.
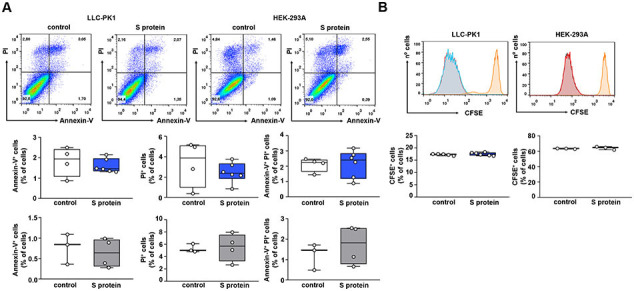
S protein does not change death and proliferation of PTECs. (A) Flow cytometry analysis of apoptosis and necrosis, accessed by Annexin V and propidium iodide (PI) staining, respectively. Analysis was performed on LLC-PK1 and HEK-293A cells treated (blue and gray bars, respectively) or not (white bars) with 5 μg/mL S protein for 16 h. Graphs representing Annexin V+ cells, PI+, and Annexin V+/PI+ cells are shown (n = 3–6, as indicated at the top of the bars). (B) Flow cytometry analysis of cell proliferation, assessed by the CellTrace CFSE dilution assay as described in the Materials and Methods section. Analysis was performed on LLC-PK1 and HEK-293A cells treated (blue and gray bars, respectively) or not (white bars) with 5 μg/mL S protein for 16 h (n = 3–6, as indicated at the top of the bars). Data are presented as means ± standard deviation.
Funding
This research was funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico (https://www.cnpq.br): 46.5656/2014-5 (to P.M.R.R. and C.C.-N.), 40.1700/2020-8 (to C.C.-N.), 304682/2015-2 (to A.A.S.P.), 303793/2015-5 (to C.C.-N.); Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro–FAPERJ (https://www.faperj.br): E-26/210.181/2020 (to P.M.R.R. and C.C.-N.), E-26/211.139/2021 (to C.C.-N.), E-26/202.950/2016 (to A.A.S.P.), E-26/202.833/2017 (to C.C.-N.); Rio Network of Innovation in Nanosystems for Health (Nanohealth/FAPERJ) - E-26/010.000983/2019 (to P.M.R.R, A.A.S.P. and C.C.-N.); Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT): 1200393 (to M.P.M.). This study was financed, in part, by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior/Brasil (CAPES) -Finance Code 001 and CAPES/PRINT-88887.508141/2020-00.
CRediT authorship contribution statement
Rodrigo P. Silva-Aguiar: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Visualization. Douglas E. Teixeira: Formal analysis, Investigation, Data curation, Writing – review & editing, Visualization. Diogo B. Peruchetti: Formal analysis, Investigation, Data curation, Writing – review & editing, Visualization. Lucas S. Florentino: Investigation, Writing – review & editing, Visualization. Rodrigo A.S. Peres: Investigation, Writing – review & editing, Visualization. Carlos P. Gomes: Formal analysis, Investigation, Writing – review & editing, Visualization. Maria-Paz Marzolo: Conceptualization, Resources, Formal analysis, Writing – review & editing, Visualization, Funding acquisition. Patricia M.R. Rocco: Conceptualization, Resources, Formal analysis, Writing – review & editing, Visualization, Funding acquisition. Ana Acacia S. Pinheiro: Validation, Formal analysis, Resources, Writing – review & editing, Visualization, Funding acquisition. Celso Caruso-Neves: Investigation, Resources, Data curation, Writing – original draft, Writing – review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank Ms. Sarah Aparecida dos Santos Alves (TCT fellowship/FAPERJ) and Ms. Giulianne Serpa (TCT fellowship/FAPERJ) for the excellent technical support.
Data availability
Data will be made available on request.
References
- 1.Lopes-Pacheco M., Silva P.L., Cruz F.F., Battaglini D., Robba C., Pelosi P., Morales M.M., Caruso Neves C., Rocco P.R.M. Pathogenesis of multiple organ injury in COVID-19 and potential therapeutic strategies. Front. Physiol. 2021;12 doi: 10.3389/fphys.2021.593223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robba C., Battaglini D., Pelosi P., Rocco P.R.M. Multiple organ dysfunction in SARS-CoV-2: MODS-CoV-2. Expert Rev Respir Med. 2020;14:865–868. doi: 10.1080/17476348.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nadim M.K., Forni L.G., Mehta R.L., Connor M.J., Jr., Liu K.D., Ostermann M., Rimmelé T., Zarbock A., Bell S., Bihorac A., Cantaluppi V., Hoste E., Husain-Syed F., Germain M.J., Goldstein S.L., Gupta S., Joannidis M., Kashani K., Koyner J.L., Legrand M., Lumlertgul N., Mohan S., Pannu N., Peng Z., Perez-Fernandez X.L., Pickkers P., Prowle J., Reis T., Srisawat N., Tolwani A., Vijayan A., Villa G., Yang L., Ronco C., Kellum J.A. COVID-19-associated acute kidney injury: consensus report of the 25th acute disease quality initiative (ADQI) workgroup. Nat Rev Nephrol. 2020;16:747–764. doi: 10.1038/s41581-020-00356-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Legrand M., Bell S., Forni L., Joannidis M., Koyner J.L., Liu K., Cantaluppi V. Pathophysiology of COVID-19-associated acute kidney injury. Nat Rev Nephrol. 2021;17:751–764. doi: 10.1038/s41581-021-00452-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batlle D., Soler M.J., Sparks M.A., Hiremath S., South A.M., Welling P.A., Swaminathan S. COVID-19 and ACE2 in cardiovascular, lung, and kidney working group. Acute kidney injury in COVID-19: emerging evidence of a distinct pathophysiology. J Am Soc Nephrol. 2020;31:1380–1383. doi: 10.1681/ASN.2020040419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Głowacka M., Lipka S., Młynarska E., Franczyk B., Rysz J. Acute kidney injury in COVID-19. Int. J. Mol. Sci. 2021;22:8081. doi: 10.3390/ijms22158081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun D.Q., Wang T.Y., Zheng K.I., Targher G., Byrne C.D., Chen Y.P., Zheng M.H. Subclinical acute kidney injury in COVID-19 patients: a retrospective cohort study. Nephron. 2020;144:347–350. doi: 10.1159/000508502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Werion A., Belkhir L., Perrot M., Schmit G., Aydin S., Chen Z., Penaloza A., De Greef J., Yildiz H., Pothen L., Yombi J.C., Dewulf J., Scohy A., Gérard L., Wittebole X., Laterre P.F., Miller S.E., Devuyst O., Jadoul M., Morelle J. Cliniques Universitaires Saint-luc (CUSL) COVID-19 research group. SARS-CoV-2 causes a specific dysfunction of the kidney proximal tubule. Kidney Int. 2020;98:1296–1307. doi: 10.1016/j.kint.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kormann R., Jacquot A., Alla A., Corbel A., Koszutski M., Voirin P., Garcia Parrilla M., Bevilacqua S., Schvoerer E., Gueant J.L., Namour F., Levy B., Frimat L., Oussalah A. Coronavirus disease 2019: acute fanconi syndrome precedes acute kidney injury. Clin. Kidney J. 2020;13(3):362–370. doi: 10.1093/ckj/sfaa109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nielsen R., Christensen E.I., Birn H. Megalin and cubilin in proximal tubule protein reabsorption: from experimental models to human disease. Kidney Int. 2016;89:58–67. doi: 10.1016/j.kint.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Marzolo M.P., Farfán P. New insights into the roles of megalin/LRP2 and the regulation of its functional expression. Biol. Res. 2011;44:89–105. doi: 10.4067/S0716-97602011000100012. [DOI] [PubMed] [Google Scholar]
- 12.Hu B., Guo H., Zhou P., et al. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T., Yang Y., Li Y., Wang Z., Ma F., Luo R., Xu X., Zhou G., Wang J., Niu J., Lv G., Crispe I.N., Tu Z. Platelets mediate inflammatory monocyte activation by SARS-CoV-2 spike protein. J. Clin. Invest. 2022;132 doi: 10.1172/JCI150101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lei Y., Zhang J., Schiavon C.R., He M., Chen L., Shen H., Zhang Y., Yin Q., Cho Y., Andrade L., Shadel G.S., Hepokoski M., Lei T., Wang H., Zhang J., Yuan J.X., Malhotra A., Manor U., Wang S., Yuan Z.Y., Shyy J.Y. SARS-CoV-2 spike protein impairs endothelial function via downregulation of ACE 2. Circ. Res. 2021;128:1323–1326. doi: 10.1161/CIRCRESAHA.121.318902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avolio E., Carrabba M., Milligan R., Kavanagh Williamson M., Beltrami A.P., Gupta K., Elvers K.T., Gamez M., Foster R.R., Gillespie K., Hamilton F., Arnold D., Berger I., Davidson A.D., Hill D., Caputo M., Madeddu P. The SARS-CoV-2 spike protein disrupts human cardiac pericytes function through CD147 receptor-mediated signalling: a potential non-infective mechanism of COVID-19 microvascular disease. Clin Sci (Lond) 2021;135:2667–2689. doi: 10.1042/CS20210735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.George S., Pal A.C., Gagnon J., Timalsina S., Singh P., Vydyam P., Munshi M., Chiu J.E., Renard I., Harden C.A., Ott I.M., Watkins A.E., Vogels C.B.F., Lu P., Tokuyama M., Venkataraman A., Casanovas-Massana A., Wyllie A.L., Rao V., Campbell M., Farhadian S.F., Grubaugh N.D., Dela Cruz C.S., Ko A.I., Perez A.Z.B., Akaho E.H., Moledina D.G., Testani J., John A.R., Ledizet M., Ben Mamoun C. Yale IMPACT team. Evidence for SARS-CoV-2 spike protein in the urine of COVID-19 patients. Kidney360. 2021;2:924–936. doi: 10.34067/KID.0002172021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LER Cunha, Stolet A.A., Strauch M.A., VAR Pereira, Dumard C.H., AMO Gomes, Monteiro F.L., Higa L.M., PNC Souza, Fonseca J.G., Pontes F.E., LGR Meirelles, JWM Albuquerque, Sacramento C.Q., Fintelman-Rodrigues N., Lima T.M., RGF Alvim, Marsili F.F., Caldeira M.M., Zingali R.B., TML Souza, Silva A.S., Muller R., DDRF Rodrigues, Jesus da Costa L., ADR Alves, Pinto M.A., Oliveira A.C., HLM Guedes, Tanuri A., Castilho L.R., Silva J.L., de Oliveira G.A.P. Polyclonal F(ab')2 fragments of equine antibodies raised against the spike protein neutralize SARS-CoV-2 variants with high potency. iScience. 2021;24(11):103315. doi: 10.1016/j.isci.2021.103315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silva-Aguiar R.P., Peruchetti D.B., Florentino L.S., Takiya C.M., Marzolo M.P., Dias W.B., Pinheiro A.A.S., Caruso-Neves C. Albumin expands albumin reabsorption capacity in proximal tubule epithelial cells through a positive feedback loop between AKT and megalin. Int. J. Mol. Sci. 2022;23:848. doi: 10.3390/ijms23020848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Augustin R., Carayannopoulos M.O., Dowd L.O., Phay J.E., Moley J.F., Moley K.H. Identification and characterization of human glucose transporter-like protein-9 (GLUT9): alternative splicing alters trafficking. J. Biol. Chem. 2004;279:16229–16236. doi: 10.1074/jbc.M312226200. [DOI] [PubMed] [Google Scholar]
- 22.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 23.Alves S.A.S., Florentino L.S., Teixeira D.E., Silva-Aguiar R.P., Peruchetti D.B., Oliveira A.C., Scharfstein J., Marzolo M.P., Pinheiro A.A.S., Caruso-Neves C. Surface megalin expression is a target to the inhibitory effect of bradykinin on the renal albumin endocytosis. Peptides. 2021;146 doi: 10.1016/j.peptides.2021.170646. [DOI] [PubMed] [Google Scholar]
- 24.Perez Bay A.E., Schreiner R., Benedicto I., Paz Marzolo M., Banfelder J., Weinstein A.M., Rodriguez-Boulan E.J. The fast-recycling receptor megalin defines the apical recycling pathway of epithelial cells. Nat. Commun. 2016;7:11550. doi: 10.1038/ncomms11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bardsley E.N., Neely O.C., Paterson D.J. Angiotensin peptide synthesis and cyclic nucleotide modulation in sympathetic stellate ganglia. J. Mol. Cell. Cardiol. 2020;138:234–243. doi: 10.1016/j.yjmcc.2019.11.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Volonte D., Sedorovitz M., Cespedes V.E., Beecher M.L., Galbiati F. Cell autonomous angiotensin II signaling controls the pleiotropic functions of oncogenic K-ras. J. Biol. Chem. 2021;296 doi: 10.1074/jbc.RA120.015188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen R., Mollet G., Esquivel E.L., Weyer K., Nielsen P.K., Antignac C., Christensen E.I. Increased lysosomal proteolysis counteracts protein accumulation in the proximal tubule during focal segmental glomerulosclerosis. Kidney Int. 2013;84:902–910. doi: 10.1038/ki.2013.218. [DOI] [PubMed] [Google Scholar]
- 28.van Beest M., Robben J.H., Savelkoul P.J., Hendriks G., Devonald M.A., Konings I.B., Lagendijk A.K., Karet F., Deen P.M. Polarisation, key to good localisation. Biochim. Biophys. Acta. 2006;1758(8):1126–1133. doi: 10.1016/j.bbamem.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 29.Silva-Aguiar R.P., Peruchetti D.B., PRM Rocco, Schmaier A.H., PMR E.Silva, Martins M.A., Carvalho V.F., AAS Pinheiro, Caruso-Neves C. Role of the renin-angiotensin system in the development of severe COVID-19 in hypertensive patients. Am. J. Physiol. Lung Cell Mol. Physiol. 2020;319:L596–L602. doi: 10.1152/ajplung.00286.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen R., Birn H., Moestrup S.K., Nielsen M., Verroust P., Christensen E.I. Characterization of a kidney proximal tubule cell line, LLC-PK1, expressing endocytotic active megalin. J Am Soc Nephrol. 1998;9:1767–1776. doi: 10.1681/ASN.V9101767. [DOI] [PubMed] [Google Scholar]
- 31.Gao Y., Zhou S., Luu S., Glowacki J. Megalin mediates 25-hydroxyvitamin D3 actions in human mesenchymal stem cells. FASEB J. 2019;33:7684–7693. doi: 10.1096/fj.201802578R. [DOI] [PubMed] [Google Scholar]
- 32.Khundmiri S.J., Chen L., Lederer E.D., Yang C.R., Knepper M.A. Transcriptomes of major proximal tubule cell culture models. J Am Soc Nephrol. 2021;32:86–97. doi: 10.1681/ASN.2020010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buzhdygan T.P., DeOre B.J., Baldwin-Leclair A., Bullock T.A., McGary H.M., Khan J.A., Razmpour R., Hale J.F., Galie P.A., Potula R., Andrews A.M., Ramirez S.H. The SARS-CoV-2 spike protein alters barrier function in 2D static and 3D microfluidic in-vitro models of the human blood-brain barrier. Neurobiol. Dis. 2020;146 doi: 10.1016/j.nbd.2020.105131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao X., Tian Y., Nguyen V., Zhang Y., Gao C., Yin R., Carver W., Fan D., Albrecht H., Cui T., Tan W. Spike protein of SARS-CoV-2 activates macrophages and contributes to induction of acute lung inflammation in male mice. FASEB J. 2021;35 doi: 10.1096/fj.202002742RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Appelbaum J., Arnold D.M., Kelton J.G., Gernsheimer T., Jevtic S.D., Ivetic N., Smith J.W., Nazy I. SARS-CoV-2 spike-dependent platelet activation in COVID-19 vaccine-induced thrombocytopenia. Blood Adv. 2022;6(7):2250–2253. doi: 10.1182/bloodadvances.2021005050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corman B., Roinel N., De Rouffignac C. Water reabsorption capacity of the proximal convoluted tubule: a microperfusion study on rat kidney. J. Physiol. 1981;316:379–392. doi: 10.1113/jphysiol.1981.sp013795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moeller M.J., Tenten V. Renal albumin filtration: alternative models to the standard physical barriers. Nat Rev Nephrol. 2013;9:266–277. doi: 10.1038/nrneph.2013.58. [DOI] [PubMed] [Google Scholar]
- 38.Diao B., Wang C., Wang R., Feng Z., Zhang J., Yang H., Tan Y., Wang H., Wang C., Liu L., Liu Y., Liu Y., Wang G., Yuan Z., Hou X., Ren L., Wu Y., Chen Y. Human kidney is a target for novel severe acute respiratory syndrome coronavirus 2 infection. Nat. Commun. 2021;12:2506. doi: 10.1038/s41467-021-22781-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peruchetti D.B., Barahuna-Filho P.F.R., Silva-Aguiar R.P., Abreu T.P., Takiya C.M., Cheng J., Pinheiro A.A.S., Cebotaru L., Guggino W.B., Caruso-Neves C. Megalin-mediated albumin endocytosis in renal proximal tubules is involved in the antiproteinuric effect of angiotensin II type 1 receptor blocker in a subclinical acute kidney injury animal model. Biochim. Biophys. Acta Gen. Subj. 2021;1865 doi: 10.1016/j.bbagen.2021.129950. [DOI] [PubMed] [Google Scholar]
- 41.Tojo A., Onozato M.L., Kurihara H., Sakai T., Goto A., Fujita T. Angiotensin II blockade restores albumin reabsorption in the proximal tubules of diabetic rats. Hypertens. Res. 2003;26:413–419. doi: 10.1291/hypres.26.413. [DOI] [PubMed] [Google Scholar]
- 42.Flemming J., Marczenke M., Rudolph I.M., Nielsen R., Storm T., Erik I.C., Diecke S., Emma F., Willnow T.E. Induced pluripotent stem cell-based disease modeling identifies ligand-induced decay of megalin as a cause of donnai-Barrow syndrome. Kidney Int. 2020;98:159–167. doi: 10.1016/j.kint.2020.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christensen E.I., Devuyst O., Dom G., Nielsen R., Van der Smissen P., Verroust P., Leruth M., Guggino W.B., Courtoy P.J. Loss of chloride channel ClC-5 impairs endocytosis by defective trafficking of megalin and cubilin in kidney proximal tubules. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8472–8477. doi: 10.1073/pnas.1432873100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peruchetti D.B., Silva-Filho J.L., Silva-Aguiar R.P., Teixeira D.E., Takiya C.M., Souza M.C., Henriques M.D.G., Pinheiro A.A.S., Caruso-Neves C. IL-4 receptor α chain protects the kidney against tubule-interstitial injury induced by albumin overload. Front. Physiol. 2020;11:172. doi: 10.3389/fphys.2020.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Navaratnarajah C.K., Pease D.R., Halfmann P.J., Taye B., Barkhymer A., Howell K.G., Charlesworth J.E., Christensen T.A., Kawaoka Y., Cattaneo R., Schneider J.W. Wanek family program for HLHS-stem cell pipeline. Highly efficient SARS-CoV-2 infection of human cardiomyocytes: spike protein-mediated cell fusion and its inhibition. J Virol. 2021;95 doi: 10.1128/JVI.01368-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.