Abstract
The outbreak of coronavirus disease 2019 (COVID-19) has now become a pandemic, and the etiologic agent is the severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2). T cell mediated immune responses play an important role in virus controlling; however, the understanding of the viral protein immunogenicity and the mechanisms of the induced responses are still limited. So, identification of specific epitopes and exploring their immunogenic properties would provide valuable information. In our study, we utilized the Immune Epitope Database and Analysis Resource and NetMHCpan to predict HLA-A2 restricted CD8+ T cell epitopes in structural proteins of SARS-CoV-2, and screened out 23 potential epitopes. Among them, 18 peptides showed strong or moderate binding with HLA-A2 with a T2A2 cell binding model. Next, the mixed peptides induced the increased expression of CD69 and highly expressed levels of IFN-γ and granzyme B in CD8+ T cells, indicating effective activation of specific CD8+ T cells. In addition, the peptide-activated CD8+ T cells showed significantly increased killing to the target cells. Furthermore, tetramer staining revealed that the activated CD8+ T cells mainly recognized seven epitopes. All together, we identified specific CD8+ T cell epitopes in SARS-CoV-2 structural proteins, which could induce the production of specific immune competent CD8+ T cells. Our work contributes to the understanding of specific immune responses and vaccine development for SARS-CoV-2.
Keywords: CD8+ T cells, epitope, SARS-CoV-2, structural protein
Graphical Abstract
Identifies CD8+ T cell epitopes in SARS-CoV-2 structural proteins, helping to further define specific immune responses that could aid in vaccine development for SARS-CoV-2.
Graphical Abstract.
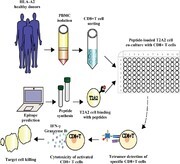
INTRODUCTION
Severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) is a single-stranded positive RNA virus, which is responsible for the coronavirus disease 2019 (COVID-19).1 SARS-CoV-2 belongs to the Coronaviridae family, and is the seventh coronavirus known to infect humans, with the former six including the human coronavirus, the SARS-CoV, and the Middle East respiratory syndrome-coronavirus (MERS-CoV).2,3 SARS-CoV-2 has caused significant mortality especially in the elderly population4 and those with comorbidities.5 Studies have shown that SARS-CoV-2 infection induces the production of specific antibodies; meanwhile T cell mediated immunity is also essential for controlling viral infection.6,7 But to date, a majority of current work has been focusing on humoral immunity, and the information on the SARS-CoV-2 specific CD8+ T cell immunity is limited. Weiskopf et al. reported that SARS-CoV-2 specific CD8+ T cells could be found in most of the intensive care unit patients, and the T cell responses appeared relatively early and increased over time.8 Wang et al. also clarified that changes in peripheral blood lymphocyte subsets were related to the clinical characteristics and treatment effects of COVID-19, and CD8+ T lymphocytes could be used as an independent predictor of disease severity and treatment effects.9 The report also suggested that CD8+ T cells from patients with moderate infection had higher clonal expansion than those from patients with severe/critical infection in the lung, and higher CD8+ T cells in the lung were associated with better control of the progression of SARS-CoV-2.10 These data shed light on a potential role of CD8+ T cell response as a function of disease remission and disease severity.
In the context of SARS-CoV, the viral infection could induce antibody responses, but SARS specific antibodies only lasted for approximately 2 yr. The titers of IgG decreased significantly in the third year, and were not detectable 6 yr postinfection.11,12 However, memory T cell responses were identified in the recovered SARS patients 6 or even 7 yr after infection.12,13 It has been reported that MERS-CoV specific antibodies were produced at low levels or absent in the patients with mild infection.14,15 Thus, these coronaviruses could trigger specific antibodies and T cell responses in infected patients, but antibody levels seemed to diminish faster than T cell responses, which shows that SARS patients might be subject to re-infection 3 yr after initial exposure. Based on current studies on SARS-CoV-2, the induced antibody responses appeared to be transient and feeble. The specific antibodies against SARS-CoV-2 might not be long lasting in patients with mild infection, who accounted for the majority of the infected population, and the IgG levels were reduced by 70% about 90 d after diagnosis.16,17 These data suggested that short-lived humoral immunity might be a common characteristic after coronavirus infection, but the rate of antibody decline was faster than that reported for SARS. On the contrary, memory T cell responses were robust in the absence or presence of circulating antibodies in convalescent COVID-19 patients.18 Thus, SARS-CoV-2 specific cellular immunity could be critical for long-term immune protection against COVID-19.
Recently, Grifoni et al. showed that SARS-CoV-2 CD8+ T cell strongly responded to S and M proteins, and at least eight SARS-CoV-2 ORFs were targeted in COVID-19 convalescent patients.19 However, the exact SARS-CoV-2 specific epitopes mediating these T cell responses have not been confirmed and described in detail. In our previous work, we identified a couple of dominant CD8+ T cell epitopes in the Spike protein (S) of SARS-CoV-2.20 Structural proteins of SARS-CoV-2, including S, membrane (M), nucleocapsid (N), and envelope (E) proteins, are the main targets that are recognized by the host immune system. In this study, we predicted the potential CD8+ T cell epitopes within the structural proteins of SARS-CoV-2, and analyzed the immune properties of these epitopes, including MHC I binding and activation of CD8+ T cells. Our work has defined a set of functional epitopes, which provides useful information for investigating the features of SARS-CoV-2 specific CD8+ T cell immunity, and contributes to the development of SARS-CoV-2 vaccines and related immunotherapy.
MATERIAL AND METHODS
Human subjects
The Institutional Review Board of the First Affiliated Hospital of Jinan University (Guangzhou, China) has approved this study. The unexposed blood donors were healthy individuals enrolled at the Guangzhou Blood Center, including four males and three females, with the ages from 24 to 42 (Table 1). All subjects were negative for detection of SARS-CoV-2 RNA by RT-PCR. The health condition of each individual was evaluated before the blood samples were used in the clinical treatment. Donors had not suffered from any systemic disease, including hepatitis B or C, HIV, syphilis, diabetes, kidney or liver diseases, malignant tumors, or autoimmune diseases. All subjects provided informed consent at the time of enrollment that their samples could be used for this study. Samples of whole blood were collected into tubes with heparin sodium and stored at room temperature.
TABLE 1.
The information of the healthy blood sample donors
| Samples | SARS-CoV-2 PCR | Gender | Age | HLA subtypes | Day 0 of tetramer staining | Day 7 of tetramer staining | Survival ratio of T2A2 cells | IFN-γ staining | Granzyme B staining |
|---|---|---|---|---|---|---|---|---|---|
| #1 | Negative | Male | 29 | HLA-A2 | ●* | ● | ● | ● | ● |
| #2 | Negative | Male | 33 | HLA-A2 | ● | ● | ● | ||
| #3 | Negative | Male | 38 | HLA-A2 | ● | ||||
| #4 | Negative | Male | 42 | HLA-A2 | ● | ||||
| #5 | Negative | Female | 25 | HLA-A2 | ● | ● | ● | ● | ● |
| #6 | Negative | Female | 30 | HLA-A2 | ● | ● | ● | ● | ● |
| #7 | Negative | Female | 35 | HLA-A2 | ● | ||||
| 1 | Negative | Female | 28 | N† | |||||
| 2 | Negative | Male | 39 | N | |||||
| 3 | Negative | Female | 44 | N | |||||
| 4 | Negative | Female | 43 | N | |||||
| 5 | Negative | Male | 27 | N | |||||
| 6 | Negative | Female | 31 | N | |||||
| 7 | Negative | Male | 22 | N | |||||
| 8 | Negative | Male | 40 | N | |||||
| 9 | Negative | Male | 34 | N | |||||
| 10 | Negative | Female | 27 | N | |||||
| 11 | Negative | Female | 23 | N | |||||
| 12 | Negative | Male | 25 | N |
Black dot means the sample was used in the experiment.
N means not HLA-A2 subtypes.
SARS-CoV-2: severe acute respiratory syndrome-coronavirus 2.
Cells
T2A2 cells are TAP-deficient T2 cells expressing HLA-A2 molecules on the cell surface.21 Cells were maintained in IMDM (Cat#SH30228.01, HyClone, Waltham, Massachusetts, USA) with 20% FBS (Cat#S711-001S, Lonsera, Uruguay) and 1% penicillin-streptomycin (Cat#SV30010, HyClone, Waltham, Massachusetts, USA). PBMCs were isolated from whole blood of healthy donors by density gradient centrifugation using Ficoll-Paque PLUS (Cat#17144003, GE, Chicago, Illinois, USA). Briefly, the blood samples were diluted by adding PBS buffer at a ratio of 1:1, gently mixed upside down. A total of 10 ml Ficoll separation solution was slowly added into the centrifuge tube, and then centrifuged for 20 min. The PBMCs were saved and washed with PBS. PBMCs were stained with FITC anti-human HLA-A2 antibody (Cat#343303, Biolegend, San Diego, California, USA) at 4°C for 30 min in the dark, and were acquired on flow cytometer FACS Canto (BD, Franklin Lakes, New Jersey, USA). Aliquots of HLA-A2+ PBMCs were cryopreserved at 90% FBS (Cat#S711-001S, Lonsera, Uruguay) and 10% DMSO (Cat#D2650, Sigma-Aldrich, Burlington, USA) and stored in liquid nitrogen or at −80°C until usage.
Antibodies and other reagents
Antibody were purchased from BioLegend, including FITC anti-human HLA-A2 antibody (Cat#343303), PE anti-human HLA-A2 antibody (Cat#343305), anti-human CD28 antibodies (Cat#302901), APC anti-human-CD69 (Cat#310909), APC anti-human CD8 (Cat#344721), PerCP anti-human IFN-γ (Cat#502524), and FITC anti-human granzyme B (Cat#515403). Flex-T monomer (Cat#280003) and PE conjugated streptavidin (Cat#405203) were purchased from BioLegend. Ficoll-Paque PLUS (Cat#17144003) was from GE and DMSO (Cat#D2650) was from Sigma-Aldrich. IMDM (Cat#SH30228.01) and penicillin-streptomycin (Cat#SV30010) were from HyClone, and FBS (Cat#S711-001S) was purchased from LONSERA. PBS (Cat#C10010500BT) was from Gibco (Waltham, Massachusetts, USA). D-biotin (Cat#2110450) was purchased from Invitrogen (Waltham, Massachusetts, USA), and Mitomycin C (Cat#50-07-7) was from Sinochem Holdings (Beijing, China). CFSE (Cat#T6802) was from Targetmol (Boston, Massachusetts, USA). EasySep Human CD8+ T Cell Isolation Kit (Cat#17953) was from Stem Cell Technologies (Vancouver, British Columbia, Canada), and IL-2 (recombinant human interleukin-2(125Ala) injection) was from SL PHARM (Beijing, China). Leuko act cktl with GolgiPlug (Cat#550583) was purchased from BD.
Prediction of HLA-A2-restricted CD8+ T cell epitopes
The S, M, N, and E protein sequences of SARS-CoV-2 Wuhan-Hu-1 strain (NC_045512.2) were used for T cell epitope prediction by using the “MHC I Binding” tool (http://www.iedb.org/). The prediction method was based on the integration of Immune Epitope Database and Analysis Resource (IEDB) recommended 2.22 and NetMHCpan EL, and these programs sorted peptides from the strongest to nonexistent any binding capacity to HLA molecules by predicting their binding capacity (half-maximal inhibitory concentration [IC50]). The selected MHC alleles were HLA-A2, the most frequent MHC class I genotype among Chinese population.22,23 Potential SARS-CoV-2 MHC-I peptides were selected and synthesized by GenScript Biotechnology Co., Ltd. (Nanjing, China) with the purity >95%, and reconstituted in DMSO at a concentration of 10 mmol/L, respectively.
Screening of peptides with T2A2 cells
The binding capability of the predicted peptides with MHC-I were assessed by an in vitro binding assay with T2A2 cells. T2A2 cells were harvested from T75 flask, washed twice with PBS and resuspended with IMDM. The cells were then seeded in 96-well plates at a density of 105 cells/well and incubated with peptides at 20 μM at 37°C for 4 h. Four groups were set: DMSO (blank control), Influenza A virus M1 peptide (GILGFVFTL, positive control), EBV peptide (IVTDFSVIK, negative control), and candidate peptides. The cells were collected and stained with PE anti-human HLA-A2 antibody at 4°C in the dark for 30 min, and samples were acquired on flow cytometer FACS Canto (BD). FlowJo v10 software was used for the analysis of flow cytometry data.
The generation of peptide-HLA-A2 tetramer
A total of 400 μM of peptide working solution was prepared by mixing 3 μl of 10 mM stock solution with 72 μl PBS (400 μM). A total of 20 μl of diluted peptide and 20 μl (1 μg/ml) Flex-T monomer were mixed well by pipetting up and down, and then added into 96-well U-bottom plates. The plates were put on ice and exposed to UV light for 30 min, and then incubated for 30 min at 37°C in the dark. A total of 30 μl of peptide-exchanged monomer was transferred into a new plate, mixed with 3.3 μl of PE conjugated streptavidin, and incubated on ice in the dark for 30 min. A total of 2.4 μl of blocking solution (1.6 μl 50 mM D-biotin in 198.4 μl PBS) was added to stop the reaction after the incubation and then incubated at 4°C to 8°C overnight.
The inhibition of T2A2 cell proliferation
To stop the proliferation of T2A2 cells during the activation of CD8+ T cells, Mitomycin C treatment was performed. T2A2 cells were set for four groups including untreated, only Mitomycin C treated (20 μg/ml), only CFSE treated (5 μmol/L), and Mitomycin C treated plus CFSE treated. All groups were detected by flow cytometer FACS Canto (BD) and data were analyzed by FlowJo v10 software.
The activation of CD8+ T cells and peptide-loaded HLA-A2 tetramer staining
CD8+ T cells were stimulated by T2A2 cells loaded with candidate peptides, and the activation marker CD69 was detected. Briefly, T2A2 cells were treated with 20 μg/ml Mitomycin C for 30 min to inhibit cell proliferation, and then stained with 5 μmol/L CFSE for an hour prior to loading with peptides. CD8+ T cells were isolated from HLA-A2+ PBMCs by using EasySep Human CD8+ T Cell Isolation Kit. A total of 5 × 105 peptide-loaded T2A2 cells were mixed with 5 × 105 CD8+ T cells, and cocultured with 1 μg/ml anti-human CD28 antibodies and 50 IU/ml IL-2 for 7 d. A total of 50 IU/ml IL-2 and 20 μM mixed peptide pool were supplemented every 2 d. The T cell activation marker CD69 was tested. On day 7, SARS-CoV-2 specific HLA-A2-restricted CD8+ T cells were detected by APC anti-human CD8 and peptide-HLA-A2 tetramer, respectively. All samples were acquired on flow cytometer FACS Canto (BD), and analyzed with FlowJo v10 software.
Cytotoxicity of CD8+ T cells by intracellular cytokine staining
On day 7, CD8+ T cells were restimulated with peptide-loaded T2A2 cells for 5 h in the presence of leuko act cktl with GolgiPlug plus 50 IU/ml IL-2, and the reactive CD8+ T cells were quantified using PerCP anti-human IFN-γ and FITC anti-human granzyme B staining. All samples were acquired on flow cytometer FACS Canto (BD), and analyzed with FlowJo v10 software.
Killing of T2A2 cells by specific CD8+ T cells
The activated CD8+ T cells and peptide-loaded T2A2 cells were cocultured for 7 d. T2A2 cells were stained with CFSE, and their survival rates were calculated with flow cytometer on days 0 and 7, respectively.
Statistical analysis
Comparison of experimental groups was performed with 1-way or 2-way ANOVA, or unpaired, 2-tailed Student's t-test. Statistical analysis was performed with the GraphPad Prism 6 and SPSS 23.0 software. P-values less than 0.05 were considered statistically significant.
RESULTS
Prediction of HLA-A2 restricted CD8+ T cell epitopes in structure proteins of SARS-CoV-2
To broadly screen the epitopes of SARS-CoV-2 and understand the mechanisms of antigen epitope recognition and activation associated with CD8+ T cells, we predicted the HLA-A2 restricted CD8+ T cell epitopes in S, M, N, and E protein of SARS-CoV-2 by using the IEDB and NetMHCpan EL 4.0. Peptides with the predicted binding capability IC50 below 100 nM were selected. Totally, 23 peptides were selected, including 14 in S protein, 3 in E protein, and 6 in M protein (Fig. 1B). The detailed information is shown in Table 2. Next, these potential epitopes were synthesized for further evaluation.
FIGURE 1.
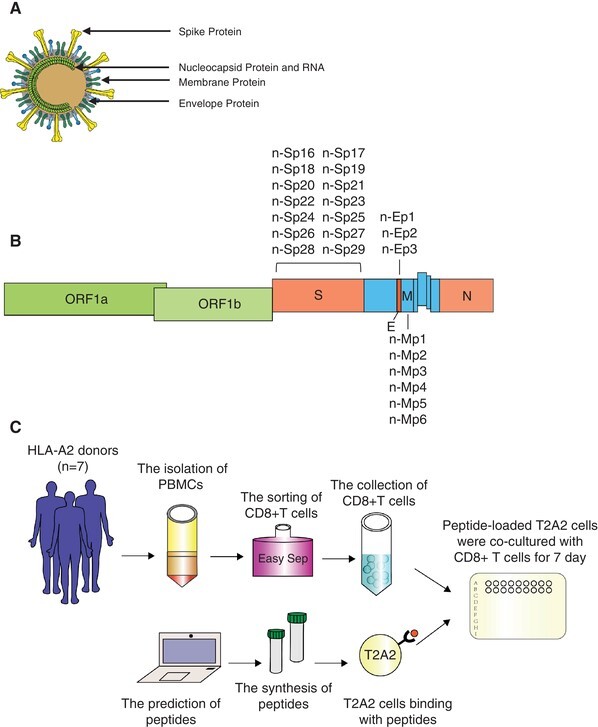
Identification of HLA-A2 restricted CD8+ T cell epitopes. (A) The schematic of severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) virion. (B) Locations of the predicted peptides in SARS-CoV-2 genome. (C) General flow chart of the experiment
TABLE 2.
| No. | HLA subtypes | Position | Sequence | Molecular weight | References |
|---|---|---|---|---|---|
| n-Sp16 | HLA-A2 | 62-70 | VTWFHAIHV | 1109.3 | |
| n-Sp17 | HLA-A2 | 424-432 | KLPDDFTGC | 995.12 | |
| n-Sp18 | HLA-A2 | 515-524 | FELLHAPATV | 1097.28 | |
| n-Sp19 | HLA-A2 | 721-729 | SVTTEILPV | 958.12 | |
| n-Sp20 | HLA-A2 | 786-794 | KQIYKTPPI | 1087.33 | |
| n-Sp21 | HLA-A2 | 817-826 | FIEDLLFNKV | 1237.46 | |
| n-Sp22 | HLA-A2 | 821-829 | LLFNKVTLA | 1018.26 | |
| n-Sp23 | HLA-A2 | 894-902 | LQIPFAMQM | 1078.35 | |
| n-Sp24 | HLA-A2 | 964-972 | KQLSSNFGA | 951.05 | |
| n-Sp25 | HLA-A2 | 976-984 | VLNDILSRL | 1042.24 | Sekine et al.18 |
| n-Sp26 | HLA-A2 | 983-991 | RLDKVEAEV | 1058.2 | |
| n-Sp27 | HLA-A2 | 1048-1056 | HLMSFPQSA | 1017.17 | |
| n-Sp28 | HLA-A2 | 1062-1070 | FLHVTYVPA | 1046.23 | |
| n-Sp29 | HLA-A2 | 1121-1129 | FVSGNCDVV | 939.05 | |
| n-Ep1 | HLA-A2 | 16-24 | SVLLFLAFV | 1008.27 | |
| n-Ep2 | HLA-A2 | 26-34 | FLLVTLAIL | 1002.31 | |
| n-Ep3 | HLA-A2 | 50-58 | SLVKPSFYV | 1039.24 | Sekine et al.18 |
| n-Mp1 | HLA-A2 | 15-23 | KLLEQWNLV | 1142.36 | Sekine et al.18 |
| n-Mp2 | HLA-A2 | 26-35 | FLFLTWICLL | 1268.62 | |
| n-Mp3 | HLA-A2 | 51-60 | LIFLWLLWPV | 1299.66 | |
| n-Mp4 | HLA-A2 | 61-70 | TLACFVLAAV | 1007.26 | |
| -Mp5 | HLA-A2 | 65-73 | FVLAAVYRI | 1051.3 | |
| n-Mp6 | HLA-A2 | 89-97 | GLMWLSYFI | 1129.38 |
Synthetic purity of peptides was >95%.
Peptides were dissolved in DMSO.
The binding of SARS-CoV-2 peptides with HLA-A2
T2A2 cells were used as a tool to analyze the HLA binding capability of the above 23 predicted peptides and screen the dominant epitopes. HLA-A2 was expressed on the T2 cell surface and the binding with suitable peptide would stabilize the pMHC complex structure and be detected with cytometry. T2A2 cells were cocultured with the predicted peptides for 4 h, and acquired on flow cytometer, respectively. Compared to the blank control and negative control, significantly increased MFI was observed in the positive control (Fig. 2A). Among candidate peptides, the MFI of Sp16, Sp19, Sp22, Sp23, Sp25, Sp27, Sp28, Ep3, Mp1, Mp5, and Mp6 were significantly increased compared to the negative control. In addition, moderately elevated signal was observed for peptides Sp18, Sp20, Sp21, Sp26, Sp29, Mp3, and Mp4. However, Sp17, Sp24, Ep1, Ep2, and Mp2 showed similar signal to that of the negative control (Fig. 2B). So, we totally identified 11 peptides with strong HLA binding capability and 7 peptides with moderate binding capability, and these peptides could potentially be presented to activate CD8+ T cells.
FIGURE 2.
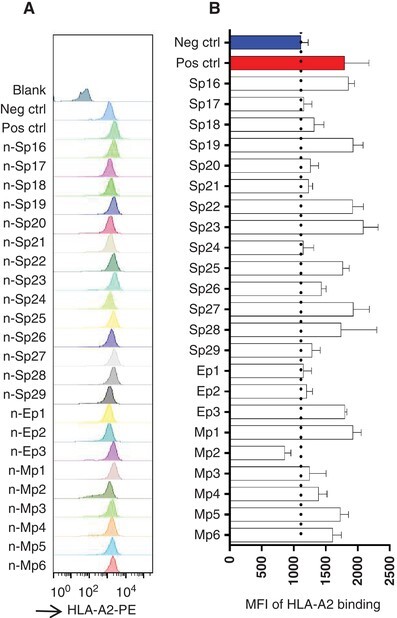
The screening of predicted peptides with T2A2 cells. (A) The binding of predicted peptides with HLA-A2 on T2A2 cells. The predicted peptides were synthesized and incubated with T2A2 cells for 4 h, and the cells were stained with FITC anti-human HLA-A2 antibody. (B) The histogram analysis of (A)
The activation of HLA-A2 restricted SARS-CoV-2 specific CD8+ T cells
We then tried to test the ability of these peptides to activate CD8+ T cells from PBMCs. Healthy adult donors were recruited for T cell isolation, including four males and three females with the ages from 24 to 42 (Table 1). All donors were identified as HLA-A2 subtypes by anti-human-HLA-A2 antibody staining. T2-A2 cells were treated with Mitomycin C to stop cell proliferation (Fig. 3A, B), and loaded with the predicted peptides. Next, the peptide-loaded T2-A2 cells were cocultured with CD8+ T cells at a ratio of 1:1. The T cell activation marker CD69 was detected after 16 h (Fig. 3C, D). The HLA-A2 restricted SARS-CoV-2 specific CD8+ T cells were detected with tetramers containing candidate peptides on day 0 and day 7, respectively. It was found that SARS-CoV-2 specific CD8+ T cells were extremely low on day 0. However, after 7 d stimulation, the proportion of CD8+ T cells specific to certain candidate peptides were significantly increased on day 7, including peptide Sp16, Sp28, Ep1, Ep2, Mp4, Mp5, and Mp6 (Fig. 3E–G).
FIGURE 3.
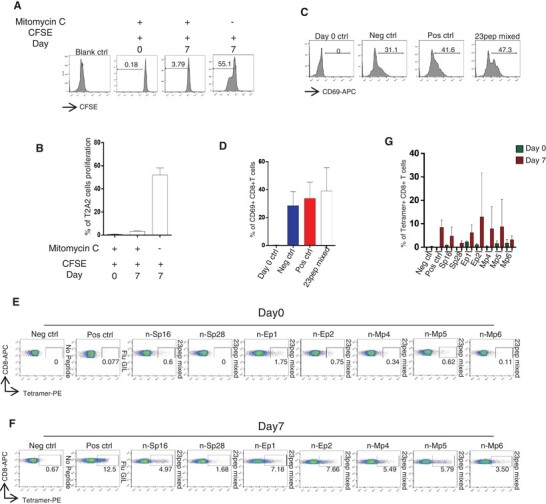
The activation of CD8+ T cells. (A) Inhibition of T2A2 cell proliferation. T2A2 cells were set for different groups, including untreated (Blank ctrl), only CFSE treated, and Mitomycin C plus CFSE treated. Cells were acquired on day 0 and day 7 to evaluate cell proliferation, respectively. (B) The histogram analysis of (A) (n = 3). The extent of T2A2 cell proliferation was presented as the intensity of CFSE. (C) The expression level of CD8+ T cell activation marker CD69. CD8+ T cells from healthy donors were cocultivated with T2A2 cells loaded with various peptides, including Neg ctrl (T2A2 cells without peptide), Pos ctrl (T2A2 cells with influenza A M1 peptide, GILGFVFTL) and T2A2 cells with mixed candidate peptides. CD69 expression was detected by using flow cytometry 16 h after cocultivation. Day 0 ctrl was set as the CD8+ cells assessed at the starting point of the mixture with T2A2 cells without peptide. (D) The histogram analysis of (C) (n = 5). (E), (F). Representative FACS plots of specific CD8+ T cells recognized by tetramers containing candidate peptides. CD8+ T cells from healthy donors were cocultivated with T2A2 cells loaded with various peptides for activation. Cells were stained with tetramers containing corresponding peptides and acquired before stimulation (day 0) and after a 7 d culture (day 7), respectively. (G) The histogram analysis of (E, F) (n = 7). Neg ctrl: T2A2 cells without peptide loading; Pos ctrl: T2A2 cells loaded with influenza A M1 peptide (GILGFVFTL)
Cytotoxicity of activated CD8+ T cells
To investigate the killing ability of the activated CD8+ T cells, we first evaluated the expression of granzyme B and IFN-γ in CD8+ T cells upon peptide stimulation. Similar to Pos ctrl, 7 d stimulation with candidate peptides induced high expression levels of granzyme B and IFN-γ in CD8+ T cells (Fig. 4A, C), and this increase was significantly higher than that in Neg ctrl group (Fig. 4B, D). This result indicated that these activated CD8+ T cells could have the function of cytotoxicity. We then cocultured the CD8+ T cells with the peptide loaded T2A2 cells, and measured the survival rates of T2A2 cells on day 0 and day 7, respectively. We found that after 7 d of cocultivation, the survival rates of T2A2 cells in the candidate peptide group were significantly decreased compared to those on day 0, indicating the killing of target cells mediated by the activated CD8+ T cells (Fig. 4E, F). Taken together, our data showed that the candidate peptides could stimulate the production of functional CD8+ T cells specific to SARS-CoV-2.
FIGURE 4.
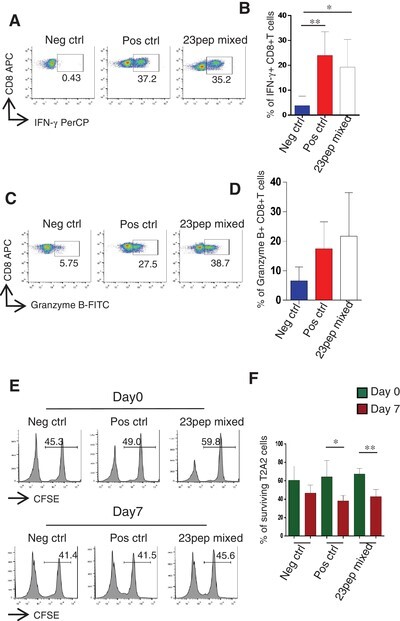
The cytotoxicity of HLA-A2 restricted severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) specific CD8+ T cells. (A) Representative FACS plots of IFN-γ staining of CD8+ T cell after stimulation with the mixed candidate peptides for 7 d. (B) The histogram analysis of (A) (n = 4). (C) Representative FACS plots of granzyme B staining of CD8+ T cells after stimulation with the mixed candidate peptides for 7 d. (D) The histogram analysis of (C) (n = 3). (E) The proportion of surviving T2A2 cells on day 0 and day 7. (F) The histogram analysis of (E) (n = 4). Statistical significance was determined with two independent sample t-tests, *P < 0.05, **P < 0.01. Neg ctrl: T2A2 cells without peptide loading; Pos ctrl: T2A2 cells loaded with influenza A M1 peptide (GILGFVFTL)
DISCUSSION
Currently, we are facing the biggest global health emergency in decades, the pandemic of COVID-19. So far, more than 100 million infections have been confirmed and more than 2.5 million people have died (WHO: https://covid19.who.int/). It has been shown that SARS-CoV-2 specific CD8+ T cell immune responses may be crucial for immune protection against COVID-19.
In order to identify more target epitopes that could elicit CD8+ T cell immunity, we predicted and screened potential MHC I-restricted CD8+ T cell epitopes within structural proteins of SARS-CoV-2 by using an in vitro epitope screening system. Based on computational tools, 23 potential epitopes were selected and tested for MHC I binding and CD8+ T cell activation. We totally identified 11 peptides with strong HLA-A2 binding, 7 peptides with moderate binding, and 5 peptides with very weak binding by T2A2 cell binding test. Next, CD8+ T cells from healthy donors were stimulated with mixed peptides and detected with tetramers prepared with candidate peptides. Specific CD8+ T cells were detected with tetramer containing Sp16, Sp28, Ep1, Ep2, Mp4, Mp5, and Mp6 after a 7 d cocultivation. For effective CD8+ cell activation, antigen epitopes need to bind with MHC I molecules on APC to provide the antigen signal, so the binding affinity between peptides and MHC I is the most important parameter for epitope prediction. Interestingly, although Ep1 and Ep2 showed very weak binding to HLA-A2, their corresponding tetramers had the highest staining of the specific CD8+ T cells. This result suggests that binding capability of peptides with MHC I molecules might not be able to directly reflect the efficiency for CD8+ T cell activation.
Furthermore, we evaluated the expression levels of granzyme B and IFN-γ in activated CD8+ T cells. Granzyme B-induced cell death has been considered to be the main mechanism used by CTL to eliminate targets like viral infected cells.24,25 IFN-γ is a biomarker of CD8+ T cells and is produced by activated CD4+ Th1 cells and CD8+ CTL.26 They are the major mechanisms for activated CD8+ T cells to kill target cells. Our study showed that activated CD8+ T cells highly expressed granzyme B and IFN-γ, which indicated a strong cytotoxic response. In addition, these CD8+ T cells mediated effective killing to target cells bearing specific peptides. Our findings suggest that SARS-CoV-2 specific CD8+ T cells may be able to differentiate into genuine memory T cells similar to influenza specific CD8+ T cells, which represent classical, fully functional memory T cells.27
At present, various approaches for screening SARS-CoV-2 specific epitopes have been reported, including immunoinformatic analysis of SARS-CoV-2 sequence,28–30 and prediction based on SARS-CoV immunologic studies.31 However, these studies have not yet been confirmed by experiments to determine the exact effective epitopes. Similar to SARS-CoV,32–34 the entry of SARS-CoV-2 into target cells is mediated by S protein through receptor-binding domain (RBD),35–37 therefore, many studies have focused on the RBD of the S protein.38–40 Other than S protein, Nelde et al. also reported HLA class I restricted T cell epitopes derived from M, E, and N proteins,41 and also showed that SARS-CoV-2 specific memory CD8+ T cells targeted S and N proteins in convalescent COVID-19 patients.42 These studies suggest that the immune responses could be triggered not only by RBD, but also epitopes from N, M, and E proteins, highlighting the importance of S, N, M, and E proteins in driving protective immune responses. In our study, tetramer staining showed that there was a small amount of reactive CD8+ T cells from unexposed donors, which is consistent with the recent reports,19,43 and this could reflect the cross reaction from common coronaviruses. Although there is another report showing no reaction in unexposed volunteers,44 this discrepancy might be due to the differences of peptides used. After a 7 d stimulation of peptides, these reactive CD8+ T cells significantly increased, indicating that these peptides could be potential immunodominant epitopes to elicit effective immune responses.
In this work, we identified specific epitopes in structural proteins of SARS-CoV-2. Besides structural proteins, nonstructure proteins of SARS-CoV-2, such as ORF1ab and ORF 8, could bear immunodominant epitopes as well. Therefore, further screening of those epitopes could provide valuable information for the understanding of SARS-CoV-2 specific cellular immunity, and contribute to the vaccine development.
AUTHORSHIP
G.C. and P.W. designed the project; J.D. performed the experiments; J.P. and M.Q. analyzed the clinical information and performed sample collection; L.M. assisted with the experiments; Z.W., G.Z., L.G., J.S., and Y.H. assisted with the clinical information and sample collection; O.J.L., P.W., and G.C. analyzed the data; and J.D., P.W., and G.C. wrote the manuscript.
Jieping Deng and Junping Pan contributed equally.
Pengcheng Wang is the lead contact.
ACKNOWLEDGMENTS
This work was supported by grants from the National Key Research and Development Program of China (2018YFC2002003), the Natural Science Foundation of China (U1801285, 81971301), the Guangzhou Planned Project of Science and Technology (201904010111, 202002020039), the Zhuhai Planned Project of Science and Technology (ZH22036302200067PWC), and the Initial Supporting Foundation of Jinan University.
Contributor Information
Jieping Deng, Department of Microbiology and Immunology; Institute of Geriatric Immunology; School of Medicine, Jinan University, Guangzhou, China; Guangdong-Hong Kong-Macau Great Bay Area Geroscience Joint Laboratory, Guangzhou, China.
Junping Pan, Department of Microbiology and Immunology; Institute of Geriatric Immunology; School of Medicine, Jinan University, Guangzhou, China; Guangdong-Hong Kong-Macau Great Bay Area Geroscience Joint Laboratory, Guangzhou, China.
Minghui Qiu, Department of Microbiology and Immunology; Institute of Geriatric Immunology; School of Medicine, Jinan University, Guangzhou, China; Guangdong-Hong Kong-Macau Great Bay Area Geroscience Joint Laboratory, Guangzhou, China.
Lipeng Mao, Department of Microbiology and Immunology; Institute of Geriatric Immunology; School of Medicine, Jinan University, Guangzhou, China; Guangdong-Hong Kong-Macau Great Bay Area Geroscience Joint Laboratory, Guangzhou, China.
Zhigang Wang, Affiliated Huaqiao Hospital, Jinan University, Guangzhou, China.
Guodong Zhu, Guangdong-Hong Kong-Macau Great Bay Area Geroscience Joint Laboratory, Guangzhou, China; Department of Geriatrics, Guangzhou First People's Hospital, School of Medicine, South China University of Technology, Guangzhou, Guangdong, China.
Lijuan Gao, Department of Microbiology and Immunology; Institute of Geriatric Immunology; School of Medicine, Jinan University, Guangzhou, China; Guangdong-Hong Kong-Macau Great Bay Area Geroscience Joint Laboratory, Guangzhou, China.
Jun Su, Affiliated Huaqiao Hospital, Jinan University, Guangzhou, China.
Yutian Hu, Meng Yi Center Limited, Macau, China.
Oscar Junhong Luo, Guangdong-Hong Kong-Macau Great Bay Area Geroscience Joint Laboratory, Guangzhou, China; Department of Systems Biomedical Sciences, School of Medicine, Jinan University, Guangzhou, China.
Guobing Chen, Department of Microbiology and Immunology; Institute of Geriatric Immunology; School of Medicine, Jinan University, Guangzhou, China; Guangdong-Hong Kong-Macau Great Bay Area Geroscience Joint Laboratory, Guangzhou, China.
Pengcheng Wang, Department of Microbiology and Immunology; Institute of Geriatric Immunology; School of Medicine, Jinan University, Guangzhou, China; Guangdong-Hong Kong-Macau Great Bay Area Geroscience Joint Laboratory, Guangzhou, China.
DISCLOSURES
The epitopes and tetramers from this study are the subjects of a patent application. The authors declare no other conflicts of interest.
REFERENCES
- 1. Lu R, Zhao X, Li J et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet. 2020;395(10224):565–574. 10.1016/s0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Su S, Wong G, Shi W et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen Y, Klein SL, Garibaldi BT et al. Aging in COVID-19: vulnerability, immunity and intervention. Ageing Res Rev. 2021;65:101205 10.1016/j.arr.2020.101205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Singh AK, Gillies CL, Singh R et al. Prevalence of co-morbidities and their association with mortality in patients with COVID -19: a systematic review and meta-analysis. Diabetes Obesity Metabol. 2020;22(10):1915–1924. 10.1111/dom.14124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rosendahl Huber S, van Beek J, de Jonge J et al. T cell responses to viral infections—opportunities for peptide vaccination. Front Immunol. 2014;5:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khan N, Best D, Bruton R, Nayak L, Rickinson AB, Moss PAH. T cell recognition patterns of immunodominant cytomegalovirus antigens in primary and persistent infection. J Immunol. 2007;178(7):4455–4465. 10.4049/jimmunol.178.7.4455. [DOI] [PubMed] [Google Scholar]
- 8. Weiskopf D, Schmitz KS, Raadsen MP et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang F, Nie J, Wang H et al. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. The Journal of Infectious Diseases. 2020;221(11):1762–1769. 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liao M, Liu Y, Yuan J et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nate Med. 2020;26(6):842–844. 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 11. Wu LP, Wang NC, Chang YH et al. Duration of antibody responses after severe acute respiratory syndrome. Emerg Infect Dis. 2007;13:1562–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tang F, Quan Y, Xin Z-T et al. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J Immunol. 2011;186(12):7264–7268. 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- 13. Ng O-W, Chia A, Tan AT, Jadi RS, Leong HN, Bertoletti A, Tan Y-J. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34(17):2008–2014. 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alshukairi AN, Khalid I, Ahmed WA et al. Antibody response and disease severity in healthcare worker MERS survivors. Emerg Infect Dis. 2016;22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Drosten C, Meyer B, Müller MA et al. Transmission of MERS-coronavirus in household contacts. N Eng J Med. 2014;371(9):828–835. 10.1056/nejmoa1405858. [DOI] [PubMed] [Google Scholar]
- 16. Long Q-X, Tang X-J, Shi Q-L et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26(8):1200–1204. 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 17. Ibarrondo FJ, Fulcher JA, Goodman-Meza D et al. Rapid decay of anti–SARS-CoV-2 antibodies in persons with mild Covid-19. N E J M. 2020;383(11):1085–1087. 10.1056/nejmc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sekine T, Perez-Potti A, Rivera-Ballesteros O et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grifoni A, Weiskopf D, Ramirez SI et al. Targets of t cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501.e15. 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qiu C, Xiao C, Wang Z et al. CD8+ T cell epitope variations suggest a potential antigen presentation deficiency for spike protein of SARS-CoV-2. bioRxiv. 2021:2021.01.22.427863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Steinle A, Schendel DJ. HLA class I alleles of LCL 721 and 174XCEM.T2 (T2). Tissue Antigens. 1994;44(4):268–270. 10.1111/j.1399-0039.1994.tb02394.x. [DOI] [PubMed] [Google Scholar]
- 22. González-Galarza FF, Takeshita LYC, Santos EJM et al. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 2015;43(D1):D784–D788. 10.1093/nar/gku1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. He Y, Li J, Mao W et al. HLA common and well-documented alleles in China. HLA. 2018;92:199–205. [DOI] [PubMed] [Google Scholar]
- 24. Lieberman J. The ABCs of granule-mediated cytotoxicity: new weapons in the arsenal. Nat Rev Immunol. 2003;3:361–370. [DOI] [PubMed] [Google Scholar]
- 25. Heusel JW, Wesselschmidt RL, Shresta S, Russell JH, Ley TJ. Cytotoxic lymphocytes require granzyme B for the rapid induction of DNA fragmentation and apoptosis in allogeneic target cells. Cell. 1994;76(6):977–987. 10.1016/0092-8674(94)90376-x. [DOI] [PubMed] [Google Scholar]
- 26. Kasahara T, Hooks JJ, Dougherty SF et al. Interleukin 2-mediated immune interferon (IFN-gamma) production by human T cells and T cell subsets. J Immunol. 1983;130:1784–1789. [PubMed] [Google Scholar]
- 27. van de Sandt CE, Hillaire ML, Geelhoed-Mieras MM et al. Human influenza A virus-specific CD8+ T-cell response is long-lived. J Infect Dis. 2015;212:81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bhattacharya M, Sharma AR, Patra P et al. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): Immunoinformatics approach. Journal of Medical Virology. 2020;92(6):618–631. 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kiyotani K, Toyoshima Y, Nemoto K, Nakamura Y. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2. Journal of Human Genetics. 2020;65 (7):569–575. 10.1038/s10038-020-0771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sanami S, Zandi M, Pourhossein B et al. Design of a multi-epitope vaccine against SARS-CoV-2 using immunoinformatics approach. Int J Biol Macromol. 2020;164:871–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ahmed SF, Quadeer AA, McKay MR. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12(3):254. 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li W, Moore MJ, Vasilieva N et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol. 2015;89(4):1954–1964. 10.1128/jvi.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li F, Li W, Farzan M et al. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. [DOI] [PubMed] [Google Scholar]
- 35. Zhou P, Yang X-L, Wang X-G et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lan J, Ge J, Yu J et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. [DOI] [PubMed] [Google Scholar]
- 37. Shang J, Wan Y, Luo C et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci. 2020;117(21):11727–11734. 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang J, Wang W, Chen Z et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586(7830):572–577. 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 39. Zost SJ, Gilchuk P, Case JB et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584(7821):443–449. 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu L, Wang P, Nair MS et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584(7821):450–456. 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 41. Nelde A, Bilich T, Heitmann JS et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat Immunol. 2021;22:74–85. [DOI] [PubMed] [Google Scholar]
- 42. Schulien I, Kemming J, Oberhardt V et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8(+) T cells. Nat Med. 2021;27:78–85. [DOI] [PubMed] [Google Scholar]
- 43. Braun J, Loyal L, Frentsch M et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. [DOI] [PubMed] [Google Scholar]
- 44. Peng Y, Mentzer AJ, Liu G et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 2020;21:1336–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]


