SUMMARY
Cytoplasmic mislocalization of the TAR-DNA binding protein of 43 kDa (TDP-43) leads to large, insoluble aggregates that are a hallmark of amyotrophic lateral sclerosis and frontotemporal dementia. Here, we study how importin α1/β recognizes TDP-43 bipartite nuclear localization signal (NLS). We find that the NLS makes extensive contacts with importin α1, especially at the minor NLS-binding site. NLS binding results in steric clashes with the C terminus of importin α1 that disrupts the TDP-43 N-terminal domain (NTD) dimerization interface. A putative phosphorylation site in the proximity of TDP-43 R83 at the minor NLS site destabilizes binding to importins by reducing the NLS backbone dynamics. Based on these data, we explain the pathogenic role of several post-translational modifications and mutations in the proximity of TDP-43 minor NLS site that are linked to disease and shed light on the chaperone activity of importin α1/β.
In brief
Doll et al. describe how the human importin α1/β heterodimer recognizes the nuclear localization signal (NLS) of TDP-43. The paper explains the pathogenic role of post-translational modifications and mutations near the TDP-43 minor NLS site that are linked to ALS and sheds light on the chaperone activity of importin α1/β.
Graphical Abstract

INTRODUCTION
In amyotrophic lateral sclerosis (ALS) and frontotemporal lobar dementia (FTLD), the principal aggregating protein is TDP-43. This RNA-binding protein is predominately nuclear (Zhang et al., 2009), although a pool of TDP-43 is constantly exported from the nucleus as part of ribonucleoprotein particles (RNPs) (Coyne et al., 2015; Majumder et al., 2016; Sephton et al., 2011). TDP-43 begins to self-oligomerize in the cytoplasm, likely when localized to stress granules (Colombrita et al., 2009), although more recent work suggests that TDP-43 may aggregate through phase separation independent of stress granules (Gasset-Rosa et al., 2019). Impairment in nuclear import of TDP-43 coupled with continued export of the protein contributes to increasing the cytoplasmic concentration of the protein, while the pool of nuclear TDP-43 is correspondingly depleted (Gasset-Rosa et al., 2019). This disruption in cellular homeostasis facilitates the growth of large TDP-43 aggregates (Yang et al., 2010).
TDP-43 function in a cell is intimately linked to RNA metabolism. The protein binds and stabilizes nascent mRNA in the nucleus (Tollervey et al., 2011) and matured mRNA in the cytoplasm (Colombrita et al., 2012). The mRNA binding activity is conferred by two RNA recognition motifs (RRMs) that occur in tandem (RRM1 and RRM2), separated by a linker sequence (Maris et al., 2005). TDP-43 contains a nuclear localization signal (NLS) immediately downstream of the N-terminal domain (NTD) (Johnson et al., 2008; Winton et al., 2008a). Classical NLSs can bind to the nuclear import adaptor importin α1 at two distinct sites, also known as the major and minor NLS binding grooves (Conti et al., 1998; Marfori et al., 2011). Association with an NLS cargo and concomitant recruitment of the receptor importin β (Cingolani et al., 1999) frees the importin β binding domain (IBB) of importin α1 (Kobe, 1999), leading to the assembly of a trimeric nuclear import complex (e.g., importin α1/β/TDP-43). The IBB functions as its own importin α1 NLS recognized with nanomolar affinity by importin β (Lott and Cingolani, 2011). Importin α1 has been shown to associate with cytoplasmic TDP-43, including phase-separated TDP-43 (Gasset-Rosa et al., 2019; Liu et al., 2021). Other importin α isoforms (Pumroy and Cingolani, 2015), such as α3 and α4, were shown to interact with TDP-43 in vitro (Nishimura et al., 2010) and based on proximity labeling studies (Chou et al., 2018). Furthermore, importin α3, alone or with importin β, was found to reverse TDP-43 aggregation mediated by arginine-rich dipeptide repeats (DPRs) in vitro (Hutten et al., 2020).
In addition to serving a role as import factors, importin α1 and β (also referred to as karyopherins) were shown to possess disaggregase activity toward TDP-43 fibrils, capable in vitro of disassociating pre-formed fibrils and returning TDP-43 to the soluble phase (Guo et al., 2018). Potent antiaggregase activity was reported for importin β2 (also known as Kap β2), which imports PY-NLS cargos without an adaptor (Lee et al., 2006). This karyopherin possesses NLS-dependent disaggregase activity toward aggregates of FUS (Guo et al., 2018; Hofweber et al., 2018; Qamar et al., 2018; Yoshizawa et al., 2018), a second aggregation-prone RNA-binding protein that contributes to a significant proportion of ALS cases. Interestingly, while Kap β2 alone is necessary and sufficient to disaggregate FUS in vitro, importin β disaggregase activity requires the adaptor importin α1 (Guo et al., 2018) or α3 (Hutten et al., 2020), as neither the receptor nor the adaptor alone is sufficient to dissolve TPD-43 aggregates (Guo et al., 2018).
Cytoplasmic TDP-43 aggregation is a complex process that is thought to depend on several concomitant events, including an imbalance in nucleocytoplasmic transport, aberrant post-translational modifications, and mutations, and that involves both the NTD and the prion-like C-terminal domain (CTD) (Prasad et al., 2019). A recent cryoelectron microscopy(cryo-EM) helical reconstruction of aggregated TDP-43 extracted from the frontal cortex of two individuals suffering from ALS with FTLD revealed an amyloid-like filament structure comprising a single protofilament spanning CTD residues 282–360 (Arseni et al., 2022). This low-complexity domain adopts a double-spiral-shaped fold consisting of 10 short β strands, distinct from TDP-43 filaments formed in vitro (Cao et al., 2019; Li et al., 2021). The stacking of TDP-43 double spirals gives rise to parallel inter-molecular β sheets that extend along the helical axis, generating amyloid-like structures, thought to be linked to TDP-43 pathogenesis (Berning and Walker, 2019). Unlike the CTD, which is intrinsically disordered in the unaggregated protein but forms pathogenic amyloid-like structures in the aggregated TDP-43, the NTD is well folded (Afroz et al., 2017; Mompeán et al., 2016; Wright et al., 2020) and likely functions as a dimer (Chang et al., 2012; Shiina et al., 2010; Wang et al., 2013), with the dimerization interface occurring within the seventh β strand of the NTD (Jiang et al., 2017). Biophysical analysis using the single-molecule fluorescence technique revealed the NTD is thermodynamically stable and undergoes reversible oligomerization that enhances the propensity of the CTD to form amyloid-like structures (Tsoi et al., 2017). Similarly, Wang et al. (2018) found the NTD enhances inter-molecular contacts between molecules promoting phase separation of the CTD and partitioning into membraneless organelles. These studies contradict previous reports that NTD-mediated oligomerization of TDP-43 represents a physiological process that may antagonize pathological aggregation (Afroz et al., 2017; Jiang et al., 2017).
Aggregated TDP-43 is subjected to heavy ubiquitination and phosphorylation (Hasegawa et al., 2008; Kametani et al., 2016). The majority of the literature focuses upon phosphorylation within the prion-like CTD (Eck et al., 2021). However, there is evidence that phosphorylation may also occur at the N terminus of TDP-43 (Kametani et al., 2016; Wang et al., 2018). Interestingly, T88, S91, and S92 within the TDP-43 NLS have been shown to be phosphorylated by casein kinase 1δ in vitro (Kametani et al., 2009). Further, S92 was phosphorylated by a truncated version of this kinase in a cellular model of TDP-43 aggregation (Nonaka et al., 2016). NLS phosphorylation is known to regulate NLS exposure and presentation to importins (Nardozzi et al., 2010), and there is evidence that phosphorylation is linked to the aggregation of RNA-binding proteins (Kurischko and Broach, 2017).
In this paper, we elucidate how importin α1 recognizes the TDP-43 NLS and determine the effect of NLS phosphorylation on the conformational dynamics of the NLS and binding affinity for the importin α1/β heterodimer.
RESULTS
The TDP-43 NLS engages importin α1 through a bipartite binding mechanism
We generated several TDP-43 constructs containing the NLS (Figure 1A) and co-expressed them in bacteria with the mammalian importin α1 lacking the IBB domain (ΔIBB-importin α1) followed by one-step purification on glutathione beads. Except for the full-length (FL) TDP-43, which is aggregation prone (Wright et al., 2020), all TDP-43 constructs containing the NLS copurified with ΔIBB-importin α1. Crystals of the NTD-NLS:-ΔIBB-importin α1 complex diffracted X-rays to 2.2 Å resolution and were used to collect a complete dataset (Table S1). The structure, solved by molecular replacement, revealed strong electron density for TDP-43 residues bound to importin α1 major (Figure 1B) and minor (Figure 1C) NLS-binding sites that correspond to TDP-43 residues 94–100 and 81–87, respectively. The linker between these two regions (residues 89–95) had weak density in a conventional difference density map but was visible in an unbiased maximum entropy map (Gull and Daniell, 1978) (Figure S1), sufficient to trace the entire NLS mainchain and identify the position of putative phosphorylation sites at T88, S91, and S92. Unexpectedly, the TDP-43 NTD (residues 1–77) was not visible in the electron density, although SDS-PAGE analysis of melted crystals indicated it was present in the crystal (Figure S2). The final model of importin α1 bound to the TDP-43 NTD-NLS was refined to an Rwork/Rfree of 18.96%/21.51% at 2.2 Å resolution (Table S1).
Figure 1. The TDP-43 NLS engages importin α1 through a bipartite mechanism.
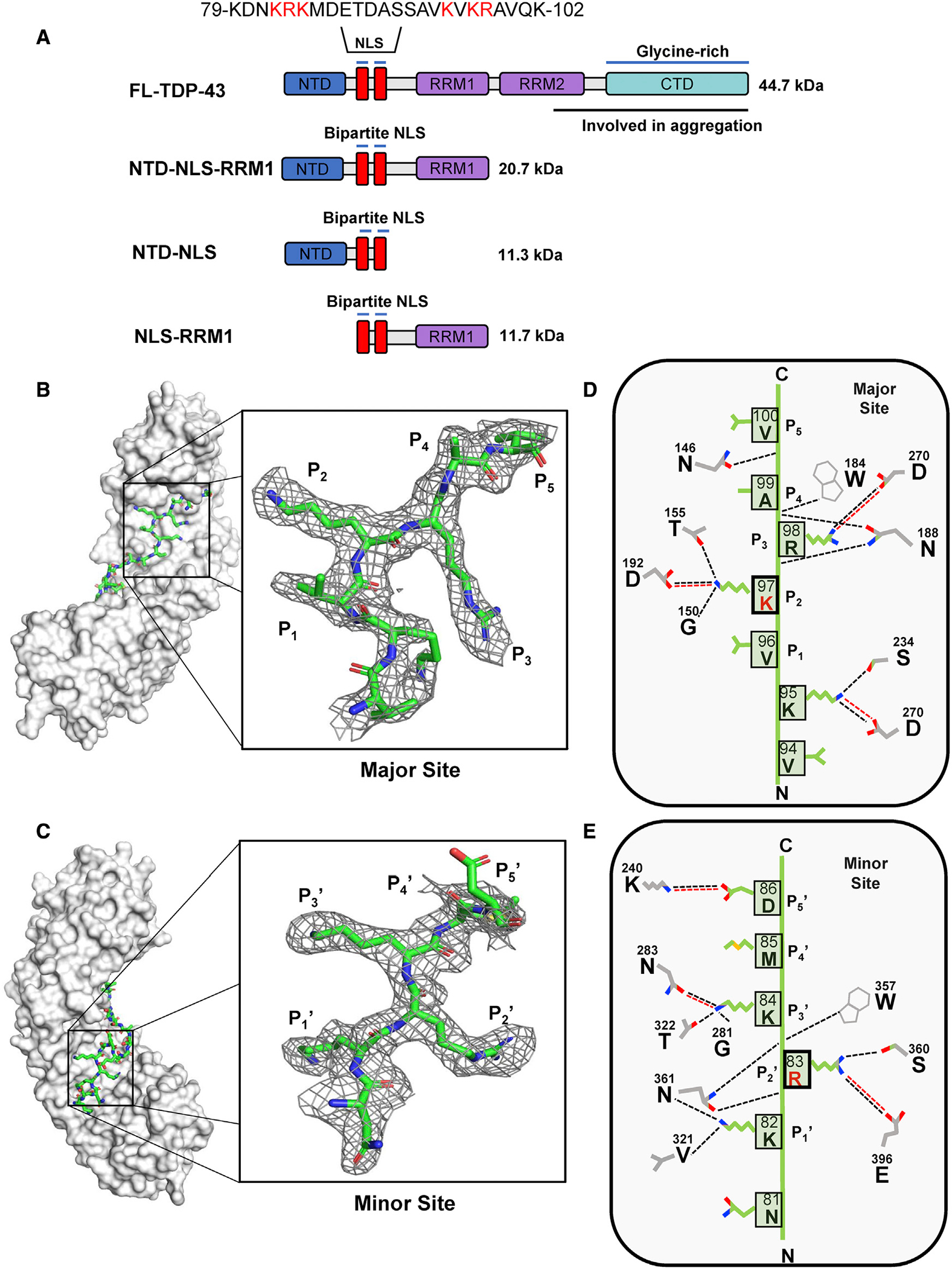
(A) Schematics of all TDP-43 constructs that were used in this work.
(B and C) Crystal structure of the TDP-43 NLS (green) bound to importin α1 (gray), major site view. The P1–P5 residues are shown in the inset, along with electron density from a Polder map contoured at 2.0 σ. (C) Rotated view of panel (B) showing the density for the TDP-43 NLS at the minor NLS site.
(D and E) Minor site view. The P1′–P5′ residues are shown in the inset, along with electron density from a Polder map contoured at 2.0 σ. Diagram of the TDP-43 NLS:importin α1 major (D) and minor (E) site interface detailing key residues of the Arm repeats within H-bonding distance of a given NLS residue. Dashed black lines indicate H-bonds, while dashed red lines indicate salt bridges.
The minor NLS pocket of importin α1 makes the majority of bonds with TDP-43 NLS
In the crystal structure, the TDP-43 NLS is oriented anti-parallel to importin α1, with the N terminus at the minor NLS binding site and the C terminus at the major NLS-binding site. K97 occupies the critical P2 position at the major NLS-pocket, with R83 at the P2′ position, as seen in other bipartite NLS (Marfori et al., 2011) (Table S2). The total buried surface area of the interface between importin α1 and the TDP-43 NLS calculated from the crystal structure is 1,317.1 Å2, and the complex is predicted to be thermodynamically stable with a positive free energy of dissociation, ΔGdiss ~1.9 kcal/mol (Krissinel and Henrick, 2007). Although the TDP-43 NLS bound to importin α1 superficially resembles the classical bipartite nucleoplasmin NLS (Conti and Kuriyan, 2000; Fontes et al., 2000), the total number and distribution of bonds differ between the two bipartite NLSs is quite different. The TDP-43 NLS makes six salt bridges, 24 H-bonds, and 214 van der Waals interactions with importin α1, as determined by PDBsum (Laskowski, 2009), versus only 14 H-bonds and 132 van der Waals interactions of the nucleoplasmin NLS. Furthermore, the TDP-43 NLS appears to have stronger bonding at the minor NLS-binding site than at the major site. Residues P1′–P5′ (Figure 1E) engage in three salt bridges, 11 H-bonds, and 67 van der Waals interactions, while residues P1–P5 (Figure 1D) form two salt bridges, eight H-bonds, and 87 van der Waals interactions. The opposite scenario was observed for the nucleoplasmin NLS (Conti and Kuriyan, 2000; Fontes et al., 2000), which makes stronger bonding at the major NLS-binding pocket (e.g., nine H-bonds and 84 van der Waals interactions) than at the minor site (e.g., five H-bonds and 21 van der Waals interactions). Like nucleoplasmin, most known bipartite NLSs make more contacts with importin α at the major NLS-binding pocket and are disrupted by a single substitution of the Lys residue at P2 position (Kralt et al., 2015; Lokareddy et al., 2015; Pumroy et al., 2015; Sankhala et al., 2017) (Table S2). Thus, TDP-43 NLS combines binding determinants seen in recognition of classical bipartite NLSs (Pumroy et al., 2015; Sankhala et al., 2017), as well as a deeper interaction at the minor NLS-binding site and particularly the P2′ position.
In cellulo pull-down identifies the P2′ as a critical determinant for importin α1/β binding
Given that TDP-43 is more bonded to importin α1 at the minor NLS site than the major, we sought to probe the effect of point mutations at P2 and P2′ positions. We introduced single and double alanine mutations at K97 (P2), and R83 (P2′) of TDP-43 NTD-NLS-RRM1 (Figure 1A), which is the largest TDP-43 fragment generated in this study proved to remain soluble when expressed in isolation. On the other hand, a significant portion of recombinant importin α and β expressed in bacteria are misfolded and/or inactive, affecting the reproducibility of biochemical experiments made using individually purified importins. The best way to purify correctly folded and active importin β is by binding it (in vitro or cellulo) to the IBB domain of importin α. Thus we devised a co-expression system where importin α1 and β were co-expressed from a polycistronic plasmid in bacteria in the presence of a second expression plasmid expressing wild-type (WT) or mutated TDP-43 constructs bearing ala-substitutions in the NLS. This system is ideal for probing the balance between importin α/β-binding and TDP-43 aggregation. As a vital control, we established that mutations in the NLS did not alter TDP-43 expression (Figure S3), allowing for a direct comparison of binding interactions with importin α1/β. The trimeric importin α1/β/TDP-43 complex was then captured on Ni beads through a histidine tag in importin α1 (Figure 2A), and the relative intensities of all three bands were quantified densitometrically (Figure 2B). Interestingly, we found that the R83A mutation at P2′ position, but not K97A at P2, disrupted binding to the importin α1/β heterodimer. Thus, the minor site of TDP-43 NLS plays a crucial role in binding to importin α1/β.
Figure 2. R83 at the P2′ site is responsible for NLS selectivity.

(A) Representative SDS-PAGE gel of the pull-down experiment in which WT TDP-43 NTD-NLS-RRM1 and variants carrying ala-mutations at P2 (K97A), P2′ (R83A), and P2+P2′ (K97A + R83A) were co-expressed with the importin α1/β heterodimer.
(B) Quantification of TDP-43 NTD-NLS-RRM1 bands from replicate SDS-PAGE gels (n = 4). Band intensity was measured through ImageJ (Abramoff et al., 2004). Histogram columns indicate the ratio of TDP-43 band intensity to normalized importin α1 intensity, with standard deviation (error bars). Individual ratios from replicate gels are shown (circles). Significant (p < 0.01) deviation from the WT TDP-43:importin α1 ratio was determined through a two-tailed Student’s t test assuming unequal variance; ***p < 0.001; n.s., not significant.
Importin α1/β binding disrupts NTD-NLS oligomerization
The 80 residues N-terminal of the TDP-43 minor NLS box form the NTD (Figure 1A). To identify a possible link between NLS recognition and TDP-43 aggregation, we investigated the oligomeric state of the TDP-43 NTD-NLS in a range of concentrations (42.5–425 μM) using analytical ultracentrifugation sedimentation velocity analysis (SV-AUC). Consistent with previous findings (Chang et al., 2012; Jiang et al., 2017; Mompean et al., 2017; Shiina et al., 2010; Wang et al., 2013), we observed that the NTD is dimeric in solution, displaying a concentration-dependent tendency to form tetramers and larger oligomers (Figures 3A and S4). Plotting the average sedimentation coefficient as a function of NTD-NLS concentration revealed a sigmoidal dependence consistent with a recent crystal structure that revealed a supramolecular helical packing built by five copies in the asymmetric unit (Wright et al., 2020). We then investigated the oligomeric state of TDP-43 NTD-NLS in complex with the importin α1/β, or just ΔIBB-importin α1. We used isoform α1, which was previously reported to exert anti-aggregase activity toward TDP-43 aggregates (Guo et al., 2018). We assembled homogeneous complexes of importin α/β and α/β/NTD-NLS by co-expression in bacteria and subjected them to sedimentation velocity analysis (Figure 3C). We found the apo-importin α/β complex exists as a dimer of ~330 kDa, in a physiological range of concentration (e.g., importin α and β concentration in a cell was estimated to be ~3 and 1 μm, respectively; Riddick and Macara, 2005). The heterodimer dimerization was likely mediated by importin α1 tendency to dimerize (Miyatake et al., 2015). In contrast, the pre-formed α/β/NTD-NLS complexes were always monomeric, consistent with a 1:1:1 binding of the three proteins (Figure 3D). The same result was obtained by performing SV-AUC analysis of the ΔIBB-importin α1:NTD-NLS complex, which also migrated as a monodisperse species of s = 2.086S corresponding to a 1:1 complex of 67.2 kDa, remarkably close to the expected mass of 66.6 kDa (Figure S5). Thus, the NLS-dependent association of importin α1 with the NTD is sufficient to disrupt TDP-43 NTD oligomerization, rendering the import cargo monomeric.
Figure 3. SV-AUC analysis of TDP-43 NTD-NLS and its complex with importin α/β.

(A) Sedimentation coefficient distributions (c(S)) derived from SV-AUC analysis of a range of concentrations for TDP-43 NTD-NLS alone (black lines, 42.5 μM, 85 μM, 153 μM, 170 μM, 256 μM, and 425 μM monomer) at 20°C. Peaks are assigned to apparent dimer (D), tetramer (T), and octamer (O) species. The corresponding experimental SV-AUC data fit with the Lamm equation, and residuals are shown in Figure S4.
(B) Plot of signal-weighted average (sw) as a function of loading concentration, in μM monomer. Values at each concentration were derived from integration of the c(S) distributions shown in (A), illustrating the concentration-dependent behavior of the NTD-NLS alone.
(C) Experimental SV-AUC data (circles) fit with the Lamm equation (lines) as implemented in the program SEDFIT (Schuck, 2000) for importin α/β alone (upper, RMSD = 0.004) and importin α/β-NTD-NLS (lower, RMSD = 0.006). Below each panel are the residuals for the fit data. For clarity, only every third data point is shown. Data were obtained with 280 nm detection at 20°C. Figures were created using the program GUSSI (Brautigam, 2015).
(D) Sedimentation coefficient distributions c(S) derived from SV-AUC data for importin α/β alone (gray dotted line, 0.95 mg/mL; gray solid line, 1.93 mg/mL) and importin α/β-NTD-NLS (black dotted line, 0.93 mg/mL; black solid line, 1.93 mg/mL) at 20°C. Peaks values are observed at 9.24 s20,w (~351 kDa) for importin α/βalone and 6.93 s20,w (~174 kDa) for importin α/β-NTD-NLS, respectively.
TDP-43 NLS is loosely connected to two small globular domains
The architecture of the TDP-43 NLS in the context of the upstream NTD and downstream RRM1 (Figure 1A) was investigated by subjecting homogeneous complexes of importin α1 bound to either domain to size-exclusion chromatography coupled with small-angle X-ray scattering (SEC-SAXS) (Acerbo et al., 2015). In a concentration range between 8 and 10 mg mL−1, pre-formed complexes of both NTD-NLS:ΔIBB-importin α1 and NLS:RRM1:ΔIBB-importin α1 gave scattering profiles suitable for biophysical analysis. From the Guinier fits of the scattering intensities, we were able to calculate a radius of gyration (Rg) for each complex. The NTD-NLS and NLS-RRM1 complexes had an Rg of 36.37 ± 0.15 Å and 38.28 ± 0.26 Å, respectively (Figures 4A and 4C; Table S3). We determined that the volume of correlation (Vc) mass for NTD-NLS and NLS-RRM1 complexes were 64.8 kDa and 67.5 kDa, respectively, which are close to the expected mass of each complex (66.6 kDa for NTD-NLS:ΔIBB-importin α1 and 67.0 kDa for NLS-RRM1:ΔIBB-importin α1). The distance distribution function P(r) calculated from data indicates a maximum diameter (Dmax) of 140.3 Å for NTD-NLS:ΔIBB-importin α1 complex and 136.7 Å for the NLS-RRM1:ΔIBB-importin α1 complex (Figures 4A and 4C). The Kratky plots of each complex suggest a degree of globularity (Figures S6A and S6B), likely provided by importin α1 and either the folded NTD or RRM1. To visualize the structural organization of the NTD-NLS and NLS-RRM1 complexes in solution, we calculated an electron density from the scattering data using Density from Solution Scattering (DENSS) (Grant, 2018). The SAXS electron densities were shaped asymmetrically (Figures 4B and 4D), allowing us to place the solenoid structure of importin α1 and the TDP-43 domains flanking the NLS. The deposited structures of the TDP-43 NTD (Mompean et al., 2016) or RRM1 (Kuo et al., 2014) domains were connected to the TDP-43 NLS described previously (Figure 1B), and the modeled TDP-43 fragments spanning NTD-NLS and NLS-RRM1 fit within the electron density generated by DENNS.
Figure 4. Solution structures of TDP-43:ΔIBB-importin α1 complexes.
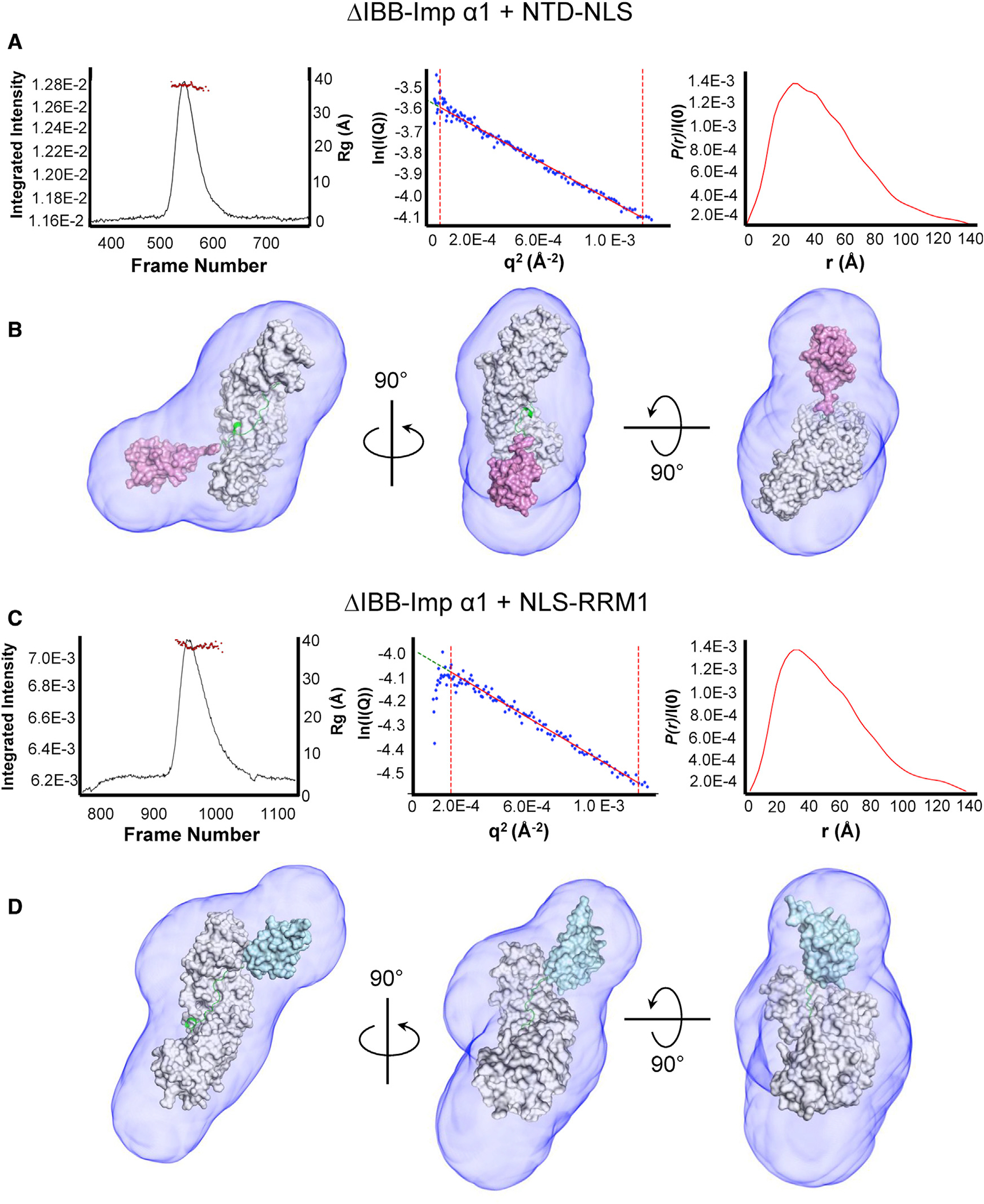
(A) Left, experimental SEC-SAXS profile of the TDP-43 NTD-NLS:ΔIBB-importin α1 complex (black trace) overlaid with Rg distribution across the peak (red circles). Center, Guinier region of the intensity I(q) to the scattering vector (q2). The qmax(Rg) cutoff was 1.3. Right, P(r) function with Dmax of 140.3 Å.
(B) Model of the TDP-43 NTD-NLS:ΔIBB-importin α1 complex fit within the electron density generated by DENSS. ΔIBB-importin α1 is shown in gray, the TDP-43 NLS in green, and the NTD in magenta. The comparison of the scattering profile predicted for the model to the empirical scattering of the complex produced a χ2 value of 1.39. (C) Left, experimental SEC-SAXS profile of the TDP-43 NLS-RRM1:ΔIBB-importin α1 complex (black trace) overlaid with Rg distribution across the peak (red circles). Center, Guinier region with qmax(Rg) cutoff of 1.3. Some repulsion was observed within the sample at low q, which was omitted from the Guinier region. Right, P(r) function with Dmax of 136.7 Å.
(D) Model of the TDP-43 NLS-RRM1:ΔIBB-importin α1 complex fit within the electron density generated by DENSS. ΔIBB-importin α1 is shown in gray, the NLS is in green and the RRM1 is colored in cyan. The comparison between predicted and observed scattering produced a χ2 value of 2.06.
To determine the reliability of our models, we generated a theoretical scattering profile for each and compared them with the observed scattering of the respective complex. We found that the NTD-NLS:ΔIBB-importin α1 model fit the experimental data with a χ2 value of 1.39, which was indicative of a strong agreement between the model and experimental data. The NLS-RRM1:ΔIBB-importin α1 model fit the experimental data with a χ2 value of 2.06, again suggesting a good agreement between our model and experimental data (Figures 4B and 4D). We further constructed a composite model of TDP-43 NTD-NLS-RRM1 (residues 1–179) bound to ΔIBB-importin α1 (shown in Figure 7) that revealed both the NTD and RRM1 are loosely connected from the importin α1 Arm core as “knots on a rope.” This is in agreement with previous SEC-SAXS data using FL TDP-43, in which the entire protein appears highly flexible if not unfolded (Wright et al., 2020). Thus, SEC-SAXS and molecular modeling revealed that the domain flanking the NTD-43 NLS does not make stable and direct contacts with importin α1. Nonetheless, importin α1 is sufficient to disrupt NTD dimerization and assembly into higher-order structures upon binding.
Figure 7. Proposed role of importins in TDP-43 NTD aggregation.
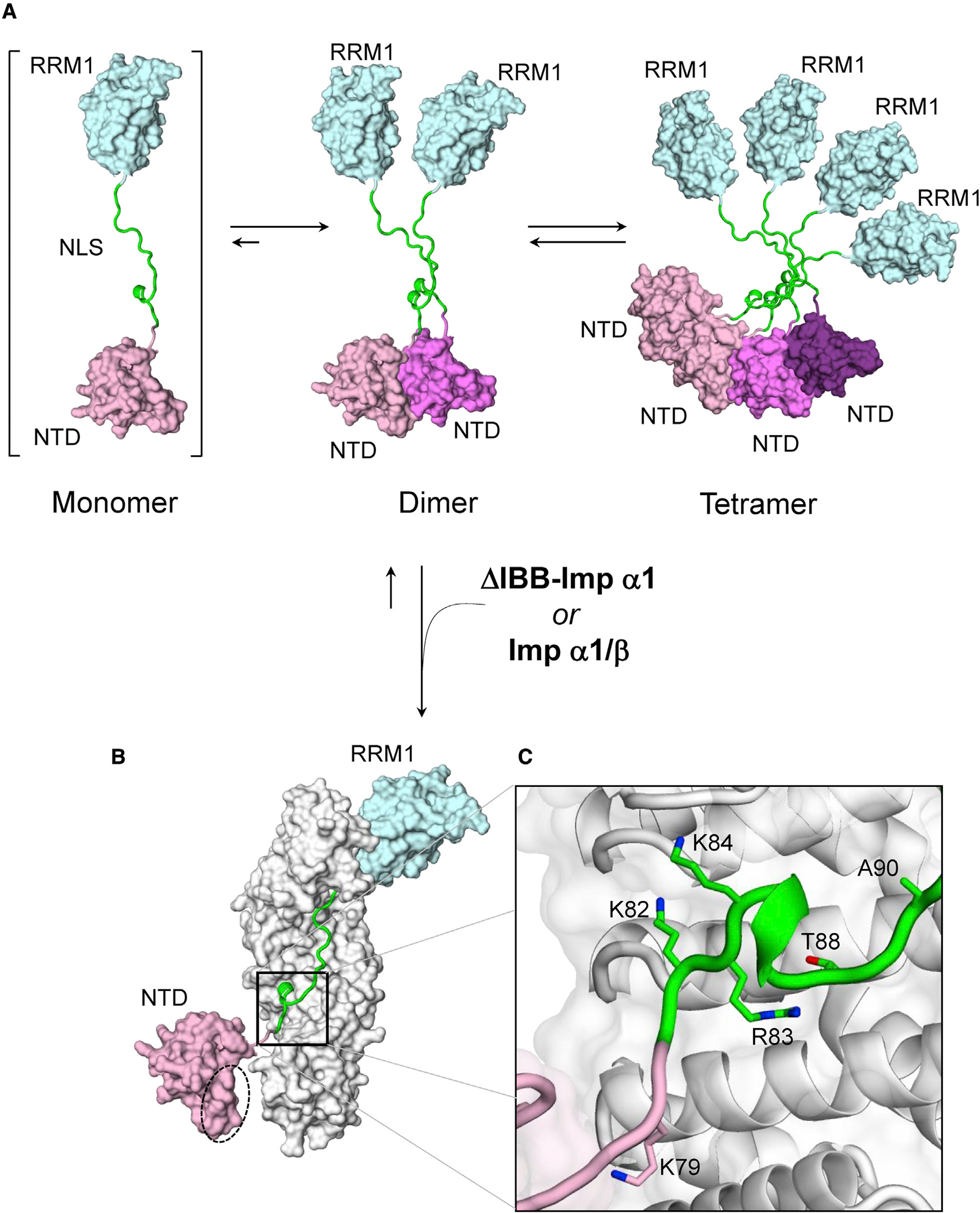
(A) TDP-43 exists in a concentration-dependent equilibrium between dimers, tetramers, and oligomers (not shown). Only the NTD-NLS-RRM1 fragment modeled from SEC-SAXS is shown.
(B) ΔIBB-importin α1, or the importin α1/β complex, breaks the NTD dimer by binding the NLS with nanomolar affinity. The NTD dimerization interface is shown by a dashed circle.
(C) A magnified view of the minor NLS-binding region highlighting all residues implicated in TDP-43 loss of function.
Phosphorylation of the TDP-43 NLS reduces affinity for importin α1
The linker sequence in TDP-43 NLS that connects the major site residues to the minor site is subject to phosphorylation at T88, S91, and S92 (Kametani et al., 2009; Nonaka et al., 2016) (Figure 5A). While the function of NLS phosphorylation is not entirely understood, the addition of this post-translational modification within the linker may inhibit association with importin α1 (Nardozzi et al., 2010). To test this hypothesis, we designed a microtiter plate binding assay to probe the affinity of the TDP-43 NTD-NLS-RRM1 WT and phosphomimetic constructs for the importin α1/β heterodimer. To generate the phosphomimetics, we individually mutated T88, S91, and S92 in the NLS linker to glutamic acid. We also generated a hyperphosphorylated phosphomimetic in which all three residues were mutated to glutamic acid. We found that all TDP-43 constructs bound the importin α1/β heterodimer with nanomolar affinity, with WT TDP-43 NTD-NLS-RRM1 having KD = 65.8 ± 14.3 nM. The pT88 mutant produced the most significant reduction in affinity for the importin α1/β heterodimer, over 3-fold relative to the WT (KD = 218.1 ± 27.6 nM) (Figure 5B). Mutations at S91 and S92 had a minor role on NLS affinity (KD = 120.7 ± 33.5 nM and 87.8 ± 8.5 nM, respectively) (Figures 5C and 5D). Interestingly, when all three mutations were introduced in the same construct, the reduction in affinity was the greatest, over 5-fold relative to the WT (KD = 350.2 ± 1.0 nM) (Figure 5E). Thus, the phosphorylation site that resides next to the critical minor NLS box dramatically affects binding affinity for the importin α1/β heterodimer; together, all phosphorylations appear to play an additive role in weakening the association to the importin α1/β heterodimer.
Figure 5. NLS phosphorylation reduces affinity for import α1/β.

(A) Ribbon diagram of the TDP-43 bipartite NLS highlights putative phosphorylation sites at T88, S91, and S92.
(B–E) Binding isotherms of importin α1/β binding to TDP-43 NTD-NLS-RRM1 containing the (B) T88E phosphomimetic (red curve), (C) S91E phosphomimetic (orange curve), (D) S92E phosphomimetic (green curve), and (E) T88E + S91E + S92E hyperphosphorylation mimetic (purple curve). The positive control, importin α1/β binding to unphosphorylated TDP-43 NTD-NLS-RRM1, is shown as a black curve in all four panels. All KD values are the average of at least two independent experiments, each with at least two independent replicates. Errors are calculated as the standard deviation from the mean.
(F–I) Carbon α root-mean-square fluctuation (RMSF) was measured across 6 μs for each isolated TDP-43 NLS system in units of Å. In each panel, NLS residues at P1–P5 (major NLS box) and P1′–P5′ (minor NLS box) are highlighted in gray. Dashed lines refer to the putative phosphorylation sites T88, S91, and S92. The RMSF curve is shown for the (F) phosphorylated T88 (red trace), (G) phosphorylated S91 (orange trace), (H) phosphorylated S92 (green trace), and (I) hyperphosphorylated T88 + S91 + S92 (purple trace), relative to the RMSF of wt TDP-43 (black trace). The color-coding of RMSF curves in (F–I) matches the binding curves in (B–E).
Phosphorylation in TDP-43 NLS modulates conformational dynamics
To rationalize the role of phosphorylation in the TDP-43 NLS linker, we performed long-timescale-all-atom molecular dynamics (MD) simulations of the unmodified TDP-43 NLS and NLS with phosphorylation at T88, S91, and S92. In each regard, we considered the NLS both independent of the importin α1 adaptor and in complex with importin α1. For the latter experiments, the relative binding energy for each system was calculated from the TDP-43:importin α1 trajectories using the Molecular Mechanics Poisson-Boltzmann Surface Area (MM/PBSA) (Homeyer and Gohlke, 2012; Miller et al., 2012) approach to explore the interaction affinity of each phosphorylated TDP-43 NLS with importin α1. To analyze the dynamical behavior of TDP-43 NLS we measured the root-mean-square fluctuation (RMSF) from simulations of free NLS, not bound to importin α1 (Figures 5F–5I). The latter data indicated that phosphorylation of the TDP-43 NLS within the linker sequence could increase the RMSF of particular NLS residues relative to reference positions determined for an unmodified NLS. Interestingly, RMSF peaks were observed at the tract of basic residues (the KRK sequence) bound at the importin α1 minor site for NLS in which T88 was phosphorylated (Figure 5F) and T88/S91/S92 were all modeled as phosphorylated (Figure 5I). Structurally, phosphorylation of T88 was found to disrupt a short helix at the minor NLS binding site. This helix is characteristic of NLS that engages the minor site (Nakada et al., 2015), and perturbing this structure through phosphorylation could explain the increase in RMSF and the consequent reduction in binding affinity (Figures 5B and 5E). A second RMSF peak was observed over the basic tract KVKR at the importin α1 major site when T88 was phosphorylated, which was not observed for any other phosphorylation event (Figures 5F–5I). Phosphorylation of S91 and S92 did not produce significant deviations in position across the entire NLS, indicating that these NLS behave similarly to the unmodified NLS (Figures 5G and 5H). For each of the complexes simulated in Figure 5, we performed cross-correlation analysis of the simulated structures against the experimental SAXS density (Figure S7). We saw consistent correlations, suggesting that alternate conformations were not observed in our simulations.
In addition to RMSF, we also calculated the change in free energy of binding for the interaction between unmodified and phosphorylated NLS with importin α1 (Figure S8). Phosphorylation caused a reduction in the free energy of binding, the most significant of which occurred for the hyperphosphorylated NLS. This observation suggested that a thermodynamic approach could not sufficiently explain the disruption of the predicted helix at the importin α1 minor binding site through conformational deviation.
Kinetic maps by Markov state modeling
To better understand how phosphorylation affects the conformational dynamics of the TDP-43 NLS, we generated kinetic maps generated by Markov state modeling. Mean first-passage time (MFPT) was derived from Markov state models (MSMs) (Bhattacharya and Lin, 2019; Huang et al., 2017), representing how phosphorylation impacts the kinetics of the TDP-43 NLS. These maps revealed that the TDP-43 NLS adopts different metastable states and distinct MFPTs between states due to phosphorylation. The arrows in Figure 6 represent the first transition time between different states for the WT NLS (Figure 6A) and each phosphorylated state (Figures 6B–6E). The lowest MFPT belongs to the WT system (<333.0 ns), suggesting the unphosphorylated NLS, which binds importin α1 with the highest affinity, also has the highest probability of transitioning to the conformation that is favored for binding to importin α1. On the other hand, the longest MFPT belongs to hyperphosphorylated TDP-43 NLS (<1862.6 ns) and indicates that the spontaneous transition between different states, relative to other phosphorylated NLS peptides, is less probable, representing a rigid structure with a low affinity for binding to importin α1(Figure 6E). Empirically, this results in a reduced affinity for importin α1/β, which in our binding experiment had over 5× higher KD for TDP-43 carrying three phosphomimetic mutations at T88/S91/S92 relative to the WT (Figure 5E). Consistent with the experiment in Figure 5, phosphorylation at T88, proximal to the critical minor NLS-binding site, stabilized a short α helix at the minor NLS site (Figure 6B), affecting the binding affinity for the importin α1/β heterodimer. Overall, these data suggest that phosphorylation in TDP-43 NLS residues not directly involved in binding to the Arm core but positioned in the proximity of the minor NLS-binding pocket destabilizes binding to importin α1/β by reducing the NLS backbone dynamics.
Figure 6. Kinetic maps from Markov state model analysis of each TDP-43 NLS system.

(A–E) The maps show peptides in secondary structure representation, with phosphorylation sites shown in licorice representation. In each map, arrows connecting states represent the mean first-passage time (MFPT) listed next to each arrow. Thicker arrows represent faster transition rates along a given direction and therefore illustrate higher transition probabilities as measured from Markov state models (MSMs). (A) Unphosphorylated NLS binds ΔIBB-importin α1 with the lowest MFPT (<333.0 ns) and has the highest probability of transitioning to the conformation favored for binding importin α1. (B) When phosphorylated at T88, the NLS had generally reduced transition times between states. (C) Phosphorylation at S91 yielded a transition map most similar to the wild-type, albeit with different NLS conformations. (D) The transition map of TDP-43 NLS with phosphorylated S92 featured a paucity of fast kinetics with different states preferred than those in panel (B). (E) The longest MFPT belongs to the hyperphosphorylated TDP-43 NLS (<1,862.6 ns), suggesting that the spontaneous transition between different states is less probable, as expected for a rigid structure with a low affinity for binding to importin α1.
DISCUSSION
In this paper, we have determined the structure and recognition of the TDP-43 NLS by the importin α1/β heterodimer. Our work investigates the interplay between TDP-43 recognition by importins and the events that lead to its aggregation.
We show that the TDP-43 NLS employs a bipartite binding mechanism to engage importin α1/β with KD = 65.8 ± 14.3 nM. The TDP-43 NLS sequences that bind at the major and minor NLS binding sites of importin α1 are in agreement with those of previously published bipartite NLSs (Marfori et al., 2011). However, unlike classical bipartite NLSs, mutation of K97 at the P2 position did not affect binding to importin α1/β, which was disrupted solely by mutation of R83 at the P2′ position. A similar phenomenon was previously reported for the NLSs of inner nuclear membrane proteins Heh1 and Heh2 (Lokareddy et al., 2015) and suggested the minor site of TDP-43 NLS plays a crucial role in binding energetics to importin α1/β. Our structural and biochemical findings agree with previous work investigating TDP-43 nuclear import in cell-based assays (Ayala et al., 2008; Pinarbasi et al., 2018; Winton et al., 2008a). These studies used TDP-43 constructs with poly-alanine mutations at the basic residues of the minor site KRK and major site KVKR sequences (Figures 1A and Table S2). In cell lines, over-expression of the ΔNLS construct in which the minor site sequence was disrupted resulted in depletion of nuclear TDP-43 and formation of cytoplasmic TDP-43 inclusions. Disruption of the major site also caused nuclear depletion, but this did not appear as substantial as when the minor site was affected (Ayala et al., 2008; Winton et al., 2008a). Similarly, in a more recent study, single K84A and K84R mutations at position P3′ prevented the nuclear import of a reporter protein consisting of the red fluorescent protein fused to TDP-43-NLS (RFP-TDP-43-NLS), whereas K95A and K95R mutants at P0, in the major binding site, were imported to the nucleus as efficiently as the WT RFP-TDP-43-NLS (Khosravi et al., 2020). Thus, recognition of the TDP-43 NLS depends chiefly on binding to the minor NLS-pocket of importin α1 and, consequentially, perturbation of the TDP-43 minor NLS tract is expected to play a critical role in slowing TDP-43 nuclear import.
In the attempt to rationalize how the NLS binding crosstalks to the TDP-43 NTD, we crystallized ΔIBB-importin α1 with the NTD-NLS construct. Although the crystals included the NTD, we found no discernable density N-terminal of the minor NLS box, suggesting the NTD is loosely connected to the NLS and does not make contact with importin α1. SEC-SAXS confirmed that both TDP-43 domains flanking the NLS (e.g., NTD and RRM1) do not directly contact importin α1. In agreement with a recent study (Wright et al., 2020), TDP-43 NTD and RRM1 can be thought of as beads on a string, independently sampling different conformations, while importin α1 stabilizes the extended bipartite NLS. Interestingly, the TDP-43 NTD has been shown to mediate dimerization of the protein and form higher-order oligomers (Afroz et al., 2017; Chang et al., 2012; Jiang et al., 2017; Mompeán et al., 2016; Shiina et al., 2010; Wang et al., 2013; Wright et al., 2020). However, we did not observe dimerization of the TDP-43 NTD-NLS construct in complex with importin α1/β, or just ΔIBB-importin α1, which lacks the autoinhibitory IBB domain (Lott and Cingolani, 2011). This is remarkable given that the NTD does not interact with importin α1 directly. Our SAXS model of the ΔIBB-importin α1:NTD-NLS complex suggests a dimeric NTD is incompatible with importin α1 association, as a second NTD protomer would clash with the C-terminal Armadillo repeats 8–10 of importin α1 (Figure 7B). The residues responsible for NTD dimerization are just upstream of the NLS (Jiang et al., 2017), and, although they do not appear to bond importin α1 directly, the protein presents a steric barrier, likely sufficient to disrupt dimerization. This observation may explain how the importin α1/β heterodimer prevents TDP-43 aggregation in vitro (Guo et al., 2018). This activity is NLS dependent and requires both the FL importin α1 and the receptor importin β, while individual importin α1 or β were not sufficient. In hindsight, importin β removes the IBB autoinhibition from importin α1, exposing the minor NLS pocket for TDP-43 binding and disrupting NTD dimerization. It is also possible that, concomitantly with importin α1-mediated disruption of TDP-43 NTD aggregation, importin β, a 97 kDa protein, also engages the rest of TDP-43, possibly shielding the CTD and further preventing its aggregation. In other words, the observed chaperone activity of importin α1/β toward TDP-43 aggregates may arise from different biochemical activities harbored in both importins.
Our data provide a reading frame to explain the deleterious effect of post-translational modifications and mutations in the proximity of the minor NLS site previously reported in the literature. Acetylation of K82, at position P1′ (Figure 7C), and ubiquitination of the nearby K79 (at position P−1′) were found in a sarkosyl-insoluble fraction of TDP-43 aggregates isolated from brains of ALS patients (Kametani et al., 2016). Acetylation of K84 at the P3′ position (Garcia Morato et al., 2022) resulted in TDP-43 cytoplasmic mislocalization, and aggregation propensity in HEK293E cells, strengthening the connection between modification of the TDP-43 minor NLS box and pathogenicity. Similarly, as previously pointed out, K84A and K84R mutations largely prevented the nuclear import of an RFP-TDP-43-NLS reporter, indicating K84 is crucial for the function of the TDP-43 NLS (Khosravi et al., 2020). C-terminal of the minor NLS box, the ALS-associated A90V mutation was found to sequester the endogenous TDP-43 into insoluble cytoplasmic aggregates (Winton et al., 2008b). This mutation is three-dimensionally close to the minor site R83, given the turn in the NLS backbone at the minor site (Figure 7C). All these modifications and mutations likely destabilize the recognition of the minor NLS box, preventing importin α1/β association that disrupts healthy nucleocytoplasmic trafficking and promotes cytoplasmic aggregation.
Beyond the phenomenological observation that a mutation or post-translational modification is linked to aggregation, in this paper, we sought to investigate how changes to just one of the many residues in TDP-43 NLS making contacts with importin α1 can have such a pleiotropic effect. To this end, we investigated three putative phosphorylation sites in the TDP-43 NLS linker at T88, S91, and S92, previously identified as targets of casein kinase 1-δ (Kametani et al., 2009; Nonaka et al., 2016). Although none of these three residues makes direct, strong contacts with importin α1 Arm core, phosphorylation of the threonine within the linker sequence (T88) produced the most significant reduction in affinity for importin α1. This was in agreement with our MD simulations of the interaction between importin α1 and phosphorylated TDP-43 NLS, in which phosphorylation at T88 produced a consistently high RMSF relative to phosphorylation of other residues within the NLS. Interestingly, T88 is close to the minor NLS binding site (Figure 7C). MD simulations indicate that phosphorylation within the NLS linker reduces the spontaneous transition of the NLS backbone to more kinetically favorable states, making binding to importin α1 less probable. Specifically, phosphorylation at T88 stabilizes a short α helix in the immediate proximity of the minor site that has a pleiotropic negative effect on binding affinity for importin α1/β. This effect was exacerbated by having three phosphorylations in the NLS linker that further stabilized secondary structure states having unfavorable binding characteristics for importin α1/β, enhancing cytoplasmic retention that leads to aggregation. This finding is particularly relevant in the disease state, in which aggregated TDP-43 becomes hyperphosphorylated, with most phosphorylation sites in the CTD (Kametani et al., 2016). Our results contrast with a recent report that phosphorylation of TDP-43 CTD suppresses condensation and aggregation (Gruijs da Silva et al., 2022) instead of enhancing it (Hasegawa et al., 2008). Notably, Gruijs da Silva et al. used in vitro phosphorylated TDP-43 lacking the three phosphorylation sites in the NLS reported by Kametani et al. (2009) that, as shown in this paper, modulate the affinity of TDP-43 for the importin α1/β complex.
We propose that, under normal conditions, importin α1/β antagonizes TDP-43 aggregation via two concomitant mechanisms: promoting its import into the nucleus and preventing NTD dimerization by steric hindrance (Figures 7A and 7B). Perturbations in the NLS that affects the minor NLS site reduce binding affinity for importin α1/β and serve two pathogenic functions: on one side, slowing down TDP-43 nuclear import, as a weaker NLS is also less likely to be imported into the cell nucleus (Fanara et al., 2000); on the other, reducing importin α1/β′s ability to stabilize monomeric TDP-43 NTD, leading to large aggregates. The literature is split on the actual role of the NTD dimerization vis à vis TDP-43 aggregation. While certain reports have suggested that TDP-43 NTD dimerization is necessary for its physiological functions like RNA splicing and may antagonize pathological aggregation (Afroz et al., 2017; Jiang et al., 2017), others have argued that the NTD dimerization is involved in TDP-43 aggregation (Shiina et al., 2010; Wang et al., 2013). Recent biophysical work has shown that NTD undergoes reversible oligomerization that enhances the propensity of the CTD to form amyloid-like structures (Tsoi et al., 2017; Wang et al., 2018). This paper supports the latter hypothesis and attempts to interpret the NTD dimerization in the context of importin α1/β recognition. NTD self-association brings TDP-43 molecules closer together, enhancing the local concentration of CTDs. This intrinsically disordered portion begins the formation of amyloid-like structures (Arseni et al., 2022), and larger and larger oligomers eventually become phase-separated aggregates. Post-translational modifications in the NLS also reduce the affinity for importins, reducing importin α1/β chaperone activity toward TDP-43 aggregates (Guo et al., 2018). Overall, the cell is overwhelmed by the disruption of TDP-43 homeostasis, promoting the formation of inexorably larger and larger inclusions that drive the disease state.
Limitations of the study
In this study, we characterize the recognition of the TDP-43 NLS by human importins α1/β or just a fragment of importin α1 lacking the IBB domain. One limitation of our work is that we limited our structural, biochemical, and computational analysis to a soluble fragment of TDP-43 spanning residues 1–177, which lacks the second RRM domain (RRM2) and the amyloid-forming CTD. This limitation was imposed by the biochemically intractable nature of the CTD that readily aggregates in solution and requires a high concentration of the ionic detergent sarkosyl to be solubilized (Wright et al., 2020). We found that sarkosyl disrupts the binding of importin α1 and β, as well as denatures ΔIBB-importin α1, making it impossible to study the association of these proteins with TDP-43 using high-resolution biophysical methods. However, we cannot exclude that the TDP-43 domains missing in our analysis, RRM2 and CTD, make additional contacts with importin α1/β that cannot be detected using our minimal TDP-43 construct (NTD-NLS-RRM1). Further studies, both in vitro and in live cells, will have to test if there exist additional binding determinants for importin α1/β in TDP-43 RRM2 and CTD capable of modulating the recognition of NLS described in this paper.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Gino Cingolani (gino.cingolani@jefferson.edu).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed materials transfer agreement.
Data and code availability
The crystallographic coordinates for the TDP-43 NTD-NLS+ΔIBB-importin α1 complex have been deposited in the Protein Data Bank and raw X-ray diffraction data are posted on Mendeley Data and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| HRP conjugated anti-GFP polyclonal primary antibody | Millipore Sigma | Cat#GERPN1236; RRID:AB_771429 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| BL21-DE3 | Millipore Sigma | Cat#CMC0016 |
| BL21-DE3 LOBSTR | Kerafast | Cat#EC1002 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| ΔIBB-importin α1 construct | This paper | Cingolani_001 |
| Importin α1/β heterodimer | This paper | Cingolani_002 |
| GST-TDP-43 NTD-NLS-RRM1 wild-type | This paper | Cingolani_003 |
| GST-TDP-43 NTD-NLS-RRM1 T88E (pT88) | This paper | Cingolani_004 |
| GST-TDP-43 NTD-NLS-RRM1 S91E (pS91) | This paper | Cingolani_005 |
| GST-TDP-43 NTD-NLS-RRM1 S92E (pS92) | This paper | Cingolani_006 |
| GST-TDP-43 NTD-NLS-RRM1 T88E, S91E, S92E (pT88+pS91+pS92) | This paper | Cingolani_007 |
| GST-TDP-43 NTD-NLS | This paper | Cingolani_008 |
| GST-TDP-43 NLS-RRM1 | This paper | Cingolani_009 |
| Ampicillin | ThermoFisher Scientific | CAS#69-53-4 Cat#11593027 |
| Kanamycin sulfate | ThermoFisher Scientific | CAS#25389-94-0 Cat#15160054 |
| Chloramphenicol | ThermoFisher Scientific | CAS#56-75-7 Cat#227925000 |
| 3,3’,5,5’-Tetramethylbenzadine (TMB) | abcam | CAS#54827-17-7 Cat#ab171523 |
| Isopropylthio-β-galactoside (IPTG), >99% | ThermoFisher Scientific | CAS#367-93-1 Cat#15529019 |
| Sodium Chloride (NaCl), >99% | ThermoFisher Scientific | CAS#7647-14-5 Cat#A12313.0I |
| Tris base, >98% | ThermoFisher Scientific | CAS#77-86-1 Cat#J22675.36 |
| 2-mercaptoethanol (BME), 14.3 M, >99% | Millipore Sigma | CAS#60-24-2 Cat#M6250 |
| Tween-20 | Millipore Sigma | CAS#9005-64-5 Cat# P1379 |
| Hydrochloric acid (HCl), 12 N | ThermoFisher Scientific | CAS#7647-01-0 Cat#NC004378 |
| Dulbecco’s Phosphate buffered saline (PBS), 10× | Millipore Sigma | CAS#7647-14-5 Cat#D1408 |
| Ethylenedinitrilotetraacetic acid disodium salt (EDTA), >99%, | Millipore Sigma | CAS#6381-92-6 Cat#E5134 |
| Imidazole, >99% | Millipore Sigma | CAS#288-32-4 Cat#814223 |
| High density nickel resin | GoldBio | Cat#H-320-5 |
| GST agarose resin | GoldBio | Cat#G-250-5 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Pierce BCA Protein Assay Kit | ThermoFisher Scientific | Cat#23227 |
| Natrix Crystallization Screen | Hampton Research | HR2-116 |
| Natrix 2 Crystalization Screen | Hampton Research | HR2-117 |
| Crystal Screen | Hampton Research | HR2-110 |
| Index Screen | Hampton Research | HR2-144 |
|
| ||
| Deposited data | ||
|
| ||
| Raw and analyzed X-ray data for TDP-43 NTD-NLS+ΔIBB-importin α1 | This paper | Mendely Data: https://data.mendeley.com/datasets/mpkdxc4hsj/ |
| TDP-43 NTD-NLS+ΔIBB-importin α1 PDB | This paper | PDB: 7N9H |
|
| ||
| Recombinant DNA | ||
|
| ||
| Vector: pET-30a | EMD biosciences/Millipore Sigma | Cat#69909 |
| Vector: pGEX-6p-1 | Amersham/Millipore Sigma | Cat# GE28-9546-48 |
| Vector: pETduet-1 | EDM biosciences/Millipore Sigma | Cat#71146 |
|
| ||
| Software and algorithms | ||
|
| ||
| ImageJ | (Abramoff et al., 2004) | N/A |
| PyMol | (DeLano, 2002) | N/A |
| HKL2000 | (Otwinowski and Minor, 1997) | N/A |
| Phaser | (McCoy, 2007) | N/A |
| Phenix.refine | (Liebschner et al., 2019) | Version 1.19.2 |
| Phenix.polder | (Liebschner et al., 2017) | N/A |
| COOT | (Emsley and Cowtan, 2004) | N/A |
| ATSAS | (Franke et al., 2017) | N/A |
| RAW | (Hopkins et al., 2017) | Version 2.1.1 |
| DENSS | (Grant, 2018) | N/A |
| Chimera | (Pettersen et al., 2004) | N/A |
| FoXS Server | (Schneidman-Duhovny et al., 2010) | N/A |
| GraphPad Prism | Non-linear regression: one-site specific binding curve | Version 9.0.0 |
| Chromlab | Bio-Rad | N/A |
| SEDFIT | (Schuck, 2000) | Center for Information Technology, NIH |
| SEDNTERP | (Laue et al., 1992) | N/A |
| GUSSI | UTSouthwestern Medical Center | N/A |
| PDBsum | (Laskowski, 2009) | N/A |
| PISA | (Krissinel and Henrick, 2007) | N/A |
| Pyemma | (Scherer et al., 2015) | Version 2.5.7 |
| Amber | (D.A. Case et al., 2018) | Version 18 |
| SHAKE algorithm | (Elber et al., 2011) | N/A |
| PCCA++ algorithm | (Roblitz et al., 2013) | N/A |
| VMD | (Humphrey et al., 1996) | N/A |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
We made use of an E. coli expression system for the synthesis of recombinant proteins. E. coli of the BL21-DE3 strain or derivations therein were transformed with one or more constructs and exposed to the corresponding selection conditions.
METHOD DETAILS
Molecular biology
Fragments of human TDP-43 (Figure 1A) were synthesized by GeneWiz and inserted into a pGEX-6p-1 vector. In the case of the TDP-43 NTD-NLS-RRM1 phosphomimetics, glutamic acid was individually substituted for each of the three residues within the NLS linker (T88, S91, and S92). An additional construct in which all of these residues were mutated to glutamic acid was also generated. Using the TDP-43 NLS:ΔIBB-importin α1 structure, we designed a second set of TDP-43 NTD-NLS-RRM1 point mutants in which the residues at the P2 (K97) and P2’ (R83) positions within the NLS were replaced with alanine; either individually or in concert. The gene encoding mouse importin α1 lacking the IBB domain was cloned in a pET30a vector (Novagen) (Sankhala et al., 2017). Human FL importin α1 and importin β were cloned in tandem into a pETDuet-1 vector for co-expression.
Protein expression and purification
The pGEX-6p-1-TDP-43 NTD-NLS and pGEX-6p-1-TDP-43 NLS-RRM1 constructs were each co-transformed with pET30a-ΔIBB importin α1 into E. coli BL21-DE3 expression strains. Protein was expressed overnight at 18°C with 400 μM IPTG. Complexes consisting of GST-TDP-43 fragments bound to ΔIBB-importin α1 were purified in a low-salt buffer (150 mM NaCl, 20 mM Tris pH 8, 5 mM BME, 1 mM EDTA, and 0.1% Tween 20) and bound to glutathione agarose resin. The GST tag was removed through overnight PreScission Protease (PPase) cleavage at 4°C on a gravity flow column, and untagged TDP-43 fragments bound to ΔIBB-importin α1 were collected in the column flowthrough. These complexes were further purified through size-exclusion chromatography using a Superdex 200 16:600 preparative column. Fractions corresponding to mono-disperse TDP-43:ΔIBB-importin α1 at a 1:1 ratio were pooled and concentrated in a 30 kDa concentrator (Vivaspin) and subjected to downstream applications.
For crystallization of the pGEX-6p-1-TDP-43 NTD-NLS-RRM1 complex with ΔIBB-importin α1, both proteins were individually transformed into BL21-DE3 E. coli expression strains. The pGEX-6–1-TDP-43 NTD-NLS-RRM1 was expressed overnight at 18°C with 400 uM IPTG. The pET30a-ΔIBB importin α1 was expressed for 5 h at 30°C with 400 uM IPTG. TDP-43 NTD-NLS-RRM1 was purified in a low-salt buffer (150 mM NaCl, 20 mM Tris pH 8, 5 mM BME, 1 mM EDTA, and 0.1% Tween 20) and bound to glutathione agarose resin. The GST tag was removed through overnight PPase cleavage at 4°C on a gravity flow column, and untagged TDP-43 NTD-NLS-RRM1 was collected in the column flowthrough. The protein was further purified by SEC using a Superdex 200 10:300 analytical column. Fractions corresponding to mono-disperse TDP-43 were used in the formation of TDP-43:ΔIBB-importin α1 complex. The ΔIBB-importin α1 was purified in a high salt buffer (600 mM NaCl, 20 mM Tris pH 8.5, 3 mM BME) and bound to high-density nickel resin. The protein was eluted in 150 mM imidazole and immediately subjected to SEC using a Superdex 200 10:300 analytical column. Fractions corresponding to mono-disperse, monomeric importin α1 were concentrated in a 30 kDa concentrator (Vivaspin) and quantified. Purified TDP-43 NTD-NLS-RRM1 was concentrated in a 10 kDa concentrator and quantified in tandem. The two proteins were pooled at a ratio in which TDP-43 NTD-NLS-RRM1 was in excess. Complex formation proceeded at 25°C for 30 min, after which the sample was applied to a Superdex 200 10:300 column and subjected to a further round of SEC. Fractions corresponding to the monodisperse complex at an apparent 1:1 rato, as determined by elution profile, were used in the downstream application, specifically hanging drop vapor diffusion.
Similar to the above, the pGEX-6p-1-TDP-43 NTD-NLS-RRM1 constructs (WT P2, P2’, and P2+P2’ mutants) were co-transformed with pETDuet-1 importin α1/β into BL21-DE3 E. coli expression strain. Proteins were expressed for 1.5 h at 30°C with 500 μM IPTG. Heterotrimeric complex consisting of TDP-43 NTD-NLS-RRM1 and importin α1/β was purified in a low-salt buffer (75 mM NaCl, 20 mM Tris pH 8, and 3 mM BME) and bound to low-density nickel beads. Complex was eluted using a low-imidazole elution buffer (75 mM imidazole, 50 mM NaCl, 20 mM NaCl, and 3 mM BME).
The TDP-43 NTD-NLS, NLS-RRM1, and NTD-NLS-RRM1 constructs could be expressed and purified individually using the same conditions employed for the TDP-43:ΔIBB-importin α1 complex. Likewise, the importin α1/β heterodimer could be expressed purified as described for the TDP-43:α1:β heterotrimeric complex.
All SEC in this paper was conducted using a BioRad NGC single wavelength FPLC unit. The Chromlab software was provided by BioRad to facilitate visualization and analysis of chromatograms. The chromatograms presented here are derivative of Chromlab.
Crystallographic methods
Crystals of ΔIBB-importin α1 bound to TDP-43 NTD-NLS were obtained using the hanging drop vapor diffusion method. A droplet containing 1 μL of gel filtration-purified complex at 5 mg mL−1 and was mixed with an equal volume of 0.1 M sodium citrate tribasic dihydrate pH 5.6, 0.7 M sodium citrate tribasic dihydrate, 10 mM BME, and equilibrated against 600 μL of the precipitant, at 18°C. Crystals were harvested in nylon cryo-loops, cryo-protected with 27% ethylene glycol, and flash-frozen in liquid nitrogen. Crystals were diffracted at the Stanford Synchrotron Radiation Lightsource (SSRL) using beamline 9–2. Data were indexed, integrated, and scaled using HKL2000 (Otwinowski and Minor, 1997). Initial phases were obtained by molecular replacement using Phaser (McCoy, 2007) and PDB 1Y2A as a search model. The TDP-43 NLS was built in Fo-Fc electron density difference maps using Coot (Emsley and Cowtan, 2004), and complete atomic models were refined using phenix.refine (Liebschner et al., 2019) using positional and isotropic B-factor refinement cycles with six Translation:Libration:Screw groups. Polder omit maps were calculated using phenix.polder (Liebschner et al., 2017). Data collection and refinement statistics are summarized in Table S1. All ribbon diagrams and surface representations were prepared using PyMol (DeLano, 2002).
Size exclusion chromatography coupled to small-angle X-ray diffraction (SEC-SAXS)
Heterodimeric complexes consisting of TDP-43 NTD-NLS:ΔIBB-importin α1 and TDP-43 NLS-RRM1:ΔIBB-importin α1 were submitted to the Cornell High Energy Synchrotron Source (CHESS) and Cornell University in Ithaca, NY. The complex was first separated on a Superdex 200 10:300 analytical column and then subjected to X-ray diffraction. Small-angle diffraction data was collected and subsequently processed using the RAW software package (version 2.1.1) (Hopkins et al., 2017), including plug-ins from ATSAS (Franke et al., 2017). Sample peak intensities were normalized to a region of the scattering baseline that satisfied the following conditions: 1. The selected baseline did not include any individual frames that differed significantly from the average intensity and 2. The selected baseline produced a distribution of radii of gyration that included a Guinier region. From the Guinier region, the molecular mass of each complex was predicted, and a P(r) plot was generated. P(r) plots corresponding to each complex were submitted to the DENsity from Solution Scattering (DENSS) (Grant, 2018) plug-in to generate low-resolution electron densities. We modeled the TDP-43 NTD-NLS and NLS-RRM1 bound to ΔIBB-importin α1 using the crystal structure of TDP-43 NLS:ΔIBB-importin α1 complex determined in this paper and previously determined structures of the TDP-43 NTD (PDB: 5MDI) and TDP-43 RRM1 (PDB: 4BS2). These complexes were fit into the DENSS density and refined against the low-resolution SAXS density using the fit-into-map command in Chimera (Pettersen et al., 2004). The scattering profiles of these models were compared to the empirically observed scattering data using the FoXS server (Schneidman-Duhovny et al., 2010), which generated a predicted-to-observed χ2 value in each case.
ELISA-based microtiter binding assay
GST-TDP-43 NTD-NLS-RRM1 WT and phosphomimetic constructs were individually purified and dialyzed overnight against 150 mM NaCl, 20 mM Tris pH 8, and 0.05% Tween 20. The importin α1/β heterodimer was purified and isolated by SEC using a Superdex 200 16:60 preparative column. The heterodimer was diluted to 2 μg:ml in adsorption buffer (approximately 50 mM sodium carbonate:sodium bicarbonate) and bound to MaxiSorp ® (Nunc) polystyrene plates at a final concentration of 200 ng per well overnight at 4°C. Plates were then blocked at 25°C for at least two hours in blocking buffer (0.1% PBS-Tween with 2% BSA). TDP-43 NTD-NLS-RRM1 ligands were prepared in dilution gradients from 0 to 500 nM in blocking buffer. Ligand dilutions were applied to the ELISA plate in 100 μL aliquots and incubated for 1 h at 25°C. Unbound ligand was removed through washing, and plates were then incubated with an HRP-conjugated anti-GST polyclonal antibody (Sigma) at a 1:5000 dilution for 1 h at 37°C. Plates were then washed to remove excess antibodies, followed by the application of 100 μL of TMB substrate. The colorimetric reaction was allowed to proceed for 15 min at 25°C before being stopped through the addition of 2 N HCl. Plates were read at 450 nm using a Tecan microplate reader (Life Sciences). Each ELISA-based assay was run in duplicate and independently reproduced at least two times. Binding isotherms were plotted in GraphPad Prism and fit by a one-site specific binding non-linear regression. Errors are given as the standard deviation across independent experiments.
Sedimentation Velocity Analytical Ultracentrifugation (SV-AUC)
SV-AUC experiments on TDP-43 NTD-NLS:ΔIBB-importin α1 heterodimer was performed at concentrations ranging 1–2 mg/mL in 20 mM Tris-HC pH 7.5, 150 mM NaCl, and 3 mM β-ME. Centrifugation was performed at 40,000 rpm at 6°C using a Beckman XLA Analytical Ultracentrifuge (Beckman-Coulter, Brea, CA). SV-AUC experiments on TDP-43 NTD-NLS and Importin α/β were performed at 20°C with an Optima analytical ultracentrifuge (Beckman-Coulter, Brea, CA) and a TiAn50 rotor with two-channel charcoal-filled Epon centerpieces and sapphire windows, using both absorbance and interference optics. Data were collected with detection at 260, 280, & 295 nm, as well as interference optics. TDP-43 NTD-NLS analyses were performed in 20 mM Tris-HCL pH 7.5, 150 mM NaCl, and 3 mM β-ME, and Importin α/β-NTD-NLS analyses were performed in 20 mM Tris-HCL pH 8.0, 50 mM NaCl, and 2 mM DTT. Complete sedimentation velocity profiles were recorded every 30 s at 40,000 rpm. Data were fit using the c(S) implementations of the Lamm equation as implemented in the program SEDFIT (Schuck, 2000). The partial specific volume (ῡ), solvent density (ρ), and viscosity (η) were derived from chemical composition by SEDNTERP (Laue et al., 1992). All S values calculated were corrected to s20,w values. Figures were created using the program GUSSI (Brautigam, 2015).
In cellulo pull-down assays
Plasmids encoding pGEX-6p-1-TDP-43 NTD-NLS-RRM1 (WT, K97A, R83A, K97A/R83A) mutants were co-expressed in E. coli BL21-DE3 expression strain with the pETDuet-1 plasmid expressing importin α1/β. Protein was expressed for 1.5 h at 30°C with 500 μM IPTG. Pre- and post-induction optical densities were measured for each co-transformant to ensure a similar rate of growth across all cultures. Trimeric complexes were purified as previously described for the importin α1/β complex and eluted from equal volumes of low-density nickel beads. Eluates were gently concentrated in 10 kDa concentrators (Vivaspin) to facilitate analysis by SDS-PAGE gel. Volumes corresponding to an equal mass of importin α1 were loaded onto a 13.7% SDS-PAGE gel alongside molecular weight marker. SDS-PAGE gels were run in triplicate (n = 3), and the average ratio of GST-TDP-43 NTD-NLS-RRM1 to importin α1 band intensity was calculated for each construct through densitometry using ImageJ (Abramoff et al., 2004). To determine statistical significance between the means of the control and experimental pull-downs, we employed a two-tailed Student’s T test assuming unequal variance. The alpha parameter had to be outside the 99th percentile for statistical significance (p < 0.01, indicated by ***). Error bars indicate the standard deviation about the mean.
Molecular dynamics (MD) simulations
The atomic structure of TDP-43 NLS bound to ΔIBB-importin α1, explained in Crystallographic methods, was used as the initial structure for MD simulations. Unphosphorylated TDP-43 NLS was utilized as the “wild-type” structure and was additionally utilized to prepare three single phosphorylated constructs, namely pT88, pS91, and pS92. An additional hyperphosphorylated construct was prepared, incorporating all three phosphorylations: pT88 + pS91 + pS92. The complex was placed in a solvent box with 12 Å of padding in positive and negative x, y, and z directions from the protein periphery. Systems employed the TIP3P water model, and all were charge neutralized and set to a NaCl concentration of 150 mM. The resulting models contained approximately 65,000 atoms, comprised of importin α1, TDP-43 NLS, ions, and water. The dimensions of each prepared system were 108 × 108 × 74 Å3. After construction, all systems were initially subjected to a minimization and thermalization procedure. During thermalization, the system was heated to 310 K gradually over 50 ps, followed by a short 1 ns simulation performed while the protein atoms were positionally restrained using a force constant of 1.0 kcal/mol Å2. Subsequently, all systems were equilibrated over 20 ns using the NPT ensemble without any restraints on the solute atoms (Table S4). Consequently, the production simulations employed five replicas, where each was run for 100 ns to generate the trajectory and be used for the Molecular Mechanics Poisson–Boltzmann Surface Area (Homeyer and Gohlke, 2012; Miller et al., 2012) MM/PBSA calculation. For each frame, the MM/PBSA was estimated, and then the mean value and standard deviation as the error bar were computed and shown in Figure S8. Minimization, thermalization, equilibration, and production steps were completed in Amber 18 (D.A. Case et al., 2018) using the ff19SB force field (Tian et al., 2020), employing a time step of 2 fs. The temperature was maintained with a Langevin heat bath, with a collision frequency of 1.0 ps−1. The SHAKE algorithm (Elber et al., 2011) was applied to restrain all covalent bonds involving hydrogen atoms. Long-range electrostatic interactions were calculated using the particle mesh Ewald (PME) method (T. Darden et al., 1993), and the nonbonded interactions were calculated with a cutoff distance of 8 Å. Our results indicated a significant difference between ΔG of WT and hyperphosphorylated structures in which the error bars from the multiple replicas of these two structures do not overlap. Therefore, WT and hyperphosphorylated TDP-43 NLS have the strongest and weakest binding affinity to ΔIBB-importin α1, respectively (Figure S8). On the other hand, the other three phosphorylated structures, pThr88, pSer91, and pSer92, have a ΔG that is placed between the ΔG values of WT and hyperphosphorylated complexes. This finding is also confirmed by the error bars definitions, which demonstrate a significant overlap between the pThr88, pSer91, and pSer92 complexes. Thus, the binding affinity of the single phosphorylated systems (pThr88, pSer91, and pSer92) are stronger than the hyperphosphorylated complex but weaker than the WT TDP-43 NLS complex. Additionally, for each of the complexes simulated in Figure 5, we performed cross-correlation analysis of the simulated structures against the experimental SAXS density (Figure S7). The MD trajectory of each system first was fitted to the experimental density map derived from SAXS data. The cross-correlation was calculated using the VMD Voltool package (Humphrey et al., 1996) to compare the SAXS data with the MD simulation trajectories, which suggested alternate conformations were not observed in our simulations.
Additionally, a second set of isolated TDP-43 NLS simulations, incorporating WT NLS and phosphorylated NLS systems, were performed. The resulting systems contained approximately 19,600 atoms each, including TDP-43 NLS, ions, and TIP3P water. Systems were neutralized, and the NaCl concentration was set to 150mM. The systems were minimized and equilibrated using Amber 18 (D.A. Case et al., 2018). We ran equilibrium simulations for 200 ns, for each phosphorylated NLS structure then selected structures at intervals of 33 ns to begin six independent production simulations for each construct. The six simulations per phosphorylated system were run for 1 μs each, totaling 6 μs for each TDP-43 variant (Scherer et al., 2015). First, the coordinates of isolated TDP-43 NLS phosphorylation simulation trajectories were transformed into features, including 1) radius of gyration, 2) root-mean-square distance, 3) distances between alpha carbon atoms (Cα) on the N- and C-terminus residues 4) distances between alpha carbon atoms on the N-terminus and Ala90 residues and 5) distances between alpha carbon atoms on the C-terminus and Ala90 residues. Subsequently, time-lagged independent component analysis (TICA) (Perez-Hernandez et al., 2013) was performed to decompose these features onto 100 slow independent components (ICs). The projected dataset was then discretized using the k-means clustering algorithm, resulting in 12 microstates. The transition probability matrix between these microstates was then computed at a lag time of 3 ns. Finally, a four-state MSM using the PCCA++ algorithm (Roblitz et al., 2013) and the mean first-passage times (MFPT) (Polizzi et al., 2016) were measured between different states for each phosphorylated TDP-43 NLS structure.
QUANTIFICATION AND STATISTICAL ANALYSIS
Band intensities on SDS-PAGE gels were calculated using ImageJ (Abramoff et al., 2004). Details regarding the statistical analysis of these data can be found within the Figure 2 legend, as well as the Method details section. Significance was defined as p < 0.01.
Signal intensity from TMB substrate conversion was measured at 450 nm using a TECAN 96-well plate reader. Details regarding the processing of the data can be found within the Method details section. Data was plotted using GraphPad Prism software and in each case was fit to a non-linear regression designed to model specific binding. We used the 95% confidence interval of each predicted KD to assess the quality of the fit and the likelihood that we had captured the true KD. Error bars represent the standard deviation of the mean. For plotted points in which error bars do not appear to be present, the spread of the error was such that it did not exceed the dimensions of the data point marker.
For the plots of the free energies of binding shown in Figure S8, error bars represent the standard deviation of the mean. Details regarding the statistical analysis of these data can be found within the Method details section.
Supplementary Material
Highlights.
The TDP-43 NLS engages importin α1 through a bipartite binding mechanism
Importin α1 minor NLS-binding site is essential for TDP-43 association
Importin α1/β disrupts the TDP-43 N-terminal domain (NTD) dimerization interface
Phosphorylation near the TDP-43 minor NLS site reduces affinity for importin α1/β
ACKNOWLEDGMENTS
This work was supported by NIH grants R01 GM100888 and R35 GM140733 to G.C. Research in this publication includes work carried out at the SKCC X-ray crystallography facility at Thomas Jefferson University, which is supported in part by NCI grant P30 CA56036 and NIH grants S10 OD017987 and S10 OD023479. AUC analyses were performed at the Johnson Foundation Structural Biology and Biophysics Core at the Perelman School of Medicine with the support of an NIH grant S10 OD018483. MacCHESS is supported by the NSF grant DMR-1829070, and the MacCHESS resource is supported by the NIH grant P30 GM124166–01A1 and NYSTAR. Atomic coordinates for ΔIBB-importin α1 bound to TDP-43 NLS have been deposited in the Protein Data Bank with accession code 7N9H.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111007.
REFERENCES
- Abramoff MD, Magelhaes PJ, and Ram SJ (2004). Image processing with ImageJ. Biophot. Int. 11, 36–42. [Google Scholar]
- Acerbo AS, Cook MJ, and Gillilan RE (2015). Upgrade of MacCHESS facility for X-ray scattering of biological macromolecules in solution. J. Synchrotron Radiat. 22, 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afroz T, Hock EM, Ernst P, Foglieni C, Jambeau M, Gilhespy LAB, Laferriere F, Maniecka Z, Pluckthun A, Mittl P, et al. (2017). Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat. Commun. 8, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arseni D, Hasegawa M, Murzin AG, Kametani F, Arai M, Yoshida M, and Ryskeldi-Falcon B (2022). Structure of pathological TDP-43 filaments from ALS with FTLD. Nature 601, 139–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala YM, Zago P, D’Ambrogio A, Xu YF, Petrucelli L, Buratti E, and Baralle FE (2008). Structural determinants of the cellular localization and shuttling of TDP-43. J. Cell Sci. 121, 3778–3785. [DOI] [PubMed] [Google Scholar]
- Berning BA, and Walker AK (2019). The pathobiology of TDP-43 C-terminal fragments in ALS and FTLD. Front. Neurosci. 13, 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, and Lin X (2019). Recent advances in computational protocols addressing intrinsically disordered proteins. Biomolecules 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam CA (2015). Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 562, 109–133. [DOI] [PubMed] [Google Scholar]
- Cao Q, Boyer DR, Sawaya MR, Ge P, and Eisenberg DS (2019). Cryo-EM structures of four polymorphic TDP-43 amyloid cores. Nat. Struct. Mol. Biol. 26, 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case DA, Ben-Shalom IY, Brozell SR, Cerutti DS, Cheatham TE III, Cruzeiro VWD, Darden TA, Duke RE, Ghoreishi D, Gilson MK, et al. (2018). AMBER 2018. University of California, San Francisco. [Google Scholar]
- Chang CK, Wu TH, Wu CY, Chiang MH, Toh EK, Hsu YC, Lin KF, Liao YH, Huang TH, and Huang JJ (2012). The N-terminus of TDP-43 promotes its oligomerization and enhances DNA binding affinity. Biochem. Biophys. Res. Commun. 425, 219–224. [DOI] [PubMed] [Google Scholar]
- Chou CC, Zhang Y, Umoh ME, Vaughan SW, Lorenzini I, Liu F, Sayegh M, Donlin-Asp PG, Chen YH, Duong DM, et al. (2018). TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci. 21, 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani G, Petosa C, Weis K, and Muller CW (1999). Structure of importin-beta bound to the IBB domain of importin-alpha. Nature 399, 221–229. [DOI] [PubMed] [Google Scholar]
- Colombrita C, Zennaro E, Fallini C, Weber M, Sommacal A, Buratti E, Silani V, and Ratti A (2009). TDP-43 is recruited to stress granules in conditions of oxidative insult. J. Neurochem. 111, 1051–1061. [DOI] [PubMed] [Google Scholar]
- Colombrita C, Onesto E, Megiorni F, Pizzuti A, Baralle FE, Buratti E, Silani V, and Ratti A (2012). TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells. J. Biol. Chem. 287, 15635–15647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E, and Kuriyan J (2000). Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure 8, 329–338. [DOI] [PubMed] [Google Scholar]
- Conti E, Uy M, Leighton L, Blobel G, and Kuriyan J (1998). Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 94, 193–204. [DOI] [PubMed] [Google Scholar]
- Coyne AN, Yamada SB, Siddegowda BB, Estes PS, Zaepfel BL, Johannesmeyer JS, Lockwood DB, Pham LT, Hart MP, Cassel JA, et al. (2015). Fragile X protein mitigates TDP-43 toxicity by remodeling RNA granules and restoring translation. Hum. Mol. Genet. 24, 6886–6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T, York D, and Pedersen L (1993). Particle mesh Ewald: an N•log(N) methodfor Ewald sums in large systems. J. Chem. Phys. 98, 10089. [Google Scholar]
- DeLano WL (2002). The PyMOL Molecular Graphics System (Schrödinger, LLC). Version 1.8. [Google Scholar]
- Eck RJ, Kraemer BC, and Liachko NF (2021). Regulation of TDP-43 phosphorylation in aging and disease. Geroscience 43, 1605–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elber R, Ruymgaart AP, and Hess B (2011). SHAKE parallelization. Eur. Phys. J. Spec. Top. 200, 211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Fanara P, Hodel MR, Corbett AH, and Hodel AE (2000). Quantitative analysis of nuclear localization signal (NLS)-importin alpha interaction through fluorescence depolarization. Evidence for auto-inhibitory regulation of NLS binding. J. Biol. Chem. 275, 21218–21223. [DOI] [PubMed] [Google Scholar]
- Fontes MR, Teh T, and Kobe B (2000). Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J. Mol. Biol. 297, 1183–1194. [DOI] [PubMed] [Google Scholar]
- Franke D, Petoukhov MV, Konarev PV, Panjkovich A, Tuukkanen A, Mertens HDT, Kikhney AG, Hajizadeh NR, Franklin JM, Jeffries CM, et al. (2017). ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 50, 1212–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Morato J, Hans F, von Zweydorf F, Feederle R, Elsasser SJ, Skodras AA, Gloeckner CJ, Buratti E, Neumann M, and Kahle PJ (2022). Sirtuin-1 sensitive lysine-136 acetylation drives phase separation and pathological aggregation of TDP-43. Nat. Commun. 13, 1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasset-Rosa F, Lu S, Yu H, Chen C, Melamed Z, Guo L, Shorter J, Da Cruz S, and Cleveland DW (2019). Cytoplasmic TDP-43 de-mixing independent of stress granules drives inhibition of nuclear import, loss of nuclear TDP-43, and cell death. Neuron 102, 339–357.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant TD (2018). Ab initio electron density determination directly from solution scattering data. Nat. Methods 15, 191–193. [DOI] [PubMed] [Google Scholar]
- Gruijs da Silva LA, Simonetti F, Hutten S, Riemenschneider H, Sternburg EL, Pietrek LM, Gebel J, Dotsch V, Edbauer D, Hummer G, et al. (2022). Disease-linked TDP-43 hyperphosphorylation suppresses TDP-43 condensation and aggregation. EMBO J. 41, e108443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gull S, and Daniell G (1978). Image reconstruction from incomplete and noisy data. Nature 272, 686–690. [Google Scholar]
- Guo L, Kim HJ, Wang H, Monaghan J, Freyermuth F, Sung JC, O’Donovan K, Fare CM, Diaz Z, Singh N, et al. (2018). Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell 173, 677–692.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Arai T, Nonaka T, Kametani F, Yoshida M, Hashizume Y, Beach TG, Buratti E, Baralle F, Morita M, et al. (2008). Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann. Neurol. 64, 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofweber M, Hutten S, Bourgeois B, Spreitzer E, Niedner-Boblenz A, Schifferer M, Ruepp MD, Simons M, Niessing D, Madl T, et al. (2018). Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173, 706–719.e13. [DOI] [PubMed] [Google Scholar]
- Homeyer N, and Gohlke H (2012). Free energy calculations by the molecular Mechanics Poisson-Boltzmann surface area method. Mol. Inform. 31, 114–122. [DOI] [PubMed] [Google Scholar]
- Hopkins JB, Gillilan RE, and Skou S (2017). BioXTAS RAW: improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J. Appl. Crystallogr. 50, 1545–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, de Groot BL, Grubmuller H, and MacKerell AD Jr. (2017). CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, and Schulten K (1996). VMD: visual molecular dynamics. J. Mol. Graph. 14, 27–38. [DOI] [PubMed] [Google Scholar]
- Hutten S, Usluer S, Bourgeois B, Simonetti F, Odeh HM, Fare CM, Czuppa M, Hruska-Plochan M, Hofweber M, Polymenidou M, et al. (2020). Nuclear import receptors directly bind to arginine-rich dipeptide repeat proteins and suppress their pathological interactions. Cell Rep. 33, 108538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang LL, Xue W, Hong JY, Zhang JT, Li MJ, Yu SN, He JH, and Hu HY (2017). The N-terminal dimerization is required for TDP-43 splicing activity. Sci. Rep. 7, 6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BS, McCaffery JM, Lindquist S, and Gitler AD (2008). A yeast TDP-43 proteinopathy model: exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc. Natl. Acad. Sci. USA 105, 6439–6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametani F, Nonaka T, Suzuki T, Arai T, Dohmae N, Akiyama H, and Hasegawa M (2009). Identification of casein kinase-1 phosphorylation sites on TDP-43. Biochem. Biophys. Res. Commun. 382, 405–409. [DOI] [PubMed] [Google Scholar]
- Kametani F, Obi T, Shishido T, Akatsu H, Murayama S, Saito Y, Yoshida M, and Hasegawa M (2016). Mass spectrometric analysis of accumulated TDP-43 in amyotrophic lateral sclerosis brains. Sci. Rep. 6, 23281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi B, LaClair KD, Riemenschneider H, Zhou Q, Frottin F, Mareljic N, Czuppa M, Farny D, Hartmann H, Michaelsen M, et al. (2020). Cell-to-cell transmission of C9orf72 poly-(Gly-Ala) triggers key features of ALS/FTD. EMBO J. 39, e102811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B (1999). Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat. Struct. Biol. 6, 388–397. [DOI] [PubMed] [Google Scholar]
- Kralt A, Jagalur NB, van den Boom V, Lokareddy RK, Steen A, Cingolani G, Fornerod M, and Veenhoff LM (2015). Conservation of inner nuclear membrane targeting sequences in mammalian Pom121 and yeast Heh2 membrane proteins. Mol. Biol. Cell 26, 3301–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, and Henrick K (2007). Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797. [DOI] [PubMed] [Google Scholar]
- Kuo PH, Chiang CH, Wang YT, Doudeva LG, and Yuan HS (2014). The crystal structure of TDP-43 RRM1-DNA complex reveals the specific recognition for UG- and TG-rich nucleic acids. Nucleic Acids Res. 42, 4712–4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurischko C, and Broach JR (2017). Phosphorylation and nuclear transit modulate the balance between normal function and terminal aggregation of the yeast RNA-binding protein Ssd1. Mol. Biol. Cell 28, 3057–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA (2009). PDBsum new things. Nucleic Acids Res. 37, D355–D359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue TM, Shah B, Ridgeway TM, and Pelletier SL (1992). Computer-aided interpretation of analytical sedimentation data for proteins. In Analytical Ultracentrifugation in Biochemistry and Polymer Science, Harding S, Rowe A, and Horton J, eds. (Royal Society of Chemistry; ), pp. 90–125. [Google Scholar]
- Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, and Chook YM (2006). Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 126, 543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Babinchak WM, and Surewicz WK (2021). Cryo-EM structure of amyloid fibrils formed by the entire low complexity domain of TDP-43. Nat. Commun. 12, 1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Moriarty NW, Poon BK, Sobolev OV, Terwilliger TC, and Adams PD (2017). Polder maps: improving OMIT maps by excluding bulk solvent. Acta Crystallogr. D Struct. Biol. 73, 148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, et al. (2019). Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu EA, Mori E, Hamasaki F, and Lieberman AP (2021). TDP-43 proteinopathy occurs independently of autophagic substrate accumulation and underlies nuclear defects in Niemann-Pick C disease. Neuropathol. Appl. Neurobiol. 7, 1019–1032. 10.1111/nan.12738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokareddy RK, Hapsari RA, van Rheenen M, Pumroy RA, Bhardwaj A, Steen A, Veenhoff LM, and Cingolani G (2015). Distinctive properties of the nuclear localization signals of inner nuclear membrane proteins Heh1 and Heh2. Structure 23, 1305–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott K, and Cingolani G (2011). The importin beta binding domain as a master regulator of nucleocytoplasmic transport. Biochim. Biophys. Acta 1813, 1578–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder P, Chu JF, Chatterjee B, Swamy KB, and Shen CJ (2016). Co-regulation of mRNA translation by TDP-43 and Fragile X Syndrome protein FMRP. Acta Neuropathol. 132, 721–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfori M, Mynott A, Ellis JJ, Mehdi AM, Saunders NF, Curmi PM, Forwood JK, Boden M, and Kobe B (2011). Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta 1813, 1562–1577. [DOI] [PubMed] [Google Scholar]
- Maris C, Dominguez C, and Allain FH (2005). The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 272, 2118–2131. [DOI] [PubMed] [Google Scholar]
- McCoy AJ (2007). Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BR 3rd, McGee TD Jr., Swails JM, Homeyer N, Gohlke H, and Roitberg AE (2012). MMPBSA.py: an efficient program for end-state free energy calculations. J. Chem. Theor. Comput. 8, 3314–3321. [DOI] [PubMed] [Google Scholar]
- Miyatake H, Sanjoh A, Unzai S, Matsuda G, Tatsumi Y, Miyamoto Y, Dohmae N, and Aida Y (2015). Crystal structure of human importin-alpha1 (Rch1), revealing a potential autoinhibition mode involving homodimerization. PLoS One 10, e0115995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mompean M, Romano V, Pantoja-Uceda D, Stuani C, Baralle FE, Buratti E, and Laurents DV (2016). The TDP-43 N-terminal domain structure at high resolution. FEBS J. 283, 1242–1260. [DOI] [PubMed] [Google Scholar]
- Mompean M, Romano V, Pantoja-Uceda D, Stuani C, Baralle FE, Buratti E, and Laurents DV (2017). Point mutations in the N-terminal domain of transactive response DNA-binding protein 43 kDa (TDP-43) compromise its stability, dimerization, and functions. J. Biol. Chem. 292, 11992–12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada R, Hirano H, and Matsuura Y (2015). Structure of importin-alpha bound to a non-classical nuclear localization signal of the influenza A virus nucleoprotein. Sci. Rep. 5, 15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardozzi JD, Lott K, and Cingolani G (2010). Phosphorylation meets nuclear import: a review. Cell Commun. Signal. 8, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura AL, Zupunski V, Troakes C, Kathe C, Fratta P, Howell M, Gallo JM, Hortobágyi T, Shaw CE, and Rogelj B (2010). Nuclear import impairment causes cytoplasmic trans-activation response DNA-binding protein accumulation and is associated with frontotemporal lobar degeneration. Brain 133, 1763–1771. [DOI] [PubMed] [Google Scholar]
- Nonaka T, Suzuki G, Tanaka Y, Kametani F, Hirai S, Okado H, Miyashita T, Saitoe M, Akiyama H, Masai H, et al. (2016). Phosphorylation of TAR DNA-binding protein of 43 kDa (TDP-43) by truncated casein kinase 1delta triggers mislocalization and accumulation of TDP-43. J. Biol. Chem. 291, 5473–5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, and Minor W (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. Macromolecular Crystallography. [DOI] [PubMed] [Google Scholar]
- Perez-Hernandez G, Paul F, Giorgino T, De Fabritiis G, and Noe F (2013). Identification of slow molecular order parameters for Markov model construction. J. Chem. Phys. 139, 015102. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Pinarbasi ES, Cağatay T, Fung HYJ, Li YC, Chook YM, and Thomas PJ (2018). Active nuclear import and passive nuclear export are the primary determinants of TDP-43 localization. Sci. Rep. 8, 7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polizzi NF, Therien MJ, and Beratan DN (2016). Mean first-passage times in Biology. Isr. J. Chem. 56, 816–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A, Bharathi V, Sivalingam V, Girdhar A, and Patel BK (2019). Molecular mechanisms of TDP-43 misfolding and pathology in amyotrophic lateral sclerosis. Front. Mol. Neurosci. 12, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumroy RA, and Cingolani G (2015). Diversification of importin-alpha isoforms in cellular trafficking and disease states. Biochem. J. 466, 13–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumroy RA, Ke S, Hart DJ, Zachariae U, and Cingolani G (2015). Molecular determinants for nuclear import of influenza A PB2 by importin alpha isoforms 3 and 7. Structure 23, 374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar S, Wang G, Randle SJ, Ruggeri FS, Varela JA, Lin JQ, Phillips EC, Miyashita A, Williams D, Strohl F, et al. (2018). FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-pi interactions. Cell 173, 720–734.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddick G, and Macara IG (2005). A systems analysis of importin-{alpha}-{beta} mediated nuclear protein import. J. Cell Biol. 168, 1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roblitz S, Stotzel C, Deuflhard P, Jones HM, Azulay DO, van der Graaf PH, and Martin SW (2013). A mathematical model of the human menstrual cycle for the administration of GnRH analogues. J. Theor. Biol. 321, 8–27. [DOI] [PubMed] [Google Scholar]
- Sankhala RS, Lokareddy RK, Begum S, Pumroy RA, Gillilan RE, and Cingolani G (2017). Three-dimensional context rather than NLS amino acid sequence determines importin alpha subtype specificity for RCC1. Nat. Commun. 8, 979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer MK, Trendelkamp-Schroer B, Paul F, Pérez-Hernández G, Hoffmann M, Plattner N, Wehmeyer C, Prinz JH, and Noé F (2015). PyEMMA 2: a software package for estimation, validation, and analysis of Markov models. J. Chem. Theor. Comput. 11, 5525–5542. [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, and Sali A (2010). FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 38, W540–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P (2000). Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sephton CF, Cenik C, Kucukural A, Dammer EB, Cenik B, Han Y, Dewey CM, Roth FP, Herz J, Peng J, et al. (2011). Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J. Biol. Chem. 286, 1204–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina Y, Arima K, Tabunoki H, and Satoh J (2010). TDP-43 dimerizes in human cells in culture. Cell. Mol. Neurobiol. 30, 641–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Kasavajhala K, Belfon KAA, Raguette L, Huang H, Migues AN, Bickel J, Wang Y, Pincay J, Wu Q, et al. (2020). ff19SB: amino-acid-specific protein backbone parameters trained against quantum Mechanics energy surfaces in solution. J. Chem. Theor. Comput. 16, 528–552. [DOI] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, König J, Hortobágyi T, Nishimura AL, Zupunski V, et al. (2011). Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 14, 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi PS, Choi KJ, Leonard PG, Sizovs A, Moosa MM, MacKenzie KR, Ferreon JC, and Ferreon ACM (2017). The N-terminal domain of ALS-linked TDP-43 assembles without misfolding. Angew. Chem. Int. Ed. Engl. 56, 12590–12593. [DOI] [PubMed] [Google Scholar]
- Wang YT, Kuo PH, Chiang CH, Liang JR, Chen YR, Wang S, Shen JC, and Yuan HS (2013). The truncated C-terminal RNA recognition motif of TDP-43 protein plays a key role in forming proteinaceous aggregates. J. Biol. Chem. 288, 9049–9057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Conicella AE, Schmidt HB, Martin EW, Rhoads SN, Reeb AN, Nourse A, Ramirez Montero D, Ryan VH, Rohatgi R, et al. (2018). A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J. 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winton MJ, Igaz LM, Wong MM, Kwong LK, Trojanowski JQ, and Lee VM (2008a). Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J. Biol. Chem. 283, 13302–13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winton MJ, Van Deerlin VM, Kwong LK, Yuan W, Wood EM, Yu CE, Schellenberg GD, Rademakers R, Caselli R, Karydas A, et al. (2008b). A90V TDP-43 variant results in the aberrant localization of TDP-43 in vitro. FEBS Lett. 582, 2252–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright GSA, Watanabe TF, Amporndanai K, Plotkin SS, Cashman NR, Antonyuk SV, and Hasnain SS (2020). Purification and structural characterization of aggregation-prone human TDP-43 involved in neurodegenerative diseases. iScience 23, 101159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Tan W, Whittle C, Qiu L, Cao L, Akbarian S, and Xu Z (2010). The C-terminal TDP-43 fragments have a high aggregation propensity and harm neurons by a dominant-negative mechanism. PLoS One 5, e15878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa T, Ali R, Jiou J, Fung HYJ, Burke KA, Kim SJ, Lin Y, Peeples WB, Saltzberg D, Soniat M, et al. (2018). Nuclear import receptor inhibits phase separation of FUS through binding to multiple sites. Cell 173, 693–705.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Xu YF, Cook C, Gendron TF, Roettges P, Link CD, Lin WL, Tong J, Castanedes-Casey M, Ash P, et al. (2009). Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc. Natl. Acad. Sci. USA 106, 7607–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The crystallographic coordinates for the TDP-43 NTD-NLS+ΔIBB-importin α1 complex have been deposited in the Protein Data Bank and raw X-ray diffraction data are posted on Mendeley Data and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| HRP conjugated anti-GFP polyclonal primary antibody | Millipore Sigma | Cat#GERPN1236; RRID:AB_771429 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| BL21-DE3 | Millipore Sigma | Cat#CMC0016 |
| BL21-DE3 LOBSTR | Kerafast | Cat#EC1002 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| ΔIBB-importin α1 construct | This paper | Cingolani_001 |
| Importin α1/β heterodimer | This paper | Cingolani_002 |
| GST-TDP-43 NTD-NLS-RRM1 wild-type | This paper | Cingolani_003 |
| GST-TDP-43 NTD-NLS-RRM1 T88E (pT88) | This paper | Cingolani_004 |
| GST-TDP-43 NTD-NLS-RRM1 S91E (pS91) | This paper | Cingolani_005 |
| GST-TDP-43 NTD-NLS-RRM1 S92E (pS92) | This paper | Cingolani_006 |
| GST-TDP-43 NTD-NLS-RRM1 T88E, S91E, S92E (pT88+pS91+pS92) | This paper | Cingolani_007 |
| GST-TDP-43 NTD-NLS | This paper | Cingolani_008 |
| GST-TDP-43 NLS-RRM1 | This paper | Cingolani_009 |
| Ampicillin | ThermoFisher Scientific | CAS#69-53-4 Cat#11593027 |
| Kanamycin sulfate | ThermoFisher Scientific | CAS#25389-94-0 Cat#15160054 |
| Chloramphenicol | ThermoFisher Scientific | CAS#56-75-7 Cat#227925000 |
| 3,3’,5,5’-Tetramethylbenzadine (TMB) | abcam | CAS#54827-17-7 Cat#ab171523 |
| Isopropylthio-β-galactoside (IPTG), >99% | ThermoFisher Scientific | CAS#367-93-1 Cat#15529019 |
| Sodium Chloride (NaCl), >99% | ThermoFisher Scientific | CAS#7647-14-5 Cat#A12313.0I |
| Tris base, >98% | ThermoFisher Scientific | CAS#77-86-1 Cat#J22675.36 |
| 2-mercaptoethanol (BME), 14.3 M, >99% | Millipore Sigma | CAS#60-24-2 Cat#M6250 |
| Tween-20 | Millipore Sigma | CAS#9005-64-5 Cat# P1379 |
| Hydrochloric acid (HCl), 12 N | ThermoFisher Scientific | CAS#7647-01-0 Cat#NC004378 |
| Dulbecco’s Phosphate buffered saline (PBS), 10× | Millipore Sigma | CAS#7647-14-5 Cat#D1408 |
| Ethylenedinitrilotetraacetic acid disodium salt (EDTA), >99%, | Millipore Sigma | CAS#6381-92-6 Cat#E5134 |
| Imidazole, >99% | Millipore Sigma | CAS#288-32-4 Cat#814223 |
| High density nickel resin | GoldBio | Cat#H-320-5 |
| GST agarose resin | GoldBio | Cat#G-250-5 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Pierce BCA Protein Assay Kit | ThermoFisher Scientific | Cat#23227 |
| Natrix Crystallization Screen | Hampton Research | HR2-116 |
| Natrix 2 Crystalization Screen | Hampton Research | HR2-117 |
| Crystal Screen | Hampton Research | HR2-110 |
| Index Screen | Hampton Research | HR2-144 |
|
| ||
| Deposited data | ||
|
| ||
| Raw and analyzed X-ray data for TDP-43 NTD-NLS+ΔIBB-importin α1 | This paper | Mendely Data: https://data.mendeley.com/datasets/mpkdxc4hsj/ |
| TDP-43 NTD-NLS+ΔIBB-importin α1 PDB | This paper | PDB: 7N9H |
|
| ||
| Recombinant DNA | ||
|
| ||
| Vector: pET-30a | EMD biosciences/Millipore Sigma | Cat#69909 |
| Vector: pGEX-6p-1 | Amersham/Millipore Sigma | Cat# GE28-9546-48 |
| Vector: pETduet-1 | EDM biosciences/Millipore Sigma | Cat#71146 |
|
| ||
| Software and algorithms | ||
|
| ||
| ImageJ | (Abramoff et al., 2004) | N/A |
| PyMol | (DeLano, 2002) | N/A |
| HKL2000 | (Otwinowski and Minor, 1997) | N/A |
| Phaser | (McCoy, 2007) | N/A |
| Phenix.refine | (Liebschner et al., 2019) | Version 1.19.2 |
| Phenix.polder | (Liebschner et al., 2017) | N/A |
| COOT | (Emsley and Cowtan, 2004) | N/A |
| ATSAS | (Franke et al., 2017) | N/A |
| RAW | (Hopkins et al., 2017) | Version 2.1.1 |
| DENSS | (Grant, 2018) | N/A |
| Chimera | (Pettersen et al., 2004) | N/A |
| FoXS Server | (Schneidman-Duhovny et al., 2010) | N/A |
| GraphPad Prism | Non-linear regression: one-site specific binding curve | Version 9.0.0 |
| Chromlab | Bio-Rad | N/A |
| SEDFIT | (Schuck, 2000) | Center for Information Technology, NIH |
| SEDNTERP | (Laue et al., 1992) | N/A |
| GUSSI | UTSouthwestern Medical Center | N/A |
| PDBsum | (Laskowski, 2009) | N/A |
| PISA | (Krissinel and Henrick, 2007) | N/A |
| Pyemma | (Scherer et al., 2015) | Version 2.5.7 |
| Amber | (D.A. Case et al., 2018) | Version 18 |
| SHAKE algorithm | (Elber et al., 2011) | N/A |
| PCCA++ algorithm | (Roblitz et al., 2013) | N/A |
| VMD | (Humphrey et al., 1996) | N/A |


