Abstract
Structural variants (SVs) are large rearrangements (>50 bp) within the genome that impact gene function and the content and structure of chromosomes. As a result, SVs are a significant source of functional genomic variation, that is, variation at genomic regions underpinning phenotype differences, that can have large effects on individual and population fitness. While there are increasing opportunities to investigate functional genomic variation in threatened species via single nucleotide polymorphism (SNP) data sets, SVs remain understudied despite their potential influence on fitness traits of conservation interest. In this future‐focused Opinion, we contend that characterizing SVs offers the conservation genomics community an exciting opportunity to complement SNP‐based approaches to enhance species recovery. We also leverage the existing literature–predominantly in human health, agriculture and ecoevolutionary biology–to identify approaches for readily characterizing SVs and consider how integrating these into the conservation genomics toolbox may transform the way we manage some of the world's most threatened species.
Keywords: conservation genetics, fitness traits, functional diversity, genome‐wide diversity, pangenomes, structural variation
1. STRUCTURAL VARIANTS: A NEW TOOL IN THE CONSERVATION GENOMICS TOOLBOX
Over the past 10 years, the field of conservation genomics has largely focused on transitioning from a handful of microsatellite markers to thousands of genome‐wide single nucleotide polymorphisms (SNPs) (Allendorf et al., 2010; Mable, 2019). Whether characterized using reduced‐representation data or whole‐genome sequence (WGS) methods, SNP‐based estimates of genome‐wide variation are being used to better inform conservation by delineating species, including cryptic species (e.g., Binks et al., 2021; Quattrini et al., 2019; but see Stanton et al., 2019), detecting hybridization and introgression (Dufresnes & Dubey, 2020; Peters et al., 2016; but see Forsdick et al., 2021 preprint; Hauser et al., 2021), identifying conservation units (Liddell et al., 2020), informing conservation translocations (Dresser et al., 2017; Glassock et al., 2021), guiding conservation breeding programmes (e.g., Galla et al., ; Wright et al., 2020) and identifying the genomic basis of adaptive traits (Duntsch et al., 2020). However, SNPs constitute only one component of genome‐wide variation (Ho et al., 2020). Structural variants (SVs) are genomic rearrangements such as insertions, deletions, duplications, inversions, and translocations, and are generally defined as ≥50 bp in length (Figure 1). Variation in chromosome structure was one of the earliest types of genetic variation studied (e.g., McClung, 1905; Sturtevant, 1921), and a recent focus on the impacts of these genomic rearrangements on sequence variation has found that SVs affect more overall genome content and intersect with genes more often than SNPs (Catanach et al., 2019; Chakraborty et al., 2019; Chiang et al., 2017; Frayling, 2014; Pang et al., 2010).
FIGURE 1.
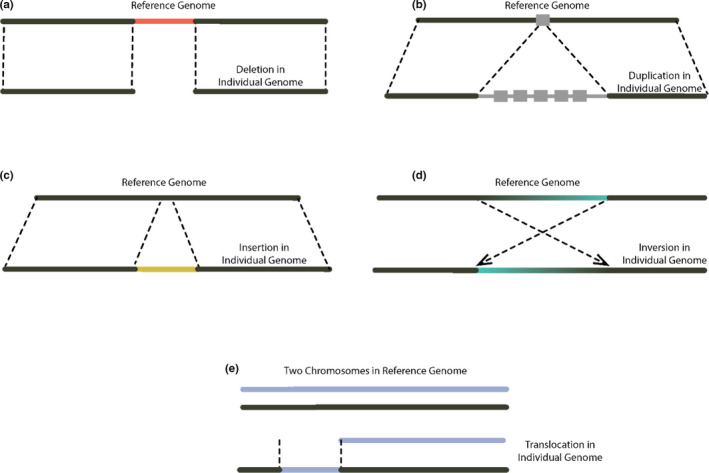
Structural variant (SV) types and how they differ from a reference genome: (a) deletion, where a segment of DNA is not present in an individual, but present in the reference; (b) duplication, a rearrangement where there is more than one copy of a particular region of the genome, often in tandem. These can be either intrachromosomal, as shown here, or interchromosomal; (c) insertion, a DNA sequence present in an individual sample, but not present in the reference; (d) inversion, a segment of the chromosome that has had a double‐stranded break at an upstream and downstream location and become reversed in order; (e) translocation, a rearrangement where a portion of one chromosome has been transposed onto another
Structural variants can have significant evolutionary impacts on species. These impacts include genomic incompatibilities between ancestral and novel variants that may influence hybridization and introgression (Weissensteiner et al., 2020) and may even lead to speciation (Davey et al., 2016; Todesco et al., 2020). In addition, emerging evidence indicates that SVs can underlay fine‐scale population structure, and may facilitate local adaptation (Cayuela et al., 2021; Dorant et al., 2020; Tigano et al., 2021). These findings–which are generally the result of integrating SNPs and SVs–highlight the opportunity for both variant types to inform conservation. Nevertheless, the broad application of SVs in conservation remains challenging in part due to the uncertainty in how best to detect and genotype all SV types at the population scale (Mérot et al., 2020). Further, although SVs are more likely to impact fitness traits than SNPs (Pang et al., 2010), the overall impacts of SVs on population persistence is relatively unexplored (Oomen et al., 2020).
Here, we leverage our cross‐sector expertise in conservation (AWS, CJH, JG, JRW, KPK, MLL, SJG, TES), human health (DE), agriculture (JG), quantitative genetics (AWS) and bioinformatics (DE, JG, JRW, MLL) to consider the significance of SVs in population persistence and species extinction risk and discuss opportunities and challenges for relating SVs to fitness traits in small populations. We identify approaches for reliably characterizing SVs with an emphasis on leveraging new and existing genomic resources for species of conservation concern. We also describe how the incorporation of genome assemblies for multiple reference individuals can be used to improve SV discovery and genotyping, even for large and/or complex SVs (Alonge et al., 2020; Eizenga et al., 2020; Gao et al., 2019; Golicz et al., 2016; Tettelin et al., 2005). We contend that the study of SVs has the potential to improve conservation outcomes by enhancing our understanding of genome‐wide variation and traits related to fitness.
2. SMALL POPULATION PARADIGM AND STRUCTURAL VARIANTS
Like SNPs, SVs vary within and among individuals, and SV variation is subject to genetic drift, selection, mutation and gene flow, and is impacted by demography, particularly effective population size (N e). Species and/or populations with large N e should harbour a higher number of SVs, whereas populations with small N e should harbour less (e.g., fewer SV counts in the critically endangered Hawaiian crow versus nonthreatened conspecifics; Weissensteiner et al., 2020). Although also influencing the mutation rates and genomic locations of SNP variants, the genome composition and architecture (e.g., overall chromosome repeat content) has a much larger impact on the number, type, location and mutation rate of SVs (Collins et al., 2020). Further, while both SNPs and SVs can be neutral, their size relative to single base substitutions mean that SVs are more likely to overlap gene and regulatory regions and impact fitness. The evolutionary dynamics of SVs in large populations will therefore be determined by the interplay between the functional effects of these SVs, if any, and variation in mutation rates of different SV sizes and types, which may lead to enrichment of SVs of particular types and/or in particular genomic regions (Conrad & Hurles, 2007).
In contrast, populations of conservation concern are likely to be fragmented or chronically small, and the evolutionary dynamics of both SNP and SV variation will be dominated by genetic drift and founder effects that interplay with the small N e to both decrease the overall level genetic variation and reduce the efficacy of natural (purifying) selection (Henn et al., 2015). Decreasing genetic variation leads to further reductions in N e which increases the probability of extinction through both the loss of adaptive variants and the increase in frequency of deleterious variants. This can lead to a downward spiral of further loss of diversity and increasing extinction risk, a process termed an “extinction vortex” (Gilpin & Soule, 1986). Further, inbreeding is unavoidable in small populations, leading to higher frequencies of deleterious recessive individuals in the population, with this inbreeding depression leading to a further fitness burden on the threatened species (Kardos et al., 2016; Stoffel et al., 2021).
While both SNPs and SVs can have deleterious fitness effects, their size and potential to overlap gene regions mean that SVs may be enriched in the pool of deleterious variants that are maintained in small populations. While SVs of very large deleterious effect may be purged from the population, small populations may selectively accumulate slightly deleterious SVs. This enrichment indicates that SVs probably underpin some negative fitness traits associated with inbreeding depression. Although there are some clues as to the evolutionary dynamics of SVs in small populations, further study is critical for resolving the significance of SVs in population persistence and species extinction risk.
3. RELATING STRUCTURAL VARIANTS TO FITNESS TRAITS
Advancements in WGS technologies coupled with increased computational capacities, have renewed interest in the role of SVs in determining trait differences (Chiang et al., 2017; Pang et al., 2010; Sadowski et al., 2019; Yi & Ju, 2018). For example, characterizing SVs in agricultural species has led to the identification of variants associated with economically significant traits in crops like grapevine (Zhou et al., 2019), maize (Yang et al., 2019), soybean (Liu et al., 2020) and tomato (Alonge et al., 2020), and identified specific genes and gene regions associated with domestication in vertebrates (Bertolotti et al., 2020; Cagan & Blass, 2016; vonHoldt et al., 2017). The impact of these SVs on trait variation may be direct or indirect. For example, SVs can lead to direct changes to gene coding regions and gene regulation that change the function or expression of proteins (Bickhart & Liu, 2014; Collins et al., 2020). In contrast, in some cases where a particular SV is associated with a trait, the SV may not be the causal variant, but rather increase the likelihood of de novo causal mutations nearby. This is the case for a relatively common 1.3 Mb inversion on the human Y‐chromosome where microdeletions accrue at inversion breakpoints, which can result in profound impacts on male fertility (Hallast et al., 2021). There is also growing evidence that SVs can further impact the gene regulatory landscape by altering the formation of topologically associating domains (TADs), genomic regions that physically interact with themselves more frequently than with regions elsewhere (Sadowski et al., 2019; Shanta et al., 2020).
There is growing evidence that SVs–in particular, the suppression of recombination and subsequent evolution of “supergenes”, or tightly linked coadapted alleles–can impact fitness traits in natural populations (Huynh et al., 2011; Jay et al., 2018; Kim et al., 2017). For example, sperm swimming speed in zebra finch is determined by inversion haplotypes, with heterokaryotypic males producing faster sperm than homokaryotypic males (Kim et al., 2017; Knief et al., 2017). In addition, a supergene resulting from an inversion has been found to determine mating strategy and morphology in the Eurasian ruff (Küpper et al., 2016). Moreover, it is notable that this inversion is probably a lethal recessive variant (Lamichhaney et al., 2016). Similarly, a large inversion resulting in a supergene underlies significant morphological and behavioural differences among white‐striped and tan‐striped morphs in White‐throated sparrows, with aggressiveness being monogenic in the white‐striped morph (Merritt et al., 2020). Although many fitness traits are likely to be polygenic, including traits of conservation interest such as disease susceptibility, reduced fertility and developmental abnormalities (e.g., Moran et al., 2021; Murchison et al., 2012; Roelke et al.,1993; Savage et al., 2020 preprint), those impacted by supergenes are likely to have relatively simple inheritance patterns which will enable their characterization and management.
To date, there are generally two approaches to investigate the genomic basis of traits, both of which require clearly defined traits and well‐curated data sets and high‐quality, well annotated, reference genomes: (1) Comparative genomics, where well‐characterized groups (individuals, populations or species) are used to identify highly differentiated genomic regions (Alonge et al., 2020; McHale et al., 2012; vonHoldt et al., 2017; Weissensteiner et al., 2020); and (2) Association studies, where associations between specific markers (i.e., SNPs and/or SVs) and traits are assessed (Chakraborty et al., 2019). For example, association studies and/or comparative approaches using SVs have revealed signatures of domestication (e.g., aquaculture salmon and dogs; Bertolotti et al., 2020; Cagan & Blass, 2016; vonHoldt et al., 2017), and the relative contribution of SVs to species evolution (e.g., Atlantic cod, Corvids, Heliconius butterflies, Sunflowers; Berg et al., 2017; Joron et al., 2006, 2011; Rieseberg et al., 1999; Weissensteiner et al., 2020).
Whole‐genome SNP‐based association studies have been extensively applied to model organisms, agriculturally significant species and humans, although they are increasingly applied to wild and non‐model species (Santure & Garant, 2018). However, for many SNP‐based association studies, significantly associated loci do not explain the vast majority of known trait heritability (i.e., the proportion of trait variation that is due to genetic rather than environmental differences between individuals; Clarke & Cooper, 2010; Eichler et al., 2010; Manolio et al., 2009). This “missing heritability” is often attributed to genetic variation not explained by genotyped loci, that is causal variants may remain uncharacterized (Manolio et al., 2009). Another hypothesis to explain this missing heritability is that SVs are a significant source of trait variation (Eichler et al., 2010; Frazer et al., 2009). Further, although a given SV is likely to be linked to a nearby SNP (Wilder et al., 2020), they may not be in strong linkage disequilibrium with each other, so that the SNP will not capture the impact of the SV on the trait (Pang et al., 2010). Structural variants may also lead to challenges in aligning reads and calling SNPs in a region, in turn preventing the identification of SVs from sequence data (Pang et al., 2010). The inclusion of SVs into association studies is therefore likely to lead to better power to detect causative variants, particularly given advances in GWAS methods, including machine‐learning approaches, that can more effectively prioritize variants in a region and account for nonadditive interactions (Brieuc et al., 2015; Ramzan et al., 2020). This is particularly promising as evidence suggests that SVs have larger phenotypic effects and may be deleterious more often than SNPs (Chakraborty et al., 2019; Conrad et al., 2010; Cridland et al., 2013; Emerson et al.,2008; Rogers et al., 2015). For example, a quantitative trait locus (QTL) study for Drosophila melanogaster found that about half of all candidate genes underpinning mapped QTL were impacted by SVs, and that a large proportion of genes contained multiple rare SVs (Chakraborty et al., 2019).
4. STRUCTURAL VARIANT DISCOVER AND GENOTYPING IN THREATENED SPECIES
4.1. A high‐quality reference genome is an invaluable investment
While linkage mapping and cytological techniques can be used to characterize SVs, here we focus on the approaches most likely to be accessible to the conservation genetics community (Deakin et al., 2019). Partnerships with global genome consortia (e.g., Vertebrate Genomes Project, Rhie et al., 2021; Earth BioGenome Project, Lewin et al., 2018; Nature and Zoonomia Consortium, 2020) are providing increased accessibility to highly contiguous, well‐annotated genome assemblies generated from both short and long read data and long‐range scaffolding approaches for many species, including those of conservation concern and their close relatives (Whibley et al., 2020). As costs of generating and analysing WGS data continue to drop, a growing number of conservation genomicists working on species beyond global genome consortia are investing in the assembly and annotation of high‐quality reference genomes. We readily recognize that not all conservation programs are able to access the resources or sample quality to generate a high‐quality, well annotated reference genome, nor do we recommend this action for all threatened species. Where there is a clear hypothesis that SVs may be impacting a particular gene or region, comparative approaches (Alonge et al., 2020; Weissensteiner et al., 2020), and target capture methods (vonHoldt et al., 2017) may be the most cost‐effective, especially if a reference genome from a closely related species can be leveraged instead. However, as described below, investment in a high‐quality reference genome is a near necessity for accurate and precise SV discovery and genotyping, particularly if the goal is to characterize a broad range of SV types for downstream functional analyses (e.g., investigating the genomic basis of traits linked to fitness). Further, if the goal is to characterize a large proportion of SVs–including large and/or complex SVs–across the genome, integration of multiple reference genomes provide a powerful tool for accurately characterizing genome‐wide variation and may facilitate genotyping across multiple variant classes (i.e., both SNPs and SVs; Figure 2).
FIGURE 2.
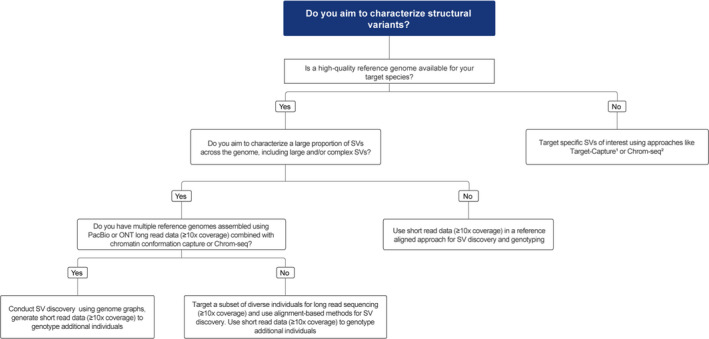
Decision tree for characterizing structural variants (SVs), including large and/or complex SVs. See text for details. Chromatin conformation capture methods include Hi‐C, Omni‐C, and Pore‐C. PacBio, Pacific Biosciences; ONT, Oxford Nanopore Technologies. 1Choi et al. (2009); 2Iannucci et al. (2021)
4.2. Alignment‐based SV discovery methods
Once a high‐quality reference genome has been assembled for a species of interest, SVs can be detected using alignment‐based methods where short‐ and/or long read sequence data for multiple individuals are aligned to the reference without prior assembly. These methods rely on identifying patterns in read mapping between a sample and the reference genome. Common algorithms include read‐pair, read depth, split‐reads and de novo or local assembly (Hajirasouliha et al., 2010; Korbel et al., 2007; Yoon et al., 2009). No single algorithm is well suited to detecting all SV types. For example, read pair‐based algorithms, where the orientation and distance between paired ends are assessed, are suitable for detecting deletions, duplications, and inversions. In the case of read depth approaches, deletions and duplications are identified through variation in mapping depth. Split‐reads identify regions where alignments map across a breakpoint, which is best suited for discerning the ends of an inversion or translocation (Figure 3). Finally, de novo and local assembly algorithms are best suited to identifying variants not present in the reference (e.g., insertions) as reads aligned to contigs may be reassembled alongside unmapped reads for pairwise comparison to a reference (Mahmoud et al., 2019). Early implementation of SV discovery programs typically relied on a single approach (e.g., BreakDancer, Pindel; Chen et al., 2009; Ye et al., 2009). Although these programs are less computationally intensive, they are limited to calling only a few SV types and tend to underperform against generalist programs that incorporate at least three algorithms for SV detection (e.g., Delly, Lumpy, Manta, SvABA; Chen et al., 2016; Layer et al., 2014; Rausch et al., 2012; Wala et al., 2018).
FIGURE 3.
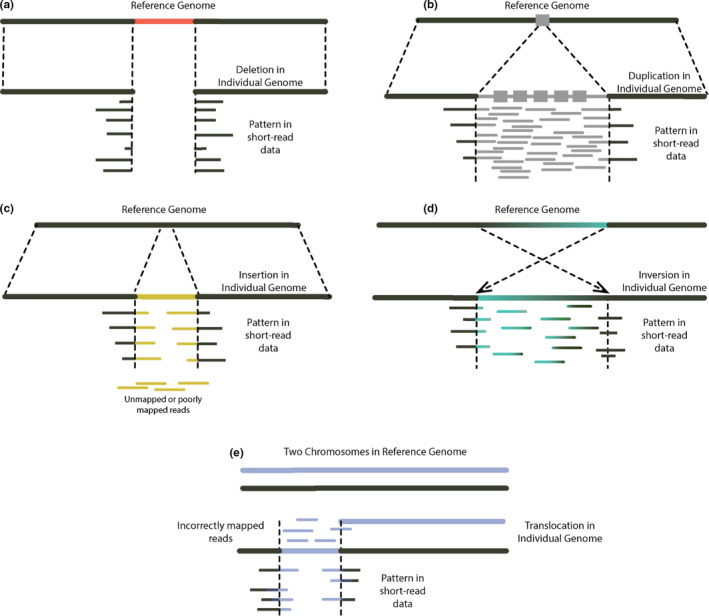
Structural variant (SV) types and common problems associated with short‐read sequence data: (a) deletion, called when reads do not map to, and/or are split across, a given region on the reference genome. Generally the most straightforward SV to detect with short‐read data, but complex rearrangements may preclude mapping and result in a false call; (b) duplication, typically identified by an increase in read depth, however reference error may result in a false call, preclude assessments of copy number variation, or miss sequence variation due to unmapped reads; (c) insertion, short‐reads may be mapped if the majority of each read aligns to the reference genome, but reads composed mostly of the insertion sequence may remain unmapped; (d) inversion, breakpoints (i.e., exact positions of double‐stranded DNA breaks) are difficult to resolve as they typically occur in highly repetitive regions; (e) translocation, breakpoints difficult to resolve as they commonly occur in highly repetitive regions, reads may also incorrectly map to the “original” chromosome from which the translocation arose. Further, it is common for more than one SV or different SV types to occur in close proximity. Resolving these complex SVs is particularly challenging with short‐read sequence data as multiple mapping signals may contradict one another (e.g., multiple deletions in close proximity, deletions that overlap with inversion/translocation breakpoints), unless large samples sizes are used (Collins et al., 2020)
Because of the challenges of identifying and classifying SVs, many SV discovery approaches have a high (and systematic) false positive rate (e.g., Cameron et al., 2019), especially when data is derived from short‐read sequencing methods (Figure 3; see more below). In order to address this error, an ensemble approach for SV characterization has been extensively applied in human data and in domesticates (Du et al., 2021; Ho et al., 2020; Zhou et al., 2019) . With an ensemble approach, multiple SV callers are integrated into a single pipeline as a means to create more certainty around SV discovery, with only variants that intersect multiple SV callers retained (Becker et al., 2018; Mohiyuddin et al., 2015; Zarate et al., 2018 preprint). Further, because validated SV call sets have been developed for humans, it is straightforward to benchmark appropriate program combinations for SV discovery (Collins et al., 2020; Ho et al., 2020; Parikh et al., 2016; Zook et al., 2020).
When validated SV call sets are lacking, we recommend using a generalist SV discovery program that combines multiple SV discovery algorithms to target a range of SVs at once. These generalist programs enable a combination of assembly‐based methods alongside read depth, read pairs and/or split‐read approaches (i.e., GRIDSS, Cameron et al., 2017; Manta, Chen et al., 2016). These programs not only perform well on their own but have the added benefit of performing well over a range of SV types (Cameron et al., 2019; Kosugi et al., 2019). Using a generalist program has an additional benefit that it overcomes the challenge faced by many ensemble methods, where distinguishing between true variants and false positives is difficult due to the significant overlap in false positives across methods (Cameron et al., 2019). However, conservation geneticists should be aware that the computational resources required to characterize SVs at the population scale using generalist SV discovery programs are substantial (e.g., for a diploid 1.15 Gb genome with 170 individuals sequenced to ≥25× coverage, 72 physical cores, 460 Gb RAM, >3 Tb storage was required to implement paired‐, split‐read and assembly‐based SV discovery algorithms; JRW personal observation).
4.3. Multiple reference genomes improve genome‐wide structural variant discovery and genotyping
A pangenome is the aggregate characterization of genomic variation present in a group of interest, including species and populations (e.g., variation between strains of tomato, Alonge et al., 2020). Pangenomes offer a straightforward solution to address the challenges associated with SV discovery and genotyping with short‐read sequence data. Although originally developed to characterize variation in bacteria (Tettelin et al., 2005), they are commonly used in studies of trait diversity in humans (Pang et al., 2010) and agriculturally significant species (e.g., cattle, goats, soybean, and maize; Bickhart et al., 2020; Della Coletta et al., 2021; Golicz et al., 2016; Liu et al., 2020; Low et al., 2020; McHale et al., 2012; Yang et al., 2019). There are two components of a pangenome: the “core” genomic regions that do not vary among individuals, and “accessory” genomic regions that vary among individuals (Bayer et al., 2020; Golicz et al., 2016; Hurgobin & Edwards, 2017; Figure 4). In a pangenomic approach, genomes of multiple individuals are assembled de novo using multiple platforms (e.g., long reads, Hi‐C, Optical mapping; Song et al., 2020; Soto et al., 2020; Weissensteiner et al., 2020; Zhou et al., 2019), followed by pairwise comparisons of whole‐genome alignments for SNP and SV discovery (e.g., Cortex, MUMmer, Minimap2; Delcher et al., 1999; Iqbal et al., 2012; Li, 2018). Once variant discovery is complete, genome graphs representing the variation in the pangenome may be constructed to efficiently represent “core” and “accessory” regions (Eizenga et al., 2020; Li, 2018; Rakocevic et al., 2019; Tettelin et al., 2005) that are encompassed in analyses of copy number variation (CNV) and presence‐absence variation (PAV) (Della Coletta et al., 2021). Genome graphs are a powerful method for population‐level genotyping and consistently outperform alignment‐based genotyping (e.g., Ebler et al., 2020 preprint; Eggertsson, 2017; Iqbal et al., 2012; Kim et al., 2019; Li, 2018). As a result, pangenomic approaches can resolve complex variants (i.e., multiple overlapping events) that may otherwise go undetected in alignment‐based approaches, hampering the discovery of causal variants (Alonge et al., 2020; McHale et al., 2012).
FIGURE 4.
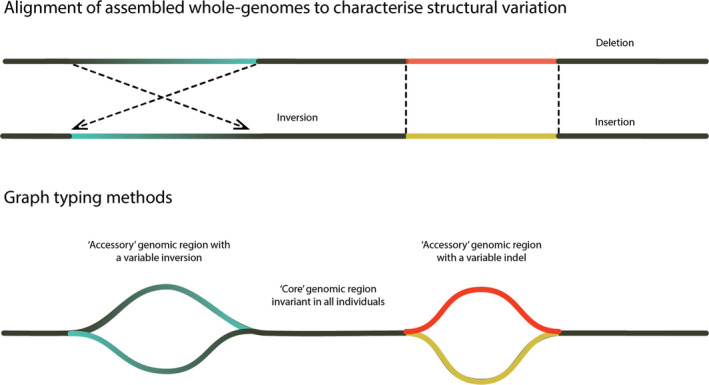
Characterization of SVs using assembled whole‐genomes and genome graphs using multiple reference genomes: (a) Schematic of structural variant (SV) discovery using alignment of assembled whole genomes. (b) Schematic of genome graphs, which can then be used to characterize a pangenome and facilitate rapid genotyping of individuals, as “core” regions (genomic regions that do not vary among individuals) are readily distinguished from “accessory” regions (genomic regions that do vary among individuals). Genome graphs can also facilitate accurate genotyping as more than one allele may be considered at once. The use of multiple reference genomes can reduce reference bias and are better able to capture insertions, inversion haplotypes and complex SVs (i.e., genomic regions where multiple SVs occur in close proximity)
For species of conservation concern, individuals selected for reference genomes may not be the most representative in that some are chosen to showcase individuals known to the public (e.g., animal ambassadors) or are the result of opportunistic sampling. Regardless, in most cases, it is unlikely that a single individual accurately represents the genomic variation within the species as a whole, even in highly inbred species (Gao et al., 2019; McHale et al., 2012). The primary advantage of a pangenomic approach is that multiple alternative alleles are assessed while read mapping, effectively removing reference bias (Paten et al., 2017). In other words, sequence reads are not precluded from mapping if they are not represented in the primary reference (e.g., insertions, Figure 3). The promise of pangenomes is exemplified in crop species such as soy, wheat and Brassica spp. where many more SVs are resolved, leading to a paradigm shift in crop improvement (Della Coletta et al., 2021; Golicz et al., 2016; Liu et al., 2020; Montenegro et al., 2017; Song et al., 2020). However, the ability of conservation programs to establish pangenomes largely depends on the availability of existing genomic resources and the ability to secure substantive funding. In lieu of a conventional pangenome, whereby all genomic variation is captured, the targeted sequencing and assembly of multiple representative individuals to construct genome graphs may provide significant improvements over the use of a single reference genome of comparable assembly quality (Figure 2). For a related approach see (Tigano et al., 2021).
4.4. SV discovery and genotyping with short‐read sequence data
Moderate coverage (e.g., ~10×) short‐read sequencing is becoming the predominant approach in conservation genomics (Cam et al., 2020; Galla et al., ; Lado et al., 2020; Lew et al., 2015; Lujan et al., 2020; Oyler‐McCance et al., 2015; Robinson et al., 2016). Moderate coverage data sets are generally cost effective and appropriate for characterizing SNPs using a single reference genome, whereas a minimum of 30x coverage is often recommended for de novo SV discovery and genotyping (Ahn et al., 2009; Kosugi et al., 2019; Sims et al., 2014; Wang et al., 2008). This is because at low coverage it is challenging to determine whether a detected variant is an artefact of sequencing/mapping error or a “true” variant (Figure 3). For example, mapping errors may occur when the relative size of a SV spans a large portion or the entire length of a read, or in the case of a complex rearrangement, prevents mapping altogether (Sedlazeck et al., 2018; Yi & Ju, 2018). In addition, read insert size (400–600 bp) may bias the discovery of insertions and deletions in some pipelines (Kosugi et al., 2019). By increasing average read depth to 30×, the likelihood of distinguishing between sequencing error, read mapping errors and true genomic variation increases as well. However, due to the increased costs, such high coverage short‐read data sets may be challenging to generate for many species of conservation concern. The question then becomes how best to fully utilize moderate coverage short‐read data sets to investigate SVs. Recent studies demonstrate that an average read depth of 10× is sufficient for population‐scale comparisons, provided sampling is representative and a high‐quality reference genome is available (Collins et al., 2020; Du et al., 2021; Zhou et al., 2019).
Regardless of coverage, it is important to note that genotypes called in short‐read SV discovery programs are prone to high error rates (Chander et al., 2019). One strategy to alleviate this is to conduct SV discovery across all individuals to establish a SV call set and then use dedicated programs for SV genotyping (e.g., BayesTyper, SVTyper, Paragraph) (Chander et al., 2019). In addition, SV genotyping programs have variable performance across a range of SV sizes and types. As such, many programs are unable to genotype across the whole range of SVs that may have been called during discovery. A final consideration is that genotyping programs are still an area of active development. Although they may alleviate some of the false discovery rates prevalent in SV discovery programs, they do not provide a definitive solution (Chander et al., 2019).
As a result, putative SVs identified using short‐read data alone should be treated as preliminary. One method that may aid in removing false variants is a trio‐binning approach, whereby SVs that fail to abide by Mendelian inheritance patterns (either due to challenges with accurate genotyping or incorrect variant calls) are removed from the SV call set (e.g., Patel et al., 2014; Pilipenko et al., 2014). This is particularly relevant for intensively managed threatened species, as pedigree relationships are often known, and suitable trios can be identified for sequencing. Alternatively, if sufficient resources are available, the sensitivity of long‐read sequence data may be leveraged across multiple reference genomes approach for SV discovery and may facilitate the use of lower coverage short‐read resequencing data for SV genotyping, but this largely remains untested. Target‐capture methods for genes or regulatory regions of interest can also provide an affordable alternative to WGS for species with large genomes and projects with tight budgets (Andermann et al., 2020). For example, vonHoldt et al. (2017) used a targeted‐sequencing approach combined with PCR validation to characterize a region on chromosome 6 under positive selection in domestic dog breeds, which significantly reduced sequence coverage requirements.
4.5. SV discovery and genotyping beyond short‐read sequence data
When characterizing genomic features, especially SVs, there are many sequencing platforms and approaches to choose from, and although they may perform well when addressing specific challenges, each has its own caveats (Table 1). Two providers prominently feature in long‐read sequencing: Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT). Since the launch of these technologies in 2011 and 2014, the long‐read sequencing space has been characterized by fast‐paced progress and innovation as demonstrated by the first telomere to telomere assembly of the human X chromosome achieved with ultra‐long‐read sequencing (Miga et al., 2020). The precise error rates between these two technologies remain somewhat contentious (Dohm et al., 2020; Lang et al., 2020 preprint), but as a general rule, ONT currently provides longer average read lengths than PacBio overall (Logsdon et al., 2020) at the cost of higher sequence error rates. Further, when PacBio sequencing providers are overseas, ONT offers the advantage that data can be generated in‐house, which may, for example, be more responsive to the needs and aspirations of Indigenous Peoples (Collier‐Robinson et al., 2019; Galla et al., 2016) and/or alleviate the need for export permits. Despite the challenges, the power of long‐read sequencing technologies to span a significant portion, if not the entire length, of complex regions of the genome in a single read provides a powerful tool for SV discovery and population‐level genotyping. When used in conjunction with a high‐quality, well annotated reference genome, this improves confidence in read mapping across the genome (Amarasinghe et al., 2020), and substantially increases precision (the proportion of variant calls that are “true”) and recall (the proportion of “true” SVs detected) rates for both SNPs and SVs (Wenger et al., 2019). In addition, platforms that directly sequence native DNA remove the amplification bias common in many short‐read sequencing approaches (Depledge et al., 2019). Furthermore, there are emerging “adaptive” sequencing approaches that have the potential to selectively sequence specific regions of the genome (Payne et al., 2020 preprint).
TABLE 1.
Comparison of short‐read sequencing, long‐read sequencing, multiple reference genomes and optical mapping approaches for characterizing structural variants (SVs)
| Feature | Short‐reads | Long‐reads | Multiple reference genomes | Optical mapping |
|---|---|---|---|---|
| Type of SV that can be assessed | Deletions, Insertions, Inversions, Copy Number Variants, and some Translocations | Deletions, insertions, inversions, translocations, duplications, complex SVs | Deletions, insertions, inversions, translocations, duplications, complex SVs | Deletions, Insertions, Inversions, Translocations, Duplications |
| High‐quality reference genome | Required | Required | Required (multiple) | Optional |
| Technology used to generate data for SV discovery and genotyping | Illumina HiSeq, NovaSeq, NextSeq | Pacific Biosciences (PacBio) Sequel; Oxford Nanopore Technologies (ONT) |
|
Bionano Genomics Saphyr |
| DNA quality | Moderate to high molecular weight | High to ultra‐high molecular weight a | High to ultra‐high molecular weight a | Ultra‐high molecular weight a |
| Length of SVs detected | 50 bp to <1 Mb | 50 bp to >1 Mb | 50 bp to >1 Mb | 500 bp to >1 Mb |
| Sequence coverage required | ≥10x | ≥10x |
|
>30× |
| Method of SV discovery | Alignment‐based, including read‐pair, read depth, split‐reads and local assembly | Alignment‐based with local assembly |
|
Alignment‐based, either to reference genome or to optical maps from different samples |
| SV genotype evidence | Multiple sources of evidence required depending on SV type (read pair, read depth, split read) | Single source of evidence (long continuous reads) usually sufficient | Two sources of evidence required (multiple references and population‐level sampling) | Order, position, and orientation of fluorescently‐labelled sequence motifs |
| Example algorithms/programs | BreakDancer; LUMPY; Manta | SMRT‐SV (PacBio); NanoVar (ONT) | Minimap2; MuMmer; Graphtyper2 | Bionano SVCaller |
| Proportion of SVs discovered across the genome | Moderate | High | High | High |
| Recall rate (proportion of “true” variants that are detected) | Low | High | High | High |
| False positive rate for detection | High | Low | Low to moderate | Low |
| SV discovery challenges |
|
Sequencing error rate can complicate read mapping and SV detection. |
|
|
| Computational resources required | High | Moderate, depending on sample number | High, depending on sample number | Moderate, depending on sample number |
| Cost per sample | Low | High | Moderate to high | Moderate |
High molecular weight DNA ≥20 Kb fragments; ultra‐high molecular weight DNA ≥100 Kb fragments
Structural variants significantly alter genome topology and impact the gene regulatory landscape (Sadowski et al., 2019; Shanta et al., 2020). In light of these impacts, the hierarchical organization of DNA within the nucleus is of particular interest when investigating the relationship of transcriptional regulation mechanisms. Chromatin conformation capture (3C) based sequencing approaches enable the investigation of the organization of chromatin across genomes (Kong & Zhang, 2019) and have identified the chromatin signature in gene expression (Lieberman‐Aiden et al., 2009; Lupiáñez et al., 2015; Shanta et al., 2020). In addition, there are emerging advancements in Nanopore sequencing methods to integrate chromatin conformation capture with long‐read sequencing (i.e., Pore‐C; Ulahannan et al., 2019 preprint). Rather than the amplification bias introduced by preparing a short read library, long‐read sequencing provides data on chromatin at a range of distances along the linear genome and enables contacts to be sequenced without amplification. This information is particularly beneficial for non‐model species where gene annotations, regulatory element annotations and gene regulatory networks are un‐ or poorly‐ characterized, leading to challenges in predicting the impacts of SVs on gene expression. For example, 3C‐based approaches combined with SV detection and analysis have revealed the molecular basis for human developmental diseases previously diagnosed through karyotypes (Melo et al., 2020) and resolved the mechanisms underlying reproductive disorders in goats (Guang‐Xin et al., 2021).
Optical mapping approaches are a useful complement to long‐read sequencing approaches, and have enhanced genome assembly outcomes by providing insights into the “big picture” of large‐scale genomic variants (as per Weissensteiner et al., 2020). Optical mapping utilizes a technique based on light‐microscopy to identify specific sequence motifs (such as restriction enzyme cut sites), which are then used to generate images of fluorescently‐labelled DNA molecules (Schwartz et al., 1993), enabling the characterization of large, complex rearrangements missed by long‐reads alone (Yuan et al., 2020). On average, optical maps span ~225 kb, providing information on the physical distance and relationship among genomic features. Besides being used to improve the scaffolding of genome assemblies (Howe & Wood, 2015; Zhang, 2015), including those of endangered species (Rhie et al., 2021), optical mapping methods directly enable the identification of both intra‐ and interspecific SVs (Levy‐Sakin et al., 2019; Zhihai et al., 2016). The primary current commercial provider of optical mapping technology is Bionano Genomics and their Saphyr instrument, which uses a nano‐channel microfluidic chip to linearize and capture images of fluorescently‐labelled ultra‐long DNA fragments to generate optical maps at a resolution of 500 bp (Yuan et al., 2020). While optical maps provide information on the physical topology of chromosomes, they do not provide sequence information on an allele. Because long‐reads and optical maps complement each other, the ideal data set for SV discovery would include both data types (e.g., Soto et al., 2020; Weissensteiner et al., 2020).
5. CONCLUDING REMARKS
Structural variants provide an exciting opportunity to complement SNP‐based approaches and expand our understanding of genome‐wide variation. However, there remains much to learn about the integration of SVs in the small population paradigm. In the meantime, mounting evidence indicates that SVs play an important role in determining fitness traits for both model and non‐model species. Whereas SNPs impact single nucleotides, SVs can capture multiple genes and gene regions simultaneously and are therefore more likely to impact fitness. Emerging approaches for SV discovery and genotyping, and relating SVs to fitness traits, are now in reach for threatened species, and we are confident that discoveries related to the role of SVs in determining fitness traits of conservation interest are imminent. Moreover, the development of SV‐specific tools for analyses of within and between population variation will further enhance the utility of SVs in species of conservation concern (e.g., SV‐Pop; Ravenhall et al., 2019).
However, developing strategies to manage fitness traits will be challenging, as selection for or against them can lead to the reduction of genome‐wide diversity (Kardos & Shafer, 2018; Kardos & Shafer, 2021 preprint). Despite this, current management practice can include such actions. For example, individuals that carry, or express, negative fitness traits can be assessed in conservation breeding programs, followed by appropriate actions or recommendations (e.g., Attwater's prairie chicken; Hammerly et al., 2013). Alternatively, when inheritance of these traits are known, individuals may be screened prior to selective pairing (e.g., California Condor; Moran et al., 2021). Regardless, as we learn more about the mechanisms underlying fitness traits, especially traits associated with negative fitness, there is a pressing need to establish comprehensive evidence‐based management strategies that mitigate the impact of these traits while minimizing the loss of genome‐wide diversity. Where we see the most promise for this is the emerging practice of integrating WGS data into individual‐based models, which will enable different management actions to be modelled and compared (Seaborn et al., 2021). Such models will allow researchers to forecast the consequences of selection‐based management of fitness traits underpinned by adaptive or deleterious SVs on genome‐wide diversity, thus better informing conservation management actions. To achieve this, strong collaboration between conservation practitioners, population ecologists (e.g., modelers), quantitative geneticists, and conservation genomicists will be required to establish the most effective models possible (Hohenlohe et al., 2020).
AUTHOR CONTRIBUTIONS
J.R.W., K.P.K., A.W.S. and T.E.S. were the lead investigators on this perspective. J.R.W. designed all figures and K.P.K. contributed Table 1. J.R.W. led the writing of the manuscript. All authors contributed to manuscript drafts, provided constructive responses to reviewer feedback and gave final approval for publication.
ACKNOWLEDGEMENTS
We wish to thank the Guest Editors of the “Whole‐genome sequences in Molecular Ecology” Special Issue for the invitation to contribute this Perspective and three anonymous reviewers for their insightful comments and constructive feedback throughout the peer review process. We are grateful to the Kākāpō125+ Consortium led by Genomics Aotearoa for providing the opportunity to explore structural variants in a conservation context. We extend our thanks to the University of Canterbury’s Conservation, Systematics and Evolution Research Team (ConSERT), including Roger Moraga at Tea Break Bioinformatics for robust dialogue–and Kylie Price and Joanna Roberts for their support and guidance–on this topic. We acknowledge the University of Canterbury, New Zealand Department of Conservation and Genomics Aotearoa for their support of JRW and NSF Track 2 EPSCoR Program (award number OIA‐1826801) for their support of SJG.
Wold, J. , Koepfli, K.‐P. , Galla, S. J. , Eccles, D. , Hogg, C. J. , Le Lec, M. F. , Guhlin, J. , Santure, A. W. , & Steeves, T. E. (2021). Expanding the conservation genomics toolbox: Incorporating structural variants to enhance genomic studies for species of conservation concern. Molecular Ecology, 30, 5949–5965. 10.1111/mec.16141
Data Availability Statement
Not applicable.
REFERENCES
- Ahn, S.‐M. , Kim, T.‐H. , Lee, S. , Kim, D. , Ghang, H. , Kim, D.‐S. , Kim, B.‐C. , Kim, S.‐Y. , Kim, W.‐Y. , Kim, C. , Park, D. , Lee, Y. S. , Kim, S. , Reja, R. , Jho, S. , Kim, C. G. , Cha, J.‐Y. , Kim, K.‐H. , Lee, B. , … Kim, S.‐J. (2009). The first Korean genome sequence and analysis: Full genome sequencing for a socio‐ethnic group. Genome Research, 19(9), 1622–1629. 10.1101/gr.092197.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allendorf, F. W. , Hohenlohe, P. A. , & Luikart, G. (2010). Genomics and the future of conservation genetics. Nature Reviews Genetics, 11(10), 697–709. 10.1038/nrg2844 [DOI] [PubMed] [Google Scholar]
- Alonge, M. , Wang, X. , Benoit, M. , Soyk, S. , Pereira, L. , Zhang, L. , Suresh, H. , Ramakrishnan, S. , Maumus, F. , Ciren, D. , Levy, Y. , Harel, T. H. , Shalev‐Schlosser, G. , Amsellem, Z. , Razifard, H. , Caicedo, A. L. , Tieman, D. M. , Klee, H. , Kirsche, M. , … Lippman, Z. B. (2020). Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell, 182(1), 145–161.e23. 10.1016/j.cell.2020.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe, S. L. , Su, S. , Dong, X. , Zappia, L. , Ritchie, M. E. , & Gouil, Q. (2020). Opportunities and challenges in long‐read sequencing data analysis. Genome Biology, 21(1), 30. 10.1186/s13059-020-1935-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andermann, T. , Torres Jiménez, M. F. , Matos‐Maraví, P. , Batista, R. , Blanco‐Pastor, J. L. , Gustafsson, A. L. S. , Kistler, L. , Liberal, I. M. , Oxelman, B. , Bacon, C. D. , & Antonelli, A. (2020). A guide to carrying out a phylogenomic target sequence capture project. Frontiers in Genetics, 10(1407), 1–20. 10.3389/fgene.2019.01407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer, P. E. , Golicz, A. A. , Scheben, A. , Batley, J. , & Edwards, D. (2020). Plant pan‐genomes are the new reference. Nature Plants, 6(8), 914–920. 10.1038/s41477-020-0733-0 [DOI] [PubMed] [Google Scholar]
- Becker, T. , Lee, W.‐P. , Leone, J. , Zhu, Q. , Zhang, C. , Liu, S. , Sargent, J. , Shanker, K. , Mil‐homens, A. , Cerveira, E. , Ryan, M. , Cha, J. , Navarro, F. C. P. , Galeev, T. , Gerstein, M. , Mills, R. E. , Shin, D.‐G. , Lee, C. , & Malhotra, A. (2018). FusorSV: An algorithm for optimally combining data from multiple structural variation detection methods. Genome Biology, 19(1), 38. 10.1186/s13059-018-1404-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg, P. R. , Star, B. , Pampoulie, C. , Bradbury, I. R. , Bentzen, P. , Hutchings, J. A. , Jentoft, S. , & Jakobsen, K. S. (2017). Trans‐oceanic genomic divergence of Atlantic cod ecotypes is associated with large inversions. Heredity, 119(6), 418–428. 10.1038/hdy.2017.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti, A. C. , Layer, R. M. , Gundappa, M. K. , Gallagher, M. D. , Pehlivanoglu, E. , Nome, T. , Robledo, D. , Kent, M. P. , Røsæg, L. L. , Holen, M. M. , Mulugeta, T. D. , Ashton, T. J. , Hindar, K. , Sægrov, H. , Florø‐Larsen, B. , Erkinaro, J. , Primmer, C. R. , Bernatchez, L. , Martin, S. A. M. , … Macqueen, D. J. (2020). The structural variation landscape in 492 Atlantic salmon genomes. Nature Communications, 11(1), 5176. 10.1038/s41467-020-18972-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickhart, D. M. , & Liu, G. E. (2014). The challenges and importance of structural variation detection in livestock. Frontiers in Genetics, 5(37), 1–14. 10.3389/fgene.2014.00037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickhart, D. M. , McClure, J. C. , Schnabel, R. D. , Rosen, B. D. , Medrano, J. F. , & Smith, T. P. L. (2020). Symposium review: Advances in sequencing technology herald a new frontier in cattle genomics and genome‐enabled selection. Journal of Dairy Science, 103(6), 5278–5290. 10.3168/jds.2019-17693 [DOI] [PubMed] [Google Scholar]
- Binks, R. M. , Steane, D. A. , & Byrne, M. (2021). Genomic divergence in sympatry indicates strong reproductive barriers and cryptic species within Eucalyptus salubris. Ecology and Evolution, 11(10), 5096–5110. 10.1002/ece3.7403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieuc, M. S. O. , Ono, K. , Drinan, D. P. , & Naish, K. A. (2015). Integration of Random Forest with population‐based outlier analyses provides insight on the genomic basis and evolution of run timing in Chinook salmon (Oncorhynchus tschawytscha). Molecular Ecology, 24, 2729–2746. [DOI] [PubMed] [Google Scholar]
- Cagan, A. , & Blass, T. (2016). Identification of genomic variants putatively targeted by selection during dog domestication. BMC Evolutionary Biology, 16(1), 10. 10.1186/s12862-015-0579-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron, D. L. , Schröder, J. , Penington, J. S. , Do, H. , Molania, R. , Dobrovic, A. , Speed, T. P. , & Papenfuss, A. T. (2017). GRIDSS: sensitive and specific genomic rearrangement detection using positional de Bruijn graph assembly. Genome Research, 27(12), 2050–2060. 10.1101/gr.222109.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam, S. L. , Daguin‐Thiébaut, C. , Bouchemousse, S. , Engelen, A. H. , Mieszkowska, N. , & Viard, F. (2020). A genome‐wide investigation of the worldwide invader Sargassum muticum shows high success albeit (almost) no genetic diversity. Evolutionary Applications, 13(3), 500–514. 10.1111/eva.12837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron, D. L. , Di Stefano, L. , & Papenfuss, A. T. (2019). Comprehensive evaluation and characterisation of short read general‐purpose structural variant calling software. Nature Communications, 10(1), 3240. 10.1038/s41467-019-11146-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanach, A. , Crowhurst, R. , Deng, C. , David, C. , Bernatchez, L. , & Wellenreuther, M. (2019). The genomic pool of standing structural variation outnumbers single nucleotide polymorphism by threefold in the marine teleost Chrysophrys auratus . Molecular Ecology, 28(6), 1210–1223. 10.1111/mec.15051 [DOI] [PubMed] [Google Scholar]
- Cayuela, H. , Dorant, Y. , Mérot, C. , Laporte, M. , Normandeau, E. , Gagnon‐Harvey, S. , Clément, M. , Sirois, P. , & Bernatchez, L. (2021). Thermal adaptation rather than demographic history drives genetic structure inferred by copy number variants in a marine fish. Molecular Ecology, 30(7), 1624–1641. 10.1111/mec.15835 [DOI] [PubMed] [Google Scholar]
- Chakraborty, M. , Emerson, J. J. , Macdonald, S. J. , & Long, A. D. (2019). Structural variants exhibit widespread allelic heterogeneity and shape variation in complex traits. Nature Communications, 10(1), 4872. 10.1038/s41467-019-12884-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chander, V. , Gibbs, R. A. , & Sedlazeck, F. J. (2019). Evaluation of computational genotyping of structural variation for clinical diagnoses. GigaScience, 8(9), giz110. 10.1093/gigascience/giz110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K. , Wallis, J. W. , McLellan, M. D. , Larson, D. E. , Kalicki, J. M. , Pohl, C. S. , McGrath, S. D. , Wendl, M. C. , Zhang, Q. , Locke, D. P. , Shi, X. , Fulton, R. S. , Ley, T. J. , Wilson, R. K. , Ding, L. I. , & Mardis, E. R. (2009). BreakDancer: An algorithm for high‐resolution mapping of genomic structural variation. Nature Methods, 6(9), 677–681. 10.1038/nmeth.1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Schulz‐Trieglaff, O. , Shaw, R. , Barnes, B. , Schlesinger, F. , Källberg, M. , Cox, A. J. , Kruglyak, S. , & Saunders, C. T. (2016). Manta: Rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics, 32(8), 1220–1222. 10.1093/bioinformatics/btv710 [DOI] [PubMed] [Google Scholar]
- Chiang, C. , Scott, A. J. , Davis, J. R. , Tsang, E. K. , Li, X. , Kim, Y. , Hadzic, T. , Damani, F. N. , Ganel, L. , Montgomery, S. B. , Battle, A. , Conrad, D. F. , & Hall, I. M. (2017). The impact of structural variation on human gene expression. Nature Genetics, 49(5), 692–699. 10.1038/ng.3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, M. , Scholl, U. I. , Ji, W. , Liu, T. , Tikhonova, I. R. , Zumbo, P. , Nayir, A. , Bakkaloğlu, A. , Özen, S. , Sanjad, S. , Nelson‐Williams, C. , Farhi, A. , Mane, S. , & Lifton, R. P. (2009). Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proceedings of the National Academy of Sciences USA, 106(45), 19096–19101. 10.1073/pnas.0910672106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, A. J. , & Cooper, D. N. (2010). GWAS: Heritability missing in action? European Journal of Human Genetics, 18(8), 859–861. 10.1038/ejhg.2010.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad, D. F. , & Hurles, M. E. (2007). The population genetics of structural variation. Nature Genetics, 39(S7), S30–S36. 10.1038/ng2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad, D. F. , Pinto, D. , Redon, R. , Feuk, L. , Gokcumen, O. , Zhang, Y. , Aerts, J. , Andrews, T. D. , Barnes, C. , Campbell, P. , Fitzgerald, T. , Hu, M. , Ihm, C. H. , Kristiansson, K. , MacArthur, D. G. , MacDonald, J. R. , Onyiah, I. , Pang, A. W. C. , Robson, S. , … Hurles, M. E. (2010). Origins and functional impact of copy number variation in the human genome. Nature, 464(7289), 704–712. 10.1038/nature08516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, R. L. , Brand, H. , Karczewski, K. J. , Zhao, X. , Alföldi, J. , Francioli, L. C. , Khera, A. V. , Lowther, C. , G., L. D. , Wang, H. , Watts, N. A. , Solomonson, M. , O’Donnell‐Luria, A. , Baumann, A. , Munshi, R. , Walker, M. , Whelan, C. W. , Huang, Y. , Brookings, T. , … Talkowski, M. E. (2020). A structural variation reference for medical and population genetics. Nature, 581(7809), 444–451. 10.1038/s41586-020-2287-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland, J. M. , Macdonald, S. J. , Long, A. D. , & Thornton, K. R. (2013). Abundance and distribution of transposable elements in two drosophila QTL mapping resources. Molecular Biology and Evolution, 30(10), 2311–2327. 10.1093/molbev/mst129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey, J. W. , Chouteau, M. , Barker, S. L. , Maroja, L. , Baxter, S. W. , Simpson, F. , Merrill, R. M. , Joron, M. , Mallet, J. , Dasmahapatra, K. K. , & Jiggins, C. D. (2016). Major Improvements to the Heliconius melpomene Genome Assembly Used to Confirm 10 Chromosome Fusion Events in 6 Million Years of Butterfly Evolution. G3 Genes|Genomes|Genetics, 6(3), 695. –708. 10.1534/g3.115.023655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher, A. L. , Kasif, S. , Fleischmann, R. D. , Peterson, J. , White, O. , & Salzberg, S. L. (1999). Alignment of whole genomes. Nucleic Acids Research, 27(11), 2369–2376. 10.1093/nar/27.11.2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin, J. E. , Potter, S. , O’Neill, R. , Ruiz‐Herrera, A. , Cioffi, M. B. , Eldridge, M. D. B. , Fukui, K. , Marshall Graves, J. A. , Griffin, D. , Grutzner, F. , Kratochvíl, L. , Miura, I. , Rovatsos, M. , Srikulnath, K. , Wapstra, E. , & Ezaz,, T. (2019). Chromosomics: Bridging the Gap between Genomes and Chromosomes. Genes, 10(8), 627. 10.3390/genes10080627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depledge, D. P. , Srinivas, K. P. , Sadaoka, T. , Bready, D. , Mori, Y. , Placantonakis, D. G. , Mohr, I. , & Wilson, A. C. (2019). Direct RNA sequencing on nanopore arrays redefines the transcriptional complexity of a viral pathogen. Nature Communications, 10(1), 754. 10.1038/s41467-019-08734-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Coletta, R. , Qiu, Y. , Ou, S. , Hufford, M. B. , & Hirsch, C. N. (2021). How the pan‐genome is changing crop genomics and improvement. Genome Biology, 22(1), 1–19. 10.1186/s13059-020-02224-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm, J. C. , Peters, P. , Stralis‐Pavese, N. , & Himmelbauer, H. (2020). Benchmarking of long‐read correction methods. NAR Genomics and Bioinformatics, 2(2), lqaa037. 10.1093/nargab/lqaa037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorant, Y. , Cayuela, H. , Wellband, K. , Laporte, M. , Rougemont, Q. , Mérot, C. , Normandeau, E. , Rochette, R. , & Bernatchez, L. (2020). Copy number variants outperform SNPs to reveal genotype–temperature association in a marine species. Molecular Ecology, 29(24), 4765–4782. 10.1111/mec.15565 [DOI] [PubMed] [Google Scholar]
- Dresser, C. M. , Ogle, R. M. , & Fitzpatrick, B. M. (2017). Genome scale assessment of a species translocation program. Conservation Genetics, 18(5), 1191–1199. 10.1007/s10592-017-0970-6 [DOI] [Google Scholar]
- Du, H. , Zheng, X. , Zhao, Q. , Hu, Z. , Wang, H. , Zhou, L. , & Liu, J. ‐F. (2021). Analysis of Structural Variants Reveal Novel Selective Regions in the Genome of Meishan Pigs by Whole Genome Sequencing. Frontiers in Genetics, 12. 10.3389/fgene.2021.550676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresnes, C. , & Dubey, S. (2020). Invasion genomics supports an old hybrid swarm of pool frogs in Western Europe. Biological Invasions, 22(2), 205–210. 10.1007/s10530-019-02112-8 [DOI] [Google Scholar]
- Duntsch, L. , Tomotani, B. M. , de Villemereuil, P. , Brekke, P. , Lee, K. D. , Ewen, J. G. , & Santure, A. W. (2020). Polygenic basis for adaptive morphological variation in a threatened Aotearoa | New Zealand bird, the hihi (Notiomystis cincta). Proceedings of the Royal Society B: Biological Sciences, 287(1933), 20200948. 10.1098/rspb.2020.0948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- E, Guang‐Xin , Zhou, D. ‐K. , Zheng, Z. ‐Q. , Yang, B. ‐G. , Li, X. ‐L. , Li, L. ‐H. , Zhou, R. ‐Y. , Nai, W. ‐H. , Jiang, X. ‐P. , Zhang, J. ‐H. , Hong, Q. ‐H. , Ma,Y. ‐H. , Chu, M. ‐X. , Gao, H. ‐J. , Zhao, Y. ‐J. , Duan, X. ‐H. , He, Y. ‐M. , Na, R. ‐S. , Han, Y. ‐G. , … Huang, Y. ‐F. (2021). Identification of a Goat Intersexuality‐Associated Novel Variant Through Genome‐Wide Resequencing and Hi‐C. Frontiers in Genetics, 11(616743), 1–11. 10.3389/fgene.2020.616743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebler, J. , Clarke, W. E. , Rausch, T. , Audano, P. A. , Houwaart, T. , Korbel, J. , Eichler, E. E. , Zody, M. C. , Dilthey, A. T. & Marschall, T. (2020). Pangenome‐based genome inference. BioRxiv, 37. 10.1101/2020.11.11.378133 [DOI] [PMC free article] [PubMed]
- Eggertsson, H. P. , Jonsson, H. , Kristmundsdottir, S. , Hjartarson, E. , Kehr, B. , Masson, G. , Zink, F. , Hjorleifsson, K. E. , Jonasdottir, A. , Jonasdottir, A. , Jonsdottir, I. , Gudbjartsson, D. F. , Melsted, P. , Stefansson, K. , & Halldorsson, B. V. (2017). Graphtyper enables population‐scale genotyping using pangenome graphs. Nature Genetics, 49(11), 11. 10.1038/ng.3964 [DOI] [PubMed] [Google Scholar]
- Eichler, E. E. , Flint, J. , Gibson, G. , Kong, A. , Leal, S. M. , Moore, J. H. , & Nadeau, J. H. (2010). Missing heritability and strategies for finding the underlying causes of complex disease. Nature Reviews Genetics, 11(6), 446–450. 10.1038/nrg2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eizenga, J. M. , Novak, A. M. , Sibbesen, J. A. , Heumos, S. , Ghaffaari, A. , Hickey, G. , Chang, X. , Seaman, J. D. , Rounthwaite, R. , Ebler, J. , Rautiainen, M. , Garg, S. , Paten, B. , Marschall, T. , Sirén, J. , & Garrison, E. (2020). Pangenome Graphs. Annual Review of Genomics and Human Genetics, 21(1), 139–162. 10.1146/annurev-genom-120219-080406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson, J. J. , Cardoso‐Moreira, M. , Borevitz, J. O. , & Long, M. (2008). Natural selection shapes genome‐wide patterns of copy‐number polymorphism in Drosophila melanogaster . Science, 320(5883), 1629–1631. 10.1126/science.1158078 [DOI] [PubMed] [Google Scholar]
- Forsdick, N. J. , Martini, D. , Brown, L. , Maloney, R. F. , Steeves, T. E. & Knapp, M. (2021). Genomic sequencing confirms absence of introgression despite past hybridisation between a common and a critically endangered bird. BioRxiv, 2020.09.28.316299. doi: 10.1101/2020.09.28.316299 [DOI]
- Frayling, T. M. (2014). Genome‐wide association studies: The good, the bad and the ugly. Clinical Medicine, 14(4), 428–431. 10.7861/clinmedicine.14-4-428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer, K. A. , Murray, S. S. , Schork, N. J. , & Topol, E. J. (2009). Human genetic variation and its contribution to complex traits. Nature Reviews Genetics, 10(4), 241–251. 10.1038/nrg2554 [DOI] [PubMed] [Google Scholar]
- Galla, S. J. , Buckley, T. R. , Elshire, R. , Hale, M. L. , Knapp, M. , McCallum, J. , Moraga, R. , Santure, A. W. , Wilcox, P. , & Steeves, T. E. (2016). Building strong relationships between conservation genetics and primary industry leads to mutually beneficial genomic advances. Molecular Ecology, 25(21), 5267–5281. 10.1111/mec.13837 [DOI] [PubMed] [Google Scholar]
- Gao, L. , Gonda, I. , Sun, H. , Ma, Q. , Bao, K. , Tieman, D. M. , Burzynski‐Chang, E. A. , Fish, T. L. , Stromberg, K. A. , Sacks, G. L. , Thannhauser, T. W. , Foolad, M. R. , Diez, M. J. , Blanca, J. , Canizares, J. , Xu, Y. , van der Knaap, E. , Huang, S. , Klee, H. J. , … Fei, Z. (2019). The tomato pan‐genome uncovers new genes and a rare allele regulating fruit flavor. Nature Genetics, 51(6), 1044–1051. 10.1038/s41588-019-0410-2 [DOI] [PubMed] [Google Scholar]
- Gilpin, M. E. , & Soulé, M. E. (1986). Minimum viable populations: processes of extinction. Conservation Biology: The Science of Scarcity and Diversity (pp. 19–34). Sunderland, MA: Sinauer Associates. [Google Scholar]
- Glassock, G. L. , Grueber, C. E. , Belov, K. , & Hogg, C. J. (2021). Reducing the Extinction Risk of Populations Threatened by Infectious Diseases. Diversity, 13(2), 63. 10.3390/d13020063 [DOI] [Google Scholar]
- Golicz, A. A. , Batley, J. , & Edwards, D. (2016). Towards plant pangenomics. Plant Biotechnology Journal, 14(4), 1099–1105. 10.1111/pbi.12499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajirasouliha, I. , Hormozdiari, F. , Alkan, C. , Kidd, J. M. , Birol, I. , Eichler, E. E. , & Sahinalp, S. C. (2010). Detection and characterization of novel sequence insertions using paired‐end next‐generation sequencing. Bioinformatics, 26(10), 1277–1283. 10.1093/bioinformatics/btq152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerly, S. C. , Morrow, M. E. , & Johnson, J. A. (2013). A comparison of pedigree‐ and DNA‐based measures for identifying inbreeding depression in the critically endangered Attwater's Prairie‐chicken. Molecular Ecology, 22(21), 5313–5328. 10.1111/mec.12482 [DOI] [PubMed] [Google Scholar]
- Hallast, P. , Kibena, L. , Punab, M. , Arciero, E. , Rootsi, S. , Grigorova, M. , Flores, R. , Jobling, M. A. , Poolamets, O. , Pomm, K. , Korrovits, P. , Rull, K. , Xue, Y. , Tyler‐Smith, C. , & Laan, M. (2021). A common 1.6 mb Y‐chromosomal inversion predisposes to subsequent deletions and severe spermatogenic failure in humans. eLife, 10, e65420. 10.7554/elife.65420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser, S. , Athrey, G. , & Leberg, P. (2021). Waste not, want not: Microsatellites remain an economical and informative technology for conservation genetics. Authorea, 10.22541/au.161175207.74315618/v1 [DOI] [PMC free article] [PubMed]
- Henn, B. M. , Botigué, L. R. , Bustamante, C. D. , Clark, A. G. , & Gravel, S. (2015). Estimating the mutation load in human genomes. Nature Reviews Genetics, 16(6), 333–343. 10.1038/nrg3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, S. S. , Urban, A. E. , & Mills, R. E. (2020). Structural variation in the sequencing era: Comprehensive discovery and integration. Nature Reviews. Genetics, 21(3), 171–189. 10.1038/s41576-019-0180-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenlohe, P. A. , Funk, W. C. , & Rajora, O. P. (2020). Population genomics for wildlife conservation and management. Molecular Ecology, 30, 62–82. 10.1111/mec.15720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, K. , & Wood, J. M. (2015). Using optical mapping data for the improvement of vertebrate genome assemblies. GigaScience, 4(1), s13742–015–0052–y. 10.1186/s13742-015-0052-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurgobin, B. , & Edwards, D. (2017). SNP Discovery Using a Pangenome: Has the Single Reference Approach Become Obsolete? Biology, 6(1), 21. 10.3390/biology6010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh, L. Y. , Maney, D. L. , & Thomas, J. W. (2011). Chromosome‐wide linkage disequilibrium caused by an inversion polymorphism in the white‐throated sparrow (Zonotrichia albicollis). Heredity, 106(4), 537–546. 10.1038/hdy.2010.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannucci, A. , Makunin, A. I. , Lisachov, A. P. , Ciofi, C. , Stanyon, R. , Svartman, M. , & Trifonov, V. A. (2021). Bridging the gap between vertebrate cytogenetics and genomics with single‐chromosome sequencing (ChromSeq). Genes, 12(1), 124. 10.3390/genes12010124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal, Z. , Caccamo, M. , Turner, I. , Flicek, P. , & McVean, G. (2012). De novo assembly and genotyping of variants using colored de Bruijn graphs. Nature Genetics, 44(2), 226–232. 10.1038/ng.1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay, P. , Whibley, A. , Frézal, L. , Rodríguez de Cara, M. Á. , Nowell, R. W. , Mallet, J. , Dasmahapatra, K. K. , & Joron, M. (2018). Supergene evolution triggered by the introgression of a chromosomal inversion. Current Biology, 28(11), 1839–1845.e3. 10.1016/j.cub.2018.04.072 [DOI] [PubMed] [Google Scholar]
- Joron, M. , Frezal, L. , Jones, R. T. , Chamberlain, N. L. , Lee, S. F. , Haag, C. R. , Whibley, A. , Becuwe, M. , Baxter, S. W. , Ferguson, L. , Wilkinson, P. A. , Salazar, C. , Davidson, C. , Clark, R. , Quail, M. A. , Beasley, H. , Glithero, R. , Lloyd, C. , Sims, S. , … ffrench‐Constant, R. H. (2011). Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature, 477(7363), 203–206. 10.1038/nature10341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joron, M. , Papa, R. , Beltrán, M. , Chamberlain, N. , Mavárez, J. , Baxter, S. , Abanto, M. , Bermingham, E. , Humphray, S. J. , Rogers, J. , Beasley, H. , Barlow, K. , H. ffrench‐Constant, R. , Mallet, J. , McMillan, W. O. , & Jiggins, C. D. (2006). A conserved supergene locus controls colour pattern diversity in heliconius butterflies. PLOS Biology, 4(10), e303. 10.1371/journal.pbio.0040303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardos, M. , Taylor, H. R. , Ellegren, H. , Luikart, G. , & Allendorf, F. W. (2016). Genomics advances the study of inbreeding depression in the wild. Evolutionary Applications, 9(10), 1205–1218. 10.1111/eva.12414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardos, M. , & Shafer, A. B. A. (2018). The peril of gene‐targeted conservation. Trends in Ecology & Evolution, 33(11), 827–839. 10.1016/j.tree.2018.08.011 [DOI] [PubMed] [Google Scholar]
- Kardos, M. , Armstrong, E. , Fitzpatrick, S. , Hauser, S. , Hedrick, P. , Miller, J. , Tallmon, D. A. , & Funk, C. W. (2021) preprint. The crucial role of genome‐wide genetic variation in conservation. bioRxiv, 1–34. 10.1101/2021.07.05.451163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Paggi, J. M. , Park, C. , Bennett, C. , & Salzberg, S. L. (2019). Graph‐based genome alignment and genotyping with HISAT2 and HISAT‐genotype. Nature Biotechnology, 37(8), 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.‐W. , Bennison, C. , Hemmings, N. , Brookes, L. , Hurley, L. L. , Griffith, S. C. , Burke, T. , Birkhead, T. R. , & Slate, J. (2017). A sex‐linked supergene controls sperm morphology and swimming speed in a songbird. Nature Ecology & Evolution, 1(8), 1168–1176. 10.1038/s41559-017-0235-2 [DOI] [PubMed] [Google Scholar]
- Knief, U. , Forstmeier, W. , Pei, Y. , Ihle, M. , Wang, D. , Martin, K. , Opatová, P. , Albrechtová, J. , Wittig, M. , Franke, A. , Albrecht, T. , & Kempenaers, B. (2017). A sex‐chromosome inversion causes strong overdominance for sperm traits that affect siring success. Nature Ecology & Evolution, 1(8), 1177–1184. 10.1038/s41559-017-0236-1 [DOI] [PubMed] [Google Scholar]
- Kong, S. , & Zhang, Y. (2019). Deciphering Hi‐C: From 3D genome to function. Cell Biology and Toxicology, 35(1), 15–32. 10.1007/s10565-018-09456-2 [DOI] [PubMed] [Google Scholar]
- Korbel, J. O. , Urban, A. E. , Affourtit, J. P. , Godwin, B. , Grubert, F. , Simons, J. F. , Kim, P. M. , Palejev, D. , Carriero, N. J. , Du, L. , Taillon, B. E. , Chen, Z. , Tanzer, A. , Saunders, A. C. E. , Chi, J. , Yang, F. , Carter, N. P. , Hurles, M. E. , Weissman, S. M. , … Snyder, M. (2007). Paired‐end mapping reveals extensive structural variation in the human genome. Science, 318(5849), 420–426. 10.1126/science.1149504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi, S. , Momozawa, Y. , Liu, X. , Terao, C. , Kubo, M. , & Kamatani, Y. (2019). Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biology, 20(1), 117. 10.1186/s13059-019-1720-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küpper, C. , Stocks, M. , Risse, J. E. , dos Remedios, N. , Farrell, L. L. , McRae, S. B. , Morgan, T. C. , Karlionova, N. , Pinchuk, P. , Verkuil, Y. I. , Kitaysky, A. S. , Wingfield, J. C. , Piersma, T. , Zeng, K. , Slate, J. , Blaxter, M. , Lank, D. B. , & Burke, T. (2016). A supergene determines highly divergent male reproductive morphs in the ruff. Nature Genetics, 48(1), 79–83. 10.1038/ng.3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lado, S. , Elbers, J. P. , Doskocil, A. , Scaglione, D. , Trucchi, E. , Banabazi, M. H. , Almathen, F. , Saitou, N. , Ciani, E. , & Burger, P. A. (2020). Genome‐wide diversity and global migration patterns in dromedaries follow ancient caravan routes. Communications Biology, 3(1), 1–8. 10.1038/s42003-020-1098-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney, S. , Fan, G. , Widemo, F. , Gunnarsson, U. , Thalmann, D. S. , Hoeppner, M. P. , Kerje, S. , Gustafson, U. , Shi, C. , Zhang, H. E. , Chen, W. , Liang, X. , Huang, L. , Wang, J. , Liang, E. , Wu, Q. , Lee, S.‐Y. , Xu, X. , Höglund, J. , … Andersson, L. (2016). Structural genomic changes underlie alternative reproductive strategies in the ruff (Philomachus pugnax). Nature Genetics, 48(1), 84–88. 10.1038/ng.3430 [DOI] [PubMed] [Google Scholar]
- Lang, D. , Zhang, S. , Ren, P. , Liang, F. , Sun, Z. , Meng, G. , … Liu, S. (2020). Comparison of the two up‐to‐date sequencing technologies for genome assembly: HiFi reads of Pacbio Sequel II system and ultralong reads of Oxford Nanopore. BioRxiv, 2020.02.13.948489. 10.1101/2020.02.13.948489 [DOI] [PMC free article] [PubMed]
- Layer, R. M. , Chiang, C. , Quinlan, A. R. , & Hall, I. M. (2014). LUMPY: A probabilistic framework for structural variant discovery. Genome Biology, 15(6), R84. 10.1186/gb-2014-15-6-r84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy‐Sakin, M. , Pastor, S. , Mostovoy, Y. , Li, L. E. , Leung, A. K. Y. , McCaffrey, J. , Young, E. , Lam, E. T. , Hastie, A. R. , Wong, K. H. Y. , Chung, C. Y. L. , Ma, W. , Sibert, J. , Rajagopalan, R. , Jin, N. , Chow, E. Y. C. , Chu, C. , Poon, A. , Lin, C. , … Kwok, P.‐Y. (2019). Genome maps across 26 human populations reveal population‐specific patterns of structural variation. Nature Communications, 10(1), 1025. 10.1038/s41467-019-08992-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew, R. M. , Finger, A. J. , Baerwald, M. R. , Goodbla, A. , May, B. , & Meek, M. H. (2015). Using next‐generation sequencing to assist a conservation hatchery: A single‐nucleotide polymorphism panel for the genetic management of endangered delta smelt. Transactions of the American Fisheries Society, 144(4), 767–779. 10.1080/00028487.2015.1037016 [DOI] [Google Scholar]
- Lewin, Harris A. , Robinson, Gene E. , Kress, W. John , Baker, William J. , Coddington, Jonathan , Crandall, Keith A. , Durbin, Richard , Edwards, Scott V. , Forest, Félix , Gilbert, M. Thomas P. , Goldstein, Melissa M. , Grigoriev, Igor V. , Hackett, Kevin J. , Haussler, David , Jarvis, Erich D. , Johnson, Warren E. , Patrinos, Aristides , Richards, Stephen , Castilla‐Rubio, Juan Carlos , … Zhang, Guojie (2018). Earth BioGenome Project: Sequencing life for the future of life. Proceedings of the National Academy of Sciences, 115(17), 4325–4333. 10.1073/pnas.1720115115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. (2018). Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics, 34(18), 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Feng, X. , & Chu, C. (2020). The design and construction of reference pangenome graphs with minigraph. Genome Biology, 21(1), 265. 10.1186/s13059-020-02168-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R. , Fu, W. , Su, R. , Tian, X. , Du, D. , Zhao, Y. , Zheng, Z. , Chen, Q. , Gao, S. , Cai, Y. , Wang, X. , Li, J. , & Jiang, Y. U. (2019). Towards the complete goat pan‐genome by recovering missing genomic segments from the reference genome. Frontiers in Genetics, 10(1169), 1–11. 10.3389/fgene.2019.01169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Li, B. O. , Cheng, C. , Xiong, Z. , Liu, Q. , Lai, J. , Carey, H. V. , Zhang, Q. , Zheng, H. , Wei, S. , Zhang, H. , Chang, L. , Liu, S. , Zhang, S. , Yu, B. , Zeng, X. , Hou, Y. , Nie, W. , Guo, Y. , … Yan, J. (2014). Genomic signatures of near‐extinction and rebirth of the crested ibis and other endangered bird species. Genome Biology, 15(12), 557. 10.1186/s13059-014-0557-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddell, E. , Cook, C. N. , Sunnucks, P. , Liddell, E. , Cook, C. N. , & Sunnucks, P. (2020). Evaluating the use of risk assessment frameworks in the identification of population units for biodiversity conservation. Wildlife Research, 47(3), 208–216. 10.1071/WR18170 [DOI] [Google Scholar]
- Lieberman‐Aiden, E. , van Berkum, N. l. , Williams, L. , Imakaev, M. , Ragoczy, T. , Telling, A. , Amit, I. , Lajoie, B. R. , Sabo, P. J. , Dorschner, M. O. , Sandstrom, R. , Bernstein, B. , Bender, M. A. , Groudine, M. , Gnirke, A. , Stamatoyannopoulos, J. , Mirny, L. A. , Lander, E. S. , Dekker, J. , & Dekker, J. (2009). Comprehensive mapping of long‐range interactions reveals folding principles of the human genome. Science, 326(5950), 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Du, H. , Li, P. , Shen, Y. , Peng, H. , Liu, S. , Zhou, G.‐A. , Zhang, H. , Liu, Z. , Shi, M. , Huang, X. , Li, Y. , Zhang, M. , Wang, Z. , Zhu, B. , Han, B. , Liang, C. , & Tian, Z. (2020). Pan‐genome of wild and cultivated soybeans. Cell, 182(1), 162–176.e13. 10.1016/j.cell.2020.05.023 [DOI] [PubMed] [Google Scholar]
- Logsdon, G. A. , Vollger, M. R. , & Eichler, E. E. (2020). Long‐read human genome sequencing and its applications. Nature Reviews Genetics, 21(10), 597–614. 10.1038/s41576-020-0236-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low, W. Y. , Tearle, R. , Liu, R. , Koren, S. , Rhie, A. , Bickhart, D. M. , Rosen, B. D. , Kronenberg, Z. N. , Kingan, S. B. , Tseng, E. , Thibaud‐Nissen, F. , Martin, F. J. , Billis, K. , Ghurye, J. , Hastie, A. R. , Lee, J. , Pang, A. W. C. , Heaton, M. P. , Phillippy, A. M. , … Williams, J. L. (2020). Haplotype‐resolved genomes provide insights into structural variation and gene content in Angus and Brahman cattle. Nature Communications, 11(1), 2071. 10.1038/s41467-020-15848-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan, N. K. , Weir, J. T. , Noonan, B. P. , Lovejoy, N. R. , & Mandrak, N. E. (2020). Is Niagara Falls a barrier to gene flow in riverine fishes? A test using genome‐wide SNP data from seven native species. Molecular Ecology, 29(7), 1235–1249. 10.1111/mec.15406 [DOI] [PubMed] [Google Scholar]
- Lupiáñez, D. G. , Kraft, K. , Heinrich, V. , Krawitz, P. , Brancati, F. , Klopocki, E. , Horn, D. , Kayserili, H. , Opitz, J. M. , Laxova, R. , Santos‐Simarro, F. , Gilbert‐Dussardier, B. , Wittler, L. , Borschiwer, M. , Haas, S. A. , Osterwalder, M. , Franke, M. , Timmermann, B. , Hecht, J. , … Mundlos, S. (2015). Disruptions of topological chromatin domains cause pathogenic rewiring of gene‐enhancer interactions. Cell, 161(5), 1012–1025. 10.1016/j.cell.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mable, B. K. (2019). Conservation of adaptive potential and functional diversity: Integrating old and new approaches. Conservation Genetics, 20(1), 89–100. 10.1007/s10592-018-1129-9 [DOI] [Google Scholar]
- Mahmoud, M. , Gobet, N. , Cruz‐Dávalos, D. I. , Mounier, N. , Dessimoz, C. , & Sedlazeck, F. J. (2019). Structural variant calling: The long and the short of it. Genome Biology, 20(1), 246. 10.1186/s13059-019-1828-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio, T. A. , Collins, F. S. , Cox, N. J. , Goldstein, D. B. , Hindorff, L. A. , Hunter, D. J. , McCarthy, M. I. , Ramos, E. M. , Cardon, L. R. , Chakravarti, A. , Cho, J. H. , Guttmacher, A. E. , Kong, A. , Kruglyak, L. , Mardis, E. , Rotimi, C. N. , Slatkin, M. , Valle, D. , Whittemore, A. S. , … Visscher, P. M. (2009). Finding the missing heritability of complex diseases. Nature, 461(7265), 747–753. 10.1038/nature08494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung, C. E. (1905). The chromosome complex of orthopteran spermatocytes. The Biological Bulletin, 9(5), 304–340. 10.2307/1535568 [DOI] [Google Scholar]
- McHale, L. K. , Haun, W. J. , Xu, W. W. , Bhaskar, P. B. , Anderson, J. E. , Hyten, D. L. , Gerhardt, D. J. , Jeddeloh, J. A. , & Stupar, R. M. (2012). Structural variants in the soybean genome localize to clusters of biotic stress‐response genes. Plant Physiology, 159(4), 1295–1308. 10.1104/pp.112.194605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo, U. S. , Schöpflin, R. , Acuna‐Hidalgo, R. , Mensah, M. A. , Fischer‐Zirnsak, B. , Holtgrewe, M. , Klever Marius‐Konstantin, Türkmen, S. , Heinrich, V. , Pluym Ilina Datkhaeva, Matoso, E. , Bernardo de Sousa, S. , Louro, P. , Hülsemann, W. , Cohen, M. , Dufke, A. , Latos‐Bieleńska, A. , Vingron, M. , Kalscheuer, V. , … Mundlos, S. (2020). Hi‐C Identifies Complex Genomic Rearrangements and TAD‐Shuffling in Developmental Diseases. The American Journal of Human Genetics, 106(6), 872–884. 10.1016/j.ajhg.2020.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt, J. R. , Grogan, K. E. , Zinzow‐Kramer, W. M. , Sun, D. , Ortlund, E. A. , Yi, S. V. , & Maney, D. L. (2020). A supergene‐linked estrogen receptor drives alternative phenotypes in a polymorphic songbird. Proceedings of the National Academy of Sciences USA, 117(35), 21673–21680. 10.1073/pnas.2011347117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérot, C. , Oomen, R. A. , Tigano, A. , & Wellenreuther, M. (2020). A Roadmap for Understanding the Evolutionary Significance of Structural Genomic Variation. Trends in Ecology & Evolution, 35(7), 561–572. 10.1016/j.tree.2020.03.002 [DOI] [PubMed] [Google Scholar]
- Miga, K. H. , Koren, S. , Rhie, A. , Vollger, M. R. , Gershman, A. , Bzikadze, A. , Brooks, S. , Howe, E. , Porubsky, D. , Logsdon, G. A. , Schneider, V. A. , Potapova, T. , Wood, J. , Chow, W. , Armstrong, J. , Fredrickson, J. , Pak, E. , Tigyi, K. , Kremitzki, M. , … Phillippy, A. M. (2020). Telomere‐to‐telomere assembly of a complete human X chromosome. Nature, 585(7823), 79–84. 10.1038/s41586-020-2547-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohiyuddin, M. , Mu, J. C. , Li, J. , Bani Asadi, N. , Gerstein, M. B. , Abyzov, A. , Wong, W. H. , & Lam, H. Y. K. (2015). MetaSV: An accurate and integrative structural‐variant caller for next generation sequencing. Bioinformatics, 31(16), 2741–2744. 10.1093/bioinformatics/btv204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro, J. D. , Golicz, A. A. , Bayer, P. E. , Hurgobin, B. , Lee, H. T. , Chan, C.‐K. , Visendi, P. , Lai, K. , Doležel, J. , Batley, J. , & Edwards, D. (2017). The pangenome of hexaploid bread wheat. The Plant Journal, 90(5), 1007–1013. 10.1111/tpj.13515 [DOI] [PubMed] [Google Scholar]
- Moran, B. M. , Thomas, S. M. , Judson, J. M. , Navarro, A. , Davis, H. , Sidak‐Loftis, L. , Korody, M. , Mace, M. , Ralls, K. , Callicrate, T. , Ryder, O. A. , Chemnick, L. G. , & Steiner, C. C. (2021). Correcting parentage relationships in the endangered California Condor: Improving mean kinship estimates for conservation management. Ornithological Applications, 123(3), duab017. 10.1093/ornithapp/duab017 [DOI] [Google Scholar]
- Murchison, E. P. , Schulz‐Trieglaff, O. B. , Ning, Z. , Alexandrov, L. B. , Bauer, M. J. , Fu, B. , Hims, M. , Ding, Z. , Ivakhno, S. , Stewart, C. , Ng, B. L. , Wong, W. , Aken, B. , White, S. , Alsop, A. , Becq, J. , Bignell, G. R. , Cheetham, R. K. , Cheng, W. , … Stratton, M. R. (2012). Genome sequencing and analysis of the tasmanian devil and its transmissible cancer. Cell, 148(4), 780–791. 10.1016/j.cell.2011.11.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oomen, R. A. , Kuparinen, A. , … Hutchings, J. A. (2020). Consequences of Single‐Locus and Tightly Linked Genomic Architectures for Evolutionary Responses to Environmental Change. Journal of Heredity, 111(4), 319–332. 10.1093/jhered/esaa020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyler‐McCance, S. J. , Cornman, R. S. , Jones, K. L. , & Fike, J. A. (2015). Genomic single‐nucleotide polymorphisms confirm that Gunnison and Greater sage‐grouse are genetically well differentiated and that the Bi‐State population is distinct. The Condor, 117(2), 217–227. 10.1650/CONDOR-14-174.1 [DOI] [Google Scholar]
- Pang, A. W. , MacDonald, J. R. , Pinto, D. , Wei, J. , Rafiq, M. A. , Conrad, D. F. , Park, H. , Hurles, M. E. , Lee, C. , Venter, J. C. , Kirkness, E. F. , Levy, S. , Feuk, L. , & Scherer, S. W. (2010). Towards a comprehensive structural variation map of an individual human genome. Genome Biology, 11(5), R52. 10.1186/gb-2010-11-5-r52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh, H. , Mohiyuddin, M. , Lam, H. Y. K. , Iyer, H. , Chen, D. , Pratt, M. , Bartha, G. , Spies, N. , Losert, W. , Zook, J. M. , & Salit, M. (2016). svclassify: A method to establish benchmark structural variant calls. BMC Genomics, 17(1), 64. 10.1186/s12864-016-2366-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paten, B. , Novak, A. M. , Eizenga, J. M. , & Garrison, E. (2017). Genome graphs and the evolution of genome inference. Genome Research, 27(5), 665–676. 10.1101/gr.214155.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne, A. , Holmes, N. , Clarke, T. , Munro, R. , Debebe, B. , & Loose, M. (2020). Nanopore adaptive sequencing for mixed samples, whole exome capture and targeted panels. BioRxiv, 2020.02.03.926956. 10.1101/2020.02.03.926956 [DOI]
- Peters, J. L. , Lavretsky, P. , DaCosta, J. M. , Bielefeld, R. R. , Feddersen, J. C. , & Sorenson, M. D. (2016). Population genomic data delineate conservation units in mottled ducks (Anas fulvigula). Biological Conservation, 203, 272–281. 10.1016/j.biocon.2016.10.003 [DOI] [Google Scholar]
- Quattrini, A. M. , Wu, T. , Soong, K. , Jeng, M.‐S. , Benayahu, Y. , & McFadden, C. S. (2019). A next generation approach to species delimitation reveals the role of hybridization in a cryptic species complex of corals. BMC Evolutionary Biology, 19(1), 116. 10.1186/s12862-019-1427-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch, T. , Zichner, T. , Schlattl, A. , Stutz, A. M. , Benes, V. , & Korbel, J. O. (2012). DELLY: structural variant discovery by integrated paired‐end and split‐read analysis. Bioinformatics, 28(18), i333–i339. 10.1093/bioinformatics/bts378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakocevic, G. , Semenyuk, V. , Lee, W. ‐P. , Spencer, J. , Browning, J. , Johnson, I. J. , Arsenijevic, V. , Nadj, J. , Ghose, K. , Suciu, M. C. , Ji, S. ‐G. , Demir, G. , Li, L. , Toptaş, B. , Dolgoborodov, A. , Pollex, B. , Spulber, I. , Glotova, I. , Kómár, P. , … Kural, D. (2019). Fast and accurate genomic analyses using genome graphs. Nature Genetics, 51(2), 354–362. 10.1038/s41588-018-0316-4 [DOI] [PubMed] [Google Scholar]
- Ravenhall, M. , Campino, S. , & Clark, T. G. (2019). SV‐Pop: population‐based structural variant analysis and visualization. BMC Bioinformatics, 20(1). 10.1186/s12859-019-2718-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramzan, F. , Gültas, M. , Bertram, H. , Cavero, D. , & Schmitt, A. O. (2020). Combining Random Forests and a Signal Detection Method Leads to the Robust Detection of Genotype‐Phenotype Associations. Genes, 11(8), 892. 10.3390/genes11080892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie, A. , McCarthy, S. A. , Fedrigo, O. , Damas, J. , Formenti, G. , Koren, S. , Uliano‐Silva, M. , Chow, W. , Fungtammasan, A. , Kim, J. , Lee, C. , Ko, B. J. , Chaisson, M. , Gedman, G. L. , Cantin, L. J. , Thibaud‐Nissen, F. , Haggerty, L. , Bista, I. , Smith, M. , … Jarvis, E. D. (2021). Towards complete and error‐free genome assemblies of all vertebrate species. Nature, 592(7856), 737–746. 10.1038/s41586-021-03451-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg, L. H. , Whitton, J. , & Gardner, K. (1999). Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics, 152(2), 713–727. 10.1093/genetics/152.2.713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J. A. , Ortega‐Del Vecchyo, D. , Fan, Z. , Kim, B. Y. , vonHoldt, B. M. , Marsden, C. D. , Lohmueller, K. E. , & Wayne, R. K. (2016). Genomic flatlining in the endangered island fox. Current Biology, 26(9), 1183–1189. 10.1016/j.cub.2016.02.062 [DOI] [PubMed] [Google Scholar]
- Robinson, J. A. , Räikkönen, J. , Vucetich, L. M. , Vucetich, J. A. , Peterson, R. O. , Lohmueller, K. E. , & Wayne, R. K. (2019). Genomic signatures of extensive inbreeding in Isle Royale wolves, a population on the threshold of extinction. Science Advances, 5(5), eaau0757. 10.1126/sciadv.aau0757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelke, M. E. , Martenson, J. S. , & O’Brien, S. J. (1993). The consequences of demographic reduction and genetic depletion in the endangered Florida panther. Current Biology, 3(6), 340–350. 10.1016/0960-9822(93)90197-V [DOI] [PubMed] [Google Scholar]
- Rogers, R. L. , Cridland, J. M. , Shao, L. , Hu, T. T. , Andolfatto, P. , & Thornton, K. R. (2015). Tandem duplications and the limits of natural selection in drosophila yakuba and Drosophila simulans . PLoS One, 10(7), e0132184. 10.1371/journal.pone.0132184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santure, A. W. , & Garant, D. (2018). Wild GWAS—association mapping in natural populations. Molecular Ecology Resources, 18(4), 729–738. 10.1111/1755-0998.12901 [DOI] [PubMed] [Google Scholar]
- Sadowski, M. , Kraft, A. , Szalaj, P. , Wlasnowolski, M. , Tang, Z. , Ruan, Y. , & Plewczynski, D. (2019). Spatial chromatin architecture alteration by structural variations in human genomes at the population scale. Genome Biology, 20(1), 148. 10.1186/s13059-019-1728-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage, J. L. , Crane, J. M. S. , Team, K. R. , & Hemmings, N. (2020). Low hatching success in the critically endangered kākāpō (Strigops habroptilus) is driven by early embryo mortality not infertility. BioRxiv, 2020.09.14.295949. 10.1101/2020.09.14.295949 [DOI]
- Schwartz, D. C. , Li, X. , Hernandez, L. I. , Ramnarain, S. P. , Huff, E. J. , & Wang, Y. K. (1993). Ordered restriction maps of Saccharomyces cerevisiae chromosomes constructed by optical mapping. Science, 262(5130), 110–114. 10.1126/science.8211116 [DOI] [PubMed] [Google Scholar]
- Sedlazeck, F. J. , Lee, H. , Darby, C. A. , & Schatz, M. C. (2018). Piercing the dark matter: Bioinformatics of long‐range sequencing and mapping. Nature Reviews Genetics, 19(6), 329–346. 10.1038/s41576-018-0003-4 [DOI] [PubMed] [Google Scholar]
- Seaborn, T. , Andrews, K. R. , Applestein, C. V. , Breech, T. M. , Garrett, M. J. , Zaiats, A. , & Caughlin T. T. (2021). Integrating genomics in population models to forecast translocation success. Restoration Ecology, 29(4), e13395. 10.1111/rec.13395 [DOI] [Google Scholar]
- Shanta, O. , Noor, A. , Chaisson, M. J. P. , Sanders, A. D. , Zhao, X. , Malhotra, A. … Human Genome Structural Variation Consortium (HGSVC) . (2020). The effects of common structural variants on 3D chromatin structure. BMC Genomics, 21(1), 95. 10.1186/s12864-020-6516-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims, D. , Sudbery, I. , Ilott, N. E. , Heger, A. , & Ponting, C. P. (2014). Sequencing depth and coverage: Key considerations in genomic analyses. Nature Reviews Genetics, 15(2), 121–132. 10.1038/nrg3642 [DOI] [PubMed] [Google Scholar]
- Song, J. ‐M. , Guan, Z. , Hu, J. , Guo, C. , Yang, Z. , Wang, S. , Liu, D. , Wang, B. , Lu, S. , Zhou, R. , Xie, W. ‐Z. , Cheng, Y. , Zhang, Y. , Liu, K. , Yang, Q. ‐Y. , Chen, L. ‐L. , & Guo, L. (2020). Eight high‐quality genomes reveal pan‐genome architecture and ecotype differentiation of Brassica napus. Nature Plants, 6(1), 34–45. 10.1038/s41477-019-0577-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto, D. C. , Shew, C. , Mastoras, M. , Schmidt, J. M. , Sahasrabudhe, R. , Kaya, G. , Andrés, A. M. , & Dennis, M. Y. (2020). Identification of structural variation in chimpanzees using optical mapping and nanopore sequencing. Genes, 11(3), 276. 10.3390/genes11030276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant, A. H. (1921). Linkage variation and chromosome maps. Proceedings of the National Academy of Sciences USA, 7(7), 181–183. 10.1073/pnas.7.7.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton, D. W. G. , Frandsen, P. , Waples, R. K. , Heller, R. , Russo, I.‐R. M. , Orozco‐terWengel, P. A. , Pedersen, C.‐E. T. , Siegismund, H. R. , & Bruford, M. W. (2019). More grist for the mill? Species delimitation in the genomic era and its implications for conservation. Conservation Genetics, 20(1), 101–113. 10.1007/s10592-019-01149-5 [DOI] [Google Scholar]
- Stoffel, M. A. , Johnston, S. E. , Pilkington, J. G. , & Pemberton, J. M. (2021). Genetic architecture and lifetime dynamics of inbreeding depression in a wild mammal. Nature Communications, 12(1). 10.1038/s41467-021-23222-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin, H. , Masignani, V. , Cieslewicz, M. J. , Donati, C. , Medini, D. , Ward, N. L. , Angiuoli, S. V. , Crabtree, J. , Jones, A. L. , Durkin, A. S. , DeBoy, R. T. , Davidsen, T. M. , Mora, M. , Scarselli, M. , Margarit y Ros, I. , Peterson, J. D. , Hauser, C. R. , Sundaram, J. P. , Nelson, W. C. , … Fraser, C. M. (2005). Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan‐genome”. Proceedings of the National Academy of Sciences USA, 102(39), 13950–13955. 10.1073/pnas.0506758102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigano, A. , Jacobs, A. , Wilder, A. P. , Nand, A. , Zhan, Y. , Dekker, J. , & Therkildsen, N. O. (2021). Chromosome‐Level Assembly of the Atlantic Silverside Genome Reveals Extreme Levels of Sequence Diversity and Structural Genetic Variation. Genome Biology and Evolution, 13(6). 10.1093/gbe/evab098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco, M. , Owens, G. L. , Bercovich, N. , Légaré, J. ‐S. , Soudi, S. , Burge, D. O. , Huang, K. , Ostevik, K. L. , Drummond, E. B. M. , Imerovski, I. , Lande, K. , Pascual‐Robles, M. A. , Nanavati, M. , Jahani, M. , Cheung, W. , Staton, S. E. , Muños, S. , Nielsen, R. , Donovan, L. A. , … Rieseberg, L. H. (2020). Massive haplotypes underlie ecotypic differentiation in sunflowers. Nature, 584(7822), 602–607. 10.1038/s41586-020-2467-6 [DOI] [PubMed] [Google Scholar]
- Ulahannan, N. , Pendleton, M. , Deshpande, A. , Schwenk, S. , Behr, J. M. , Dai, X. , … Imielinski, M. (2019). Nanopore sequencing of DNA concatemers reveals higher‐order features of chromatin structure. BioRxiv, 833590, 10.1101/833590 [DOI]
- vonHoldt, B. M. , Shuldiner, E. , Koch, I. J. , Kartzinel, R. Y. , Hogan, A. , Brubaker, L. , Wanser, S. , Stahler, D. , Wynne, C. D. L. , Ostrander, E. A. , Sinsheimer, J. S. , & Udell, M. A. R. (2017). Structural variants in genes associated with human Williams‐Beuren syndrome underlie stereotypical hypersociability in domestic dogs. Science Advances, 3(7), e1700398. 10.1126/sciadv.1700398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wala, J. A. , Bandopadhayay, P. , Greenwald, N. F. , O'Rourke, R. , Sharpe, T. , Stewart, C. , Schumacher, S. , Li, Y. , Weischenfeldt, J. , Yao, X. , Nusbaum, C. , Campbell, P. , Getz, G. , Meyerson, M. , Zhang, C.‐Z. , Imielinski, M. , & Beroukhim, R. (2018). SvABA: Genome‐wide detection of structural variants and indels by local assembly. Genome Research, 28(4), 581–591. 10.1101/gr.221028.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Wang, W. , Li, R. , Li, Y. , Tian, G. , Goodman, L. , Fan, W. , Zhang, J. , Li, J. , Zhang, J. , Guo, Y. , Feng, B. , Li, H. , Lu, Y. , Fang, X. , Liang, H. , Du, Z. , Li, D. , Zhao, Y. , … Wang, J. (2008). The diploid genome sequence of an Asian individual. Nature, 456(7218), 60–65. 10.1038/nature07484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissensteiner, M. H. , Bunikis, I. , Catalán, A. , Francoijs, K.‐J. , Knief, U. , Heim, W. , Peona, V. , Pophaly, S. D. , Sedlazeck, F. J. , Suh, A. , Warmuth, V. M. , & Wolf, J. B. W. (2020). Discovery and population genomics of structural variation in a songbird genus. Nature Communications, 11(1), 3403. 10.1038/s41467-020-17195-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger, A. M. , Peluso, P. , Rowell, W. J. , Chang, P.‐C. , Hall, R. J. , Concepcion, G. T. , Ebler, J. , Fungtammasan, A. , Kolesnikov, A. , Olson, N. D. , Töpfer, A. , Alonge, M. , Mahmoud, M. , Qian, Y. , Chin, C.‐S. , Phillippy, A. M. , Schatz, M. C. , Myers, G. , DePristo, M. A. , … Hunkapiller, M. W. (2019). Accurate circular consensus long‐read sequencing improves variant detection and assembly of a human genome. Nature Biotechnology, 37(10), 1155–1162. 10.1038/s41587-019-0217-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whibley, A. , Kelley, J. L. , & Narum, S. R. (2020). The changing face of genome assemblies: Guidance on achieving high‐quality reference genomes. Molecular Ecology Resources, 21(3), 641–652. 10.1111/1755-0998.13312 [DOI] [PubMed] [Google Scholar]
- Wilder, A. P. , Palumbi, S. R. , Conover, D. O. , & Therkildsen, N. O. (2020). Footprints of local adaptation span hundreds of linked genes in the Atlantic silverside genome. Evolution Letters, 4(5), 430–443. 10.1002/evl3.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, B. R. , Farquharson, K. A. , McLennan, E. A. , Belov, K. , Hogg, C. J. , & Grueber, C. E. (2020). A demonstration of conservation genomics for threatened species management. Molecular Ecology Resources, 20(6), 1526–1541. 10.1111/1755-0998.13211 [DOI] [PubMed] [Google Scholar]
- Yang, N. , Liu, J. , Gao, Q. , Gui, S. , Chen, L. U. , Yang, L. , Huang, J. , Deng, T. , Luo, J. , He, L. , Wang, Y. , Xu, P. , Peng, Y. , Shi, Z. , Lan, L. , Ma, Z. , Yang, X. , Zhang, Q. , Bai, M. , … Yan, J. (2019). Genome assembly of a tropical maize inbred line provides insights into structural variation and crop improvement. Nature Genetics, 51(6), 1052–1059. 10.1038/s41588-019-0427-6 [DOI] [PubMed] [Google Scholar]
- Ye, K. , Schulz, M. H. , Long, Q. , Apweiler, R. , & Ning, Z. (2009). Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired‐end short reads. Bioinformatics, 25(21), 2865–2871. 10.1093/bioinformatics/btp394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, K. , & Ju, Y. S. (2018). Patterns and mechanisms of structural variations in human cancer. Experimental & Molecular Medicine, 50(8), 1–11. 10.1038/s12276-018-0112-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, S. , Xuan, Z. , Makarov, V. , Ye, K. , & Sebat, J. (2009). Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Research, 19(9), 1586–1592. 10.1101/gr.092981.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Y. , Chung, C.‐ Y.‐L. , & Chan, T.‐F. (2020). Advances in optical mapping for genomic research. Computational and Structural Biotechnology Journal, 18, 2051–2062. 10.1016/j.csbj.2020.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarate, S. , Carroll, A. , Krashenina, O. , Sedlazeck, F. J. , Jun, G. , Salerno, W. , & Gibbs, R. (2018). Parliament2: Fast structural variant calling using optimized combinations of callers. BioRxiv, 424267, 10.1101/424267 [DOI]
- Zhang, G. (2015). Bird sequencing project takes off. Nature, 522(7554), 34. 10.1038/522034d [DOI] [PubMed] [Google Scholar]
- Zhihai, H. , Jiang, X. , Shuiming, X. , Baosheng, L. , Yuan, G. , Chaochao, Z. , Xiaohui, Q. , Wen, X. , & Shilin, C. (2016). Comparative optical genome analysis of two pangolin species: Manis pentadactyla and Manis javanica. GigaScience, 5(1), giw001. 10.1093/gigascience/giw001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y. , Minio, A. , Massonnet, M. , Solares, E. , Lv, Y. , Beridze, T. , Cantu, D. , & Gaut, B. S. (2019). The population genetics of structural variants in grapevine domestication. Nature Plants, 5(9), 965–979. 10.1038/s41477-019-0507-8 [DOI] [PubMed] [Google Scholar]
- Zook, J. M. , Hansen, N. F. , Olson, N. D. , Chapman, L. , Mullikin, J. C. , Xiao, C. , Sherry, S. , Koren, S. , Phillippy, A. M. , Boutros, P. C. , Sahraeian, S. M. E. , Huang, V. , Rouette, A. , Alexander, N. , Mason, C. E. , Hajirasouliha, I. , Ricketts, C. , Lee, J. , Tearle, R. , … Salit, M. (2020). A robust benchmark for detection of germline large deletions and insertions. Nature Biotechnology, 38(11), 1347–1355. 10.1038/s41587-020-0538-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoonomia Consortium (2020). A comparative genomics multitool for scientific discovery and conservation. Nature, 587(7833), 240–245. https://dx.doi.org/10.1038%2Fs41586‐020‐2876‐6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


