Summary
Liver vascular network is patterned by sinusoidal and hepatocyte co-zonation. How intra-liver vessels acquire their hierarchical specialized functions is unknown. We study heterogeneity of hepatic vascular cells during mouse development through functional and single-cell RNA sequencing. The acquisition of sinusoidal endothelial cell identity is initiated during early development and is completed postnatally, originating from a pool of undifferentiated vascular progenitors at E12. The peri-natal induction of the transcription factor c-Maf is a critical switch for sinusoidal identity determination. Endothelium restricted deletion of c-Maf disrupts liver sinusoidal development, aberrantly expands postnatal liver hematopoiesis, promotes excessive postnatal sinusoidal proliferation, and aggravates liver pro-fibrotic sensitivity to chemical insult. Enforced c-Maf overexpression in generic human endothelial cells switches on a liver sinusoidal transcriptional program that maintains hepatocyte function. c-Maf represents an inducible intra-organotypic and niche responsive molecular determinant of hepatic sinusoidal cell identity and lays the foundation for strategies for vasculature-driven liver repair.
Keywords: Vascular heterogeneity, endothelial cell specification, liver, development, single-cell molecular profiling, fibrosis, c-Maf
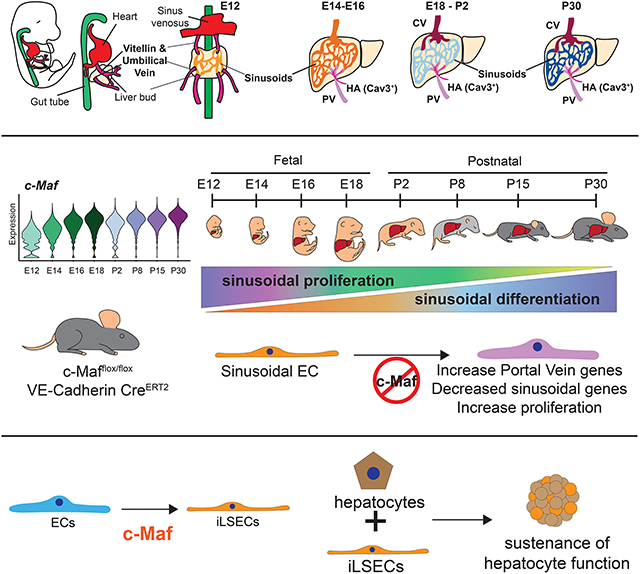
Graphical Abstract
eTOC blurb
Blood vessels adapt to the requirements of each organ. In the liver, this adaptation is a progressive transition from fetal to postnatal development, that is facilitated by the transcription factor c-Maf. In vitro c-Maf induces a liver like vascular phenotype allowing long term hepatocyte co-culture.
Introduction
The vascular system encompasses a heterogeneous pool of specialized endothelial cells (ECs) that anastomose arteries to veins and establishes functional and structural organotypic diversity (Harvey and Oliver, 2004). This angiodiversity is achieved by the unique capacity of ECs to customize and meet the cellular and metabolic demands of each organ. In addition to inter-organ specific heterogeneity, endothelial progenitor cells acquire remarkable intra-organ diversity with certain organs, such as the liver, ultimately arborizing with differentiated specialized ECs (Augustin and Koh 2017; Gomez-Salinero and Rafii 2018; Jakab and Augustin 2020; Rafii et al. 2016). Tissue-specific microvascular ECs also instruct neighboring cells and adapt to metabolic demands during homeostasis, regeneration and aging (Augustin and Koh, 2017; Rafii et al., 2016). The capacity of ECs to meet these tissue-specific diversified functions is achieved by actively co-adapting to their microenvironment by deploying distinct angiocrine factors to maintain homeostasis and regeneration, or mal-adapt to support tumorigenesis (Cao et al. 2017; Dejana et al. 2017; Ding et al. 2010; Ding et al. 2011; Tavora et al. 2020). While the transcriptional networks regulating arterial-venous specification have been extensively described, the molecular determinants of intra-organ microvascular EC diversity, such as zonation within liver capillaries (termed sinusoids) remain to be fully characterized.
Liver vasculature adapt to various developmental, homeostatic and pathophysiological processes, including liver regeneration, fibrosis and cancer (Cao et al. 2017; Ding et al. 2010; de Haan et al. 2020; Halpern et al. 2018; Lotto et al. 2020; MacParland et al. 2018; Rafii et al. 2016; Sharma et al. 2020). Furthermore, liver endothelium is organized in a patterned hepatocyte co-zonation gradient that regulates liver functionalization in part by angiocrine release of Rspo3 and Wnt9b from the Central Vein (Halpern et al., 2018; Planas-Paz et al., 2016; Rocha et al., 2015). Similarly, the liver sinusoids secrete Wnt2, Hgf, and other angiocrine factors that modulate liver homeostasis and regeneration (Ding et al., 2010, 2014; Rafii et al., 2016). While Gata4 and Bmp9 have been identified as regulators of the sinusoidal signature (Desroches-Castan et al., 2019a; Géraud et al., 2017; Winkler et al., 2021), the mechanism by which differentiated liver vasculature acquires its diverse attributes is still undefined.
Here, we resolve intra-organ vascular heterogeneity by performing single-cell RNA sequencing (scRNA-seq) of liver endothelium from early fetal to late postnatal development. We show that acquisition of the sinusoidal identity is initiated during early development and is fully established postnatally. We identify induction of the transcription factor c-Maf as a critical driver of liver sinusoidal maturation. Absence of c-Maf impairs hepatic sinusoidal specification, aberrantly expands postnatal liver hematopoiesis, and increases the fibrotic damage induced by chemical insult in adult mouse liver. Notably, c-Maf overexpression induces sinusoidal identity in human generic endothelium. Thus, c-Maf represent an inducible specification factor for liver sinusoids and set the stage to devise future approaches for therapeutic liver repair.
Results
Diversification of liver vasculature is developmentally specified
Upon differentiation, liver endothelial progenitors acquire their specialization by expressing defined surface receptors and cytokines to meet the demands of the co-localized liver parenchyma. While fetal and adult liver sinusoids are phenotypically distinct (Bankston and Pino, 1980; Barberá-Guillem et al., 1986; Nonaka et al., 2007), it is unclear how this heterogeneity is progressively established and maintained across fetal and postnatal development. To address this, we performed scRNA-seq on sorted ECs defined as CD45negCD31+ cells every two days from embryonic day 12 to 18 (E12-E18) and at postnatal day 2, 8, 15 and 30 (P2, P8, P15 and P30, respectively) of development (Figure 1A, Figure S1A-B and Table S1). Our analysis identified a total of 17 EC clusters (Figure 1B), with contamination of hematopoietic cells and a small fraction of parenchymal cells (Figure S1C-E).
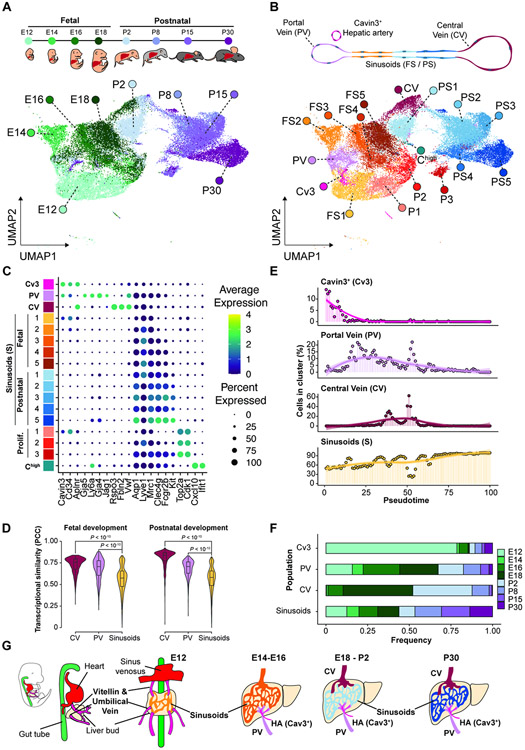
Figure 1. Liver vasculature progenitor diversification is acquired during transition from fetal to postnatal development.
(A) Uniform manifold approximation (UMAP) from ECs identified by scRNA-seq of sorted liver ECs (CD45negCD31+) at fetal and postnatal time points E12, E14,E16, E18, P2, P8, P15, and P30. Colors are assigned based on the sample time point as indicated in the upper panel. (B) UMAP labeling of the different EC populations identified from the scRNA-seq analysis from (A). Cv3, Cavin3+; PV, portal vein; CV, central vein; FS1-5, fetal sinusoidal EC populations; PS1-5, postnatal sinusoidal EC populations; P1-3, proliferating ECs; Chigh, Cxcl10 high expressing ECs. (C) Identification of specific markers associated to the EC populations identified from the scRNA-seq. (D) Transcriptional similarity analysis within portal vein, central vein, and sinusoids cell groups during fetal and postnatal development, as calculated by Pearson’s correlation coefficient (PCC) in the principal component space. (E) Vascular populations identified from the scRNA-seq ordered using pseudotime and plotted based on their predicted order. (F) Proportion of the contribution of Cavin3+ (Cv3), portal vein (PV), central vein (CV), or all sinusoids (S) per time point. Colors indicate the time point. (G) Diagram of the development of the liver vascular system following the observations from scRNA-seq. During early development (E12), the liver bud is infiltrated by the vitellin vein, umbilical vein, and sinus venosus. After this infiltration, the endothelium differentiates into the portal vein and the sinusoids. The central vein population is differentiated later in development, starting primarily at E18. During the progressive development of the liver, the sinusoidal transcriptome transitions from a fetal to an adult state. Colors are based on the identified populations in (B).
The EC populations were identified as CD31+CDH5+CD45neg (Figure S1E,F), and cell cluster identity was defined based on marker gene expression: Cavin3+ (Cav3+) by expression of Cavin3, Portal Vein (PV) by expression of Gja5, Cd34, Ly6a, Gja4, Jag1 and Vwf, Central Vein (CV) by expression of Rspo3, Fbln2, Vwf, sinusoids (S) expressing Cd34, Mrc1, Fcgr2b, Clec4g and Kit, a proliferative cell cluster expressing Top2a and Cdk1, and a Cxcl10 high (Chigh) cluster with expression of Cxcl10 and Ifit1 (Figure 1C and Figure S1E-I). Within the sinusoidal group, we observed a unique expression pattern of undifferentiated markers such as: Cd34, Pgk1 and Mif compared to adult differentiated markers, such as Aqp1, Mrc1, Fcgr2b, Clec4g and Kit (Figure 1C and Figure S1E,G,H). Notably, the sinusoidal EC population displays the lowest degree of internal transcriptional similarity within the developmental stages (Fig 1D), rendering sinusoids the most transcriptionally diverse cell type across the landscape of vascular capillary development compared with large vessel clusters Portal Vein and Central Vein.
Angiogenic signals arising from the liver bud at E10-E12 induce its vascularization from the progenitors within the Vitellin Vein, the Umbilical Vein and Sinus Venosus (DeSesso, 2017), facilitating contribution from the endocardium to liver endothelium (Zhang et al., 2016). We noted a decrease in the frequency of the Cavin3+ cluster at pseudotimes corresponding to later developmental stages (Figure 1E-F), although a small percentage was present at postnatal stages and these cells were identified as Hepatic artery cells based on Cavin3 staining in the Protein Atlas (Figure S1I). The Portal Vein cluster increases in frequency shortly after the decrease of the Cavin3+ cluster (Figure 1E-F). Notably, the establishment of the Central Vein cluster occurs during late fetal and early postnatal development (E18-P2; Figure 1E-F). While the Central Vein and Portal Vein signatures occurs during restricted developmental windows, sinusoid frequency steadily increases over time (Figure 1E-F).
Based on protein and RNA expression patterns, we characterized the developmental changes of the Portal Vein and adult sinusoidal marker Aqp1, the fetal enriched marker Lyve1 and the Central Vein marker Emcn (Figure S2A-C). scRNA-seq analyses revealed specific vascular populations during restricted developmental times with Aqp1 induction by the Portal Vein at E14 and Endomucin (Emcn) by the Central Vein at E18 (Figure S2A-C). Moreover, establishment of Central Vein induces the hepatocyte co-zonation as measured by CYP2E1 at stage P2 (Figure S2D). This is in agreement with the requirement of Rspo3 angiocrine secretion from Central Vein regulating early liver postnatal function (Boj et al., 2012; Planas-Paz et al., 2016; Rocha et al., 2015). These data suggest a temporal specification of each liver vascular component and defines sinusoids as the most transcriptionally diverse EC type during liver development (Figure 1G).
Temporal dynamics of liver sinusoidal specialization
To uncover transcriptional dynamics during liver vascular differentiation, we performed RNA velocity analysis (Bergen et al., 2020) on EC fractions (Figure 2A). Estimation of transition confidence between developmental stages using graph abstraction (Wolf et al., 2019) recapitulated the expected in vivo development (Figure S3A and Table S2). Ordering of the cells along RNA velocity-inferred pseudotime revealed two main groups of driver genes (fit likelihood > 0.17), associated with either fetal or postnatal development (Figure 2B and Figure S3B). At early timepoints (E12), undifferentiated EC progenitors express Lyve1 and Cd34 as observed by scRNA-seq and immunofluorescence analysis, although Cd34 becomes restricted to the Portal Vein (PV) at E14 (Figure S3C-E). We show that sinusoids acquire the capacity to express angiocrine factors Igf2r, Dpp4, Bmp2, Wnt2 and Ptprb during development (Figure 2B and Figure S3F-T). The genes driving the fetal developmental process were enriched for markers of cell proliferation, active protein turnover, and cell migration, among others (Figure 2C). Postnatal development driver genes were enriched for vascular maturation markers, including regulation of blood pressure, immune system, blood vessel remodeling, and secretion of cytokines involved in angiogenesis and cell proliferation (Figure 2C). Active transcription of adult sinusoidal markers, such as Mrc1 and Fcgr2b measured by RNA velocity was observed at early time points during fetal development and increased during postnatal stages (Figure 2D). Thus, an initial colonization of the liver bud by CD34+ endothelial progenitors lead to a progressive differentiation into the liver vasculature.
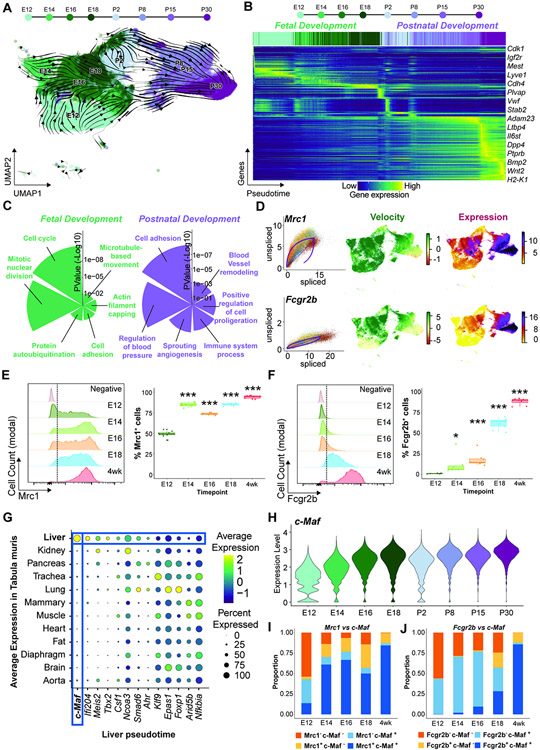
Figure 2. Liver vascular development is associated with a bi-sequential specification within a fetal to postnatal transition.
(A) Pseudotime analysis of the EC populations. Arrows show the directionality of changes associated with the transition from fetal to postnatal development. (B) Identification of the top 300 genes contributing to the changes over pseudotime. Colors correspond to an increase in change. (C) Gene ontology analysis of the genes contributing to fetal or postnatal pseudotime transition. (D) Analysis of the expression velocity of Mrc1 and Fcgr2b, measured as the ratio of spliced versus unspliced RNA over time, and the expression levels of these genes, derived from scRNA-seq. (E and F) Analysis of the percentage of ECs positive for Mrc1 (E) and Fcgr2b (F), of the ECs across the fetal E12, E14, E16, E18, and 4 weeks. Data represents n ≥ 7. Student’s t test analysis was performed comparing E12 to each individual time point, *p < 0.05, ***p < 0.001. (G) Average expression of the transcription factors driving the postnatal vascular transition from liver pseudotime in (B) within the EC of each organ obtained from the Tabula Muris database. Colors represent the average expression while size of the dot represents the percentage of cells where the expression was detected. (H) Violin plot showing expression of c-Maf within the EC clusters per time point from the scRNA-seq. (I and J) Flow cytometry analysis of the expression levels of c-Maf and Mrc1 (I) and c-Maf and Fcgr2b (J) in the liver ECs (n ≥ 5). Colors indicate the quadrant from the flow cytometry gating.
To verify the progressive acquisition of sinusoidal identity, we performed fluorescence cytometry analysis of the sinusoidal markers Mrc1 and Fcgr2b during fetal development and compared it at 4 weeks postnatal stage. Mrc1 was detected early at E12 (Figure 2E) and precedes the Fcgr2b appearance at E18 (Figure 2F). Therefore, liver sinusoidal EC fate is specified in a dynamic temporal sequence initiated during development and completed postnatally.
Identification of c-Maf as a liver sinusoidal specific transcription factor
To elucidate the molecular determinants of sinusoids, we analyzed transcription factors identified as drivers (fit likelihood > 0.17) in our RNA velocity analysis. To define those transcription factors specific to liver endothelium, we performed differential gene expression analysis across tissues from the Tabula Muris database (Tabula Muris Consortium et al., 2018). We found c-Maf to be the top-enriched (FDR<10−10; fold change = 3.6) transcription factor in liver ECs (Figure 2G), with a continuous increase over time (Figure 2H), being the most highly expressed member of the Maf family within liver sinusoids (Figure S4A). Notably, while Gata4 also mediates liver sinusoidal development (Géraud et al., 2017); Gata4 widespread homogenous expression across liver EC types and developmental stages suggests a wider functionality as compared to c-Maf (Figure S4B,C).
Expression of Bmp9 (Desroches-Castan et al., 2019a), LIF and IL6 (Giordano et al., 2015; Yang et al., 2005) have been associated with c-Maf expression. Treatment of liver EC cultures with Bmp9 in vitro, but not with LIF or IL6 increased c-Maf expression (Figure S4D), along with other liver endothelial genes (Figure S4E). To uncover the contribution of c-Maf to liver vascular development, we performed flow cytometry analysis of c-Maf and the sinusoidal markers Mrc1 and Fcgr2b from fetal E12.5 to E18.5 and in 4 weeks postnatal stage (Figure S4F). We observed an increase in co-expression of c-Maf, Mrc1 (Figure 2I and Figure S4G,H) and c-Maf with Fcgr2b (Figure 2J and Figure S4I,J) through the developmental stages. Hence, c-Maf is well-positioned to mandate liver sinusoidal identity.
c-Maf is a key determinant of liver sinusoidal signature
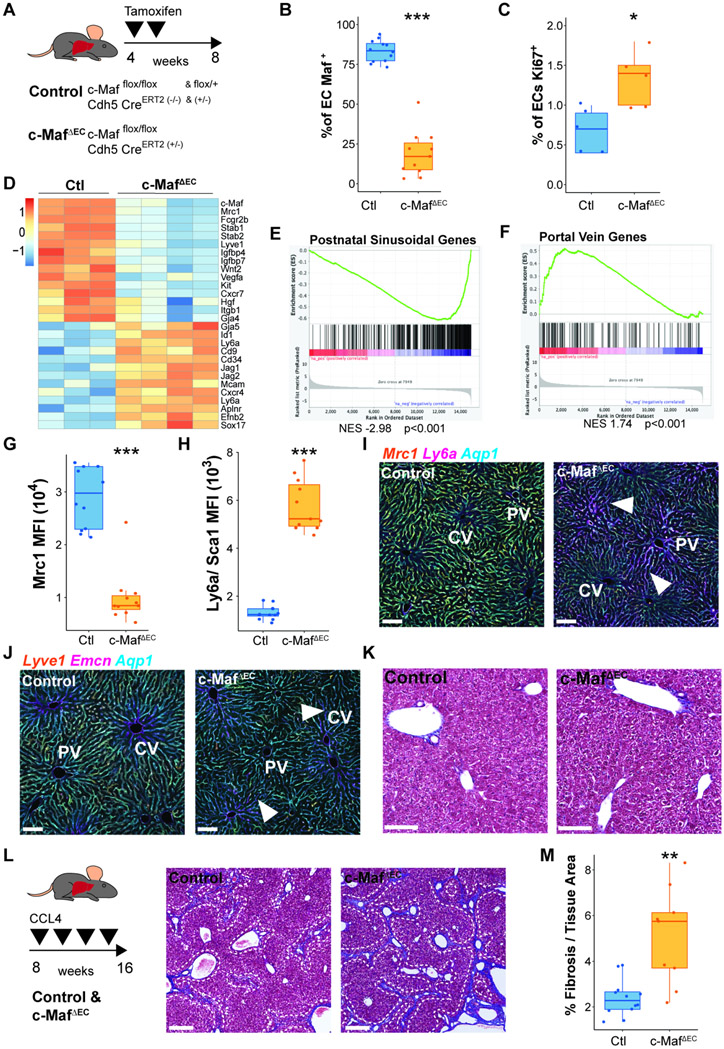
Developmental deletion of c-Maf is embryonically lethal phenotype due to a lack of erythropoiesis associated with aberrant alteration of macrophages in the blood islands of the liver, and reduced liver size (Kusakabe et al., 2011). To circumvent this impediment and study the contribution of endothelial c-Maf in regulating sinusoidal differentiation, we generated an inducible and tissue-specific mouse model, in which upon tamoxifen treatment c-Maf is selectively and conditionally deleted in ECs (VEcadherin-CreErt2/c-Mafflox/flox) to create c-MafΔEC mice (Wende et al., 2012). To prevent recombination in the hematopoietic lineage, we deleted c-Maf in the VEcadherin-CreErt2/c-Mafflox/flox mice by treating the pregnant mice with tamoxifen at E12 to E14 and analyzed the mice at E16 (Figure 3A, left panel). As opposed to constitutive deletion of c-Maf, (Kusakabe et al., 2011), embryos from c-MafΔEC mice did not show macroscopic abnormalities, including anemia or reduced liver size (Figure 3A, right panel), however CD45negCD31+ ECs (Figure S4K) showed a reduction in c-Maf+ ECs (Figure 3B), whereas CD45+CD68+ hematopoietic myeloid cells remained unaffected (Figure S4L).
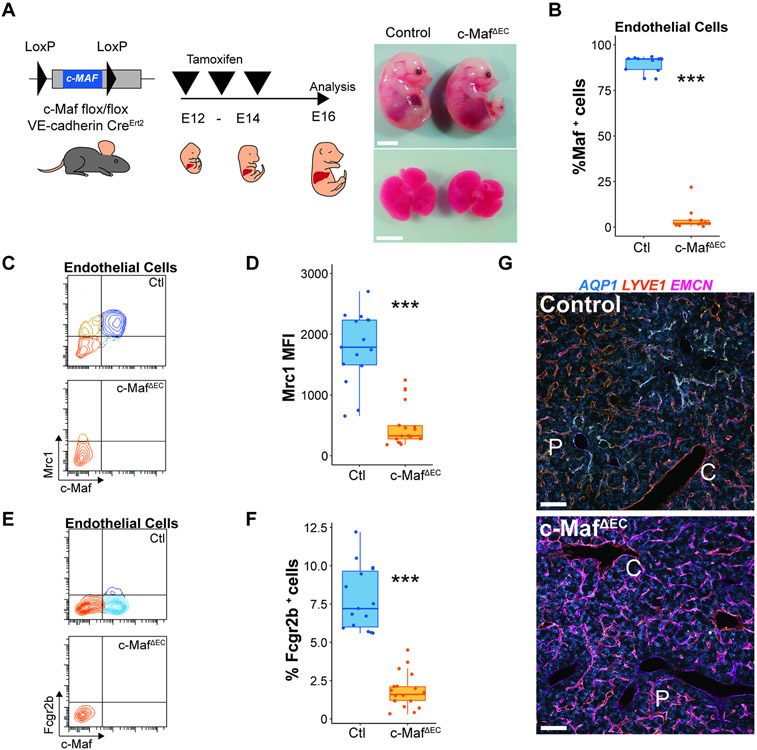
Figure 3. c-Maf choreographs the acquisition of sinusoidal attributes during maturation of immature liver capillaries.
(A) c-Mafflox/flox mice were crossed with VEcadherin(Cdh5)-CreErt2 mice, induced with tamoxifen from E12 to E14, to generate c-MafDEC mice and analyzed at the E16 developmental time point. Fetal embryos and livers from control and c-MafΔEC mice are shown. Scale bars 500 μm. (B) Flow cytometry analysis of the vasculature showing the deletion specificity of c-Maf within the ECs. Student’s t test analysis ***p < 0.001 (n = 12). (C) Flow cytometry analysis of the expression of c-Maf and Mrc1 at E16.5 in control and c-MafΔEC mice. (D) Analysis of the medium fluorescence intensity of Mrc1 expression in ECs in the control and c-MafΔEC mice. Student’s t test analysis ***p < 0.001 (n ≥ 15). (E) Flow cytometry analysis of the expression of c-Maf and Fcgr2b at E16.5 in control and c-MafΔEC mice. (F) Analysis of the percentage of ECs positive for Fcgr2b in the control and c-MafΔEC mice. 5Student’s t test analysis ***p < 0.001 (n ≥ 15). (G) Immunofluorescence analysis of the vascular markers Aqp1, Lyve1, and Emcn in control and c-MafΔEC mice at E16.5 of development, induced as shown in (A) from n = 5 mice. “C” represents central vein and “P” represents portal vein. Scale bars: 100 μm.
Embryos from EC-specific c-Maf deletion showed a reduction of sinusoidal markers Mrc1 (Figure 3C,D) and Fcgr2b (Figure 3E,F), consistent with a critical role of c-Maf in sinusoidal identity determination. This was coupled with an increase in the venous marker Emcn (Figure 3G). Cell proliferation as measured by Ki67 did not show significant changes in the endothelial compartment (Figure S4M). A decrease in Fractal dimension indicated that deletion of c-Maf disrupts the patterning of zonated sinusoids, (Figure S4N). Thus, c-Maf is required during development for the phenotypic specification and formation of sinusoidal network.
Deletion of c-Maf expression prevents sinusoidal differentiation and aberrantly expands postnatal liver hematopoiesis
To further explore the role of c-Maf during postnatal liver development, we induced its deletion by administrating 4-hydroxytamoxifen to mouse pups and analyzing them prior to P15 (Figure 4A, upper panel). We performed scRNA-seq analysis of cells from the endothelial (CD31+CD45neg) and hematopoietic (CD31negCD45+) compartments (Figure 4A, bottom panel). Cells from two animals, male and female, per condition, control and c-MafΔEC, were sorted and combined, and separated analytically based on the expression of sexual dimorphism genes (Figure S5A). We identified a total of 23 populations, subdivided into 7 vascular endothelial sub-clusters, 14 hematopoietic sub-clusters, and two clusters of contaminant doublets and hepatocytes (Figure 4B, Figure S5B and Table S3). Within the ECs, c-Maf expression was reduced in c-MafΔEC cells compared to controls, but not in Kupffer cells (Figure 4C and Figure S5C,D). In c-MafΔEC mice, only a small fraction of ECs (~17.7%) retained c-Maf expression, in line with a deletion efficiency of 92.8% calculated by flow cytometry (Figure S5E). We observed EC clusters in the c-Maf deficient mice that were absent in the WT counterparts (clusters KO-S[1-3]; Figure 4D) suggestive of a sinusoidal differentiation defect. Notably, within the EC clusters, there was an increased proportion of proliferative Ki67+ ECs in the c-MafΔEC mice (Figure 4E).
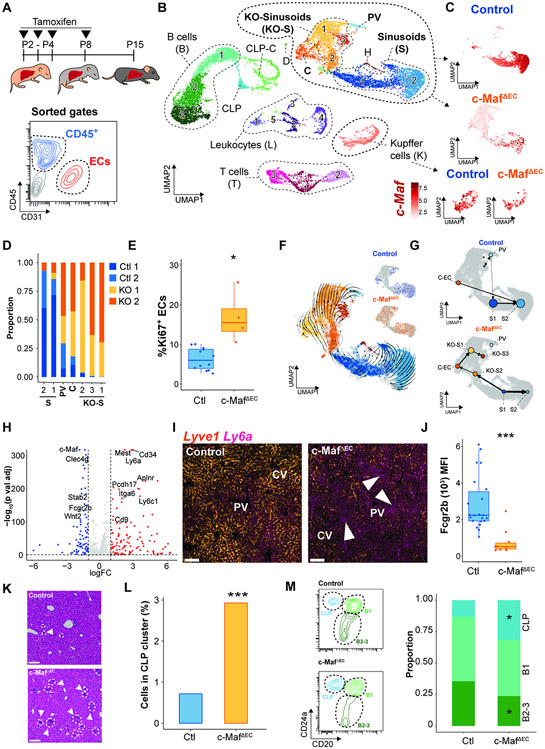
Figure 4. c-Maf postnatal deficiency overextends the restricted hematopoietic liver sojourn through an aberrant vascular arterialization.
(A) Analysis of the influence of c-Maf on postnatal liver vascular development was performed by 4-hydroxytamoxifen administration from P2 to P4 and at P8 and analyzed at P15 in control and c-MafΔEC mice. Flow-assisted cell sorting (FACS) was performed for the gated CD45+ CD31neg hematopoietic, and CD45negCD31+ endothelial cell populations, as indicated on the flow chart, from control and c-MafΔEC mice. (B) ScRNA-seq of the hematopoietic and EC-sorted populations from (A) was performed. UMAP labeling of the different endothelial and hematopoietic populations identified: endothelial populations: sinusoids (S 1–2), KO-sinusoids (KO-S 1–3), cycling (C), and portal vein (PV); hematopoietic populations: CLPs, cycling – CLP (CLP-C), B cells (B 1–3), T cells (T 1–3), leukocytes (L 1–5), and Kupffer cells (K); contaminant cells were identified as: doublets (D) and hepatocytes (H). (C) Expression of c-Maf in the endothelial and Kupffer cell clusters of control and c-MafΔEC mice. (D) Proportion analysis shows identification of the generation of a unique clusters of cells within the c-MafΔEC mice associated with the c-Maf-deficient cells. (E) Flow cytometry analysis of the percentage of Ki67 positive cells within the ECs. Student’s t test analysis *p < 0.05 (n ≥ 4). (F) Pseudotime analysis of the EC populations. Arrows show the directionality of changes associated with the transition between different cells. The top right panel shows the associated pseudotime per sample. (G) Partition-based graph abstraction analysis of transition confidence between EC clusters in either control or c-MafΔEC mice. (H) Volcano plot of the differentially expressed genes between the control and c-MafΔEC mice. Several genes associated with the sinusoidal cell and portal vein populations are shown. Colors indicate fold change in expression: red – increased and blue – decreased. (I) Immunofluorescence staining of Sca1 and Lyve1 in control and c-MafΔEC mice. Representative image of n = 5, scale bars 100 μm. Arrows indicate loss of Lyve1 and presence of Sca1 in sinusoids. “PV” represents portal vein and “CV” represents central vein. (J) Quantification by flow cytometry of the expression levels of Fcgr2b in control versus c-MafΔEC mice. Student’s t test analysis ***p < 0.001 (n ≥ 10). (K) Hematoxylin and eosin staining of control and c-MafΔEC livers. Arrows and circles show areas with increased deposition of hematopoietic cells. Representative images of n = 5. Scale bars: 100 μm. (L) Percentage of cells in the cluster of CLPs identified from the scRNA-seq in (B). Fisher test ***p<0.001. (M) Flow cytometry analysis previously gated on CD45+CD45RA+ cells positive for CD24a and CD20 were defined as CLPs and B cells (B1, B2-3). Student’s t test analysis *p < 0.05 (n ≥ 5).
To estimate the differentiation trajectories of ECs, we performed RNA-velocity analysis (Bergen et al., 2020) (Figure 4F). We observed a distinct trajectory in c-MafΔEC ECs towards KO-S[1-3] cell clusters (Figure 4F, inset). These differentiation trajectories were also observed when performing graph abstraction analysis to estimate transition confidence (Figure 4G). Consistent with a differentiation defect in c-MafΔEC liver ECs, RNA velocity-derived pseudotime analysis show an enrichment of c-MafΔEC cells at early pseudotime points (Figure S5F). Analysis of the differentially expressed genes between the Control and c-MafΔEC ECs showed decreased expression of sinusoidal genes, such as Fcgr2b, Stab2, Clec4g and Wnt2, and an increase in arterial genes, including Cd34, Ly6a, Aplnr and Cd9 (FDR < 0.05, log2FC > 1; Figure 4H). The changes in the expression of sinusoidal and arterial signatures were further confirmed by immunofluorescence analysis of Ly6a and Lyve1 staining, showing an increase in Ly6a and decrease in Lyve1 staining (Figure 4I). Furthermore, we showed a decrease in Fcgr2b (Figure 4J) and Mrc1 (Figure S5G). These results are consistent with scRNA-seq analysis of an unscheduled appearance of a unique cell cluster of ECs within the c-MafΔEC sinusoids with higher expression of arterial genes.
The expression of an arterial phenotype is associated with emergence of hematopoietic progenitors in the fetal liver and adult bone marrow (Guo et al. 2017; Khan et al. 2016; Poulos et al. 2013). Histological analysis of the liver by H&E staining showed an increase in hematopoietic cells, specifically common lymphoid progenitor (CLP) cell cluster and decrease of differentiated B2-B3 cells in c-MafΔEC mice livers (Figure 4K,L,M). Matrix cellulose colony-forming assays also confirmed an increase in hematopoietic progenitors as shown by a significant increase in colony forming units in the c-MafΔEC mice (Figure S5H). Hence, postnatal vascular c-Maf deficiency impairs the acquisition of the sinusoidal identity, promotes an arterial phenotype and pathophysiological retention of liver hematopoiesis.
c-Maf orchestrates liver sinusoidal program in adult mice
To investigate the role of c-Maf in the adult liver endothelium, we induced its deletion in ECs at postnatal week 4 and analyzed them at postnatal week 8 (Figure 5A). We observed c-Maf deletion in 81.3% of the ECs (Figure 5B), with no significant effect in Cd45+Cd68+ myeloid cells (Figure S6A), nor in Cd45+Cd68+F4/80+Cdh5+ Kupffer cells (Figure S6B). Similar to what we observed at P15, there was an increase proliferative capacity of the ECs in the c-MafΔEC mice (Figure 5C). RNA-seq analysis show an overall decrease in sinusoidal signature, such as Mrc1, Fcgr2b, Stab1, Stab2, and Lyve1 and angiocrine genes, including Wnt2, Hgf, Itgb1, Igfbp4, and Igfbp7 in c-MafΔEC mice (Figure 5D). Conversely, expression of genes involved in the Portal Vein transcriptional program was increased, including Ly6a, Cd34, Cd9, Ephb2, Gja5 and Sox17 (Figure 5D). Employing gene set enrichment analysis (GSEA) also showed a depletion of postnatal sinusoidal genes (Figure 5E) and an increase in Portal Vein genes (Figure 5F).
Figure 5. c-Maf adult vascular deficiency aberrantly induces a Portal Vein signature of the capillaries and facilitates stress-induced liver fibrosis.
(A) Induction of c-Maf deficiency in c-Mafflox/floxVEcadherin(Cdh5)-CreERT2 mice specifically in vascular endothelium was performed by tamoxifen administration after P30 using 3 days on, 3 days off, and 3 days on protocol, to generate c-MafΔEC mice. (B) Analysis of vascular ECs shows c-Maf deletion within the endothelium. Student’s t test analysis ***p < 0.001 (n ≥ 11). (C) Flow cytometry analysis of the percentage of Ki67 positive cells within the ECs. Student’s t test analysis *p < 0.05 (n = 5). (D) Bulk RNA-seq analysis of adult liver ECs isolated from control and c-MafΔEC mice, showing the differential expression signatures between them. (E) GSEA analysis of the postnatal sinusoidal gene list from population PS5 identified in Figure 1B. The analysis shows a decrease in the expression of sinusoidal genes associated within the c-MafΔEC mice. (F) GSEA analysis of the portal vein gene list from population PV identified in Figure 1B. The analysis shows an increase in the expression of these genes within the c-MafΔEC mice. (G and H) Flow cytometry quantification of the medium fluorescence intensity of sinusoidal marker Mrc1 (G) and Ly6a/Sca1 (H) expression in control and c-MafΔEC mice. Student’s t test analysis ***p < 0.001 (n ≥ 11). (I and J) Immunofluorescence analysis of the expression of liver vascular markers Mrc1, Ly6a/Sca1, Lyve1, Emcn, and Aqp1 in control and c-MafΔEC mice. Representative image from n = 5. Scale bars, 100 μm. “CV” represents central vein, “PV” represents portal vein. Arrows show expression of Ly6a and Emcn within the sinusoids in I and J, respectively. (K) Representative images of Masson’s trichrome from n = 3 of control and c-MafΔEC mice under basal conditions. A small fraction of fibrosis deposition could be observed surrounding the portal vein, both in the control and the c-MafΔEC mice. Scale bars: 100 μm. (L) Induction of fibrosis using CCl4 treatment in control and c-MafΔEC mice 1 month after deletion was induced. Representative images of Masson’s trichrome from n ≥ 9 of control and c-MafΔEC mice, showing the fibrotic deposition in blue. Image shows a zoom region from a tile scan of the whole liver. The white dashed line follows the fibrosis area. Scale bars 200 μm. (M) Quantification of the percentage of fibrosis over the total tissue area of control and c-MafΔEC mice. Student’s t test analysis **p < 0.01 (n ≥ 9).
To unravel c-Maf regulatory landscape, we defined the position of the c-Maf DNA binding motifs within open chromatin regions of liver ECs using available ATAC-seq data (Winkler et al., 2021). We next intersected the list of genes with associated c-Maf motifs, and the sinusoidal gene list derived from our scRNA-seq data. Gene set enrichment analysis showed a decreased signature of postnatal sinusoidal genes with associated c-Maf motif in c-MafΔEC mice (Figure S6C). We also show a decrease in sinusoidal Mrc1 (Figure 5G) and Fcgr2b (Figure S6D), and increase in arterial Ly6a (Figure 5H) and Cd9 (Figure S6E). upon c-Maf deletion immunofluorescence analysis indicated the decrease of Mrc1 and concomitant increase of the arterial marker Ly6a in liver sinusoids (Figure 5I). Moreover, expression of other adult sinusoidal markers, including Lyve1 were disrupted, along with increased expression of the venous marker Emcn in the vicinity of the Central Vein (Figure 5J). However, deletion of c-Maf in adults with already established vascular networks did not alter fractal dimension patterning (Figure S6F).
Analysis of the microstructure of the sinusoids by electron microscopy did not show absence of fenestrations after c-Maf deletion, suggesting that other pathways might control this unique structural remodeling (Figure S6G). Notably, analysis of the zonation markers E-cadherin and Cyp2E1 also did not reveal changes in the portal to centro-lobular zonation gradient (Figure S6H). However, we observed an expansion of glutamine synthetase-expressing hepatocytes within the c-MafΔEC mice, with an increase staining of the venous marker Emcn (Figure S6I), suggestive of mild disruption of liver co-zonation. Loss of sinusoidal identity can potentially arise from two distinct scenarios: i) expansion of the Portal Vein or ii) activation of arterial signatures within the sinusoids. Our previous results in postnatal development suggest the latter scenario, together with increased in the proliferative phenotype. To test this hypothesis, we generated confetti c-MafΔEC mice to examine clonal expansions in the liver. We observed an increased size of sinusoidal clones, supporting our previous observations (Figure S6J). Thus, c-Maf deletion in adult liver sinusoids results in the loss of sinusoidal identity with acquisition of Portal Vein markers and a mild disruption of liver co-zonation.
Loss of both liver co-zonation and sinusoidal identity triggers liver fibrosis (Desroches-Castan et al., 2019a). Hence, we studied whether induction of fibrosis results in loss of c-Maf positive cells. We induced liver fibrosis using biweekly doses of 25% carbon tetrachloride (CCl4) for 1 month. In line with c-Maf capable of determining sinusoidal identity, induction of fibrosis resulted in an increase of Mrc1neg/c-Mafneg ECs (Figure S6K). Although no fibrosis was observed in basal conditions in the c-MafΔEC mice (Figure 5K, and Figure S6L), CCl4 treatment increased fibrotic area compared to controls (Figure 5L, M). Therefore, loss of sinusoidal identity postnatally is not sufficient for induction of liver fibrosis in adult mice, but increases predisposition towards healing by fibrosis upon chemical insult. Hence, c-Maf is required for the maintenance of liver sinusoidal identity and for restoring fibrosis-free homeostasis upon chemical insult in adult mice.
scRNA-seq of human liver ECs informs of signature-based isolation strategy
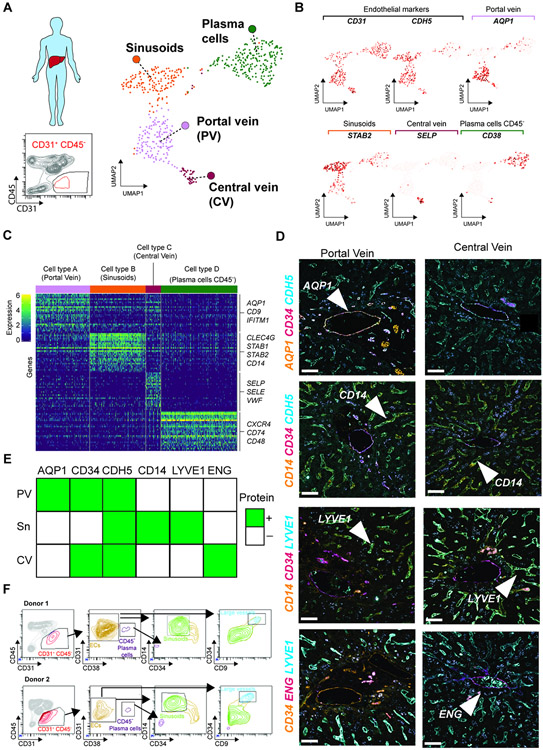
scRNA-seq of liver sinusoids demonstrate co-zonation with hepatocytes (Halpern et al., 2018; MacParland et al., 2018). Here, we performed scRNA-seq analysis of the CD45negCD31+ population in human adult liver to resolve cell heterogeneity. We observed four distinct cell clusters, of which the clusters representing Sinusoids, Portal Vein and Central Vein were assigned to the EC compartment based on the expression of CD31, CDH5, AQP1, STAB2 and SELP (Figure 6A,B and Table S4). Notably, we identified plasma cells as CD45negCD31+ and CD38 (MacParland et al., 2018) (Figure 6A,B and Table S4). The Sinusoid and Portal Vein clusters expressed well-established marker genes (Figure 6C) that enabled their classification based on prior studies (Halpern et al., 2018; MacParland et al., 2018). To resolve the identity of the Central vein cluster, we performed differential expression between Central and Portal Vein clusters (Figure S7A), which warranted its classification based on ENG and RSPO3 angiocrine expression. Furthermore, while large vessels are CD34+, the Portal Vein is the only AQP1+ human vascular population (Figure 6D,E). Notably, the expression of CD14 and LYVE1 was associated with sinusoidal ECs, while endoglin (ENG) was observed exclusively in Central Vein (Figure 6D,E). Accordingly, we developed a flow cytometry strategy that allowed us to identify human liver ECs as CD45negCD31+CD38neg cells, of which the sinusoids were phenotypically defined as CD45negCD31+CD38negCD14+CD34low, and large vessels (Portal and Central Veins) as CD45negCD31+CD38negCD14+CD34highCD9+ ECs (Figure 6F). These results expand the previous description of the molecular features of the human liver endothelium and can potentially serve as a reference dataset for future analysis.
Figure 6. Human liver EC subpopulations can be identified by the expression of unique differential markers acquired by immunofluorescence and flow cytometry.
(A) Human liver CD45negCD31+ cells were sorted and analyzed by scRNA-seq analysis (n = 1). UMAP analysis of the EC compartment from the human liver sorted as CD45negCD31+ cells. Colors represent each population identified, based on their differentially expressed markers. (B) ScRNA-seq analysis of human liver CD45negCD31+ cells identified four clusters based on the expression of specific markers such as CD31, CDH5, AQP1, STAB2, SELP, or CD38. We used the co-expression of both CD31 and CDH5 to identify the ECs. These four clusters were associated with the portal vein, sinusoids, central vein, and putative hematopoietic plasma cells (CD45negCD38+). (C) Identification of the expression of cluster-associated specific markers between each subpopulation represented as a heatmap. Colors show the varying expression levels of each gene per cell. (D) Immunofluorescence validation of the vascular markers identified by single-cell RNA-seq analysis allows the identification of specific markers associated with the portal vein, sinusoids, or central vein based on the expression of Aqp1, Cd34, Cdh5, Cd14, Lyve1, and Eng. Arrows represent the labeling of the gene in a particular population. Representative images from 3 patients. Scale bars, 100 μm. (E) Table representing the identification of protein expression by immunofluorescence on the portal vein (PV), sinusoids (S), or central vein (CV) in the human liver. Green color indicates positive identification of expression in (D). (F) Identification by flow cytometry of the different vascular subpopulations found in the scRNA-seq, based on the expression of the surface markers CD45, CD31,CD38, CD14, CD34, and CD9, performed in two different human liver samples.
Importantly, analysis of a human fetal liver at 17 weeks of development showed a different pattern from the adult. Similar to mouse development, the Portal Vein was phenotypically differentiated early in development, with the absence of the sinusoidal marker LYVE1, while the Central Vein remained LYVE1+ (Figure S7B). We also defined the sinusoids as CD14+ in the human fetal liver at 17 weeks, recapitulating previous observations at week 20 (Couvelard et al., 1996) (Figure S7C). Unlike in adult liver CD31/CD45 gating strategy, no plasma cells marked by CD45negCD31+CD38+, were observed during fetal development (Figure S7C). Therefore, scRNA-seq analysis of sorted CD31+CD45neg revealed specific markers for precise gating of adult liver ECs. Our proposed strategy prevents inclusion of plasma cells into the EC gate, improving the purity of the EC population for downstream analysis.
c-Maf overexpression induces the human sinusoidal transcriptional program in vitro
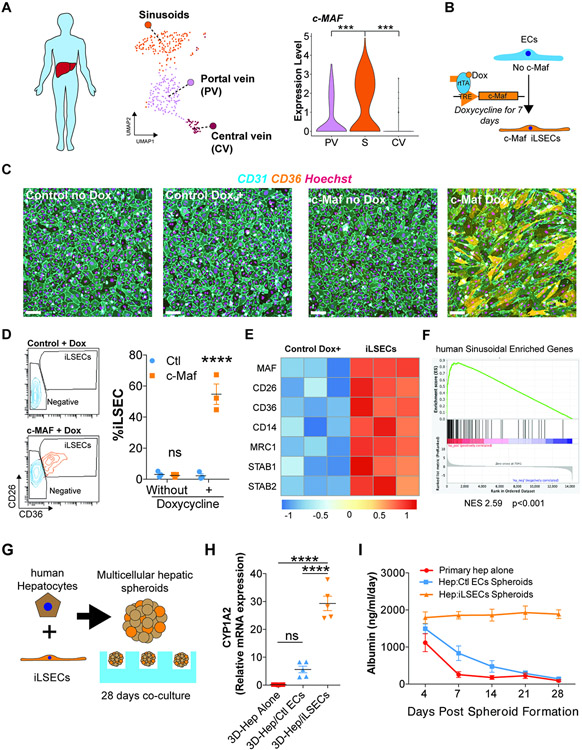
We next leveraged our human scRNA-seq data to explore the role of c-Maf in human ECs enforced specification into sinusoidal signature. Notably, human liver sinusoids showed higher expression levels of c-Maf compared to the Portal or Central Veins (Figure 7A). To test whether c-Maf has the capacity to induce a sinusoidal program in human ECs, we used a doxycycline inducible lentiviral system to overexpress c-Maf in human Umbilical Vein ECs (HUVECs) that represent generic vessels (Figure 7B).
Figure 7. c-Maf induces a pro-regenerative liver signature in vascular ECs, enabling long-term sustenance of co-cultured hepatocytes.
(A) Expression of c-Maf in the EC subpopulations from the human single-cell analysis. UMAP represents the different EC populations identified. The results show an enrichment in the sinusoidal population (S) compared with the portal vein (PV) and central vein (CV). Student’s t test comparison between total cells per cluster from n = 1 sample, ***p < 0.001. (B) Diagram representing the induction of c-Maf overexpression in human ECs in vitro using an inducible doxycycline system for 7 days. (C) Immunofluorescence analysis of the expression of CD31 and CD36 in ECs transduced with a lentivirus control or lentivirus overexpressing c-Maf under the control of doxycycline. Nuclei are stained with Hoechst. Analysis of cell cultures of control and c-Maf cells, with or without doxycycline, for 7 days. Representative image of n = 3. Scale bars, 100 μm. (D) Flow cytometry identification and quantification of induced LSECs (iLSECs) after the administration of doxycycline. Cells overexpressing c-Maf acquire elevated expression of CD26 and CD36. Representative flow of n = 3 (different donors). Data are presented as mean ± SEM. Two-way ANOVA ****p < 0.0001 (n = 3). (E) Bulk RNA-seq analysis of control and iLSECs treated for 7 days with doxycycline. Heatmap shows differential expression analysis between the iLSEC sorted population overexpressing c-Maf and control cells from n = 3 donor cells. (F) GSEA analysis of the expression values of the human sinusoidal enriched genes from Figure 6A identified an upregulation of the sinusoidal signature in the iLSECs. (G) Representation of the co-culture of human hepatocytes and iLSECs to generate spheroids. (H) qRT-PCR results indicating relative CYP1A2 expression at day 28 of the co-culture of human hepatocytes (Hep) with ECs (control or iLSECs) or hepatocytes alone. Data are presented as mean ± SEM. One-way ANOVA, ****p < 0.0001 (n = 5). (I) ELISA quantification of the expression of albumin over the 28-day co-culture of hepatocytes (Hep) with ECs (control or iLSECs). Data are presented as mean ± SEM (n = 5).
Human liver sinusoids are known to co-express microvascular markers, including CD36, CD26 (Fukui et al., 1990; Strauss et al., 2017), and CD14 (Couvelard et al. 1996; MacParland et al. 2018; Strauss et al. 2017). Overexpression of c-Maf in HUVECs lead to increase expression of CD36 by immunofluorescence and morphological changes (Figure 7C and Figure S7D,E), and emergence of double positive CD36+/CD26+ population comprising 54.76% ± 9.36 (mean ± standard deviation) of EC population(Figure 7D). We termed the CD36+CD26+ population as Induced Liver Sinusoidal ECs (iLSECs). RNA-seq analysis of the iLSECs showed an enhanced expression of multiple liver sinusoidal markers (FDR < 0.05) compared to control cells (Figure 7E), which was further supported by gene set enrichment analysis (Figure 7F), although not all sinusoidal genes, such as Fcgr2b, were induced. Remarkably, human liver sinusoidal markers CD14 and MRC1 (Figure S7F,G) were also induced in iLSECs overexpressing c-Maf (Figure S7H). Thus, enforced c-Maf expression is sufficient to induce activation of a pro-sinusoidal transcriptional program and sinusoid phenotype in human ECs in vitro.
To determine if iLSECs support hepatocyte function, we designed a co-culture system of iLSECs with human adult primary hepatocyte aggregates (Song et al., 2015) (Figure 7G). Electron microscopy showed ECs from control and iLSECs interacting with the hepatocyte aggregates at day 7, displaying cytoplasmic fenestration gaps in the iLSECs, but not in the controls (Figure S7I). In order to assess hepatocyte function, we quantified the expression of CYP1A2, a key enzyme in liver metabolism, and the secretion of albumin during 28 days of co-culture. Contrary to mock-transduced control ECs, iLSECs sustained long-term hepatocyte function, as demonstrated by CYP1A2 expression (Figure 7H) and maintenance of albumin production (Figure 7I). Thus, c-Maf overexpression is sufficient for induction of liver sinusoidal identity and enables long-term sustainability of hepatocyte function. Therefore, c-Maf plays a key role in specifying sinusoidal identity and function.
Discussion
During development, EC progenitors by differentiating into specialized blood vessels adapt to the organotypic metabolic, immunological, and homeostatic demands (Augustin and Koh 2017; Gomez-Salinero and Rafii 2018; Jakab and Augustin 2020; Rafii et al. 2016). In particular, the liver is arborized by variegated vasculature with remarkable intra-organ vascular heterogeneity, and complex regional co-zonation with hepatocytes (Halpern et al., 2018; MacParland et al., 2018; Planas-Paz et al., 2016). Transcription factor Gata4 is enriched in liver endothelium compared to other organs, supporting hepatic functions (Géraud et al., 2010, 2017; Winkler et al., 2021). However, by performing scRNA-seq of liver endothelium at multiple stages of development, we found Gata4 to be homogeneously and non-specifically expressed across all EC types in the liver, including arterial, venous and sinusoidal liver ECs. By contrast, we found c-Maf, another putative but poorly studied liver EC specific transcription factor, to be developmentally induced exclusively in the hepatic sinusoids, but not arterial or venous liver ECs. Employing a genetic model of c-Maf deletion in the endothelium across multiple developmental stages, we show the critical requirement of c-Maf for the establishment of sinusoidal identity. These results extend the previous view of how liver endothelium specializes over time, identifying c-Maf as a unique central player in determining sinusoidal identity and function.
Leveraging scRNA-seq analysis, we resolved liver vasculature heterogeneity during fetal to postnatal development, being specified with signatures of large vessels and sinusoids. We uncovered an early undifferentiated population of sinusoidal EC progenitors at E12 with robust Cd34 expression that acquire their differentiation markers through development. While a small percentage of liver ECs originate from Csf1r+ erythro-myeloid progenitor cell (Plein et al., 2018), we could not detect these cells, probably due to the silencing of this gene before EC differentiation. Notably, while the Portal Vein is specified early in fetal development, we found that the Central Vein program is specified late in fetal development, coinciding with the requirement of early postnatal hepatocyte co-zonation (Boj et al., 2012; Planas-Paz et al., 2016; Rocha et al., 2015). Our results concur with recent finding that scRNA-seq analysis does not always match protein expression in ECs (Inverso et al., 2021), as observed for Emcn. We unraveled a high degree of transcriptional heterogeneity within the sinusoidal compartment during development and identified the transcription factor c-Maf as a main regulator of this process. Deletion of c-Maf results in the erasure of postnatal sinusoidal program and activation of an arterial phenotype in the sinusoids that resembles Portal Vein ECs. Upon c-Maf deletion, we observed an aberrant expansion of the postnatal hematopoietic cells, consistent with the findings that arterial endothelial fate during liver development orchestrates fetal and adult bone marrow hematopoiesis (Guo et al., 2017; Khan et al., 2016; Poulos et al., 2013).
Abnormal sinusoidal arterialization has been associated with liver fibrosis (Desroches-Castan et al., 2019a); however, it remains to be determined whether arterialization is sufficient for development of liver fibrosis. Our results suggest that acquisition of Portal Vein markers by the sinusoids in c-Maf deficient mice is not sufficient to cause fibrosis in adult stages. However, c-Maf deletion enhanced the sensitivity towards CCl4-induced fibrosis. These results differ from previous reports, in which an increased fibrotic response was observed under basal conditions in Gata4 or Bmp9 deficient mice, (Desroches-Castan et al., 2019a; Winkler et al., 2021). These differences could be attributed to an earlier activation of the Cre as reported for Gata4 (Géraud et al., 2017; Winkler et al., 2021) or to differences in the background as observed for Bmp9 between C57/Bl6 and 129/Ola mice (Desroches-Castan et al., 2019b). Understanding these differential responses between postnatal to adult induction and the genetic background of the mice may authorize the development of improved anti-fibrotic therapies for mitigating liver fibrosis.
The transcription factors Gata4, c-Maf, and Meis2 might represent a complementary transcriptional core for liver sinusoidal specification (Géraud et al., 2010; de Haan et al., 2020). Furthermore, their overexpression in human ECs induce a liver signature at the RNA level (Géraud et al., 2017; de Haan et al., 2020), and Gata4 induction during the differentiation of liver ECs from pluripotent stem cells (Gage et al., 2020). We show that overexpression of c-Maf alone induces most sinusoidal gene signatures, enforcing the long-term sustenance of hepatocytes aggregates in vitro. Although the combined expression of Gata4, c-Maf, and Meis2 could regulate sinusoidal transcriptional program on its entirety, expression of c-Maf is sufficient for the acquisition of sinusoidal identity and certain essential specialized functions. Similar observations have also been made in other organs, including kidney, in which the sole expression of the transcription factor Tbx3 sustain glomerulus EC signatures (Barry et al., 2019), while co-expression of Pbx1 and Gata5 complement Tbx3 glomeruli specification. Deciphering the cooperative interactions among various transcription factors favors the implementation and maintenance of angiodiversity will warrant the development of a vascular zone-specific transcription factor code that could be used for guiding organ regeneration.
Thus, acquisition of the different vascular cell identities is a dynamic process initiated during fetal development and completed postnatally. Our results shed light on the temporal acquisition of EC organotypic attributes in the liver, identifying c-Maf as a critical transcription factor for sinusoidal cell identity. Deletion of c-Maf expression disrupts vascular development, promotes acquisition of Portal Vein traits in the sinusoid compartment, abnormal hematopoietic potential and increased sensitivity to fibrosis upon liver insult. However, the mechanism by which liver sinusoids interact with their post-natal microenvironment to acquire c-Maf dependent specialization requires further investigation. Notwithstanding of these caveats, the present study resolves intra-organ vascular cell heterogeneity in the liver and defines the role of c-Maf in the establishment of sinusoidal identity, laying the foundation for the design of vascular-driven liver repair.
Limitations of the study
We have identified c-Maf as liver sinusoidal specification transcription factor, apart from Gata4 and potentially another candidate Meis2. We have used a VEcadherin-CreERT2 to selectively delete c-Maf post-natally in 4 weeks old mice, resulting in erasure of sinusoidal signature but not structural defects, such as fibrosis-although we observed increase fibrosis after CCl4 treatment. Whether earlier deletion of c-Maf during fetal stages or perinatally could lead to more severe loss of sinusoidal identity with capillarization and fibrosis need to be further investigated. Moreover, as VEcadherin-CreERT2 is not 100% efficient in deleting c-Maf, and as lymphatic vessels, also express c-Maf (Hansen et al., 2010), other liver specific Cre, such as Clec4g-iCre (Wohlfeil et al., 2019) or longer tamoxifen treatment might achieve more efficient c-Maf deletion specifically in liver vessels. Nonetheless, due to the small contribution of lymphatics in the liver at steady state; loss of c-Maf in these cells is most likely inconsequential for the described liver phenotype.
We show that c-Maf overexpression, induced liver sinusoidal gene signatures, except for a few genes, including Fcgr2b. It is plausible that use of a combination of cooperative transcription factors, including Gata4, Meis2 and c-Maf among others along with signals from microenvironmental and metabolic cues, might be required to implement full sinusoidal signatures. Alternatively, co-culture with hepatocytes, stellate cells and Kupffer cells could also be essential to provide with the appropriate inductive signaling pathways required for their adequate specialization.
Finally, we show that LSEC within the stretch of each sinusoidal vessel extending from portal vein to the central vein are also zonated. While c-Maf seems to re-establish the LSEC signature the mechanism by which LSEC acquire their hierarchical fenestrated zonation and co-zonation with hepatocytes might be driven by biochemical and biophysical cues. Employing models in which human ECs are engineered as adaptable endothelial cells, such as Reset-Vascular Endothelial Cells (R-VECs) (Palikuqi et al., 2020) could set stage for uncovering the determinants of LSEC zonation as well.
STAR Methods:
Resource availability
Lead contact
Any request related to the resources and reagents generated should be directed to the lead contact: Shahin Rafii (srafii@cornell.med.edu)
Material availability
Plasmids and other resources generated on this study are available upon request to the lead contact.
Data and code availability
The RNA and single cell RNAseq data presented in this study have been deposited at GEO: GSE172360, GSE172361, GSE172362. Accession numbers are provided in the Key resource table. Code for analysis of the single cell RNAseq has been deposited at GitHub and is publicly available. Access information for raw files can be found in the Key resource table.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PE Mouse anti-human CD31 (clone WM59) | BD Bioscience | Cat 555446 |
| BV421 Mouse anti-human CD45 (clone 2D1) | Biolegend | Cat 368522 |
| BV421 mouse anti-human CD31 (clone WM59) | Biolegend | Cat 303124 |
| Fitc mouse anti-human CD14 (clone 63D3) | Biolegend | Cat 367116 |
| Pe mouse anti-human CD26 (clone BA5b) | Biolegend | Cat 302706 |
| PE/Cyanine 7 mouse anti-human CD36 (clone 5-271) | Biolegend | Cat 336222 |
| Goat anti-mouse MRC1/CD206 (polyclonal) | R&D systems | Cat AF2535 |
| PE mouse anti-human/mouse MAF (clone symOF1) | ThermoFisher Scientific | Cat 12-9855-42 |
| APC/Cyanine 7 anti-human CD45 (clone HI30) | Biolegend | Cat 304014 |
| PE/Cyanine 7 mouse anti-human CD34 (clone 581) | Biolegend | Cat 343416 |
| BV421 mouse anti-human CD38 (clone HB-7) | Biolegend | Cat 356618 |
| Human TruStain FC Blocker | Biolegend | Cat 422302 |
| Pe/Cyanine 7 rat anti-mouse CD31 (clone 390) | Biolegend | Cat 102418 |
| Alexa 488 rat anti-mouse CD45 (clone 30-F11) | Biolegend | Cat 103122 |
| BV421 rat anti-mouse CD45 (clone 30-F11) | Biolegend | Cat 103134 |
| APC/Cyanine 7 rat anti-mouse CD45 (clone 30-F11) | Biolegend | Cat 103116 |
| PE mouse anti-mouse CD32B (clone AT130-2) | ThermoFisher Scientific | Cat 12-0321082 |
| BV421 rat anti-mouse MRC1/CD206 (clone C068C2) | Biolegend | Cat 141717 |
| PE/Cyanine 7 rat anti-mouse Ly6A/E (clone D7) | Biolegend | Cat 108114 |
| APC mouse anti-mouse CD32B (clone AT130-2) | ThermoFisher Scientific | Cat 17-0321-82 |
| APC rat anti-mouse CD68 (clone FA-11) | Biolegend | Cat 137008 |
| Alexa 488 rat anti-mouse CD31 (clone 390) | Biolegend | Cat 102414 |
| TruStain FcX rat anti-mouse CD16/32 (Clone 93) | Biolegend | Cat 101320 |
| APC rat anti-mouse CD45R/B220 (clone RA3-6B2) | Biolegend | Cat 103212 |
| Alexa 488 rat anti-mouse CD24 (clone M1/69) | Biolegend | Cat 101816 |
| PE/Cyanine 7 rat anti-mouse CD45 (clone 30-F11) | Biolegend | Cat 103114 |
| PE rat anti-mouse CD20 (clone SA275A11) | Biolegend | Cat 150409 |
| PE/Cyanine 7 rat anti-mouse F4/80 (clone BM8) | Biolegend | Cat 123113 |
| BV421 rat anti-mouse CD68 (clone FA-11) | Biolegend | Cat 137017 |
| Alexa 700 rat anti-mouse CD45 (clone 30-F11) | Biolegend | Cat 103128 |
| Alexa 647 rat anti-mouse Cdh5 (clone11D4.1) | BD Bioscience | Cat 562242 |
| Alexa 647 rat anti-mouse Cdh5 (clone BV13) | Biolegend | Cat 138005 |
| Rabbit anti-human CD34 (polyclonal) | Millipore Sigma | Cat HPA036722 |
| Mouse anti-human CD34 (clone QBEnd 10) | Dako | Cat M716501-2 |
| Goat anti-mouse Lyve1 (polyclonal) | R&D systems | Cat AF2125 |
| Goat anti-human Lyve1 (polyclonal) | R&D systems | Cat AF2089 |
| Goat anti-human VE-Cadherin (polyclonal) | R&D systems | Cat AF938 |
| Rabbit anti-human CD14 (polyclonal) | Millipore Sigma | Cat HPA001887 |
| Mouse anti-human ENG (clone CL1912) | Millipore Sigma | Cat AMAB90925 |
| Rat anti-mouse Endomucin (clone V.7C7) | Santa Cruz | Cat sc-65495 |
| Rabbit anti-human/mouse Aqp1 (polyclonal) | Millipore Sigma | Cat HPA019206 |
| Mouse anti-human MRC1/CD206 (clone 685645) | R&D systems | Cat MAB25341 |
| Mouse anti-human/mouse Glutamine Synthetase | BD Bioscience | Cat 610518; RRID AB_397880 |
| Rabbit anti-human/mouse Cyp2E1 (polyclonal) | Millipore Sigma | Cat AB1252 |
| Alexa 594 Rat anti-mouse E-cadherin (clone DEMCA-1) | Biolegend | Cat 147306 |
| Alexa 488 donkey Anti-rabbit (polyclonal) | ThermoFisher Scientific | Cat A21206 |
| Alexa 594 donkey anti-rat (polyclonal) | ThermoFisher Scientific | Cat A21209 |
| Alexa 555 donkey anti-mouse (polyclonal) | ThermoFisher Scientific | Cat A31570 |
| Alexa 647 donkey anti-goat (polyclonal) | Jackson Immuno Research Laboratories | Cat 705-605-147; RRID: AB_2340437 |
| Deposited data | ||
| Single cell of temporal analysis of mouse liver endothelium | GEO | GSE174209 |
| Single cell of hematopoietic and endothelial cells at postnatal day P15 of Control and c-Maf deficient endothelial mice | GEO | GSE174208 |
| Bulk RNA-seq from adult mouse endothelium | GEO | GSE172360 |
| Single cell from human CD31+ CD45neg sorted liver cells | GEO | GSE172361 |
| Experimental models: Cell lines | ||
| Primary Human Vascular Umbilical Endothelial Cells | Laboratory of Shahin Rafii | NA |
| Lentivirus packaging cell line: 293T cells | ATCC | Cat CRL-3216 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratories | Cat 000664 |
| Mouse: VE-cadherin Cre/ERT2 | Laboratory of Ralf H Adams | MGI: 3848982 |
| Mouse: Maf flox/flox | Laboratory of Carmen Birchmeier | MGI: 5316775 |
| Oligonucleotides | ||
| Table S5 | NA | NA |
| Recombinant DNA | ||
| Tamoxifen inducible human Maf (c-Maf) | Vectorbuilder | Cat VB170606-1099bva |
| Software and algorithms | ||
| R v4.0.2 | R | NA |
| Prism v.5 | GraphPad | RRRID: SCR_002798 |
| FACS Diva | BD Bioscience | NA |
| Zen blue | Nikon | NA |
| Seurat (v 3.2.3) | Butler et al., 2018; Stuart et al., 2019 | NA |
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
Experimental model and subject details
Animal husbandry
All animal experiments were performed under the approval of Weill Cornell Medicine Animal Care and Use Committee. The c-Mafflox/flox mice were obtained from Professor Carmen Birchmeier (Max Delbrück Center for Molecular Medicine). These mice have been crossed more than 10 times with C57/Bl6 mice and were crossed with the VEcadherin-CreERT2 transgenic mice donated from Ralph Adams, also in C57/Bl6 background. For the developmental analysis, female c-Mafflox/flox were crossed with male c-Mafflox/flox VEcadherin-CreERT2 heterozygotes. The induction of the Cre was performed by the administration of a daily injection of 40mg/kg of tamoxifen from E12.5 to E14.5. Tamoxifen was prepared in sunflower seed oil. Embryos were isolated at E16.5 and analyzed by flow cytometry, immunofluorescence, and RNA-seq.
The analysis of liver sinusoidal postnatal development was performed by induction of Cre with a solution of 2mg/mL 4-hyidroxytamoxifen from P2 to P4 and at P8. Mice were inoculated with 25ul of this solution at P2 and 50ul the remainder of the days. Littermate mice were analyzed at P15 when livers were processed for flow cytometry, sorting, or imaging as previously described.
The analysis of liver sinusoidal markers in Figure 1A was performed in C57/Bl6 mice obtained from Jackson Laboratories and analyze at 8 weeks of age. Adult mice were sacrifice in a CO2 chamber and perfused with 15mL of PBS and 10mL of 4% paraformaldehyde (PFA). Livers were subsequent isolated and fixed overnight on 4% PFA. For the developmental analysis females were sacrificed at the indicated timepoint and the embryonic livers were isolated and fixed overnight with 4% PFA or processed for flow cytometry. The postnatal timepoint analysis was performed as indicated for the developmental timepoints, with the exception that mice were perfused with PBS and PFA from P8 onwards, similarly to the adults.
The analysis of adult mice was performed both for male and female mice, including c-Mafflox/flox (controls) and c-Mafflox/flox–VEcadherin-CreERT2+/−, originating from the same litters. The induction of the deletion was performed by the administration of 6 doses of 40mg/kg of tamoxifen over 9 days – 3 days on, 3 days off, and 3 days on – after the mice were 4 weeks old. The mice were analyzed 4 weeks after the Tamoxifen administration, at 2 months of age.
Mice used for RNAseq analysis were intravitally label using a VEcadherin Alexa-647 antibody. Mice were euthanized using a CO2 chamber and processed immediately thereafter. All mice were perfused with 10 mL of PBS. From the mice used for RNA-seq experiments, livers were extracted and stained for flow cytometry as described above. From the mice used for flow cytometry and staining, samples from the larger lobe of the liver were extracted and processed for flow cytometry, and after those mice were perfused with 10 mL of 4% PFA.
The development of liver fibrosis was induced by administrating CCl4 twice per week at 25% dilution in sunflower oil. For the study of CCl4 in control mice we used C57/Bl6 mice from Jackson Laboratories at 8 weeks of age, inoculated with CCl4 or only oil as a control. For the study of CCL4 in c-MafΔEC mice, we treated the mice with tamoxifen at week 4 as previously indicated and posteriorly with CCl4 at week 8. The administration of CCl4 was started at 2 months of age and performed for 4 weeks. Mice were euthanized 48 hours after the last injection was performed. Mice were perfused with 10 mL of PBS and 10 mL of 4% PFA solution. Livers were kept in 4% PFA solution overnight and transferred to 70% ethanol or 30% sucrose as described above.
Cell cultures
Human umbilical vein ECs (HUVECs) were extracted from umbilical cords obtained from NewYork-Presbyterian Hospital as previously described (Zhang et al., 2003, 2004). Briefly, the umbilical cords were flashed with Hanks’ Balanced Salt Solution (HBSS) and the inner side of the umbilical vein was incubated with 0.5% collagenase for 20 minutes as previously described. Cells were flashed out using HBSS and spun down in a centrifuge. Afterwards, cells were cultured on plates coated with 1% collagen using the following medium formulation: M199 basal medium (SH30253FS Fisher Scientific) supplemented with 10% fetal calf serum (Corning), 10% human serum (100-512 Gemini Bio-Products), 1X Corning glutagro Supplement (25-015-CI Corning), 1X Hepes (25-060-Cl Corning), 1X antimicrobial antifungal (2020/11 Corning), 100 μg/mL heparin (H3393 – Sigma), 20 mg/mL bFGF (100-18B Peprotech), 10mg/mL EGF (AF-100-15 Peprotech), and 10mg/mL Vegf (400-32 Peprotech). The medium was changed every 2 to 3 days.
HUVECs were transduced with control or c-Maf lentivirus at passage 2 and selected using 1μg/mL of puromycin for 5 days, having a negative control on the plate. The induction of iLSECs was performed between passages 4 and 6 using the following protocol. Cells were grown until 50% confluence before administration of doxycycline on induction medium containing: M199 basal medium (SH30253FS Fisher Scientific) supplemented with 10% fetal calf serum (Corning), 10% human serum (100-512 Gemini Bio-Products), 1X Corning glutagro Supplement (25-015-CI Corning) , 1X Hepes (25-060-Cl Corning) , 1X Antimicrobial antifungal (2020/11 Corning), 100 μg/mL heparin (H3393 – Sigma), 20 mg/mL bFGF (100-18B Peprotech), 10mg/mL EGF (AF-100-15 Peprotech), 10mg/mL Vegf (400-32 Peprotech), 10mg/mL IL6 (200-06 Peprotech), and 1μg/mL doxycycline (04-0016 Stemolecule). The medium was changed every 2 days. Cells were induced over 7 days, starting on day 0, and analyzed by flow cytometry and immunofluorescence as explained above.
The study of the expression of c-Maf in mouse liver ECs in culture was performed using freshly isolated cells cultured for 48 hours in the presence of Bmp9, I16, or Lif. Mouse livers from healthy C57/Bl6 mice at 8 weeks of age were isolated, minced in a solution of collagenase/dispase for 15 minutes, and cultured with anti-mouse CD31 magnetic beads for 1-2 hours. ECs were isolated using a magnet, washed five times with 1X MACS solution, and rinsed once in cold PBS. After that, cells were cultured in a medium containing: M199 basal medium (SH30253FS Fisher Scientific) supplemented with 2% fetal calf serum (Corning), 1X Corning glutagro Supplement (25-015-CI Corning), 1X Hepes (25-060-Cl Corning), 1X antimicrobial antifungal (2020/11 Corning), 100 μg/mL heparin (H3393 – Sigma), 20 ng/mL bFGF (100-18B Peprotech), and 10ng/mL Vegf (400-32 Peprotech). For stimulation with we used 10ng/mL of BMP9, Il6 or Lif.
For the hepatocyte co-culture, c-Maf (CD31+ CD26+ CD36+) were sorted on an Aria II Flow sorter and co-cultured with the hepatocytes using a previously described co-culture system (Song et al., 2015). Briefly, human adult hepatocytes were obtained from Bioreclamation IVT. Hepatocytes and ECs were seeded on microwell plates together with lethally irradiated fibroblasts to promote their aggregation. The co-cultures were maintained for 28 days, with changes of medium every 2 to 3 days.
The production of lentivirus was performed on 293T cells grown on DMEM, 10% FBS, 1X L-glutamax, 1X Hepes, and 1X antimicrobial antifungal, similarly to previously described protocols (Lis et al., 2017). Cells were transduced with the following plasmids: VSVG, REV, and RES from Cyagen, Inc., and the lentiviral plasmids pLenti empty control and pLenti c-Maf. Viral titers were calculated using a p24 assay (632200 - Clontech).
Method details
Flow cytometry and Cell Sorting
Isolation of cells to be used in flow cytometry and cell sorting analysis were performed as follows. Liver samples were isolated from the mice, minced, and incubated with collagenase A (25mg/ml) and dispase II (25mg/mL) at 37°C for 10 to 15 minutes. Cells were filtered using a 100 μm filter and spun down at 300g for 5 minutes. Samples were RBC lysed for 5 minutes on ice using RBC lysis solution (Biolegend). After 5 minutes, samples were rinsed with PBS and spun down. Liver samples were stained using 1X MACS buffer solution. Cells were first incubated with the FC-quenching antibody before staining.
From the human livers, the non-parenchymal cell (NPC) fraction was obtained from the laboratory of Dr. Robert Schwartz. Samples were processed similarly to mouse liver samples after the RBC lysis step. The human fetal liver sample was processed similarly to the mouse livers from the beginning of the protocol.
10X Chromium scRNA-seq analysis
Developmental trajectory experiment:
Mouse endothelial cells (ECs) were sorted as indicated above under the Flow cytometry and Sorter description. The sorted CD45neg CD31+ cells were transfer to the Genomics Core from Weill Cornell Medicine to proceed with the Chromium Single Cell 3’ Reagent Kit v2 (10x Genomics, product code # 120267) cell protocol using 10X Genomics’ Chromium Controller. A total of 8,000 to 10,000 cells were loaded into each channel of the Single-Cell A Chip from each sample. Samples were sequenced at the recommended depth of 50,000 reads per cell in an Illumina HiSeq 4000 sequencer. Sequencing output was de-multiplexed and post-processed following the 10X genomics custom pipelines using the cellRanger (v4.0.0.) software. Raw base calls were de-multiplexed with the mkfastq command, followed by alignment to the mm 10 reference genome. Barcode and unique molecular identifier (UMI) counting was performed using the cellranger command with default parameters. Cell barcodes with UMI > 500 and mitochondrial reads < 20% were retained for downstream analysis. After filtering, 5,081 +/− 1,014 (mean +/− standard deviation) cells per sample were obtained, with a total of 40,655 cells recovered.
Samples were analyzed using the software Seurat (v 3.2.3) (Butler et al., 2018; Stuart et al., 2019). Normalization was performed using the SCTransform function, regressing out the following variables: total number of UMIs per cell, percentage of mitochondrial UMIs, S phase score and G2M score. Following normalization, principal component (PC) analysis was performed. We next verified the amount of variance explained by the top 30 principal components. The initial 24 PCs showed above 0.5% of variance explained. Visual inspection of PCs above 25 showed a more homogeneous expression of the composing genes with no clear separation and a decrease of the variance explained. Therefore, PCs up to PC24, were retained for downstream analysis. Clusters were defined using the FindNeighbors function in the PC space, followed by the FindClusters function with a resolution of 0.8, as it resulted in biological relevant clusters with clear marker genes that were manually curated for cell type annotation. Cluster marker genes were identified using the FindAllMarkers function with the following parameters: log.fc.threshold = 0.25, min.pct = 0.1 and only.pos = TRUE.
For RNA velocity analysis, loom files were generated using velocyto (v0.17.17) (La Manno et al., 2018) using the run10X function. RNA velocity calculation was performed using scVelo (v0.2.2) (Bergen et al., 2020). Genes were filtered and normalized using the filter_and_normalize function with the following parameters: min_shared_counts = 100 and n_top_genes = 2000. First and second order moments (means and uncentered variances) were computed using the moments function, with n_pcs = 30 and n_neighbors = 30. Gene dynamics were recovered using the recover_dynamics function and RNA velocities were calculated with the velocity function setting mode = ‘dynamical’. For downstream analysis of vascular endothelial cells, we selected endothelial cell clusters based on the expression of the markers Cdh5, Pecam1 and the absence of Ptprc as indicated in: Figure 1 and Supplementary Figure 2 for the developmental trajectory. RNA velocity vector field was estimated using the velocity_embedding_stream with basis = ‘umap’ and min_mass = 0. Estimation of RNA velocity-based pseudotime was performed using the velocity_pseudotime function with default parameters. For estimation of transition probabilities across developmental time points, we performed PAGA graph abstraction (Wolf et al., 2019) using the paga function, setting the developmental time points as the ‘groups’ parameter.
The identification of the transcription factors from the pseudotime analysis in Figure 2 was performed based on the TTRUST version2 mouse transcription factor list (Han et al., 2018). Comparison of all liver endothelial transcription factors was performed by downloading the Tabula muris database and calculation of the liver enriched genes compared to endothelial cells from other organs. The liver enriched list of genes was compared to the transcription factors identified from the pseudotime analysis using TTRUST version2.
P15 control and c-MafΔEC experiment:
For the analysis of P15 samples from Control and c-MafΔEC mice was performed over a mixed sample containing male and female cells coming from each sample. Count matrices were generated using the cellRanger software (v4.0.0.) as described above, and Seurat (v3.2.3) was used for downstream analysis. Samples were processed and split based on the expression of the following sexual dimorphism genes: male: Ddx3y, Eif2s3y, Gm29650, Kdm5d and Uty; and female: Xist. These genes were excluded from further analysis to avoid enrichment based on sexual dimorphism. Data was split based on the sex of the mice and normalized using the SCTransform function. In order to perform integration, we selected 3,000 integration features using the SelectIntegrationFeatures function and excluding the sexual dimorphism genes. Next, we applied the PrepSCTintegration function setting assay=‘SCT’, and integration anchors were defined using the FindIntegrationAnchors with dims = 1:40. Final integration was performed using the IntegrateData function with normalization.method = ‘SCT’. Following integration, PCA analysis was performed using the RunPCA function, and 20 PCs were retained for downstream analysis. Cell clustering was performed with the FindNeighbors function followed by the FindClusters function with resolution = 0.5. Uniform manifold approximation (UMAP) dimensionality reduction for visualization was performed using the RunUMAP function.
For RNA velocity analysis, loom files were generated using (v0.17.17) (La Manno et al., 2018) as described above, and scVelo (v0.2.2) (Bergen et al., 2020) was used for downstream analysis. Endothelial cells were retained based on expression of the markers Cdh5, Pecam1 and the absence of Ptprc and downstream RNA velocity analysis was performed as described above.
ddSeq scRNA-seq analysis
Human liver ECs were sorted as indicated above and transferred to the Genomics Core Facility at Weill Cornell Medicine to proceed with Illumina Bio-Rad SureCell WTA 3’ Library Prep kit using the Bio-Rad ddSEQ Single-Cell Isolator system. Briefly, according to manufacturer’s instructions (Illumina, cat # 20014280), the sorted cells were washed with 1x PBS + 0.1% BSA, counted by Bio-Rad TC20 Cell Counter, and cell viability was assessed. A total of 12,000 cells and barcode mixes were loaded into each channel of the cartridge to generate the droplets on the ddSeq Single Cell Isolator, from which we recovered a total of 769 cells. After the first strand was synthesized in droplets, individual droplets were disrupted. The second strand cDNA synthesis was carried out and the RNA template was removed. Using the Illumina Nextera SureCell transposome kit, cDNA was fragmented simultaneously and tagged with adapter sequences in a single step. Following PCR amplification, cDNA libraries were assessed using the Agilent Technology 2100 Bioanalyzer and sequenced on the Illumina NextSeq 500 sequencer using the high output mode with 150 cycle kit. FASTQ files were then generated in the Illumina BaseSpace SureCell Single-Cell System.
Cell barcodes with UMI < 500 and UMI > 15,000 were filtered out. Mitochondrial genes were removed as a potential confounder of clustering. Data was analyzed using the Seurat R package (v 3.2.3) (Butler et al., 2018; Stuart et al., 2019). Raw UMI counts were log-normalized using the NormalizaData function with the following parameters: normalization.method = “LogNormalize” and scale.factor = 1,000. Variable genes were selected by using the FindVariableGenes function with the following parameters: mean.function = ExpMean, dispersion.function = LogVMR, x.low.cutoff = 0.3; x.high.cutoff = 3; y.cutoff = 0.5. Data was then scaled and UMI number was regressed out as potential confounders of clustering using the ScaleData function. Following normalization, PC analysis was performed followed by JackStraw analysis to determine the significant PCs to be used in downstream analysis. A total of 11 significant PCs were retained for downstream analysis. Clusters were defined using the FindNeighbors function in the PC space, followed by the FindClusters function with a resolution of 0.4.
We identified a total of four clusters. We identified the genes enriched in the four different clusters of ECs using the FindAllMarkers function with the following parameters: only.pos = TRUE; min.pct = 0.25; thresh.use = 0.25 (Supplementary Table 1). One of the clusters corresponded to contamination of CD45− CD31+ CD38+ SCD1+ plasma cells, which were removed for analysis of ECs in the main figures, but indicated in the Supplement. UMAP plots were generated using the RunUMAP function with the significant PCs.
Bulk RNA sequencing
Mouse ECs from control and MafΔEC were sorted as indicated above using the markers CD45− CD31+ VEcadherin+. Human cells overexpressing c-Maf were isolated by sorting the fraction of cells positive for CD31, CD26, and CD36. Control cells were sorted based on the expression of CD31, since these cells do not express either CD26 or CD36. RNA was isolated using the RNA isolation Mini Kit from Qiagen (74104 - Qiagen), following the instructions from the manufacturer. At least 100 ng of total RNA from freshly harvested cells was isolated and purified using Qiagen’s RNeasy Mini Kit. RNA quality was verified using an Agilent Technologies 2100 Bioanalyzer prior to sequencing. RNA library preps were generated and multiplexed using Illumina’s TruSeq RNA Library Preparation Kit v2 (non-stranded and poly-A selection). 10 nM of cDNA was used as input for high-throughput sequencing via Illumina’s HiSeq 4000, producing 51 bp paired-end reads.
NGS data processing and statistical analysis.
Sequencing reads were de-multiplexed (bcl2fastq v2.17), checked for quality (FastQC v0.11.5), and trimmed/filtered when appropriate (Trimmomatic v0.36). The resultant high quality reads were mapped (TopHat2 v2.1.0; Bowtie2 v2.2.6) to the transcriptome sequence reference of the UCSC mm10 genome build. Gene counts were quantified using the Python package HTSeq (v0.11.1). Transcript abundance measures (FPKM values) were quantified using Cufflinks (v2.2.1). Gene-level differential expression analysis was performed using the Bioconductor R package DESeq2 (v1.22.2).
Transcriptome Data Analysis.
Transcriptomic data analysis was summarized in the forms of heatmaps and gene set enrichment plots. Heatmaps were generated using the CRAN R package pheatmap (v1.0.12). GSEA plots were generated using the R scripts available from the Broad Institute (GSEA v1.0).
Processing of ATACseq files
We download liver endothelial cell ATACseq from GSE154828 (Winkler et al., 2021). Fastq files were checked for quality using FastQC and processed using adapters trimmed with Cutadapt wrapper Trim Galore. Reads were aligned with bowtie2 and duplicated reads were marked with Picard. Reads with multimap, duplicates and low-quality reads were removed. Remaining reads were shifted on positive/negative strand and peaks were identified using MACS2. Motif analysis was performed using HOMER and scan against c-Maf motif locations. c-Maf peaks were extracted as peaks that overlap a c-Maf motif region after removing those in blacklisted regions. We determined genes flancking the peaks using a 10kb window from the peak with Bedtools. From this list of genes, we look at which were proximal to up and down regulated differentially expressed genes.
Immunofluorescence protocol
Mouse tissues embedded in OCT compound were section on a cryostat at 20 μm thickness and kept at −80°C. Sections were thawed at room temperature and washed three times with a PBS solution for 5 minutes. Tissues were permeabilized using 0.1% Triton solution for 10 minutes and washed with PBS 3 times for 5 minutes. Afterwards samples were incubated for 30 minutes with blocking solution (1X PBS, 5% donkey Serum, 0.1% Triton). Tissues were incubated for two days at 4°C with the primary antibodies at a dilution of 1:100 in blocking solution. Primary antibodies were wash three times with PBS for 5 minutes. Secondary antibody staining was performed for three hours at room temperature in blocking solution. Secondary antibodies were washed three times in PBS for 10 minutes. Samples were mounted on Fluoroshield with Dapi (F6057, Sigma) and imaged using a Zeiss 710 confocal microscope.
Human adult livers were obtained from the histopathology department at Weill Cornell Medicine. All samples were deparaffinized following this procedure: 2x 3 minutes methanol, 1 minute 50% methanol 50% ethanol, 1 minute 95% ethanol, 1 minute 80% ethanol, 1 minute 75% ethanol, 1 minute 50% ethanol, 1 minute 25% ethanol, 3 minutes PBS. After that, samples were incubated with 1X Buffer B from Electron Microscopy Science (62706-11) and posteriorly boiled on a Retriever Thermal Slide Processor (Electron Microscopy Science) for 1 hour. After cooling down samples were washed three times for 5 minutes with PBS and incubated with 0.1% PBS-Triton for 10 minutes. Samples were washed three times for 5minutes with PBS and incubated with blocking solution for 30 minutes. Primary antibodies were incubated with blocking solution at a concentration of 1:100 overnight. After washing three times for 5 minutes with PBS, secondary antibody staining was performed at 1:500 dilution with blocking solution for 3 hours at room temperature. Samples were mounted on Fluoroshield with Dapi (F6057 – Sigma) and imaged using a Zeiss 710 Confocal Microscope.
Human fetal liver was obtained from Advanced Bioscience Resources, Inc. Small pieces of this sample were incubated on 4% PFA overnight, washed with PBS twice for 5 minutes, and kept in a 30% sucrose solution for 3 days before being included in OCT. The staining for this sample proceeded similarly to the ones from the mouse tissues.
Histology
All organs were fixed overnight in 4% PFA at 4°C, and cleaned three times with 1X PBS for 10 minutes each time. After this step, the organs used for paraffin histology were kept in 70% Ethanol and were sent to Histoserv, Inc. to be processed for: hematoxyline, eosin, and Masson’s trichrome staining. The livers used for immunofluorescence were maintained for 3 days in a 30% Sucrose solution at 4°C. After this time organs were included in OCT. Histopathology analysis was conducted to evaluate the degree of liver damage and a pathologist blindly analyzed necrosis. Histological images were acquired on a Zeiss Axio Observer Z.1 microscope with a 20X/0.8NA objective and a AxioCam 305 color camera. Quantification of fibrosis deposition was performed using the Zeiss Zen 2.6 image analysis package, using tiled images of whole liver sections. The amount of liver fibrosis was divided by the surface are of the liver quantified for all images.
Lacunarity and Fractal analysis
The branching and space-filling of the liver structure display a hierarchical self-similarity quantifiable with fractal mathematics, in particular the parameters of fractal dimension (D) and lacunarity (Λ) (Adelson et al., 2021). D is a ratio indicating the change in complexity of the structural pattern with scale, while Λ quantifies the size and spatial distribution of gaps between branches in the pattern. Liver images were analyzed in ImageJ in order to quantify these fractal properties. Color-binarized two-dimensional confocal microscopy images were loaded into ImageJ using the FracLac plug-in.
D of each liver image was quantified using a box-counting method, with a grid of known box dimension (r) overlaid on each image. The number of boxes containing structural pixels (N) was counted. This procedure was repeated for all images for each of 100 grids of varying r (5 to 1,742 pixels wide), with N decreasing exponentially as box size increased.
Plotting r versus N reveals D, which approximately equals to the negation of the slope of the best-fit regression line. The relevant equation is:
Λ was measured using a similar method, a sliding box scan algorithm, which involves sliding boxes over the image in an overlapping pattern. The mean (μ) and standard deviation (σ) of pixels per box was calculated for each r, and Λ was approximated using the following equation (an average Λ was then calculated based on all r used):
Matrix cellulose assay
Analysis of the hemato-progenitor cells contained in the livers from control and c-Maf endothelial deficient mice were performed at postnatal day P15. Livers were processes as previously indicated for flow assisted cytometry sorting analysis. Hematopoietic cells were identified as a CD45+ CD31− population, from which we sorted 90.000 cells. Hematopoietic cells were transfer to 3mL matrix cellulose assay tubes with MethoCult GF m3434 (#03444 StemCell Technologies), from which 2mL were plated and cultured for 14 days. After 2 weeks, the number of colonies was counted for all plates and the average of the two plates per mouse were calculated, providing the total colony forming units per 30,000 cells.
Cloning
c-Maf was PCR amplified from the plasmid (VB170606-1099bva) from Vectorbuilder, Inc (Cyagen Bioscience) and cloned into a pZip lentiviral plasmid carrying a Puromycin resistance and RtTa. The PCR product and Lentiviral plasmid were digested with the enzymes BamHI and AsisI and Ligated. Individual clones were validated by sequencing.
Quantitative real time reverse transcription polymerase chain reaction (qRT-PCR):
Spheroid aggregates of iLSECs and hepatocytes were washed with PBS three times and then collected into Eppendorf tubes. After centrifugation at 1000 rpm for 5 minutes, the pellets were lysed with RLT buffer and total RNA was isolated using an RNAease Kit (Qiagen). Isolated RNA was then treated with DNase (NEB) and RNA cleaned up with the RNA Clean and Concentrator kit (Zymo Research). 2 μg of RNA was used for cDNA synthesis (iSCRIPT cDNA synthesis kit, Biorad). qRT-PCR was performed using SYBR Green PCR (Biorad) on a Biorad CFX PCR machine. The gene expression data was normalized using β-actin as a housekeeping gene. The following primers were used:
Albumin secretion:
Albumin secretion was measured in culture medium. The medium was collected and replaced with fresh medium every 2 days. The collected medium was centrifuged at 1000 rpm for 5 minutes and the supernatant was stored at −20°C. Secreted albumin was quantified by an enzyme-linked immunosorbent assay (ELISA) kit using sheep anti-human albumin antibodies (Bethy Labs) and horseradish peroxidase detection (3,3’,5,5’-tetramethylbenzidine, Invitrogen).
Quantification and Statistical analysis:
Statistics were calculated using R software, the R package “ggpubr” and Graph Pad. Groups of 2 different conditions were compared using T-test. Statistical significance is shown as: *p<0.05, **p<0.01, ***p<0.001. Boxplots indicate the medium, data within the boxplot shows the interquartile range, whisker shows the minimum and maximum values used for the boxplot, values outside the whiskers were consider as outlawyers by R. Barplots represent medium of the data ± SEM. All information related to the analysis of each panel can be found in the figure legend. Quantification of liver fibrosis was performed using Zeiss Zen 2.6 image analysis package. Total liver fibrosis was divided by the total surface are of the liver for all images.
Supplementary Material
Table S1. Mouse liver vascular population markers through development, related to Figure 1A and 1B. Identification of mouse liver endothelial cell markers enriched on each population from the timeseries from fetal to postnatal development.
Table S2. Pseudotime prediction of gene expression transition from the endothelial cell populations, related to Figure 2A and Figure S3A. Numbers indicate the pseudotime transition prediction between different timepoints.
Table S3. Mouse liver vascular and hematopoietic markers at P15 postnatal development, related to Figure 4B. Identification of genes enriched in endothelial and hematopoietic cells sorted at P15 from control and c-MafΔEC mice.
Table S4. Human liver vascular markers, related to Figure 6A. Identification of genes enriched in the cell populations identified from sorted human liver CD45− CD31+ cells.
Table S5. List of primers used for quantitative RT-PCR analysis and cloning, related to Star Methods.
Highlights.
Liver large vessel identity is developmentally acquired.
Sinusoids are the most versatile population with continuous adaptation until adult.
The transcription factor c-Maf choreographs liver sinusoidal program.
c-Maf induces a liver sinusoidal like state in human generic endothelial cells.
Acknowledgements
We acknowledge Dr. Carmen Birchmeier (Max Delbrück Center for Molecular Medicine) for kindly providing us with the c-Mafflox/flox mice. We acknowledge Dr. Rui Benedito (CNIC) for kindly sharing his expertise in genetic deletion during postnatal development.
Funding:
JM Gomez-Salinero was a New York Stem Cell Foundation – Druckenmiller Fellow, and is currently supported by Weill Cornell Medicine Fund for the Future program. F Izzo is supported by the American Society of Hematology Fellow-to-Faculty Scholar Award. S Rafii is funded by Hartman Institute for Therapeutic Organ Regeneration, Ansary Stem Cell Institute, grants from the National Institute of health (NIH), R35 HL150809, RC2 DK114777, U01AI138329, the Empire State Stem Cell Board NYSTEM) (C030160), Daedalus Fund for Innovation and iSelma and Lawrence Ruben Science to Industry Bridge Fund from Weill Cornell Medicine, the Starr Foundation stem cell core project and initiatives TRI-SCI #2019-029.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest:
S Rafii is the co-founder and non-paid consultant to Angiocrine Bioscience, San Diego CA.
R.E.S. is on the scientific advisory board of Miromatrix Inc. and is a speaker and consultant for Alnylam Inc.
References
- Adelson RP, Palikuqi B, Weiss Z, Checco A, Schreiner R, Rafii S, and Rabbany SY (2021). Morphological characterization of Etv2 vascular explants using fractal analysis and atomic force microscopy. Microvasc. Res 138, 104205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin HG, and Koh GY (2017). Organotypic vasculature: From descriptive heterogeneity to functional pathophysiology. Science 357. [DOI] [PubMed] [Google Scholar]
- Bankston PW, and Pino RM (1980). The development of the sinusoids of fetal rat liver: morphology of endothelial cells, Kupffer cells, and the transmural migration of blood cells into the sinusoids. Am. J. Anat 159, 1–15. [DOI] [PubMed] [Google Scholar]
- Barberá-Guillem E, Arrue JM, Ballesteros J, and Vidal-Vanaclocha F (1986). Structural changes in endothelial cells of developing rat liver in the transition from fetal to postnatal life. J. Ultrastruct. Mol. Struct. Res 97, 197–206. [DOI] [PubMed] [Google Scholar]
- Barry DM, McMillan EA, Kunar B, Lis R, Zhang T, Lu T, Daniel E, Yokoyama M, Gomez-Salinero JM, Sureshbabu A, et al. (2019). Molecular determinants of nephron vascular specialization in the kidney. Nat. Commun 10, 5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen V, Lange M, Peidli S, Wolf FA, and Theis FJ (2020). Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol 38, 1408–1414. [DOI] [PubMed] [Google Scholar]
- Boj SF, van Es JH, Huch M, Li VSW, José A, Hatzis P, Mokry M, Haegebarth A, van den Born M, Chambon P, et al. (2012). Diabetes risk gene and Wnt effector Tcf7l2/TCF4 controls hepatic response to perinatal and adult metabolic demand. Cell 151, 1595–1607. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Ye T, Sun Y, Ji G, Shido K, Chen Y, Luo L, Na F, Li X, Huang Z, et al. (2017). Targeting the vascular and perivascular niches as a regenerative therapy for lung and liver fibrosis. Sci. Transl. Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvelard A, Scoazec JY, Dauge MC, Bringuier AF, Potet F, and Feldmann G (1996). Structural and functional differentiation of sinusoidal endothelial cells during liver organogenesis in humans. Blood 87, 4568–4580. [PubMed] [Google Scholar]
- Dejana E, Hirschi KK, and Simons M (2017). The molecular basis of endothelial cell plasticity. Nat. Commun 8, 14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSesso JM (2017). Vascular ontogeny within selected thoracoabdominal organs and the limbs. Reprod. Toxicol 70, 3–20. [DOI] [PubMed] [Google Scholar]
- Desroches-Castan A, Tillet E, Ricard N, Ouarné M, Mallet C, Belmudes L, Couté Y, Boillot O, Scoazec J-Y, Bailly S, et al. (2019a). Bone morphogenetic protein 9 is a paracrine factor controlling liver sinusoidal endothelial cell fenestration and protecting against hepatic fibrosis. Hepatology 70, 1392–1408. [DOI] [PubMed] [Google Scholar]
- Desroches-Castan A, Tillet E, Ricard N, Ouarné M, Mallet C, Feige J-J, and Bailly S (2019b). Differential consequences of bmp9 deletion on sinusoidal endothelial cell differentiation and liver fibrosis in 129/ola and C57BL/6 mice. Cells 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B-S, Nolan DJ, Butler JM, James D, Babazadeh AO, Rosenwaks Z, Mittal V, Kobayashi H, Shido K, Lyden D, et al. (2010). Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature 468, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B-S, Nolan DJ, Guo P, Babazadeh AO, Cao Z, Rosenwaks Z, Crystal RG, Simons M, Sato TN, Worgall S, et al. (2011). Endothelial-derived angiocrine signals induce and sustain regenerative lung alveolarization. Cell 147, 539–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B-S, Cao Z, Lis R, Nolan DJ, Guo P, Simons M, Penfold ME, Shido K, Rabbany SY, and Rafii S (2014). Divergent angiocrine signals from vascular niche balance liver regeneration and fibrosis. Nature 505, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y, Yamamoto A, Kyoden T, Kato K, and Tashiro Y (1990). Quantitative immunogold localization of dipeptidyl peptidase IV (DPP IV) in rat liver cells. Cell Struct. Funct 15, 117–125. [DOI] [PubMed] [Google Scholar]
- Gage BK, Liu JC, Innes BT, MacParland SA, McGilvray ID, Bader GD, and Keller GM (2020). Generation of Functional Liver Sinusoidal Endothelial Cells from Human Pluripotent Stem-Cell-Derived Venous Angioblasts. Cell Stem Cell 27, 254–269.e9. [DOI] [PubMed] [Google Scholar]
- Géraud C, Schledzewski K, Demory A, Klein D, Kaus M, Peyre F, Sticht C, Evdokimov K, Lu S, Schmieder A, et al. (2010). Liver sinusoidal endothelium: a microenvironment-dependent differentiation program in rat including the novel junctional protein liver endothelial differentiation-associated protein-1. Hepatology 52, 313–326. [DOI] [PubMed] [Google Scholar]
- Géraud C, Koch P-S, Zierow J, Klapproth K, Busch K, Olsavszky V, Leibing T, Demory A, Ulbrich F, Diett M, et al. (2017). GATA4-dependent organ-specific endothelial differentiation controls liver development and embryonic hematopoiesis. J. Clin. Invest 127, 1099–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano M, Henin C, Maurizio J, Imbratta C, Bourdely P, Buferne M, Baitsch L, Vanhille L, Sieweke MH, Speiser DE, et al. (2015). Molecular profiling of CD8 T cells in autochthonous melanoma identifies Maf as driver of exhaustion. EMBO J. 34, 2042–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Salinero JM, and Rafii S (2018). Endothelial cell adaptation in regeneration. Science 362, 1116–1117. [DOI] [PubMed] [Google Scholar]
- Guo P, Poulos MG, Palikuqi B, Badwe CR, Lis R, Kunar B, Ding B-S, Rabbany SY, Shido K, Butler JM, et al. (2017). Endothelial jagged-2 sustains hematopoietic stem and progenitor reconstitution after myelosuppression. J. Clin. Invest 127, 4242–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan W, Øie C, Benkheil M, Dheedene W, Vinckier S, Coppiello G, Aranguren XL, Beerens M, Jaekers J, Topal B, et al. (2020). Unraveling the transcriptional determinants of liver sinusoidal endothelial cell specialization. Am. J. Physiol. Gastrointest. Liver Physiol 318, G803–G815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern KB, Shenhav R, Massalha H, Toth B, Egozi A, Massasa EE, Medgalia C, David E, Giladi A, Moor AE, et al. (2018). Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat. Biotechnol 36, 962–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A, Henderson S, Lagos D, Nikitenko L, Coulter E, Roberts S, Gratrix F, Plaisance K, Renne R, Bower M, et al. (2010). KSHV-encoded miRNAs target MAF to induce endothelial cell reprogramming. Genes Dev. 24, 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Cho J-W, Lee S, Yun A, Kim H, Bae D, Yang S, Kim CY, Lee M, Kim E, et al. (2018). TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 46, D380–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey NL, and Oliver G (2004). Choose your fate: artery, vein or lymphatic vessel? Curr. Opin. Genet. Dev 14, 499–505. [DOI] [PubMed] [Google Scholar]
- Inverso D, Shi J, Lee KH, Jakab M, Ben-Moshe S, Kulkarni SR, Schneider M, Wang G, Komeili M, Vélez PA, et al. (2021). A spatial vascular transcriptomic, proteomic, and phosphoproteomic atlas unveils an angiocrine Tie-Wnt signaling axis in the liver. Dev. Cell 56, 1677–1693.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakab M, and Augustin HG (2020). Understanding angiodiversity: insights from single cell biology. Development 147. [DOI] [PubMed] [Google Scholar]
- Khan JA, Mendelson A, Kunisaki Y, Birbrair A, Kou Y, Arnal-Estapé A, Pinho S, Ciero P, Nakahara F, Ma’ayan A, et al. (2016). Fetal liver hematopoietic stem cell niches associate with portal vessels. Science 351, 176–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakabe M, Hasegawa K, Hamada M, Nakamura M, Ohsumi T, Suzuki H, Tran MTN, Kudo T, Uchida K, Ninomiya H, et al. (2011). c-Maf plays a crucial role for the definitive erythropoiesis that accompanies erythroblastic island formation in the fetal liver. Blood 118, 1374–1385. [DOI] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis R, Karrasch CC, Poulos MG, Kunar B, Redmond D, Duran JGB, Badwe CR, Schachterle W, Ginsberg M, Xiang J, et al. (2017). Conversion of adult endothelium to immunocompetent haematopoietic stem cells. Nature 545, 439–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotto J, Drissler S, Cullum R, Wei W, Setty M, Bell EM, Boutet SC, Nowotschin S, Kuo Y-Y, Garg V, et al. (2020). Single-Cell Transcriptomics Reveals Early Emergence of Liver Parenchymal and Non-parenchymal Cell Lineages. Cell 183, 702–716.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacParland SA, Liu JC, Ma X-Z, Innes BT, Bartczak AM, Gage BK, Manuel J, Khuu N, Echeverri J, Linares I, et al. (2018). Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun 9, 4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka H, Tanaka M, Suzuki K, and Miyajima A (2007). Development of murine hepatic sinusoidal endothelial cells characterized by the expression of hyaluronan receptors. Dev. Dyn 236, 2258–2267. [DOI] [PubMed] [Google Scholar]
- Palikuqi B, Nguyen D-HT, Li G, Schreiner R, Pellegata AF, Liu Y, Redmond D, Geng F, Lin Y, Gómez-Salinero JM, et al. (2020). Adaptable haemodynamic endothelial cells for organogenesis and tumorigenesis. Nature 585, 426–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas-Paz L, Orsini V, Boulter L, Calabrese D, Pikiolek M, Nigsch F, Xie Y, Roma G, Donovan A, Marti P, et al. (2016). The RSPO-LGR4/5-ZNRF3/RNF43 module controls liver zonation and size. Nat. Cell Biol 18, 467–479. [DOI] [PubMed] [Google Scholar]
- Plein A, Fantin A, Denti L, Pollard JW, and Ruhrberg C (2018). Erythro-myeloid progenitors contribute endothelial cells to blood vessels. Nature 562, 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos MG, Guo P, Kofler NM, Pinho S, Gutkin MC, Tikhonova A, Aifantis I, Frenette PS, Kitajewski J, Rafii S, et al. (2013). Endothelial Jagged-1 is necessary for homeostatic and regenerative hematopoiesis. Cell Rep. 4, 1022–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafii S, Butler JM, and Ding B-S (2016). Angiocrine functions of organ-specific endothelial cells. Nature 529, 316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha AS, Vidal V, Mertz M, Kendall TJ, Charlet A, Okamoto H, and Schedl A (2015). The angiocrine factor rspondin3 is a key determinant of liver zonation. Cell Rep. 13, 1757–1764. [DOI] [PubMed] [Google Scholar]
- Sharma A, Seow JJW, Dutertre C-A, Pai R, Blériot C, Mishra A, Wong RMM, Singh GSN, Sudhagar S, Khalilnezhad S, et al. (2020). Onco-fetal Reprogramming of Endothelial Cells Drives Immunosuppressive Macrophages in Hepatocellular Carcinoma. Cell 183, 377–394.e21. [DOI] [PubMed] [Google Scholar]
- Song W, Lu Y-C, Frankel AS, An D, Schwartz RE, and Ma M (2015). Engraftment of human induced pluripotent stem cell-derived hepatocytes in immunocompetent mice via 3D co-aggregation and encapsulation. Sci. Rep 5, 16884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss O, Phillips A, Ruggiero K, Bartlett A, and Dunbar PR (2017). Immunofluorescence identifies distinct subsets of endothelial cells in the human liver. Sci. Rep 7, 44356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabula Muris Consortium, Overall coordination, Logistical coordination, Organ collection and processing, Library preparation and sequencing, Computational data analysis, Cell type annotation, Writing group, Supplemental text writing group, and Principal investigators (2018). Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavora B, Mederer T, Wessel KJ, Ruffing S, Sadjadi M, Missmahl M, Ostendorf BN, Liu X, Kim J-Y, Olsen O, et al. (2020). Tumoural activation of TLR3-SLIT2 axis in endothelium drives metastasis. Nature 586, 299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wende H, Lechner SG, Cheret C, Bourane S, Kolanczyk ME, Pattyn A, Reuter K, Munier FL, Carroll P, Lewin GR, et al. (2012). The transcription factor c-Maf controls touch receptor development and function. Science 335, 1373–1376. [DOI] [PubMed] [Google Scholar]
- Winkler M, Staniczek T, Kürschner SW, Schmid CD, Schönhaber H, Cordero J, Kessler L, Mathes A, Sticht C, Neßling M, et al. (2021). Endothelial GATA4 controls liver fibrosis and regeneration by preventing a pathogenic switch in angiocrine signaling. J. Hepatol 74, 380–393. [DOI] [PubMed] [Google Scholar]
- Wohlfeil SA, Häfele V, Dietsch B, Schledzewski K, Winkler M, Zierow J, Leibing T, Mohammadi MM, Heineke J, Sticht C, et al. (2019). Hepatic Endothelial Notch Activation Protects against Liver Metastasis by Regulating Endothelial-Tumor Cell Adhesion Independent of Angiocrine Signaling. Cancer Res. 79, 598–610. [DOI] [PubMed] [Google Scholar]
- Wolf FA, Hamey FK, Plass M, Solana J, Dahlin JS, Göttgens B, Rajewsky N, Simon L, and Theis FJ (2019). PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 20, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ochando J, Yopp A, Bromberg JS, and Ding Y (2005). IL-6 plays a unique role in initiating c-Maf expression during early stage of CD4 T cell activation. J. Immunol 174, 2720–2729. [DOI] [PubMed] [Google Scholar]
- Zhang F, Hackett NR, Lam G, Cheng J, Pergolizzi R, Luo L, Shmelkov SV, Edelberg J, Crystal RG, and Rafii S (2003). Green fluorescent protein selectively induces HSP70-mediated up-regulation of COX-2 expression in endothelial cells. Blood 102, 2115–2121. [DOI] [PubMed] [Google Scholar]
- Zhang F, Cheng J, Hackett NR, Lam G, Shido K, Pergolizzi R, Jin DK, Crystal RG, and Rafii S (2004). Adenovirus E4 gene promotes selective endothelial cell survival and angiogenesis via activation of the vascular endothelial-cadherin/Akt signaling pathway. J. Biol. Chem 279, 11760–11766. [DOI] [PubMed] [Google Scholar]
- Zhang H, Pu W, Tian X, Huang X, He L, Liu Q, Li Y, Zhang L, He L, Liu K, et al. (2016). Genetic lineage tracing identifies endocardial origin of liver vasculature. Nat. Genet 48, 537–543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Mouse liver vascular population markers through development, related to Figure 1A and 1B. Identification of mouse liver endothelial cell markers enriched on each population from the timeseries from fetal to postnatal development.
Table S2. Pseudotime prediction of gene expression transition from the endothelial cell populations, related to Figure 2A and Figure S3A. Numbers indicate the pseudotime transition prediction between different timepoints.
Table S3. Mouse liver vascular and hematopoietic markers at P15 postnatal development, related to Figure 4B. Identification of genes enriched in endothelial and hematopoietic cells sorted at P15 from control and c-MafΔEC mice.
Table S4. Human liver vascular markers, related to Figure 6A. Identification of genes enriched in the cell populations identified from sorted human liver CD45− CD31+ cells.
Table S5. List of primers used for quantitative RT-PCR analysis and cloning, related to Star Methods.
Data Availability Statement
The RNA and single cell RNAseq data presented in this study have been deposited at GEO: GSE172360, GSE172361, GSE172362. Accession numbers are provided in the Key resource table. Code for analysis of the single cell RNAseq has been deposited at GitHub and is publicly available. Access information for raw files can be found in the Key resource table.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PE Mouse anti-human CD31 (clone WM59) | BD Bioscience | Cat 555446 |
| BV421 Mouse anti-human CD45 (clone 2D1) | Biolegend | Cat 368522 |
| BV421 mouse anti-human CD31 (clone WM59) | Biolegend | Cat 303124 |
| Fitc mouse anti-human CD14 (clone 63D3) | Biolegend | Cat 367116 |
| Pe mouse anti-human CD26 (clone BA5b) | Biolegend | Cat 302706 |
| PE/Cyanine 7 mouse anti-human CD36 (clone 5-271) | Biolegend | Cat 336222 |
| Goat anti-mouse MRC1/CD206 (polyclonal) | R&D systems | Cat AF2535 |
| PE mouse anti-human/mouse MAF (clone symOF1) | ThermoFisher Scientific | Cat 12-9855-42 |
| APC/Cyanine 7 anti-human CD45 (clone HI30) | Biolegend | Cat 304014 |
| PE/Cyanine 7 mouse anti-human CD34 (clone 581) | Biolegend | Cat 343416 |
| BV421 mouse anti-human CD38 (clone HB-7) | Biolegend | Cat 356618 |
| Human TruStain FC Blocker | Biolegend | Cat 422302 |
| Pe/Cyanine 7 rat anti-mouse CD31 (clone 390) | Biolegend | Cat 102418 |
| Alexa 488 rat anti-mouse CD45 (clone 30-F11) | Biolegend | Cat 103122 |
| BV421 rat anti-mouse CD45 (clone 30-F11) | Biolegend | Cat 103134 |
| APC/Cyanine 7 rat anti-mouse CD45 (clone 30-F11) | Biolegend | Cat 103116 |
| PE mouse anti-mouse CD32B (clone AT130-2) | ThermoFisher Scientific | Cat 12-0321082 |
| BV421 rat anti-mouse MRC1/CD206 (clone C068C2) | Biolegend | Cat 141717 |
| PE/Cyanine 7 rat anti-mouse Ly6A/E (clone D7) | Biolegend | Cat 108114 |
| APC mouse anti-mouse CD32B (clone AT130-2) | ThermoFisher Scientific | Cat 17-0321-82 |
| APC rat anti-mouse CD68 (clone FA-11) | Biolegend | Cat 137008 |
| Alexa 488 rat anti-mouse CD31 (clone 390) | Biolegend | Cat 102414 |
| TruStain FcX rat anti-mouse CD16/32 (Clone 93) | Biolegend | Cat 101320 |
| APC rat anti-mouse CD45R/B220 (clone RA3-6B2) | Biolegend | Cat 103212 |
| Alexa 488 rat anti-mouse CD24 (clone M1/69) | Biolegend | Cat 101816 |
| PE/Cyanine 7 rat anti-mouse CD45 (clone 30-F11) | Biolegend | Cat 103114 |
| PE rat anti-mouse CD20 (clone SA275A11) | Biolegend | Cat 150409 |
| PE/Cyanine 7 rat anti-mouse F4/80 (clone BM8) | Biolegend | Cat 123113 |
| BV421 rat anti-mouse CD68 (clone FA-11) | Biolegend | Cat 137017 |
| Alexa 700 rat anti-mouse CD45 (clone 30-F11) | Biolegend | Cat 103128 |
| Alexa 647 rat anti-mouse Cdh5 (clone11D4.1) | BD Bioscience | Cat 562242 |
| Alexa 647 rat anti-mouse Cdh5 (clone BV13) | Biolegend | Cat 138005 |
| Rabbit anti-human CD34 (polyclonal) | Millipore Sigma | Cat HPA036722 |
| Mouse anti-human CD34 (clone QBEnd 10) | Dako | Cat M716501-2 |
| Goat anti-mouse Lyve1 (polyclonal) | R&D systems | Cat AF2125 |
| Goat anti-human Lyve1 (polyclonal) | R&D systems | Cat AF2089 |
| Goat anti-human VE-Cadherin (polyclonal) | R&D systems | Cat AF938 |
| Rabbit anti-human CD14 (polyclonal) | Millipore Sigma | Cat HPA001887 |
| Mouse anti-human ENG (clone CL1912) | Millipore Sigma | Cat AMAB90925 |
| Rat anti-mouse Endomucin (clone V.7C7) | Santa Cruz | Cat sc-65495 |
| Rabbit anti-human/mouse Aqp1 (polyclonal) | Millipore Sigma | Cat HPA019206 |
| Mouse anti-human MRC1/CD206 (clone 685645) | R&D systems | Cat MAB25341 |
| Mouse anti-human/mouse Glutamine Synthetase | BD Bioscience | Cat 610518; RRID AB_397880 |
| Rabbit anti-human/mouse Cyp2E1 (polyclonal) | Millipore Sigma | Cat AB1252 |
| Alexa 594 Rat anti-mouse E-cadherin (clone DEMCA-1) | Biolegend | Cat 147306 |
| Alexa 488 donkey Anti-rabbit (polyclonal) | ThermoFisher Scientific | Cat A21206 |
| Alexa 594 donkey anti-rat (polyclonal) | ThermoFisher Scientific | Cat A21209 |
| Alexa 555 donkey anti-mouse (polyclonal) | ThermoFisher Scientific | Cat A31570 |
| Alexa 647 donkey anti-goat (polyclonal) | Jackson Immuno Research Laboratories | Cat 705-605-147; RRID: AB_2340437 |
| Deposited data | ||
| Single cell of temporal analysis of mouse liver endothelium | GEO | GSE174209 |
| Single cell of hematopoietic and endothelial cells at postnatal day P15 of Control and c-Maf deficient endothelial mice | GEO | GSE174208 |
| Bulk RNA-seq from adult mouse endothelium | GEO | GSE172360 |
| Single cell from human CD31+ CD45neg sorted liver cells | GEO | GSE172361 |
| Experimental models: Cell lines | ||
| Primary Human Vascular Umbilical Endothelial Cells | Laboratory of Shahin Rafii | NA |
| Lentivirus packaging cell line: 293T cells | ATCC | Cat CRL-3216 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratories | Cat 000664 |
| Mouse: VE-cadherin Cre/ERT2 | Laboratory of Ralf H Adams | MGI: 3848982 |
| Mouse: Maf flox/flox | Laboratory of Carmen Birchmeier | MGI: 5316775 |
| Oligonucleotides | ||
| Table S5 | NA | NA |
| Recombinant DNA | ||
| Tamoxifen inducible human Maf (c-Maf) | Vectorbuilder | Cat VB170606-1099bva |
| Software and algorithms | ||
| R v4.0.2 | R | NA |
| Prism v.5 | GraphPad | RRRID: SCR_002798 |
| FACS Diva | BD Bioscience | NA |
| Zen blue | Nikon | NA |
| Seurat (v 3.2.3) | Butler et al., 2018; Stuart et al., 2019 | NA |
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.