Abstract
Background:
MC1R polymorphisms interact with CDKN2A mutations modulating melanoma risk and contribute to a less suspicious clinical and dermoscopic appearance of melanomas. Different strategies, including dermoscopic comparative approach and digital monitoring, are used for the melanoma diagnosis in this context.
Objective:
To analyse the diagnostic accuracy of the morphologic approach and comparative approach in dermoscopy, to detect melanoma in familial melanoma (FamMM) patients according to different genetic backgrounds.
Methods:
Two independent readers evaluated 415 lesions belonging to 25 FamMM: 26 melanomas (62% in situ, 36% early invasive) and 389 nevi, blinded for dermoscopic and histopathologic diagnosis, following two different steps. First step-Randomized: all lesions were randomly located in one single folder. Second step-Comparative approach: the lesions were clustered by patient. Sensitivity, specificity, and number needed to excise (NNE) for melanoma diagnosis were calculated for both diagnostic strategies. Sensitivity and specificity were also assessed regarding the genetic background.
Results:
The comparative approach showed lower sensitivity compared to the morphologic approach (69.2 and 73.1 vs. 76.9 both readers) but better specificity (95.9 and 95.1 vs. 84.3 and 90.2, respectively). NNE was better in the comparative approach. The readers had more difficulties diagnosing lesions from CDKN2A mutation carriers with red hair colour (RHC) MC1R variants.
Conclusion:
The comparative approach can be useful in high risk patients to decrease the NNE. Early melanomas in CDKN2A carriers with RHC polymorphisms are more difficult to diagnose even with the comparative approach and benefit from the detection of changes during digital dermoscopy monitoring for early diagnosis.
Keywords: melanoma, genetics, dermoscopy, familial melanoma, nevi
Introduction
Melanoma is characterized by a complex aetiology, involving both genetic and environmental risk factors. It is estimated that 10% of cases of melanomas occur in a familial setting as an autosomal dominant trait with incomplete penetrance. In approximately 20–40% of these familial melanoma (FamMM) cases, germline mutations in high-risk genes can be found, with Cyclin Dependent Kinase Inhibitor 2A (CDKN2A) being the major susceptibility gene.1 High-risk germline mutations can also be found in around 10% of sporadic patients with multiple primary melanomas.2–4 Polymorphisms in the Melanocortin 1 Receptor (MC1R) gene are considered as medium susceptibility variants for melanoma development, increasing melanoma risk by up to 10 times compared with wild-type.5
Dermoscopy represents the gold standard for the management of patients with skin tumours because it improves the diagnostic accuracy and early diagnosis of melanoma compared with the unaided eye and permits the differentiation of melanomas from the myriads of nevi.6–9
The diagnosis of melanomas in a given patient is based on two different strategies: i) morphology-based recognition which implies the assessment of the clinical and dermoscopic features of a given lesion;6 ii) the comparative approach in which individual lesions (i.e. nevi) are evaluated in the context of a patient’s overall nevus profile (i.e. signature pattern) and this is especially valuable for patients with multiple nevi.10–13
It is well known that the interaction between medium-risk MC1R gene variants among FamMM cases can increase the genetic risk in CDKN2A mutation carriers by up to 14 times and then contribute to a less suspicious clinical and dermoscopic appearance of melanomas14–16 and larger benign nevi.17 Thus, a correct management of these patients will include more diagnostic strategies although little is known about the value of the comparative approach in diagnosing all melanomas and the avoidance of unnecessary surgical excisions of nevi.
The aim of our study was to determine whether the comparative approach is a useful method to detect melanoma in patients belonging to FamMM compared to the dermoscopic evaluation without knowing the patient nevus type. Moreover, the predominant dermoscopic patterns of nevi and melanoma in individual FamMM patients were also analysed.
Materials and Methods
In this study 25 FamMM patients (according to the rule of 2)18 referred to the Melanoma Unit of Barcelona, Spain, were included. Demographic data (sex, age, skin phototype) as well as the mutation status of CDKN2A and MC1R were reported. DNA obtained from peripheral blood lymphocytes was available for all FamMM patients. Exons 1 alpha, 1 beta, 2 and 3, intronic variant IVS2–105 and the c.−34G>T variant of CDKN2A, and exon 2 from CDK4 were sequenced as previously referenced.19 MC1R was sequenced following the protocol previously described.17 Dermoscopic images had been captured with Dermlite Foto Equipment (3Gen, LLC, Dana Point, California, USA) at 10-fold magnification.
Dermoscopic images of all melanomas and nevi belonging to each patient were included. Two independent Readers (C.L, A.M), dermatologists with 10 years-training in dermoscopy and not involved in the health care of these patients, evaluated all lesions following two different steps:
First step-Randomized (morphologic approach): the Readers, blinded for the dermoscopic and histopathologic diagnosis, evaluated all lesions, melanomas and nevi from all 25 FamMM patients randomly located in one single folder.
Second step-Comparative approach: the Readers, blinded for the dermoscopic and histopathologic diagnosis, evaluated the lesions that were clustered patient by patient (each folder contained all nevi and melanomas belonging to a given patient) for a total of 25 folders belonging to 25 patients. The Readers evaluated the series of images individually, with an interval of 8 weeks between the 2 steps of the study to avoid recall bias.
For each step, the Readers were asked to formulate a diagnosis (melanoma: yes or no) and to give management recommendations (excision or follow-up).
Dermoscopic characteristics of melanomas and the main nevus pattern (reticular; globular; homogeneous; complex pattern) for each patient were evaluated.
The local Institutional Review Board approved the study and all clinical investigation was conducted according to the Declaration of Helsinki Principles and was performed following Good Clinical Practice standards. Patients were given an Informed Consent Document concerning their agreement.
Statistics
All statistical calculations were made with the SPSS 19.0 (Statistical Package for Social Sciences, SPSS Inc., Chicago, Ill.). Sensitivity, specificity, Receiver Operating characteristic (ROC) curves and area under the curve (AUC), positive predictive value, negative predictive value for each Reader and for each step was calculated. Absolute and relative frequencies of dermoscopic pattern were provided. Number needed to excise (NNE) was also calculated as: (TOTAL lesions excised)/(melanoma diagnosis). Additionally, sensitivity, specificity, ROC and AUC were calculated for each reader and for each step according to the genetic status of CDKN2A and MC1R.
Results
Study population
Seven males and 18 females with a mean age of 51.25 years old (range 25–85) at the study inclusion were included in our study. Table 1 lists the main clinical and genetic data.
Table 1.
Demographic and genetic mutational status of patients.
| Patient | Sex | Age | CDKN2A | MC1R* | Skin phototype | Number of primary melanomas# | 1st or 2nd degree relatives with melanoma |
|---|---|---|---|---|---|---|---|
| 1 | female | 30 | wild-type | R163Q | 2 | 1 | 1 |
| 2 | male | 68 | wild-type | R160W | 1 | 1 | 1 |
| 3 | female | 85 | wild-type | M128T | 2 | 1 | 1 |
| 4 | male | 25 | wild-type | D294H | 2 | 1 | 1 |
| 5 | female | 56 | wild-type | V60L | 2 | 1 | 1 |
| 6 | female | 59 | G101W | D294H | 3 | 2 | 1 |
| 7 | female | 55 | wild-type | D294H | 1 | 4 | 0 |
| 8 | male | 40 | G101W | V60L/R160W | 2 | 4 | 3 |
| 9 | male | 39 | wild-type | R142H | 3 | 2 | 2 |
| 10 | female | 63 | wild-type | wild-type | 2 | 1 | 2 |
| 11 | female | 44 | wild-type | V60L | 3 | 1 | 1 |
| 12 | male | 46 | wild-type | V60L | 3 | 1 | 1 |
| 13 | female | 59 | wild-type | D294H | 2 | 2 | 2 |
| 14 | female | 56 | V59G | wild-type | 2 | 3 | 1 |
| 15 | male | 83 | wild-type | R163Q | 2 | 6 | 1 |
| 16 | female | 38 | wild-type | D294H | 2 | 4 | 1 |
| 17 | female | 33 | G101W | D294H | 2 | 7 | 1 |
| 18 | female | 76 | G101W | wild-type | 3 | 4 | 2 |
| 19 | female | 41 | G101W | R163Q | 3 | 2 | 2 |
| 20 | female | 57 | wild-type | V92M | 2 | 2 | 1 |
| 21 | female | 29 | G101W | R151C/R160W | 2 | 8 | 1 |
| 22 | female | 52 | G101W | R151C/D294H | 2 | 9 | 1 |
| 23 | male | 61 | G101W | R151C | 2 | 4 | 2 |
| 24 | female | 34 | Q50R | V60L | 3 | 1 | 2 |
| 25 | female | 54 | wild-type | D294H | 2 | 1 | 1 |
Synonymous variants are not listed. All variants were found in heterozygosis.
This number corresponds to all melanomas diagnosed to date
Patients 6 and 17, 18 and 19, and 21 and 22 belong to the same melanoma family, respectively.
A total of 415 lesions including 26 melanomas and 389 nevi belonging to 25 FamMM patients (mean number of images of 16.6 per patient) were analysed. Example images of nevi and melanoma from three patients are included in figures 1–3. Sixty-two percent of the melanomas included were in situ, the mean Breslow thickness of invasive melanomas was 0.83 mm (range 0.21–1.9). Patients were followed-up for 10 years after the study. A total of 18 new melanomas were diagnosed in 11 of the patients. The histopathology characteristics of all melanomas are found in Table S1. The nevi included in the study remained stable throughout 10 years of follow-up.
Figure 1.
Dermoscopy of 9 of the 11 lesions included from patient 4, CDKN2A wild-type and carrier of one MC1R RHC variant. Lesion e is the melanoma. Both experts diagnosed it correctly as melanoma.
Figure 3.
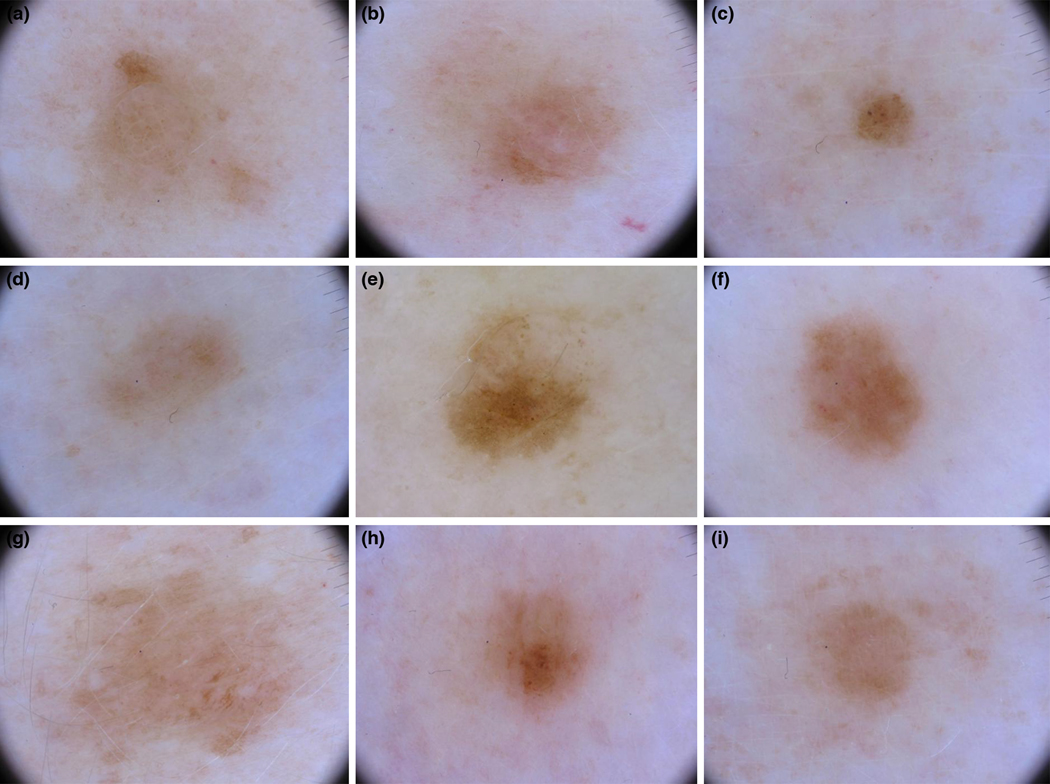
Dermoscopy of 9 of the 17 lesions included from patient 22, carrier of a CDKN2A mutation and two MC1R RHC variants. Lesion e is melanoma. None of the experts diagnosed it as melanoma with a morphological approach but R2 identified it as suspicious of melanoma with the comparative approach.
Dermoscopically, melanomas were frequently typified by the presence of reticular pattern in 46% of cases, reticulo-homogeneous in 19% of cases, multicomponent in 15% of cases, globular in 8% of cases and reticular-globular or homogeneous or structureless in one case each (4%). Additional clues were the presence of red structureless areas or dotted vessels in four cases (15%). Patterns in nevi were similar with predominance of reticular pattern present in 96% of patients, presence of homogeneous pattern in 32% of patients and globular pattern in 12% of patients. Only one patient had no reticular pattern nevi, presenting mainly homogeneous nevi (Fig. S1). Multicomponent pattern was not a predominant nevus pattern in any patient.
With the morphologic approach, both Readers showed a sensitivity of 76.9% and a specificity of 84.3% and 90.2%, respectively, for excision of melanomas. The positive predictive value was 24.7 and 34.5 and the negative predictive value was 98.2 and 98.3, for readers 1 and 2 respectively. When using the comparative approach, specificity was higher for both Readers compared with the morphologic approach, while sensitivity was almost similar (Table 2, Fig. S2) for melanoma excision. The positive predictive value was higher 52.9 and 50.0 and the negative predictive value was 97.9 and 98.1, for readers 1 and 2 respectively. Notably, the NNE improved when using the comparative approach compared with the morphologic approach for both Readers, improving from 1:4.1 to 1:1.9 for Reader 1 and from 1:2.9 to 1:2 for Reader 2, respectively.
Table 2.
Diagnostic accuracy of Readers 1 and 2 when using the morphologic and comparative approach, respectively, and according to the germline genetic status of the patients. SN = Sensitivity; SP = Specificity; AUC = Area Under the Curve (in ROC analysis). MC1R alleles: R = variant strongly associated with the red hair colour phenotype or RHC; r = variant low-associated with RHC; - = wild-type.
| MORPHOLOGY | COMPARISON | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GOLD STANDARD | READER 1 | READER 2 | READER 1 | READER 2 | |||||||||||
| Melanoma | Nevi | SN | SP | AUC | SN | SP | AUC | SN | SP | AUC | SN | SP | AUC | ||
| ALL lesions | 26 | 389 | 76.9% | 84.3% | 0.806 | 76.9% | 90.2% | 0.826 | 69.2% | 95.9% | 0.836 | 73.1% | 95.1% | 0.841 | |
| CDKN2A status | MC1R genotype | Melanoma | Nevi | SN | SP | AUC | SN | SP | AUC | SN | SP | AUC | SN | SP | AUC |
| Mutated | ALL | 11 | 173 | 81.8% | 80.9% | 0.814 | 72.7% | 90.2% | 0.815 | 72.7% | 96.5% | 0.846 | 72.7% | 94.8% | 0.838 |
| Wild-type | ALL | 15 | 216 | 73.3% | 87.0% | 0.802 | 80.0% | 90.3% | 0.851 | 66.7% | 95.4% | 0.810 | 73.3% | 95.4% | 0.844 |
| ALL | RR, Rr,R- | 14 | 225 | 78.6% | 84.0% | 0.813 | 64.4% | 89.8% | 0.770 | 64.3% | 96.9% | 0.806 | 57.1% | 95.1% | 0.761 |
| ALL | rr, r-,-- | 12 | 164 | 75.0% | 84.8% | 0.799 | 91.7% | 90.9% | 0.913 | 75.0% | 94.5% | 0.848 | 91.7% | 95.1% | 0.934 |
| Mutated | RR, Rr,R- | 7 | 104 | 71.4% | 77.9% | 0.747 | 57.1% | 87.5% | 0.723 | 57.1% | 97.1% | 0.771 | 57.1% | 94.2% | 0.757 |
| Mutated | rr, r-,-- | 4 | 69 | 100% | 85.5% | 0.928 | 100% | 94.2% | 0.971 | 100% | 95.7% | 0.978 | 100% | 95.7% | 0.978 |
| Wild-type | RR, Rr,R- | 7 | 121 | 85.7% | 89.3% | 0.875 | 71.4% | 91.7% | 0.816 | 71.4% | 96.7% | 0.841 | 57.1% | 95.9% | 0.765 |
| Wild-type | rr, r-,-- | 8 | 95 | 62.5% | 84.2% | 0.734 | 87.5% | 88.4% | 0.880 | 62.5% | 93.7% | 0.781 | 87.5% | 94.7% | 0.911 |
Sensitivity and specificity varied according to the genetic background of the patients assessed (Table 2). Both readers had more difficulties correctly diagnosing lesions from CDKN2A mutation carriers with MC1R red hair colour (RHC) associated variants. No differences were observed in CDKN2A mutation carriers and wild-type patients when not considering MC1R status. However, we observed that Reader 2 also had more difficulties correctly diagnosing lesions from MC1R RHC variant carriers, independently of the CDKN2A mutation status.
Discussion
Diagnosing melanoma is always a complex and multifactorial process that is further challenging when dealing with individuals at increased risk of developing cancer such as FamMM. The optimal management of these patients relies on the identification of all melanomas while avoiding the excision of benign nevi. However, FamMM patients harbour distinct phenotypic traits that render them a special subset of individuals with a peculiar morphology of nevi and melanoma.14–17, 20 Strategies to recognize a given melanoma in high-risk patients should consider the variability of nevi and the deviation from the common nevus pattern usually encountered when screening general population. Herein, we assessed the value of a “comparative” approach in which a comparison of equivocal lesions with a patient’s other nevi is confronted with the baseline purely morphologic strategy.
In our study, the comparative approach dramatically increased the specificity for both Readers showing 95% value. However, sensitivity was lower for the comparative approach.
Argenziano and colleagues have tested the use of the comparative approach in patients with multiple nevi but not genetic susceptibility.10 In their study, the authors found that the overall NNE for 6 dermoscopists decreased from 52.3 using the morphologic approach (single lesion) to 13.4 using the comparative approach in patients with multiple nevi.
Our data, analysing FamMM patients, confirmed that the systematic use of the comparative approach would translate into a reduction of surgical excision of benign nevi because a higher specificity can be reached. However, sensitivity could decrease when using a comparative approach. This could be explained by the fact that, in these patients, melanoma are quite inconspicuous and often display dermoscopic features that overlap with nevi, such as the presence of delicate network and structureless areas. Also, the fact of having a high proportion of in situ and very early melanoma may influence the diagnostic sensitivity.
Thus, in the context of high-risk patients, the use of the comparative approach, although valid and powerful, should be paralleled with the systematic use of digital dermoscopic monitoring that might reveal subtle changes occurring over time that aid the recognition of melanoma that are featureless at the baseline visit. The use of reflectance confocal microscopy, 21–23 when available, should be encouraged for any atypical lesion/s identified by means of the comparative approach or during digital monitoring.
In a previous study we observed that suspicious melanoma lesions from CDKN2A mutation carriers with MC1R RHC variants had lower ABCD total dermoscopy score.16 In this study, we also confirmed the difficulties to correctly diagnose melanomas in CDKN2A carriers with MC1R RHC variants. Moreover, one reader had more difficulties in lesions from RHC carriers, independently of the CDKN2A status. This could be partially explained by the different geographic area of origin of this reader where RHC variants are less prevalent.
Furthermore, multiple new melanomas have been diagnosed in the following 10 years after the last lesion was evaluated. Those melanomas were not previously included as nevi in our study but detected as new lesions. Thus, in these patients it is also necessary to perform total body photography for map comparison and not only focus on the digital follow-up of single lesions.24 The limitations of our study include the retrospective design and the relatively small number of patients, although it refers to a special cohort of subjects with 10 years follow-up. Furthermore, another limitation was the fact that analysis was carried out by highly experienced readers although it is well known that FamMM patients require a high level of expertise and dedicated physicians for their management. Additionally, in single lesion teleconsultation in this group of patients the accuracy is limited.
To conclude, our findings demonstrate that even the comparative approach is preferable to the sole use of morphologic analysis in the context of high-risk patients, especially FamMM patients, digital dermoscopy, total body photography and total body examination to not miss melanoma. Red hair polymorphisms in MC1R increase the difficulties in detecting melanomas. Further imaging devices such as in vivo confocal microscopy need to be tested in this highly selected narrow population to improve melanoma diagnostic accuracy.
Supplementary Material
Supplementary Figure 1. Dermoscopy images of 4 of the 4 lesions included from patient 10, wild type for CDKN2A and MC1R. Lesion a was a melanoma. None of the experts diagnosed it as melanoma with a morphological approach but R2 identified it as suspicious of melanoma with the comparative approach.
Supplementary Figure 2. ROC curves showing sensitivity and specificity of melanoma identification according to readers 1 and 2 (R1, R2), both approaches (morphology and comparison) and germline genetic status of patient (CDKN2A and MC1R).
Figure 2.
Dermoscopy of 9 of the 19 lesions included from patient 7, CDKN2A wild-type and carrier of one MC1R RHC variant. Lesion e is melanoma. None of the experts diagnosed it as melanoma.
Acknowledgements
Thanks to our patients and their families who are the main reason for our studies; to nurses from the Melanoma Unit of Hospital Clínic of Barcelona, Daniel Gabriel, Pablo Iglesias, Mireia Domínguez and Maria E Moliner for helping to collect patient data, and to Judit Mateu from the “Melanoma: image, genetics and immunology” group at IDIBAPS for her technical assistance.
Funding source:
The research at the Melanoma Unit in Barcelona is partially funded by Spanish Fondo de Investigaciones Sanitarias grants PI15/00716 and PI15/00956; by the grant AC16/00081, integrated in the Plan Estatal I+D+I, IMMUSPHINX – Transcan-2; CIBER de Enfermedades Raras of the Instituto de Salud Carlos III, Spain, co-funded by ISCIII-Subdirección General de Evaluación and European Regional Development Fund (ERDF), “a way to build Europe”; AGAUR 2017_SGR_1134 of the Catalan Government, Spain; European Commission under the 6th Framework Programme, Contract No. LSHC-CT-2006–018702 (GenoMEL) and by the European Commission under the 7th Framework Programme, Diagnoptics; The National Cancer Institute (NCI) of the US National Institute of Health (NIH) (CA83115); a grant from “Fundación Científica de la Asociación Española Contra el Cáncer” GCB15152978SOEN, Spain, and CERCA Programme / Generalitat de Catalunya. Part of the work was developed at the Centro Esther Koplowitz building, Barcelona.
Footnotes
Conflict of interest: none
References
- 1.Potrony M, Badenas C, Aguilera P, et al. Update in genetic susceptibility in melanoma. Ann Transl Med 2015;3:210doi: 10.3978/j.issn.2305-5839.2015.08.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puig S, Malvehy J, Badenas C, et al. Role of the CDKN2A locus in patients with multiple primary melanomas. J Clin Oncol 2005;23:3043–51doi: 10.1200/JCO.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 3.Puig S, Potrony M, Cuellar F, et al. Characterization of individuals at high risk of developing melanoma in Latin America: bases for genetic counseling in melanoma. Genet Med 2016;18:727–36doi: 10.1038/gim.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auroy S, Avril MF, Chompret A, et al. Sporadic multiple primary melanoma cases: CDKN2A germline mutations with a founder effect. Genes Chromosomes Cancer 2001;32:195–202http://www.ncbi.nlm.nih.gov/pubmed/11579459. Accessed 3 Jul 2019. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein AM, Chaudru V, Ghiorzo P, et al. Cutaneous phenotype and MC1R variants as modifying factors for the development of melanoma in CDKN2A G101W mutation carriers from 4 countries. Int J cancer 2007;121:825–31doi: 10.1002/ijc.22712. [DOI] [PubMed] [Google Scholar]
- 6.Argenziano G, Soyer HP, Chimenti S, et al. Dermoscopy of pigmented skin lesions: results of a consensus meeting via the Internet. J Am Acad Dermatol 2003;48:679–93doi: 10.1067/mjd.2003.281. [DOI] [PubMed] [Google Scholar]
- 7.Thomas L, Puig S. Dermoscopy, Digital Dermoscopy and Other Diagnostic Tools in the Early Detection of Melanoma and Follow-up of High-risk Skin Cancer Patients. Acta Derm Venereol 2017;Suppl 218:14–21doi: 10.2340/00015555-2719. [DOI] [PubMed] [Google Scholar]
- 8.Kittler H, Marghoob AA, Argenziano G, et al. Standardization of terminology in dermoscopy/dermatoscopy: Results of the third consensus conference of the International Society of Dermoscopy. J Am Acad Dermatol 2016;74:1093–106doi: 10.1016/j.jaad.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrera C, Marchetti MA, Dusza SW, et al. Validity and Reliability of Dermoscopic Criteria Used to Differentiate Nevi From Melanoma: A Web-Based International Dermoscopy Society Study. JAMA dermatology 2016;152:798–806doi: 10.1001/jamadermatol.2016.0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Argenziano G, Catricalà C, Ardigo M, et al. Dermoscopy of patients with multiple nevi: Improved management recommendations using a comparative diagnostic approach. Arch Dermatol 2011;147:46–9doi: 10.1001/archdermatol.2010.389. [DOI] [PubMed] [Google Scholar]
- 11.Gaudy-Marqueste C, Wazaefi Y, Bruneu Y, et al. Ugly duckling sign as a major factor of efficiency in melanoma detection. JAMA Dermatology 2017;153:279–84. [DOI] [PubMed] [Google Scholar]
- 12.Grob JJ. The “Ugly Duckling” Sign: Identification of the Common Characteristics of Nevi in an Individual as a Basis for Melanoma Screening. Arch Dermatol 1998;134:103-a-104doi: 10.1001/archderm.134.1.103-a. [DOI] [PubMed] [Google Scholar]
- 13.Scope A, Dusza SW, Halpern AC, et al. The “ugly duckling” sign: Agreement between observers. Arch Dermatol 2008;144:58–64. [DOI] [PubMed] [Google Scholar]
- 14.Carrera C, Palou J, Malvehy J, et al. Early stages of melanoma on the limbs of high-risk patients: clinical, dermoscopic, reflectance confocal microscopy and histopathological characterization for improved recognition. Acta Derm Venereol 2011;91:137–46doi: 10.2340/00015555-1021. [DOI] [PubMed] [Google Scholar]
- 15.Bassoli S, Maurichi A, Rodolfo M, et al. CDKN2A and MC1R variants influence dermoscopic and confocal features of benign melanocytic lesions in multiple melanoma patients. Exp Dermatol 2013;22:411–6doi: 10.1111/exd.12168. [DOI] [PubMed] [Google Scholar]
- 16.Cuéllar F, Puig S, Kolm I, et al. Dermoscopic features of melanomas associated with MC1R variants in Spanish CDKN2A mutation carriers. Br J Dermatol 2009;160:48–53doi: 10.1111/j.1365-2133.2008.08826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vallone MG, Tell-Marti G, Potrony M, et al. Melanocortin 1 receptor (MC1R) polymorphisms’ influence on size and dermoscopic features of nevi. Pigment Cell Melanoma Res 2018;31:39–50doi: 10.1111/pcmr.12646. [DOI] [PubMed] [Google Scholar]
- 18.Leachman SA, Carucci J, Kohlmann W, et al. Selection criteria for genetic assessment of patients with familial melanoma. J Am Acad Dermatol 2009;61:677.e1–14doi: 10.1016/j.jaad.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Potrony M, Puig-Butillé JA, Aguilera P, et al. Increased prevalence of lung, breast, and pancreatic cancers in addition to melanoma risk in families bearing the cyclin-dependent kinase inhibitor 2A mutation: implications for genetic counseling. J Am Acad Dermatol 2014;71:888–95doi: 10.1016/j.jaad.2014.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fargnoli MC, Sera F, Suppa M, et al. Dermoscopic features of cutaneous melanoma are associated with clinical characteristics of patients and tumours and with MC1R genotype. J Eur Acad Dermatol Venereol 2014;28:1768–75doi: 10.1111/jdv.12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alarcon I, Carrera C, Palou J, Alos L, Malvehy J, Puig S. Impact of in vivo reflectance confocal microscopy on the number needed to treat melanoma in doubtful lesions. Br J Dermatol 2014;170:802–8doi: 10.1111/bjd.12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pellacani G, Pepe P, Casari A, Longo C. Reflectance confocal microscopy as a second-level examination in skin oncology improves diagnostic accuracy and saves unnecessary excisions: a longitudinal prospective study. Br J Dermatol 2014;171:1044–51doi: 10.1111/bjd.13148. [DOI] [PubMed] [Google Scholar]
- 23.Borsari S, Pampena R, Lallas A, et al. Clinical Indications for Use of Reflectance Confocal Microscopy for Skin Cancer Diagnosis. JAMA dermatology 2016;152:1093–8doi: 10.1001/jamadermatol.2016.1188. [DOI] [PubMed] [Google Scholar]
- 24.Salerni G, Carrera C, Lovatto L, et al. Benefits of total body photography and digital dermatoscopy ("two-step method of digital follow-up") in the early diagnosis of melanoma in patients at high risk for melanoma. J Am Acad Dermatol 2012;67:e17–27doi: 10.1016/j.jaad.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Dermoscopy images of 4 of the 4 lesions included from patient 10, wild type for CDKN2A and MC1R. Lesion a was a melanoma. None of the experts diagnosed it as melanoma with a morphological approach but R2 identified it as suspicious of melanoma with the comparative approach.
Supplementary Figure 2. ROC curves showing sensitivity and specificity of melanoma identification according to readers 1 and 2 (R1, R2), both approaches (morphology and comparison) and germline genetic status of patient (CDKN2A and MC1R).