Abstract
Methods for detecting microorganisms on surfaces are needed to locate biocontamination sources and to relate surface and airborne concentrations. Research was conducted in an experimental room to evaluate surface sampling methods and quantitative PCR (QPCR) for enhanced detection of a target biocontaminant present on flooring materials. QPCR and culture analyses were used to quantitate Bacillus subtilis (Bacillus globigii) endospores on vinyl tile, commercial carpet, and new and soiled residential carpet with samples obtained by four surface sampling methods: a swab kit, a sponge swipe, a cotton swab, and a bulk method. The initial data showed that greater overall sensitivity was obtained with the QPCR than with culture analysis; however, the QPCR results for bulk samples from residential carpet were negative. The swab kit and the sponge swipe methods were then tested with two levels of background biological contamination consisting of Penicillium chrysogenum spores. The B. subtilis values obtained by the QPCR method were greater than those obtained by culture analysis. The differences between the QPCR and culture data were significant for the samples obtained with the swab kit for all flooring materials except soiled residential carpet and with the sponge swipe for commercial carpet. The QPCR data showed that there were no significant differences between the swab kit and sponge swipe sampling methods for any of the flooring materials. Inhibition of QPCR due solely to biological contamination of flooring materials was not evident. However, some degree of inhibition was observed with the soiled residential carpet, which may have been caused by the presence of abiotic contaminants, alone or in combination with biological contaminants. The results of this research demonstrate the ability of QPCR to enhance detection and enumeration of biocontaminants on surface materials and provide information concerning the comparability of currently available surface sampling methods.
Human exposure to bioaerosols has been associated with a variety of diseases, and dissemination of microbial contaminants in indoor environments has been linked to the development of a cluster of symptoms that include eye and sinus irritation, sore throat, headache, fatigue, and dizziness (11). Contamination and subsequent dispersal of biocontaminants in the workplace and living quarters provide an environment that increases the possibility of occupant exposure and adverse health effects ranging from lost productivity to severe illness. Detection and measurement of biocontaminants in indoor environments are needed to assess contamination levels and to estimate the resulting exposure of occupants. However, monitoring is hampered by a lack of methods that provide precise, accurate, and representative exposure estimates for bioaerosols and microbe-contaminated surfaces (3). Conventional biocontaminant monitoring relies on collection of airborne and surface microorganisms and analysis of samples either by culturing on artificial growth media or by microscopic assay. Culture analysis methods underestimate concentrations because only culturable cells are enumerated and identified, while nonculturable organisms go undetected (5). Microscopic assays are laborious and imprecise, and identification is generally limited to the genus level. The inaccuracy of conventional methods and the long analytical time required to characterize bioaerosol and surface contaminant concentrations underscore the need to develop new analytical techniques that can provide rapid, reliable data for bioaerosol exposure monitoring.
PCR (8) has been shown to enhance detection of target microorganisms in bioaerosol samples (1, 2, 6, 7, 9, 10). Methods for detecting microorganisms on surfaces are also needed to locate biocontamination sources and to relate surface and airborne concentrations. The purpose of this research was to evaluate surface sampling methods and quantitative PCR (QPCR) for enhanced detection of a target biocontaminant present on flooring materials. Endospores of Bacillus subtilis subsp. niger (also designated Bacillus globigii) were used for this study. Development and optimization of a rapid, reliable, quantitative method for indoor environmental monitoring involved both laboratory experiments and the use of an experimental room designed for bioaerosol studies. Protocols for B. subtilis subsp. niger DNA extraction and sample processing methods compatible with PCR analysis were developed. B. subtilis subsp. niger-contaminated flooring materials were sampled in the experimental room, and the concentrations of the target organism were measured. Interference with QPCR as a result of background biological contamination and abiotic material was also determined. This research established analytical methods for characterization of a specific microorganism on flooring materials that are quantitative, rapid, sensitive, and useful for monitoring organisms in indoor environments.
MATERIALS AND METHODS
Test organisms and culture media.
The bacterium B. subtilis subsp. niger was used as the test organism in this study. B. subtilis subsp. niger endospores were obtained from the U.S. Army Dugway Proving Ground in Utah. Spore concentration standards were prepared from a liquid purified spore suspension, and the B. subtilis subsp. niger spores aerosolized into the experimental room were from a dry, purified spore culture. B. subtilis subsp. niger was cultured on tryptic soy agar amended with 100 μg of cycloheximide per ml (TSAC) (pH 7.0) (Difco Laboratories, Detroit, Mich.), which was incubated at 28°C for 1 to 2 days. Spores of the fungus Penicillium chrysogenum were used to determine the effect of background biological contamination on B. subtilis subsp. niger culture and QPCR analysis. To obtain spores for the experiments, P. chrysogenum was cultured on malt extract agar (pH 4.7) (Difco), which was incubated at 23°C for 30 days. Spores were harvested from the agar plates and stored dry at 4°C until they were needed. For analysis of surface samples, spores were cultured on malt extract agar that was incubated at 23°C for 3 to 5 days.
Experimental room.
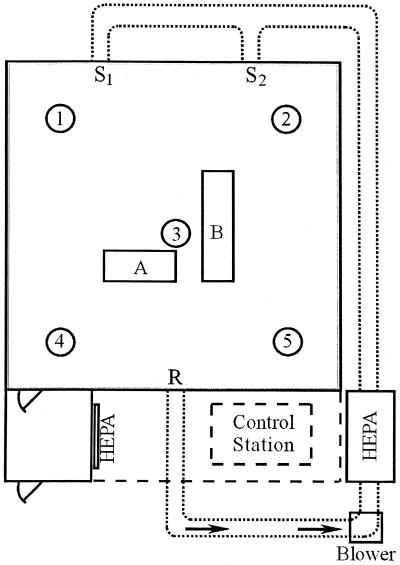
An experimental room designed to resemble a residential indoor environment was used in this study (Fig. 1) (4). The room, which is 4.0 by 4.0 m by 2.2 m high, has a sheet vinyl tile floor. The interior walls, exterior walls, and ceiling are covered with sheetrock and coated with interior latex paint. The room is equipped with a heating, ventilation, and air conditioning system that is sized to simulate a residential system and has 13- by 20-cm rectangular bare metal ductwork. The room is a closed system with two registers (10 by 20 cm) located 1.8 m off the floor and 1.8 m apart, which supply HEPA-filtered (99.97% efficiency) air, and one return vent (25 by 30 cm) located 30 cm off the floor on the opposite wall. During aerosolization experiments, the heating, ventilation, and air conditioning system was operated with an airflow rate of 4.2 m3/min, which resulted in approximately seven room air volume exchanges per hour. The duct velocity was approximately 2.8 m/s. The room was maintained with positive static pressure (0.02 in. of water) during operation to minimize movement of contaminants from the surrounding area into the room. An anteroom equipped with a HEPA-filtered air shower attached to the room entrance reduces mixing of air resulting from entering and leaving the room during experiments. The temperature was monitored with 20 type T thermocouples (Thermo Electric Co., Saddle Brook, N.J.), and the relative humidity was monitored with five relative humidity probes (Hy-Cal Engineering, El Monte, Calif.) located in the room. During all activities in the experimental room, technicians wore full-face respirators and nonwoven protective clothing. Upon completion of each series of experimental room trials, contaminated flooring materials were removed and the interior surfaces of the room were disinfected.
FIG. 1.
Diagram of the experimental room. HEPA-filtered air is delivered into the room via two supply registers (S1 and S2) and exits through the return register (R). Sampling stands 1 to 5 support temperature and relative humidity probes. A, location of APS; B, location of flooring material sections.
Contamination of flooring materials.
Surface sampling was performed with four types of flooring materials: (i) sheet vinyl tile (Armstrong Possibilities [corlon]; catalog no. 86106); (ii) residential cut-pile carpet (Aladdin Silhouette; 34-oz. textured Stainmaster Xtra Life, with 7/16 6-lb. rebond); (iii) commercial loop carpet (Mannington Holbrook; vinyl back; Antron Legacy fiber); and (iv) soiled, worn, 5-year-old residential cut-pile carpet. The flooring materials were cut into 5.7- by 5.7-cm sections for surface sampling studies.
For laboratory studies liquid suspensions of B. subtilis subsp. niger spores were applied to flooring material sections. The sections were placed individually in sterile containers, and each section was inoculated with the desired concentration of B. subtilis subsp. niger spores in a biological safety cabinet. For trials in the experimental room, the flooring materials were contaminated by introducing B. subtilis subsp. niger or P. chrysogenum spores into the room via the air supply duct at one of the supply registers with a Pitt 3 dry-aerosol generator (4, 12). A dry aerosol of spores was generated inside a sealed Plexiglas column by acoustic vibration. Filtered, dry CO2 gas conveyed the spores from the Pitt 3 aerosol generator to the supply duct at a rate of approximately 10 liters/min. The airborne spore concentration in the room was measured with a laser-based aerodynamic particle sizer (APS) (TSI, Inc., St. Paul, Minn.) located inside the room. The APS is capable of obtaining real-time measurements for particles in the 0.5- to 30-μm size range and operates at an airflow rate of 5 liters/min. The background levels of particles were measured with the APS before spores were released. The number of B. subtilis subsp. niger spores per cubic meter was determined by enumerating particles in the 0.7- to 2.5-μm size range. The number of P. chrysogenum spores per cubic meter was determined by enumerating particles in the 1.8- to 3.5-μm size range. After aerosolization of the spores, the room air-handling system was turned off to allow the spores to settle onto the flooring materials. The levels of contamination were determined by removing replicate sections of each flooring material and performing a culture analysis by the bulk sampling method, as described below.
Measurement of spore suspension concentrations.
B. subtilis subsp. niger spore suspensions whose concentrations were known were required for preparation of quantitation standards for QPCR and for laboratory studies involving contamination of flooring materials. The total concentrations in B. subtilis subsp. niger spore suspensions were determined with a Coulter Multisizer II electronic particle counter (Beckman Coulter, Inc., Miami, Fla.). A sample of spores was placed in 0.01 M potassium phosphate buffer with 0.05% Tween 20 (PBT) (pH 7.0), vortexed for 1 min, and then sonicated for 10 min with a Branson 1200 sonicator (Branson Ultrasonics Corp., Danbury, Conn.). The spore suspension was then diluted with filtered Isoton II solution (Beckman Coulter, Inc.), and the spores were enumerated with the Coulter Multisizer II counter. Ten 50-μl aliquots of each sample were counted. The data were averaged, and the total number of spores per milliliter of spore suspension was determined. To determine the number of viable spores per milliliter, serial dilution and spread plating of the spore suspension onto triplicate agar plates were performed. After incubation of the plates and enumeration of the colonies, the average for the triplicate plates was determined and used to calculate the concentration of viable spores in the suspension.
Surface sampling methods.
Four surface sampling methods were tested in laboratory and experimental room studies: a swab kit method (Surface Sampler; Truetech Inc., Long Island, N.Y.), a cotton swab method (Fisher Scientific, Pittsburgh, Pa.), a sponge swipe method (New Horizons Diagnostics Corp., Columbia, Md.), and a bulk sampling method. The protocol developed for the swab kit and cotton swab sampling methods consisted of moistening each swab with PBT and sampling the flooring material sections by swabbing the entire area. The swab was resuspended in 9 ml of PBT and vortexed for 1 min. An aliquot of the sample was reserved for culture analysis, and 6 ml was concentrated by filtration through a 13-mm-diameter, 0.45-μm-pore-size HA filter membrane (mixed cellulose acetate and nitrate; Millipore Corp., Bedford, Mass.) for subsequent DNA extraction. For the sponge swipe method, a sponge was moistened in a sterile stomacher bag (Tekmar Co., Cincinnati, Ohio) containing 30 ml of PBT. The sponge was squeezed to remove the excess buffer and used to sample flooring material sections by wiping the entire area. The sponge was returned to the stomacher bag, and the sample was hand mixed for 1 min. An aliquot of the sample was reserved for culture analysis, and 20 ml was concentrated by filtration, as described above, for subsequent DNA extraction. The bulk sampling method consisted of placing an entire section of contaminated flooring material in a sterile stomacher bag. The sample was removed from the experimental room and stomached for 1 min in 30 ml of PBT by using a Stomacher 80 mixer (Tekmar Co.). An aliquot of the sample was reserved for culture analysis. For vinyl tile, 20 ml of the processed sample was concentrated by filtration for subsequent DNA extraction. For carpet materials, 20 ml was prefiltered through a no. 25 glass fiber filter (Schleicher & Schuell, Keene, N.H.) and the filter was rinsed with 5 ml of PBT. The filtrate was then concentrated by filtration, as described above, for DNA extraction.
Surface sampling efficiency.
Laboratory experiments were performed to determine the sampling efficiencies of three methods, the swab kit, cotton swab, and sponge swipe methods, for removal of B. subtilis subsp. niger spores from surfaces. Three sterile glass petri dishes were inoculated with 10-μl portions of a B. subtilis subsp. niger spore suspension containing 7.4 × 106 CFU of B. subtilis subsp. niger spores. The B. subtilis subsp. niger spores were distributed over an area of approximately 5 cm2 and allowed to dry. For each sampling method, the entire area containing the B. subtilis subsp. niger inoculum was sampled. The swab kit and cotton swab samples were placed in 9 ml of PBT, vortexed for 1 min, and cultured on TSAC. The sponge swipe samples were placed in a stomacher bag containing 30 ml of PBT, hand stomached for 1 min, and cultured on TSAC. The concentrations of B. subtilis subsp. niger spores remaining in the petri dishes were determined by pipetting 10 ml of PBT into each of the dishes to rehydrate the remaining dried inoculum. The resulting suspensions were cultured on TSAC. The overall efficiencies, sampling losses, and sample processing losses were calculated from the data by using the following equations: (i) percent sampling loss = (CFU remaining in petri dish/CFU applied) × 100; (ii) percent sample processing loss = [(CFU applied − CFU per sample − CFU remaining in petri dish)/CFU applied] × 100; and (iii) percent overall efficiency = (CFU per sample/CFU applied) × 100.
Experimental room trials.
A series of experimental room trials was designed to test the protocols that were developed in the laboratory. A total of seven experiments were conducted. Thirty 5.7- by 5.7-cm sections of each flooring material were placed on a sampling bench in the experimental room (Fig. 1) and contaminated with dry B. subtilis subsp. niger spores by aerosolization of spores into the experimental room for 10 min, as described above. The room air-handling system was turned off to allow settling of the B. subtilis subsp. niger spores onto the test materials. The concentrations of B. subtilis subsp. niger spores on the flooring material sections were determined by using the bulk sampling method and culture analysis, as described above. In the first three trials, four sampling methods (the swab kit, sponge swipe, cotton swab, and bulk methods) were compared for detection of B. subtilis subsp. niger by QPCR and culture analysis. Each trial consisted of sampling one section of each of the four types of flooring material with each method (a total of 16 samples). Two sampling methods (the swab kit and sponge swipe methods) were then used for further comparisons to determine the effects of background biological contamination on sample analysis. Trials 4 and 5 were conducted after the flooring materials were contaminated with approximately 102 CFU of P. chrysogenum spores per cm2. P. chrysogenum spores were aerosolized into the experimental room as described above, and the room air-handling system was turned off to allow the spores to settle onto the B. subtilis subsp. niger-contaminated test materials. Trials 6 and 7 were conducted in order to compare the two sampling methods when approximately 104 CFU of P. chrysogenum spores per cm2 were used. To obtain this level of contamination, sequential P. chrysogenum aerosol releases into the experimental room were performed.
DNA extraction and purification.
A DNA extraction and concentration protocol was developed for quantitation standards and all surface sampling methods. Processed samples were filtered through a 0.45-μm-pore-size HA filter membrane (Millipore Corp.), and the membrane was suspended in 0.5 ml of PBT. Each concentrated sample was pretreated with sodium dodecyl sulfate (final concentration, 0.5%) and proteinase K (final concentration, 20 μg/ml), incubated at 50°C for 5 min, and then boiled for 15 min. The sample was chilled on ice for 2 min, and bovine serum albumin (final concentration, 0.05%) was added to block nonspecific binding of DNA to the membrane. The sample was then incubated for 5 min at 37°C in a rotary shaker at 225 rpm. The membrane was removed from the sample, the DNA was purified by using the Pellet Paint protocol (Novagen, Madison, Wis.), and the purified DNA was resuspended in 50 μl of TE buffer. PCR quantitation standards were prepared from a purified B. subtilis subsp. niger spore suspension. B. subtilis subsp. niger spore suspensions were enumerated electronically with a Coulter Multisizer II counter and also by culturing on spread plates, as described above. The electronic particle counts for spore suspensions were greater than the culture measurements because of the nonculturability of some spores. Therefore, electronic measurements were used to determine the concentrations of spore suspensions used as PCR quantitation standards. PCR quantitation standards were prepared from B. subtilis subsp. niger spore suspensions by using the DNA extraction and purification methods used to process samples.
QPCR.
The ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, Calif.) was used for QPCR analysis. A segment of the B. subtilis subsp. niger recA gene was amplified by using primer sequences obtained from the Naval Medical Research Center and a fluorescently labeled probe (Synthetic Genetics, San Diego, Calif.) designed by using Primer Express software (Applied Biosystems), which produced a 131-bp amplicon. The primer sequences were ACCAGACAATGCTCGACGTT (forward) and CCCTCTTGAAATTCCCGAAT (reverse). The probe sequence was 6-FAM-5′TGCGCCCATTTTTCAAGCTGCG3′-TAMRA. Primers were obtained from Operon Technologies (Alameda, Calif.). The amplification conditions specified by the Naval Medical Research Center for use with the Perkin-Elmer reagents were as follows: B. subtilis subsp. niger DNA template, 1× TaqMan buffer A, 5 mM MgCl2, 0.1 mM dATP, 0.1 mM dCTP, 0.1 mM dGTP, 0.2 mM dUTP, 2.5 U of AmpliTaq Gold, 0.5 U of AmpErase uracyl N-glycosylase, each primer at a concentration of 0.2 μM, 0.2 μM probe, and a total reaction volume of 50 μl. The TaqMan cycling conditions were as follows: 2 min at 50°C; 10 min at 95°C; and 40 cycles of 15s at 95°C followed by 1 min at 60°C.
Quantitation with the ABI Prism 7700 sequence detection system was accomplished by amplifying standards of known concentrations that were processed in the same manner as the unknown samples. Standards (100 to 105 templates/reaction mixture) were amplified in duplicate at the same time and under the same conditions as the replicate unknown samples. After amplification, the data were analyzed by using the software provided with the ABI Prism 7700 sequence detection system. The concentrations of the standards were determined based on electronic particle counts, and the software constructed a standard curve for Ct value versus concentration. Ct refers to the PCR cycle in which detectable amplification product was measured. The Ct value was inversely proportional to the initial DNA template concentration. Concentrations for the unknown samples were extrapolated from the standard curve by the software and are reported below as means based on two replicates.
Data and statistical analyses.
The numbers of B. subtilis subsp. niger CFU per milliliter determined for duplicate samples were averaged and converted to the number of CFU per sample. The concentrations of initial target DNA in PCR amplification mixtures were averaged for sample replicates and converted to the number of B. subtilis subsp. niger templates per sample of flooring material. All culture and PCR data were converted to number of CFU/per square centimeter and number of templates per square centimeter of flooring material, respectively. The data were log10 transformed, and analysis of variance and Student t tests were performed to compare data sets. Lower detection limits were determined based on detection of 1 B. subtilis subsp. niger CFU per sample and 1 B. subtilis subsp. niger template per PCR amplification mixture and conversion of the data to number of CFU per square centimeter and number of templates per square centimeter, respectively, as described above.
RESULTS
Sampling efficiency.
The overall process efficiencies of sampling and analysis were compared in the laboratory for the swab kit, cotton swab, and sponge swipe sampling methods by using culture analysis. The overall efficiency was affected by both the efficiency of removal of B. subtilis subsp. niger from surfaces by the sampling method and losses during sample processing and analysis. Glass petri dishes seeded with liquid suspensions containing known concentrations of B. subtilis subsp. niger spores and allowed to dry were sampled. The amount of B. subtilis subsp. niger remaining in each dish was determined, as was the culturable concentration of the sample (Table 1). The results showed that there were no significant differences (P = 0.351) among the swab kit, cotton swab, and sponge swipe methods for sampling a smooth glass surface and that the majority of losses occurred during the processing steps (i.e., vortexing or hand mixing).
TABLE 1.
Sampling efficiencies of three methods for removing culturable B. subtilis subsp. niger spores from surfacesa
| Sampling method | Trial | Sampling loss (%) | Processing loss (%) | Overall efficiency (%) |
|---|---|---|---|---|
| Swab kit | 1 | 1.3 | 29.8 | 68.9 |
| 2 | 2.5 | 18.3 | 79.2 | |
| 3 | 2.6 | 25.0 | 72.4 | |
| Mean ± SE | 2.1 ± 0.4 | 24.4 ± 3.3 | 73.5 ± 3.0 | |
| Cotton swab | 1 | 5.9 | 26.9 | 67.2 |
| 2 | 3.3 | 23.8 | 72.9 | |
| 3 | 12.5 | 21.7 | 65.8 | |
| Mean ± SE | 7.2 ± 2.7 | 24.1 ± 1.5 | 68.6 ± 2.2 | |
| Sponge swipe | 1 | 0.30 | 30.8 | 68.9 |
| 2 | 0.04 | 25.0 | 75.0 | |
| 3 | 0.01 | 21.1 | 78.9 | |
| Mean ± SE | 0.12 ± 0.09 | 25.6 ± 2.8 | 74.3 ± 2.9 |
Sterile glass petri dishes were each inoculated with 10 μl of a B. subtilis subsp. niger spore suspension containing 7.4 × 106 CFU, and the preparations were allowed to dry. The inoculated surfaces of the dishes were sampled by each method (n = 3), and the samples were processed and cultured on TSAC to determine the culturable concentrations of B. subtilis subsp. niger. The petri dishes were rinsed with buffer, and samples were cultured to determine the B. subtilis subsp. niger concentrations remaining in the dishes after surface sampling. Percent loss and overall efficiency values were calculated from the data (see text).
Experimental room trials.
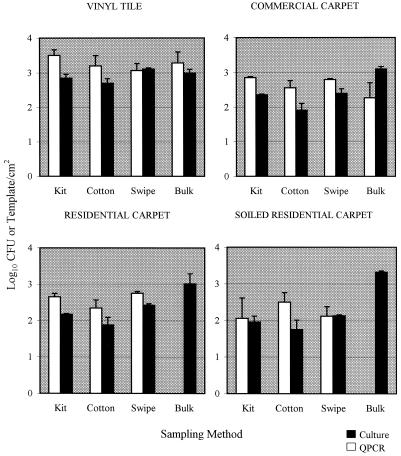
The average airborne concentration of B. subtilis subsp. niger in the experimental room during aerosolization was 1.94 × 106 particles/m3/min during the 10-min release. Culture analysis of flooring material sections in the room after settling of the bioaerosol indicated that the concentration was approximately 102 to 103 B. subtilis subsp. niger CFU/cm2, depending on the flooring material analyzed. In the first three trials, four surface sampling methods for detection of B. subtilis subsp. niger on contaminated flooring materials were compared by using QPCR and culture analysis (Fig. 2). The concentrations of B. subtilis subsp. niger retrieved from surfaces and determined by QPCR were generally greater than or equivalent to the concentrations determined by the culture analysis. However, the PCR results for bulk samples of new and soiled residential carpets were negative (lower detection limit, 0.5 template copy/cm2). If the the bulk samples were excluded, there was no significant difference among the other sampling methods for QPCR detection of B. subtilis subsp. niger on any of the flooring materials (Table 2). When culture analysis was used, significantly greater concentrations were determined with the bulk sampling method for the three carpet materials, while no significant differences between sampling methods were observed for vinyl tile (Table 2).
FIG. 2.
Comparison of culture data (log10 CFU per square centimeter) and QPCR data (log10 templates per square centimeter) obtained from samples of B. subtilis subsp. niger-contaminated flooring materials obtained with four sampling methods. The bar heights represent means based on three samples; the error bars indicate standard errors. Kit, swab kit; Cotton, cotton swab; Swipe, sponge swipe.
TABLE 2.
Statistical comparison of four sampling methods (swab kit, sponge swipe, cotton swab, and bulk methods) for experimental room trials with B. subtilis subsp. niger-contaminated flooring materialsa
| Flooring material |
P value
|
|
|---|---|---|
| PCR | Culture | |
| Vinyl tile | 0.674 | 0.102 |
| Commercial carpet | 0.364 | 0.001b |
| Residential carpet | 0.194c | 0.011b |
| Soiled residential carpet | 0.770c | 0.000b |
Analysis of variance tests were performed with QPCR and culture data (n = 3).
P values less than 0.05 indicate that there was a significant difference between sampling methods.
The results for bulk samples analyzed by QPCR were negative and therefore excluded from the analysis.
Two sampling methods, the swab kit and sponge swipe methods, were selected for further testing. Two trials were conducted with 102 CFU of P. chrysogenum per cm2 as background contamination on the flooring material sections, and this was followed by two additional trials conducted with 104 CFU of P. chrysogenum per cm2 as background contamination. Addition of 102 CFU P. chrysogenum per cm2 to the flooring materials was accomplished by aerosolization of 4.1 × 105 spores/m3/min (average concentration) for 15 min. A higher level of P. chrysogenum contamination (104 CFU/cm2) was obtained by performing four consecutive aerosol releases of spores (average concentration, 8 × 106 particles/m3/min) for 10 min, with a settling period between aerosolizations. The mean B. subtilis subsp. niger concentrations determined by QPCR analysis were greater than the concentrations determined by the culture method for both levels of P. chrysogenum background contamination with one exception, sponge swipe samples from residential carpet at the 102-CFU/cm2 P. chrysogenum level (Table 3). No apparent inhibitory effects on QPCR analysis were observed when P. chrysogenum was added as a background biological contaminant. A comparison of B. subtilis subsp. niger values obtained in performance trials with no P. chrysogenum contamination and the two levels of P. chrysogenum contamination showed that there were no significant differences among the data sets obtained by QPCR analysis (Table 4). For the culture data, one significant difference was observed with the sponge swipe sampling method and residential carpet; in this case the values obtained in the 102-CFU/cm2 P. chrysogenum contamination trials were significantly greater than the values obtained in the other two trials (P = 0.045) (Table 4).
TABLE 3.
Comparison of B. subtilis subsp. niger measurements obtained for flooring materials with two levels of background P. chrysogenum contaminationa
| Flooring material | Background P. chrysogenum level (CFU/cm2) | Log10B. subtilis subsp. niger concn (SE)
|
|||
|---|---|---|---|---|---|
| Swab kit
|
Sponge swipe
|
||||
| PCR (templates/cm2) | Culture (CFU/cm2) | PCR (templates/cm2) | Culture (CFU/cm2) | ||
| Vinyl tile | 102 | 3.531 (0.014) | 2.842 (0.077) | 3.347 (0.073) | 2.981 (0.102) |
| Commercial carpet | 102 | 3.007 (0.001) | 2.468 (0.057) | 2.948 (0.038) | 2.506 (0.083) |
| Residential carpet | 102 | 2.755 (0.092) | 2.210 (0.071) | 2.599 (0.599) | 2.696 (0.108) |
| Soiled residential carpet | 102 | 2.587 (0.160) | 2.173 (0.139) | 2.428 (0.090) | 2.083 (0.134) |
| Vinyl tile | 104 | 3.721 (0.251) | 3.005 (0.300) | 3.555 (0.128) | 2.755 (0.392) |
| Commercial carpet | 104 | 3.119 (0.272) | 2.394 (0.260) | 3.018 (0.099) | 2.516 (0.111) |
| Residential carpet | 104 | 2.732 (0.125) | 2.079 (0.070) | 2.856 (0.033) | 2.369 (0.002) |
| Soiled residential carpet | 104 | 2.576 (0.190) | 1.897 (0.149) | 2.554 (0.086) | 2.172 (0.127) |
The swab kit and sponge swipe sampling methods were used to sample four flooring materials for culture and QPCR analysis (n = 2).
TABLE 4.
Statistical comparison of data collected from B. subtilis subsp. niger-contaminated flooring materials with no P. chrysogenum background contamination (n = 3), 102 CFU of P. chrysogenum per cm2 (n = 2), and 104 CFU of P. chrysogenum per cm2 (n = 2)a
| Sampling method | Flooring material |
P value
|
|
|---|---|---|---|
| PCR | Culture | ||
| Swab kit | Vinyl tile | 0.663 | 0.769 |
| Commercial carpet | 0.408 | 0.813 | |
| Residential carpet | 0.773 | 0.296 | |
| Soiled residential carpet | 0.652 | 0.543 | |
| Sponge swipe | Vinyl tile | 0.241 | 0.482 |
| Commercial carpet | 0.091 | 0.746 | |
| Residential carpet | 0.839 | 0.045b | |
| Soiled residential carpet | 0.395 | 0.812 | |
Analysis of variance tests were performed with PCR and culture data for the swab kit and sponge swipe sampling methods.
The P value less than 0.05 indicates that there was a significant difference between data for different P. chrysogenum contamination levels.
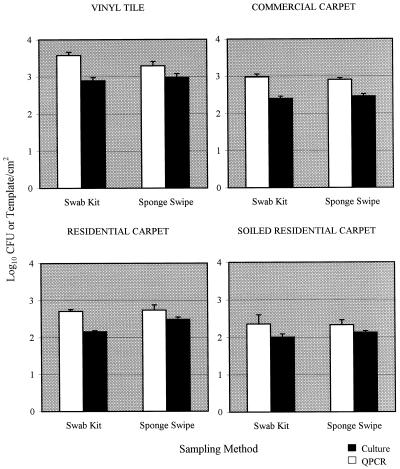
The swab kit and sponge swipe data from the seven trials were pooled for further analysis. Higher mean levels of B. subtilis subsp. niger on flooring materials were obtained with QPCR than with culture analysis in all cases (Fig. 3). The values obtained by QPCR when the swab kit sampling method was used were significantly greater than those obtained by the culture technique for all flooring materials except the soiled residential carpet (Table 5). For the sponge swipe method, the values obtained by QPCR were significantly greater than the culture values only for commercial carpet. A comparison of the swab kit and sponge swipe sampling methods showed that there were no significant differences between these sampling methods for any of the flooring materials when QPCR analysis was used (Table 6; Fig. 3). With culture analysis, the values obtained with the sponge swipe sampling method for residential carpet were significantly greater than those obtained with the swab kit method (Table 6). A comparison of the values for the different flooring materials indicated that the QPCR and culture values were significantly greater with vinyl tile than with the three carpet materials for both sampling methods (P = 0.000).
FIG. 3.
Comparison of culture data (log10 CFU per square centimeter) and QPCR data (log10 templates per square centimeter) obtained from all trials with B. subtilis subsp. niger-contaminated flooring materials when the swab kit and sponge swipe sampling methods were used. The bar heights represent means based on seven samples; the error bars indicate standard errors.
TABLE 5.
Statistical comparison of QPCR and culture analysis methods in which pooled data from experimental room trials (n = 7) were useda
| Sampling method | Flooring material | P value |
|---|---|---|
| Swab kit | Vinyl tile | 0.000b |
| Commercial carpet | 0.000b | |
| Residential carpet | 0.000b | |
| Soiled residential carpet | 0.210 | |
| Sponge swipe | Vinyl tile | 0.074 |
| Commercial carpet | 0.000b | |
| Residential carpet | 0.130 | |
| Soiled residential carpet | 0.180 |
Student t tests were performed for each sampling method and flooring material.
P values less than 0.05 indicate that there was a significant difference between the QPCR and culture analysis methods.
TABLE 6.
Statistical comparison of the swab kit and sponge swipe sampling methods in which pooled data from experimental room trials (n = 7) were useda
| Flooring material |
P value
|
|
|---|---|---|
| PCR | Culture | |
| Vinyl tile | 0.078 | 0.570 |
| Commercial carpet | 0.450 | 0.470 |
| Residential carpet | 0.830 | 0.002b |
| Soiled residential carpet | 0.930 | 0.230 |
Student t tests were performed with QPCR and culture data for each flooring material.
The P values less than 0.05 indicates that there was a significant difference between sampling methods.
Residential carpet data were compared with soiled residential carpet data to determine whether soiling was a significant factor in inhibiting measurement of B. subtilis subsp. niger in surface samples. With QPCR data, no significant differences were observed between the two materials (swab kit method, P = 0.210; sponge swipe method, P = 0.054), although the mean values for residential carpet were greater than those for soiled residential carpet for both sampling methods. With culture analysis, the residential carpet values were significantly greater than the values for soiled residential carpet for the sponge swipe sampling method (swab kit method, P = 0.140; sponge swipe method, P = 0.001.)
DISCUSSION
QPCR detection of B. subtilis subsp. niger in surface samples was more sensitive than culture analysis in these experiments. In addition, QPCR analysis by the working protocol developed in this study can be performed on the day that a sample is collected compared with a delay of 24 to 48 h for culture analysis. Inhibitors of QPCR that may be present in environmental samples remain a concern. In the present study, inhibition due solely to background biological contamination consisting of P. chrysogenum spores on flooring materials was not evident. However, some degree of inhibition likely occurred during amplification of samples from soiled residential carpet, as indicated by the lower mean template concentrations measured for those samples compared to the concentrations for new residential carpet samples (Fig. 3; Table 3). The differences were observed with both the swab kit and sponge swipe sampling methods but were not statistically significant based on the mean data from seven experimental trials. There may have been some inhibition of the QPCR caused by the presence of biological contaminants other than P. chrysogenum. Culture analysis of soiled residential carpet samples using the swab kit and sponge swipe sampling methods revealed non-B. subtilis subsp. niger background bacterial concentrations of approximately 102 CFU/cm2 and fungal concentrations of 101 CFU/cm2 (data not shown). Inhibition may also have been caused by the presence of nonbiological contaminants on both new and soiled residential carpet samples. Negative results were obtained for PCR analysis of bulk samples from both new residential carpet and soiled residential carpet, but not bulk samples from commercial carpet (Fig. 2). One explanation for these results is that stomaching of the carpet section may have released chemical inhibitors from the residential carpet that were not present in samples collected by the other sampling methods. The negative PCR results obtained for residential carpet but not for commercial carpet may have been due to differences in the compositions of the two carpet materials. In addition, more fiber debris was observed in the stomacher buffer of the residential carpet samples. This debris made filtration of these samples more difficult than filtration of the commercial carpet samples, even after prefiltration. PCR inhibition by environmental contaminants can be reduced by further purification of the sample DNA. However, the sensitivity may be reduced due to loss of sample. Because the success of a purification protocol depends on the type of inhibitors present, further testing of the PCR analysis protocol should be conducted with field samples in order to determine the degree of inhibition compared with inhibition in control samples and the most effective means to overcome inhibition.
The swab kit, sponge swipe, and cotton swab sampling methods were more effective with the relatively smooth surface of vinyl tile than with carpet (Fig. 2 and 3; Table 3), where sampling losses likely occurred due to settling of B. subtilis subsp. niger spores into the depths of the carpet. Analysis of the QPCR data from the first three trials revealed no significant differences among the sampling methods tested for any of the flooring materials (Table 2). According to the experimental design, two sampling methods were selected for further testing with background biological contamination. The swab kit and sponge swipe methods were chosen, while the bulk and cotton swab protocols were eliminated from further study. The bulk method is a destructive sampling method with limited field practicality that was included primarily to provide a baseline of culture data for comparison with QPCR data. In addition, inhibition of PCR by carpet material extracts was observed with bulk samples. The cotton swab method was eliminated because the principle of sampling was similar to that of the swab kit method and it was less sensitive. The swab kit and sponge swipe methods were effective for detection and quantitation of B. subtilis subsp. niger on flooring materials, and there were no significant differences between these methods in terms of the PCR data obtained in seven trials. Therefore, selection of a sampling method may depend on the field situation. The sponge swipe method is probably the most practical method for large sampling areas, and the swab kit method was the most efficient for smooth vinyl tile surfaces.
The results of this research demonstrate the ability of QPCR to enhance detection and enumeration of biocontaminants on surface materials and provide information concerning the comparability of currently available sampling methods. Additional studies designed to minimize biotic and abiotic interference would increase the field applicability of surface sampling for detection of biocontaminants in indoor environments.
REFERENCES
- 1.Alvarez A J, Buttner M P, Stetzenbach L D. PCR for bioaerosol monitoring: sensitivity and environmental interference. Appl Environ Microbiol. 1995;61:3639–3644. doi: 10.1128/aem.61.10.3639-3644.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez A J, Buttner M P, Toranzos G A, Dvorsky E A, Toro A, Heikes T B, Mertikas L E, Stetzenbach L D. The use of solid-phase polymerase chain reaction for the enhanced detection of airborne microorganisms. Appl Environ Microbiol. 1994;60:374–376. doi: 10.1128/aem.60.1.374-376.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Conference of Governmental and Industrial Hygienists. Bioaerosols: assessment and control. Cincinnati, Ohio: American Conference of Governmental and Industrial Hygienists; 1999. [Google Scholar]
- 4.Buttner M P, Stetzenbach L D. Monitoring of fungal spores in an experimental indoor environment to evaluate sampling methods and the effects of human activity on air sampling. Appl Environ Microbiol. 1993;59:219–226. doi: 10.1128/aem.59.1.219-226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox C S. Airborne bacteria and viruses. Sci Prog Oxford. 1989;73:469–500. [PubMed] [Google Scholar]
- 6.Haugland R A, Vesper S J, Wymer L J. Quantitative measurement of Stachybotrys chartarum conidia using real time detection of PCR products with the TaqManTM fluorogenic probe system. Mol Cell Probes. 1999;13:329–340. doi: 10.1006/mcpr.1999.0258. [DOI] [PubMed] [Google Scholar]
- 7.Mukoda T, Todd L, Sobsey M. PCR and gene probes for detecting bioaerosols. J Aerosol Sci. 1994;25:1523–1532. [Google Scholar]
- 8.Saiki R K, Scharf S, Faloona F, Mullis K B, Horn G T, Erlich H, Arnheim N. Enzymatic amplification of β-globin genomic sequence and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 9.Sawyer M H, Chamberlin C J, Wu Y N, Aintablian N, Wallace M R. Detection of varicella-zoster virus DNA in air samples from hospital rooms. J Infect Dis. 1994;169:91–94. doi: 10.1093/infdis/169.1.91. [DOI] [PubMed] [Google Scholar]
- 10.Stärk K D C, Nicolet J, Frey J. Detection of Mycoplasma hypopneumoniae by air sampling with a nested PCR assay. Appl Environ Microbiol. 1998;64:543–548. doi: 10.1128/aem.64.2.543-548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stetzenbach L D. Introduction to aerobiology. In: Hurst C J, editor. Manual of environmental microbiology. Washington, D.C.: ASM Press; 1997. pp. 619–628. [Google Scholar]
- 12.Weyel D A, Ellakkani M, Alarie Y, Karol M. An aerosol generator for the resuspension of cotton dust. Toxicol Appl Pharmacol. 1984;76:544–547. doi: 10.1016/0041-008x(84)90359-4. [DOI] [PubMed] [Google Scholar]