Abstract
The genotypic diversity that occurs in natural populations of antagonistic microorganisms provides an enormous resource for improving biological control of plant diseases. In this study, we determined the diversity of indigenous 2,4-diacetylphloroglucinol (DAPG)-producing Pseudomonas spp. occurring on roots of wheat grown in a soil naturally suppressive to take-all disease of wheat. Among 101 isolates, 16 different groups were identified by random amplified polymorphic DNA (RAPD) analysis. One RAPD group made up 50% of the total population of DAPG-producing Pseudomonas spp. Both short- and long-term studies indicated that this dominant genotype, exemplified by P. fluorescens Q8r1-96, is highly adapted to the wheat rhizosphere. Q8r1-96 requires a much lower dose (only 10 to 100 CFU seed−1 or soil−1) to establish high rhizosphere population densities (107 CFU g of root−1) than Q2-87 and 1M1-96, two genotypically different, DAPG-producing P. fluorescens strains. Q8r1-96 maintained a rhizosphere population density of approximately 105 CFU g of root−1 after eight successive growth cycles of wheat in three different, raw virgin soils, whereas populations of Q2-87 and 1M1-96 dropped relatively quickly after five cycles and were not detectable after seven cycles. In short-term studies, strains Q8r1-96, Q2-87, and 1M1-96 did not differ in their ability to suppress take-all. After eight successive growth cycles, however, Q8r1-96 still provided control of take-all to the same level as obtained in the take-all suppressive soil, whereas Q2-87 and 1M1-96 gave no control anymore. Biochemical analyses indicated that the superior rhizosphere competence of Q8r1-96 is not related to in situ DAPG production levels. We postulate that certain rhizobacterial genotypes have evolved a preference for colonization of specific crops. By exploiting diversity of antagonistic rhizobacteria that share a common trait, biological control can be improved significantly.
Biological control of soil-borne plant pathogens by application of specific microorganisms to seeds or planting material has been studied intensively over the past three decades. Notable among biocontrol agents are antibiotic-producing fluorescent Pseudomonas spp. (3, 13, 47). In evaluating the last decade of research on biological control, it is clear that most biocontrol agents, including strains of antibiotic-producing Pseudomonas spp., are still too variable in their performance to be successfully used as a common practice in agriculture and horticulture. This inconsistency has been attributed to a number of factors, including the variable expression of genes involved in disease suppression and poor root colonization by the applied biocontrol agent. Consequently, research has focused on studying gene expression in rhizosphere environments (27) and traits involved in rhizosphere competence (8).
Rhizosphere competence comprises the ability of biocontrol agents to distribute along growing plant roots, to propagate, and to survive over a considerable time period in the presence of the indigenous microflora (32, 52, 55). The significance of the rhizosphere competence of biocontrol agents for disease suppression has been emphasized by various studies (5, 18, 19, 23, 35, 38, 44). Collectively, these studies have demonstrated that biocontrol agents must establish and maintain a certain threshold population density to preempt or limit infection by the pathogen or induce host defenses. Population densities of introduced Pseudomonas strains can vary among root systems of different plants or among roots of single plants by several orders of magnitude, a pattern referred to as lognormal distribution (1, 26). Moreover, their rhizosphere population densities tend to decline substantially over a prolonged period of time and with increasing distance from the inoculum source (2, 17, 26, 36, 51).
Given the importance of root colonization in biological control, the selection of strains that are rhizosphere competent will significantly contribute to improve the efficacy of biocontrol agents. Two approaches have been widely used to select for potential biocontrol agents (55). The first approach consists of isolating antagonistic microorganisms from the intended environment of use, such as soils, seeds, or roots, whereas the second approach comprises the isolation of antagonists from soils that are naturally suppressive to a particular pathogen. Both selection procedures are based on the assumption that antagonistic microorganisms will be better adapted to the environment or host-pathogen system from which they were originally isolated. Although many microorganisms have been randomly isolated by both procedures and subsequently tested in greenhouse and field experiments, few of these biocontrol agents have been effective over a long period of time and a broad range of conditions.
The genotypic diversity that occurs in natural populations of biocontrol agents provides a tremendous resource for improving biological control of plant diseases (13, 47). This approach has been widely used to select for better biocontrol agents of insects and to improve the use of microorganisms in the production of fermented foods and in the biodegradation of xenobiotic compounds (45). However, the exploitation of genotypic diversity among biocontrol agents of soil-borne fungi, so far, has received much less attention. Therefore, knowledge of the diversity within a group of strains that share a common biocontrol trait may provide a new approach for identifying biocontrol strains that are superior with respect to rhizosphere competence and ability to suppress soil-borne pathogens.
We have focused on the role of the antifungal metabolite 2,4-diacetylphlorglucinol (DAPG) in biological control of soil-borne pathogens by fluorescent Pseudomonas spp. (47). Genetic studies, modeled after Koch's postulates, demonstrated unequivocally that DAPG plays a major role in the suppression of a variety of soil-borne plant pathogens by fluorescent Pseudomonas strains (9, 20, 41, 49). Moreover, DAPG-producing fluorescent Pseudomonas spp. were shown to be highly enriched in take-all suppressive soils (37) and key components of the natural biocontrol that operates in these soils (38, 39). In this study, we begin to explore the relationship between the genotype of a DAPG producer and the ability to colonize wheat roots and to suppress take-all. We hypothesize that certain genotypes will have evolved a preference for the colonization of specific crops or an enhanced activity against a particular disease. We show that Pseudomonas fluorescens strain Q8r1-96, which is representative of a genotypic group of DAPG-producers common on roots of wheat grown in take-all decline (TAD) soils, has a root-colonizing ability far superior to that of two other, genotypically different, DAPG-producing P. fluorescens strains.
MATERIALS AND METHODS
Bacterial strains and growth media.
The Pseudomonas strains used in this study are listed in Table 1. The reference strains are well-characterized biocontrol agents of a variety of plant pathogenic fungi. Rhizosphere competence and biocontrol assays were performed with strains Q2-87, 1M1-96, and Q8r1-96. Strains Q2-87 (34) and Q8r1-96 (38) were isolated in 1987 and 1996, respectively, from roots of wheat grown in a take-all suppressive soil collected from an agricultural field near the city of Quincy, Wash. Strain 1M1-96 was isolated in 1996 from roots of wheat grown in a pea-wilt suppressive soil from a field near the city of Mount Vernon, Wash. Spontaneous rifampin-resistant derivatives of strains Q2-87, 1M1-96, and Q8r1-96 were used in the experiments. All three strains harbor the phlD gene, one of the key genes in the biosynthesis of the antibiotic DAPG. In vitro DAPG production by strains Q2-87, 1M1-96, and Q8r1-96 was confirmed by C18 reverse-phase high-performance liquid chromatography (HPLC) followed by photodiode array spectroscopy (4, 39). Pseudomonas strains were routinely grown on King's medium B agar (KMB) (22) at 25°C. Naturally occurring fluorescent Pseudomonas spp. were isolated from wheat roots on KMB agar supplemented with cycloheximide (100 μg ml−1), chloramphenicol (13 μg ml−1), and ampicillin (40 μg ml−1) (KMB+) (43). Rhizosphere population densities of the rifampin-resistant derivatives of Q2-87, 1M1-96, and Q8r1-96 were determined on KMB+ supplemented with rifampin (100 μg ml−1) (KMB+Rif).
TABLE 1.
Characteristics of Pseudomonas spp. used in this study
| Strain | DAPG productiona | Target of biocontrol activityb | Origin | Source and/ or reference |
|---|---|---|---|---|
| P. fluorescens | ||||
| Q2-87 | + | G. graminis on wheat | Wheat, Washington | 49; this study |
| 1M1-96 | + | G. graminis on wheat | Wheat, Washington | This study |
| Q8r1-96 | + | G. graminis on wheat | Wheat, Washington | Tthis study |
| 2-79RN10 | − | G. graminis on wheat | Wheat, Washington | 50 |
| Q69c-80 | − | G. graminis on wheat | Wheat, Washington | 14 |
| CHAO | + | G. graminis on wheat, T. basicola on tobacco, P. ultimum on cucumber | Tobacco, Switzerland | 20 |
| Pf-1 | + | T. basicola on tobacco, P. ultimum on cucumber | Tobacco, Switzerland | 21 |
| F113 | + | P. ultimum on sugar beet | Sugar beet, Ireland | 41 |
| PFM2 | + | S. tritici on wheat | Wheat, Oklahoma | 25 |
| Pf5 | + | P. ultimum and R. solani on cotton | Cotton, Texas | 16 |
| Other Pseudomonas sp. | ||||
| PINR2 | + | P. ultimum on cucumber, F. oxysporum on tomato | Tobacco, Italy | 21 |
| PILH1 | + | P. ultimum on cucumber, F. oxysporum on tomato | Tomato, Italy | 21 |
| PGNR1 | + | P. ultimum on cucumber, F. oxysporum on tomato | Tobacco, Ghana | 21 |
+, Production of DAPG; −, no DAPG- production.
The full names of the strains indicated are Gaeumannomyces graminis var. tritici, Thielaviopsis basicola, Pythium ultimum, Septoria tritici, Rhizoctonia solani, and Fusarium oxysporum f. sp. radicis-lycopersici. For many strains, the data on biocontrol activity come from unpublished experiments.
Soils.
Soils were obtained from fields near the cities of Quincy, Lind, and Moses Lake Wash. Soil from the agricultural field near Quincy, designated Quincy TAD, is suppressive to take-all of wheat. In 1980, the Quincy TAD field had been cropped continuously to wheat for 22 years; between 1980 and 1995 other crops besides wheat also were grown. The virgin soils from Quincy, Lind, and Moses Lake, designated Quincy Virgin, Lind Virgin, and Moses Lake Virgin, respectively, were covered by native vegetation such as sagebrush and bunchgrass. The Quincy Virgin soil was located near the corresponding Quincy TAD field. The virgin soils are conducive to take-all of wheat. All soils were collected in March 1995 from the upper 30 cm of the soil profile, air dried for 1 week, and passed through a 0.5-cm-mesh screen prior to use. Their physical and chemical properties were determined by the Analytical Sciences Laboratory, University of Idaho, and were described previously (37, 54).
Isolation of indigenous DAPG-producing Pseudomonas spp. from roots of wheat.
Twelve wheat seeds were sown in square polyvinyl chloride pots (8 cm high, 7.5 cm wide) containing 200 g of sieved raw Quincy TAD soil and 50 ml of water supplemented with metalaxyl (Novartis, Greensboro, N.C.) at 2.5 mg ml−1 as the active ingredient to control Pythium root rot. A 1-cm layer of soil was spread on top of the seeds. Plants were grown in a controlled-environment chamber at 16°C with a 12-h photoperiod. Pots received 50 ml of dilute (2:3 [vol/vol]) Hoaglund's solution (macro-elements only) twice a week. After 3 weeks of growth, four to six randomly selected plants were harvested from each replicate, and root samples were prepared to determine the population size of indigenous DAPG-producing Pseudomonas spp. The shoots of the remaining plants were excised, and the soil and associated root system were decanted into a plastic bag and shaken vigorously to aerate and mix. This cultivated soil was stored for 1 week at 15°C, returned to the same pot, and then replanted with 12 seeds. This process of plant growth, harvesting, and determination of population sizes was repeated for a total of eight cycles. To determine the population size of indigenous DAPG-producing Pseudomonas spp., 1 g of roots and associated rhizosphere soil was suspended in 5.0 ml of sterile water and shaken vigorously for 1 min on a Vortex mixer. The samples were subsequently sonicated in an ultrasonic cleaner for 1 min, and then serial dilutions of the root washes were plated onto KMB+. Plates were incubated at 25°C, and colonies were enumerated after 48 h. The number of fluorescent Pseudomonas spp. that harbor the phlD gene was determined by colony hybridization followed by PCR analysis (37).
Genotypic diversity of indigenous DAPG-producing Pseudomonas spp.
A PCR-based fingerprinting method with randomly amplified polymorphic DNA (RAPD) markers was used for genotypic characterization of indigenous DAPG-producing Pseudomonas spp. and reference strains. Two 10-mer primers, M13 and D7, were used and were obtained from Operon Technologies, Inc. (Alameda, Calif.). Primers M13 and D7 were tested by Keel et al. (21) on a wide variety of DAPG-producing Pseudomonas spp. and were selected from a total of 64 primers because they both produced distinct and consistent banding patterns with polymorphic markers. PCR-RAPD amplifications were carried out in a 25-μl reaction mixture which contained 5 μl of a diluted heat-lysed cell suspension (37), 1× GeneAmp PCR buffer (Perkin-Elmer Corp., Norwalk, Conn.); 200 μM concentrations of dATP, DTTP, dGTP, and dCTP (Perkin-Elmer); 80 pmol of M13 or D7 primer; and 2.0 U of AmpliTaq DNA polymerase (Perkin-Elmer). Each reaction mixture was covered with 1 drop of mineral oil. The cycler used was a Perkin-Elmer 480. The PCR program consisted of an initial denaturation at 94°C for 120 s, followed by 2 cycles of 94°C for 30 s, 36°C for 30 s, and 72°C for 120 s; 30 cycles of 94°C for 20 s, 36°C for 15 s, 45°C for 15 s, and 72°C for 90 s; and a final incubation at 72°C for 10 min. The amplification products were separated on a 2.5% agarose gel in 1× TBE (90 mM Tris-borate, 2 mM EDTA [pH 8.3]) at 75 V for 3 h. The gel was stained with ethidium bromide, and the amplification products were visualized with a UV transilluminator. All PCR-RAPD reactions were repeated at least three times, and only the RAPD bands which appeared consistently were evaluated. Band sizes were determined with the Phoretix_1D software (version 3.0; Phoretix International). Calculations of the pairwise coefficients of similarity (Dice) were based on the presence or absence of bands, and cluster analysis with the UPGMA algorithm were performed with the NTSYS-pc numerical taxonomy and multivariate analysis system (40).
Biochemical identification and characterization.
Pseudomonas strains Q2-87, 1M1-96, and Q8r1-96 were identified and characterized in detail by the American Type Culture Collection (ATCC, Rockville, Md). Characterizations included cellular and colonial morphology, fatty acid profiles (fatty acid methyl ester [FAME] analysis), growth at different temperatures and on diverse agar media, pigment, acid and enzyme production, and the ability to utilize 53 substrates as a sole carbon source.
Seed treatment.
Wheat seeds (cv. Penewawa) were coated with 1% methylcellulose (Sigma) (control) or with suspensions of the rifampin-resistant derivatives of strains Q2-87, 1M1-96, or Q8r1-96 in 1% methylcellulose. The coated seeds were air dried for 5 h in a laminar flow cabinet. The final densities of the strains were approximately 101, 102, 103, 104, 105, 106, or 107 CFU per seed as determined by dilution plating on KMB+Rif.
Determination of rhizosphere competence.
In short-term experiments, nine treated wheat seeds were sown in square polyvinyl chloride pots (8 cm high, 7.5 cm wide) containing 200 g of sieved raw Quincy virgin soil and 50 ml of water supplemented with metalaxyl at 2.5 mg ml−1 as the active ingredient to control Pythium root rot. A 1-cm layer of soil was spread on top of the seeds. Plants were grown as described above. After 3 weeks of growth, plants were harvested, and loosely adhering soil was removed from the roots by gently shaking. Root samples were prepared from six randomly selected plants, and the population sizes of the introduced Pseudomonas strains were determined by dilution plating root washes, prepared as described above, onto KMB+Rif. Three replicates were used for each treatment. In long-term experiments, 12 nontreated wheat seeds were sown in square polyvinyl chloride pots containing 200 g of sieved raw Quincy virgin, Lind virgin, or Moses Lake virgin soils into which suspensions of Q2-87, 1M1-96, or Q8r1-96 were introduced to obtain a final density of approximately 100 CFU per g of soil fresh weight. Plants were grown for eight successive cycles as described above; Q2-87, 1M1-96, and Q8r1-96 were introduced into the soils only at the beginning of the first cycle and not in the successive cycles. Every cycle, four to six randomly selected plants were harvested from each replicate, root samples were prepared, and population sizes of introduced strains were determined by dilution plating as described above. For each treatment, three replicates were used. Each soil was studied independently. The shoots of the remaining plants were excised, and the soil and associated root system were decanted into a plastic bag and shaken vigorously to aerate and mix. The soils were returned to the same pot and then replanted with 12 nontreated seeds.
Take-all pathogen and disease ratings.
R3-111a-1 is a virulent isolate of Gaeumannomyces graminis (Sacc.) Arx & D. Olivier var. tritici J. Walker, originally isolated from wheat near Moses Lake (7), and was routinely cultured on potato dextrose agar at room temperature. In both short-term and long-term experiments, soil was amended with 0.1 to 0.2% (wt/wt) of an oat grain inoculum of R3-111a-1 (53). From each replicate, six randomly selected plants were harvested and washed, and the disease severity was determined on a 0-to-8 scale, where “0” indicates no disease and “8” indicates a dead plant (46). In both short-term and long-term experiments with the Quincy virgin soil, nontreated Quincy virgin and Quincy TAD soils were included in order to compare the take-all suppressiveness of the soils.
Antibiotic production on roots of wheat.
DAPG was isolated from roots of wheat according to the method described by Bonsall et al. (4). A 30-g portion of wheat roots with adhering rhizosphere soil, but without remnants of seeds, was mixed in a 250-ml flask with 40 ml of 80% acetone acidified to pH 2.0 with 10% trifluoroacetic acid (TFA) and then shaken (200 rpm) for 2 h at room temperature. Samples were subsequently filtered (Whatman no. 1) through a Buchner funnel, and the filtrate was centrifuged at 12,400 × g for 30 min at 4°C to remove small soil particles. The supernatant was evaporated to a volume of 8 ml, acidified to pH 2.0 with 10% TFA, extracted twice with 20 ml of ethyl acetate, and evaporated to dryness. Extracts were suspended in 1 ml of 35% acetonitrile (ACN) and 0.1% TFA, and then centrifuged in an Eppendorf 5415 centrifuge at 16,000 × g for 20 min at room temperature prior to separation and identification by HPLC. The extraction efficiency of DAPG was approximately 60% (4). The Waters HPLC system consisted of a 717 Plus autosampler, a 600E solvent delivery system, a 600 controller, and a 996 photodiode array detector. Root extracts were fractionated by C18 reverse-phase HPLC (Waters symmetry column, 3.9 by 150 mm) with 50- to 200-μl sample injections. Solvent conditions included a flow rate of 0.5 ml/min with a 2-min initialization at 10% ACN–0.1% TFA followed by a 20-min gradient to 100% ACN–0.1% TFA using curve profile 5. HPLC gradient profiles were monitored at 270 and 330 nm, values which represent the peak maxima of DAPG in the designated solvent system. The seven-point standard curve used for quantification was generated by spiking known concentrations of pure DAPG into root samples (30 g), collected from wheat grown in Quincy virgin soil, prior to the extraction procedure described above. A highly significant linear relationship was found for the standard curve (DAPG = 0.00156 × A, r2 = 0.99, P < 0.0001), in which DAPG represents the total amount of DAPG (in nanograms) and A represents the area of the DAPG peak.
Statistical analysis.
Nonlinear regression analyses (SPSS, Inc., release 7.5) were performed to determine the relationship between the initial density of Q2-87, 1M1-96, or Q8r1-96 on the seed and their final density in the rhizosphere of 3-week-old wheat plants. The initial and final population densities were transformed to log10(CFU + 1) prior to the regression analyses. The equation used in the nonlinear regression analyses is based on the Michaelis-Menten kinetics (saturation function). This equation is Y = α∗X/(β + X), where Y represents the final density (log CFU gram of root−1), X is the initial density (log CFU seed−1), α is the maximum final density, and β is the initial density necessary to reach half of the maximum final density. In biocontrol assays, differences between rhizosphere population densities and shoot height were determined by analysis of variance followed by Tukey's studentized range test (SAS Institute, Inc., Cary N.C.). For shoot height, the distance between the stem base and the tip of the longest leaf was used. Differences in disease severity were analyzed by using the Wilcoxon rank-sum test (α = 0.05). Each experiment was performed at least twice.
RESULTS
Genotypic diversity.
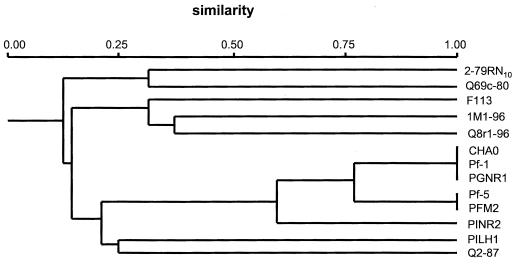
Wheat was grown in Quincy TAD soil for eight successive cycles of four weeks each and DAPG-producing Pseudomonas spp. were isolated by colony hybridization followed by PCR. The population density of DAPG producers was below the level of detection in the first cycle, increased during the second, third, and fourth cycles to a density of 106 CFU g of root−1 and thereafter stabilized at a density of approximately 2 × 105 CFU g of root−1. A detailed representation of the population dynamics of DAPG-producing Pseudomonas spp. in the Quincy TAD soil was described previously (38). From this cycling experiment, 101 DAPG-producing isolates were selected for RAPD analysis with primers M13 and D7. Amplification with primers M13 and D7 gave rise to 51 and 52 bands, respectively, which ranged in size from approximately 50 to 2,300 bp. The reproducibility of the amplification patterns was confirmed in three independent experiments. Sixteen different groups with a unique RAPD profile were identified among the 101 isolates. One RAPD group made up 49.9% of the selected DAPG isolates and represented, for cycles 2 through 8, an average population density of 2.2 × 105 CFU g of root−1. Pseudomonas sp. strain Q8r1-96, isolated after eight successive cycles, was selected as the representative isolate of this dominant RAPD group. Calculations of the pairwise coefficients of similarity showed that strain Q8r1-96 gave a RAPD fingerprint that was different from those obtained from other DAPG-producing Pseudomonas strains, including F113, CHA0, Pf-1, Pf-5, PINR2, Q2-87, and 1M1-96 (Fig. 1). Strains Q8r1-96, Q2-87, and 1M1-96, all isolated from wheat rhizosphere, were selected to determine whether different genotypes have different root-colonizing and biocontrol abilities.
FIG. 1.
Cluster dendrogram of DAPG-producing Pseudomonas strains, isolated from different crops and soils worldwide, based on RAPD analyses with two 10-mer primers. The pairwise coefficients of similarity (Dice) were clustered with the UPGMA algorithm of the NTSYS numerical taxonomy and multivariate analysis system (41). Characteristics of the strains can be found in Table 1. P. fluorescens strains 2-79RN10 and Q69c-80 do not produce DAPG and were included as controls.
Characterization of strains Q8r1-96, Q2-87, and 1M1-96.
Q8r1-96, Q2-87, and 1M1-96 were identified as P. fluorescens biovar II by both gas chromatography (GC)-FAME analysis and classical bacteriological tests (Table 2). Denitrification is a discriminatory characteristic between biovar II and biovar I of P. fluorescens and was observed for all three strains. All three strains had very similar substrate utilization spectra. Differential responses among the strains were observed for only seven substrates: threhalose, ethanol, benzoate, 2-ketogluconate, valerate, dl-norleucine, and l-proline (Table 2). Q8r1-96 was able to utilize trehalose, benzoate, and valerate as a sole carbon source, whereas Q2-87 and 1M1-96 could not. There also were differences among the three strains in casein hydrolysis and gelatinase activity. All three strains produced DAPG in vitro and in the rhizosphere of wheat (Table 2).
TABLE 2.
Biochemical characteristics of Pseudomonas strains Q2-87, 1M1-96, and Q8r1-96
| Characteristic | Q2-87 | 1M1-96 | Q8r1-96 |
|---|---|---|---|
| GC-FAME analysis | P. fluorescens biovar II | P. fluorescens biovar II | P. fluorescens biovar II |
| Growth with SCSa | |||
| Trehalose | − | − | + |
| Ethanol | − | + | + |
| Benzoate | − | − | + |
| 2-Ketogluconate | + | − | + |
| Valerate | − | − | + |
| dl-Norleucine | + | − | − |
| l-Proline | − | + | + |
| Casein hydrolysis | + | − | + |
| Gelatinase (plate) | + | − | − |
| In situ DAPG productionb | |||
| ng/g of root (SE) | 25.0 (8.3) | 141.5 (38.3) | 128.1 (90.5) |
| ng/105 CFU (SE) | 0.8 (0.5) | 2.0 (0.3) | 0.3 (0.2) |
Growth (+) or no growth (−) on various substrates as a sole carbon source (SCS). A total of 53 carbon sources were tested. Assays were performed in Stanier's basal medium. Those carbon sources that showed differential responses by the three isolates are presented here. The other carbon sources for which strains Q2-87, 1M1-96, and Q8r1-96 gave the same response were l-arabinose (+), cellobiose (−), d-fructose (+), d-glucose (+), lactose (−), maltose (−), d-mannitol (+), l-rhamnose (−), d-ribose (+), d-sorbitol (+), sucrose (+), d-xylose (+), adonitol (−), erythritol (−), glycerol (+), geraniol (−), i-inositol (+), sebacic acid (−), acetamide (−), adipate (−), butyrate (−), citraconate (−), d-gluconate (+), M-hydroxybenzoate (−), dl-lactate (+), L-malate (+), pelargonate (+), propionate (+), quinate (+), succinate (+), L-tartrate (−), B-alanine (+), d-alanine (+), betaine (+), glycine (−), l-histidine (+), d-tryptophan (−), l-valine (+), dl-arginine (+), benzylamine (−), butylamine (−), putrescine (+), mesaconate (−), dl-glycerate (+), l-tryptophan (−), and methanol (−).
Production of DAPG on the roots of wheat grown in raw Quincy virgin soil for 4 weeks. Wheat seeds were treated with Q2-87, 1M1-96, or Q8r1-96 at a density of approximately 104 CFU seed−1. Mean values of three replicates are presented. For every replicate, five pots with 25 plants each were used. DAPG production was determined by HPLC and is expressed as the total amount of DAPG produced per gram of root fresh weight (ng g of root−1) and as the amount of DAPG produced per population unit (105 CFU) of the introduced strains. For both parameters, no statistically significant differences were observed between the three strains (Tukey's studentized range test, α = 0.05).
Short-term rhizosphere competence and biocontrol studies.
To test strains Q8r1-96, Q2-87, and 1M1-96 for their ability to control take-all of wheat in Quincy virgin soil, seeds were treated with each of these strains at a density of approximately 106 CFU seed−1. This initial dose was selected because it is on the low end of what is commonly used in biocontrol studies. After 4 weeks of plant growth, all three strains reduced take-all severity to the same extent and to a level similar to that obtained in the Quincy TAD soil (Table 3). Rhizosphere population densities of introduced or naturally occurring DAPG producers were greater than 107 CFU g of root−1 in all treatments except for the Quincy virgin control.
TABLE 3.
“Short-term” bioassay: control of take-all of wheat by DAPG-producing Pseudomonas strains Q2-87, 1M1-96, and Q8r1-96a
| Treatmentb | Population density of DAPG-producing Pseudomonas (CFU g of root−1)c | Disease severityd |
|---|---|---|
| Quincy virgin | ND | 5.5 a |
| Quincy virgin plus Q2-87 | 1.7 × 107 a | 3.7 b |
| Quincy virgin plus 1M1-96 | 5.6 × 107 b | 3.8 b |
| Quincy virgin plus Q8r1-96 | 4.1 × 107 b | 3.4 b |
| Quincy TAD | 2.1 × 107 ab | 2.7 b |
Wheat seeds were treated with Q2-87, 1M1-96, or Q8r1-96 at a density of approximately 106 CFU seed−1 and sown in raw Quincy virgin soil amended with 0.1% (wt/wt) of an oat grain inoculum of the take-all fungus. Untreated seeds sown in Quincy virgin and Quincy TAD soil served as controls. Disease severity and rhizosphere population densities of introduced or naturally occurring DAPG-producing Pseudomonas strains were determined after 4 weeks of plant growth.
The Quincy virgin soil is conducive to take-all of wheat, whereas the complementary Quincy TAD soil is suppressive to take-all.
Mean values of three replicates of six plants each are presented. Population densities of introduced strains Q2-87, 1M1-96, and Q8r1-96 were determined by dilution plating onto KMB+ Rif. Population densities of DAPG-producing Pseudomonas spp. occurring naturally on roots of wheat grown in the Quincy virgin or TAD soil were determined by colony hybridization followed by PCR. Different lowercase letters indicate a statistically significant difference (Tukey's studentized range test, α = 0.05). ND, not detected (below the lower limit of detection of 104 CFU g of root−1).
Mean values of six replicates of two plants each are presented. The severity of take-all was rated on a 0-to-8 scale (0 = no disease; 8 = dead plant). Differences between treatments were analyzed by using the Wilcoxon rank-sum test. Different lowercase letters indicate a statistically significant difference (α = 0.05).
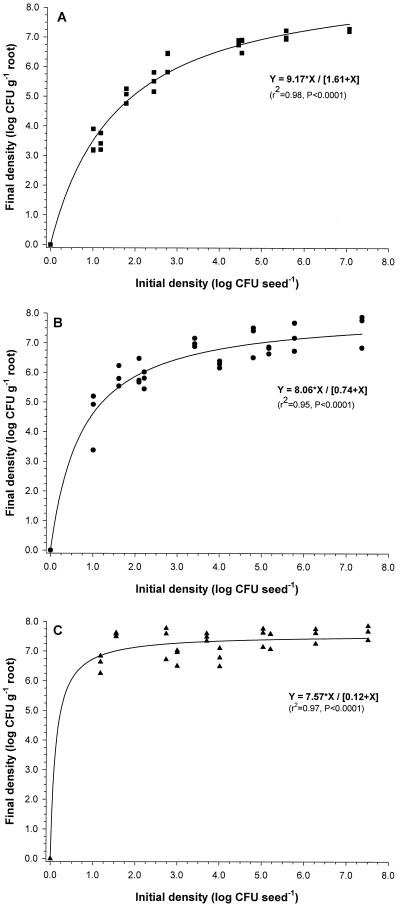
Dose-response studies were performed with the three strains to determine the relationships between the initial density on the seed and the final density in the rhizosphere of wheat grown for 3 weeks in raw Quincy virgin soil. At initial densities of approximately 10 to 100 CFU seed−1, strain Q8r1-96 established rhizosphere population densities of 107 CFU g of root−1 (Fig. 2). In contrast, strains 1M1-96 and Q2-87 required initial densities of approximately 104 and 105 CFU seed−1, respectively, to establish rhizosphere population densities of 107 CFU g of root−1 (Fig. 2). For all three strains, nonlinear regression analyses showed highly significant asymptotic relationships between the initial density on the seed and the rhizosphere population density after 3 weeks of plant growth. The equation used in the nonlinear regression analyses was based on the Michaelis-Menten kinetics (saturation function), initially used to describe substrate-limited growth of bacteria.
FIG. 2.
Relationship between the initial density (log CFU seed−1) and rhizosphere population density (log CFU g of root−1) of P. fluorescens strains Q2-87 (A), 1M1-96 (B), and Q8r1-96 (C). Wheat seeds were treated with each of the strains to obtain final densities of approximately 0, 101, 102, 103, 104, 105, 106, or 107 CFU per seed. Plants were grown for 3 weeks in raw Quincy virgin soil. For each initial density, three replicates were used. The equation used in nonlinear regression analysis is Y = α∗ X/[β + X], where Y represents the final density (log CFU per gram of root), X is the initial density (log CFU seed−1), α is the maximum final density, and β is the initial density necessary to reach half of the maximum final density.
Long-term rhizosphere competence and biocontrol studies.
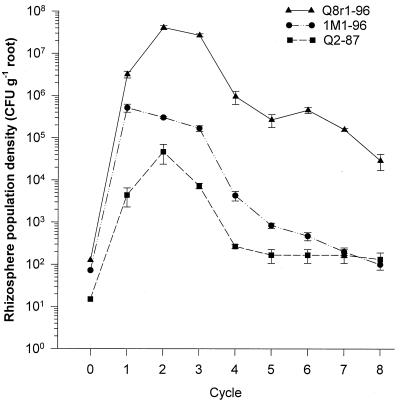
The ability of strains Q8r1-96, 1M1-96, and Q2-87 to colonize the rhizosphere of wheat over an extended period of time was tested in experiments where wheat was grown for successive cycles. All three strains were introduced into raw virgin soils only once (cycle 0) at densities of approximately 10 to 100 CFU g of soil−1. The rhizosphere competence of strain Q8r1-96 was substantially greater than that of the other two strains (Fig. 3). In Quincy virgin soil, strain Q8r1-96 established rhizosphere population densities ranging from 107 to 5 × 107 CFU g of root−1 during the first three growth cycles of wheat, and thereafter its population density decreased relatively slowly to a density of approximately 105 CFU g of root−1 in cycles seven and eight. In contrast, during the first three growth cycles, strains Q2-87 and 1M1-96 established rhizosphere population densities that were 100- to 1,000-fold lower than those established by Q8r1-96. Furthermore, the populations of Q2-87 and 1M1-96 dropped relatively quickly to densities of approximately 2 × 102 and 5 × 102 CFU g of root−1, respectively, after five successive growth cycles of wheat and were not detectable (lower limit of detection was 102 CFU g of root−1) after seven cycles. When Q8r1-96, Q2-87, and 1M1-96 were introduced into Lind and Moses Lake virgin soils, the population dynamics during successive cycling were very similar to those in the Quincy virgin soil. After eight successive growth cycles, the take-all fungus was introduced into bacterium-treated soils and nontreated Quincy virgin and TAD soils, which served as controls. Strain Q8r1-96 maintained a rhizosphere population density of 1.9 × 105 CFU g of root−1 and provided significant control of take-all of wheat (Table 4; Fig. 4). No control of take-all occurred in soils treated with strains Q2-87 and 1M1-96, and the populations of both strains remained undetectable. The suppression provided by Q8r1-96 was similar to the level of control obtained in the Quincy TAD soil. The difference in shoot height between plants grown in the Quincy TAD soil and Quincy virgin soil treated with Q8r1-96 (Fig. 4) seems not to be related to take-all severity but may have been due to differences in the nutrient status between the two soils.
FIG. 3.
Population dynamics of P. fluorescens strains Q2-87, 1M1-96, and Q8r1-96 on the roots of wheat grown in raw Quincy virgin soil for eight successive cycles of 3 weeks each. Strains Q2-87, 1M1-96, and Q8r1-96 were introduced into raw Quincy virgin soil to a final density of approximately 10 to 100 CFU g of soil−1 (cycle 0). At the end of each growth cycle, the rhizosphere population densities of the introduced strains were determined by dilution plating onto rifampin-amended medium. For each strain, three replicates were used. Mean values and standard errors are presented. Strains Q2-87 and 1M1-96 were not detected anymore in the seventh and eighth cycles and were assigned a population size of 102 CFU g of root−1, which is the lower limit of detection.
TABLE 4.
“Long-term” bioassay: control of take-all of wheat by DAPG-producing Pseudomonas strains Q2-87, 1M1-96, and Q8r1-96a
| Treatment | Population density of DAPG producers (CFU g of root−1)b | Shoot ht (cm)c | Disease severityd |
|---|---|---|---|
| Quincy virgin | ND | 15.5 a | 6.0 a |
| Quincy virgin plus Q2-87 | ND* | 15.6 a | 6.1 a |
| Quincy virgin plus 1M1-96 | ND* | 15.0 a | 6.4 a |
| Quincy virgin plus Q8r1-96 | 1.9 × 105 a | 20.9 b | 3.3 b |
| Quincy TAD | 6.3 × 107 b | 26.2 c | 3.8 b |
Strains were introduced into raw Quincy virgin soil at a final density of approximately 100 CFU g of soil−1 (dry weight). After eight successive growth cycles of wheat of 3 weeks each (see Fig. 3), the soils were amended with 0.2% (wt/wt) of an oat grain inoculum of the take-all fungus. Wheat grown in Quincy virgin and Quincy TAD soils for eight successive growth cycles of 3 weeks each served as controls. Disease severity and rhizosphere population densities of introduced or naturally occurring DAPG-producing Pseudomonas strains were determined after 3 weeks of plant growth.
Mean values of three replicates of six plants each are presented. Population densities of introduced strains Q2-87, 1M1-96, and Q8r1-96 were determined by dilution plating onto KMB+ Rif. Population densities of DAPG-producing Pseudomonas spp. occurring naturally on roots of wheat grown in the Quincy virgin or TAD soil were determined by colony hybridization followed by PCR. Different lowercase letters indicate a statistically significant difference (Tukey's studentized range test, α = 0.05). ND, not detected (below lower limit of detection of 104 CFU g of root−1); ND*, not detected (below lower limit of detection of 102 CFU g of root−1).
Mean values of six replicates of two plants each are presented. The severity of take-all was rated on a 0-to-8 scale (0 = no disease; 8 = dead plant). Differences between treatments in shoot height were analyzed by analysis of variance followed by Tukey's studentized range test, whereas differences in disease severity were analyzed by using the Wilcoxon rank-sum test. Different lowercase letters indicate a statistically significant difference (α = 0.05).
FIG. 4.
Long-term bioassay: biological control of take-all of wheat by P. fluorescens strains Q2-87, 1M1-96, and Q8r1-96. Strains Q2-87, 1M1-96, and Q8r1-96 were introduced into raw Quincy virgin soil to a final density of approximately 100 CFU g of soil−1, and wheat was grown for eight successive cycles of 3 weeks each. The population dynamics of each of the strains is shown in Fig. 3. After eight successive cycles, an oat grain inoculum of the take-all pathogen was introduced to a final density of 0.2% (wt/wt). Wheat plants were grown for 3 weeks in the infested soils, after which take-all severity was rated on a 0-to-8 scale, and the shoot height was determined (Table 4). Wheat grown in Quincy virgin (conducive to take-all) and Quincy TAD (suppressive to take-all) soils for eight successive growth cycles of 3 weeks each served as controls. 1, Quincy virgin; 2, Quincy virgin plus Q2-87; 3, Quincy virgin plus 1M1-96; 4, Quincy virgin plus Q8r1-96; 5, Quincy TAD.
DISCUSSION
TAD is a natural biological control of the wheat root disease take-all, which develops in response to a severe outbreak of the disease during extended monoculture of wheat or barley (15). Root-associated fluorescent Pseudomonas spp., which produce the antibiotic DAPG, are highly enriched in various TAD soils (37) and are key components of the natural biological control that operates in these soils (38, 39). Identification and analysis of the genotypic diversity of DAPG producers from wheat-growing regions in the United States and The Netherlands have been conducted recently using a combination of phenotypic and PCR-based molecular techniques (31). A significant amount of diversity occurs within this group of fluorescent pseudomonads isolated from wheat rhizosphere. For example, in the Quincy TAD soil, the subject of this and several other studies (34, 37, 38), three and four distinct genotypes were identified on the basis of whole-cell BOX-PCR and ERIC-PCR, respectively (31). In our study of 101 isolates collected during 8 months of successive growth cycles of wheat in the Quincy TAD soil, 16 groups were identified by RAPD analysis with M13 and D7, two 10-mer primers that give consistent and reproducible banding patterns. We opted for the use of M13 and D7 in our study, because extended cycling of wheat might cause only subtle changes in the genotypic diversity of DAPG producers and two primers would increase our ability to detect changes in diversity within the population. The level of genotypic diversity in the Quincy TAD soil may reflect the cropping history of the field prior to the collection of the soil in 1995 for this study. Between 1958 and 1980, the Quincy TAD field had been cropped only to wheat; however, from 1981 to 1995 other crops (sweet corn, field corn, lima beans, or dry beans) were rotated with wheat.
One key question has been whether DAPG-producing isolates from monoculture wheat field soils contribute equally to the phenomenon of TAD. Knowledge of the diversity within these populations may implicate specific subsets of DAPG-producing Pseudomonas spp. in TAD and may allow the identification of strains that have different abilities to colonize the rhizosphere and suppress soil-borne pathogens of wheat. Different genotypes of DAPG-producing Pseudomonas spp. have been reported to differ in their ability to suppress Fusarium crown and root rot and Pythium root rot (42), to produce other antibiotics in addition to DAPG (21, 42), and to colonize roots of maize plants of different growth stages (33). Our results indicate that wheat exerts a specific selection pressure on a certain DAPG-producing genotype, and this genotype is highly adapted to the wheat rhizosphere. Of the 16 RAPD groups identified on roots of wheat grown successively in the Quincy TAD soil, one group made up 50% of the total population of DAPG-producing Pseudomonas spp., representing, on average, a population density of approximately 2 × 105 CFU g of root−1. This genotype, exemplified by P. fluorescens Q8r1-96 and corresponding BOX-PCR-group D, is genotypically different from other well-known DAPG-producing Pseudomonas strains, including F113, CHA0, Pf-1, Pf-5, PINR2, and Q2-87 (Fig. 1) and is common in some monoculture wheat field soils in the northern United States (31).
Evidence that Q8r1-96 is highly adapted to the wheat rhizosphere was provided by our colonization studies with three different genotypes Q8r1-96, 1M1-96, and Q2-87. Short-term studies clearly showed that Q8r1-96 requires a much lower dose (only 10 to 100 CFU seed−1 or soil−1) to establish rhizosphere population densities of 107 CFU g of root−1 than does Q2-87 or 1M1-96. The model used in the nonlinear regression analyses to describe the relationship between initial dose and final rhizosphere population densities of Q8r1-96, Q2-87, and 1M1-96 (Fig. 2) was initially developed to describe substrate-limited growth by bacteria. Earlier studies have applied linear regression analyses to describe the relationship between a limited range of initial and final densities of introduced strains (5, 35). However, the asymptotic nature of the model used in this study makes it much more appropriate for describing the relationship for a wide range of initial densities. Furthermore, the model parameter β, which represents the initial density required to reach half of the maximal attainable rhizosphere population density, provides a quantitative measure for rhizosphere competence of a particular Pseudomonas strain in a given set of abiotic and biotic conditions. For strain Q8r1-96, the β value was 0.12, which means that, in raw Quincy virgin soil, Q8r1-96 needs only 0.12 log CFU seed−1 to reach a rhizosphere population density of 3.79 log CFU g of root−1 (half of 7.57 log CFU g of root−1) after 3 weeks of plant growth. For strains Q2-87 and 1M1-96, the β values were 1.61 and 0.74, respectively, illustrating again that strain Q2-87 and 1M1-96 are less rhizosphere competent than Q8r1-96 (Fig. 2). It should be emphasized, however, that spatial colonization patterns of the introduced strains as well as their performance and persistence on different wheat cultivars grown under field conditions were not taken into account in this study. These aspects of rhizosphere competence will be addressed in future studies.
The strongest evidence that Q8r1-96 is highly adapted to the wheat rhizosphere was provided in our long-term cycling experiments. After successive growth cycles of wheat, Q8r1-96 maintained a population density of approximately 105 CFU g of root−1 (Fig. 3), a density that is very similar to the density at which this strain occurs naturally on roots of wheat grown in the Quincy TAD soil. The observation that Q8r1-96 showed the same population dynamics relative to Q2-87 and 1M1-96 during successive cycling of wheat in the Lind and Moses Lake virgin soils, both of which have different physical-chemical properties, suggests that its superior rhizosphere competence is not soil specific. To our knowledge, this superior ability to establish and maintain high rhizosphere population densities over an extended period of plant growth is quite unique among rhizobacteria. During the last two decades, studies on the population dynamics of other Pseudomonas strains have shown that, in general, population densities declined substantially over a prolonged period of time, often to levels below the detection limit (2, 17, 26, 30, 36, 51). In an attempt to maintain population densities above the threshold required for pathogen suppression or growth promotion, higher initial doses have been applied to seed or soil (5). Nevertheless, variable colonization (52) and the cost of applying large doses remain a major impediment to the widespread use of rhizobacteria in commercial agriculture.
Because microorganisms in the rhizosphere depend on substrates liberated from the root for their growth, the host plant profoundly influences the quantity and composition of indigenous microorganisms as well as the population dynamics of introduced strains. Qualitative and quantitative differences have been observed between the rhizosphere microflora of crop cultivars that were either susceptible or resistant to a given soil-borne pathogen (12). Lemanceau et al. (24) have demonstrated crop-specific influences on the population structure of fluorescent Pseudomonas spp. from a silty loam soil and from the rhizosphere, rhizoplane, or root interior of flax and tomato plants grown in that soil. A much higher proportion of tomato isolates than flax isolates could assimilate inositol, ribose, saccharose, trehalose, erythritol, m-hydroxybenzoate, and 5-cetogluconate. Similarly, Mavingui et al. (29) found that among many strains of Bacillus polymyxa from bulk soil and the rhizosphere environment of wheat, all the rhizoplane isolates could metabolize sorbitol and shared identical restriction fragment length polymorphism patterns. To begin to identify the mechanism(s) responsible for the superior rhizosphere competence of strain Q8r1-96, it seems reasonable to first consider its ability to utilize trehalose, valerate, and/or benzoate. These three carbon sources were the only ones, among the 53 substrates tested, that Q8r1-96 could utilize but both Q2-87 and 1M1-96 could not (Table 2). Interestingly, trehalose has been implicated in osmotolerance and, additionally, as an inducer of antagonism toward Pythium debaryanum in P. fluorescens ATCC 17400 (11). Furthermore, Frey et al. (10) suggested that several fungi, and in particular the ectomycorrhizal fungus Laccaria bicolor, release trehalose and thereby may exert a selection on fluorescent Pseudomonas spp. that are able to efficiently utilize this substrate. Benzoate has been shown to act as a chemo-attractant for Azospirillum spp. (28) and may provide Q8r1-96 with a competitive advantage in the first steps of root colonization. Certain substrates contained in root exudates are also known to act as specific inducers of gene expression in plant-associated bacteria. In this context, Van Overbeek and Van Elsas (48) screened several transcriptional fusion mutants for their response to wheat root exudates and identified one mutant in which gene expression was specifically induced by proline. Both strain Q8r1-96 and 1M1-96 can utilize proline as a sole carbon source, whereas the least rhizosphere competent strain Q2-87 could not (Table 2). Further investigation will be necessary to conclusively identify the role of these and possibly other substrates. The superior rhizosphere competence of Q8r1-96 seems not to be related to in situ DAPG production levels. No significant differences were observed between the three strains with respect to the total amount of DAPG produced in the rhizosphere of wheat and the amount of DAPG produced per population unit (Table 2). These production levels are similar to those reported earlier for Q2-87 and Q8r1-96 (4, 39). In addition, Carroll et al. (6) reported that loss of DAPG production did not reduce the ecological fitness of P. fluorescens F113 in the rhizosphere of sugar beets.
No difference in the ability of strains Q8r1-96, Q2-87, and 1M1-96 to suppress take-all were observed in short-term studies (Table 3). This is not surprising given that all three strains produced similar amounts of DAPG in situ and established rhizosphere population densities above the threshold density (105 CFU g of root−1 [38]) required to control take-all of wheat. In contrast, Q8r1-96 differed significantly from the other two strains in the suppression of take-all in the long-term cycling experiment (Table 4; Fig. 4), reflecting the fact that only Q8r1-96 has sufficient rhizosphere competence to maintain its density above the threshold density for an extended period of time. This experiment highlights the key role of root colonization in the suppression of take-all. The relationship between take-all suppression and the rhizosphere population density of DAPG-producing Pseudomonas spp. (38) also showed that there is no significant increase in the level of suppression at densities above the threshold density. This observation explains why the level of suppression provided by Q8r1-96 was similar to that obtained in the Quincy TAD soil, in spite of significantly higher rhizosphere population densities of indigenous DAPG-producers in the TAD soil (Table 4; Fig. 4). The results of this study suggest that one particular genotype of DAPG-producing Pseudomonas spp., found on roots of wheat grown in the Quincy TAD soil, is responsible for a major part, if not all, of the natural suppression that operates in this particular Quincy TAD soil. McSpadden-Gardener et al. (31) found that nearly one-third of the DAPG isolates obtained from different wheat-growing areas in the United States were genotypically similar to Q8r1-96. Furthermore, preliminary results have indicated that these isolates have the same root-colonizing ability (B. B. McSpadden-Gardener and D. M. Weller, unpublished data). Collectively, these results suggest that Q8r1-like strains are able to successfully adapt to the wheat rhizosphere regardless of the prevailing biotic and abiotic conditions. More genotypes of DAPG-producing Pseudomonas spp. naturally present in TAD soils, including those that represent other RAPD groups, should be evaluated to support these hypotheses. Root colonization studies with crops other than wheat should be performed to evaluate the rhizosphere competence and biocontrol efficacy of strain Q8r1-96 in other host-pathogen systems.
In conclusion, the present study demonstrated that genotypic diversity within a group of antagonistic microorganisms that share a common biocontrol trait has great potential for improving biological control. This approach capitalizes on existing knowledge concerning mechanisms, while exploiting the differences among strains to face the challenges of diverse soil and rhizosphere environments. By matching rhizobacterial genotypes with crops or varieties for which they have a colonization preference, root colonization and biocontrol can be increased without increasing the amount of inoculum.
ACKNOWLEDGMENTS
This research was supported by grant 94-37107-0439 from the U.S. Department of Agriculture, Office of Grants and Program Systems, National Research Initiative, Competitive Grants Program.
We thank K. Hays, O. Tak-Wong, S. E. Kalloger, and K. L. Schroeder for their technical support. We are grateful to I. A. Lamour for her advice on the nonlinear regression analyses, to R. F. Bonsall for HPLC analysis, and to L. S. Thomashow for her insightful comments.
REFERENCES
- 1.Bahme J B, Schroth M N. Spatial-temporal colonization patterns of a rhizobacterium on underground organs of potatoes. Phytopathology. 1987;77:1093–1100. [Google Scholar]
- 2.Bakker P A H M, Lamers J G, Bakker A W, Marugg J D, Weisbeek P J, Schippers B. The role of siderophores in potato growth stimulation by Pseudomonas putida in a short rotation of potato. Neth J Plant Pathol. 1986;92:249–256. [Google Scholar]
- 3.Bender C L, Rangaswamy V, Loper J E. Polyketide production by plant-associated pseudomonads. Annu Rev Phytopathol. 1999;37:175–196. doi: 10.1146/annurev.phyto.37.1.175. [DOI] [PubMed] [Google Scholar]
- 4.Bonsall R F, Weller D M, Thomashow L S. Quantification of 2,4-diacetylphloroglucinol produced by fluorescent Pseudomonas spp. in vitro and in the rhizosphere of wheat. Appl Environ Microbiol. 1997;63:951–955. doi: 10.1128/aem.63.3.951-955.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bull C T, Weller D M, Thomashow L S. Relationship between root colonization and suppression of Gaeumannomyces graminis var. tritici by Pseudomonas fluorescens strain 2-79. Phytopathology. 1991;81:954–959. [Google Scholar]
- 6.Carroll H, Moenne-Loccoz Y, Dowling D N, O'Gara F. Mutational disruption of the biosynthesis genes coding for the antifungal metabolite 2,4-diacetylphloroglucinol does not influence the ecological fitness of Pseudomonas fluorescens F113 in the rhizosphere of sugarbeets. Appl Environ Microbiol. 1995;61:3002–3007. doi: 10.1128/aem.61.8.3002-3007.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook R J, Weller D M. Management of take-all in consecutive crops of wheat or barley. In: Chet I, editor. Innovative approaches to plant disease. New York, N.Y: Wiley Interscience; 1987. pp. 41–76. [Google Scholar]
- 8.Dekkers L C, Poehlich C C, van der Fits L, Lugtenberg B J J. A site-specific recombinase is required for competitive root colonization by Pseudomonas fluorescens WCS365. Proc Natl Acad Sci USA. 1998;95:7051–7056. doi: 10.1073/pnas.95.12.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenton A M, Stephens P M, Crowley J, O'Callaghan M, O'Gara F. Exploitation of gene(s) involved in 2,4-diacetylphloroglucinol biosynthesis to confer a new biocontrol capability to a Pseudomonas strain. Appl Environ Microbiol. 1992;58:3873–3878. doi: 10.1128/aem.58.12.3873-3878.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frey P, Frey-Klett P, Garbaye J, Berge O, Heulin T. Metabolic and genotypic fingerprinting of fluorescent pseudomonads associated with the Douglas fir-Laccaria bicolor mycorrhizosphere. Appl Environ Microbiol. 1997;63:1852–1860. doi: 10.1128/aem.63.5.1852-1860.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaballa A, Abeysinghe P D, Urich G, Matthijs S, de Greve H, Cornelis P, Koedam N. Trehalose induces antagonism towards Pythium debaryanum in Pseudomonas fluorescens ATCC 17400. Appl Environ Microbiol. 1997;63:4340–4345. doi: 10.1128/aem.63.11.4340-4345.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert S G, Handelsman J, Parke J L. Root camouflage and disease control. Phytopathology. 1994;84:222–225. [Google Scholar]
- 13.Handelsman J, Stabb E V. Biocontrol of soilborne plant pathogens. Plant Cell. 1996;8:1855–1869. doi: 10.1105/tpc.8.10.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison L A, Letendre L, Kovacevich P, Pierson E A, Weller D M. Purification of an antibiotic effective against Geaumannomyces graminis var. tritici produced by a biocontrol agent, Pseudomonas aureofaciens. Soil Biol Biochem. 1993;25:215–221. [Google Scholar]
- 15.Hornby D. Suppressive soils. Annu Rev Phytopathol. 1983;21:65–85. [Google Scholar]
- 16.Howell C R, Stipanovic R D. Control of Rhizoctonia solani on cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium. Phytopathology. 1979;69:480–482. [Google Scholar]
- 17.Howie W J, Cook R J, Weller D M. Effects of soil matric potential and cell motility on wheat root colonization by fluorescent pseudomonads suppressive to take-all. Phytopathology. 1987;77:286–292. [Google Scholar]
- 18.Johnson K B. Dose-response relationships and inundative biological control. Phytopathology. 1994;84:780–784. [Google Scholar]
- 19.Johnson K B, DiLeone J A. Effect of antibiosis on antagonist dose-plant disease response relationships for the biological control of crown gall of tomato and cherry. Phytopathology. 1999;89:974–980. doi: 10.1094/PHYTO.1999.89.10.974. [DOI] [PubMed] [Google Scholar]
- 20.Keel C, Schnider U, Maurhofer M, Voisard C, Laville J, Burger P, Wirthner P, Haas D, Défago G. Suppression of root diseases of by Pseudomonas fluorescens CHA0: importance of the secondary metabolite 2,4-diacetylphloroglucinol. Mol Plant-Microbe Interact. 1992;5:4–13. [Google Scholar]
- 21.Keel C, Weller D M, Natsch A, Défago G, Cook R J, Thomashow L S. Conservation of the 2,4-diacetylphloroglucinol biosynthesis locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl Environ Microbiol. 1996;62:552–563. doi: 10.1128/aem.62.2.552-563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King E O, Ward M K, Raney D E. Two simple media for demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 23.Larkin R P, Fravel D R. Mechanisms of action and dose-response relationships governing biological control of Fusarium wilt of tomato by nonpathogenic Fusarium spp. Phytopathology. 1999;89:1152–1161. doi: 10.1094/PHYTO.1999.89.12.1152. [DOI] [PubMed] [Google Scholar]
- 24.Lemanceau P, Corberand T, Gardan L, Latour X, Laguerre G, Boeufgras J M, Alabouvette C. Effect of two plant species, flax (Linum usitatissinum L.) and tomato (Lycopersicon esculentum Mill.), on the diversity of soilborne populations of fluorescent pseudomonads. Appl Environ Microbiol. 1995;61:1004–1012. doi: 10.1128/aem.61.3.1004-1012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levy E, Gough F J, Berlin K D, Guiana P W, Smith J T. Inhibition of Septoria tritici and other phytopathogenic fungi and bacteria by Pseudomonas fluorescens and its antibiotics. Plant Pathol. 1992;41:335–341. [Google Scholar]
- 26.Loper J E, Suslow T V, Schroth M N. Lognormal distribution of bacterial populations in the rhizosphere. Phytopathology. 1984;74:1454–1460. [Google Scholar]
- 27.Loper J E, Lindow S E. Reporter gene systems useful in evaluating in situ gene expression by soil- and plant-associated bacteria. In: Hurst C J, Knudsen G R, McInerney M J, Stetzenbach L D, Walter M V, editors. Manual of environmental microbiology. Washington, D.C.: ASM Press; 1997. pp. 482–492. [Google Scholar]
- 28.Lopez-de-Victoria G, Lovell C R. Chemotaxis of Azospirillum species to aromatic compounds. Appl Environ Microbiol. 1993;59:2951–2955. doi: 10.1128/aem.59.9.2951-2955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mavingui P, Laguerre G, Berge O. Genetic and phenotypic diversity of Bacillus polymyxa in soil and in the wheat rhizosphere. Appl Environ Microbiol. 1992;58:1894–1903. doi: 10.1128/aem.58.6.1894-1903.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazzola M, Cook R J. Effects of fungal root pathogens on the population dynamics of biocontrol, strains of fluorescent pseudomonads in the wheat rhizosphere. Appl Environ Microbiol. 1991;57:2171–2178. doi: 10.1128/aem.57.8.2171-2178.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McSpadden-Gardener B B, Schroeder K L, Kalloger S E, Raaijmakers J M, Thomashow L S, Weller D M. Genotypic and phenotypic diversity of phlD-containing Pseudomonas isolated from the rhizosphere of wheat. Appl Environ Microbiol. 2000;66:1939–1946. doi: 10.1128/aem.66.5.1939-1946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parke J L. Root colonization by indigenous and introduced microorganisms. In: Keister D L, Gregan P B, editors. The rhizosphere and plant growth. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1991. pp. 33–42. [Google Scholar]
- 33.Picard C, di Cello F, Ventura M, Fani R, Guckert A. Frequency and diversity of 2,4-diacetylphloroglucinol-producing bacteria isolated from the maize rhizosphere at different stages of growth. Appl Environ Microbiol. 2000;66:948–955. doi: 10.1128/aem.66.3.948-955.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierson E A, Weller D M. Use of mixtures of fluorescent pseudomonads to suppress take-all and improve growth of wheat. Phytopathology. 1994;84:940–947. [Google Scholar]
- 35.Raaijmakers J M, Leeman M, van Oorschot M M P, van der Sluis I, Schippers B, Bakker P A H M. Dose-response relationships in biological control of fusarium wilt of radish by Pseudomonas spp. Phytopathology. 1995;85:1075–1081. [Google Scholar]
- 36.Raaijmakers J M, van der Sluis I, Koster M, Bakker P A H M, Weisbeek P J, Schippers B. Utilization of heterologous siderophores and rhizosphere competence of fluorescent Pseudomonas spp. Can J Microbiol. 1995;41:126–135. [Google Scholar]
- 37.Raaijmakers J M, Weller D M, Thomashow L S. Frequency of antibiotic-producing Pseudomonas spp. in natural environments. Appl Environ Microbiol. 1997;63:881–887. doi: 10.1128/aem.63.3.881-887.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raaijmakers J M, Weller D M. Natural plant protection by 2,4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. Mol Plant-Microbe Interact. 1998;11:144–152. [Google Scholar]
- 39.Raaijmakers J M, Bonsall R F, Weller D M. Effect of population density of Pseudomonas fluorescens on production of 2,4-diacetylphloroglucinol in the rhizosphere of wheat. Phytopathology. 1999;89:470–475. doi: 10.1094/PHYTO.1999.89.6.470. [DOI] [PubMed] [Google Scholar]
- 40.Rohlf E J. NTSYS-pc: numerical taxonomy and multivariate analysis system for IBM microcomputer, version 1.8. Setauket, N.Y: Applied Biostatistics, Inc.; 1994. [Google Scholar]
- 41.Shanahan P, O'Sullivan D J, Simpson P, Glennon J D, O'Gara F. Isolation of 2,4-diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Appl Environ Microbiol. 1992;58:353–358. doi: 10.1128/aem.58.1.353-358.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharifi-Tehrani A, Zala M, Natsch A, Moënne-Loccoz Y, Défago G. Biocontrol of soil-borne fungal plant diseases by 2,4-diacetylphloroglucinol-producing fluorescent pseudomonads with different restriction profiles of amplified 16S rDNA. Eur J Plant Pathol. 1998;104:631–643. [Google Scholar]
- 43.Simon A, Ridge E H. The use of ampicillin in a simplified selective medium for the isolation of fluorescent pseudomonads. J Appl Bacteriol. 1974;37:459–460. doi: 10.1111/j.1365-2672.1974.tb00464.x. [DOI] [PubMed] [Google Scholar]
- 44.Smith K P, Handelsman J, Goodman R. Modeling dose-response relationships in biological control: partitioning host responses to the pathogen and the biocontrol agent. Phytopathology. 1997;87:720–729. doi: 10.1094/PHYTO.1997.87.7.720. [DOI] [PubMed] [Google Scholar]
- 45.Stabb E V, Jacobson L M, Handelsman J. Zwittermycin A-producing strains of Bacillus cereus from diverse soils. Appl Environ Microbiol. 1994;60:4404–4412. doi: 10.1128/aem.60.12.4404-4412.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomashow L S, Weller D M. Role of phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J Bacteriol. 1988;170:3499–3508. doi: 10.1128/jb.170.8.3499-3508.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomashow L S, Weller D M. Current concepts in the use of introduced bacteria for biological disease control: mechanisms and antifungal metabolites. In: Stacey G, Keen N T, editors. Plant-microbe interactions. Vol. 1. New York, N.Y: Chapman & Hall; 1996. pp. 187–236. [Google Scholar]
- 48.Van Overbeek L S, van Elsas J D. Root exudate induced promoter activity in Pseudomonas fluorescens mutants in the rhizosphere of wheat. Appl Environm Microbiol. 1995;61:890–898. doi: 10.1128/aem.61.3.890-898.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vincent M N, Harrison L A, Brackin J M, Kovacevich P A, Murkerji P, Weller D M, Pierson E A. Genetic analysis of the antifungal activity of a soilborne Pseudomonas aureofaciens strain. Appl Environ Microbiol. 1991;57:2928–2934. doi: 10.1128/aem.57.10.2928-2934.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weller D M. Colonization of wheat roots by a fluorescent pseudomonad suppressive to take-all. Phytopathology. 1983;73:1548–1553. [Google Scholar]
- 51.Weller D M. Distribution of a take-all suppressive strain of Pseudomonas fluorescens on seminal roots of winter wheat. Appl Environ Microbiol. 1984;48:897–899. doi: 10.1128/aem.48.4.897-899.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weller D M. Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu Rev Phytopathol. 1988;26:379–407. [Google Scholar]
- 53.Weller D M, Zhang B-X, Cook R J. Application of a rapid screening test for selection of bacteria suppressive to take-all of wheat. Plant Dis. 1985;69:710–713. [Google Scholar]
- 54.Weller D M, Raaijmakers J M, Thomashow L S. The rhizosphere ecology of antibiotic-producing pseudomonads and their role in take-all decline. In: Ogoshi A, Kobayashi K, Homma Y, Kodama F, Kondo N, Akino S, editors. Plant growth-promoting rhizobacteria: present status and future prospects. Sapporo, Japan: Kakanishi Printing; 1997. pp. 58–64. [Google Scholar]
- 55.Whipps J M. Developments in the biological control of soil-borne plant pathogens. Adv Bot Res. 1997;26:1–133. [Google Scholar]