Abstract
Purpose
Until now, 1H MRSI of the prostate has been performed with suppression of the large water signal to avoid distortions of metabolite signals. However, this signal can be used for absolute quantification and spectral corrections. We investigated the feasibility of water‐unsuppressed MRSI in patients with prostate cancer for water signal–mediated spectral quality improvement and determination of absolute tissue levels of choline.
Methods
Eight prostate cancer patients scheduled for radical prostatectomy underwent multi‐parametric MRI at 3 T, including 3D water‐unsuppressed semi‐LASER MRSI. A postprocessing algorithm was developed to remove the water signal and its artifacts and use the extracted water signal as intravoxel reference for phase and frequency correction of metabolite signals and for absolute metabolite quantification.
Results
Water‐unsuppressed MRSI with dedicated postprocessing produced water signal and artifact‐free MR spectra throughout the prostate. In all patients, the absolute choline tissue concentration was significantly higher in tumorous than in benign tissue areas (mean ± SD: 7.2 ± 1.4 vs 3.8 ± 0.7 mM), facilitating tumor localization by choline mapping. Tumor tissue levels of choline correlated better with the commonly used (choline + spermine + creatine)/citrate ratio (r = 0.78 ± 0.1) than that of citrate (r = 0.21 ± 0.06). The highest maximum choline concentrations occurred in high‐risk cancer foci.
Conclusion
This report presents the first successful water‐unsuppressed MRSI of the whole prostate. The water signal enabled amelioration of spectral quality and absolute metabolite quantification. In this way, choline tissue levels were identified as tumor biomarker. Choline mapping may serve as a tool in prostate cancer localization and risk scoring in multi‐parametric MRI for diagnosis and biopsy procedures.
Keywords: absolute quantification, choline, 1H MR spectroscopic imaging, prostate cancer, water signal
1. INTRODUCTION
Prostate cancer (PCa) is the second‐most commonly occurring cancer in men. 1 In the diagnosis of PCa, multi‐parametric MRI (mpMRI) together with scoring by the Prostate Imaging Reporting and Data System is now widely applied. 2 Among the MR techniques that can detect cancer in the prostate and assess its aggressiveness is 1H MRSI by which the distribution of metabolites in prostate tissue can be measured. 3 , 4 , 5 Typical metabolites assessed by MRSI are choline, spermine, creatine, and citrate. Although MRSI was originally included in the mpMRI and Prostate Imaging Reporting and Data System approach, it is used only occasionally primarily because of limited practicality and robustness. However, as multiple studies have demonstrated that MRSI has added clinical value, 3 , 5 it is worthwhile to investigate acquisition and postprocessing methods that go beyond what is standardly available on commercial MR systems, to overcome these limitations and extend the scope of prostate MRSI, such as improved volume selection by semi‐LASER sequences. 6 , 7 , 8
Proton MRSI is commonly performed with suppression of the large signal of water to avoid that it distorts the much smaller signals of metabolites. However, this water signal can be very useful for referencing purposes, such as estimating absolute metabolite values and correcting for artifacts such as line shape or phase deformations of metabolite peaks. Therefore, often a separate MRS data set without suppression of the signal of water is acquired to obtain this signal, 9 but this is prone to movement artifacts and at the expense of extra time, precluding its application in MRSI, and thus rarely used in a clinical setting. Applying additional RF pulses for water signal suppression may require time for their tuning, increasing the amount of RF power deposition, and may suppress signals of compound protons with a chemical shift close to that of the water. Moreover, RF pulses targeting water proton spins may also result in magnetization transfer effects on signals of metabolites, which can cause errors in metabolite quantification. For all of these reasons, it remains an attractive option to acquire MRSI data without water suppression. 10
Water signal unsuppressed 1H‐MR spectra suffer from spurious peaks, which are symmetrically present in the up and downfield range of the water signal and have opposite phase. 10 , 11 , 12 , 13 , 14 These so‐called sideband artifacts arise due to mechanical vibrations of the gradient coils inducing a time‐dependent magnetic field, leading to frequency modulations of the FID. Their magnitude depends on a number of factors, and their phases are coherent with those of the gradients. 12 , 15 Gradient switching also induces magnetic field (B0) oscillations, mostly in the magnet cryostat, but these so‐called eddy currents are largely compensated by shielded gradients. 9 As the sideband artifacts are in the order of 0.01% or less, 12 , 15 they are negligible for metabolite peaks. However, if arising from the large water signal, they may significantly contribute to the spectral region with metabolite peaks, with similar magnitude as these peaks, and therefore have to be removed or circumvented to exploit the full advantages of water‐unsuppressed MRS. 10 An additional potential problem is the tail of the water resonance extending into this spectral region, which may hamper quantification of metabolite signals, and therefore needs to be properly amended.
Several studies have explored non‐water‐suppressed 1H MRSI of the brain using either special pulse sequences to circumvent the spectral presence of water and its sidebands or using postprocessing methods to extract and remove the water signal and eliminate its artifacts. 10 , 11 , 12 , 13 , 14 , 16 , 17 , 18 , 19 As the acquisition methods suffer from long measurement times, long TE, and/or movement artifacts, they are not attractive for application to prostate MRSI. Most postprocessing methods rely on the selection of the modulus signal to remove sidebands. 13 , 14
The principle aim of this study was to develop water‐unsuppressed 3D MRSI of the prostate with post‐acquisition removal of the water signal and its artifacts (sidebands and baseline) and to validate its value in a number of prostate cancer patients. The water signal extracted from the raw data was used to improve the quality of the spectra and as an internal reference for absolute metabolite quantification. Subsequently tissue concentration maps of choline, a positive cancer biomarker, were generated for localization and characterization of prostate tumors. The diagnostic value of these choline concentrations was compared with that of the conventionally used signal ratio (choline + spermine + creatine)/citrate.
2. METHODS
2.1. Subjects and histopathology
In this study, 8 patients were included who were scheduled for a radical prostatectomy procedure. Ethical approval was obtained from the “Concernstaf Kwaliteit en Veiligheid, Commissie mensgebonden Onderzoek” Regio Arnhem‐Nijmegen (Netherlands). All participants provided prior written informed consent. After the MRI and MRSI examination, the prostate of the patients was removed by radical prostatectomy. Thereafter, the prostate was fixated in formalin and serially sectioned at 4‐mm intervals into full axial slices for hematoxylin and eosin staining. A pathologist delineated the regions of tumor and performed major and minor grading for Gleason scores. 20 , 21 The histopathology data were used as a gold standard for interpretation of the MRS data. Tumor foci with Gleason score ≤ 3 + 3 were classified as low risk, with Gleason score 3 + 4 as intermediate risk, and with Gleason score 3 + 4 with a grade 5 tertiary component or with Gleason score ≥ 4 + 3 as high risk.
2.2. Magnetic resonance data acquisition
All measurements were performed on a 3T MR system (MAGNETOM Trio; Siemens Healthcare, Erlangen, Germany) with a body coil for transmission and an endorectal coil (MEDRAD, Pittsburgh, PA) for signal reception. The patients were subjected to a standard mpMRI exam including T2‐weighted imaging, DWI, and dynamic contrast‐enhanced MRI.
For water‐unsuppressed 3D MRSI of all patients, we used a semi‐LASER sequence with MEGA pulses, in which the water‐suppression pulses were removed to allow detection of metabolite peaks simultaneous with the water signal, but still suppressing the lipid peaks. 22 , 23 , 24 The MRSI measurements were performed with a TE = 88 ms, TR = 1.35‐2.1 seconds, vector size = 2048, and spectral width = 2400 Hz. In the three orthogonal directions, between 10 and 14 phase‐encoding steps were applied to each. Data were acquired with weighted k‐space sampling and a 100% Hamming filter. After spatial zero filling to a 16 × 16 × 16 k‐space matrix, the nominal voxel dimension was 6 × 6 × 6 mm3.
2.3. Voxel selection
For prostate delineation and voxel annotation, an in‐house graphical user interface called “voxel viewer” was developed in MATLAB (version 2014b; The MathWorks, Natick, MA) and used to create masks of prostate histopathological slices with the delineated tumor area overlaying the T2‐weighted MR images (Supporting Information Figure S1). The voxel grids of each of the 1H‐MRSI data sets were also overlaid on the T2‐weighted MR images without showing the MRS data. This overlay was used to select voxels covering the prostate by visual comparison to prostatectomy slices and ADC maps derived from the DWI measurements. 4 These voxels were carefully delineated to be entirely within the prostate and not touching seminal vesicles, the urethra, or ejaculatory ducts. The quality of the MR spectra was evaluated with an automatic method that reproduces the decisions of expert spectroscopists 25 removing spectra with broad resonances, high lipid signal contamination, and poor SNR from further analysis. The quality control step was performed before the spectral fitting step at the end of the processing pipeline (vide infra). Because of the Hamming‐filtered weighted acquisition, the effective voxel volume was a sphere of about 0.7 mm3 (with a diameter for a spatial response function at 64% full height of the main lobe), which was also taken into consideration in the voxel selection.
2.4. Processing of water‐unsuppressed 1H MRSI of the prostate
To process water signal–unsuppressed 3D 1H‐MRSI data of the prostate, an algorithm was developed including previously reported procedures such as applying modulus signal selection to cancel the acoustic water sidebands and wavelet transformations in the baseline correction. 13 , 14 , 16
The flow diagram in Figure 1 gives an outline of the pipeline applied to obtain metabolite concentrations from the raw spectral data. This pipeline divides into three phases: (1) preprocessing and wavelet selection, (2) water‐signal modeling, and (3) metabolite signal quantification. Preprocessing of the spectra involved phase correction and frequency alignment 26 , 27 of the water signals (steps 1 and 2). Then, as FIDs, the spectral signals were filtered using maximum overlap discrete wavelet transforms (step 3). Filtered components were restructured and combined to make a low‐pass‐filtered FID of the water signal and an FID containing this water signal and the metabolite signals. This separate “water” FID is then used for eddy current and phase correction 27 , 28 , 29 of the “water and metabolites” FID (step 4). Subsequently, the metabolite signals are fitted with model‐metabolite signals, assuming the water line shape (step 5). Subtracting these fitted metabolite signals and the separate water signal from the modulus of the water and metabolites spectra provides a baseline (step 6) that is smoothed with a discrete wavelet transform. This baseline is combined with the separate water signal to provide a second, new model of the dominant water reference signal (“full water” signal) including its spectral extension in the region of the metabolite resonances as a baseline (step 7). In parallel, the metabolite resonances are frequency‐aligned (step 8). Subsequently, phase and eddy current correction is then reapplied 27 , 28 , 29 using the full water signal (step 9). Then this water signal is subtracted from the phase‐corrected and frequency‐corrected data set, and the modulus signal is selected (step 10). The modulus spectrum halves the amplitude of the metabolite signals but cancels the symmetrical acoustic ringing from the water signal leaving just the metabolite resonances. After automatic spectral quality control, 25 the metabolite peaks are fitted using the line shape of the full water signal (step 11). The integral of this signal is then used as a concentration reference to calculate in vivo metabolite concentrations (step 12). A more detailed description of the postprocessing pipeline is provided in Supporting Information.
FIGURE 1.
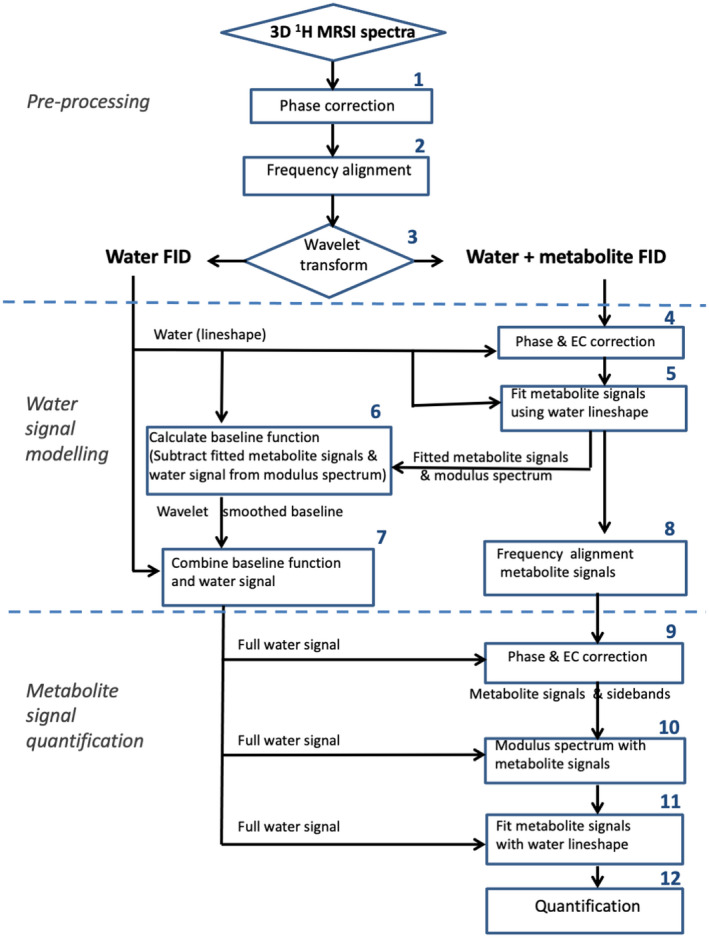
Flow of data from water signal–unsuppressed 3D‐MRSI exams along the postprocessing steps, including preprocessing with peak phasing and frequency alignment, wavelet water filtering, artifact removal, and metabolite quantification using the extracted and reconstructed water signal. For details, see the Supporting Information. Abbreviation: EC, eddy current
2.5. Determination of absolute choline tissue concentration with T1 and T2 relaxation correction
Absolute choline tissue concentrations were determined from the fitted choline resonance area, taking the water signal area as a reference, assuming a tissue‐water content in the prostate of 39.4 mM/g wet weight. 30 With a prostate tissue density of 1.02 kg/L, 31 this equals a tissue‐water concentration of 40.2 mM. Signal attenuation due to T1 and T2 relaxation was corrected using the following relaxation times reported for 3 T: citrate protons: T1: 0.47 seconds and T2: 0.17 seconds; choline methyl protons: T1: 1.1 seconds and T2: 0.220 seconds 22 ; water protons in benign peripheral zone: T1: 1.6 seconds and T2: 0.142 seconds 32 , 33 ; and water protons in tumor tissue: 0.109 seconds. 32 , 33
With TE = 88 ms and TR = 1.35 seconds, we obtained the following relaxation correction factor for choline protons in benign peripheral zone:
and for tumor tissue this factor is 0.539. If TR = 2.1 seconds, these relaxation factors only increase by 6%.
2.6. Choline mapping, voxel selection of tumor and normal prostate tissue, and statistical analysis
Absolute choline tissue‐concentration maps were generated from the 1H‐MRSI data set of each patient. Subsequently, MR spectra were selected from voxels completely positioned in tumor or normal‐appearing tissue, and the choline tissue concentrations were evaluated for each. Between 7 and 21 voxels were selected from tumorous areas and between 99 and 368 voxels for benign areas. The (choline + spermine + creatine) over citrate ratio (CSC/C) was also calculated from the fitted MR spectra of each of the voxels. Correlation analysis was performed to evaluate the strength of the relationship between absolute choline and citrate concentration to CSC/C ratio. Pearson correlation coefficients and Spearman’s rank correlation coefficients were calculated for choline to CSC/C ratio and citrate to CSC/C ratio, respectively. A two‐tailed unpaired Welch t‐test was performed to compare the average total choline concentrations between tumor and benign tissue.
3. RESULTS
Proton MR spectra of the prostate, obtained with a semi‐LASER 3D‐MRSI sequence without water signal suppression, are dominated by the resonance of water, as demonstrated for voxels in a transversal slice (Figure 2A‐D). After zooming in on the spectral range from 2.3 to 3.3 ppm and expanding the vertical scale 250 times, the signals of citrate protons at 2.6 ppm and of choline methyl protons at 3.2 ppm become visible (Figure 2E). The quality of the data is illustrated in Figure 2F, showing the water resonance (top panels) and further expansions of the 2.5‐3.5‐ppm region (bottom panels) for voxels from the peripheral zone (blue) and transition zone (red, orange).
FIGURE 2.
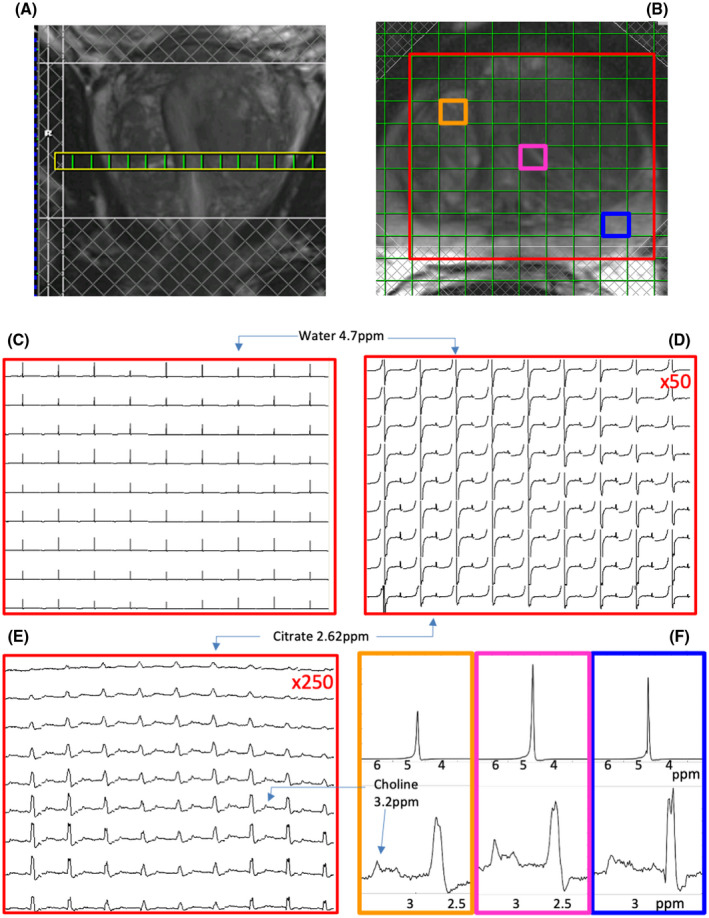
Magnetic resonance spectra in a transversal slice of a 3D 1H‐MRSI data set acquired with a semi‐LASER sequence without water‐signal suppression. (A) Coronal T2‐weighted MRI slice showing position of transversal slice. (B) Transversal T2‐weighted MRI slice with MRSI grid and selected volume of interest (VOI). The lower row in the VOI is partly covered by a saturation slab (hatched bar); hence, the signal intensities in this row are decreased. In addition, three voxels are indicted, from which the spectra are displayed in (F). The orange voxel is positioned toward the ventral side of the central gland. The magenta voxel is also in the central gland, and the blue voxel is in the peripheral zone. (C) Full spectra with the water signal indicated. (D) Magnetic resonance spectra in the range of 5.8 ppm to 2.3 ppm. The water signal and citrate signals are indicated. The vertical scale is enhanced by a factor of 50. (E) Magnetic resonance spectra in the range of 3.3 ppm to 2.3 ppm. The vertical scale is enhanced by a factor of 250. The signals of choline and citrate are indicated. (F) Details of MR spectra from the water‐unsuppressed data set showing the region around the water signal (top row, 6.5‐3 ppm) and around the metabolites of interest (3.5‐2.5 ppm). The spectra shown are from the voxels indicated in (B) according to the colors and demonstrate a range of line widths and line shapes across the data set
To remove the water‐signal contributions from these MR spectra, which were acquired without water‐suppression pulses, we developed a postprocessing protocol as described in section 2 and illustrated in Figure 1. Examples of MR spectra from a single voxel obtained along this processing pipeline are shown in Figure 3. A spectrum obtained after zero‐order phase correction of a raw spectrum, corresponding to preprocessing phase step 1, is shown in Figure 3A (black line). Then, frequency alignment (step 2) was applied, and subsequently, wavelet filtering was performed on the spectra (step 3) to obtain an FID for a separate water signal (Figure 3A, blue line) and an FID containing the signals of water and the metabolites. After phase and eddy current correction (step 4), the latter was transformed in a modulus spectrum, removing the side bands, and the metabolite signals fitted to be used in the spectral baseline estimation. A modulus spectrum obtained after step 5 for step 6 in the water signal modeling phase is shown in Figure 3B (red line). Thereafter, the estimated model baseline and water signal were combined (Figure 3C, green line) and removed from the final modulus spectrum (step 10) leaving only the metabolite signals (Figure 3D and Supporting Information Figure S2) for metabolite signal fitting and quantification in steps 11 and 12. Of the MR spectra from eligible voxels, 88.4 ± 5.5% (averaged over all patients, ±SD) passed the quality control procedure. In the final baseline‐corrected MR spectra, the proton signals of the metabolites choline, spermine, creatine, and citrate were fitted with model resonances for the corresponding protons convolved with the full water line shape (Figure 3D).
FIGURE 3.
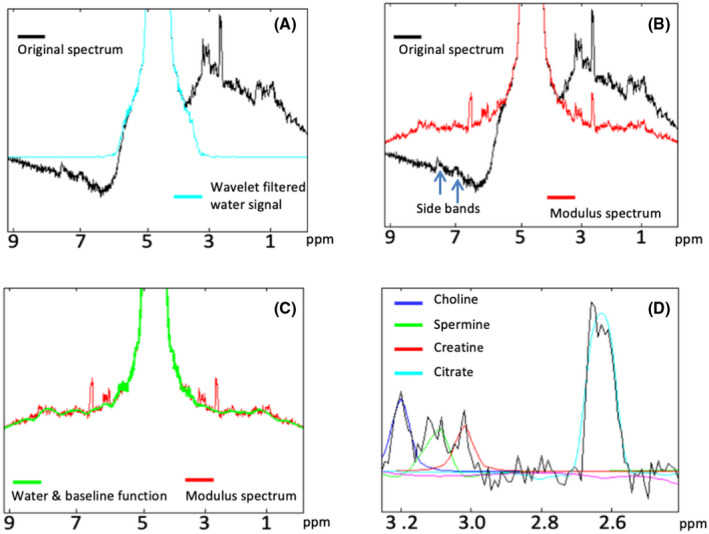
Examples of spectra from a single voxel obtained along the processing pipeline, shown in Figure 1. (A) Original raw spectrum after zero‐order phasing of the water signal (black line, step 1) and spectrum after frequency alignment and wavelet filtering of the water signal (blue, step 3). (B) After phase and EC correction, the modulus spectrum is taken, canceling the water side bands (red, steps 5 and 6). (C) Next, a full water signal is generated from a model baseline function and the water signal (green, step 7), which is then subtracted from the modulus spectrum (red, step 10). (D) The subtraction results in the water and baseline free spectrum that can be fitted and quantified (steps 11 and 12)
In the spectra of the 3D‐MRSI data of all patients, we observed increased choline and decreased citrate signals in prostate areas identified as tumor lesions from a comparison with the matching histopathological slices. The variation of absolute choline levels in different locations in the prostate is illustrated in Figure 4 for a patient with a Gleason score 5 tumor. From the MRSI voxel grid, overlaid on the T2‐weighted image of a transversal slice through the prostate, three separate voxels were selected: a voxel in benign tissue (yellow), one at the edge of the tumor (red), and one in the tumor (blue). The corresponding MR spectra of these voxels with fits of the methyl proton resonances of choline and creatine compounds, polyamine protons, and citrate protons are shown in Figures 4A (benign), 4B (edge of the tumor), and 4C (tumor). As outlined in section 2, we calculated absolute tissue concentrations of choline from the integral of its fitted resonance and that of the water signal of the same voxel as a reference. This resulted in an absolute choline concentration for the voxel in benign tissue of 2.9 mM, for the voxel at the edge of the tumor of 3.7 mM and for the voxel in the tumor of 5.4 mM.
FIGURE 4.
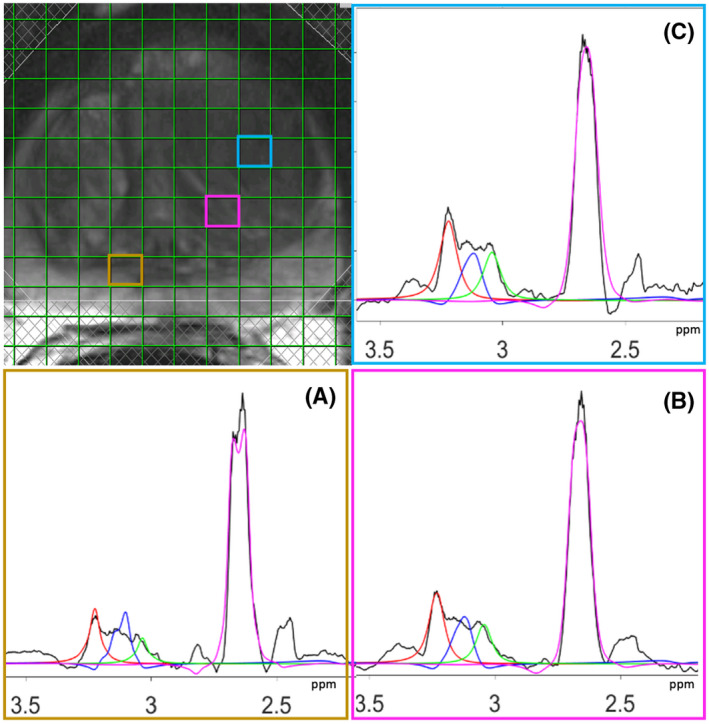
Magnetic resonance spectra from three separate voxels in the prostate from a patient with a Gleason score 5 tumor lesion. The baseline‐corrected MR spectra obtained after step 11 in Figure 1 are depicted in black. For each spectrum, the fits are shown for the resonances of methyl protons in choline compounds (red), protons in polyamines (blue), and methyl protons in total creatine (green) and protons in citrate (magenta). The different spectra correspond to benign tissue (A), edge of the tumor (B), and tumor (C)
From the absolute tissue levels of choline in each voxel, we reconstructed choline maps and compared these with T2‐weighted MRIs and ADC maps from DWIs from the same prostate position, along with a corresponding axial slice through the prostatectomy specimen. The results for the prostates of 3 patients with high, intermediate, and low‐risk cancer lesions are shown in Figure 5 with the location of the tumor foci indicated on the histopathological slices. In all cases, the voxels with increased choline tissue levels colocalized with a significant tumor focus as identified by histopathology and guided by mpMRI. Interestingly, we observed that the highest maximum absolute choline concentrations occurred in high‐risk cancer foci (Figure 5).
FIGURE 5.
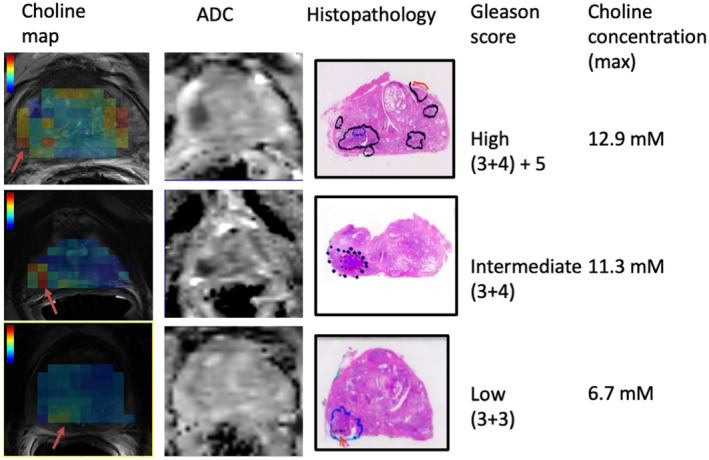
Representative transversal slices with 1H‐MRS choline maps on top of T2‐weighted MR images (left‐hand side) and ADC maps and corresponding prostate prostatectomy histopathology slices with tumor delineation from 3 patients with high, intermediate, and low‐risk cancer lesions. Areas with low ADC values seen on the ADC maps of intermediate and high‐risk lesions co‐localize with tumor foci identified by histopathology. For all lesions, these tumor foci correspond with increased choline levels. The maximum choline tissue concentrations in the tumor areas (from voxels with red arrow) are presented on the right‐hand side and increase with Gleason score–based risk category. The color bars indicate choline concentrations in the range of 2 to 14 mM
In the prostates of each patient included in this study, the mean choline concentration of the voxels from tumorous areas was significantly higher than the mean of the voxels from benign areas (Table 1). Taking the data of all patients together, the average choline tissue concentration in benign tissue (3.8 ± 0.7 mM) was significantly different (p < .0001) from that in tumor tissue (7.2 ± 1.4 mM). For the same areas we also calculated average CSC/C ratios (Table 1). This ratio is commonly used as MRS biomarker for cancer tissue in 1H‐MRSI data of prostates. All tumor areas had a higher CSC/C ratio than benign areas, and compiling the data of all patients, the average CSC/C ratio in benign tissue (0.38 ± 0.06) was significantly different (p = .0002) from that in tumor tissue (0.88 ± 0.22). Furthermore, the average choline content of low‐risk lesions (Gleason < 3 + 4) was significantly lower (p = .046) than that of the intermediate + high risk lesions (Gleason ≥ 3 + 4), but the CSC/C ratio did not differ between these lesion groups (p = .9).
TABLE 1.
Absolute tissue levels of choline (mM) and the (choline + spermine + creatine)/citrate ratio in benign and tumor areas for all patients (P1‐P8)
| Patient | Choline (mM; mean ± SD) | CSC/Cit (mean ± SD) | Number of voxels | Gleason score | |||
|---|---|---|---|---|---|---|---|
| Benign | Tumor | Benign | Tumor | Benign | Tumor | ||
| P_1 | 3.6 ± 0.1 | 6.2 ± 0.1 | 0.34 ± 0.06 | 0.66 ± 0.03 | 149 | 7 | 2 + 3 = 5 |
| P_2 | 2.4 ± 0.2 | 9.3 ± 0.6 | 0.35 ± 0.18 | 0.8 ± 0.34 | 368 | 21 | 3 + 4 = 7 |
| P_3 | 2.9 ± 0.2 | 4.8 ± 0.4 | 0.41 ± 0.09 | 1.23 ± 0.29 | 210 | 10 | 3 + 3 = 6 |
| P_4 | 4.5 ± 0.3 | 8.6 ± 0.5 | 0.39 ± 0.12 | 1.2 ± 0.49 | 259 | 8 | 3 + 4 = 7 |
| P_5 | 3.6 ± 0.2 | 7.1 ± 0.4 | 0.35 ± 0.05 | 0.89 ± 0.09 | 99 | 10 | 4 + 3(+5) = 7 + 5 |
| P_6 | 3.9 ± 0.2 | 7.4 ± 0.3 | 0.3 ± 0.05 | 0.84 ± 0.08 | 185 | 16 | 3 + 4(+5) |
| P_7 | 4.4 ± 0.2 | 7.6 ± 0.2 | 0.36 ± 0.06 | 0.66 ± 0.08 | 336 | 8 | 4 + 3(+5) = 7 + 5 |
| P_8 | 3.6 ± 0.2 | 6.8 ± 0.2 | 0.5 ± 0.16 | 0.79 ± 0.1 | 316 | 20 | 3 + 3 = 6 |
| All patients | 3.6 ± 0.7 | 7.2 ± 1.4 | 0.38 ± 0.06 | 0.88 ± 0.22 | |||
Cancer in the prostates of most patients was multifocal. For each patient, the index tumor lesion with highest Gleason score was chosen for inclusion in this table. The number of voxels selected for the calculation of these levels in benign tissue and tumor foci as well as Gleason score are also presented. Cancer lesions in patients 5, 6, and 7 contained a tertiary grade 5 component. For each patient, the p‐value comparing mean benign and tumor choline values is < .0001, according to a Welch unpaired two‐tailed t‐test, except for CSC/C ratio in P4, for which the p‐value = .0023.
Abbreviation: CSC/C, (choline + spermine + creatine)/citrate signal ratio.
Finally, we compared the CSC/C ratio with the absolute tissue levels of choline and citrate. In voxels that colocalize with tumor, the estimated choline concentration strongly correlates with the CSC/C ratio with an average coefficient (±SD) of 0.78 ± 0.08, whereas citrate correlates with a coefficient of −0.57 ± 0.08. In healthy tissue, these correlations with the CSC/C ratio are 0.21 ± 0.06 for choline and 0.57 ± 0.04 for citrate (Table 2).
TABLE 2.
Correlation analysis of absolute choline and citrate tissue levels to the CSC/C signal ratio in tumor and benign regions in all patients (P1‐P8)
| Patient | Tumor | Benign | ||
|---|---|---|---|---|
| Pearson correlation coefficient | Spearman correlation coefficient | Pearson correlation coefficient | Spearman correlation coefficient | |
| Cho to CSC/C | Cit to CSC/C | Cho to CSC/C | Cit to CSC/C | |
| P1 | 0.78 | −0.56 | 0.21 | −0.59 |
| P2 | 0.74 | −0.61 | 0.23 | −0.6 |
| P3 | 0.77 | −0.46 | 0.29 | −0.54 |
| P4 | 0.79 | −0.67 | 0.2 | −0.47 |
| P5 | 0.94 | −0.63 | 0.27 | −0.58 |
| P6 | 0.65 | −0.48 | 0.22 | −0.61 |
| P7 | 0.76 | −0.51 | 0.16 | −0.56 |
| P8 | 0.81 | −0.61 | 0.11 | −0.57 |
| Mean ± SD | 0.78 ± 0.1 | −0.57 ± 0.08 | 0.21 ± 0.06 | −0.57 ± 0.05 |
Pearson correlation coefficients and Spearman’s rank correlation coefficients are calculated for choline (Cho) to CSC/C and citrate (Cit) to CSC/C ratio, respectively.
4. DISCUSSION
In this paper we report the first successful performance of 3D 1H MRSI of the prostate without using water signal suppression or circumventing pulses. To remove the water signal and its sidebands from the MR spectra, we developed an effective post‐acquisition pipeline including wavelet transform, peak alignment, and phasing and modulus selection. The quality of spectra from voxels was improved by convolving the spectra with the line shape of the water signal in the same voxels. In this way, we obtained MR spectra throughout the prostate, showing signals for choline, spermine, creatine, and citrate with sufficient resolution to be analyzed separately. Because the extracted water signal can be used as a voxel‐selective internal reference, it was possible to derive absolute tissue levels of these compounds for each MRSI voxel of the prostate. Water‐unsuppressed 3D MRSI was applied to patients with prostate cancer, using a semi‐LASER sequence for volume selection, and the results were compared with whole‐mount histopathology of their prostates. We focused on the signal of choline compounds, as currently it is the only in vivo spectral component that positively correlates with the presence of cancer in the prostate. The results demonstrate that in the prostate of each patient, the mean choline level significantly discriminates cancer tissue from benign tissue with a performance that is at least comparable to that of the CSC/C ratio, which is classically used for this discrimination. In addition, our preliminary data indicate that the maximum choline level of a tumor lesion is related to Gleason score.
Up until now, proton MRS and MRSI without water suppression essentially has only been implemented for the brain using either acquisition or postprocessing methods. 10 Acquisition methods to eliminate the water signal and its side bands include J‐resolved MRS, 11 long‐TE MRSI, 18 and two MRS scans. 17 , 19 , 34 , 35 We have refrained from these methods, as their MRSI application in the prostate may suffer from long measurement times, too‐long TE, and/or movement artifacts. Alternatively, postprocessing methods have been proposed to remove the water signal and its artifacts from water‐unsuppressed MRS and MRSI data. A key step in the pipeline of several of these postprocessing methods was the selection of the modulus signal to remove side band artifacts of the water signal. 10 , 13 , 14 , 36 In our postprocessing, we also made use of modulus signal selection for this purpose next to the use of wavelet transforms to select and smooth the water signal. 16 The used TE of 88 ms in semi‐LASER MRSI acquisition is optimal for the in‐phase detection of the citrate signal 34 ; in a non‐water version, this TE leads to attenuated and broader acoustic sidebands further away from the water signal than occurring at shorter TEs. 12 A potential drawback of the modulus selection is the theoretical loss in SNR of the metabolite signals up to a factor of √2. However, as shown in previous studies, there is little if any loss in SNR due to the noise distribution in the modulus signal and other corrective effects of the modulus operation such as on eddy current artifacts. 13 , 14 , 36 The modulus operation may cause mirroring of downfield signals around the water line, interfering with the metabolite signals of interest. However, 1H‐MR spectra of the prostate at TE = 88 ms do not show resonances downfield of the water signal, and if such signals would appear at shorter TE, this mirroring problem can be mitigated by the procedure described in Le Fur and Cozzone. 36 Recently, the fast MRSI method SPICE has been presented in a version without water‐signal suppression, which requires acquisition of a training set of data for proper water‐signal removal and makes no provisions for removing acoustic sidebands of the water signal. 37
A benefit of water‐unsuppressed MRSI is that a water signal is obtained without the need for any additional measurement. This water signal can be used as an internal reference for various purposes such as metabolite line‐shape distortion correction, absolute metabolite quantification, multicoil signal corrections, and in the case of single voxel, to correct for motion‐induced phase fluctuations between individual scans. 36 Moreover, in a non‐water‐suppressed 1H‐MRSI approach with SPICE, the water signal was used for susceptibility mapping. 38 In our study we used the water line to correct for metabolite line‐shape distortions and for absolute quantification of choline content in the prostate. Acquiring the water signal also provides a reference for removal of B0 inhomogeneities between voxels, allowing for more proscriptive restriction of the frequencies for the fitting of model metabolite signals. Nevertheless, the overlap between the signals of spermine residues and cholines cannot always be removed, so that the quantification of one will influence the other. The referencing will improve the choline peak fitting, but for low concentrations there will be some correlated error with the spermine quantification. Despite this, the voxels of interest, in which choline is high and spermine is low, will be easily identifiable from these low concentration voxels by this fitting method.
In the metabolite quantification, we applied relaxation‐time corrections using literature values for the T1 and T2 of the choline methyl and water spins 22 , 32 , 33 and a reported value for the prostate water content. 30 , 31 For quantification in tumorous areas, we assumed that these values are not different from those in benign prostate tissue, except for the T2 of the water spins, which are lower in tumor tissue. 32 It has been reported that the T1 values of water spins are also lower for tumor tissue compared with benign tissue 39 ; however, if we apply these lower T1 values, it had little effect on the calculated choline tissue concentrations at the TR used in data acquisition. If a lower T1 value is taken into account, the combined relaxation correction factor for tumor tissue approaches that of benign tissue. It has been argued that the water content varies only by a few percent between benign and pathological prostate tissue, and thus has little effect on the calculation of absolute metabolite content from MRS data. 40
Water‐signal referencing in MRS of the prostate has an advantage compared with that in brain. Absolute quantification of brain metabolites using the water signal may be hampered by a combination of CSF presence and point spread function. Any contribution of this fluid in a voxel (overlap with a ventricle) may considerably decrease the apparent concentration of metabolites if not corrected for. On the contrary, proton density images of the normal prostate show little signal variation and are featureless, and no large differences in the water content of different regions of the prostate have been reported. 30 , 39
The levels of choline compounds that we derived from the 3.2‐ppm peak for benign prostate tissue (3.8 ± 0.7 mM) is comparable to what has been reported by others using MRS, which is in the range of 2.6‐7 mM in healthy subjects. 5 In MRS of cancer tissue, it is common that the signals of choline compounds are increased; therefore, these are often used as a diagnostic marker. 41 Also in 1H MRS of prostate cancer, the intensity of the methyl peak at about 3.2 ppm, which is composed primarily of signals of free choline, phosphocholine and glycerophosphocholine, 42 , 43 has been used as an in vivo diagnostic tumor marker. In most cases the choline signal was used as a ratio to other prostate metabolites, such as in the commonly used CSC/C ratio, but also as choline/creatine. 3 , 5 , 8 , 44 More detailed analysis of prostate biopsies identified phosphocholine as a major component responsible for this increase. 42 However, the only report we are aware of on the assessment of the absolute level of choline compounds in prostate tumor tissue measured by in vivo MRS did not detect an increase. 45 The authors used single‐voxel PRESS for volume selection (TE = 32 ms; voxel size between 1.5 and 4 cc) and suggested that this finding may be due to bad depiction of the choline signal. In contrast, our study demonstrates that mapping of absolute choline tissue concentrations derived from water‐unsuppressed 1H MRSI may serve as an excellent tool in the detection and localization of prostate tumors, in particular for clinically relevant cancer foci of ≥ 0.5 cc and therefore can be of value in mpMRI examinations for diagnosis and for MR‐guided biopsy procedures. Moreover, our data suggest that the choline content is associated with cancer‐risk groups. Several studies have shown that choline compound signal ratios are correlated with Gleason score in tumors 3 , 7 , 8 , 44 , 46 , 47 and are complementary to DWI in mpMRI as quantitative indicators of tumor aggressiveness. 3 , 4 , 5 , 44
In our study, the correlation with the CSC/C ratio, classically used to identify tumor tissue by MRS, was better for choline than citrate, indicating that the absolute choline level may be a better biomarker than that of citrate. In healthy tissue, the CSC/C ratio has a stronger correlation to citrate than choline, suggesting that the ratio is more dependent on the former metabolites concentrations, whereas in tumor tissue the two metabolites have an equally strong influence on this ratio. This suggests that in tumors, the CSC/C is equally influenced by the displacement of benign tissue as it is on the increase of proliferation among tumor cells. Separating these metabolic biomarkers out allows for a choline concentration map that may act as a more specific marker for proliferative growth.
Although this is a small data set, and there may be a dependence on tumor volume, we hypothesize that the maximum absolute tissue choline concentration could be a marker for tumor lesions with a Gleason grade 5 component, making choline maps a useful tool to assess the most aggressive tumor foci in the prostate. With a larger data set than collected here, it can be validated if the choline tissue concentration can serve in the clinic to discriminate between low aggressive tumor lesions and the more malignant.
Supporting information
FIGURE S1 Screenshot of an in‐house‐built graphical user interface (GUI) tool to delineate voxels in prostate (left) and tumor regions (right). The GUI provides a clickable MRSI grid to select voxels while viewing corresponding DWI (ADC), T2‐weighted MRI, and histopathology images
FIGURE S2 A, Example of the final subtraction of the model water signal from the original data and conversion to a modulus spectrum. The errors in subtraction of the water signal are seen as a ringing around 4.7 ppm, but beyond this is a largely flat baseline of 0 amplitude (the 0 amplitude position is indicated on the y‐axis). B, Detail of this modulus spectrum without water signal or its sidebands show a largely flat baseline with some errors of water‐signal subtraction above 4 ppm, the metabolites of interest, and some residual lipid signals around 1 ppm that were not fully suppressed by the sequence. The region of the spectrum that is used for fitting the final model metabolite signals is indicated by the red lines
ACKNOWLEDGMENT
We thank Sjaak J.A. van Asten and Eline K. Vos for their valuable expert contributions to the research work.
Tayari N, Wright AJ, Heerschap A. Absolute choline tissue concentration mapping for prostate cancer localization and characterization using 3D 1H MRSI without water‐signal suppression. Magn Reson Med. 2021;87:561–573. 10.1002/mrm.29012
Nassim Tayari and Alan J. Wright contributed equally to this work.
Funding information
This work was funded by the Marie Curie FP7‐PEOPLE‐2012‐ITN project TRANSACT (PITN‐GA‐2012‐316679)
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Hamoen EHJ, De Rooij M, Witjes JA, Barentsz JO, Rovers MM. Use of the prostate imaging reporting and data system (PI‐RADS) for prostate cancer detection with multiparametric magnetic resonance imaging: a diagnostic meta‐analysis. Eur Urol. 2015;67:1112‐1121. [DOI] [PubMed] [Google Scholar]
- 3. Kurhanewicz J, Vigneron DB. Magnetic resonance spectroscopy of prostate cancer. eMagRes. 2016;1:923‐944. [Google Scholar]
- 4. Kobus T, Vos PC, Hambrock T, et al. Prostate cancer aggressiveness: in vivo assessment of MR spectroscopy and diffusion‐weighted imaging at 3 T. Radiology. 2012;265:457‐467. [DOI] [PubMed] [Google Scholar]
- 5. Tayari N, Heerschap A, Scheenen TWJ, Kobus T. In vivo MR spectroscopic imaging of the prostate, from application to interpretation. Anal Biochem. 2017;529:158‐170. [DOI] [PubMed] [Google Scholar]
- 6. Steinseifer IK, Philips BWJ, Gagoski B, Weiland E, Scheenen TWJ, Heerschap A. Flexible proton 3D MR spectroscopic imaging of the prostate with low‐power adiabatic pulses for volume selection and spiral readout. Magn Reson Med. 2017;77:928‐935. [DOI] [PubMed] [Google Scholar]
- 7. Tayari N, Steinseifer IK, Selnæs KM, Bathen TF, Maas MC, Heerschap A. High‐quality 3‐dimensional 1H magnetic resonance spectroscopic imaging of the prostate without endorectal receive coil using a semi‐LASER sequence. Invest Radiol. 2017;52:640‐646. [DOI] [PubMed] [Google Scholar]
- 8. Gholizadeh N, Greer PB, Simpson J, et al. Supervised risk predictor of central gland lesions in prostate cancer using 1H MR spectroscopic imaging with gradient offset‐independent adiabaticity pulses. J Magn Reson Imaging. 2019;50:1926‐1936. [DOI] [PubMed] [Google Scholar]
- 9. de Graaf RA. Vivo NMR Spectroscopy: Principles and Techniques. 3rd ed. John Wiley & Sons Inc. 2019. [Google Scholar]
- 10. Dong Z. Proton MRS and MRSI of the brain without water suppression. Prog Nucl Magn Reson Spectrosc. 2015;86‐87:65‐79. [DOI] [PubMed] [Google Scholar]
- 11. Hurd RE, Gurr D, Sailasuta N. Proton spectroscopy without water suppression: the oversampled J‐resolved experiment. Magn Reson Med. 1998;40:343‐347. [DOI] [PubMed] [Google Scholar]
- 12. Clayton DB, Elliott MA, Leigh JS, Lenkinski RE. 1H spectroscopy without solvent suppression: characterization of signal modulations at short echo times. J Magn Reson. 2001;153:203‐209. [DOI] [PubMed] [Google Scholar]
- 13. Serrai H, Clayton DB, Senhadji L, Zuo C, Lenkinski RE. Localized proton spectroscopy without water suppression: removal of gradient induced frequency modulations by modulus signal selection. J Magn Reson. 2002;154:53‐59. [DOI] [PubMed] [Google Scholar]
- 14. Le Fur Y, Cozzone PJ. FID modulus: a simple and efficient technique to phase and align MR spectra. MAGMA. 2014;27:131‐148. [DOI] [PubMed] [Google Scholar]
- 15. Wu Y, Chronik BA, Bowen C, Mechefske CK, Rutt BK. Gradient‐induced acoustic and magnetic field fluctuations in a 4T whole‐body MR imager. Magn Reson Med. 2000;44:532‐536. [DOI] [PubMed] [Google Scholar]
- 16. Dong Z, Dreher W, Leibfritz D. Toward quantitative short‐echo‐time in vivo proton MR spectroscopy without water suppression. Magn Reson Med. 2006;55:1441‐1446. [DOI] [PubMed] [Google Scholar]
- 17. Kreis R, Boesch C. Localized 1H‐MRS without water saturation: techniques and initial results for human brain and muscle. In: Proceedings of the 24th Annual Meeting of ISMRM, Sydney, Australia, 1998:0024. [Google Scholar]
- 18. Van der Veen JWC, Weinberger DR, Tedeschi G, Frank JA, Duyn JF. Proton MR spectroscopic imaging without water suppression. Radiology. 2000;217:296‐300. [DOI] [PubMed] [Google Scholar]
- 19. MacMillan EL, Chong DGQ, Dreher W, Henning A, Boesch C, Kreis R. Magnetization exchange with water and T1 relaxation of the downfield resonances in human brain spectra at 3.0 T. Magn Reson Med. 2011;65:1239‐1246. [DOI] [PubMed] [Google Scholar]
- 20. Epstein JI, Allsbrook WC, Amin MB, et al. The 2005 International Society of Urological Pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma. Am J Surg Pathol. 2005;29:1228‐1242. [DOI] [PubMed] [Google Scholar]
- 21. Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason grading of prostatic carcinoma. Am J Surg Pathol. 2016;40:244‐252.26492179 [Google Scholar]
- 22. Scheenen TWJ, Gambarota G, Weiland E, et al. Optimal timing for in vivo 1H‐MR spectroscopic imaging of the human prostate at 3T. Magn Reson Med. 2005;53:1268‐1274. [DOI] [PubMed] [Google Scholar]
- 23. Scheenen TWJ, Klomp DWJ, Wijnen JP, Heerschap A. Short echo time 1H‐MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses. Magn Reson Med. 2008;59:1‐6. [DOI] [PubMed] [Google Scholar]
- 24. Klomp D, Scheenen T, Hambrock T, Heerschap A. A new method for optimum MRSI of the prostate at 3T using adiabatic RF pulses and internal water referencing. In: Proceedings of the Joint Annual Meeting of ISMRM‐ESMRMB, Berlin, Germany, 2007. Abstract #2885. [Google Scholar]
- 25. Wright AJ, Kobus T, Selnaes KM. Quality control of prostate 1 H MRSI data. NMR Biomed. 2013;26:193‐203. [DOI] [PubMed] [Google Scholar]
- 26. Ernst RR. Numerical Hilbert transform and automatic phase correction in magnetic resonance spectroscopy. J Magn Reson. 1969;1:7‐26. [Google Scholar]
- 27. Klose U. In vivo proton spectroscopy in presence of eddy currents. Magn Reson Med. 1990;14:26‐30. [DOI] [PubMed] [Google Scholar]
- 28. Simonetti AW, Melssen WJ, Van der Graaf M, Heerschap A, Buydens LMC. Automated correction of unwanted phase jumps in reference signals which corrupt MRSI spectra after eddy current correction. J Magn Reson. 2002;159:151‐157. [DOI] [PubMed] [Google Scholar]
- 29. Wright AJ, Buydens LMC, Heerschap A. A phase and frequency alignment protocol for 1H MRSI data of the prostate. NMR Biomed. 2012;25:755‐765. [DOI] [PubMed] [Google Scholar]
- 30. Lowry M, Liney GP, Turnbull LW, Manton DJ, Blackband SJ, Horsman A. Quantification of citrate concentration in the prostate by proton magnetic resonance spectroscopy: zonal and age‐related differences. Magn Reson Med. 1996;36:352‐358. [DOI] [PubMed] [Google Scholar]
- 31. “Report of the Task Group on the Reference Man, International Commission on Radiological Protection, no 23.” Pergamon Press Inc. 1981. [Google Scholar]
- 32. Gibbs P, Liney GP, Pickles MD, Zelhof B, Rodrigues G, Turnbull LW. Correlation of ADC and T2 measurements with cell density in prostate cancer at 3.0 Tesla. Invest Radiol. 2009;44:572‐576. [DOI] [PubMed] [Google Scholar]
- 33. de Bazelaire CMJ, Duhamel GD, Rofsky NM, Alsop DC. MR imaging relaxation times of abdominal and pelvic tissues measured in vivo at 3.0 T: preliminary results. Radiology. 2004;230:652‐659. [DOI] [PubMed] [Google Scholar]
- 34. Dreher W, Leibfritz D. New method for the simultaneous detection of metabolites and water in localized in vivo1H nuclear magnetic resonance spectroscopy. Magn Reson Med. 2005;54:190‐195. [DOI] [PubMed] [Google Scholar]
- 35. Giapitzakis IA, Shao T, Avdievich N, Mekle R, Kreis R, Henning A. Metabolite‐cycled STEAM and semi‐LASER localization for MR spectroscopy of the human brain at 9.4T. Magn Reson Med. 2018;79:1841‐1850. [DOI] [PubMed] [Google Scholar]
- 36. Le Fur Y, Cozzone PJ. Hemi‐spectrum substitution after water signal fitting (HESWAF): an improvement of the modulus post‐processing of MR spectra. Magn Reson Mater Phys Biol Med. 2015;28:67‐85. [DOI] [PubMed] [Google Scholar]
- 37. Lam F, Li Y, Guo R, Clifford B, Liang Z‐P. Ultrafast magnetic resonance spectroscopic imaging using SPICE with learned subspaces. Magn Reson Med. 2020;83:377‐390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guo R, Zhao Y, Li Y, et al. Simultaneous QSM and metabolic imaging of the brain using SPICE: Further improvements in data acquisition and processing. Magn Reson Med. 2021;85:970‐977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baur ADJ, Hansen CM, Rogasch J, et al. Evaluation of T1 relaxation time in prostate cancer and benign prostate tissue using a Modified Look‐Locker inversion recovery sequence. Sci Rep. 2020;10:3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liney GP, Turnbull LW, Lowry M, Turnbull LS, Knowles AJ, Horsman A. In vivo quantification of citrate concentration and water T2 relaxation time of the pathologic prostate gland using 1H MRS and MRI. Magn Reson Imaging. 1997;15:1177‐1186. [DOI] [PubMed] [Google Scholar]
- 41. Glunde K, Bhujwalla ZM, Ronen SM. Choline metabolism in malignant transformation. Nat Rev Cancer. 2011;11:835‐848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Swanson MG, Keshari KR, Tabatabai ZL, et al. Quantification of choline‐ and ethanolamine‐containing metabolites in human prostate tissues using 1H HR‐MAS total correlation spectroscopy. Magn Reson Med. 2008;60:33‐40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Decelle EA, Cheng LL. High‐resolution magic angle spinning 1H MRS in prostate cancer. NMR Biomed. 2014;27:90‐99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Starobinets O, Simko JP, Kuchinsky K, et al. Characterization and stratification of prostate lesions based on comprehensive multiparametric MRI using detailed whole‐mount histopathology as a reference standard. NMR Biomed. 2017;30:e3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Basharat M, Payne GS, Morgan VA, Parker C, Dearnaley D, deSouza NM. TE = 32 ms vs TE = 100 ms echo‐time (1)H‐magnetic resonance spectroscopy in prostate cancer: tumor metabolite depiction and absolute concentrations in tumors and adjacent tissues. J Magn Reson Imaging. 2015;42:1086‐1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van Asten JJA, Cuijpers V, Hulsbergen‐van de Kaa C, et al. High resolution magic angle spinning NMR spectroscopy for metabolic assessment of cancer presence and Gleason score in human prostate needle biopsies. Magn Reson Mater Phys Biol Med. 2008;21:435‐442. [DOI] [PubMed] [Google Scholar]
- 47. Keshari KR, Tsachres H, Iman R, et al. Correlation of phospholipid metabolites with prostate cancer pathologic grade, proliferative status and surgical stage—impact of tissue environment. NMR Biomed. 2011;24:691‐699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Screenshot of an in‐house‐built graphical user interface (GUI) tool to delineate voxels in prostate (left) and tumor regions (right). The GUI provides a clickable MRSI grid to select voxels while viewing corresponding DWI (ADC), T2‐weighted MRI, and histopathology images
FIGURE S2 A, Example of the final subtraction of the model water signal from the original data and conversion to a modulus spectrum. The errors in subtraction of the water signal are seen as a ringing around 4.7 ppm, but beyond this is a largely flat baseline of 0 amplitude (the 0 amplitude position is indicated on the y‐axis). B, Detail of this modulus spectrum without water signal or its sidebands show a largely flat baseline with some errors of water‐signal subtraction above 4 ppm, the metabolites of interest, and some residual lipid signals around 1 ppm that were not fully suppressed by the sequence. The region of the spectrum that is used for fitting the final model metabolite signals is indicated by the red lines


