Abstract
The structure, stability, and function of various coding and noncoding RNAs are influenced by chemical modifications. Pseudouridine (Ψ) is one of the most abundant post‐transcriptional RNA base modifications and has been detected at individual positions in tRNAs, rRNAs, mRNAs, and snRNAs, which are referred to as Ψ‐sites. By allowing formation of additional bonds with neighboring atoms, Ψ strengthens RNA–RNA and RNA–protein interactions. Although many aspects of the underlying modification reactions remain unclear, the advent of new transcriptome‐wide methods to quantitatively detect Ψ‐sites has recently changed our perception of the functional roles and importance of Ψ. For instance, it is now clear that the occurrence of Ψs appears to be directly linked to the lifetime and the translation efficiency of a given mRNA molecule. Furthermore, the administration of Ψ‐containing RNAs reduces innate immune responses, which appears strikingly advantageous for the development of generations of mRNA‐based vaccines. In this review, we aim to comprehensively summarize recent discoveries that highlight the impact of Ψ on various types of RNAs and outline possible novel biomedical applications of Ψ.
Keywords: epitranscriptome, mRNA, mRNA vaccine, pseudouridine, RNA modification, rRNA, snoRNA, snRNA, translation, tRNA
Abbreviations
A, adenine
ASL, anticodon stem and loop
C, cytosine
CDS, coding sequence
CMC, N‐cyclohexyl‐N′‐β‐(4‐methylmorpholinium) ethylcarbodiimide p‐tosylate
DKC1, dyskeratosis congenita 1
G, guanine
LSU, large subunit
miRNA, microRNA
MRT55, mental retardation
mt‐tRNA, mitochondrial tRNAs
NAP57, nucleolar‐associated protein 57
Prp5, pre‐mRNA‐processing ATP‐dependent RNA helicase PRP5
PUS, pseudouridine synthase
RluA, ribosomal large subunit pseudouridine synthase A
RsuA, ribosomal small subunit pseudouridine synthase A
SECIS, selenocysteine insertion sequence
snRNA, small nuclear RNA
SSU, small subunit
TruA, tRNA pseudouridine synthase A
TruB, tRNA pseudouridine synthase B
TruD, tRNA pseudouridine synthase D
U, uracil
Ψ, pseudouridine
Although RNA molecules consist of only four standard nucleotides, namely adenine (A), cytosine (C), guanine (G), and uracil (U), the incorporation of chemical RNA base modifications greatly expands their structural diversity [1]. To date, over 170 different RNA modifications have been identified and pseudouridine (Ψ), an isoform of U, is the most abundant decoration on RNAs [2] in all three domains of life—it is sometimes even referred to as the fifth RNA nucleoside [3, 4]. Ψ was the first discovered modification in RNA [5], and it is the product of the post‐transcriptional isomerization of incorporated uridines. In detail, the uracil base is repositioned, and therefore, the canonical carbon–nitrogen glycosidic bond (C1–N1) connecting it to the ribose is replaced by a carbon–carbon bond (C1–C5) [6, 7] (Fig. 1). The free N1‐H position of Ψ expands base stacking possibilities at the ‘Hoogsteen’ edge to facilitate folding, proper secondary structure formation, rigidity, and stability in single‐stranded and duplex regions [8, 9, 10]. Though recent structural simulations suggest that the base stacking effect could also be context dependent and governed by neighboring sequences [11], many Ψ‐sites in structured RNAs are conserved across species to stabilize essential RNA duplexes by forming Ψ‐A, Ψ‐G, Ψ‐U, and Ψ‐C pairs and enhance RNA–protein interactions.
Fig. 1.
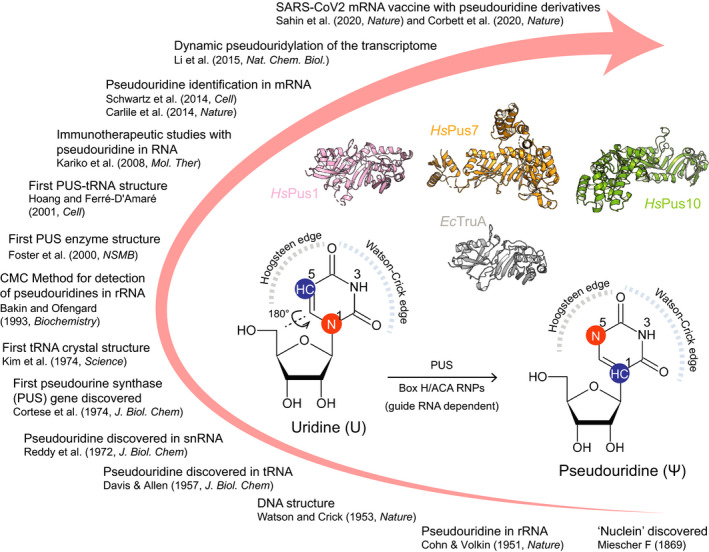
Timeline of selected key events in the history of Ψ research. The inset shows details of conversion reaction from uridine to Ψ. Representative crystal structures of PUS enzymes [EcTruA (PDB: 2NQP); HsPus1 (PDB: 4ITS); HsPus7 (PDB: 5KKP); HsPus10 (PDB: 1V9K)] are shown in cartoon representation.
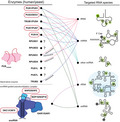
The formation of Ψ is catalyzed by a group of enzymes called pseudouridine synthases (PUS). This functional family of enzymes can be further subcategorized into two separate classes, which promote the pseudouridylation reaction in completely different fashions. (a) RNA guide‐dependent enzymes rely on a guide RNAs that are complimentary to the specific target site, whereas (b) stand‐alone enzymes do not require a guide RNA and act like canonical enzymes [12] (Fig. 2). Guide RNAs are also known as H/ACA snoRNAs and together with H/ACA core proteins, namely NAP57/dyskerin (DKC1 in the mammalian system), NOP10, NHP2, and GAR1, form a small nucleolar ribonucleoprotein complex to target rRNAs for pseudouridylation [13, 14]. Stand‐alone PUS enzymes comprise six ancient subfamilies, including TruA, TruB, TruD, RsuA, RluA, and PUS10. These proteins modify the target sites by directly recognizing specific sequences and/or secondary structural motifs of substrates. TruA is distinct from other subfamilies as it modifies multiple tRNAs without distinct sequence specificity in the region of the modification sites. The remaining five enzyme subfamilies show higher specificity and selectivity toward their substrates and target sequences. For instance, TruB specifically catalyzes reactions on Ψ55 in the T‐stem loop of tRNAs [15, 16, 17]. Despite the rather divergent amino acid sequences of Pus enzymes, their catalytic domains fold into a very similar and characteristic core structure [18, 19, 20] (Fig. 1). In addition, they utilize a highly conserved catalytic aspartate residue, which despite the low primary sequence similarities suggests a common ancestor and an evolutionarily conserved catalytic mechanism [21]. Surprisingly, PUS10 and the pseudouridylation complex DKC1/NHP2/NOP10/GAR1 have been associated with additional cellular functions. For instance, they participate in miRNA processing and form a complex with the telomerase RNA component to regulate telomerase activity, respectively [22, 23].
Fig. 2.

List of human pseudouridine synthases and their substrates. The names of yeast homologs are mentioned, and disease‐associated enzymes are highlighted in red circles. The substrates of RPUSD1, RPUSD2, PUS1L, and PUS7L are currently unknown.
In many model organisms, the loss of Ψs by mutations in the pseudouridine synthases or genetic engineering leads to overall growth retardation [24, 25]. The direct causative pathogenic association between decreased Ψ levels and human disorders, particularly neuronal dysfunctions and resulting behavior defects, has been confirmed [26] (Fig. 2). The list of Ψ‐linked diseases includes mitochondrial myopathy and sideroblastic anemia caused by mutations in the PUS1 gene that impairs pseudouridylation of specific tRNAs and PUS1‐dependent mRNA expression [27, 28, 29]. Autosomal recessive mental retardation (MRT55) is related to PUS3 mutations while intellectual disorders are associated with missense mutations in PUS7 [26, 30]. Together with other factors, the PUS10 gene was identified as a risk locus in Crohn's disease, an inflammatory disease [31]. Recently, clinical‐relevant mutations in DKC1 and NOP10 have been proven to reduce pseudouridylation in rRNAs and cause nephrotic syndromes in fish [24]. In addition, aberrant pseudouridylation profiles have been linked to cancer formation [32].
De novo protein synthesis is a highly complex mechanism that requires the precise coordination of multiple processes and numerous macromolecular entities, including mRNAs, tRNAs, and ribosomes. Only the properly timed and well‐orchestrated action of all involved players allows to decode and translate the genetic code into functional enzymes and to maintain all cellular functions and activities. Basically, all RNAs that have either direct or indirect roles in modulating protein synthesis are known to carry Ψs in multiple places (Fig. 3). Here, we will focus on the most recent findings about pseudouridylated tRNAs, rRNA, noncoding RNAs, and mRNAs in the cytoplasm as well as in mitochondria. We will discuss the functional consequences at the cellular level and highlight the incorporation of Ψ in the synthetic mRNAs for medical therapies. For more detailed information on Pus enzymes and their mechanisms of action, we refer the interested reader to the other several excellent reviews [12, 33].
Fig. 3.
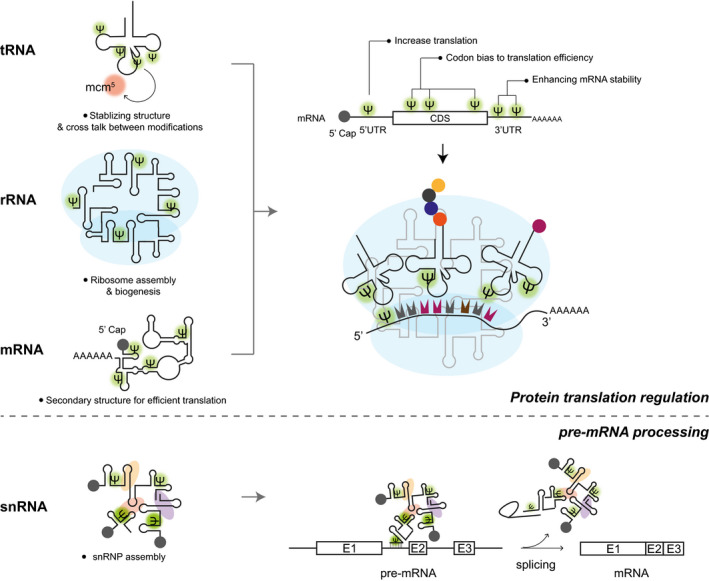
Scheme of pseudouridylated noncoding and coding RNAs and their impact on cellular functions. The left side illustrates the different types of pseudouridylated RNA molecules, while the right side highlights the biological processes that they regulate, including protein translation and pre‐mRNA processing. The small nuclear ribonucleoproteins (snRNPs) are presented as colored ovoid shapes in the snRNP assembly.
Ψs in tRNAs and their functions
In general, tRNA molecules are heavily modified and Ψs account for around 2–5% of all identified tRNA modifications [4]. On average, each tRNA molecule harbors 2–3 Ψ‐sites, which not only represents the most abundant type of tRNA modification [34], but also serves several biological roles. For instance, Ψs are known to affect tRNA structure, tRNA stability, tRNA modification circuitry, and protein translation rates. As all tRNAs need to be accommodated in the tight A‐, P‐, and E‐sites of the ribosome, the rigid tRNA molecules are a highly conserved and structurally almost identical. The characteristic L‐shaped tertiary structure is mainly created by the ‘elbow’ region, which is anchored via the interaction between D‐loop and TΨC‐loop. In detail, Ψ55, located at TΨC loop, pairs with G18 in the D‐loop that mediates the stabilization of the elbow conformation [35]. In addition, Ψ55 is introduced prior to other modifications in the elbow region, which also affects the modification cascades acting on the neighboring nucleotides [36], like m5U54. Uridines in numerous positions of the anticodon stem and loop (ASL) region of eukaryotic tRNAs, namely U26, U27, U28, U30, U31, U32, U38, and U39, can undergo isomerization and are found as Ψ. Most of them are known to confer structural rigidity, which is highlighted by the base pairing between Ψ39 and A31 and the noncanonical base pairing between Ψ38 and position 32 at the transition point between stem and loop region [37, 38]. A functional crosstalk between Ψ38/39 and other modifications in the ASL of tRNAs was recently reported [25]. In detail, Ψ38/39 appears in tandem with other modifications at U34, including mcm5U or s2U, which contributes to Glu/Gln‐rich protein translation. Moreover, the absence of Ψ38/39 has an unexpected influence on the mutation frequency and wide spectrum of proteome changes in Pseudomonas spp [39]. Apart from these universally highly conserved Ψ‐sites, Ψs directly in the anticodon (positions 34, 35, and 36) are found only in certain tRNAs [40], such as Ψ35 in tRNATyr, or Ψ34/36 in tRNAIle. Ψ35 has been demonstrated to regulate the base pairing between tRNA and mRNA that are rich in tyrosine codons in the bacterial system [41], while the role of Ψ34/36 still remains elusive. It can be speculated that evolutionary processes have counterselected against the presence of Ψs in the codon–anticodon region to reduce decoding mistakes due to the possibilities of alternative base pairing and/or avoid overly stable nucleotide pairs [42].
Ψs in rRNAs and their functions
The ribosome is a large ribozyme that consists of a small subunit (SSU) and a large subunit (LSU) that are both built from four rRNAs and around 80 ribosomal proteins. rRNAs are highly complex macromolecules with an extremely complex tertiary fold. Conserved Ψ sites in rRNAs are found across different species [43], and Ψ‐clusters are located at the interface of the two subunits, which place them also close to the peptidyl transferase center in the LSU and the decoding site in the SSU. In particular, 1‐methyl‐3‐α‐amino‐α‐carboxyl‐propyl pseudouridine (m1acp3Ψ) is a hypermodified site that seems to directly interact with P‐site tRNA (m1acp3Ψ1191 in yeast SSU and the equivalent to m1acp3Ψ1248 in the human 18S rRNA). Its absence strongly delays rRNA maturation [44] and blocks other modifications at the decoding center, which can lead to elevated misincorporation rates of amino acids during translation elongation. Recently, the decrease in m1acp3Ψ1248 modification was shown to drive cancer formation in 46% of colorectal carcinoma patients [32].
It is currently estimated that budding yeast rRNAs carry around 45 Ψs and human rRNAs harbor approximately 100 Ψ‐sites. Those Ψs are found to stabilize various local motifs, such as single‐stranded tracts or local helical systems, which are involved in mediating long‐range interactions inside the complex rRNA structure. The presence of Ψ generally stabilizes the structure of the functionally important areas and thus tunes ribosome functions for efficient and accurate protein translation. Moreover, a single particle cryo‐electron microscopy study has recently managed to visualize the human ribosome structures in three‐dimensions with an overall resolution of approximately 2.5 Å [45]. The local resolution in the central regions was even higher and allowed to directly map 25 Ψ‐sites (e.g., the conserved Ψ1677 and Ψ1683 in helix H37) in the structure by recognizing their distinct pattern of hydrogen bonds. Moreover, m3Ψ3762 at the tip of helix H69, an intersubunit bridge in the 60S ribosomal subunit [46, 47], is visualized and shown to mediate the interaction via a coordinated water molecule. Due to the isomeric nature of the structure of U and Ψ, other predicted Ψ‐sites still remain to be confirmed structurally even at this high spatial resolution. Although the overall structure and functional regulation of ribosomes is conserved, the global pseudouridylation profile of rRNA varies among different cell types, different stages of cell differentiation [20, 48], and after exposure to different external stimuli, such as starvation or stress. For instance, the level of Ψ3371 within the E‐site of the 28s rRNA is dynamically regulated during differentiation, which is believed to impact on translation fidelity [20]. Therefore, Ψ modifications constitute an additional source to dynamically shape ribosome heterogeneity and create specialized translational units that allow to locally tune protein translation.
Ψs in snoRNAs and their functions
snoRNAs are a group of noncoding RNAs with a length between 60 and 300 nucleotides and strong secondary structures. snoRNA can be divided into two classes, namely (a) C/D box snoRNAs and (b) H/ACA box snoRNAs [49]. Both types of snoRNAs are mainly responsible for guiding post‐transcriptional RNA modifications. Whereas C/D box snoRNAs allow the specific recognition of sites that should receive a 2′‐O‐ribose methylation, H/ACA box snoRNAs mark RNA sequences for pseudouridylation. snoRNA themselves is known carry Ψs for a long time as well, but the precise sites were mapped only recently [50]. Interestingly, Ψs are found mostly in the regions involved in direct base pairing with target sites. For instance, they occur in the C/D box snoRNA at the 5′ terminus of the guiding sequence and in H/ACA box snoRNAs at the targeting hairpin or the 5′ arm of the guiding sequence. Little is known about the targeting mechanisms that guide the modifications in snoRNAs, their dynamic regulation, and their functional consequences.
Ψs in snRNAs and their functions
The splicing of pre‐mRNAs is a key step during mRNA maturation and requires five major snRNPs, namely U1, U2, U4, U5, and U6. These snRNPs consist of small nuclear RNAs (snRNAs) and auxiliary proteins and are ultimately necessary to remove introns and rejoin exons in the correct order. In addition, another set of less known spliceosomal snRNA species, namely U11, U12, U4atac, and U6atac, participates in the splicing reaction of an atypical class of U12 introns. All spliceosomal snRNAs, including major and minor species, are predominantly modified with Ψs, and the modification sites are mainly located in the functionally important regions. The strategic location and enrichment of Ψ is conserved between species. Over the years, Ψs in snRNAs have been extensively studied in the yeast model system, where some of the Ψ modifications have been shown to be essential for spliceosome assembly and efficient pre‐mRNA splicing [33]. Most of the Ψs in snRNAs are mediating RNA–RNA interaction to enhance folding and maintain structure. Interestingly, it was recently shown that Ψ35, Ψ42, and Ψ44 in the U2 snRNA are key for the interaction with Prp5, an RNA‐dependent ATPase [51]. The U2–Prp5 interaction modulates the ATPase activity of Prp5 is crucial for the subsequent spliceosome assembly.
Most snRNAs are constitutive modified whereas U2 snRNA in yeast, as a first example, has shown to contain conditionally inducible pseudouridylation sites, including positions 56 and 93, which occur upon nutrient deprivation or heat‐shock stimulations [51]. The U56 and U93 locate in the conformationally dynamic stem II of U2 snRNA, and the pseudouridylation on the two sites affects the stem dynamics, including the interaction with metal ion cofactors. For example, the presence of Ψ93 of U2 snRNA introduces conformational flexibility which may be the cause to downregulated pre‐mRNA splicing efficiency [51, 52]. In addition, it has been shown that the snR81 RNP‐mediated Ψ93 formation is controlled by the TOR signaling pathway which regulates cell entry into stationary phase [51].
Impact of Ψ on mitochondrial protein expression
Mitochondrial protein translation takes place within mitochondria and requires a dedicated set of tRNAs, rRNAs, and mRNAs. Mitochondrial tRNAs (mt‐tRNAs) such as cytosolic tRNAs are also extensively pseudouridylated at conserved sites. A recent study has mapped Ψ sites in human mt‐tRNAs and identified new Ψ sites at positions 66 and 67 in the mt‐tRNAPro [53]. Surprisingly, Ψ35 was also identified in the human mt‐tRNAHis GUG. Although biochemical properties of Ψ in mt‐tRNAs are less characterized compared with cytosolic ones, it can be speculated that they serve similar roles in maintaining mt‐tRNA structure stability and thus contribute to the mitochondrial protein translation regulation [54]. The detection of Ψ in mitochondrial transcripts has been performed using human 143B cells, and it revealed the targets of several mitochondria‐specific pseudouridine synthases, including TRUB2, RPUSD3, and RPUSD4. The respective targets include 16S rRNA (Ψ3069) [55], mt‐tRNALeu (Ψ3259), COXI mRNA (Ψ6294), and COXIII mRNA (Ψ9904–9906). Depending on the targeting site, they are found to be essential for 16s rRNA stability and the assembly of functional mitochondrial ribosomes. Furthermore, mt‐Ψs affect mitochondrial protein synthesis, oxidative phosphorylation, and overall cell survival.
Ψs in mRNAs and their functions
mRNAs typically carry a coding sequence (CDS) that contains the transcribed genetic information for protein synthesis, common regulatory sequences (e.g., 5′‐UTR, 3′‐UTR, or polyA‐tail), and short mRNA‐specific signaling motifs (e.g., SECIS) [56]. The level of protein synthesis from a given mRNA molecule depends on the translation initiation rates, translation elongation speed, mRNA half‐life time, re‐initiation efficiency, and to a lesser extent on termination rates. Post‐transcriptional modifications of mRNA, also known as ‘epitranscriptomic’ marks, have been studied intensively over the last couple of years [57]. The interest in mRNA marks has been sparked by their involvement in human diseases and their potential use as diagnostic markers [58]. While the focus of the field was directed toward base methylations, several recent transcriptome‐wide studies have mapped at least 260 Ψs in 238 mRNAs in Saccharomyces cerevisiae and 96 Ψs in 89 human mRNAs [8, 50]. Another study in HEK293 cells presented over two thousand (2084) Ψ sites in human mRNAs (1889) and a mass spectrometry approach reported that 0.3% of all Us are converted into Ψ in human cell lines [59]. In addition, the extensive pseudouridylation of mRNAs also occurs in human pathogens, such as the Toxoplasma gondii parasite [60]. Ψs seem to be distributed unevenly within the CDS as well as the 5′‐ and 3′‐UTRs [8, 50, 59, 61]. Among codons, GUA codons (encoding for valine) seem to be the most frequently modified. However, a comparison of all these datasets has pointed out that a small fraction of candidates were consistently identified and considered across these studies [62]. Interestingly, the profile of Ψ sites in mRNAs is highly dynamic and this suggests that Ψ at a given site occurs in response to changing cellular growth conditions [50]. Although the functional role of each specific Ψs in mRNAs is not clear, it was proposed that Ψ could alter protein synthesis through various ways, for instance by increasing mRNA stability, by altering secondary structures, or by affecting the interaction of mRNAs with the translating ribosomes [8].
As it is difficult to uncouple the influence of Ψ in mRNA from other Ψ‐containing entities in protein synthesis, most studies address this question using artificial mRNAs with Ψs [63, 64, 65, 66]. This approach allows introducing Ψs in mRNAs either globally or site specifically and thus separates these confounding relationships. The presence of Ψ in codons was shown to reduce rate constants for amino acid incorporation and overall protein synthesis. Interestingly, the higher protein expression rate from Ψ‐containing mRNAs is more pronounced in in vivo models than in cell culture systems [65]. Moreover, when the uridine is replaced with a Ψ in a stop codon, it results in nonsense suppression at the translational level [63] and this Ψ‐mediated read‐through is independent of the sequence context [67]. However, this translational termination suppression is not likely to happen under normal conditions [66]. In addition, although Ψ‐containing codons exhibit higher rate of amino acid substitution [63], and it seems to be a rare event in unstressed cells [66]. Of note, other studies have approached these questions differently using site‐specific or completely pseudouridylated GFP reporter constructs. Strikingly, they have not observed amino acid substitutions or decreasing protein production rates [64, 65, 68]. Therefore, the question remains whether the impact of Ψ in mRNA is limited to specific sequences, affects local structures context dependently, or controls general aspects of ribosomal protein synthesis [65, 68].
Other potential factors contributing to the efficiency of protein synthesis include the mRNA sequence itself and the presence of secondary structure motifs. Interestingly, global changes in mRNA secondary structure are related to the introduction of Ψ and N1‐methyl‐Ψ. Like for other types of RNAs, the introduction of Ψ seems to impact the formation of secondary structure elements, which are obviously less common in mRNAs than in short structured noncoding RNA, but the impact of short structural elements for the regulation of the whole mRNA molecule cannot be underestimated. The reduction in secondary structures in the 5′‐UTR reduces translation initiation efficiency, but in the remaining parts of the mRNA (e.g., CDS, 3′‐UTR) induces high protein expression. Ψs also occur in these regions; however, their roles in the secondary structural control are still unclear. It is speculated that Ψs regulates the secondary structure in UTRs (i.e., more constrained structure) and thus affects RNA binding protein affinities at these areas [69]. In addition, the occurrence of Ψs in these regions is also dynamically regulated upon external stimulation, such as starvation or heat shock [50, 61]. Nonetheless, the introduction of a single Ψ at a specific site in a specific mRNA might have manifold roles for the regulation of a respective mRNA molecule.
Ψ in therapeutic applications
The quick response to the COVID‐19 pandemic through mRNA vaccines has brought mRNA therapeutics to front pages worldwide and to the attention of the lay population. Nonetheless, mRNA‐based technologies have been considered a valuable tool and promising technology for the treatment of a broad range of human illnesses for more than a decade [70]. Numerous clinical trials of different classes of RNA‐based therapeutics, and particularly mRNA‐based therapies, are underway [71, 72]. RNA therapeutics include antisense oligonucleotides, siRNAs, miRNAs, RNA aptamers, and mRNA therapeutics [73] and can be subclassified into (a) mRNA‐based cell therapies and (b) mRNA vaccines.
Despite the recent success stories, the translation of these new type of drugs and vaccines into clinical applications presented the scientific community with novel and unique technological challenges. In addition to many other ‘minor’ issues, the establishment of RNA‐based therapies required to solve two major issues inherently related to the use an active RNA compound—(a) the widespread degradation of exogenous RNA by ubiquitous RNases and (b) the immunogenic nature of exogenous RNA leading to cell toxicity and impaired therapeutic protein translation. One of the earliest propositions to increase the efficacy and suitability of RNA molecules for therapeutic usage was the incorporation of widely abundant naturally occurring RNA base modifications [74, 75]. As mentioned above, most of the known 172 RNA modifications are quite rare and affect only certain positions in specific types of RNAs. Ψ is by far the most abundant modification and present in almost all types of RNA molecules. Hence, the idea was born to tweak therapeutic RNA molecules by incorporating Ψs and its derivatives, thus fully replacing uridines.
Since the formulation of the basic concept, it has been shown that incorporation of Ψ and its derivatives indeed enhance the stability of the parent mRNA and lead to strongly increased (approx. 10‐fold) protein translation of the encoded protein as compared to unmodified mRNAs delivered by the same application route [65, 76, 77]. Furthermore, it has been shown that incorporation of the synthetic derivative N1‐methypseudouridine can further improve translation and still evade our innate immune response [78, 79]. The increased stability and half‐life time of the modified transcripts are likely to be the factors contributing to the increased translation of modified mRNAs compared with their corresponding unmodified version. The additional stability of Ψ‐containing mRNAs is attributed to the positive influence of the modification on the phospho‐di‐ester backbone and the stronger binding parameters of A‐Ψ base pairing over a conventional A‐U pair. Ψ‐containing mRNA transcripts have also been shown to be more stable by subverting the NMD decay pathway [63]. Despite the lack of sufficient experimental evidence, it appears very likely that the presence of Ψ also influences splicing, translation, the formation of secondary structure elements and thus the overall lifetime of an mRNA molecule.
The presence of Ψ also has a profound effect on the immunogenicity of the mRNA molecule. Strikingly, it has been shown that the exchange of U by Ψ makes the transcripts much less immunogenic in comparison with unmodified transcripts, which typically induce dendritic cells (DC) to secrete IFN‐α and TNF‐α. The modified transcripts also did not activate the innate immune response, as basically no pro‐inflammatory cytokines were found in the serum postinoculation [76, 77]. However, it was reported that unmodified mRNA transcripts can also serve as nonimmunogenic therapeutic molecules [63, 66]. A more extensive study evaluated the effects of Ψ incorporation by profiling more than 30 cytokines simultaneously. The results show that both immunogenicity and efficacy are comparable between the unmodified and modified therapeutic transcripts after application in liver tissue [80]. Therefore, the exact mechanism of the effect and influence on the therapeutic outcome of the mRNA with Ψs are not yet fully understood and might need to be adjusted to the specific application and targeted cell/tissue type.
Though, the small advantageous effects of Ψ collectively can have a huge impact on the clinical performance of the RNA therapy and thus can be crucial for its successful outcome. For instance, we recently learned that a mRNA vaccine candidate for SARS‐CoV‐2 that did not use modified RNA bases showed strongly reduced efficiency, 47% in a Phase 2b/3 study, and did not meet the prescribed statistical success criteria in avoiding the associated disease. It is worth mentioning, however, that overall effectiveness of a particular candidate vaccine depends on multiple factors such as the dose of the vaccine, the lipid encapsulation, and the mRNA sequence used as well. Thus, more detailed studies are needed to find out the exclusive contribution of the modifications to successful mRNA vaccine candidates. Last but not least, it should be re‐emphasized that, although the spotlight on mRNA vaccines [81, 82, 83] has been brought about by the response to COVID‐19, mRNA vaccine candidates have been in the pipeline for many type of cancers, viral infections, and physiological conditions such as ischemic heart disease, myasthenia gravis, and methylmalonic aciduria [71, 84, 85].
Conclusions and perspectives
Although Ψ was discovered as the first RNA modification more than 70 years ago (Box 1), its functional significance has been recognized only recently. In the past few years, we have learned that Ψs can not only be found in various noncoding RNAs but are also vastly present in mRNAs. The development of a global detection method using CMC‐coupled mapping by sequencing has provided both deep insight into the global distribution of this unique modification and has also revealed a completely new dynamic layer of regulation of gene expression and protein translation (Box 2). Moreover, a crosstalk among ribosome, tRNA, and mRNA via their modification sites during translation process has emerged. In the coming years, we will need to verify the functional impact of the Ψ sites detected in different RNAs, investigate their contribution to specific regulatory pathways and understand their dynamic regulation. For instance, it is not clear whether Ψs are present in introns of pre‐mRNAs as the previous detection methods mainly focused on the mature mRNAs. The interest in Ψ was recently boosted by its broad use and application in the production of mRNA vaccines. The incorporation of Ψ and various derivatives have been shown to reduce immunogenicity and to increase translation efficiency and efficacy. After all, Ψ is not ‘pseudo’ but could turn out to be even more useful than regular uridine. Due to the specific features of Ψ, there is light at the end of the tunnel that the word ‘undruggable’ could become obsolete in the near future.
Box 1. The history of Ψ‐research continues.
The discovery of the constituents of the nucleus, the so called ‘nuclein’ [86], represents the prelude to modern molecular biology. The first scientific report of Ψ being present in rRNA was published seven decades ago [5]. Just after the discovery of the ‘double helix structure’ of DNA [87], Ψ was detected in tRNA [88]. After a gap of 15 years, Ψ was reported to be present in another type of RNA, namely snRNAs [89]. The mapping of the first Ψ synthase gene [90] revealed the enzymatic players that are involved in the underlying modification reaction. The same year the first tRNA crystal structures were reported, which also represent the first structures of complex RNA molecules [91, 92]. After another gap of more than 20 years, a unique probing method was developed that allows the specific detection of the conversion from U to Ψ [93]. In the following years, the structure of a stand‐alone Ψ synthase (PUS) was determined by crystallography [94], and the next year, the first PUS‐tRNA complex structure revealed mechanistic insights into substrate recognition and catalysis [17]. While the efforts to understand Ψ, its synthesis, and implications started to show noticeable results, the development of RNA therapeutics lead to testing of the naturally occurring RNA base modifications, including Ψs, for their use and suitability in clinical applications [74, 76]. Subsequently, it was shown that the incorporation of Ψs and the use of modified RNAs indeed leads to a desirable reduction of immunogenic responses [76]. In parallel, technological developments in genomics, transcriptomics paved the way for transcriptome‐wide Ψ identification in mRNAs and ncRNAs [8, 50]. In the following year, a novel method for the dynamic identification of Ψ in mammalian transcripts was reported [59]. Finally, the SARS‐CoV‐2 pandemic and the timely response with highly efficient mRNA vaccines [81, 83] that are based Ψ and its derivatives have led to an unprecedented interest in RNA modifications and in particular Ψs [65, 66, 81, 83]. We believe that the recent technological developments for Ψ detection and its successful application in mRNA vaccines will further boost the interest in these unique modifications.
Box 2. Methods for Ψ identification.
The initial method of detecting Ψ was based on alkaline hydrolysis followed by TLC detection of isotope‐labeled nucleotides. Later, selective chemical labels of Ψ, like cyanoethylation or N‐cyclohexyl‐N′‐β‐(4‐methylmorpholinium) ethylcarbodiimide p‐tosylate (CMC) conjugation, were developed and fully established. The labeled Ψs created by the respective chemical reactions can be identified using mass spectrometry, allowing the precise localization of modified sites, respectively. As CMC gets conjugated at the N3 position of Ψ within the Watson‐Crick edge, the CMC‐Ψ conjugation site specifically terminates reverse transcription reactions and the resulting cDNA products can be analyzed by gel electrophoresis methods. This approach allows the direct analysis of the specific size of the aborted transcript and the identification of modification sites in individual candidate RNAs. These common identification methods allowed the hypothesis‐driven identification of Ψ in selected structured noncoding RNAs, especially in tRNAs and rRNAs [95, 96]. Recently, several groups have successfully coupled the classical CMC‐Ψ protocol to next‐generation sequencing (NGS), enabling the identification of Ψ sites at the transcriptome‐wide level. Simultaneously and independently, these groups revealed that the Ψ modifications also occur in mRNAs. In detail, Ψ can commonly be found in the 5ʹ‐UTR, CDS, and 3ʹ‐UTR regions at several conserved sites that can be modified under different conditions [8, 50, 59]. These findings implicate an additional layer of dynamic gene regulation based at an unprecedented level.
Acknowledgements
We thank the members of the Glatt lab for vivid discussion and suggestions. This work was supported by the Sonata Grant (2019/35/D/NZ1/02397; T‐YL and RM) from the National Science Centre. This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (Grant agreement No 101001394).
Ting‐Yu Lin and Rahul Mehta contributed equally to this article.
Edited by Claus Azzalin
Contributor Information
Ting‐Yu Lin, Email: ting-yu.lin@uj.edu.pl.
Sebastian Glatt, Email: sebastian.glatt@uj.edu.pl.
References
- 1. Lane BG (1998) Historical perspectives on RNA nucleoside modifications. In Modification and Editing of RNA (Henri G, Rob B, eds), pp. 1–20. American Society of Microbiology Press, Washinton, DC. [Google Scholar]
- 2. Hall RH (1965) A general procedure for the isolation of “minor” nucleosides from ribonucleic acid hydrolysates. Biochemistry 4 :661–670. [DOI] [PubMed] [Google Scholar]
- 3. Zaringhalam M and Papavasiliou FN (2016) Pseudouridylation meets next‐generation sequencing. Methods 107, 63–72. [DOI] [PubMed] [Google Scholar]
- 4. Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crécy‐Lagard V, Ross R, Limbach PA, Kotter A et al. (2018) MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46, D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cohn WE, Volkin E (1951) Nucleoside‐5′‐phosphates from ribonucleic acid. Nature 167, 483–484. [Google Scholar]
- 6. Veerareddygari GR, Singh SK and Mueller EG (2016) The pseudouridine synthases proceed through a glycal intermediate. J Am Chem Soc 138, 7852–7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Motorin Y and Marchand V (2021) Analysis of RNA modifications by second‐and third‐generation deep sequencing: 2020 update. Genes (Basel) 12, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schwartz S, Bernstein D, Mumbach M, Jovanovic M, Herbst R, León‐Ricardo B, Engreitz J, Guttman M, Satija R, Lander E et al. (2014) Transcriptome‐wide mapping reveals widespread dynamic‐regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kierzek E, Malgowska M, Lisowiec J, Turner DH, Gdaniec Z and Kierzek R (2014) The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res 42, 3492–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lorenz C, Lünse CE and Mörl M (2017) tRNA modifications: impact on structure and thermal adaptation. Biomolecules 7, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deb I, Popenda Ł, Sarzyńska J, Małgowska M, Lahiri A, Gdaniec Z and Kierzek R (2019) Computational and NMR studies of RNA duplexes with an internal pseudouridine‐adenosine base pair. Sci Rep 9, 16278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rintala‐Dempsey AC and Kothe U (2017) Eukaryotic stand‐alone pseudouridine synthases – RNA modifying enzymes and emerging regulators of gene expression? RNA Biol 14, 1185–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ni J, Tien AL and Fournier MJ (1997) Small nucleolar RNAs direct site‐specific synthesis of pseudouridine in ribosomal RNA. Cell 89, 565–573. [DOI] [PubMed] [Google Scholar]
- 14. Ganot P, Bortolin ML and Kiss T (1997) Site‐specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89, 799–809. [DOI] [PubMed] [Google Scholar]
- 15. Sprinzl M and Vassilenko KS (2005) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 33, D139–D140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gu X, Yu M, Ivanetich KM and Santi DV (1998) Molecular recognition of tRNA by tRNA pseudouridine 55 synthase. Biochemistry 37, 339–343. [DOI] [PubMed] [Google Scholar]
- 17. Hoang C and Ferré‐D'Amaré AR (2001) Cocrystal structure of a tRNA Ψ55 pseudouridine synthase: nucleotide flipping by an RNA‐modifying enzyme. Cell 107 :929–939. [DOI] [PubMed] [Google Scholar]
- 18. Czudnochowski N, Wang AL, Finer‐Moore J and Stroud RM (2013) In human pseudouridine synthase 1 (hPus1), a C‐terminal helical insert blocks tRNA from binding in the same orientation as in the Pus1 bacterial homologue TruA, consistent with their different target selectivities. J Mol Biol 425, 3875–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hur S and Stroud RM (2007) How U38, 39, and 40 of many tRNAs become the targets for pseudouridylation by TruA. Mol Cell 26, 189–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCleverty CJ, Hornsby M, Spraggon G and Kreusch A (2007) Crystal structure of human Pus10, a novel pseudouridine synthase. J Mol Biol 373, 1243–1254. [DOI] [PubMed] [Google Scholar]
- 21. Huang L, Pookanjanatavip M, Gu X and Santi DV (1998) A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. Biochemistry 37, 344–351. [DOI] [PubMed] [Google Scholar]
- 22. Song J, Zhuang Y, Zhu C, Meng H, Lu B, Xie B, Peng J, Li M and Yi C (2020) Differential roles of human PUS10 in miRNA processing and tRNA pseudouridylation. Nat Chem Biol 16, 160–169. [DOI] [PubMed] [Google Scholar]
- 23. Nguyen THD, Tam J, Wu RA, Greber BJ, Toso D, Nogales E and Collins K (2018) Cryo‐EM structure of substrate‐bound human telomerase holoenzyme. Nature 557, 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Balogh E, Chandler JC, Varga M, Tahoun M, Menyhárd DK, Schay G, Goncalves T, Hamar R, Légrádi R, Szekeres Á et al. (2020) A pseudouridylation defect due to DKC1 and NOP10 mutations causes nephrotic syndrome with cataracts, hearing impairment, and enterocolitis. Proc Natl Acad Sci USA 117, 15137–15147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Han L, Kon Y and Phizicky EM (2015) Functional importance of Ψ38 and Ψ39 in distinct tRNAs, amplified for tRNAGln(UUG) by unexpected temperature sensitivity of the s2U modification in yeast. RNA 21, 188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Brouwer APM, Abou Jamra R, Körtel N, Soyris C, Polla DL, Safra M, Zisso A, Powell CA, Rebelo‐Guiomar P, Dinges N et al. (2018) Variants in PUS7 cause intellectual disability with speech delay, microcephaly, short stature, and aggressive behavior. Am J Hum Genet 103, 1045–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carlile TM, Martinez NM, Schaening C, Su A, Bell TA, Zinshteyn B and Gilbert WV (2019) mRNA structure determines modification by pseudouridine synthase 1. Nat Chem Biol 15, 966–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bykhovskaya Y, Casas K, Mengesha E, Inbal A and Fischel‐Ghodsian N (2004) Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA). Am J Hum Genet 74, 1303–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mangum JE, Hardee JP, Fix DK, Puppa MJ, Elkes J, Altomare D, Bykhovskaya Y, Campagna DR, Schmidt PJ, Sendamarai AK et al. (2016) Pseudouridine synthase 1 deficient mice, a model for mitochondrial myopathy with sideroblastic anemia, exhibit muscle morphology and physiology alterations. Sci Rep 6, 26202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shaheen R, Han L, Faqeih E, Ewida N, Alobeid E, Phizicky EM and Alkuraya FS (2016) A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum Genet 135, 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Festen EAM, Goyette P, Green T, Boucher G, Beauchamp C, Trynka G, Dubois PC, Lagacé C, Stokkers PCF, Hommes DW et al. (2011) A meta‐analysis of genome‐wide association scans identifies IL18RAP, PTPN2, TAGAP, and PUS10 as shared risk loci for Crohn's disease and celiac disease. PLoS Genet 7, e1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Babaian A, Rothe K, Girodat D, Minia I, Djondovic S, Milek M, Spencer Miko SE, Wieden HJ, Landthaler M, Morin GB et al. (2020) Loss of m1acp3Ψ ribosomal RNA modification is a major feature of cancer. Cell Rep 31, 107611. [DOI] [PubMed] [Google Scholar]
- 33. Morais P, Adachi H and Yu YT (2021) Spliceosomal snRNA epitranscriptomics. Front Genet 12, 652129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miura K‐I (1967) Specificity in the structure of transfer RNA. Prog Nucleic Acid Res Mol Biol 6, 39–82. [DOI] [PubMed] [Google Scholar]
- 35. Kimura S and Waldor MK (2019) The RNA degradosome promotes tRNA quality control through clearance of hypomodified tRNA. Proc Natl Acad Sci USA 116, 1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kai‐Leigh Schultz S and Kothe U (2020) tRNA elbow modifications affect the tRNA pseudouridine synthase TruB and the methyltransferase TrmA. RNA 26, 1131–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krutyhołowa R, Zakrzewski K and Glatt S (2019) Charging the code—tRNA modification complexes. Curr Opin Struct Biol 55, 138–146. [DOI] [PubMed] [Google Scholar]
- 38. Durant PC and Davis DR (1999) Stabilization of the anticodon stem‐loop of tRNA(Lys,3) by an A+‐C base‐pair and by pseudouridine. J Mol Biol 285, 115–131. [DOI] [PubMed] [Google Scholar]
- 39. Tagel M, Ilves H, Leppik M, Jürgenstein K, Remme J and Kivisaar M (2021) Pseudouridines of TRNA anticodon stem‐loop have unexpected role in mutagenesis in Pseudomonas sp. Microorganisms 9, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Szweykowska‐Kulinska Z, Senger B, Keith G, Fasiolo F and Grosjean H (1994) Intron‐dependent formation of pseudouridines in the anticodon of Saccharomyces cerevisiae minor tRNA(Ile). EMBO J 13, 4636–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Addepalli B and Limbach PA (2016) Pseudouridine in the anticodon of Escherichia coli tRNATyr(QΨA) is catalyzed by the dual specificity enzyme RluF. J Biol Chem 291, 22327–22337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grosjean H and Westhof E (2016) An integrated, structure‐ and energy‐based view of the genetic code. Nucleic Acids Res 44, 8020–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sharma S and Lafontaine DLJ (2015) “View from a bridge”: a new perspective on eukaryotic rRNA base modification. Trends Biochem Sci 40, 560–575. [DOI] [PubMed] [Google Scholar]
- 44. Liang XH, Liu Q and Fournier MJ (2009) Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre‐rRNA processing. RNA 15, 1716–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Natchiar SK, Myasnikov AG, Kratzat H, Hazemann I and Klaholz BP (2017) Visualization of chemical modifications in the human 80S ribosome structure. Nature 551, 472–477. [DOI] [PubMed] [Google Scholar]
- 46. Liang X, Liu Q and Fournier MJ (2007) rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol Cell 28, 965–977. [DOI] [PubMed] [Google Scholar]
- 47. Chikne V, Doniger T, Rajan KS, Bartok O, Eliaz D, Cohen‐Chalamish S, Tschudi C, Unger R, Hashem Y, Kadener S et al. (2016) A pseudouridylation switch in rRNA is implicated in ribosome function during the life cycle of Trypanosoma brucei . Sci Rep 6, 25296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marchand V, Pichot F, Neybecker P, Ayadi L, Bourguignon‐Igel V, Wacheul L, Lafontaine DLJ, Pinzano A, Helm M and Motorin Y (2020) HydraPsiSeq: a method for systematic and quantitative mapping of pseudouridines in RNA. Nucleic Acids Res 48, e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kiss T (2002) Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109, 145–148. [DOI] [PubMed] [Google Scholar]
- 50. Carlile TM, Rojas‐Duran MF, Zinshteyn B, Shin H, Bartoli KM and Gilbert WV (2014) Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu G, Adachi H, Ge J, Stephenson D, Query CC and Yu Y (2016) Pseudouridines in U2 snRNA stimulate the ATPase activity of Prp5 during spliceosome assembly. EMBO J 35, 654–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. van der Feltz C, DeHaven AC and Hoskins AA (2018) Stress‐induced pseudouridylation alters the structural equilibrium of yeast U2 snRNA stem II. J Mol Biol 430, 524–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Suzuki T, Yashiro Y, Kikuchi I, Ishigami Y, Saito H, Matsuzawa I, Okada S, Mito M, Iwasaki S, Ma D et al. (2020) Complete chemical structures of human mitochondrial tRNAs. Nat Commun 11, 4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang M, Liu H, Zheng J, Chen B, Zhou M, Fan W, Wang H, Liang X, Zhou X, Eriani G et al. (2016) A deafness‐and diabetes‐associated tRNA mutation causes deficient pseudouridinylation at position 55 in tRNAGlu and mitochondrial dysfunction. J Biol Chem 291, 21029–21041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Antonicka H, Choquet K, Lin Z, Gingras A, Kleinman CL and Shoubridge EA (2017) A pseudouridine synthase module is essential for mitochondrial protein synthesis and cell viability. EMBO Rep 18, 28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thanbichler M and Böck A (2002) The function of SECIS RNA in translational control of gene expression in Escherichia coli . EMBO J 21, 6925–6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roundtree IA, Evans ME, Pan T and He C (2017) Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Song H, Liu D, Dong S, Zeng L, Wu Z, Zhao P, Zhang L, Chen ZS and Zou C (2020) Epitranscriptomics and epiproteomics in cancer drug resistance: therapeutic implications. Signal Transduct Target Ther 5, 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li X, Zhu P, Ma S, Song J, Bai J, Sun F and Yi C (2015) Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol 11, 592–597. [DOI] [PubMed] [Google Scholar]
- 60. Nakamoto MA, Lovejoy AF, Cygan AM and Boothroyd JC (2017) mRNA pseudouridylation affects RNA metabolism in the parasite Toxoplasma gondii . RNA 23, 1834–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lovejoy AF, Riordan DP and Brown PO (2014) Transcriptome‐wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae . PLoS One 9, e110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen W and Liu K (2019) Analysis and comparison of RNA pseudouridine site prediction tools. Curr Bioinform 15, 279–286. [Google Scholar]
- 63. Karijolich J and Yu Y‐T (2011) Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 474, 395–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hoernes TP, Clementi N, Faserl K, Glasner H, Breuker K, Lindner H, Hüttenhofer A and Erlacher MD (2016) Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res 44, 852–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mauger DM, Cabral BJ, Presnyak V, Su SV, Reid DW, Goodman B, Link K, Khatwani N, Reynders J, Moore MJ et al. (2019) mRNA structure regulates protein expression through changes in functional half‐life. Proc Natl Acad Sci USA 116, 24075–24083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Eyler DE, Franco MK, Batool Z, Wu MZ, Dubuke ML, Dobosz‐Bartoszek M, Jones JD, Polikanov YS, Roy B and Koutmou KS (2019) Pseudouridinylation of mRNA coding sequences alters translation. Proc Natl Acad Sci USA 116, 23068–23074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Adachi H and Yu Y‐T (2020) Pseudouridine‐mediated stop codon readthrough in S. cerevisiae is sequence context‐independent. RNA 26, 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hoernes TP, Heimdörfer D, Köstner D, Faserl K, Nußbaumer F, Plangger R, Kreutz C, Lindner H and Erlacher MD (2019) Eukaryotic translation elongation is modulated by single natural nucleotide derivatives in the coding sequences of mRNAs. Genes (Basel) 10, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Delorimier E, Hinman MN, Copperman J, Datta K, Guenza M and Berglund JA (2017) Pseudouridine modification inhibits muscleblind‐like 1 (MBNL1) binding to CCUG repeats and minimally structured RNA through reduced RNA flexibility. J Biol Chem 292, 4350–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Vogel FR and Sarver N (1995) Nucleic acid vaccines. Clin Microbiol Rev 8, 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhu X, Yin L, Theisen M, Zhuo J, Siddiqui S, Levy B, Presnyak V, Frassetto A, Milton J, Salerno T et al. (2019) Systemic mRNA therapy for the treatment of Fabry disease: preclinical studies in wild‐type mice, Fabry mouse model, and wild‐type non‐human primates. Am J Hum Genet 104, 625–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Krienke C, Kolb L, Diken E, Streuber M, Kirchhoff S, Bukur T, Akilli‐Öztürk Ö, Kranz LM, Berger H, Petschenka J et al. (2021) A noninflammatory mRNA vaccine for treatment of experimental autoimmune encephalomyelitis. Science 371, 145–153. [DOI] [PubMed] [Google Scholar]
- 73. Damase TR, Sukhovershin R, Boada C, Taraballi F, Pettigrew RI and Cooke JP (2021) The limitless future of RNA therapeutics. Front Bioeng Biotechnol 9, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Karikó K, Buckstein M, Ni H and Weissman D (2005) Suppression of RNA recognition by Toll‐like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175. [DOI] [PubMed] [Google Scholar]
- 75. Lockhart J, Canfield J, Mong EF, VanWye J and Totary‐Jain H (2019) Nucleotide modification alters microRNA‐dependent silencing of microRNA switches. Mol Ther Nucleic Acids 14, 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Karikó K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S and Weissman D (2008) Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther 16, 1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Karikó K, Muramatsu H, Ludwig J and Weissman D (2011) Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside‐modified, protein‐encoding mRNA. Nucleic Acids Res 39, e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Andries O, Mc Cafferty S, De Smedt SC, Weiss R, Sanders NN and Kitada T (2015) N1‐methylpseudouridine‐incorporated mRNA outperforms pseudouridine‐incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J Control Release 217, 337–344. [DOI] [PubMed] [Google Scholar]
- 79. Svitkin YV, Cheng YM, Chakraborty T, Presnyak V, John M and Sonenberg N (2017) N1‐methyl‐pseudouridine in mRNA enhances translation through eIF2α‐dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res 45, 6023–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kauffman KJ, Mir FF, Jhunjhunwala S, Kaczmarek JC, Hurtado JE, Yang JH, Webber MJ, Kowalski PS, Heartlein MW, DeRosa F et al. (2016) Efficacy and immunogenicity of unmodified and pseudouridine‐modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials 109, 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sahin U, Muik A, Vogler I, Derhovanessian E, Kranz LM, Vormehr M, Quandt J, Bidmon N, Ulges A, Baum A et al. (2021) BNT162b2 vaccine induces neutralizing antibodies and poly‐specific T cells in humans. Nature 595, 572–577. [DOI] [PubMed] [Google Scholar]
- 82. Vogel AB, Kanevsky I, Che YE, Swanson KA, Muik A, Vormehr M, Kranz LM, Walzer KC, Hein S, Güler A et al. (2021) BNT162b vaccines protect rhesus macaques from SARS‐CoV‐2. Nature 592, 283–289. [DOI] [PubMed] [Google Scholar]
- 83. Corbett KS, Edwards DK, Leist SR, Abiona OM, Boyoglu‐Barnum S, Gillespie RA, Himansu S, Schäfer A, Ziwawo CT, DiPiazza AT et al. (2020) SARS‐CoV‐2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 586, 567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. An D, Schneller JL, Frassetto A, Liang S, Zhu X, Park J‐S, Theisen M, Hong S‐J, Zhou J, Rajendran R et al. (2017) Systemic messenger RNA therapy as a treatment for methylmalonic acidemia. Cell Rep 21, 3548–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Carlsson L, Clarke JC, Yen C, Gregoire F, Albery T, Billger M, Egnell A‐C, Gan L‐M, Jennbacken K, Johansson E et al. (2018) Biocompatible, purified VEGF‐A mRNA improves cardiac function after intracardiac injection 1 week post‐myocardial infarction in swine. Mol Ther Clin Dev 9, 330–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Miescher F (1869) Letter I; to Wilhelm His; Tübingen, February 26th, 1869. In: Die Histochem und Physiologischen Arbeiten von Friedrich Miescher‐Aus dem wissenschaftlichen Briefwechsel von F Miescher 1, 33–38.
- 87. Watson JD and Crick FHC (1953) Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature 171 :737–738. [DOI] [PubMed] [Google Scholar]
- 88. Davis FF and Allen FW (1957) Ribonucleic acids from yeast which contain a fifth nucleotide. J Biol Chem 227, 907–915. [PubMed] [Google Scholar]
- 89. Reddy R, Ro‐Choi TS, Henning D, Shibata H, Choi YC and Busch H (1972) Modified nucleosides of nuclear and nucleolar low molecular weight ribonucleic acid. J Biol Chem 247, 7245–7250. [PubMed] [Google Scholar]
- 90. Cortese R, Kammen HO, Spengler SJ and Ames BN (1974) Biosynthesis of pseudouridine in transfer ribonucleic acid. J Biol Chem 249, 1103–1108. [PubMed] [Google Scholar]
- 91. Kim SH, Suddath FL, Quigley GJ, McPherson A, Sussman JL, Wang AHJ, Seeman NC and Rich A (1974) Three‐dimensional tertiary structure of yeast phenylalanine transfer RNA. Science 185, 435. [DOI] [PubMed] [Google Scholar]
- 92. Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BFC and Klug A (1974) Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature 250 :546–551. [DOI] [PubMed] [Google Scholar]
- 93. Bakin A and Ofengand J (1993) Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyl transferase center: analysis by the application of a new sequencing technique. Biochemistry 32 :9754–9762. [DOI] [PubMed] [Google Scholar]
- 94. Foster PG, Huang L, Santi DV and Stroud RM (2000) The structural basis for tRNA recognition and pseudouridine formation by pseudouridine synthase I. Nat Struct Biol 7, 23–27. [DOI] [PubMed] [Google Scholar]
- 95. Charette M and Gray MW (2000) Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49, 341–351. [DOI] [PubMed] [Google Scholar]
- 96. Agris PF, Eruysal ER, Narendran A, Väre VYP, Vangaveti S and Ranganathan SV (2018) Celebrating wobble decoding: half a century and still much is new. RNA Biol 15, 537–553. [DOI] [PMC free article] [PubMed] [Google Scholar]


