Abstract
Because oral transmission of SARS-CoV-2 is 3–5 orders of magnitude higher than nasal transmission, we investigated debulking of oral viruses using viral trap proteins (CTB-ACE2, FRIL) expressed in plant cells, delivered through the chewing gum. In omicron nasopharyngeal (NP) samples, the microbubble count (based on N-antigen) was significantly reduced by 20 μg of FRIL (p < 0.0001) and 0.925 μg of CTB-ACE2 (p = 0.0001). Among 20 delta or omicron NP samples, 17 had virus load reduced below the detection level of spike protein in the RAPID assay, after incubation with the CTB-ACE2 gum powder. A dose-dependent 50% plaque reduction with 50–100 ng FRIL or 600–800 μg FRIL gum against Influenza strains H1N1, H3N2, and Coronavirus HCoV-OC43 was observed with both purified FRIL, lablab bean powder or gum. In electron micrographs, large/densely packed clumps of overlapping influenza particles and FRIL protein were observed. Chewing simulator studies revealed that CTB-ACE2 release was time/dose-dependent and release was linear up to 20 min chewing. Phase I/II placebo-controlled, double-blinded clinical trial (IND 154897) is in progress to evaluate viral load in saliva before or after chewing CTB-ACE2/placebo gum. Collectively, this study advances the concept of chewing gum to deliver proteins to debulk oral viruses and decrease infection/transmission.
Abbreviations: SARS-CoV-2, COVID-19; Topical drug delivery, Chewing gum; Omicron, Delta strains, Influenza virus; Plant cells, Virus trap proteins; Oral viruses, Debulking
Graphical abstract
Twitter headline: Chewing gum neutralizing both SARS-CoV-2 and influenza viruses is advanced to clinical testing.
1. Introduction
Oral diseases caused by microbial infections afflict 3.5 billion people worldwide. Bacteria and fungi colonize tooth surfaces forming tenacious and intractable biofilms resulting in severe dental caries, while saliva is a major source of pathogens that are transmitted as droplets or aerosolized particles [1]. Of recent concern is COVID-19, in which the salivary glands are the primary replication site of SARS-CoV-2, leading to the loss of taste and smell [[2], [3], [4], [5]]. In addition, Influenza, HPV, HSV1, EBV and KSHV viruses are also transmitted orally and their life cycle in the oral epithelium is well characterized [[6], [7], [8], [9], [10], [11]].
Although SARS-CoV-2 transmission between unvaccinated individuals is the primary cause of continued spread, fully vaccinated individuals with breakthrough infections have peak viral loads similar to unvaccinated individuals and efficiently transmit virus in household settings [12]. Equally important is the pattern of SARS-CoV-2 evolution that strengthen viral infectivity through antibody-resistant mutations [13]. Although SARS-CoV-2 is transmitted nasally and orally, oral transmission is 3–5 orders of magnitude higher than nasal transmission [[14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28]]. Airborne-lifetime-weighed volume of saliva droplets in healthy subjects is 3–5 orders of magnitude higher than breath droplets; speaking four words releases more virus particles than an entire hour of breathing [29], suggesting that a decrease in oral viral load could have substantial effect on virus transmission [[14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28]]. Therefore, new methods are proposed to debulk pathogens in the oral cavity and minimize transmission.
Clinical evaluation of mouth rinses in COVID-19 patients reveal no statistically significant changes in saliva viral load after rinse, up to 2 h [30]. It is possible that qPCR detects non-viable viral particles, as evidenced by detection of SARS-CoV-2 several weeks after disappearance of symptoms [[31], [32], [33]] and subsequent CDC guidelines to not perform qPCR testing up to ninety days after the onset of infection. Daniell lab has expressed ACE2 enzyme in chloroplasts to treat pulmonary hypertension, which is now advancing to the clinic to treat COVID-19 patients [34]. In addition, CTB-ACE2 chewing gum was able to markedly debulk SARS-CoV-2 (>95%) in COVID-19 patient saliva or swab samples as measured by microbubbles or qPCR [35] by direct binding of the spike protein to soluble ACE2 (Fig. 1 A). Blocking engagement of spike protein to ACE2 and GM1 receptors by CTB-ACE2 (Fig. 1A) was evaluated by inhibiting uptake of lentivirus or VSV pseudo-type virions into Vero cells [35]. In addition to the native human ACE2 enzyme used in this study, several mutated versions with much higher affinity to the SARS-CoV-2 have been developed and could be utilized as viral trap proteins [36,37].
Fig. 1.
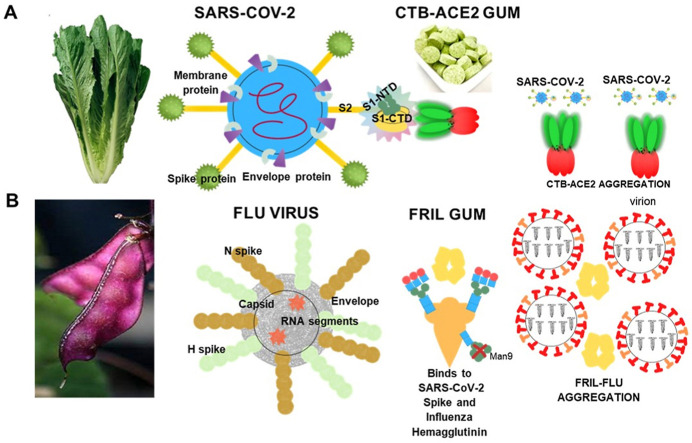
Blocking and neutralization mechanisms of SARS-CoV-2 BY PLANT-MADE viral trap proteins - CTB-ACE2 and FRIL: (A) The pentameric insoluble microparticles of the lettuce chloroplast CTB-ACE2 in the chewing gum tablet effectively binds to the viral spike proteins and sediments the SARS-CoV-2 in the chewing gum or blocks the viral entry into human cells by binding to ACE2/GM1 receptors. (B) The homo-tetramer, FRIL a plant lectin (from Lablab bean) binds to complex-type-N glycans present on the surface of SARS-CoV-2 and influenza viruses forming aggregates and also sequesters virions at the late endosomal stage, thereby preventing entry into the nucleus of human cells.
The foremost viruses with the strongest capabilities for spread by aerosol transmission and causing illness and death to the widest population are SARS-CoV-2 and Influenza [38]. The significance of the plant lectin FRIL is its preferential entrapment of viruses that express complex-type N-glycans (Fig. 1B) on their outer envelopes [39,40]. Of note enveloped viruses express either high mannose, complex N-linked glycans, or a non-complex-hybrid polysaccharides. Both SARS-CoV-2 and Influenza (Flu) viruses have been shown to contain complex-type N-glycans on their envelopes [[39], [40], [41], [42]]. Influenza (Flu) is an especially significant respiratory virus that affects the worldwide population [43]. The infections are seasonal and mainly endemic with a high rate of morbidity and mortality [43]. Globally, influenza causes half a million deaths annually [41]. Each year a new Flu vaccine is deployed based on emerging strains with small and gradual variations (Genetic Drift) [43]. Flu is especially life-threatening when the segmented genomic strands that separately encode HA and NA of human Flu become mixed upon infection with a different species e.g., bird or pig (Genetic Shift) [43]. Several major pandemics have occurred in the past 150 years with 1918 being the worst case, resulting in 50 million deaths and infecting one-third of the world's population [41,43]. A potential global outbreak of Avian H5N1 Bird Flu appeared in China in 2013; the Bird Flu directly infected humans with the potential to spread worldwide [44].
Based on the success of the COVID gum to debulk SARS-CoV-2 using the native human protein ACE2 [35], in this study we explored the entrapment efficacy in different strains of SARS-CoV-2 using microbubbling (N-antigen) or RAPID (spike protein) assays. In addition, we investigated the virus trap plant protein lectin (FRIL) for its potential to neutralize SARS-CoV-2 and influenza virus by plaque reduction assay and mechanism of entrapment using electron micrography. For five decades protein drugs have been delivered as sterile injections, requiring cold storage/transportation, thereby decreasing patient affordability and compliance. In order to address some of these challenges, the Daniell lab has developed oral delivery systems through encapsulation of protein drugs in plant cells [[45], [46], [47], [48], [49], [50]]. Plant cell bioencapsulation eliminates cold storage/transportation challenges [[51], [52], [53], [54]]. Plant cells are now being developed as a novel strategy to deliver protein drugs against pathogens that colonize the oral cavity by disrupting biofilm to kill pathogens that cause dental caries [51] or debulking SARS-CoV-2 in saliva to decrease reinfection and transmission [35]. This approach is especially suitable for reducing viral load in saliva or clearing the throat surface, where most viral infections originate. While chewing gums have been used since 1928 [55] to deliver small molecules like aspirin [56] caffeine [57], calcium carbonate [58], chlorhexidine [59], nicotine [60,61], and Xylitol [62], delivering protein via gums pose additional challenges in their stability and release kinetics. For example, insulin in chewing gum tablets was mostly degraded in the gastric juice and was not validated by animal testing [63] but oral delivery of insulin bioencapsulated in plant cells is feasible [64]. Similarly, when proteins are bioencapsulated in plant cells, they are not degraded during the gum manufacturing process (requiring high temperature) and are stable for several years in chewing gum [35,51]. In this study, we optimize protein release kinetics from the chewing gum to initiate clinical trials of proteins in the oral cavity.
2. Materials and methods
2.1. Preparation of clinical grade CTB-ACE2 protein drug in lettuce
CTB-ACE2 lettuce plant material was grown, washed, and lyophilized at Fraunhofer according to the procedure described previously [35]. Grinding of plant material was performed on a pre-disinfected bench in a clean room facility using a steel grinder (BioloMix Mill Grinder, Swing −700 g) which was washed and rinsed thoroughly to remove any traces of detergent on the surface. All washed equipment's were disinfected using 70% Isopropyl Alcohol (IPA)/Ethanol to remove any bio-load attached to the surface. Forceps, aluminum foil sheets and sieve (USA standard sieve – ASTM E11 specifications; No.25; 710 μm) were autoclaved at 121 °C for 20 min. The lyophilized leaves were placed on the sterile aluminum foil sheet and placed on the clean bench and mid-ribs were removed using the pre-sterilized forceps. Ten grams of the plant material was weighed on a pre-disinfected weighing scale and transferred to the grinder. Plant material was ground for 12 s. Time for grinding was carefully monitored using Traceable Nano Timer (Fisher Scientific). The milled powder was aseptically sieved through onto sterile aluminum foil using ASTM E11 No.25,710 μm pore size sieve and transferred to presterilized Uline black container. All leftover materials on the sieve were discarded. The Uline containers containing material were stored in a steel cabinet at room temperature. An aliquot of 100 mg ground sample was aseptically removed in sterile container for bioburden assessment to evaluate total microbial and yeast/mold counts as per USP <61> and <62>. Moisture content of the plant material used for gum preparation was determined by the protocol described previously [35] (Fig. S1).
2.2. Preparation of CTB-ACE2 or FRIL chewing gum tablets
Chewing gum tablets containing ground CTB-ACE2 lettuce plant powder or Lablab purpureus bean powder were produced by Per Os Biosciences (Hunt Valley, MD) by a compression process that preserves the efficacy of the active ingredient instead of the traditional gum manufacturing technique which routinely involves extrusion/rolling at high temperatures that can introduce variability in the protein concentration and degrade its efficacy. The CTB-ACE2 gum tablets were prepared with the following excipients – gum base (24.46%), magnesium stearate (3.00%), maltitol (15.98%), Xylitol (1.98%), sorbitol (20.93%), silicon dioxide (0.40%), isomalt (10.00%), stevia 99% (0.45%), natural flavoring agents (maltodextrin, dextrose, gum arabic, essential oils) to make the gum tablets flavorful, and conducive to compression. The gums thus manufactured containing 50 mg plant powder (2 g weight/tablet) performs exactly like conventional gums available in the market with respect to physical characteristics. The gum tablets received from Per Os Biosciences were stored in the mylar bags to avoid moisture absorption. Few tablets were kept in the Uline black containers for routine examination viz; bioburden, moisture content, drug dose quantitation and release (Fig. S1).
2.3. Drug dosage determination in CTB-ACE2 gum tablets
The total dose of CTB-ACE2 in the 2 g gum tablet was examined by western blot technique. 100 mg of the crushed gum powder was suspended in 500 μL of plant extraction buffer (PEB) (100 mM NaCl; 10 mM EDTA; 200 mM Tris-HCl, pH 8.0; 0.05% (v/v) Tween-20; 1 x protease inhibitor cocktail (PIC); 0.1% SDS; 14 mM β -Mercapto-ethanol; 400 mM sucrose; and 2 mM Phenyl-methylsulfonyl fluoride (PMSF)) and incubated for 1 h at 4 °C on a vortex. This was followed by sonication for 9 cycles at 80% amplitude for 10s on and 15s off using a sonicator 3000 (Misonix, Farmingdale, NY). Bradford assay for total protein quantitation followed by immunoblot analysis for total CTB-ACE2 dose quantitation were performed following protocols developed by Daniell lab [35].
For evaluation of the CTB-ACE2 release, 100 mg of the crushed gum tablet was suspended in 500 μL of PEB and incubated for 30 min while vortexing at 4 °C. This was followed by centrifugation at 750g for 5 min at 4 °C. The supernatant fraction was stored on ice until analysis. The remaining pellet fraction was resuspended in PEB and sonicated for 3 cycles at 80% amplitude for 5s on and 10s off using a sonicator 3000 (Misonix, Farmingdale, NY). Bradford assay for total protein quantitation followed by immunoblot analysis for CTB-ACE2 release were performed following protocols developed by Daniell lab [35].
2.4. Nasopharyngeal patient swab sample preparation
Oropharyngeal (OP) and nasopharyngeal (NP) swabs samples were collected from patients admitted to the Hospital of the University of Pennsylvania with clinically-confirmed COVID-19 infection using flocked nylon swabs (Copan Diagnostics). The OP and NP swabs were eluted together in 1.5 mL of viral transport media (VTM) as previously described [65], aliquoted and frozen (−80 °C) prior to analysis. Informed consent was provided by all study participants under protocols approved by the University of Pennsylvania IRB (protocol #823392). Virus quantification was carried out by qPCR using N1 primers as previously described [65]. Virus lineage assignment (Table 1 ) was based on whole genome sequencing using the POLAR protocol [66,67] and assignment using Pangolin lineage as described [68], or based on S gene target failure in the patient's clinical diagnostic sample RT-PCR, which is a marker for the Omicron lineage [69,70]; in some cases, Omicron lineage assignment was based on near-100% Omicron circulation within the local community at time of sampling.
Table 1.
Viral titer information and virus lineage assignment through whole genome sequencing [[66], [67], [68]] of SARS-CoV-2 NP/OP samples treated with FRIL and CTB-ACE2 antiviral trap proteins and assessed by microbubble SARS-CoV-2 antigen assay and RAPID. In order to access SARS-CoV-2 genome sequence, please use link as an example- https://www.ncbi.nlm.nih.gov/nuccore/ON495672.1/
| Sample | Actual sample type | Virus titer per uL RNA (N1 qPCR) | Virus titer per uL neat sample * | Variant of Concern | Assignment method | VSP ID | NCBI Accession numbers |
|---|---|---|---|---|---|---|---|
| 620 | NP/OP | 8.33E+02 | 2.98E+02 | Omicron | WGS | VSP3590 | ON495672 |
| 613 | NP/OP | 2.78E+04 | 9.93E+03 | Omicron | WGS | VSP3583 | ON480514 |
| 614 | NP/OP | 1.40E+01 | 5.00E+00 | Omicron | SGTF | ||
| 615 | NP/OP | 1.17E+01 | 4.18E+00 | Omicron | SGTF | ||
| 593 | NP/OP | 8.70E+02 | 3.11E+02 | No | WGS | VSP3562 | OM570126 |
| 595 | NP/OP | 9.63E+02 | 3.44E+02 | Delta | WGS | VSP3563 | OM570127 |
| 596 | NP/OP | 8.84E+03 | 3.16E+03 | Delta | WGS | VSP3564 | OM570128 |
| 597 | NP/OP | 5.30E+04 | 1.89E+04 | Delta | WGS | VSP3565 | OM570129 |
| 600 | NP/OP | 2.59E+05 | 9.25E+04 | Delta | WGS | VSP3566 | OM570130 |
| 601 | NP/OP | 1.19E+04 | 4.25E+03 | Delta | WGS | VSP3567 | OM570131 |
| 603 | NP/OP | NA | NA | Delta | Date | ||
| 605 | NP/OP | NA | NA | Delta | Date | ||
| 607 | NP/OP | NA | NA | Delta | Date | ||
| 609 | NP/OP | 5.17E+03 | 1.85E+03 | Delta | WGS | VSP3572 | OM570132 |
| 613 | NP/OP | 2.78E+04 | 9.93E+03 | Omicron | WGS | VSP3583 | ON480514 |
| 615 | NP/OP | 1.17E+01 | 4.18E+00 | Omicron | SGTF | ||
| 619 | NP/OP | 1.02E+01 | 3.64E+00 | Omicron | SGTF | ||
| 620 | NP/OP | 8.33E+02 | 2.98E+02 | Omicron | WGS | VSP3590 | ON495672 |
| 624 | ETA | 6.22E+01 | 2.22E+01 | Omicron | SGTF | ||
| 626 | ETA | 1.55E+03 | 5.54E+02 | Omicron | WGS | VSP3600 | ON495673 |
| 628 | NP/OP | NA | NA | Omicron | Date | ||
| 629 | NP/OP | NA | NA | Omicron | Date | ||
| 630 | NP/OP | NA | NA | Omicron | Date | ||
| 631 | NP/OP | NA | NA | Omicron | Date | ||
| 476 | NP/OP | 5.92E+05 | 2.11E+05 | Unknown | |||
| 482 | NP/OP | 6.32E+06 | 2.26E+06 | Unknown | |||
| 505 | NP/OP | 1.17E+06 | 4.18E+05 | Unknown |
WGS - whole genome sequencing (Polar Protocol and ARTIC primers) SGTF - S gene target failure by PCR, presumed Omicron Date - circulation at time of complete or near-complete variant dominance * adjusting for the fact that RNA from 140 μl of neat sample was eluted in 50 μL buffer NA: not available.
2.5. Microbubbling SARS-CoV-2 antigen assay
The microbubbling SARS-CoV-2 antigen assay was performed with the clinical NP swab samples as described previously [35]. Out of the available four patient NP/OP samples of the omicron variant of SARS-CoV-2, two were tested with the FRIL bean powder and the remaining with CTB-ACE2 gum powder. Briefly, patient samples (150 μL) were incubated on the vortex machine with different doses of FRIL bean powder (5, 10, 25 mg containing 20, 40, 100 μg protein respectively) and CTB-ACE2 gum (10, 25, 50 mg containing 0.18, 0.46, 0.92 μg respectively) at 4 °C for 30 min. This was followed by centrifugation at 14000 rpm for 20 min at 4 °C. The supernatant thus recovered (120 μL) was carefully collected in separate tubes without disrupting the pellet. The supernatant was first treated with the lysis buffer of 10% Tween 20 (100x, 1.2 μL) and 100x protease inhibitor cocktail (1.2 μL) and incubated for 30 min at room temperature. 100 μL of the lysed samples were incubated with suspensions of 500,000 capture antibody functionalized magnetic beads in a 96 well-plate and secured on a rotator (12 rpm) for 30 min at room temperature. The 96-well plate was then placed on a magnet which separates the magnetic beads and the wells were washed thrice using washing buffer (0.05% Tween 20 in PBS buffer, pH 7.4) followed by resuspension in 100 μL of 250 ng/mL biotinylated detection antibody in PBS containing 1% BSA. After incubation for 30 min at room temperature, the beads were similarly washed thrice followed by resuspension in 100 μL of 1 μg/mL NeutrAvidin functionalized Pt nanoparticles at room temperature for 30 min. The beads were then washed thrice and finally resuspended in 100 μL of 30% H2O2. The magnetic bead mix containing immunosandwich complexes formed between magnetic beads/target protein/PtNps were transferred to the previously designed [71,72] microbubbling microchips containing microwell arrays (14 μm × 14 μm, 7 μm depth, 100 × 100). These microchips were then placed on a neodymium disk magnet subjecting it to an external magnetic field for 9 min to pull down the beads to the microwells. Microbubbles formed as a result of the accumulation of oxygen in the microwells catalyzed by PtNPs through H2O2 decomposition were imaged using an iPad with the uHandy mobile phone microscope (9× magnification, 5 mm focusing length; Aidmics Biotechnology, Taipei, Taiwan).
2.6. RAPID assay
The electrochemical sensors were prepared as described previously [73]. A SquidStat Plus (Admiral Instruments) potentiostat and Electrochemical impedance spectroscopy (EIS) were used for viral detection. The EIS measurements were performed as described previously [73]. NP/OP patient swab samples of SARS-CoV-2 delta and omicron variants were heat inactivated for 1 h at 56 °C. For RAPID assays, 150 μL of each sample was treated with 20 mg of CTB-ACE2 crushed gum powder for 1 h under stirring at 4 °C. After incubation, samples were vortexed again and spun down at 14,000 rpm for 20 min at 4 °C. First, 10 μL of VTM (blank) were added to the working electrode and left for 2 min, followed by addition of 200 μL redox probe to cover all the electrodes (counter, reference, and working electrodes), and EIS experiments were performed to obtain the blank signal. After the blank was analyzed, the sensor was washed with PBS, pH 7.4 and 10 μL aliquots of the resulting supernatant of the samples before and after incubation and centrifugation of the CTB-ACE2 chewing gum were placed directly onto the working electrode of the biosensor. The sample was removed after 2 min of exposure, cleaned carefully with PBS and after addition of 200 μL of the redox probe EIS analyses were performed.
2.7. Cell lines
African green monkey kidney epithelial Vero E6 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Invitrogen), supplemented with 5% heat-treated fetal bovine serum (FBS, Sigma), 100 units/mL penicillin, 2 mM l-glutamine, 50 μg/mL gentamicin, 100 μg/mL streptomycin, 1.25 μg/mL of amphotericin B (Fungizone), and 10 mM HEPES (4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid, pH 7.2). Cells were incubated with 5% CO2 at 37 °C. Madin-Darby Canine Kidney (MDCK) cells were cultured in Minimum Essential Media Alpha (MEM α, Gibco), supplemented with 5% heat-treated fetal bovine serum (FBS, Sigma), 100 units/mL penicillin, 2 mM l-glutamine, 50 μg/mL gentamicin, 100 μg/mL streptomycin, and 1.25 μg/mL of amphotericin B (Fungizone), and 10 mM HEPES (4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid, pH 7.2).
2.8. FRIL protein purification
Lablab purpureus bean powder was extracted in PBS buffer and dialyzed with reduced levels of salt concentrations overnight. Next, the sediment was resuspended in 20 mM phosphate buffer (pH 8.0) and transferred onto an Unosphare Q column (BioRad, Hercules, California). Bonded proteins were extracted with different gradients of NaCl (0–0.5 M). The fractions with the highest neutralization titer were pooled, concentrated, and loaded onto a Superdex s200 10/300 GL size exclusion column (GE, Boston, Massachusetts). The fractions with the highest neutralization titers were then pooled and concentrated. Finally, the bands representing FRIL were isolated through cibardon blue affinity chromatography (Affi-Gel, BioRad) flow-through and separated from nonspecific bands at ∼30 and 40 kDa.
2.9. Plaque reduction assay
Influenza Virus: Purified FRIL as well as lablab bean powder, as the source of FRIL, were evaluated for their abilities to prevent infection of influenza virus strains H1N1 (A/California/7/2009-X181) and H3N2 (A/Singapore/INFMH-16/0019/2016) using a quantitative viral plaque reduction assay. The assay was conducted in 100 μL by co-incubating the Flu strains (80 pfu) with increasing amounts of purified FRIL (0–3.2 μg) in serum-free medium or protein extract of Bean Powder (0–2 mg) in PBS at 37 °C for 1 h. The virus plus FRIL and powder, respectively, were then added onto MDCK cells (at ∼90% confluence) in 48-well plates for infection. Following 1 h adsorption at 37 °C, the virus mixtures were aspirated and washed to eliminate unabsorbed Flu. The cells were overlaid with Avicel/methylcellulose, incubated at 37 °C for 28 h, fixed and immune-stained with anti-Flu nucleoprotein antibody. Viral plaques were microscopically counted and used to generate dose response curves.
Coronavirus: OC43 was co-incubated with increasing amounts of purified FRIL (0–3.2 μg) in serum-free medium or protein extract of lablab bean powder (0–2 mg) in PBS for 1 h. Vero cells were then infected by adsorbing 100 μL of the OC43 FRIL mixtures at 34 °C for 1 h in serum free medium and then placed in culture medium containing heat-treated serum for 5 days at 37 °C after which cells were fixed and stained in 4% formaldehyde and 0.2% crystal violet. Viral plaques were microscopically counted and used to generate dose response curves.
2.10. Negative staining electron microscopy
H1N1 virus culture: H1N1 (A/California/7/2009-X181) viruses and purified FRIL protein (10 μg/mL and 150 μg/mL) were co-incubated in HEPES buffer (50 nM, pH = 8.0) at 37 °C for 60 min. The H1N1 virus was diluted from a stock concentration of 4 × 107 pfu/mL using HEPES buffer to a titer of 1 × 106 pfu/mL. The virus and purified FRIL protein were pre-treated with centrifugation at 15,000 rpm for 5 min. After incubation, samples were applied to a glow-discharged carbon-coated 400 mesh copper grid. The carbon-coated grid is stained with 2% uranyl acetate and washed twice with 5 μL of diH2O. The viruses were then observed using transmission electron microscope (FEI Tecnai T12) operating at 100 kV with a CMOS camera (Gatan Oneview, Pleasanton, California). Images were captured at the magnification of 42K using Gatan Digital Gatan Digital Micrographic software.
Purified H1N1 virus: Sucrose-gradient purified viruses were used for microscopic visualization. H1N1 (A/California/7/2009-X181) viruses and 150 μg/mL of FRIL were co-incubated at 37 °C for 30 min. Glow-discharge treatment of the carbon-coated 400 mesh copper grid was performed using PELCO easiGlow™ 91000 Glow discharge cleaning system (Ted Pella Inc., USA). The grids were glow discharged with 15 mA current for 30 s. After incubation, samples were diluted with PBS buffer and loaded onto the carbon-coated grid. The grid is negatively stained with Nano-W (Nanoprobes, Yaphank, New York). The viruses were observed using electron microscope (JEM-1400. JEOL, Peabody, Massachusetts) operating at 120 kV with a CCD camera (Gatan 895. Gatan, Pleasanton, California). Images were captured at the magnifications of 2.6K and 5K by Gatan Digital Micrographic software.
2.11. Protein release from chewing gum tablets
The feasibility of topical drug delivery was tested on two different drug-containing gums: the green fluorescent protein (GFP) and the angiotensin converting enzyme-2 (ACE2). After lyophilization, plant cells expressing GFP or CTB-ACE2 were incorporated into chewing gums, and release kinetics after mechanical simulation were quantified via fluorescence intensity for GFP or by western blot quantitation for CTB-ACE2. The GFP gum contained 448 μg GFP, and two doses of CTB-ACE2 gum were comprised of 365.8 and 752.7 μg CTB-ACE2.
GFP gum: A 2-g gum tablet with 25 mg (mg) of plant powder was placed in 6mL of PEB [51]. Chewing of the gum was simulated using the cyclic loading mode of a Universal Mechanical Testing Machine with Merlin software (Instron Model 5564) for 0, 10, 20, and 30 min . A minimum compressive load of −1200 N and a maximum extension of 0.7 mm ( at a speed of 300 mm/min was used to mimic human chewing habits. Utilizing these parameters, 0, 286, 470, and 645 simulated chewing cycles were achieved at 0, 10, 20, and 30 min, respectively. After grinding at each time point, 100 μL of sample were removed and stored on ice for analysis. Each sample was centrifuged at 13,550 rpm at 4 °C for 10 min. Afterwards, the homogenate fraction (HMG) was collected and stored on ice. The release of GFP from the HMG was quantified using known concentrations of the recombinant GFP protein (ab119740, abcam). The HMG was diluted 1:10 with phosphate-buffered saline (PBS). The diluted HMG (200 μL) was loaded into the wells of an assay plate (Corning Incorporated, Costar Assay Plate 3916, ME USA) and read on a microplate reader (BioTek, Synergy H1, VT USA). The fluorescence intensity was detected at 485 nm (excitation) and 538 nm (emission) [51].
CTB-ACE2 gum: Two different doses of CTB-ACE2 chewing gum (2 g), namely a 365.8 and 752.7 μg CTB-ACE2 gum tablet, were analyzed using the same machine at 0, 10, and 20 min and CTB-ACE2 was quantified via Western Blot. A minimum compressive load of −1200 N and a maximum extension of 0.7 mm at a speed of 300 mm/min were used for both doses, and 0, 140, and 385 chewing cycles were achieved for the 365.8 μg CTB-ACE2 gum and 0, 192, and 464 simulated chewing cycles were obtained for the 752.7 μg CTB-ACE2 gum at 0, 10, and 20 min, respectively. PEB composition as well as the volumes removed after each time point for both doses of CTB-ACE2 gum were identical to the GFP gum experiment. Each 100 μL sample was centrifuged at 750 g at 4 °C for 5 min. After centrifugation, the HMG was collected and stored on ice. The HMG was analyzed for total protein concentration by Bradford assay [74] (Bio-rad Laboratories, Hercules, CA) using known concentrations of bovine serum albumin (BSA) standards that ranged from 0.025 to 0.80 μg/μL. The homogenate gum samples were diluted 1:5 with Milli-Q water, and 10 μL of the diluted sample were loaded into the wells of an assay plate 96 wells (Corning Incorporated, Costar Assay Plate 9017, ME USA). This was followed by quantitation of CTB-ACE2 by western blot analysis as previously described, with appropriate modifications [35].
3. Results and discussion
As pointed out in the introduction, oral transmission of SARS-CoV-2 is 3–5 orders of magnitude higher than nasal transmission [[14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28]]. After two years of the current pandemic, there is still no FDA approved quantitative detection method to end the quarantine or return of employees back to workplaces after testing positive for SARS-CoV-2. It is possible that qPCR detects non-viable viral particles, as evidenced by detection of SARS-CoV-2 several weeks after disappearance of symptoms [[31], [32], [33]] and CDC guidelines do not support qPCR testing up to ninety days after the onset of infection. In order to develop a quantitative antigen test and study the changes in viral load in COVID-19 samples in response to viral trap proteins, this study utilizes two different approaches. Microbubbling (N antigen) and RAPID (spike protein) assays are used to evaluate CTB-ACE2 gum or the lablab bean powder containing the viral trap protein – FRIL. In addition, the impact of FRIL on influenza virus strains is evaluated in plaque reduction assays. Mechanism of FRIL entrapment is further investigated using electron microscopic studies. Various steps involved in the process of creation of plants expressing CTB-ACE2, production of seeds, biomass, sterilization, lyophilization, grinding/sieving, production of chewing gum with lyophilized plant powder, evaluation of FDA requirements (moisture content, bioburden, drug dosage, stability, etc.), functional characterization, toxicology/pharmacokinetic studies, filing of IND and clinical trial design are summarized in Fig. 8 , Fig. S1.
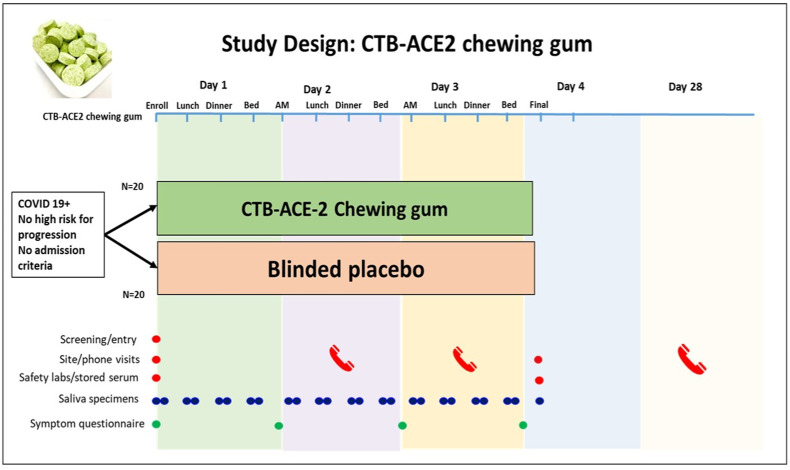
Fig. 8.
CTB-ACE2 chewing gum Phase I/II clinical study design. Based on FDA approval of IND (154897) Phase I/II placebo-controlled, double-blind randomized CTB-ACE2 or placebo gum study is in progress. Study duration is four days with 13 gums total, four gums each in days 1–3. An unstimulated whole saliva sample will be collected before eating or drinking or brushing teeth. Subjects will chew the CTB-ACE2 chewing gum/placebo gum (study product containing 2 g of CTB-ACE2 or placebo) for 10 min and then immediately provide a post-treatment 2–5 mL saliva sample in pre-labeled salivary collection tubes. Viral load will be quantified by qPCR or protein (N or spike) quantitation.
3.1. CTB-ACE2 dose quantitation and release from gum tablets during incubation
Western blot assessment of gum tablet revealed the total dose of CTB-ACE2 to be 365.8 ± 18.0 μg/2 g tablet (Fig. S2 A, B). CTB-ACE2 release without sonication was found to be 37 ± 1.0 μg/tablet (2 g) (Fig. S2 C, D) under optimized experimental conditions performed in the lab. The grinding conditions were optimized to maximize the release of protein conducive for chewing gum drug delivery system. This is different from the orally delivered protein drugs [50,64] that are bioencapsulated and hence protected from degradation in the stomach by gastric juices and subsequently released in the gut by microbial digestion of the plant cell wall.
Bioburden assessment for plant material used for preparation of gum and the gum tablets revealed no microbial or fungal growth adhering compliance to the FDA parameters. Moisture content of clinical grade CTB-ACE2 lettuce powder was found to be 5.5% which meets the FDA requirements for orally delivered plant powder. The gum tablets prepared thus, comply for the FDA specifications and ready to be used for clinical trials.
3.2. Evaluation of SARS-CoV-2 in clinical swab samples by microbubbling after treatment with CTB-ACE2 gum and FRIL bean protein
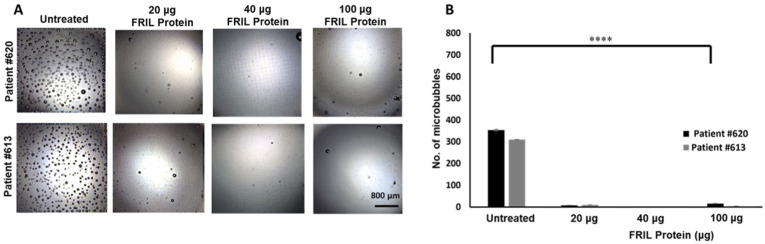
The microbubbling SARS-CoV-2 antigen assay is designed to detect the nucleocapsid (N) antigen at femtomolar concentrations [71]. Due to limited sample volume availability, dose dependent analysis was restricted to three doses. Omicron variant NP samples from patients #620 and #613 were tested for the debulking potential of FRIL bean protein at doses 20, 40 and 100 μg each and patients #614 and #615 were tested for the same with CTB-ACE2 gum at doses 0.18, 0.46 and 0.92 μg. The viral titers for all four omicron variant NP samples are shown in.
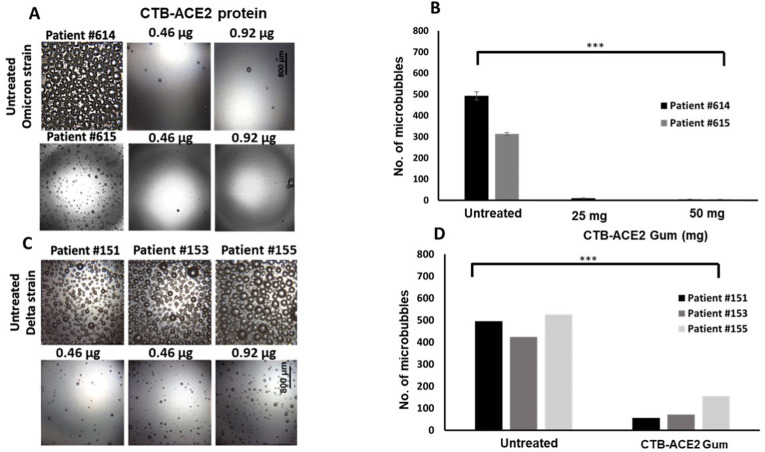
As seen in Fig. 2A and B FRIL protein is capable of significantly reducing the microbubble count (p < 0.0001) even at the lowest dose of 20 μg. A dose dependent reduction in microbubbles was seen with CTB-ACE2 gum, with least number of microbubbles seen in the 0.92 μg treated sample (p = 0.0001) (Fig. 3A and B).
Fig. 2.
Neutralization of SARS-CoV-2 Omicron variant in NP swab samples with FRIL from Lablab bean powder: (A) Images from clinical NP/OP swab samples of Patients #620 and #613 treated with FRIL bean powder (20,40,100 μg FRIL protein in 5,10 and 25 mg of bean powder respectively). (B) Number of microbubbles are quantified. All images share the same scale bar. Data were analyzed by One-way ANOVA test. There is a significant difference in the microbubble count between the Untreated vs FRIL bean powder at all concentrations. (***p-value < 0.0001). Omicron SARS-CoV-2 strain identification is based on whole genome sequencing or S gene target failure or date of collection. Please refer to Table 1 for other details on patient IDs.
Fig. 3.
Neutralization of SARS-CoV-2 Delta and Omicron variants in NP swab samples by CTB-ACE2 gum (A) Images from clinical NP/OP swab samples of Patients #614, #615 treated with CTB-ACE2 gum (0.46 or 0.92 μg CTB-ACE2 protein in 25 and 50 mg of gum respectively) (B) Quantitation of microbubbles show a significant difference between the untreated Omicron strain vs CTB-ACE2 gum at both concentrations. (***p-value = 0.0001). All images shown share the same scale bar. (C) Images from clinical NP/OP swab samples of Patients #151, #153 and #155 treated with ACE2 gum (0.46 or 0.92 μg CTB-ACE2 protein in 25 and 50 mg of gum respectively). (D) Quantitation of microbubbles show a significant difference between the untreated Delta strain vs CTB-ACE2 gum at both concentrations. (***p-value = 0.0009). All data shown were analyzed by One-way ANOVA test. Omicron and Delta SARS-CoV-2 strain identification is based on whole genome sequencing or S gene target failure or date of collection. Please refer to Table 1 for other details on patient IDs.
As we compare the doses between both plant-based anti-viral trap proteins, it is interesting to note that CTB-ACE2 is effective at a dose of 0.46 μg while almost similar debulking potential was seen with 20 μg of FRIL. In our recently published paper [35] we have shown the debulking efficacy of CTB-ACE2 against the delta variant of the virus as seen in Fig. 3C and D. The neutralization potential of CTB-ACE2 is effective across both variants of the SARS-CoV-2. These results also confirm that the SARS-CoV-2 debulking efficiency of FRIL, a plant lectin, is distinct from that of CTB-ACE2. While ACE2 binds directly to the SARS-CoV-2 spike protein and entrapping the virus in the pentameric insoluble microparticle structure of CTB-ACE2 [34], FRIL preferentially binds to complex-typeN-glycans on viral glycoproteins forming aggregates [39]. Both these distinct mechanisms facilitate removal of entrapped viral particles and hence could be an effective preventive measure in limiting viral transmission. In all four patient swab samples, we do not observe a positive correlation between the viral load titer as determined by RT-PCR and the microbubble count that depends on the presence of N antigen (Table 1). Patients (#620, #613) with higher viral load have lower microbubble count, which suggests that RT-PCR may be detecting N1 gene fragments that do not produce intact N1 antigens with epitopes for antibody binding [72].
3.3. Evaluation of SARS-CoV-2 in clinical swab samples by RAPID after treatment with CTB-ACE2 gum
Our previously developed method RAPID (Real-time Accurate Portable Impedimetric Detection) [73,75], which uses electrochemical impedance spectroscopy (EIS) to detect the binding between spike protein and human receptor ACE2, was used here to validate the trapping effect of the CTB-ACE2 chewing gum [[76], [77], [78], [79], [80], [81], [82]]. Based on our prior work with hundreds of clinical samples in order to elucidate the optimal analytical conditions for the assay [73,75], we used the normalized RCT response, defined by the following equation:
where Z is the RCT of the sample group and Z0 is the RCT of the blank group (virus transportation medium, VTM). RAPID presents low limits of detection (LOD, 6.29 fg mL−1 SP) and quantification (LOQ, 20.96 fg mL−1 SP) in VTM medium based on the signal to noise ratio (S/N = 3) and (S/N = 10), respectively [70].
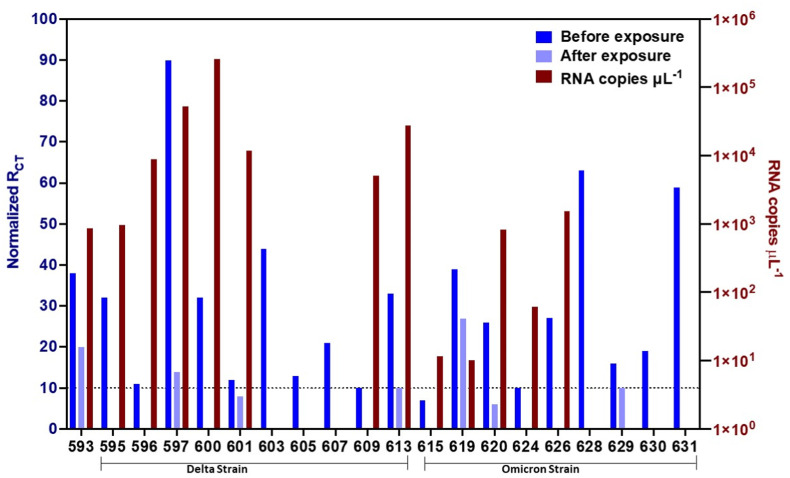
With the exception of one sample (ID 593), the samples were genotyped based on whole genome sequencing or S gene target failure or date of collection (Table 1, Fig. 4). Samples 595–609 are Delta and 613–631 are Omicron strains of SARS-CoV-2. RNA extraction for all samples were from equal patient sample volume (140 μL) and RNA extracted was resuspended and quantified by qPCR under similar conditions. There is no unique change observed in viral load between Delta and Omicron samples, but viral load varied significantly among patients. Patient samples (150 μL of sample) were incubated with the CTB-ACE2 chewing gum and RAPID was performed in 10 μl aliquots of the patient sample (Fig. 4) and RCT values varies significantly among patient samples. The CTB-ACE2 chewing gum was able to trap viral particles efficiently, decreasing viral concentration in all the 20 samples analyzed (Fig. 4). For 17 of the samples, no viral particles were detected by RAPID after the incubation with the CTB-ACE2 chewing gum, underscoring the ability of this material to trap the viral particles up to very low SARS-CoV-2 SP concentrations levels (below the LOD of RAPID).
Fig. 4.
CTB-ACE2 chewing gum trapping efficacy assessed through diagnosis with RAPID. Normalized charge-transfer resistance values for the electrochemical impedance spectroscopy measurements of 20 SARS-CoV-2 clinical samples (left y-axis) before and after exposure to 20 mg of ACE2 chewing gum. RNA copies μL −1 from qPCR of infected samples prior to exposure to the ACE2 chewing gum (right y-axis). Numbers below samples are patient IDs. Please refer to Table 1 for other details. Sample ID 593 was not genotyped. Samples 595–609 are Delta and 613–631 are Omicron SARS-CoV-2 strain identification is based on whole genome sequencing or S gene target failure or date of collection. RNA extraction was from 140 μL patient sample (1/50 μL shown) and RAPID was performed in 100 μL patient sample (10 μL Rct value shown).
3.4. FRIL exhibits potent neutralization of influenza virus and coronavirus
As FRIL has binding affinity to complex-type N-linked glycans, it could be effective against a wide spectrum of enveloped viruses that express complex-type N-glycans, such as Influenza virus, HBV, and HSV [83,84]. First, we examined the antiviral activities of purified FRIL protein and lablab bean powder against the Influenza H1N1 (A/California/7/2009-X181), and H3N2 (A/Singapore/INFMH-16/0019/2016), as well as Coronavirus HCoV-OC43 using plaque reduction assays (Fig. 5, Fig. S3).
Fig. 5.
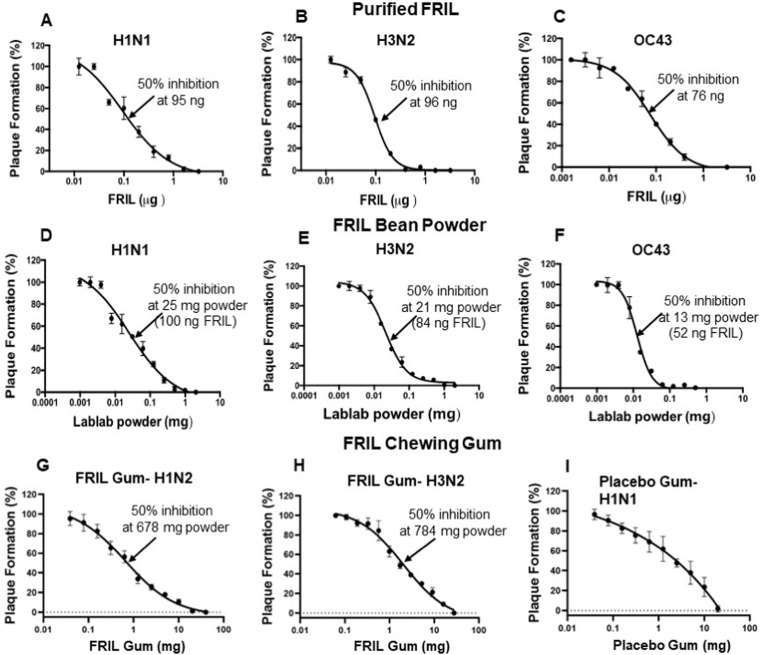
FRIL Plaque Reduction Neutralization Assay: H1N1 (A/California/7/2009-X181), H3N2 (A/Singapore/INFMH-16/0019/2016), and HCoV-OC43 viruses were pre-incubated with increasing amounts of purified FRIL protein or soluble extract of lablab bean powder or soluble extract of FRIL chewing gum powder at 37 °C for 1 h in 100 μL. The pretreated viruses were then added onto cells for the plaque reduction assay and plaque numbers were quantified at 28 h post infection for the two Flu viruses and 5 days post infection for HCoV-OC43 virus. The data represents Mean ± SD of plaque numbers from two independent experiments in duplicate. One mg of bean powder contains 4 μg FRIL. Dosage for 50% inhibition is shown in nanograms (ng) for purified FRIL and milligrams (mg) for bean powder and gum.
In the Influenza plaque reduction assays, purified FRIL protein exhibited a midpoint inhibition at 95 ng (in 100 μL treatment volume) against H1N1 (Figs. 5A) and 96 ng against H3N2 (Fig. 5B). Interestingly, FRIL's antiviral potency of 76 ng against OC43 was experimentally close to that of the Influenza viruses (Fig. 5C), taking into consideration possible minor differences in viral titers and plaque counts. These results correspond to the previously reported 50% plaque reduction neutralization test (PRNT50) value against H1N1, H3N2, and hCoV-19/Taiwan/NTU04/2020 [39].
Compared to purified FRIL, lablab bean powder is a better candidate for anti-viral chewing gum with advantages related to affordability, accessibility, and stability of enzymes. Therefore, we examined the neutralization ability of lablab bean powder, which contains ∼4 μg FRIL protein per 1 mg of bean powder. Plaque assays with soluble lablab bean powder exhibited mid-point inhibition values of 25 μg (estimated to release 100 ng FRIL) against H1N1 (Fig. 5D), and 21 μg (84 ng FRIL) against H3N2 (Fig. 5E) and 13 μg (52 ng FRIL) against OC43 (Fig. 5F). More importantly, the estimated amount of solubilized FRIL released at midpoint inhibition is experimentally close to the amount of purified FRIL used for midpoint inhibition, taking into consideration possible minor differences in viral titers and plaque counts. These data show that purification of FRIL from lablab bean powder is not required for its antiviral potency, which lays the logical foundation for manufacturing chewing gum with lablab bean powder. In addition, for both Influenza stains and the OC43 Coronavirus, less than 1 mg/mL of lablab bean powder (which releases 4 μg/mL of FRIL) can effectively inhibit all infections. Such high potency of FRIL bean powder allows for the manufacturing of chewing gum containing FRIL that could effectively reduce the transmission risk of both Influenza and Coronavirus.
Based on the outstanding efficacy of lablab bean powder, FRIL chewing gum was manufactured by Per Os Biosciences using Lablab Purpureus bean powder. FRIL chewing gum demonstrated strong and specific anti-viral protection against the H1N1 and H3N2 viruses. Plaque assays with soluble FRIL chewing gum powder exhibited mid-point inhibition values of 678 μg against H1N1 (Figs. 5G), and 784 μg against H3N2 (Fig. 5H) in 100 μL treatment volume. The differences between mid-point inhibition values of H1N1 and H3N2 is not significant considering variations in the viral load, viral activity, and experimental variations (Fig. S3). Moreover, 100% inhibition of plaque formation is achieved at 20 mg of FRIL gum powder. Considering that the volume of stimulated saliva during early chewing is 4–5 mL [[85], [86], [87]], we estimate dosage of FRIL in the chewing gum is more than adequate to effectively debulk different Influenza virus strains and Coronaviruses in saliva, thereby preventing viral infection/transmission.
It is important to note that we observed inhibitory effect against H1N1 and H3N2 in the placebo gum powder as well (Fig. 5I). The inhibitory effect of placebo gum is more significant at higher concentrations, while FRIL gum powder exhibits stronger anti-viral capacity at all doses (Fig S3, S4). Interestingly, the plaque reduction of placebo gum does not fit into Hill equation, which is typically suitable for inhibitory effect based on protein-protein interactions [88]. The marginal reduction of plaque does not decrease as dosage of placebo gum increases, suggesting that the inhibitory effect of placebo gum is not based on specific interactions with the influenza virus. We suspect the inhibitory effect of the placebo gum is due to maltitol (15.98% in chewing gum), Xylitol (1.98%), sorbitol (20.93%) added to the gum base. Sorbitol and xylitol has been reported to have anti-viral activity against human respiratory syncytial virus (hRSV), Influenza virus, and SARS-CoV-2 virus in animal models or cell culture [[90], [91]] but not evaluated in the clinic. The combination of sugar alcohol (non-specific antiviral) and highly specific anti-viral proteins make anti-viral chewing gums even more attractive tool for prevention and treatment of SARS-CoV-2 and Influenza virus.
3.5. Mechanism of virus entrapment by FRIL bean protein
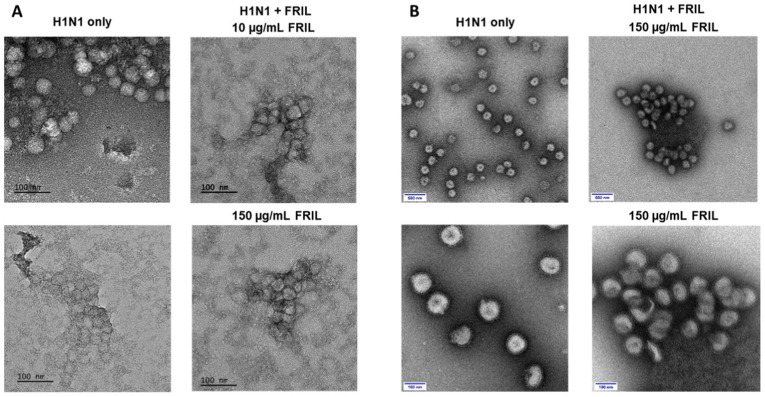
Different from the debulking mechanism of CTB-ACE2, FRIL entraps virions through its binding affinity to the complex-type N-linked glycans on virus envelopes. Assemblies of entrapped virus particles were observed at 10 μg/mL FRIL with unpurified Influenza virus using negative staining EM (Fig. 6A). Influenza particles were bond at much closer proximity to one another by FRIL compared to untreated virus particles, with visible FRIL proteins surrounding the viral aggregates. Large and densely packed clumps of overlapping influenza particles and FRIL protein were observed at 150 μg/mL FRIL concentrations but not in untreated virus particles (Fig. 6A).
Fig. 6.
FRIL entraps influenza virus by aggregation: (A) Negative-stain EM images of H1N1 (A/California/7/2009-X181) cell culture virions alone (left panels) and aggregated virus particles with FRIL after incubating with 10 μg/mL and 150 μg/mL FRIL (right panels). Data are representative of 2 independent experiments. (B) Negative-stain EM images of sucrose-gradient purified X181 virions alone (left panels) and aggregated X181 virus particles after mixing with 150 μg/mL FRIL (right panels). Scale bar: 100 and 500 nm, respectively.
To better observe the aggregation of Influenza particles, we then conducted the protocol with sucrose-gradient purified viruses and 150 μg/mL purified FRIL. Aggregates of Influenza virus particles can be observed surrounding FRIL aggregates (Fig. 6B). The amount of isolated Influenza virus, which can be observed abundantly in untreated Influenza virus, is greatly decreased with the addition of FRIL.
These images suggest that FRIL's carbohydrate-binding domain (CBD) on each of its four monomers could connect multiple virus particles, thereby entrapping virions in large aggregates at 10–150 μg/mL of FRIL protein (Fig. 6). The clustering of virus particles could explain FRIL's debulking mechanisms: the entrapped Influenza virus particles would be neutralized and removed from the sample during centrifugation. Furthermore, other virus-trapping lectins MBL and SP-D have been documented to promote viral clearance with the immune system [92], and FRIL could have a similar effect. Since the spike (S) protein present on the SARS-CoV-2 virus envelope encodes 22 complex-type N-linked glycan sites per monomer [41], we hypothesize that FRIL could entrap and debulk SARS-CoV-2 through binding to the N-glycan sites on spike protein in the microbubble assay. Mainly, when the patient sample was incubated with lablab bean powder, FRIL protein released from the lablab bean powder could entrap SARS-CoV-2 virus particles to form into clumps. The entrapped virus particles are then removed during the centrifugation, leading to a lower microbubble count.
3.6. Protein release from GFP-Protegrin and CTB-ACE2 chewing gum tablets by simulation of human chewing
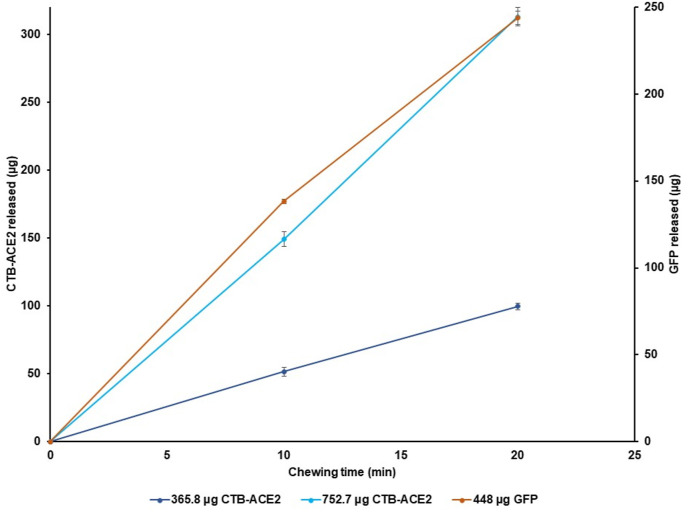
In this study, we optimized in vitro release kinetics by simulating human chewing of gum tablets containing different doses of target proteins. Data on time versus load and extension versus load were also collected at all time points, and cyclic reproducibility is observed (Fig. S5). The percentage of GFP-protegrin released in the HMG was calculated, with the amount of GFP-protegrin released increasing with chewing time. GFP release from gum tablets was linear up to 30 min of mechanical chewing. GFP-protegrin concentration in PEB increased to 312.6 μg after 30 min, which is 70% of the total GFP present in the gum tablet. Additionally, there was a significant difference (****p < 0.00001) between the GFP released at 0 min (0 μg) and 20 min (238.7 μg). Similarly, the percentage of CTB-ACE2 released in the HMG was quantified, with the amount of CTB-ACE2 released from the pellet increasing linearly with time of mechanical bite force. CTB-ACE2 release from both dosages of gum tablets were linear until 20 min of mechanical chewing, with most of the protein release occurring during early chewing. After 20 min of simulated chewing, 99.6 ± 4.9 μg (27%) and 313.0 ± 6.8 μg (42%) CTB-ACE2 release was observed for the 365.8 μg and 752.7 μg ACE2 tablets, respectively, which amounts to an excess of 213.4 μg CTB-ACE2 with the clinical gum. Therefore, in future clinical studies, a much lower dose of CTB-ACE2 gum tablet could be evaluated. Again, there was a significant difference (****p < 0.00001) between the CTB-ACE2 released at 0 min and 20 min for both doses of CTB-ACE2 chewing gums (Fig. 7). Therefore, protein release was both time and dose-dependent.
Fig. 7.
Release of proteins from the non-clinical and clinical CTB-ACE2 chewing gums and GFP chewing gum. Protein release was evaluated in the homogenate at 0, 10, and 20 min. After 20 min of simulated chewing, 99.6 ± 4.9 μg CTB-ACE2 (27% release) and 313.0 ± 6.8 μg (42% release) was released from the 365.8 ± 18.0 μg and 752.7 ± 3.4 μg tablet, respectively. The 448 μg GFP gum released 243.9 ± 7.4 μg (54% release) after 20 min of simulated chewing. There is a significant difference between the CTB-ACE2 released at 0 and 20 min for both doses of chewing gum (****p-value < 0.00001). Similarly, there is significant difference of GFP released from the GFP tablet at 0 and 20 min (****p-value < 0.00001); data were analyzed by a one-way ANOVA test. The data represents mean ± standard deviation (SD) (n = 4).
Chewing patterns and bite force greatly vary by individual and do not show a linear pattern as a function of age [93], with bite forces ranging from 113 to 1692 N [94]. Bite forces of 777.7 ± 78.7 N and 481.6 ± 190.4 N (mean ± SD) have been reported among adult males and females, respectively [95]. Therefore, the cyclic loading parameters were strategically chosen to account for the variability in chewing habits while also not exceeding a load of 1500 N. When comparing the varying doses of CTB-ACE2 chewing gum, the lower-dosed gum released less CTB-ACE2 at each timepoint than the higher-dosed gum. Therefore, the dose of the chewing gum is the most critical criterion. Moreover, as both the CTB-ACE2 and GFP chewing gums showed sustained release of the plant protein, longer chewing ensures optimal results. As shown above, complete debulking of the SARS-CoV-2 virus is achieved with 0.46 μg CTB-ACE2 in 120 μL patient samples. Therefore, in 4–5 mL volume of initial stimulated saliva [[85], [86], [87]], we expect complete debulking after 20 min of chewing either dose of the CTB-ACE2 gum. Finally, drug dosage was more critical than time of mechanical chewing, and increased levels of protein release were observed with higher dose of gums.
3.7. Advancing chewing gum delivery of viral trap proteins from laboratory to the clinic
While ex vivo SARS-CoV-2 debulking is effective for various SARS-CoV-2 strains, including the highly transmissible omicron strain, the time required for repopulation of saliva by SARS-CoV-2 is still unknown. While current CTB-ACE2 gum could be used for short duration (in dental clinics, public transportation, gatherings, restaurants etc.), therapeutic applications of the CTB-ACE2 gum for successful reduction of viral load in COVID-19 patients would require data on viral load kinetics in saliva. Therefore, a phase I/II placebo-controlled, double-blind study of CTB-ACE2 chewing gum is now conducted as detailed in the study plan (Fig. 8).
Investigational New Drug application on CTB-ACE2 chewing gum has been approved by the FDA (IND 154897) and Phase I/II placebo-controlled, double-blinded clinical trial is underway to assess viral load in saliva before and after chewing of CTB-ACE2/placebo gum. Because Lablab bean is consumed globally, used as dry powder in protein enriched diet [96,97], this would be Generally Regarded As Safe by FDA, thereby decreasing hurdles in the regulatory approval process. Recruitment and informed consent, those meeting enrollment criteria will be randomized 1:1 (CTB-ACE2 chewing gum/placebo gum). A stratified blocked randomization with randomly permuted blocks will be performed based on age (<60, ≥60) given the strong impact of age factors on outcomes in COVID-19. Participants will be evaluated in the Clinical Trials Unit (CTU) of the University of Pennsylvania. An unstimulated whole saliva sample before eating or drinking or brushing teeth will be collected. Subjects will then chew CTB-ACE2 chewing gum/placebo gum (study product containing 2 g of CTB-ACE2 or placebo) for 10 min and then immediately provide a post-treatment 2–5 mL saliva sample in pre-labeled salivary collection tubes. In addition, the subjects will be asked to chew a piece of CTB-ACE2 gum/placebo gum for 10 min at home in the AM (not on the day of enrollment), before lunch, in the late afternoon and before sleeping on days 1, 2 and 3, and collect a saliva sample before eating or drinking or brushing teeth. Subjects will then chew CTB-ACE2 chewing gum/placebo gum (study product) for 10 min and collect before and after treatment 2–5 mL saliva samples, in pre-labeled salivary collection tubes. On Day 4, one final unstimulated saliva sample will be collected in the morning before eating or drinking or brushing teeth at home and the specimens (from day 1–4) will be brought to the CTU for a final clinical visit. Participants will be contacted by phone on day 28 for a final status update.
This study will use the infrastructure of the funded CTU at the University of Pennsylvania (Pablo Tebas, PI). Study participants will be recruited from both University of Pennsylvania's Infectious Disease Clinic as well as Penn Community Practice at Penn Presbyterian Medical Center by way of provider referrals, self-referrals, and identification of eligible patients using the PennChart (EMR of the Hospital of the University of Pennsylvania). The Investigative Team will also leverage extensive School of Medicine resources to support early phase trials, including the Office of Clinical Research, the Penn Medicine Academic Computing Service (PMACS), the Investigational Drug Service (IDS), and our CTSA-supported Center for Human Phenomic Sciences (CHPS). The Office of Clinical Research (OCR) is a central office in the Perelman School of Medicine (PSOM) designed to support the management and conduct of clinical research while promoting compliance.
The primary safety analyses will be conducted on all participants receiving study product and presented by age strata, all solicited and unsolicited AEs will be summarized by frequency per arm for the whole group, and stratified by age, as percentages, along with associated exact 95% Clopper-Pearson confidence intervals. For the virology endpoints, levels of SARS-CoV-2 RNA on days 1,2,3,4 will be compared between arms using non-parametric Wilcoxon rank-sum tests and descriptive statistics, separately at each scheduled measurement time. In addition, viral antigens (N or spike protein) will be quantified in saliva samples using microbubble or RAPID assays shown above. For the evaluation of the clinical evolution of COVID 19, the severity ranking will be based on the area under the curve AUC of the daily total symptom score associated with COVID-19 over time.
4. Conclusion and future directions
Approximately one fourth of American population chew gum 2–3 times a week [98] mostly for pleasure, although delivery of small molecules is used currently or in the past for delivering aspirin, nicotine, caffeine or for enhancing oral hygiene or health. Attempts to deliver therapeutic proteins like insulin using chewing gum have failed so far, primarily because of degradation in the digestive system but is well suited for neutralizing pathogens in the oral cavity or in the throat surface to reduce transmission of infection. Most importantly, proteins produced in plants have shown incredible stability in the gum during production at high temperature and long-term storage at ambient temperature. This study illustrates the power of delivering viral trap proteins delivered via chewing gum to reduce infection and transmission of SARS-CoV-2 or influenza viruses and the potential to advance this platform to various other orally transmitted viral or bacterial diseases.
4.1. Statistical analyses
CTB-ACE2 total dose quantitation and release data are presented by means ± SD. Microbubbling SARS-CoV-2 Antigen Assay data are presented by means ± SD. Statistical significance was determined using by One-Way Electrochemical impedimetric measurements are presented as an average of 3 replicates for each condition. Graphs were created and statistical tests were conducted in GraphPad Prism 9.2. In plaque reduction assay, the plaque numbers were quantified under a dissecting microscope. Half-maximal EC50 values were obtained by nonlinear regression fitting to a variable slope, four parameter dose-response model using the GraphPad Prism 6 software (GraphPad Software, LaJolla, CA).
Author contributions
HD conceived this project, designed experiments, created materials, identified collaborators and coordinated projects with different groups or institutions, interpreted data, wrote/edited several versions of this manuscript. SKN quantified CTB-ACE2 in chewing gum and performed microbubble assays, interpreted data and wrote corresponding sections. HG and YG performed plaque reduction assays in different viral strains. YG assisted in electron microscopy microbubble and plaque reduction assays, MSM, YM and CM prepared purified FRIL protein and lablab bean powder and quantified FRIL dosage, as well as the negative-stain images of influenza virus aggregated by FRIL. JGW and RGC contributed NP/OP samples and ADM, FDB quantified viral load and identified virus genotype through sequencing shown in Table 1. WZ and PW provided microbubbling assay reagents and instruments and assisted in experiments. GW characterized the CTB-ACE2 chewing gum with SKN and RJK. RJK quantified CTB-ACE2 and GFP in chewing gums in addition to performing release kinetic studies in chewing gums, interpreting data, and writing corresponding sections with SKN, with guidance from FM. RJK assisted in preparing Fig. 1 and Fig. S1. WRA manufactured the electrochemical devices, MDTT performed electrochemical experiments designed by CFN. SKM prepared grids and generated electron micrographs using cultured influenza viruses. WRT is the PI on phase I/II CTB-ACE2 gum clinical trial, PT is sub-investigator contributing to study design and KBM is the medical director. RPR advised and interpreted influenza virus data and wrote corresponding sections. MSW contributed technical discussions on release studies and historical applications of the chewing gum.
Funding
Research in the Daniell laboratory is supported by funding from NIH grant R01 HL 107904, R01 HL 109442, R01 HL 133191. Commonwealth of Pennsylvania, Department of Community and Economic Development grant to HD on “COVID-19 Pennsylvania Discoveries: Responding to SARS-COV-2 Through Innovation & Commercialization” funded purchase of freeze dryers, toxicology studies on ACE2 produced at Fraunhofer USA/AeroFarms and production of the chewing gum. NP/OP sample collection in the Collman lab is supported by R33-HL137063 and the Penn Center for Coronavirus Research. Microbubbling assay in the Wang laboratory is supported by funding from the Penn Center for Precision Medicine, Penn Health-Tech, Penn Center for Innovation and Precision Dentistry, NIH RADx and BARDA. RAPID assay is supported by the Nemirovsky Prize and the Dean's Innovation Fund from the Perelman School of Medicine at the University of Pennsylvania. Research in Ma laboratory is supported by Taiwan Ministry of Science and Technology (MOST 110-2113-M-001-034-MY3).
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Henry Daniell has patent #USA patent 11,246,819 issued to University of Pennsylvania. Henry Daniell has patent #USA patent 10,806,775 issued to University of Pennsylvania. Henry Daniell has patent #USA patent 10,314,893 issued to University of Pennsylvania.
Acknowledgements
Authors thank Lisa Bachman and Robert Estey for preparation of chewing gums, Admiral Instruments for donating the potentiostats used in the electrochemical experiments for RAPID assays and Naila Shahid for quantification of proteins in lablab bean powder. Authors thank Steven Szewczyk and Michelle Paolicelli for assisting with chewing gum protein release studies and acknowledge use of facilities and instrumentation supported by the Materials Science and Engineering Departmental Laboratory at the University of Pennsylvania and NSF through the University of Pennsylvania Materials Research Science and Engineering Center (MRSEC) (DMR-1720530).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.biomaterials.2022.121671.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Bowen W.H., Burne R.A., Wu H., Koo H. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 2018;26:229–242. doi: 10.1016/j.tim.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang N., Pérez P., Kato T., Mikami Y., Okuda K., Gilmore R.C., Dominguez Conde C., Gasmi B., Stein S., Beach M., et al. SARS-CoV-2 infection of the oral cavity and saliva. Nat. Med. 2021;27:892–903. doi: 10.1038/s41591-021-01296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y., Ren B., Peng X., Hu T., Li J., Cong T., Tang B., Xu X., Zhou X. Saliva is a non‐negligible factor in the spread of COVID‐19. Mol. Oral Microbiol. 2020;35:141–145. doi: 10.1111/omi.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan J.F.-W., Yip C.C., To K.K., Tang T.H., Wong S.C., Leung K., Fung A.Y., Ng A.C., Zou Z., Tsoi H., et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58:10–20. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 6.Atyeo N., Rodriguez M.D., Papp B., Toth Z. Clinical manifestations and epigenetic regulation of oral herpesvirus infections. Viruses. 2021;13(681):2–17. doi: 10.3390/v13040681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tewari P., Banka P., Kernan N., Reynolds S., White C., Pilkington L., O'Toole S., Sharp L., D'Arcy T., Cliona M., et al. Prevalence and concordance of oral HPV infections with cervical HPV infections in women referred to colposcopy with abnormal cytology. J. Oral Pathol. Med. 2021;50(7):692–699. doi: 10.1111/jop.13172. [DOI] [PubMed] [Google Scholar]
- 8.Paules C., Subbarao K. Influenza. Lancet. 2017;390:697–708. doi: 10.1016/S0140-6736(17)30129-0. [DOI] [PubMed] [Google Scholar]
- 9.Jang Y.H., Seong B.L. The quest for a truly universal influenza vaccine. Front. Cell. Infect. Microbiol. 2019;9:344. doi: 10.3389/fcimb.2019.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W.J., Xiao H., Dai L., Liu D., Chen J., Qi X., Bi Y., Shi Y., Gao G.F., Liu Y. Avian influenza A (H7N9) virus: from low pathogenic to highly pathogenic. Front. Med. 2021;15(4):507–527. doi: 10.1007/s11684-020-0814-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Y., Wu Y., Tefsen B., Shi Y., Gao G.F. Bat-derived influenza-like viruses H17N10 and H18N11. Trends Microbiol. 2014;22:183–191. doi: 10.1016/j.tim.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singanayagam A., Hakki S., Dunning J., Madon K.J., Crone M.A., Koycheva A., Derqui-Fernandez N., Barnett J.L., Whitfield M.G., Varro R., et al. Community transmission and viral load kinetics of the SARS-CoV-2 delta (B.1.617.2) variant in vaccinated and unvaccinated individuals in the U.K.: a prospective, longitudinal, cohort study. Lancet Infect. Dis. 2022;22(2):183–195. doi: 10.1016/S1473-3099(21)00648-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang R., Chen J., Wei G.-W. Mechanisms of SARS-CoV-2 evolution revealing vaccine resistant mutations in Europe and America. J. Phys. Chem. Lett. 2021;12:11850–11857. doi: 10.1021/acs.jpclett.1c03380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stadnytskyi V., Anfinrud P., Bax A. Breathing, speaking, coughing or sneezing: what drives transmission of SARS-CoV-2? J. Intern. Med. 2021;290:1010–1027. doi: 10.1111/joim.13326. https://onlinelibrary.wiley.com/doi/full/10.1111/joim.133260.1007/s00784-020-03413-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anfinrud P., Stadnytskyi V., Bax C.E., Bax A. Visualizing speech-generated oral fluid droplets with laser light scattering. N. Engl. J. Med. 2020;382:2061–2063. doi: 10.1056/NEJMc2007800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bax A., Bax C.E., Stadnytskyi V., Anfinrud P. SARS-CoV-2 transmission via speech-generated respiratory droplets. Lancet Infect. Dis. 2021;21(3):318. doi: 10.1016/S1473-3099(20)30726-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bahl P., de Silva C., Bhattacharjee S., Stone H., Doolan C., Chughtai A.A., Raina Maclntyre C.R. Droplets and Aerosols generated by singing and the risk of COVID-19 for choirs. Clin. Infect. Dis. 2020;72:639–641. doi: 10.1093/cid/ciaa1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alsved M., Matamis A., Bohlin R., Richter M., Bentsson P.E., Fraenkel C.-J., Medstrand P., Löndahl J. Exhaled respiratory particles during singing and talking. Aerosol Sci. Technol. 2020;54:1245–1248. [Google Scholar]
- 19.Smith S.H., Somsen G.A., Rijn C.V., Kooij S., van der Hoek L., Bem R.A., Bonn D. Aerosol persistence in relation to possible transmission of SARS-CoV-2. Phys. Fluids. 2020;32 doi: 10.1063/5.0027844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stadnytskyi V., Bax C.E., Bax A., Anfinrud P. The airborne lifetime of small speech droplets and their potential importance in SARS-CoV-2 transmission. Proc. Natl. Acad. Sci. U.S.A. 2020;117(22):11875–11877. doi: 10.1073/pnas.2006874117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregson F.K.A., Watson N.A., Orton C.M., Haddrell A.E., McCarthy L.P., Finnie T.J.R., Gent N., Donaldson G.C., Shah P.L., Calder J.D. Comparing aerosol concentrations and particle size distributions generated by singing, speaking and breathing. Aerosol Sci. Technol. 2021;55(6):681–691. [Google Scholar]
- 22.Lam-Hine T., McCurdy S.A., Santora L., Duncan L., Corbett-Detig R., Kapusinszky B., Willis M., et al. Outbreak associated with SARS-CoV-2 B.1.617.2 (delta) variant in an elementary School - marin county, California, may-june 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70(35):1214–1219. doi: 10.15585/mmwr.mm7035e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.CDC . 2021. Scientific Brief: SARS-CoV-2 Transmission.https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/sars-cov-2-transmission.html [Google Scholar]
- 24.Asadi S., Wexler A.S., Cappa C.D., Barreda S., Bouvier N.M., Ristenpart W.D. Effect of voicing and articulation manner on aerosol particle emission during human speech. PLoS One. 2020;15(1) doi: 10.1371/journal.pone.0227699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majra D., Benson J., Pitts J., Stebbing J. SARS-CoV-2 (COVID-19) superspreader events. J. Infect. 2021;82(1):36–40. doi: 10.1016/j.jinf.2020.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asadi S., Wexler A.S., Cappa C.D., Barreda S., Bouvier N.M., Ristenpart W.D. Aerosol emission and superemission during human speech increase with voice loudness. Sci. Rep. 2019;9:2348. doi: 10.1038/s41598-019-38808-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chao C.Y.H., Wan M.P., Morawska L., Johnson G.R., Ristovski Z.D., Hargreaves M., Mengersen K., Corbett S., Li Y., Xie X., Katoshevski D. Characterization of expiration air jets and droplet size distributions immediately at the mouth opening. J. Aerosol Sci. 2009;40(2):122–133. doi: 10.1016/j.jaerosci.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kushalnagar P., Chow C.C., Bax A. Self-infection with speech aerosol may contribute to COVID-19 severity. J. Intern. Med. 2021;290(6):1275–1277. doi: 10.1111/joim.13370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen Y., Courtney J.M., Anfinrud P., Bax A. Hybrid measurement of respiratory aerosol reveals a dominant coarse fraction resulting from speech that remains airborne for minutes. Proc. Natl. Acad. Sci. USA. 2022;119(26) doi: 10.1073/pnas.2202086119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferrer M.D., Barrueco A.S., Martinez-Beneyto Y., Mateos-Moreno M.V., Ausina-Marquez V., Garcìa-Vázquez E., Puche-Torres M., Forner Giner M.J., González A.C., Coello J.M.S., et al. Clinical evaluation of antiseptic mouth rinses to reduce salivary load of SARS-CoV-2. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-03461-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Y., Li X., Zhu B., Liang H., Fanf C., Gong Y., Guo Q., Sun X., Zhao D., Shen J., Zhang H., Liu H., et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26(4):502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrera D., Serrano J., Roldan S., Sanz M. Is the oral cavity relevant in SARS-CoV-2 pandemic? Clin. Oral Invest. 2020;24(8):2925–2930. doi: 10.1007/s00784-020-03413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katz M.H. Challenges in testing for SARS-CoV-2 among patients who recovered from COVID-19. JAMA Intern. Med. 2021;181(5):704–705. doi: 10.1001/jamainternmed.2020.7575. [DOI] [PubMed] [Google Scholar]
- 34.Daniell H., Mangu V., Yakubov B., Park J., Habibi P., Shi Y., Gonnella P.A., Fisher A., Cook T., Zeng L., et al. Investigational new drug enabling angiotensin oral-delivery studies to attenuate pulmonary hypertension. Biomaterials. 2020;233 doi: 10.1016/j.biomaterials.2019.119750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daniell H., Nair S.K., Esmaeili N., Wakade G., Shahid N., Ganesan P.K., Islam MdR., Shepley-Mctaggart A., Feng S., Gary E.N., et al. Debulking SARS-CoV-2 in saliva using angiotensin converting enzyme 2 in chewing gum to decrease oral virus transmission and infection. Mol. Ther. 2022;30:1966–1978. doi: 10.1016/j.ymthe.2021.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bracken C.J., Lim S.A., Solomon P., Rettko N.J., Nguyen D.P., Zha B.S., Schaefer K., Byrnes J.R., Zhou J., Lui I., Liu J., Pance K. In: Bi-paratopic and Multivalent VH Domains Block ACE2 Binding and Neutralize SARS-CoV 2. Zhou X.X., Leung K.K., Wells J.A., editors. vol. 17. 2021. QCRG structural biology consortium; pp. 113–121. (Nat Chem Biol). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glasgowa A., Glasgowb J., Limontac D., Solomonb P., Luib I., Zhang Y., Nixe M.A., Rettkob N.J., Zhaf S., Yaming R., Kao K., Rosenberg O.S., Ravetchg J.V., Wiita A.P., Leung K.K., Limb S.A., Zhou X.X., Hobman T.C., Kortemme T., Wells J.A. Engineered ACE2 receptor traps potently neutralize SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 2020;117:28046–28055. doi: 10.1073/pnas.2016093117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lv J., Gao J., Wu B., Yao M., Yang Y., Chai T., Li N. Aerosol transmission of Coronavirus and influenza virus of animal origin. Front. Vet. Sci. 2021;8 doi: 10.3389/fvets.2021.572012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y., Shahed-Al-Mahmud M., Chen X., Chen T., Liao K., Lo J.M., Wu Y., Ho M., Wu C., Wong C., Jan J., Ma C. A carbohydrate-binding protein from the edible lablab beans effectively blocks the infections of influenza viruses and SARS-CoV-2. Cell Rep. 2020;32(6):108016–108020. doi: 10.1016/j.celrep.2020.108016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bagdonaite I., Wandall H.H. Global aspects of viral glycosylation. Glycobiology. 2018;28:443–467. doi: 10.1093/glycob/cwy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krammer F., Smith G.J.D., Fouchier R.A.M., Peiris M., Kedzierska K., Doherty P.C., Palese P., Shaw M.L., Treanor J., Webster R.G., Garcìa-Sastre A. Influenza. Nat. Rev. Dis. Prim. 2018;4(1):3. doi: 10.1038/s41572-018-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369(6501):330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taubenberger J.K., Moren D.M. The pathology of influenza virus infections. Annu. Rev. Pathol. 2008;3:499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsieh Y.H., Wu J., Fang J., Yang Y., Lou J. Quantification of bird-to-bird and bird-to-human infections during 2013 novel H7N9 avian influenza outbreak in China. PLoS One. 2014;9(2) doi: 10.1371/journal.pone.0111834.eCollection2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shenoy V., Kwon K.C., Rathinasabapathy A., Lin S., Jin G., Song C., Shil P., Nair A., Qi Y., Li Q., et al. Oral delivery of angiotensin-converting enzyme 2 and angiotensin-(1-7) bioencapsulated in plant cells attenuates pulmonary hypertension. Hypertension. 2014;64:1248–1259. doi: 10.1161/HYPERTENSIONAHA.114.03871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shil P., Kwon K.C., Zhu P., Verma A., Daniell H., Li Q. Oral delivery of ACE2/ang-(1–7) bioencapsulated in plant cells protects against experimental uveitis and autoimmune uveoretinitis. Mol. Ther. 2014;22:2069–2082. doi: 10.1038/mt.2014.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khan I., Daniell H. Oral delivery of therapeutic proteins bioencapsulated in plant cells: preclinical and clinical advances. Curr. Opin. Colloid Interface Sci. 2021;54 doi: 10.1016/j.cocis.2021.101452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park J., Yan G., Kwon K.C., Liu M., Gonnella P.A., Yang S., Daniell H. Oral delivery of novel human IGF-1 bioencapsulated in lettuce cells promotes musculoskeletal cell proliferation, differentiation, and diabetic fracture healing. Biomaterials. 2020;233 doi: 10.1016/j.biomaterials.2019.119591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He W., Baysal C., Lobato Gómez M., Huang X., Alvarez D., Zhu C., ArmarioNajera V., Blanco Perera A., Cerda Bennaser P., Saba-Mayoral A., et al. Contributions of international plant science community to fight against infectious diseases in human—part 2: affordable drugs in edible plants for endemic and reemerging diseases. Plant Biotechnol. J. 2021;19:1921–1936. doi: 10.1111/pbi.13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwon C., Daniell H. Oral delivery of protein drugs bioencapsulated in plant cells. Mol. Ther. 2016;24(8):1342–1350. doi: 10.1038/mt.2016.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh R., Ren Z., Shi Y., Lin S., Kwon K.C., Balamurugan S., Rai V., Mante F., Koo H., Daniell H. Affordable oral health care: dental biofilm disruption using chloroplast made enzymes with chewing gum delivery. Plant Biotechnol. J. 2021;19(10):2113–2125. doi: 10.1111/pbi.13643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daniell H., Kulis M., Herzog R.W. Plant cell-made protein antigens for induction of Oral tolerance. Biotechnol. Adv. 2019;37 doi: 10.1016/j.biotechadv.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Daniell H., Jin S., Zhu X., Gitzendanner M.A., Soltis D.E., Soltis P.S. Green giant—a tiny chloroplast genome with mighty power to produce high‐value proteins: history and phylogeny. Plant Biotechnol. J. 2021;19:430–447. doi: 10.1111/pbi.13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Srinivasan A., Herzog R.W., Khan I., Sherman A., Bertolini T., Wynn T., Daniell H., et al. Preclinical development of plant‐based oral immune modulatory therapy for hemophilia B. Plant Biotechnol. J. 2021;19(10):1952–1966. doi: 10.1111/pbi.13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaushik P., Kaushik D. Medicated chewing gums: recent patents and patented technology platforms. Recent Pat. Drug Deliv. Formulation. 2019;13(3):184–191. doi: 10.2174/1872211313666191010093148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greene T., Rogers S., Franzen A., Gentry R. A critical review of the literature to conduct a toxicity assessment for oral exposure to methyl salicylate. Critical Rev. Toxiciol. 2017;47(2):98–120. doi: 10.1080/10408444.2016.1236071. [DOI] [PubMed] [Google Scholar]
- 57.Ryan E.J., Kim C., Fickes E.J., Williamson M., Muller M.D., Barkley J.E., Gunstad J., Glickman E.L. Caffeine gum and cycling performance: a timing study. J. Strength Condit Res. 2013;27(1):259–264. doi: 10.1519/JSC.0b013e3182541d03. [DOI] [PubMed] [Google Scholar]
- 58.Brown R., Sam C., Green T., Wood S., Effect of GutsyGumTM A novel gum, on subjective ratings of gatsro esophageal reflux following A refluxogenic meal. J. Diet. Suppl. 2015;12(2):138–145. doi: 10.3109/19390211.2014.950783. [DOI] [PubMed] [Google Scholar]
- 59.Simons D., Brailsford S., Kidd E.A., Beighton D. The effect of chlorhexidine acetate/xylitol chewing gum on the plaque ad gingival indices of elderly occupants in residential homes. J. Clin. Periodontol. 2001;28(11):1010–1015. doi: 10.1034/j.1600-051x.2001.281104.x. [DOI] [PubMed] [Google Scholar]
- 60.Wennike P., Danielsson T., Landfeldt B., Westin A., Tonnesen P. Smoking reduction promotes smoking cessation: results from a double blind, randomized, placebo-controlled trial of nicotine gum with 2-year follow-up. Addiction. 2003;98(10):1395–1402. doi: 10.1046/j.1360-0443.2003.00489.x. [DOI] [PubMed] [Google Scholar]
- 61.Round E.K., Chen P., Taylor A.K., Schmidt E. Biomarkers of tobacco exposure decrease after smokers switch to an E-cigarette or nicotine gum. Nicotine Tob. Res. 2019;21(9):1239–1247. doi: 10.1093/ntr/nty140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Söderling E., Pienihäkkinen K. Effects of xylitol chewing gum and candies on the accumulation of dental plaque: a systematic review. Clin. Oral Invest. 2022;26:119–129. doi: 10.1007/s00784-021-04225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Freitas A.A.R., Ribeiro A.J., Santos A.C., Veiga F., Nunes L.C.C., Silva D.A., Soares-Sobrinho J.L., Silva-Filho E.C. Sterculia striata gum as a potential oral delivery system for protein drugs. Int. J. Biol. Macromol. 2020;164:1683–1692. doi: 10.1016/j.ijbiomac.2020.07.276. [DOI] [PubMed] [Google Scholar]
- 64.Boyhan D., Daniell H. Low-cost production of proinsulin in tobacco and lettuce chloroplasts for injectable or oral delivery of functional insulin and C-peptide. Plant Biotechnol. J. 2011;9(5):585–598. doi: 10.1111/j.1467-7652.2010.00582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Everett J., Hokama P., Roche A.M., Reddy S., Hwang Y., Kessller L., Glaascock A., Li Y., Whelan J.N., Weiss S.R. SARS-CoV-2 genomic variation in space and time in hospitalized patients in philadelphia. mBio. 2021;12(1) doi: 10.1128/mBio.03456-20. e03456-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marques A.D., Sherrill-Mix S., Everett J.K., Reddy S., Hokama P., Roche A.M., Hwang Y., Glascock A., Whiteside S.A., Graham-Wooten G. SARS-CoV-2 variants associated with vaccine breakthrough in the Delaware valley through summer 2021. mBio. 2022;13(1) doi: 10.1128/mbio.03788-21. e03788-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hilaire B.G.S., Durand N.C., Mitra N., Pulido S.G., Mahajan R., Blackburn A., Colaric Z.L., Theisen J.W.M., Weisz D., Dudchenko O. A rapid, low cost and highly sensitive SARS-CoV-2 diagnostic based on whole genome sequencing. 2020. bioRxiv preprint. [DOI] [PMC free article] [PubMed]
- 68.Toole A., Scher E., Underwood A., Jackson B., Hill V., McCrone J., Colquhoun R., Ruiis C., Abu-Dahab K., Taylor B., et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021;7 doi: 10.1093/ve/veab064. veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith B.F., Graven P.F., Yang D.Y., Downs S.M., Hansel D.E., Faan G., Qin X. Using spike gene target failure to estimate growth rate of the Alpha and omicron variants of SARS-CoV-2. J. Clin. Microbiol. 2022;60(4) doi: 10.1128/jcm.02573-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brown K.A., Gubbay J., Hopkins J., Patel S., Buchan S.A.A., Daneman N., Goneau L.W. S-gene target failure as a marker of variant B.1.1.7 among SARS-CoV-2 isolates in the greater toronto area, december 2020 to march 2021. JAMA. 2021;325(20):2115–2116. doi: 10.1001/jama.2021.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen H., Li Z., Zhang L., Sawaya P., Shi J., Wang P. Quantitation of femtomolar-level protein biomarkers using a simple microbubbling digital assay and bright-field smartphone imaging. Angew. Chem. Int. Ed. 2019;58(39):13922–13928. doi: 10.1002/anie.201906856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen H., Li Z., Feng S., Richard-Greenblatt M., Hutson E., Andrianus S., Glaser L.J., Rodino K.G., Qian J., Jayaraman D., Collman R.G., Glascock A., Bushman F.D., Lee J.S., Cherry S., Fausto A., Weiss S.R., Koo H., Corby P.M., O'Doherty U., Garfall A.L., Vogl D.T., Stadtmauer E.A., Wang P. Femtomolar SARS-CoV-2 antigen detection using the microbubbling digital assay with smartphone readout enables antigen burden quantitation and tracking. Clin. Chem. 2022;68(1):230–239. doi: 10.1093/clinchem/hvab158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Torres M.D.T., de Araujo W.R., de Lima L.F., Ferreira A.L., de la Fuente-Nunez C. Low-Cost biosensor for rapid detection of SARS-CoV-2 at the point of care. Matter. 2021;4(7):2403–2416. doi: 10.1016/j.matt.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 75.Torres M.D.T., de Lima L.F., Ferreira A.L., de Araujo W.R., Callahan P., Dávila A., Abella B.S., de la Fuente-Nunez C. Detection of SARS-CoV-2 with RAPID: a prospective cohort study. iScience. 2022;25(4) doi: 10.1016/j.isci.2022.104055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang J., Petitjean S.J.L., Koehler M., Zhang Q., Dumitru A.C., Chen W., Derclaye S., Vincent S.P., Soumillion P., Alsteens D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020;11:4541. doi: 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muñoz J., Montes R., Baeza M. Trends in electrochemical impedance spectroscopy involving nanocomposite transducers: characterization, architecture surface and bio-sensing. TrAC, Trends Anal. Chem. 2017;97:201–215. doi: 10.1016/j.trac.2017.08.012. [DOI] [Google Scholar]
- 79.Pereira A.R., Sedenho G.C., Souza J.C.P.D.E., Crespilho F.N. Advances in enzyme bioelectrochemistry. An. Acad. Bras. Cienc. 2018;90(1 suppl 1):825–857. doi: 10.1590/0001-3765201820170514. [DOI] [PubMed] [Google Scholar]
- 80.Mauritz K.A., Moore R.B. State of understanding of nafion. Chem. Rev. 2004;104(10):4535–4586. doi: 10.1021/cr0207123. [DOI] [PubMed] [Google Scholar]
- 81.Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 82.Rashed M.Z., Kopechek J.A., Priddy M.C., Hamorsky K.T., Palmer K.E., Mittal N., Valdez J., Flynn J., Williams S.J. Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li Y., Liu D., Wang Y., Su W., Liu G., Dong W. The importance of glycans of viral and host proteins in enveloped virus infection. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.638573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Luo S., Hu K., He S., Wang P., Zhang M., Huang X., Du T., Zheng C., Liu Y., Hu Q. Contribution of N-linked glycans on HSV-2 gB to cell-cell fusion and viral entry. Virology. 2015;483:72–82. doi: 10.1016/j.virol.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 85.Dawes C., Macpherson L.M. Effects of nine different chewing-gums and lozenges on salivary flow rate and pH. Caries Res. 1992;26:176–182. doi: 10.1159/000261439. [DOI] [PubMed] [Google Scholar]
- 86.Iorgulescu G. Saliva between normal and pathological. Important factors in determining systemic and oral health. J. Med. Life. 2009;2:303–307. [PMC free article] [PubMed] [Google Scholar]
- 87.Dawes C., Macpherson L.M.D. The distribution of saliva and sucrose around the mouth during the use of chewing gum and the implications for the site-specificity of caries and calculus deposition. J. Dent. Res. 1993;72:852–857. doi: 10.1177/00220345930720050401. [DOI] [PubMed] [Google Scholar]
- 88.Goutelle S., Maurin M., Rougier F., Barbaut X., Bourguignon L., Ducher M., Maire P. The Hill equation: a review of its capabilities in pharmacological modelling. Fundam. Clin. Pharmacol. 2008;22(6):633–648. doi: 10.1111/j.1472-8206.2008.00633.x. [DOI] [PubMed] [Google Scholar]
- 90.Xu M.L., Wi G.R., Kim H.J., Kim H.J. Ameliorating effect of dietary xylitol on human respiratory syncytial virus (hRSV) infection. Biol. Pharm. Bull. 2016;39(4):540–546. doi: 10.1248/bpb.b15-00773. [DOI] [PubMed] [Google Scholar]
- 91.Bansal, S., Jonsson, C.B., Taylor, S.L., Figueroa, J.M., Dugour, A.V., Palacios, C., Vega, J.C. Iota carrageenan and xylitol inhibit SARS-CoV-2 in Vero cell culture. PLoS One. 16:e0259943. doi: 10.1371/journl.pone.0259943. [DOI] [PMC free article] [PubMed]
- 92.Hartshorn K.L., White M.R., Shepherd V., Reid K., Jensenius J.C., Crouch E.C. Mechanisms of anti-influenza activity of surfactant proteins A and D: comparison with serum collections. Am. J. Physiol. 1997;273(6):1156–1166. doi: 10.1152/ajplung.1997.273.6.L1156. [DOI] [PubMed] [Google Scholar]
- 93.Takaki P., Vieira M., Bommarito S. Maximum bite force analysis indifferent age groups. Int. Arch. Otorhinolaryngol. 2014;18:272–276. doi: 10.1055/s-0034-1374647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Waltimo A., Könönen M. A novel bite force recorder and maximal isometric bite force values for healthy young adults. Scand. J. Dent. Res. 1993;101:171–175. doi: 10.1111/j.1600-0722.1993.tb01658.x. [DOI] [PubMed] [Google Scholar]
- 95.Varga S., Spalj S., Lapter Varga M., Anic Milosevic S., Mestrovic S., Slaj M. Maximum voluntary molar bite force in subjects with normal occlusion. Eur. J. Orthod. 2011;33:427–433. doi: 10.1093/ejo/cjq097. [DOI] [PubMed] [Google Scholar]
- 96.Letting F.K., Venkataramana P.B., Ndakidemi P.A. Breeding potential of lablab (Lablab purpureus): a review on characterization and bruchid studies towards improved production and utilization in Africa. Genet. Resour. Crop Evol. 2021;68:3081–3101. doi: 10.1007/s10722-021-01271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hossain S., Ahmed R., Bhowmick S., Sarkar M., Nahar T., Uddin B., Basunia M., Mamun A., Hashimoto M., Shido O. Lallab purpureus bean flour ameliorates plasma proteins and accretion of docosahexaenoic acid in the plasma, liver and brain of malnourished rats. Legume Sci. 2020;1–13 doi: 10.1002/leg3.34. [DOI] [Google Scholar]
- 98.Martyn D.M., Lau A. Chewing gum consumption in the United States among children, adolescents and adults. Food Addit. Contam. 2019;36(3):350–358. doi: 10.1080/19440049.2019.1567944. [DOI] [PubMed] [Google Scholar]
Further reading
- 89.Mathew S., Thani A., Yaassine H.M. Computational screening of known broad-spectrum antiviral small organic molecules for potential influenza HA stem inhibitors. PLoS One. 2018;13 doi: 10.1371/journal.pone.0203148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.