Abstract
Key points
Maximal endurance performance is greater in the early daytime.
Timed exercise differentially alters the muscle transcriptome and (phospho)‐proteome.
Early daytime exercise triggers energy provisioning and tissue regeneration.
Early night‐time exercise activates stress‐related and catabolic pathways.
Scheduled training has limited effects on the muscle and liver circadian clocks.
Abstract
Timed physical activity might potentiate the health benefits of training. The underlying signalling events triggered by exercise at different times of day are, however, poorly understood. Here, we found that time‐dependent variations in maximal treadmill exercise capacity of naïve mice were associated with energy stores, mostly hepatic glycogen levels. Importantly, running at different times of day resulted in a vastly different activation of signalling pathways, e.g. related to stress response, vesicular trafficking, repair and regeneration. Second, voluntary wheel running at the opposite phase of the dark, feeding period surprisingly revealed a minimal zeitgeber (i.e. phase‐shifting) effect of training on the muscle clock. This integrated study provides important insights into the circadian regulation of endurance performance and the control of the circadian clock by exercise. In future studies, these results could contribute to better understanding circadian aspects of training design in athletes and the application of chrono‐exercise‐based interventions in patients.
Keywords: circadian clock, energy homeostasis, exercise, metabolism, proteomics, skeletal muscle, transcriptomics, zeitgeber
Introduction
Almost all aspects of mammalian physiology undergo changes relative to the time of day. Many of these variations are, directly or indirectly, driven by the circadian clock, an evolutionarily conserved time‐keeping mechanism that is present in virtually all cells of the body (Buhr & Takahashi, 2013). Adequate timing of physical activity might have been of particular evolutionary importance to synchronize movement with predator–prey interactions, implying a strong control by the circadian clock, both at the central level to control behaviour and in skeletal muscle for adequate functionality (Albrecht & Eichele, 2003). Inversely, adaptation of clock‐controlled physiological parameters, for example, anticipation and execution of metabolic pathways important for foraging, optimally can be influenced by activity patterns to adapt to changes in the external environment (Hughes & Piggins, 2012). Thus, in addition to potentially being downstream of clock control, muscle activity has also been proposed as an upstream zeitgeber to synchronize peripheral clocks, and the application of chrono‐exercise put forward in various diseases characterized by abnormal circadian rhythmicity (Gabriel & Zierath, 2019; Gutierrez‐Monreal et al. 2020).
Consistent with the circadian modulation of muscle physiology, hundreds of transcripts in skeletal muscle oscillate with a 24‐h period in humans and mice (McCarthy et al. 2007; Perrin et al. 2018). Moreover, insulin sensitivity, mitochondrial respiration, glucose and lipid‐related metabolites likewise follow similar patterns in muscle tissue (Loizides‐Mangold et al. 2017; Dyar et al. 2018; Sato et al. 2018). In line with this, daily variations in resistance and endurance exercise peak performance have been reported during the normal active phase in humans (Mirizio et al. 2020) and rodents (Ezagouri et al. 2019) in most, but not all (Knaier et al. 2019; Mirizio et al. 2020) trials. The robustness and timing of such performance peaks seem highly variable, depending on a multitude of parameters including chronotype, time from awakening, muscle and liver glycogen levels, nutritional status, and temperature (Facer‐Childs & Brandstaetter, 2015; Hearris et al. 2018). It thus is unknown whether and how the intrinsic muscle clock machinery influences the physiological and molecular responses of skeletal muscle to exercise and ultimately physical performance. Inversely, a potential zeitgeber activity of training also is unclear, i.e. whether repeated bouts of exercise affect phase and amplitude of the molecular clock in muscle or other tissues. Training studies at different times of day suffer from confounding aspects such as light‐mediated inhibition of voluntary locomotion in animals, or restricted analysis of reporter gene‐based approaches. Therefore, whether the putative enhanced effects of timed exercise training on health parameters in both clinical and preclinical contexts (Yamanaka et al. 2015; Savikj et al. 2019) depend on the zeitgeber properties of exercise remains to be investigated.
To address open questions about the cross‐regulation of exercise and the circadian clock, we first evaluated effects of timed exercise at the systemic and muscle cellular levels by assessing maximal treadmill exercise capacity across the 24‐h light–dark (LD) cycle. We furthermore dissected the transcriptome and (phospho‐)proteome responses of working muscles at two distinct phases of the LD cycle. Second, to investigate the potential zeitgeber effects of exercise, we used a skeleton photoperiod (SPP) in combination with restricted wheel‐running access to interrogate the consequences of scheduled daytime voluntary training on skeletal muscle gene and protein regulation.
Methods
Animals
Eight‐week‐old C57BL/6JRj male mice (Janvier Labs, Le Genest‐Saint‐Isle, France) were housed in standard cages under 12:12 light:dark (LD) conditions, with light onset at 06.00 h (zeitgeber time 0; ZT0) or entrained to a skeleton photoperiod (SPP) as described below and in Fig. 7, unless otherwise stated. The mice had ad libitum access to a standard chow diet (Maintenance 3432, KLIBA NAFAG, Kaiseraugst, Switzerland) and water, unless otherwise stated. All experiments were performed in agreement with the principles of the Basel Declaration, with Federal and Cantonal Laws regulating the care and use of experimental animals in Switzerland, and the institutional guidelines of the Biozentrum and the University of Basel. The protocol with all methods described here was approved by the ‘Kantonales Veterinäramt’ of the Kanton Basel‐Stadt, under consideration of the well‐being of the animals and the 3R principle.
Figure 7. Phospho‐/proteomic analyses reveal time‐dependent differences in muscle response to early daytime vs. early night‐time exercise.
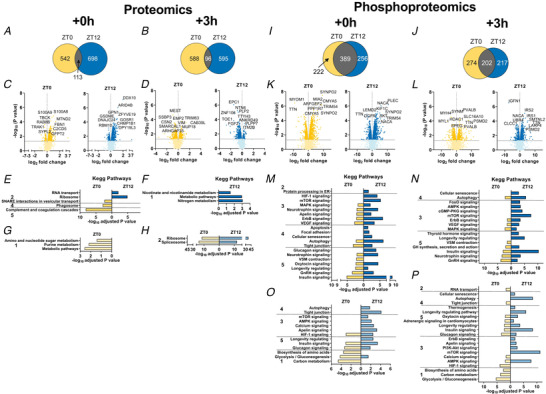
A and B, Venn diagrams displaying the number of differentially regulated proteins immediately (A; Ex + 0 h) and 3 h (B; Ex + 3 h) after early daytime (ZT0) or early night‐time (ZT12) exercise, and resulting overlap. C and D, volcano plots showing the differentially regulated proteins (with top 10 indicated) as above. E–H, KEGG analysis of up‐regulated (E and F) and down‐regulated (G and H) proteins by exercise as described above. I–L, Venn diagrams (I and J) and volcano plots (K and L) displaying the differentially phosphorylated proteins in the same groups as above. M–P, KEGG analysis (top 5 terms per category) for increased (M and N) and decreased (O and P) protein phosphorylation in Ex + 0 h and Ex + 3 h groups. KEGG pathway categories: 1, metabolism; 2, genetic information processing; 3, environmental information processing; 4, cellular processes; 5, organismal systems. [Colour figure can be viewed at wileyonlinelibrary.com]
Forced high‐intensity exercise performance across the day
Group‐housed sedentary mice were acclimated to the treadmill equipped with a shock grid (Columbus Instruments, Columbus, OH, USA) on three consecutive days prior to the experiment. The accommodation period consisted of: Day 1, placing the mice in the treadmill for 10 min without shock and belt movement followed by 5 min at 5 m min−1; Day 2, running at 5, 7 and 10 m min−1 for 5 min each, without shock; Day 3, running at 8, 10 and 12 m min−1 for 5 min each, with shock. After one resting day, a maximal exercise capacity test was performed by 3 min at 8 m min−1 increasing treadmill speed by 2 m every 2 min, at a 15° slope, until exhaustion. To provide additional motivation to avoid the lower portion of the treadmill and thus the shock grid, we gently scratched the back of the animal during accommodation and maximal exercise capacity test. Exhaustion was met if an animal remained on the electrical grid (providing a mild electrical stimulus of 0.5 mA, 200 ms pulse, 1 Hz) for more than 5 s. Tail blood glucose (Accu‐Chek, Roche, Basel, Switzerland) and lactate (Lactate Plus meter, Nova Biomedical, Waltham, MA, USA) values were determined immediately prior to treadmill exercise and within 1 min after physical exhaustion. Mice were either sacrificed immediately after exhaustion (≤5 min of time delay; Ex + 0 h) or 3 h after exercise (Ex + 3 h). For the latter group, mice were returned to their home cages without access to food. This experiment was repeated every 4 h for 24 h starting from ZT0 (06.00 h; scheme in Fig. 1A ). A non‐exercised group (Sedentary; Sed) of mice was always sacrificed at a similar ZT (≤30 min of time delay) as the exercised mice (Ex + 0 h and Ex + 3 h). Note that control Sed mice were placed in new cages, with new bedding but no food access 60 min prior to sacrifice. Importantly, all mice had a free access to food prior to an exercise bout, that is, were not fasted. Lastly, to take into account changes in basal gene expression over time, gene expression data obtained from Ex + 0 h and Ex + 3 h mouse groups running, e.g. at ZT0, were compared to sedentary controls sacrificed at ZT0 and ZT4, respectively.
Figure 1. Time‐of‐day‐dependent variations in mouse treadmill exercise performance and physiological responses to exercise.
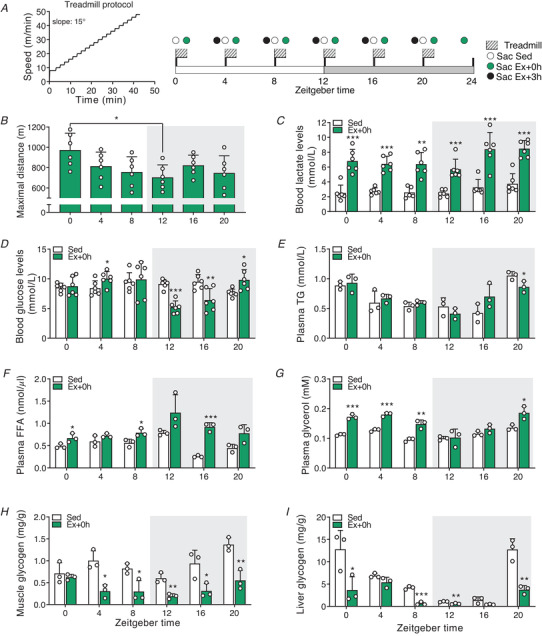
A, treadmill protocol and experimental scheme. Mice were divided into two groups, sedentary (Sed) and exercise (Ex). The latter group was further divided into two groups: sacrificed immediately (+ 0 h) or 3 h after exhaustion (+ 3 h). B, maximal distance reached at exhaustion. C–G, blood lactate (C), glucose (D), plasma triglyceride (TG) (E), free fatty acid (FAA) (F), and glycerol (G) levels. H and I, muscle (H) and liver (I) glycogen levels. Light and dark periods are depicted by white and grey background, respectively. Data are shown as the means ± SD (n = 3). * P < 0.05, ** P < 0.01, *** P < 0.001, one‐way ANOVA (B) and unpaired Student's t‐test (C–I). [Colour figure can be viewed at wileyonlinelibrary.com]
Daytime scheduled wheel‐running activity
Mice were single‐housed in standard cages, within an environment‐controlled cabinet (UniProtect Air Flow Cabinet, Bioscape, Castrop‐Rauxel, Germany), with the temperature set to 23°C. The mice had access to a wheel with rods (∅ 11.5 cm, Starr Life Sciences, Oakmont, PA, USA) under constant 12:12 LD conditions for 3 weeks prior to exposure to a SPP. The SPP consisted of two repeated light‐pulses (LP): 1 h LP at the beginning of the resting period and 1 h LP at the end of the resting period, interrupted by 10 h of darkness (Fig. 7A ). After 3 weeks’ acclimatization to the SPP, 3 weeks of intervention followed. During the intervention, one group of mice (control, CTRL) had ad libitum access to food, water and free access to running wheels. Another group of mice (daytime activity, DA) was food‐restricted to the longer dark phase (active phase) and had access to a wheel only during the shorter dark period (resting phase) (see Fig. 7). Wheel and food access were controlled manually, without opening the cage to not disturb the animals. Importantly, the use of the wheel (light phase: 309 turns vs. dark phase: 17,732 turns) and food intake (light phase: 0.4 g vs. dark phase: 4.7 g) during the resting/inactive period was virtually absent in the CTRL group. Lastly, we only compared daytime and night‐time wheel active animals in our study, trained for a similar length of time; the comparison of sedentary mice with mice given free access to a running wheel is well documented elsewhere (Holloszy, 1967; Allen et al. 2001; McKie et al. 2019).
Body temperature and locomotor activity recordings
General locomotor activity and core body temperature data were acquired with the E‐Mitter Telemetry System (Starr Life Sciences) from single‐caged animals placed in an environment‐controlled cabinet (UniProtect Air Flow Cabinet, Bioscape). Briefly, small transponders (G2 E‐Mitter, Starr Life Sciences) were implanted into the abdominal cavity of mice under isoflurane anaesthesia (2% isoflurane + O2). Mice were treated with meloxicam (1 mg kg−1) pre‐ and post‐operatively and allowed to recover for 3 weeks. The abovementioned parameters together with the wheel‐running activity were recorded with a PC‐based acquisition system connected to ER4000 Receivers (VitalView, Starr Life Sciences).
Muscle tissue preparation and blood collection
Mice from the different experiments were sacrificed by short exposure to CO2 and immediate exsanguination. Blood was collected in tubes containing lithium heparin (Microvette 500 LH, Sarstedt, Munich, Germany) centrifuged at 2000 g for 5 min at RT and stored at −80°C. The glycolytic quadriceps and gastrocnemius muscles, as well as liver samples, were quickly snap frozen in liquid nitrogen and stored at −80°C until further analysis.
Quantitative real‐time PCR
Total RNA from muscle tissues was extracted using a hybrid method combining TRI‐Reagent (T9424, Sigma‐Aldrich, St Louis, MO, USA) and RNeasy Mini Kit (74104, Qiagen, Hilden, Germany). RNA quantity and purity were measured with a NanoDrop OneC (Thermo Fisher Scientific, Waltham, MA, USA). High‐Capacity cDNA Reverse Transcript Kit (4368814, Thermo Fisher Scientific, Waltham, MA, USA) was used for cDNA synthesis with 1 μg of total RNA. Quantitative real‐time PCR was performed with Fast SYBR Green Master Mix (4385612, Thermo Fisher Scientific, Waltham, MA, USA) in a RT‐PCR System (StepOnePlus, Thermo Fisher Scientific, Waltham, MA, USA). PCR reactions were done in duplicate with the addition of negative controls (i.e. no reverse transcription and no template controls). Relative expression levels were determined using the comparative ΔΔC T method to normalize target gene mRNA to hypoxanthine guanine phosphoribosyltransferase (Hprt). Primer sequences are summarized in Supplementary Table 10. Rhythmicity and differential rhythmicity were assessed using the methods RAIN (Thaben & Westermark, 2014) and DODR (Thaben & Westermark, 2016), respectively.
Blood parameters analysis
Quantification of plasma triglyceride was done with the Cobas c111 analyser (Roche, Basel, Switzerland). Plasma free fatty acids were analysed using the Free Fatty Acid Quantification Assay Kit (ab65341, Abcam, Cambridge, UK) following manufacturer's recommendations. Muscle and liver glycogen levels were measured with a Glycogen Assay Kit (Abcam, ab65620).
RNA sequencing and data analysis
RNA quality was determined on the Bioanalyzer instrument (Agilent Technologies, Santa Clara, CA, USA) using the RNA 6000 Nano Chip (Agilent, cat. no. 5067‐1511) and quantified by spectrophotometry using the NanoDrop ND‐1000 Instrument (NanoDrop Technologies, Wilmington, DE, USA). Library preparation was performed with 1 μg total RNA using the TruSeq Stranded mRNA Library Prep Kit High Throughput (cat. no. RS‐122‐2103, Illumina, San Diego, CA, USA). Libraries were quality‐checked on the Fragment Analyzer (Advanced Analytical, Ames, IA, USA) using the Standard Sensitivity NGS Fragment Analysis Kit (cat. no. DNF‐473, Advanced Analytical) revealing excellent quality of libraries (average concentration was 152 ± 9 nmol l−1 and average library size was 374 ± 4 base pairs). Samples were pooled to equal molarity. Each pool was quantified by PicoGreen Fluorometric measurement in order to be adjusted to 1.8 pM and used for clustering on the NextSeq 500 instrument (Illumina). Samples were sequenced single reads of 76 bases using the NextSeq 500 High Output Kit 75‐cycles (Illumina, cat. no. FC‐404‐1005), and primary data analysis was performed with the Illumina RTA version 2.4.11 and Basecalling Version bcl2fastq‐2.20.0.422.
To quantify mRNA expression levels, kallisto version 0.46.0 (Bray et al. 2016) was used. To build the index for kallisto, the GRCm38.p6 (mm10) genome assembly and the ncbiRefSeqCurated transcript annotation of the UCSC genome browser were used (Karolchik et al. 2014; Pruitt et al. 2014). MicroRNAs (miRbase Version 19; Kozomara & Griffiths‐Jones, 2011; translated to RefSeq IDs through BioMart; Durinck et al. 2009) were excluded. Only one transcript was retained if several had both identical start and end coordinates, slightly flattening the annotation, and preference was given to transcripts with IDs starting with ‘NM_’. Transcripts mapping to more than one chromosome or to random or chrUn contigs were discarded. tximport version 1.14.0 (Soneson et al. 2015) was used to transform expression levels to flattened gene level pseudo‐counts, using the ‘lengthScaledTPM’ option. For this, RefSeq IDs were mapped to Entrez gene IDs using the org.Mm.eg.db database of R/Bioconductor version 3.10 (Gentleman et al. 2004). DESeq2 version 1.26.0 (Love et al. 2014) was used for statistical analysis of gene level differential expression. Here, log2 fold changes were estimated by the DESeq2 shrinkage estimator.
Proteomics and data analysis
Phosphopeptide enrichment and liquid chromatography–tandem mass spectrometry analysis
Tissue was lysed in 8 M Urea, 0.1 M ammonium bicarbonate, phosphatase inhibitors (Sigma‐Aldrich, P5726 and P0044) by sonication (Bioruptor, 10 cycles, 30 s on/off, Diagenode, Liège, Belgium) and proteins were digested as described previously (Ahrne et al. 2016). Shortly, proteins were reduced with 5 mm tris(2‐carboxyethyl)phosphine (TCEP) for 60 min at 37°C and alkylated with 10 mm chloroacetamide for 30 min at 37°C. After diluting samples with 100 mm ammonium bicarbonate buffer to a final urea concentration of 1.6 M, proteins were digested by incubation with sequencing‐grade modified trypsin (1/50, w/w; Promega, Madison, WI, USA) for 12 h at 37°C. After acidification using 5% trifluoroacetic acid (TFA), peptides were desalted using C18 reverse‐phase spin columns (Macrospin, Harvard Apparatus, Holliston, MA, USA) according to the manufacturer's instructions, dried under vacuum and stored at −20°C until further use.
Peptide samples were enriched for phosphorylated peptides using Fe(III)‐IMAC cartridges on an AssayMAP Bravo (Agilent Technologies, Basel, Switzerland) platform as recently described (Post et al. 2017). Unmodified peptides (‘flowthrough’) were subsequently used for tandem mass tag (TMT) analysis.
Phospho‐enriched peptides were resuspended in 0.1% aqueous formic acid and subjected to liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis using a Q Exactive HF Mass Spectrometer or an Orbitrap Fusion Lumos Mass Spectrometer fitted with an EASY‐nLC 1000 or an EASY‐nLC 1200, respectively (both Thermo Fisher Scientific) and a custom‐made column heater set to 60°C. Peptides were resolved using a reversed phase (RP)‐HPLC column (75 μm × 30 cm or 75 μm × 36 cm) packed in‐house with C18 resin (ReproSil‐Pur C18‐AQ, 1.9 μm resin; Dr Maisch GmbH, Ammerbuch, Germany) at a flow rate of 0.2 μl min−1. The following gradient was used for peptide separation: Q Exactive HF from 5% B to 8% B over 5 min to 20% B over 45 min to 25% B over 15 min to 30% B over 10 min to 35% B over 7 min to 42% B over 5 min to 50% B over 3 min to 95% B over 2 min followed by 18 min at 95% B, Orbitrap Fusion Lumos from 5% B to 8% B over 5 min to 20% B over 45 min to 25% B over 15 min to 30% B over 10 min to 35% B over 7 min to 42% B over 5 min to 50% B over 3min to 95% B over 2 min followed by 18 min at 95% B. Buffer A was 0.1% formic acid in water and buffer B was 80% acetonitrile–0.1% formic acid in water.
The Q Exactive HF mass spectrometer was operated in DDA mode with a total cycle time of approximately 1 s. Each MS1 scan was followed by high‐collision dissociation (HCD) of the 10 most abundant precursor ions with dynamic exclusion set to 45 s. For MS1, 3 × 106 ions were accumulated in the Orbitrap over a maximum time of 100 ms and scanned at a resolution of 120,000 full width at half‐maximum (FWHM; at 200 m/z). MS2 scans were acquired at a target setting of 1 × 105 ions, maximum accumulation time of 100 ms and a resolution of 30,000 FWHM (at 200 m/z). Singly charged ions and ions with unassigned charge state were excluded from triggering MS2 events. The normalized collision energy was set to 28%, the mass isolation window was set to 1.4 m/z and one microscan was acquired for each spectrum. The Orbitrap Fusion Lumos mass spectrometer was operated in DDA mode with a cycle time of 3 s between master scans. Each master scan was acquired in the Orbitrap at a resolution of 120,000 FWHM (at 200 m/z) and a scan range from 375 to 1600 m/z followed by MS2 scans of the most intense precursors in the Orbitrap at a resolution of 30,000 FWHM (at 200 m/z) with isolation width of the quadrupole set to 1.4 m/z. Maximum ion injection time was set to 50 ms (MS1) and 54 ms (MS2) with an AGC target set to 1 × 106 and 5 × 104, respectively. Only peptides with charge state 2–5 were included in the analysis. Monoisotopic precursor selection (MIPS) was set to Peptide, and the intensity threshold was set to 2.5 × 104. Peptides were fragmented by HCD with collision energy set to 30%, and one microscan was acquired for each spectrum. The dynamic exclusion duration was set to 30 s.
The acquired raw‐files were imported into Progenesis QI software (v2.0, Nonlinear Dynamics Ltd, Newcastle upon Tyne, UK), which was used to extract peptide precursor ion intensities across all samples applying the default parameters. The generated mgf‐file was searched using MASCOT against a murine database (consisting of 34,026 forward and reverse protein sequences downloaded from Uniprot on 20,190,129) and 392 commonly observed contaminants using the following search criteria: full tryptic specificity was required (cleavage after lysine or arginine residues, unless followed by proline); three missed cleavages were allowed; carbamidomethylation (C) was set as fixed modification; oxidation (M) and phosphorylation (STY) were applied as variable modifications; mass tolerance of 10 ppm (precursor) and 0.02 Da (fragments). The database search results were filtered using the ion score to set the false discovery rate (FDR) to 1% on the peptide and protein level, respectively, based on the number of reverse protein sequence hits in the datasets. Exported peptide intensities were normalized based on the protein regulations observed in the corresponding TMT experiments in order to account for changes in protein abundance. Only peptides corresponding to proteins which were regulated significantly with a P value ≤5% in the TMT analysis were normalized. Quantitative analysis results from label‐free quantification were processed using the SafeQuant R package v.2.3.2 (Ahrne et al. 2016; https://github.com/eahrne/SafeQuant/) to obtain peptide relative abundances. This analysis included global data normalization by equalizing the total peak/reporter areas across all LC‐MS runs, data imputation using the knn algorithm, summation of peak areas per peptide and LC‐MS/MS run, followed by calculation of peptide abundance ratios. Only isoform specific peptide ion signals were considered for quantification. The summarized peptide expression values were used for statistical testing of between‐condition differentially abundant peptides. Here, empirical Bayes‐moderated t‐tests were applied, as implemented in the R/Bioconductor limma package (http://bioconductor.org/packages/release/bioc/html/limma.html).
TMT labelling and LC‐MS/MS analysis
Tryptic peptides were labelled with isobaric tandem mass tags (TMT10plex or TMTpro 16plex, Thermo Fisher Scientific). Peptides were resuspended in labelling buffer (2 m urea, 0.2 m Hepes, pH 8.3) by sonication and TMT reagents were added to the individual peptide samples followed by a 1 h incubation at 25°C shaking at 500 rpm. To quench the labelling reaction, aqueous 1.5 m hydroxylamine solution was added and samples were incubated for another 5 min at 25°C shaking at 500 rpm followed by pooling of all samples. The pH of the sample pool was increased to 11.9 by adding 1 m phosphate buffer (pH 12) with incubation for 20 min at 25°C shaking at 500 rpm to remove TMT labels linked to peptide hydroxyl groups. Subsequently, the reaction was stopped by adding 2 m hydrochloric acid until a pH <2 was reached. Finally, peptide samples were further acidified using 5% TFA, desalted using Sep‐Pak Vac 1cc (50 mg) C18 cartridges (Waters, Milford, MA, USA) according to the manufacturer's instructions and dried under vacuum. For TMTpro 16plex analysis, four peptide samples were prepared from C2C12 cells, TMT labelled and included in the analysis to boost protein coverage.
TMT‐labelled peptides were fractionated by high‐pH reversed phase separation using a XBridge Peptide BEH C18 column (3.5 μm, 130 Å, 1 mm × 150 mm, Waters) on an Agilent 1260 Infinity HPLC system. Peptides were loaded on column in buffer A (20 mM ammonium formate in water, pH 10) and eluted using a two‐step linear gradient from 2% to 10% in 5 min and then to 50% buffer B (20 mM ammonium formate in 90% acetonitrile, pH 10) over 55 min at a flow rate of 42 μl min−1. Elution of peptides was monitored with a UV detector (215 nm, 254 nm) and a total of 36 fractions were collected, pooled into 12 fractions using a post‐concatenation strategy as previously described (Wang et al. 2011) and dried under vacuum.
Dried peptides were resuspended in 0.1% aqueous formic acid and subjected to LC‐MS/MS analysis using a Q Exactive HF Mass Spectrometer or an Orbitrap Fusion Lumos Mass Spectrometer fitted with an EASY‐nLC 1000 or an EASY‐nLC 1200, respectively (both Thermo Fisher Scientific) and a custom‐made column heater set to 60°C. Peptides were resolved using a RP‐HPLC column (75 μm × 30 cm or 75 μm × 36 cm, respectively) packed in‐house with C18 resin (ReproSil‐Pur C18–AQ, 1.9 μm resin; Dr Maisch GmbH) at a flow rate of 0.2 μl min−1. The following gradient was used for peptide separation: Q Exactive HF from 5% B to 15% B over 10 min to 30% B over 60 min to 45% B over 20 min to 95% B over 2 min followed by 18 min at 95% B, Orbitrap Fusion Lumos from 5% B to 15% B over 9 min to 30% B over 90 min to 45% B over 21 min to 95% B over 2 min followed by 18 min at 95% B. Buffer A was 0.1% formic acid in water and buffer B was 80% acetonitrile, 0.1% formic acid in water.
The Q Exactive HF mass spectrometer was operated in DDA mode with a total cycle time of approximately 1 s. Each MS1 scan was followed by HCD of the 10 most abundant precursor ions with dynamic exclusion set to 30 s. For MS1, 3e6 ions were accumulated in the Orbitrap over a maximum time of 100 ms and scanned at a resolution of 120,000 FWHM (at 200 m/z). MS2 scans were acquired at a target setting of 1e5 ions, maximum accumulation time of 100 ms and a resolution of 30 000 FWHM (at 200 m/z). Singly charged ions and ions with unassigned charge state were excluded from triggering MS2 events. The normalized collision energy was set to 30%, the mass isolation window was set to 1.1 m/z and one microscan was acquired for each spectrum. The Orbitrap Fusion Lumos mass spectrometer was operated in DDA mode with a cycle time of 3 s between master scans. Each master scan was acquired in the Orbitrap at a resolution of 120,000 FWHM (at 200 m/z) and a scan range from 375 to 1600 m/z followed by MS2 scans of the most intense precursors in the Orbitrap at a resolution of 30,000 FWHM (at 200 m/z) with isolation width of the quadrupole set to 1.1 m/z. Maximum ion injection time was set to 50 ms (MS1) and 54 ms (MS2) with an AGC target set to 1e6 and 1e5, respectively. MIPS was set to Peptide, and the intensity threshold was set to 5e4. Peptides were fragmented by HCD with collision energy set to 38%, and one microscan was acquired for each spectrum. The dynamic exclusion duration was set to 45 s.
The acquired raw‐files were analysed using SpectroMine software (Biognosis AG, Schlieren, Switzerland). Spectra were searched against a murine database consisting of 17,013 protein sequences (downloaded from Uniprot on 20,190,307) and 392 commonly observed contaminants. Standard Pulsar search settings for TMT10plex (‘TMT10plex Quantification’) and TMTpro 16plex (‘TMTpro Quantification’) were used and resulting identifications and corresponding quantitative values were exported on the PSM level using the ‘Export Report’ function. Acquired reporter ion intensities in the experiments were employed for automated quantification and statistical analysis using our in‐house‐developed SafeQuant R script v2.3.2 (Ahrne et al. 2016). This analysis included adjustment of reporter ion intensities, global data normalization by equalizing the total reporter ion intensity across all channels, summation of reporter ion intensities per protein and channel, calculation of protein abundance ratios and testing for differential abundance using empirical Bayes‐moderated t‐statistics.
Pathway enrichment analysis
Pathway enrichment analysis was performed using g:Profiler (Reimand et al. 2016). Note that our input lists contained all transcripts or (phospho‐)proteins of a given mouse group at a given time point meeting our statistical thresholds.
Statistics
The n‐number used per genotype for each experiment is indicated in the figure legend. Data are presented as means ± SD and statistically analysed with GraphPad Prism 8 (GraphPad Software Inc., La Jolla, CA, USA). A two‐tailed, unpaired Student's t‐test was performed to evaluate statistical difference between two groups. For multiple comparisons, data were analysed using one‐way and two‐way ANOVAs followed by Sidak's multiple comparisons test. Corresponding symbols to highlight statistical significance are as followed: * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001. A statistical report is available online with this manuscript. All raw data are available upon demand.
Results
Time‐of‐day‐dependent variations in mouse treadmill exercise performance
Differences in low‐ and moderate‐intensity treadmill exercise performance were previously reported within the active phase of wild‐type mice (Ezagouri et al. 2019). It is, however, unclear whether mice show broader variations in maximal exercise performance between the light (i.e. resting/inactive) and the dark (i.e. feeding/active) periods. To investigate how the time of day affects exercise capacity, we challenged different groups of untrained C57BL/6J mice to an acute bout of high‐intensity exercise every 4 h across the whole 24‐h LD cycle and sacrificed them immediately (Ex + 0 h) or 3 h (Ex + 3 h) after exhaustion was reached (experimental design and protocol, Fig. 1A ). We found a significant variation of maximal treadmill running capacity, with a peak and trough of performance at zeitgeber time 0 (ZT0, light‐on; hereafter, early daytime) and ZT12 (light‐off; hereafter, early night‐time), respectively (Fig. 1B and 2A and B ). Importantly, all mice reached exhaustion and displayed an elevation of blood lactate, yet without significant relation to the time of day (Figs 1C and 2C ). Conversely, we observed that blood glucose levels significantly dropped at ZT12 and ZT16 upon exercise (Figs 1D and 2D ). Lastly, higher serum corticosterone levels were observed in all exercised groups regardless of time (Fig. 2E ).
Figure 2. Time‐of‐day‐dependent fluctuations in exercise performance and exercise‐induced corticosterone release.
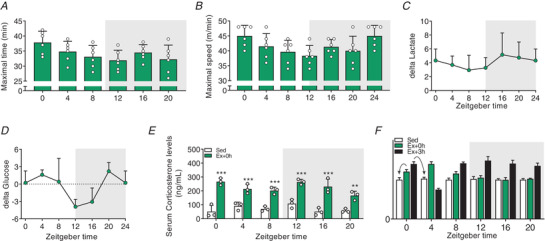
A, maximal time reached at exhaustion. B, maximal speed reached at exhaustion. C and D, delta blood lactate (C) and delta glucose levels (D) (resting values subtracted from values at exhaustion). E, serum corticosterone levels in Sed and at exhaustion. Light and dark periods are depicted by white and grey background, respectively. Data are shown as the mean ± SD (n = 3). ** P < 0.01, *** P < 0.001, one‐way ANOVA (D). F, mock dataset to illustrate the analysis of the gene expression data depicted in Figs 3 and 4. Fold‐change (FC) values are plotted according to time at which exercise was started. FC estimation was performed according to the time of sacrifice (i.e. as indicated by the arrows: gene expression data of mice exercised from ZT0 and sacrificed directly at exhaustion are expressed relative to Sed ZT0 plotted at ZT0; and the gene expression of mice exercised at ZT0 and sacrificed 3 h after exhaustion are expressed relative to Sed ZT4, but plotted at ZT0). [Colour figure can be viewed at wileyonlinelibrary.com]
To further evaluate the metabolic outcome of maximal treadmill exercise across the day, we measured circulating energy substrates, muscle and liver glycogen levels immediately after exercise (Ex + 0 h). We observed that plasma triglyceride (TG) levels were mainly unchanged at exhaustion (Fig. 1E ), which is consistent with circulating TG not being the primary source of energy during moderate to high‐intensity exercise in untrained animals (Hargreaves & Spriet, 2018). Conversely, plasma free fatty acids (FFA) and glycerol levels, a lipolytic marker, were affected by treadmill exercise in a time‐dependent manner (Fig. 1F and G ). Lastly, muscle glycogen stores were consistently reduced by exercise, except for ZT0, the time at which mice show their greatest performance (Fig. 1H ). In contrast, hepatic glycogen stores, an essential source of glucose during exercise (Richter & Hargreaves, 2013), were significantly impacted by exercise across the LD cycle except for ZT4 (Fig. 1I ).
Altogether, these data demonstrate that mice are better at performing a maximal running test in the early daytime. Furthermore, we show that when basal hepatic glycogen stores are low (i.e. in the early dark phase), mice are unable to sustain a prolonged workout and to maintain homeostatic blood glucose levels.
Exercise around the clock induces broad and time‐dependent gene responses in skeletal muscle
Using semiquantitative polymerase chain reaction (qPCR), we next evaluated the expression of genes that are part of the immediate, early response of skeletal muscle to exercise across the day (see Methods and Fig. 2F for details of fold‐change determination and statistical analysis; for each transcript, normalized data are provided as bar graphs, while raw data are provided as daily line graphs). We found that nuclear receptor subfamily 4 group A member 3 (Nr4a3) and activating transcription factor 3 (Atf3), both exercise‐responsive genes (Fernandez‐Verdejo et al. 2017; Pillon et al. 2020), were induced immediately after exercise and remained elevated 3 h post‐exercise (Figs 3A and 4A ). Conversely, while the expression of the peroxisome proliferator‐activated receptor γ coactivator 1α 4 (Ppargc‐1α4) isoform induced by resistance exercise (Ruas et al. 2012) was up‐regulated 3 h post‐exercise (Fig. 4A ), the Ppargc‐1α1 isoform particularly involved in oxidative muscle remodelling (Lin et al. 2002) was mainly induced directly after exercise (Fig. 4A ). Moreover, interleukin‐6 (Il‐6), a myokine implicated in muscle glycolysis and adipose fat lipolysis (Pedersen & Febbraio, 2008), was increased immediately post‐exercise, especially in the late part of the day and at night (i.e. from ZT8 to 20) (Fig. 3A ). Finally, vascular endothelial growth factor A (Vegfa), an important regulator of muscle regeneration, exercise adaptation and angiogenesis (Delavar et al. 2014), was only induced when exercise was performed during the daytime (Fig. 3A ).
Figure 3. Exercise around the clock induces broad and time‐dependent gene responses in skeletal muscle.
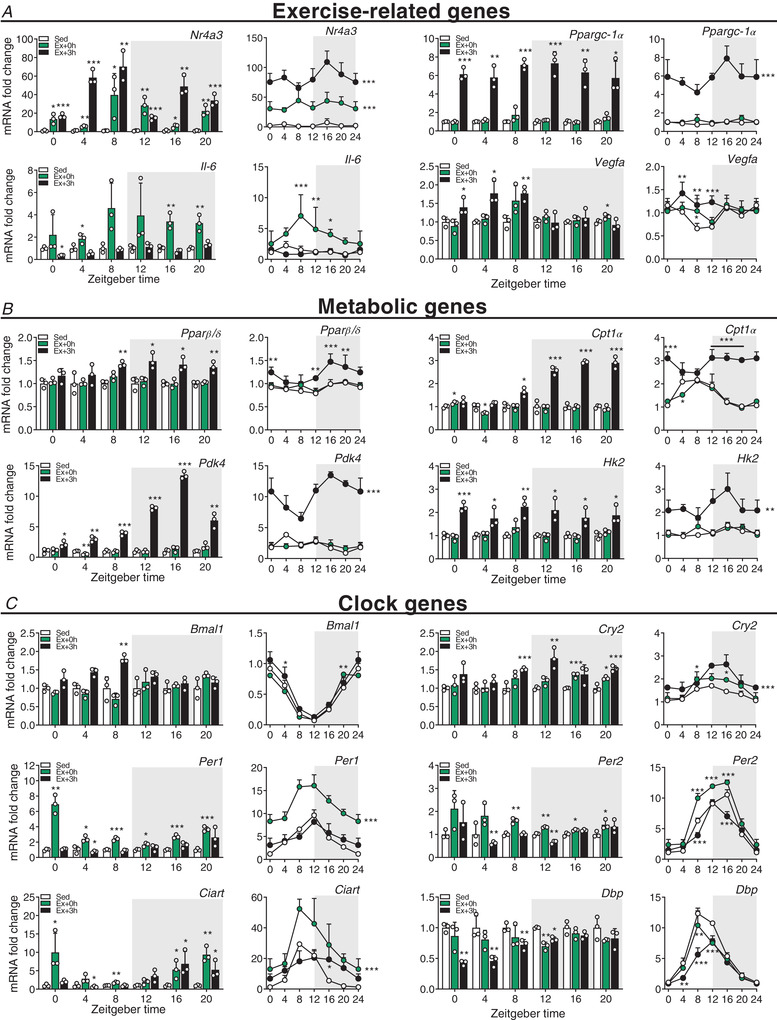
Gene expression in sedentary (Sed) and exercised mice at (Ex + 0 h) and 3 h (Ex + 3 h) after exhaustion. A, exercise‐related genes. B, metabolic genes. C, clock genes. Expression values were determined by qPCR and normalized to Hprt. Light and dark periods are depicted by white and grey background, respectively. Data in bar graphs are shown as the mean fold‐change ± SD (n = 3) relative to the expression in Sed set to 1 (see Methods for details on normalization). Data in line graph are shown as the mean fold‐change ± SD (n = 3) relative to the expression in the Sed ZT0 group set to 1. * P < 0.05, ** P < 0.01, *** P < 0.001, unpaired Student's t‐test (bar graphs) and one‐way ANOVA (line graphs). Group significance in line graphs is indicated on the right side of the group line. [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 4. Scheduled treadmill induces broad and time‐dependent transcriptional responses in skeletal muscle.
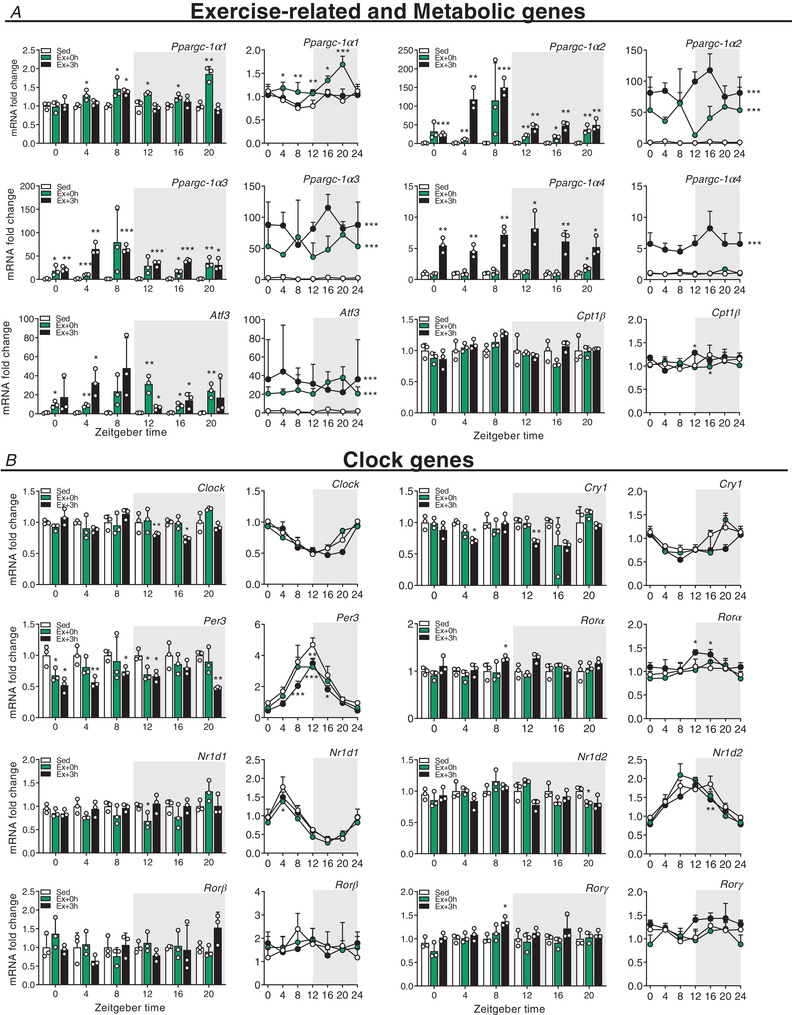
Gene expression in sedentary mice (Sed) and exercised mice at (Ex + 0 h) and 3 h (Ex + 3 h) after exhaustion. A, exercise‐related genes. B, metabolic genes. C, clock genes. Expression values were determined by qPCR and normalized to Hprt. Light and dark periods are depicted by white and grey background, respectively. Data in bar graphs are shown as the mean fold‐change ± SD (n = 3) relative to the expression in Sed set to 1 (see Methods for details on normalization). Data in line graph are shown as the mean fold‐change ± SD (n = 3) relative to the expression in the Sed ZT0 group set to 1. * P < 0.05, ** P < 0.01, *** P < 0.001, unpaired Student's t‐test (bar graphs) and one‐way ANOVA (line graphs). Group significance in line graphs is indicated on the right side of the group line. [Colour figure can be viewed at wileyonlinelibrary.com]
We likewise measured genes involved in the regulation of lipid and glucose metabolism: some were induced at the day–night transition (e.g. peroxisome proliferator‐activated receptor β/δ (Pparβ/δ); Fig. 3B ), induced at night (e.g. the liver‐specific isoform carnitine palmitoyltransferase‐1α (Cpt1α) and pyruvate dehydrogenase kinase 4 (Pdk4); Fig. 3B ), broadly induced (e.g. hexokinase 2 (Hk2); Fig. 3B ), or unchanged by exercise (e.g. the muscle‐specific isoform (Cpt1β); Fig. 4A ).
Finally, we assessed the expression of core circadian clock genes. We observed changes in the expression of brain and muscle arnt‐like (Bmal)1, circadian locomotor output cycles kaput (Clock), cryptochrome (Cry)1, Cry2, period (Per)1, Per2, Per3, circadian associated repressor of transcription (Ciart), retinoic acid receptor‐related receptor α (Rorα), Rorγ, nuclear receptor subfamily 1 group D member 1 (Nr1d1, Rev‐Erbα), Nr1d2 (Rev‐Erbβ) and albumin D box‐binding protein (Dbp), while only Rorβ transcript levels remained unaffected (Figs 3C and 4B ). The transcription of most of these clock genes was positively affected by exercise, with some exhibiting a bimodal regulation, e.g. a repression following an induction for Per1, Per2 or Ciart at specific ZTs (Fig. 3C ). Overall, these data demonstrate that the regulation of prototypical exercise response and metabolic genes is only in part influenced by the time of day, which, for example, seems completely irrelevant for Ppargc‐1α. Moreover, acute endurance exercise bouts extensively affect the expression of core clock genes in the immediate response after fatigue is reached.
Daytime vs. night‐time treadmill exercise elicits distinct gene signatures in skeletal muscle
In light of our qPCR results, we further explored the transcriptional signatures of working muscles at distinct phases of the LD cycle. We performed RNA‐sequencing (RNA‐seq) gene expression profiling of muscles harvested at times when treadmill exercise performance differed the most, namely ZT0 (i.e. light onset) and ZT12 (i.e. light offset).
The overall extent of the transcriptional response shifted from the immediate time point (Ex + 0) at the early daytime exercise to the late time point (Ex + 3) at the early night‐time exercise (Figs 5A and B and 6). Surprisingly, only about 25% of the differentially expressed genes (DEGs; FDR ≤ 0.05) were shared between ZT0 and ZT12, regardless of the time of sacrifice (i.e. Ex + 0 h vs. Ex + 3 h). Examples representing this ‘core program’ are the MAF transcription factors Maff and Mafk, implicated in cellular stress response and detoxification (Katsuoka et al. 2005), and metallothionein 1/2 (Mt1/2), involved in oxidative stress protection and regulation of hypertrophy via the Akt pathway (Di Foggia et al. 2014) (Fig. 5C and D , and Supplementary Table 1). The expression of most genes, however, differs qualitatively or at least temporally between the two ZTs. For instance, in line with the qPCR data, Per1 and Ciart transcripts were significantly induced immediately after exercise when performed at ZT0, but not at ZT12 (Fig. 5C ).
Figure 5. Daytime vs. night‐time treadmill exercise elicit distinct transcriptomic signatures.
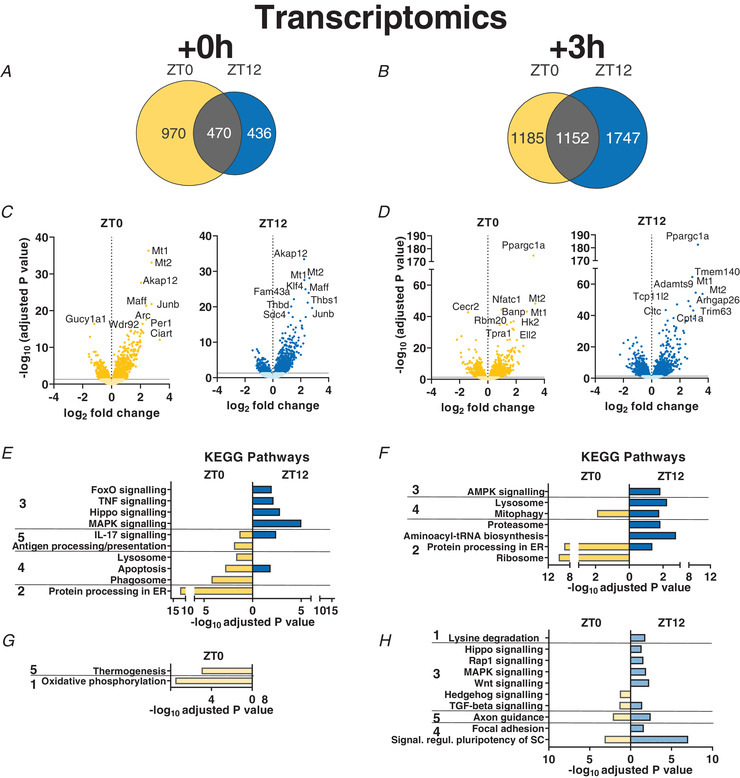
A and B, Venn diagrams displaying the number of DEGs immediately (A; Ex + 0 h) and 3 h (B; Ex + 3 h) after early daytime (ZT0) or early night‐time (ZT12) exercise, and resulting overlap. C and D, volcano plots displaying DEGs (with top 10 indicated) as described above. E–H, KEGG analysis (top 5 terms per category) of up‐regulated (E and F) and down‐regulated (G and H) genes by exercise as above. KEGG pathway categories: 1, metabolism; 2, genetic information processing; 3, environmental information processing; 4, cellular processes; 5, organismal systems. [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 6. Distinct muscle gene expression signatures after early daytime vs. early night‐time treadmill exercise.
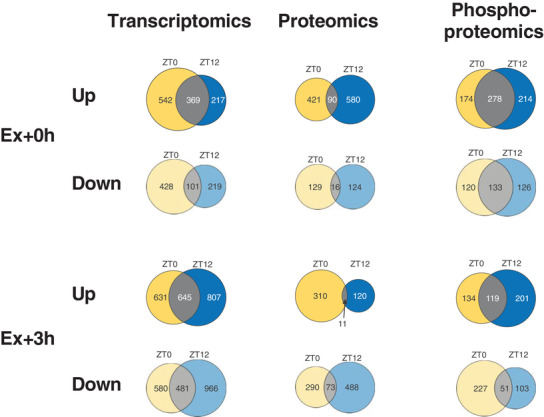
Venn diagrams displaying the number of differentially regulated genes, proteins and phosphosites immediately (A; Ex + 0 h) or 3 h (B; Ex + 3 h) after early daytime (ZT0), early night‐time (ZT12) exercise, and the overlap. [Colour figure can be viewed at wileyonlinelibrary.com]
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis using g:Profiler (Reimand et al. 2016) revealed that exercise at both ZTs activated mitophagy, inflammation and apoptotic pathways (Fig. 5E and F , and Supplementary Table 1). Conversely, exercise at early daytime is linked with a robust immediate increase in genes linked to protein processing, lysosome and phagosome, associated with a down‐regulation of oxidative phosphorylation (Fig. 5G ), followed by a further increase in protein processing and ribosome‐related genes 3 h later. In contrast, early night‐time exercise triggers a broad immediate induction in genes related to mitogen‐activated protein kinase (MAPK) and other stress, muscle wasting pathways (i.e. Hippo and FoxO), with an ensuing activation of catabolic signals linked to AMP‐activated protein kinase (AMPK), lysosome and proteasome 3 h later. At this time point, some of the genes related to the stress signalling pathways that were activated earlier are mostly down‐regulated (Fig. 5H ).
Taken together, the time of day markedly affects the transcriptional response to an acute bout of endurance exercise. Moreover, broadly speaking, daytime exercise positively regulates transcriptional processes associated with protein synthesis and stability, while night‐time exercise triggers responses associated with energy stress.
Proteome and secretome changes associated with daytime vs. night‐time treadmill exercise
To investigate whether the exercise‐induced transcriptional changes were accompanied by significant modifications at the protein level, we performed mass spectrometric analyses of muscles from mouse groups exercised at ZT0 and ZT12, and identified a total of about 5300 proteins using SpectroMine, specifying a false discovery rate of 1% at the peptide and protein level.
Similar to the transcriptomic data, only a relatively small overlap between the two ZTs regarding the levels of differentially affected proteins was observed at either time point (Figs 6A and 7A–D ). The control of specific programs was underlined by the small overlap in KEGG terms between ZT0 and ZT12 (Fig. 7E–H ), indicating differential regulation of protein expression and pathway activation between time points. For example, the up‐regulation of soluble N‐ethylmaleimide‐sensitive factor attachment protein receptors (SNARE) interactions in vesicular transport at exhaustion following ZT0 exercise indicates the increase in a class of membrane‐associated proteins, which, besides their involvement in neurotransmitter release (Dunant & Israel, 2000; Kasai et al. 2012), regulate GLUT4‐containing vesicle trafficking (Cheatham, 2000). In line with this, vesicle‐associated membrane protein 3 and 8 (VAMP3, 8), syntaxin 6 and 8 (STX6, 8), synaptobrevin homolog YKT6 (YKT6) and synaptosomal‐associated protein 23 (SNAP23), which are all key mediators of GLUT4 translocation to the cell surface, were elevated at this time point (Bryant & Gould, 2011; Zong et al. 2011; Morris et al. 2020) (Supplementary Table 2). Moreover, the Rho family GTPase RAC1, an essential regulator of glucose transport in contracting muscles (Sylow et al. 2017), its upstream regulator protein tyrosine phosphatase α (PTPα; PTPRA) (Sun et al. 2012), together with the GTPase Ras‐related protein (RALA), a known mediator of insulin‐dependent glucose uptake (Takenaka et al. 2015), were all consistently elevated (Supplementary Table 2). Finally, we observed increased expression of the Rho GTPases RHOA, B, and C, in favour of enhanced GLUT4 intracellular trafficking (Duong & Chun, 2019) (Supplementary Table 2). Collectively, these changes imply an activation of glucose uptake into skeletal muscle that seems more specific to the adaptation in early daytime exercise. Moreover, early daytime treadmill exercise robustly initiated the ‘complement system’ (Fig. 7E ). Complement activation triggers tissue regeneration in response to muscle injury and inflammation (Zhang et al. 2017). Accordingly, complement C3a, C4b, C8a, C8b, C9, CFD, and CFB proteins, were all up‐regulated (Supplementary Table 2). In contrast, the levels of proteins linked to ribosome function were only elevated in early night‐time exercise at 0 h, followed by a subsequent decrease at 3 h, associated with a strong induction in proteins involved in nicotinamide and nitrogen metabolism (Fig. 7F and H ). Hence, proteostasis could be more affected at this time point compared to early daytime running.
Skeletal muscle tissue is an endocrine organ, releasing small molecules, so‐called myokines, in response to exercise (Delezie & Handschin, 2018). We thus used predictive tools to identify putatively secreted proteins that could influence muscle performance and metabolism at distinct phases of the LD cycle. Of the top 100 proteins exclusively induced in the immediate response to daytime exercise (Ex + 0 h), we retrieved 25 candidates as potential secreted proteins using SignalP 5.0, and 12 additional candidates using SecretomeP 2.0 (Bendtsen et al. 2004; Nielsen, 2017) (Supplementary Table 3). For instance, the major urinary proteins (MUP) 3, 17 and 18 were all up‐regulated at ZT0 and could potentially be associated with the regulation of systemic blood glucose, as well as liver and skeletal muscle metabolism (Zhou et al. 2009). We, moreover, identified fibrillin‐1 (FBN1, also called asprosin), a glucogenic protein hormone that is secreted by adipose cells and recruited at the surface of hepatocytes to increase plasma glucose level (Romere et al. 2016). Night‐time exercise resulted in a smaller number of potentially secreted proteins using the same analysis tools (Supplementary Table 3). The seven predicted proteins included the SPARC‐like protein 1 (SPARCL1), a member of the SPARC family of proteins that also includes SPARC/osteonectin, an exercise‐regulated myokine with a potential effect on myogenic differentiation (Lee & Jun, 2019).
Overall, these proteomic results highlight the robust activation of time‐dependent cellular responses; enhancing glucose metabolism in the early daytime (supported by the predicted secretome) and, conversely, alterations in protein homeostasis when exercise is performed in the early night.
The phosphorylome of daytime vs. night‐time working muscles
Many proteins are regulated by phosphorylation independently of their expression (Huttlin et al. 2010), in particular upon exercise (Hoffman et al. 2015). To identify signalling pathways that are modulated by exercise at different times of day, we performed a comprehensive phosphoproteomics analysis, in which approximately 7000 sequence ions carrying post‐translational modifications in response to exercise mapping to about 1600 unique proteins were identified (Figs 6 and 7I–L ).
A larger enrichment of concurrent phosphorylation and dephosphorylation of proteins in stress‐related and catabolic pathways was detected in night‐time exercise, both at Ex + 0 h and Ex + 3 h (Fig. 7M–P ). While the α2‐subunit of AMPK, the predominant form in the muscle (Garcia & Shaw, 2017), was consistently dephosphorylated on Ser377 by exercise regardless of ZT, only exercise at night induced further changes in the phosphorylation status of both α1 and α2 AMPK subunits. Night‐time exercise rapidly phosphorylated α1/α2 at Ser496/491, and we found increased phosphorylation of the critical regulator of autophagy ULK1 on Ser637, which is dependent on AMPK (Mack et al. 2012). The mechanistic target of rapamycin (mTOR) protein was similarly phosphorylated on Ser1162/1261 upon exercise at both ZTs, yet both TSC1 and TSC2 proteins, important integrators of different signalling pathways to control mTOR signalling, were predominantly regulated by night‐time exercise (Supplementary Table 4). Emblematic of the activation of the ‘mTOR pathway’, there were robust post‐translational changes of, for example, lipin1 (LPIN1), eukaryotic translation initiation factor 4 (EIF4) B, EIF4E binding protein and ribosomal protein S6 (RPS6). Similarly, key components of the ‘HIF‐1 signalling’ pathway, e.g. the glycolytic enzymes enolase 1 (ENO1), phosphofructokinase (PFK) L‐M, and aldolase A (ALDOA), displayed unique and/or distinct phosphorylation profiles at night (Fig. 7M , and Supplementary Table 4). In line with the decrease in muscle glycogen content upon exercise at ZT12, the muscle‐specific isoform of glycogen phosphorylase (PYGM), a key enzyme in the first step of glycogenolysis, was specifically phosphorylated on Ser2 and on Ser15; the latter residue is particularly known to enhance phosphorylase activity and the degradation of glycogen (Johnson, 1992). In contrast, these two sites were not phosphorylated in response to early daytime exercise, consistent with muscle glycogen sparing (Fig. 1H ). Moreover, daytime exercise exclusively enhanced phosphorylation on Ser3/616 residues of the TBC1 domain family member 4 (TBC1D4 or AS160), a strong regulator of GLUT4 trafficking in skeletal muscle (Sakamoto & Holman, 2008; Cartee, 2015). Finally, besides the increased abundance of asprosin in daytime contracting muscles, we observed active post‐translational modifications on Ser2566 and Ser2711 (Supplementary Table 4), which could warrant secretory processing to boost hepatic glucose production. We furthermore detected post‐translational modifications of hormone receptors that play a key role in energy metabolism, e.g. the phosphorylation of the glucocorticoid receptor (Nr3c1) on T152 and S275 residues only at night‐time (Supplementary Table 4), even though plasma corticosterone levels were similarly elevated by exercise at ZT0 and ZT12 (Fig. 2E ).
Broad changes in the phosphorylation of proteins associated with modulation of muscle contractile properties (i.e. titin; myosin heavy chain 3, 4 and 9; sarcoplasmic reticulum calcium‐ATPase 1 and 2) were similarly observed independent of the time of day (Fig. 7K and L ). However, the ‘vascular smooth muscle contraction’ KEGG pathways were exclusively enriched upon daytime exercise, particularly at exhaustion (Ex + 0 h), indicating a higher engagement of calcium and cAMP signalling pathways. In line, phosphorylation of the calcium voltage‐gated channel subunit α 1 S (CACNA1S) protein, a regulator of contractile force in response to stress, fear and exercise, the so‐called ‘fight‐or‐flight’ response (Emrick et al. 2010; Catterall, 2015), was specific to residue Ser5 and Ser1617/1640 and T700 (Supplementary Table 4). These sites are not yet characterized but could indicate an up‐regulation of calcium channel activity, leading to increased force production. In support of this, expression of the calcium release‐activated calcium channel protein 1 (ORAI1), a crucial calcium regulator limiting muscle fatigue (Wei‐Lapierre et al. 2013), was exclusively up‐regulated in daytime exercised muscles (Supplementary Table 2).
Taken together, the phosphoproteomics data mirror the transcriptomics and proteomics inasmuch as clear distinctions between different time points were observed, mainly in regard to stress and catabolism. It thus is clear that besides a robust core exercise programme, the time of day has a major impact on gene expression, protein levels and post‐translational modifications triggered by acute physical activity.
Mice with restricted wheel access spontaneously run during daytime under a skeleton photoperiod
While the studies have so far pertained to the consequences of the time of day on the acute exercise response, we next assessed how training at different times of day affects the clock. Due to the masking (i.e. suppressive) effect of light on physical activity and other nocturnal behaviour of mice, voluntary use of the running wheel under normal conditions exclusively spreads throughout the dark period, with a peak in the early hours of the night, with intermittent multiple eating events (Yasumoto et al. 2015). These and other limitations have precluded a comprehensive analysis of prolonged training as a potential zeitgeber under physiological conditions so far. We therefore tested the use of a skeleton photoperiod (SPP) in combination with time‐restricted wheel and food access to evaluate whether mice would spontaneously run in a wheel during their normal resting phase (see Methods and Fig. 8A ). SPPs maintain central clock oscillation (Oishi et al. 2002), reducing the strong inhibition of light on physical activity in mice (Delezie et al. 2016).
Figure 8. Scheduled daytime wheel‐running activity in mice exposed to a skeleton photoperiod.
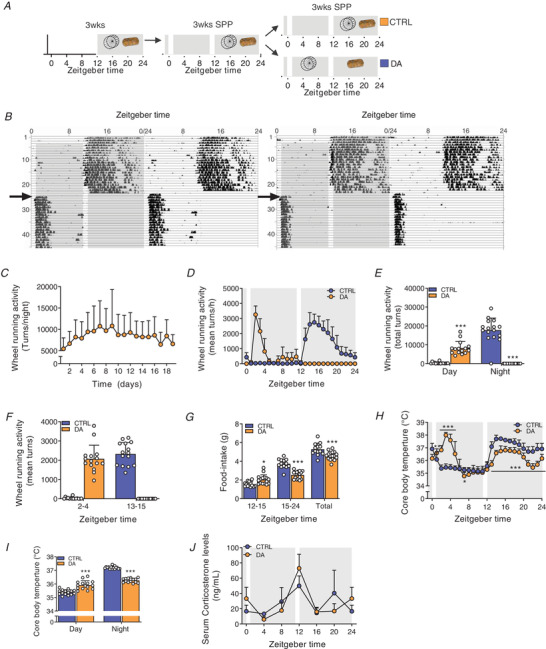
A, experimental scheme. Mice had free access to a wheel for 3 weeks under 12:12 L:D before transfer to a skeleton photoperiod (SPP). The SPP consists of 1 h light pulse from ZT0 to ZT1 and from ZT11 to ZT12. After 3 weeks under SPP conditions, mice were separated into two groups: one with free access to wheel and food, the other with exclusive access to wheel during day and food during night. B, representative double‐plotted actograms of DA mice. The grey background indicates light off, and the arrow indicates the time shift in wheel access. C, daily wheel‐running activity from the first day of restriction of the DA mice. D and E, 24‐h activity levels of CTRL and DA mice (D), and resulting average of day vs. night locomotor activity (E). F, quantified wheel‐running activity during the first 2 h of wheel excess. G, food intake during the active, feeding period and average over the full food access period. H and I, 24‐h core body temperature levels of CTRL and DA mice (H), and resulting day and night values (I). J, serum corticosterone levels. Light and dark periods are depicted by white and grey background, respectively. Data are shown as the mean ± SD (n = 24). * P < 0.05, ** P < 0.01, *** P < 0.001, unpaired Student's t‐test (E, G, I) and one‐way ANOVA (H). [Colour figure can be viewed at wileyonlinelibrary.com]
Under SPP conditions, DA mice used the wheel from the first day of daytime‐restricted access and maintained a stable onset and level of activity during the following days (Fig. 8B and C ). DA mice almost exclusively ran within the first 3 h of daytime wheel access (Fig. 8D ). Even though total activity only represented 50% of the control group (Fig. 8E ), running intensity was comparable between the two groups during the first 2 h of wheel access (Fig. 8D and F ). Daytime‐restricted wheel access promoted food intake in the first few hours of the night‐time period but significantly decreased the overall amount of food consumed (Fig. 8G ). As expected, there was an elevation of daytime core body temperature paralleling the increase in wheel‐running activity of DA mice (Fig. 8H and I ). The temporal organization of night‐time core body temperature values in DA mice, however, closely resembled those of CTRL, night‐time active mice, likely caused by comparable feeding bout activities (Fig. 8G ). To evaluate whether the animal handling or the running at an abnormal time point caused a severe physiological stress, we measured serum corticosterone levels, but did not observe significant changes (Fig. 8J ). This also demonstrates that daytime training does not affect the daily rhythm of corticosterone and, moreover, that voluntary wheel running is not associated with a stress response in mice, in stark contrast to treadmill exercise (Fig. 2E ). This stress affects the clock and thereby confounds the analysis of the zeitgeber activity of exercise (Tahara et al. 2017).
Daytime wheel running profoundly affects skeletal muscle gene expression but not the core clock
Having established a robust system of voluntary, stress‐free daytime activity uncoupled from feeding behaviour, we evaluated the impact of day‐ and night‐time activity on gene expression. To do so, we collected muscle tissues after 3 weeks of daytime wheel access, when running performance already plateaued in particular in DA mice (Fig. 8C ), without depriving mice of wheel access. First, we studied the impact of training on the daily expression of clock genes, which would reveal potential zeitgeber activity. We have shown that acute bouts of treadmill exercise cause the rapid expression of some of the core clock transcriptional (co‐)regulators (Figs 3C and 4B ). Moreover, night‐time wheel running exercise modulates the daily amplitude of core clock gene expression (Yasumoto et al. 2015). Prolonged daytime wheel running resulted in a marked effect on the amplitude, but much less on the phase, of most core clock genes. For example, the relative amplitude (peak‐trough amplitude divided by mean) of Bmal1, Clock, Cry1‐2, Per1‐3, RORα and RORγ was dampened in muscles of the DA group (Figs 9A and 10A , and Supplementary Table 5).
Figure 9. Daytime wheel running profoundly affects skeletal muscle gene expression but not the core clockwork.
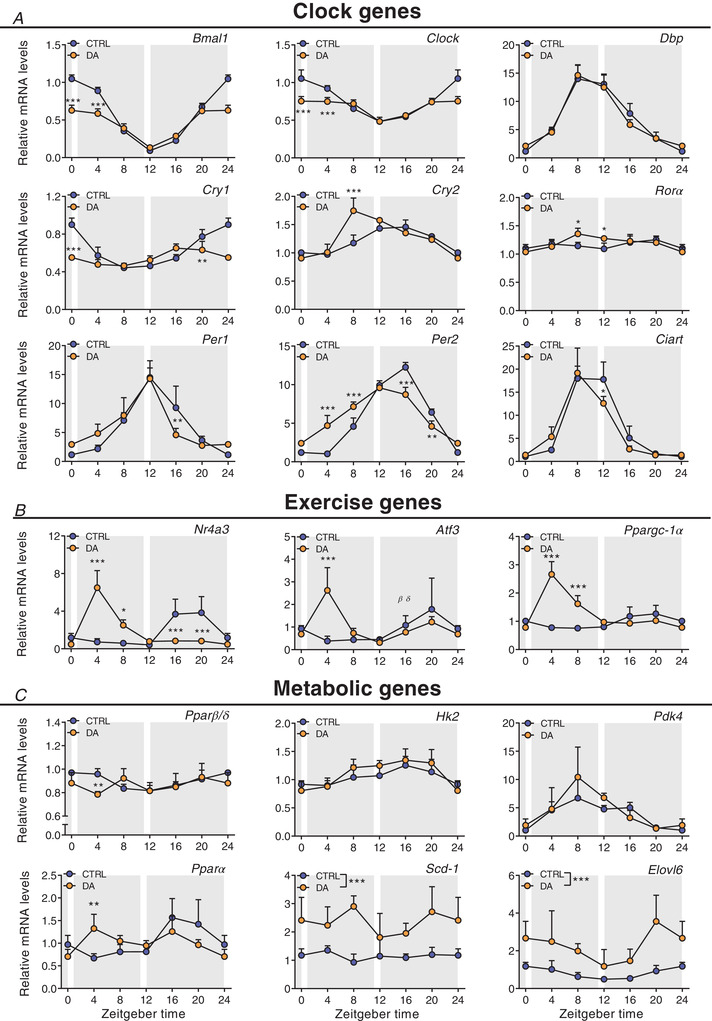
Gene expression in control (CTRL) and daytime activity (DA) mice. A, clock genes. B, exercise‐related genes. C, metabolic genes. Expression values were determined by qPCR and normalized to Hprt. Light and dark periods are depicted by white and grey background, respectively. Data are shown as the mean fold‐change ± SD (n = 4) relative to the expression in CTRL ZT0 set to 1. * P < 0.05, ** P < 0.01, *** P < 0.001, one‐way ANOVA. [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 10. Distinct muscle gene expression signatures after daytime vs. night‐time wheel running training.
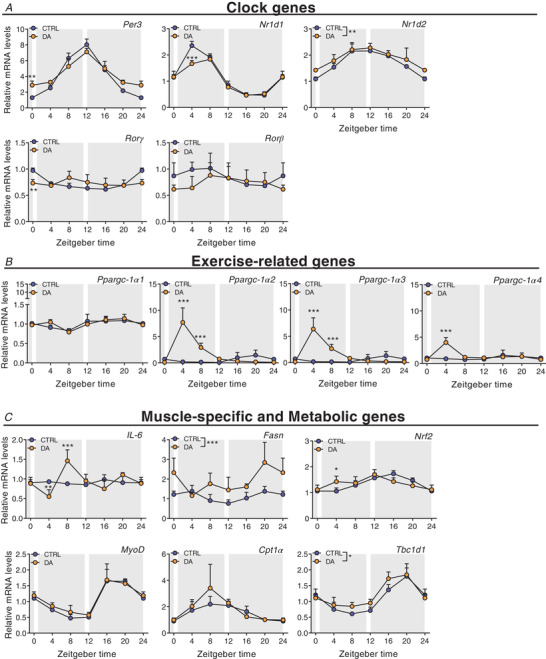
Gene expression in control (CTRL) and daytime activity (DA) mice. A, clock genes. B, exercise‐related genes. C, muscle‐specific and metabolic genes. Expression values were determined by qPCR and normalized to Hprt. Light and dark periods are depicted by white and grey background, respectively. Data are shown as the mean fold‐change ± SD (n = 4) relative to the expression in CTRL ZT0 set to 1. * P < 0.05, ** P < 0.01, *** P < 0.001, one‐way ANOVA. [Colour figure can be viewed at wileyonlinelibrary.com]
To elucidate whether daytime training affects the clock of other peripheral tissues, we also measured clock genes in the liver, without observing robust changes (Fig. 11A ). Thus, to compare these effects to an established zeitgeber, we finally determined clock expression in liver and muscle of mice undergoing daytime feeding (DF). As established (Damiola et al. 2000), this zeitgeber paradigm resulted in a complete reversal of the liver clock (Fig. 11B ). Interestingly, however, DF exerted an effect on the skeletal muscle clock that resembles that of daytime activity, with a blunting of the amplitude of several genes, and minor phase shifts in others (Reznick et al. 2013) (Fig. 11C ). Thus, collectively, the skeletal muscle clock seems much more refractory to external perturbations compared to the liver clock, even though training resulted in clear differences in the regulation of selected exercise (e.g. Ppargc‐1α) and metabolic transcripts (e.g. Pparα) in muscles of DA mice (Figs 9B and C and 10B and C ).
Figure 11. Distinct liver clock gene expression signatures after daytime vs. night‐time wheel running training.
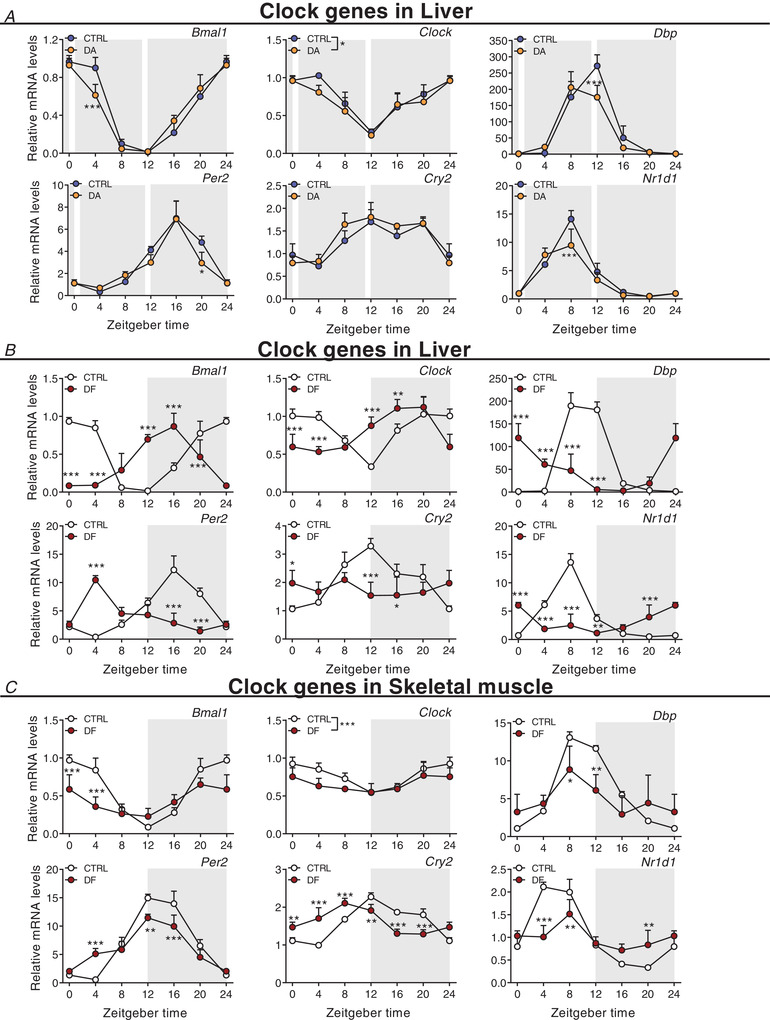
Gene expression in control (CTRL) and daytime activity (DA) mice. Clock genes in liver of DA mice (A), of daytime feeding (DF) mice (B), and in skeletal muscle of DF mice (C). Expression values were determined by qPCR and normalized to Hprt. Light and dark periods are depicted by white and grey background, respectively. Data are shown as the mean fold‐change ± SD (n = 4) relative to the expression in CTRL ZT0 set to 1. * P < 0.05, ** P < 0.01, *** P < 0.001, one‐way ANOVA. [Colour figure can be viewed at wileyonlinelibrary.com]
Characterization of the muscle transcriptome, proteome and phosphoproteome responses of daytime wheel active mice
We next dissected the global changes elicited by day‐ compared to night‐time activity on the transcriptome, proteome and phosphoproteome at ZT4 and ZT16, near the peak of wheel running activity in DA and CTRL mice, respectively. Daytime training led to the differential expression of 2094 genes at ZT4, with a similar proportion of down‐ and up‐regulated transcripts (Fig. 12A ). In comparison, training at night‐time was a weaker modifier of gene expression: we only found 368 transcripts for which the expression was different between CTRL, night‐time‐trained muscles and those obtained from daytime‐trained animals at ZT16 (Fig. 12A ). The overlap between the day‐ and night‐time activity groups describes known exercise‐regulated genes, including members of the KLF transcription factor gene family (i.e. Klf4, 5 and 15), the MAF transcription factors and Nr4a3. Incidentally, Atf3, Per2 and Ppargc‐1α were among the top induced genes only in daytime working muscles (Fig. 12B ). The nuclear receptor Pparα, a regulator of fatty acid oxidation (Muoio et al. 2002), was likewise specifically induced by daytime wheel running. Lastly, the KEGG pathways of DA mice reflected some of those that were activated by acute exercise bouts at early daytime, indicating a hardwiring of the pathways related to protein processing and mitophagy in chronic activity (Fig. 12C and D , Supplemental Table 6).
Figure 12. Characterization of the muscle transcriptome, proteome and phosphoproteome responses of daytime wheel active mice.
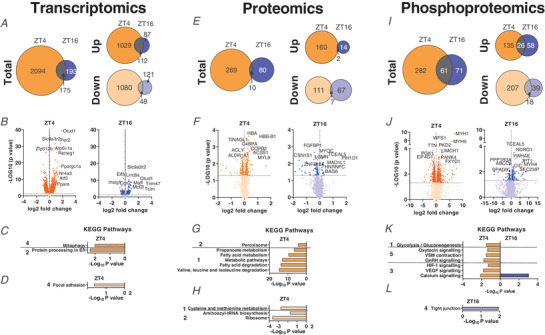
A, Venn diagrams with number of DEGs in DA mice at ZT4 and CTRL mice at ZT16, and resulting overlap. B, volcano plots displaying the DEGs (top 10 shown) as identified in the same groups as above. C and D, KEGG analysis (top 5) of up‐regulated (C) and down‐regulated (D) genes as above. E, Venn diagrams displaying the number of differentially regulated proteins for night‐time, and daytime wheel running and the overlap. F, volcano plot showing the differentially regulated proteins (top 10 only) by night‐time and daytime wheel running. G and H, KEGG analysis (top 5) for proteins induced (G) or decreased (H) by night‐time and daytime wheel running. I, Venn diagram displaying the number of differentially phosphorylated proteins as described above. J, volcano plot showing the differentially phosphorylated proteins (top 10 shown). K and L, KEGG analysis (top 5) for phosphorylated (K) and dephosphorylated (L) proteins. KEGG pathway categories: 1, metabolism; 2, genetic information processing; 3, environmental information processing; 4, cellular processes; 5, organismal systems. [Colour figure can be viewed at wileyonlinelibrary.com]
Up to 3800 proteins were detected in skeletal muscles of DA and CTRL mice, including 246 for which the expression was exclusively altered by daytime wheel running (Fig. 12E and F ). The majority of up‐regulated proteins were linked to metabolic functions (Fig. 12G and H Supplemental Table 7), including an important cluster regulating fatty acid metabolism (e.g. ATP‐citrate synthase (ACLY), mitochondrial trifunctional enzyme subunit (HADH)α, FASN and PDK4) and the citrate cycle (e.g. isocitrate dehydrogenase (IDH)2, succinate dehydrogenase (SDH)b) (Fig. 12F ), most of which are direct targets of PPARα (Boergesen et al. 2012; Gan et al. 2018). In line with the transcriptomic data, exclusive night‐time running‐induced proteins are scarcer, and not attributable to specific pathways by KEGG analysis (Supplemental Table 7). Moreover, in silico prediction of the secretome of both groups did not return notable myokine candidates (Supplementary Table 8). In regard to post‐translational modifications, we observed the differential phosphorylation of 344 and 135 proteins in response to daytime and night‐time training, respectively, with an overlap of 62 protein‐specific post‐translational modifications, some of which associated with ‘calcium signalling’ (Fig. 12I ). However, most of the proteins shared between ZT4 and ZT16 were differentially phosphorylated by wheel running exercise. For example, CACNA1s was phosphorylated on Ser700 in the early daytime and dephosphorylated on Ser1617 in the early night‐time. Similar results were found for RYR1 and STIM1, with the identification of previously uncharacterized sites (Supplementary Table 9). Notably, there was a strong daytime‐dependent post‐translational regulation of PYGM on Ser15 (Fig. 12J ; Supplementary Table 9), potentially promoting the degradation of glycogen (Johnson, 1992). Accordingly, ‘glycolysis’ was enriched in the DA group (Fig. 12K and L), and ALDOA, a key enzyme in the fourth step of glycolysis, was one of the top phosphorylated enzymes (Supplementary Table 9).
Overall, marked differences in gene expression, protein levels and phosphorylation patterns were observed in mice undergoing daytime compared to night‐time activity, implying a strong impact of the time of training on muscle physiology and function in absence of a major change in the regulation of the skeletal muscle clock.
Discussion
Physical activity is a crucial behaviour for which strong evolutionary pressure exists to ensure critical timing with availability of food and evasion of predators. Thus, a strong mutual interaction between skeletal muscle, the circadian timing system and exercise has been proposed (Gutierrez‐Monreal et al. 2020). First, the daily variations in metabolic and functional properties, ultimately influenced by the circadian system, will affect endurance and strength capacities. For example, depletion of energy stores after the resting phase will influence muscle metabolism and performance. Second, repeated aberrations from normal behaviour, e.g. temporal changes in activity levels, could exert zeitgeber activity to modulate and synchronize peripheral clocks according to altered environmental constraints. However, for both of these hypotheses, strong influence of time of day on mouse muscle cellular responses and performance, and the zeitgeber effects of exercise training, remain poorly described. Herein, we further evaluated these aspects in controlled animal studies, e.g. using SPPs as a technical intervention to reduce confounding factors.
First, collectively, our results strongly suggest that early daytime voluntary wheel running training has very poor zeitgeber activity on the skeletal muscle clock, barely affecting the phase of circadian gene expression in muscle, very different from the strong and paradigmatic effect of daytime feeding on the liver clock (Damiola et al. 2000; Fig. 12B ). In stark contrast, early daytime training leads to a dramatically altered profile in terms of transcription, protein levels and phosphoproteomics. In most aspects, the exercise response was amplified in the daytime activity group, and in part resembled that following an acute exercise bout. For example, normal night‐time running wheel activity is linked to only a very moderate transcriptional response of Ppargc‐1α gene expression. This gene responds very strongly to an acute exercise bout in naïve mice, qualitatively and quantitatively comparable to what we have observed in the daytime running wheel group, which could be interpreted as a lack of training adaptation, or habituation, in this group of mice (Perry et al. 2010). Alternatively, it is conceivable that the difference in exercise response in daytime compared to night‐time running wheel activity is at least in part related to the difference in feeding state, since food access in both groups was restricted to the dark period. In particular, the difference in proteins related to fatty acid metabolism, citrate cycle and glucose metabolism indicates that the metabolic state in these two groups might instruct the response to a metabolic stimulus such as physical activity. It will be interesting to further dissect these variables by the addition of different time‐restricted feeding interventions to the physical activity paradigm in future studies.
The running capacity of untrained mice at different times of day, hence the influence of the clock on muscle function, will have to be interpreted in a similar manner. Metabolic constraints most likely contribute significantly to the observed differences in endurance observed in our present study as well as previously reported (Ezagouri et al. 2019). For example, we observed a strong correlation between performance and liver, and to a lesser extent also skeletal muscle, glycogen. Depletion of glycogen stores is a major determinant of muscle fatigue in mice and humans, arguably liver glycogen even more in rodents than the skeletal muscle counterpart (Richter & Hargreaves, 2013; Hargreaves & Spriet, 2018). ‘Train low’, exercise under low glycogen conditions, is an established training paradigm in humans that, despite lower performance capacity in the individual endurance training bout, elicits a higher overall training adaptation (Hawley et al. 2018). Whether this is also the case in the mice running at ZT12 (lowest endurance performance, low liver and muscle glycogen stores) compared to those running at ZT0 (highest endurance performance, high liver and muscle glycogen stores) remains to be shown. Intriguingly, the transcriptomics, proteomics and phosphoproteomics analyses of these two different groups indicated that besides a robust, similar response of muscle to one acute exercise bout, very different pathways are activated depending on the time of day. In the high performing mice running at ZT0, the immediate activation of pathways related to inflammation, the complement system and autophagy, followed by a later increase in ribosomes suggests an efficient repair and regeneration process. In parallel, a coordinated regulation of proteins involved in muscle contraction and energy provisioning, e.g. those involved in calcium signalling and GLUT4 vesicle transport, is observed. Finally, the increased expression of MUPs and asprosin could be interpreted as a muscle‐derived signal to coordinate glycogenolysis and glucose release from liver. In stark contrast, the early response of skeletal muscle of the mice running at ZT12 indicates a broad activation of cellular stress, followed by an engagement of catabolic processes to match energetic demands in the absence of high abundance of storage of energy substrates, e.g. indicated by activation of AMPK and the glucocorticoid receptor, which could be interpreted as a signal for muscle protein degradation to fuel liver gluconeogenesis in this context. Accordingly, FoxO signalling is only observed in these mice. Differential control of proteostasis is furthermore underlined by the selective modulation of mTOR and related pathways.
The metabolic constraints that could explain the differences in performance depending on the time of day are at least secondarily under the control of the intrinsic muscle clock, e.g. in regards to energy level. Future experiments with inducible, skeletal muscle‐specific knockout models for clock components might thus help to clarify the contribution of this system to muscle metabolism and functionality if not confounded by other consequences of such dysregulations (Dyar et al. 2014). Our data demonstrate clear effects of acute exercise bouts and of prolonged voluntary daytime activity on several components of the clock mechanism, indicating that at least some of the affected genes are regulated by exercise. It is not clear, however, how some of these proteins will impart such changes in level or activity on the clock, whether such perturbations are mitigated by other, unperturbed clock control mechanisms, and to what extent such proteins affect other cellular processes besides clock oscillation. For example, the Period 1–2 genes, which we found to be robustly induced by exercise at different times of day, have been described as integrators of external and internal perturbations at the intersection of modulating the molecular clock and other cellular functions (Ripperger & Albrecht, 2012). Based on our data, these systems can become unlinked in certain settings and contexts, even though mutual regulation has been found, e.g. also by AMPK, HIF‐1α or mTOR on the clock (Panda, 2016; Reinke & Asher, 2019).
Our data also indicate that appropriate timing of exercise bouts might facilitate repair and regeneration, important to speed up recovery and potentially allow higher intensity and/or volume. The best time of training, however, will not be deducible from experiments performed in nocturnal, in‐bred laboratory mouse strains that do not recapitulate the complexity of the situation in humans with genetic variations, chronotypes, difference in eating habits and sleeping patterns, and other factors. Our comprehensive transcriptome, proteome and phosphoproteome analyses might provide a starting point to identify and validate markers for such a stratification in humans, contributing to personalized training design (Facer‐Childs et al. 2018) (Fig. 13). Lastly, our data indicate that scheduled training has very little or no phase‐shifting properties in a healthy mouse muscle, but, if chronically performed at the wrong time of day, might lead to perturbations of the muscle clock which resemble those of other peripheral clocks in pathological states and in ageing, characterized by a general dampening of oscillation amplitudes (Chellappa et al. 2019; Rijo‐Ferreira & Takahashi, 2019). Our experiments, however, will not allow any conclusions about a potential therapeutic effect of timed training (chrono‐exercise) on muscle and other peripheral clocks in pathologies that are linked to abnormal circadian rhythmicity (Cederroth et al. 2019).
Figure 13. Graphical summary of key findings of exercise studies in the mouse.
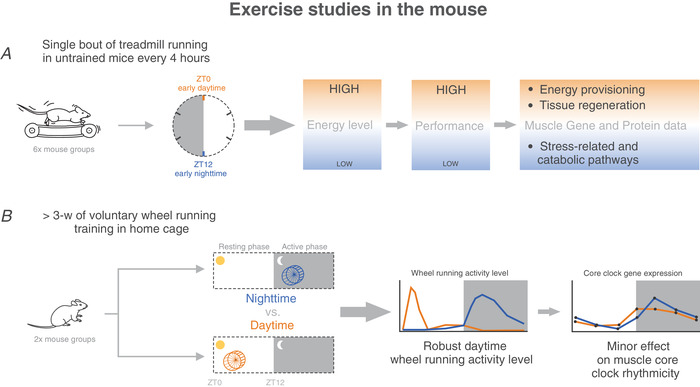
High energy levels, high performance and tissue regeneration‐related pathways were observed when treadmill exercise was performed at ZT0. In contrast, low energy levels decreased performance, and the activation of stress‐related and catabolic processes was observed when treadmill exercise was performed at ZT12. Moreover, chronic daytime wheel running has minor effects on muscle core clock gene expression. [Colour figure can be viewed at wileyonlinelibrary.com]
Additional information
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author contributions
Conceptualization and supervision: J.D. and C.H. Methodology: G.M., J.D., P.O.W. and D.R. Investigation: G.M., J.D. and G.S. Formal analysis and visualization: G.M., J.D., P.O.W. and D.R. Writing – original draft: G.M. and J.D. Writing – review and editing, G.M., J.D., P.O.W. and C.H. Funding acquisition: C.H. All authors have read and approved the final version of this manuscript and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All persons designated as authors qualify for authorship, and all those who qualify for authorship are listed.
Funding
The work in the laboratory of C.H. is funded by the Swiss National Science Foundation, the European Research Council (ERC) Consolidator grant 616830‐MUSCLE_NET, Swiss Cancer Research grant KFS‐3733‐08‐2015, the Swiss Society for Research on Muscle Diseases (SSEM), SystemsX.ch, the Novartis Stiftung für Medizinisch‐Biologische Forschung and the University of Basel.
Supporting information
Peer Review History
Supplementary Table 1. Treadmill Transcriptomics.
Supplementary Table 2. Treadmill Proteomics.
Supplementary Table 3. Treadmill Secretome Prediction.
Supplementary Table 4. Treadmill Phosphoproteomics.
Supplementary Table 5. DA Amplitude and Phase analysis.
Supplementary Table 6. DA Transcriptomics.
Supplementary Table 7. DA Proteomics.
Supplementary Table 8. DA Secretome Prediction.
Supplementary Table 9. DA Phosphoproteomics.
Supplementary Table 10. Primer sequences.
Statistical Summary Document
Acknowledgements
We are very grateful to the Quantitative Genomics Facility, in particular Phillippe Demougin for preparing cDNA libraries. We thank Katja Lamia, Scripps Research, for helpful comments on our manuscript. We also acknowledge the support of the Center for Scientific Computing (sciCORE) of the University of Basel.
Open access funding provided by Universitat Basel.
Biographies
Geraldine Maier completed her PhD in Biomedical Research in 2020 at the Biozentrum, University of Basel (Switzerland).

Julien Delezie completed his PhD in Neurobiology in 2012 at the University of Strasbourg (France). Both are interested in exercise and circadian biology and, in particular, how the circadian clock contributes to skeletal muscle function and overall health.

Edited by: Michael Hogan & Audrey Bergouignan
Linked articles: This article is highlighted in a Perspectives article by Macêdo et al and a Journal Club article by Chaffer et al. To read these articles, visit https://doi.org/10.1113/JP282005 and https://doi.org/10.1113/JP282134.
The peer review history is available in the Supporting Information section of this article (https://doi.org/10.1113/JP281535#support‐information‐section).
[Correction added on May 25, 2022, after first online publication: CSAL funding statement has been added.]
G. Maier and J. Delezie are equal first authors.
This article was first published as a preprint. Maier G, Delezie J, Westermark PO, Santos G, Ritz D. 2020. Transcriptomic, proteomic and phosphoproteomic underpinnings of daily exercise performance and Zeitgeber activity of endurance training. bioRxiv. https://doi.org/10.1101/2020.10.19.345686.
This is an Editor's Choice article from the 15 February 2022 issue.
Data availability statement
A statistical report is available online with this manuscript. Sequencing data sets associated with this work are available from GEO, accession code GSE178262. All proteomics datasets generated during the current study are available from ProteomeXchange, accession code PXD026461. All other data supporting the findings of this study are available from the corresponding author upon request.
References
- Ahrne E, Glatter T, Vigano C, Schubert C, Nigg EA & Schmidt A (2016). Evaluation and improvement of quantification accuracy in isobaric mass tag‐based protein quantification experiments. J Proteome Res 15, 2537–2547. [DOI] [PubMed] [Google Scholar]
- Albrecht U & Eichele G (2003). The mammalian circadian clock. Curr Opin Genet Dev 13, 271–277. [DOI] [PubMed] [Google Scholar]
- Allen DL, Harrison BC, Maass A, Bell ML, Byrnes WC & Leinwand LA (2001). Cardiac and skeletal muscle adaptations to voluntary wheel running in the mouse. J Appl Physiol 90, 1900–1908. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Jensen LJ, Blom N, Von Heijne G & Brunak S (2004). Feature‐based prediction of non‐classical and leaderless protein secretion. Protein Eng Des Sel 17, 349–356. [DOI] [PubMed] [Google Scholar]
- Boergesen M, Pedersen TA, Gross B, van Heeringen SJ, Hagenbeek D, Bindesboll C, Caron S, Lalloyer F, Steffensen KR, Nebb HI, Gustafsson JA, Stunnenberg HG, Staels B & Mandrup S (2012). Genome‐wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator‐activated receptor alpha in mouse liver reveals extensive sharing of binding sites. Mol Cell Biol 32, 852–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P & Pachter L (2016). Near‐optimal probabilistic RNA‐seq quantification. Nat Biotechnol 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Bryant NJ & Gould GW (2011). SNARE proteins underpin insulin‐regulated GLUT4 traffic. Traffic 12, 657–664. [DOI] [PubMed] [Google Scholar]
- Buhr ED & Takahashi JS (2013). Molecular components of the mammalian circadian clock. Handb Exp Pharmacol, 3–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartee GD (2015). Roles of TBC1D1 and TBC1D4 in insulin‐ and exercise‐stimulated glucose transport of skeletal muscle. Diabetologia 58, 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA (2015). Regulation of cardiac calcium channels in the fight‐or‐flight response. Curr Mol Pharmacol 8, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederroth CR, Albrecht U, Bass J, Brown SA, Dyhrfjeld‐Johnsen J, Gachon F, Green CB, Hastings MH, Helfrich‐Forster C, Hogenesch JB, Levi F, Loudon A, Lundkvist GB, Meijer JH, Rosbash M, Takahashi JS, Young M & Canlon B (2019). Medicine in the fourth dimension. Cell Metab 30, 238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheatham B (2000). GLUT4 and company: SNAREing roles in insulin‐regulated glucose uptake. Trends Endocrinol Metab 11, 356–361. [DOI] [PubMed] [Google Scholar]
- Chellappa SL, Vujovic N, Williams JS & Scheer F (2019). Impact of circadian disruption on cardiovascular function and disease. Trends Endocrinol Metab 30, 767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury‐Olela F & Schibler U (2000). Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14, 2950–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delavar H, Nogueira L, Wagner PD, Hogan MC, Metzger D & Breen EC (2014). Skeletal myofiber VEGF is essential for the exercise training response in adult mice. Am J Physiol Regul Integr Comp Physiol 306, R586–R595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delezie J, Dumont S, Sandu C, Reibel S, Pevet P & Challet E (2016). Rev‐erbα in the brain is essential for circadian food entrainment. Scientific Reports 6, 29386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delezie J & Handschin C (2018). Endocrine crosstalk between skeletal muscle and the brain. Front Neurol 9, 698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Foggia V, Zhang X, Licastro D, Gerli MF, Phadke R, Muntoni F, Mourikis P, Tajbakhsh S, Ellis M, Greaves LC, Taylor RW, Cossu G, Robson LG & Marino S (2014). Bmi1 enhances skeletal muscle regeneration through MT1‐mediated oxidative stress protection in a mouse model of dystrophinopathy. J Exp Med 211, 2617–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunant Y & Israel M (2000). Neurotransmitter release at rapid synapses. Biochimie 82, 289–302. [DOI] [PubMed] [Google Scholar]
- Duong KHM & Chun KH (2019). Regulation of glucose transport by RhoA in 3T3‐L1 adipocytes and L6 myoblasts. Biochem Biophys Res Commun 519, 880–886. [DOI] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E & Huber W (2009). Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 4, 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyar KA, Ciciliot S, Wright LE, Bienso RS, Tagliazucchi GM, Patel VR, Forcato M, Paz MI, Gudiksen A, Solagna F, Albiero M, Moretti I, Eckel‐Mahan KL, Baldi P, Sassone‐Corsi P, Rizzuto R, Bicciato S, Pilegaard H, Blaauw B & Schiaffino S (2014). Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol Metab 3, 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyar KA, Lutter D, Artati A, Ceglia NJ, Liu Y, Armenta D, Jastroch M, Schneider S, de Mateo S, Cervantes M, Abbondante S, Tognini P, Orozco‐Solis R, Kinouchi K, Wang C, Swerdloff R, Nadeef S, Masri S, Magistretti P, Orlando V, Borrelli E, Uhlenhaut NH, Baldi P, Adamski J, Tschop MH, Eckel‐Mahan K & Sassone‐Corsi P (2018). Atlas of circadian metabolism reveals system‐wide coordination and communication between clocks. Cell 174, 1571–1585.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emrick MA, Sadilek M, Konoki K & Catterall WA (2010). Beta‐adrenergic‐regulated phosphorylation of the skeletal muscle CaV1.1 channel in the fight‐or‐flight response. Proc Natl Acad Sci U S A 107, 18712–18717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezagouri S, Zwighaft Z, Sobel J, Baillieul S, Doutreleau S, Ladeuix B, Golik M, Verges S & Asher G (2019). Physiological and molecular dissection of daily variance in exercise capacity. Cell Metabolism 30, 78–91.e4, [DOI] [PubMed] [Google Scholar]
- Facer‐Childs ER, Boiling S & Balanos GM (2018). The effects of time of day and chronotype on cognitive and physical performance in healthy volunteers. Sports Med Open 4, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facer‐Childs E & Brandstaetter R (2015). The impact of circadian phenotype and time since awakening on diurnal performance in athletes. Curr Biol 25, 518–522. [DOI] [PubMed] [Google Scholar]
- Fernandez‐Verdejo R, Vanwynsberghe AM, Essaghir A, Demoulin JB, Hai T, Deldicque L & Francaux M (2017). Activating transcription factor 3 attenuates chemokine and cytokine expression in mouse skeletal muscle after exercise and facilitates molecular adaptation to endurance training. FASEB J 31, 840–851. [DOI] [PubMed] [Google Scholar]
- Gabriel BM & Zierath JR (2019). Circadian rhythms and exercise—re‐setting the clock in metabolic disease. Nat Rev Endocrinol 15, 197–206. [DOI] [PubMed] [Google Scholar]
- Gan Z, Fu T, Kelly DP & Vega RB (2018). Skeletal muscle mitochondrial remodeling in exercise and diseases. Cell Res 28, 969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D & Shaw RJ (2017). AMPK: mechanisms of cellular energy sensing and restoration of metabolic balance. Mol Cell 66, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY & Zhang J (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez‐Monreal MA, Harmsen JF, Schrauwen P & Esser KA (2020). Ticking for metabolic health: The skeletal‐muscle clocks. Obesity 28 (Suppl 1), S46–S54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves M & Spriet LL (2018). Exercise metabolism: Fuels for the fire. Cold Spring Harb Perspect Med 8, a029744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley JA, Lundby C, Cotter JD & Burke LM (2018). Maximizing cellular adaptation to endurance exercise in skeletal muscle. Cell Metab 27, 962–976. [DOI] [PubMed] [Google Scholar]
- Hearris MA, Hammond KM, Fell JM & Morton JP (2018). Regulation of muscle glycogen metabolism during exercise: implications for endurance performance and training adaptations. Nutrients 10, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman NJ, Parker BL, Chaudhuri R, Fisher‐Wellman KH, Kleinert M, Humphrey SJ, Yang P, Holliday M, Trefely S, Fazakerley DJ, Stockli J, Burchfield JG, Jensen TE, Jothi R, Kiens B, Wojtaszewski JF, Richter EA & James DE (2015). Global phosphoproteomic analysis of human skeletal muscle reveals a network of exercise‐regulated kinases and AMPK substrates. Cell Metab 22, 922–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloszy JO (1967). Biochemical adaptations in muscle. Effects of exercise on mitochondrial oxygen uptake and respiratory enzyme activity in skeletal muscle. J Biol Chem 242, 2278–2282. [PubMed] [Google Scholar]
- Hughes ATL & Piggins HD (2012). Feedback actions of locomotor activity to the circadian clock. Prog Brain Res 199, 305–336. [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME & Gygi SP (2010). A tissue‐specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LN (1992). Glycogen phosphorylase: control by phosphorylation and allosteric effectors. FASEB J 6, 2274–2282. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, Harte RA, Heitner S, Hinrichs AS, Learned K, Lee BT, Li CH, Raney BJ, Rhead B, Rosenbloom KR, Sloan CA, Speir ML, Zweig AS, Haussler D, Kuhn RM & Kent WJ (2014). The UCSC genome browser database: 2014 update. Nucleic Acids Res 42, D764–D770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai H, Takahashi N & Tokumaru H (2012). Distinct initial SNARE configurations underlying the diversity of exocytosis. Physiol Rev 92, 1915–1964. [DOI] [PubMed] [Google Scholar]
- Katsuoka F, Motohashi H, Ishii T, Aburatani H, Engel JD & Yamamoto M (2005). Genetic evidence that small maf proteins are essential for the activation of antioxidant response element‐dependent genes. Mol Cell Biol 25, 8044–8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaier R, Infanger D, Niemeyer M, Cajochen C & Schmidt‐Trucksass A (2019). In athletes, the diurnal variations in maximum oxygen uptake are more than twice as large as the day‐to‐day variations. Front Physiol 10, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A & Griffiths‐Jones S (2011). miRBase: integrating microRNA annotation and deep‐sequencing data. Nucleic Acids Res 39, D152–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH & Jun HS (2019). Role of myokines in regulating skeletal muscle mass and function. Front Physiol 10, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel‐Duby R & Spiegelman BM (2002). Transcriptional co‐activator PGC‐1α drives the formation of slow‐twitch muscle fibres. Nature 418, 797–801. [DOI] [PubMed] [Google Scholar]
- Loizides‐Mangold U, Perrin L, Vandereycken B, Betts JA, Walhin JP, Templeman I, Chanon S, Weger BD, Durand C, Robert M, Paz Montoya J, Moniatte M, Karagounis LG, Johnston JD, Gachon F, Lefai E, Riezman H & Dibner C (2017). Lipidomics reveals diurnal lipid oscillations in human skeletal muscle persisting in cellular myotubes cultured in vitro. Proc Natl Acad Sci U S A 114, E8565–E8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W & Anders S (2014). Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack HI, Zheng B, Asara JM & Thomas SM (2012). AMPK‐dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy 8, 1197–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JJ, Andrews JL, McDearmon EL, Campbell KS, Barber BK, Miller BH, Walker JR, Hogenesch JB, Takahashi JS & Esser KA (2007). Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics 31, 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKie GL, Medak KD, Knuth CM, Shamshoum H, Townsend LK, Peppler WT & Wright DC (2019). Housing temperature affects the acute and chronic metabolic adaptations to exercise in mice. J Physiol 597, 4581–4600. [DOI] [PubMed] [Google Scholar]
- Mirizio GG, Nunes RSM, Vargas DA, Foster C & Vieira E (2020). Time‐of‐day effects on short‐duration maximal exercise performance. Sci Rep 10, 9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris S, Geoghegan ND, Sadler JBA, Koester AM, Black HL, Laub M, Miller L, Heffernan L, Simpson JC, Mastick CC, Cooper J, Gadegaard N, Bryant NJ & Gould GW (2020). Characterisation of GLUT4 trafficking in HeLa cells: comparable kinetics and orthologous trafficking mechanisms to 3T3‐L1 adipocytes. PeerJ 8, e8751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muoio DM, MacLean PS, Lang DB, Li S, Houmard JA, Way JM, Winegar DA, Corton JC, Dohm GL & Kraus WE (2002). Fatty acid homeostasis and induction of lipid regulatory genes in skeletal muscles of peroxisome proliferator‐activated receptor (PPAR) α knock‐out mice. Evidence for compensatory regulation by PPARδ. J Biol Chem 277, 26089–26097. [DOI] [PubMed] [Google Scholar]
- Nielsen H (2017). Predicting secretory proteins with signalP. Methods Mol Biol 1611, 59–73. [DOI] [PubMed] [Google Scholar]
- Oishi K, Fukui H, Sakamoto K, Miyazaki K, Kobayashi H & Ishida N (2002). Differential expressions of mPer1 and mPer2 mRNAs under a skeleton photoperiod and a complete light–dark cycle. Brain Res Mol Brain Res 109, 11–17. [DOI] [PubMed] [Google Scholar]
- Panda S (2016). Circadian physiology of metabolism. Science 354, 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen BK & Febbraio MA (2008). Muscle as an endocrine organ: Focus on muscle‐derived interleukin‐6. Physiological Reviews 88, 1379–1406. [DOI] [PubMed] [Google Scholar]
- Perrin L, Loizides‐Mangold U, Chanon S, Gobet C, Hulo N, Isenegger L, Weger BD, Migliavacca E, Charpagne A, Betts JA, Walhin JP, Templeman I, Stokes K, Thompson D, Tsintzas K, Robert M, Howald C, Riezman H, Feige JN, Karagounis LG, Johnston JD, Dermitzakis ET, Gachon F, Lefai E & Dibner C (2018). Transcriptomic analyses reveal rhythmic and CLOCK‐driven pathways in human skeletal muscle. eLife 7, e34114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry CG, Lally J, Holloway GP, Heigenhauser GJ, Bonen A & Spriet LL (2010). Repeated transient mRNA bursts precede increases in transcriptional and mitochondrial proteins during training in human skeletal muscle. J Physiol 588, 4795–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon NJ, Gabriel BM, Dollet L, Smith JAB, Sardon Puig L, Botella J, Bishop DJ, Krook A & Zierath JR (2020). Transcriptomic profiling of skeletal muscle adaptations to exercise and inactivity. Nat Commun 11, 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post H, Penning R, Fitzpatrick MA, Garrigues LB, Wu W, MacGillavry HD, Hoogenraad CC, Heck AJ & Altelaar AF (2017). Robust, sensitive, and automated phosphopeptide enrichment optimized for low sample amounts applied to primary hippocampal neurons. J Proteome Res 16, 728–737. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Brown GR, Hiatt SM, Thibaud‐Nissen F, Astashyn A, Ermolaeva O, Farrell CM, Hart J, Landrum MJ, McGarvey KM, Murphy MR, O'Leary NA, Pujar S, Rajput B, Rangwala SH, Riddick LD, Shkeda A, Sun H, Tamez P, Tully RE, Wallin C, Webb D, Weber J, Wu W, DiCuccio M, Kitts P, Maglott DR, Murphy TD & Ostell JM (2014). RefSeq: an update on mammalian reference sequences. Nucleic Acids Res 42, D756–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H & Vilo J (2016). g:Profiler‐a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res 44, W83–W89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke H & Asher G (2019). Crosstalk between metabolism and circadian clocks. Nat Rev Mol Cell Biol 20, 227–241. [DOI] [PubMed] [Google Scholar]
- Reznick J, Preston E, Wilks DL, Beale SM, Turner N & Cooney GJ (2013). Altered feeding differentially regulates circadian rhythms and energy metabolism in liver and muscle of rats. Biochim Biophys Acta 1832, 228–238. [DOI] [PubMed] [Google Scholar]
- Richter EA & Hargreaves M (2013). Exercise, GLUT4, and skeletal muscle glucose uptake. Physiol Rev 93, 993–1017. [DOI] [PubMed] [Google Scholar]
- Rijo‐Ferreira F & Takahashi JS (2019). Genomics of circadian rhythms in health and disease. Genome Med 11, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger JA & Albrecht U (2012). The circadian clock component PERIOD2: from molecular to cerebral functions. Prog Brain Res 199, 233–245. [DOI] [PubMed] [Google Scholar]
- Romere C, Duerrschmid C, Bournat J, Constable P, Jain M, Xia F, Saha PK, Del Solar M, Zhu B, York B, Sarkar P, Rendon DA, Gaber MW, LeMaire SA, Coselli JS, Milewicz DM, Sutton VR, Butte NF, Moore DD & Chopra AR (2016). Asprosin, a fasting‐induced glucogenic protein hormone. Cell 165, 566–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruas JL, White JP, Rao RR, Kleiner S, Brannan KT, Harrison BC, Greene NP, Wu J, Estall JL, Irving BA, Lanza IR, Rasbach KA, Okutsu M, Nair KS, Yan Z, Leinwand LA & Spiegelman BM (2012). A PGC‐1α isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell 151, 1319–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K & Holman GD (2008). Emerging role for AS160/TBC1D4 and TBC1D1 in the regulation of GLUT4 traffic. Am J Physiol Endocrinol Metab 295, E29–E37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Parr EB, Devlin BL, Hawley JA & Sassone‐Corsi P (2018). Human metabolomics reveal daily variations under nutritional challenges specific to serum and skeletal muscle. Mol Metab 16, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savikj M, Gabriel BM, Alm PS, Smith J, Caidahl K, Bjornholm M, Fritz T, Krook A, Zierath JR & Wallberg‐Henriksson H (2019). Afternoon exercise is more efficacious than morning exercise at improving blood glucose levels in individuals with type 2 diabetes: a randomised crossover trial. Diabetologia 62, 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Love MI & Robinson MD (2015). Differential analyses for RNA‐seq: transcript‐level estimates improve gene‐level inferences. F1000Res 4, 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Cheng SY, Chen M, Lim CJ & Pallen CJ (2012). Protein tyrosine phosphatase α phosphotyrosyl‐789 binds BCAR3 to position Cas for activation at integrin‐mediated focal adhesions. Mol Cell Biol 32, 3776–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylow L, Kleinert M, Richter EA & Jensen TE (2017). Exercise‐stimulated glucose uptake—regulation and implications for glycaemic control. Nat Rev Endocrinol 13, 133–148. [DOI] [PubMed] [Google Scholar]
- Tahara Y, Aoyama S & Shibata S (2017). The mammalian circadian clock and its entrainment by stress and exercise. J Physiol Sci 67, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka N, Sumi Y, Matsuda K, Fujita J, Hosooka T, Noguchi T, Aiba A & Satoh T (2015). Role for RalA downstream of Rac1 in skeletal muscle insulin signalling. Biochem J 469, 445–454. [DOI] [PubMed] [Google Scholar]
- Thaben PF & Westermark PO (2014). Detecting rhythms in time series with RAIN. J Biol Rhythms 29, 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaben PF & Westermark PO (2016). Differential rhythmicity: detecting altered rhythmicity in biological data. Bioinformatics 32, 2800–2808. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yang F, Gritsenko MA, Wang Y, Clauss T, Liu T, Shen Y, Monroe ME, Lopez‐Ferrer D, Reno T, Moore RJ, Klemke RL, Camp DG 2nd & Smith RD (2011). Reversed‐phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics 11, 2019–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei‐Lapierre L, Carrell EM, Boncompagni S, Protasi F & Dirksen RT (2013). Orai1‐dependent calcium entry promotes skeletal muscle growth and limits fatigue. Nat Commun 4, 2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka Y, Hashimoto S, Takasu NN, Tanahashi Y, Nishide SY, Honma S & Honma K (2015). Morning and evening physical exercise differentially regulate the autonomic nervous system during nocturnal sleep in humans. Am J Physiol Regul Integr Comp Physiol 309, R1112–R1121. [DOI] [PubMed] [Google Scholar]
- Yasumoto Y, Nakao R & Oishi K (2015). Free access to a running‐wheel advances the phase of behavioral and physiological circadian rhythms and peripheral molecular clocks in mice. PLoS One 10, e0116476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Wang C, Li Y, Miwa T, Liu C, Cui W, Song WC & Du J (2017). Complement C3a signaling facilitates skeletal muscle regeneration by regulating monocyte function and trafficking. Nat Commun 8, 2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Jiang L & Rui L (2009). Identification of MUP1 as a regulator for glucose and lipid metabolism in mice. J Biol Chem 284, 11152–11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong H, Wang CC, Vaitheesvaran B, Kurland IJ, Hong W & Pessin JE (2011). Enhanced energy expenditure, glucose utilization, and insulin sensitivity in VAMP8 null mice. Diabetes 60, 30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Peer Review History
Supplementary Table 1. Treadmill Transcriptomics.
Supplementary Table 2. Treadmill Proteomics.
Supplementary Table 3. Treadmill Secretome Prediction.
Supplementary Table 4. Treadmill Phosphoproteomics.
Supplementary Table 5. DA Amplitude and Phase analysis.
Supplementary Table 6. DA Transcriptomics.
Supplementary Table 7. DA Proteomics.
Supplementary Table 8. DA Secretome Prediction.
Supplementary Table 9. DA Phosphoproteomics.
Supplementary Table 10. Primer sequences.
Statistical Summary Document
Data Availability Statement
A statistical report is available online with this manuscript. Sequencing data sets associated with this work are available from GEO, accession code GSE178262. All proteomics datasets generated during the current study are available from ProteomeXchange, accession code PXD026461. All other data supporting the findings of this study are available from the corresponding author upon request.


