Transposable elements (TEs) in plants are best known for their ability to inflate genome size and their potential effects on host phenotypes. In this essay, we suggest that many TEs do none of these things, but survive and replicate inconspicuously in the host genome. Transposable elements are frequently depicted as “invasive” sequences with a tendency to replicate in “bursts” as soon as the silencing mechanisms keeping them in check are relaxed. While massive amplifications do occur and have intriguing consequences, this way of thinking about TEs, guided by analogies from horizontally transmitted pathogens, can be misleading. By means of the example of Alesia elements—a retrotransposon lineage present at low copy numbers throughout angiosperms—we propose a scenario of vertical descent in which TEs are maintained in evolution not because of their ability to invade and amplify, but because they have evolved strategies to persist at low copy numbers. Studying the adaptive traits of rare TEs across species promises intriguing insights into the world of intragenomic conflict and a more nuanced view of transposition dynamics in plants.
THE PHENOMENON OF RARE TE LINEAGES
To illustrate the varying abundance of TEs in plants, we annotated long‐terminal repeat retrotransposons (LTR‐RTs) in the genomes of 20 angiosperm species (Fig. 1A). These elements are the most prominent TEs in plants and can be classified into major evolutionary lineages based on their reverse transcriptase (RT) sequence (Fig. 1B; Neumann et al., 2019). Retrotransposon lineages share basic structural features and can be thought of as “genera”: groups of elements with a similar biology due to common descent, yet in some cases comprising considerable heterogeneity as they have diversified into numerous sublineages. This classification relies on the presence of a RT and does therefore not capture degraded copies and the sometimes large populations of non‐autonomous elements.
Figure 1.
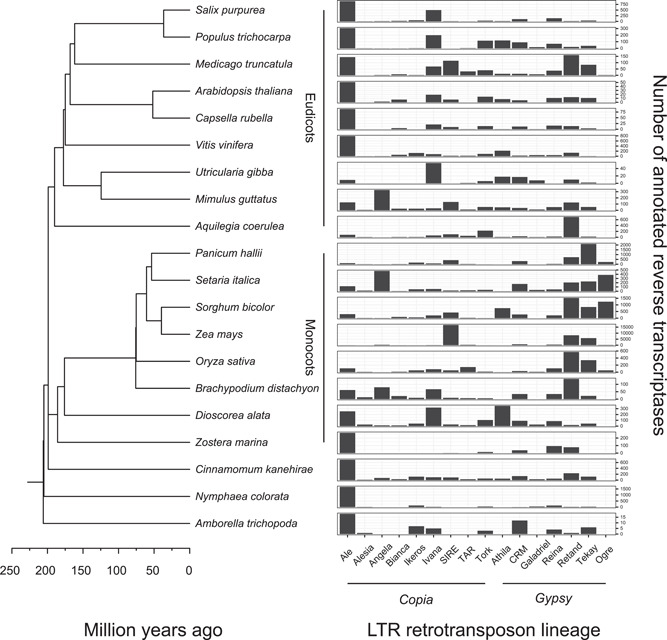
Long‐terminal repeat retrotransposons (LTR‐RT) in the genomes of 20 diverse angiosperm species (phylogeny modified from Janssens et al., 2020). Barplots show the abundance of annotated reverse transcriptases, a key enzyme in the self‐replication of these elements. The de novo annotation of the 20 genomes was done using EDTA (Ou et al., 2019), and lineages were assigned by aligning the RT amino acid sequence of each intact element against the RTs of previously identified lineages from the RepeatExplorer data base (Neumann et al., 2019)
Most genomes are dominated by one to three LTR‐RT lineages, while the other lineages are comparatively rare (Fig. 1B). In nine genomes, including the three basal angiosperms, Ale elements are the most abundant LTR‐RTs. This lineage has been noted for its diversity and tendency to split into numerous low‐copy families (Wicker and Keller, 2007), possibly reflecting arms race dynamics. Other peaks in the TE landscape are due to amplifications in the recent past, for example of Tekay elements in Panicum hallii (2108 copies) or SIRE elements in Zea mays (16,729 copies). These are the “bursts” that have received much attention in the past, on the one hand because they explain the old riddle of genome size variation (Elliott and Gregory, 2015), on the other because they have complicated the genomics of economically important crops (Vitte et al., 2014).
The peaks in the TE landscape, however, distract from a pattern no less intriguing: several TE lineages are present yet rare throughout the phylogeny (Fig. 1). Alesia elements are present in 15 of the 20 species and have one (Amborella trichopoda) to 28 (Dioscorea alata) copies (Fig. 2). The TE and the host phylogeny agree poorly, reflecting distinct sublineages present in most plant species (see next section). Strikingly, while Alesia copies are present in species that diverged up to 200 million years ago, the copies themselves are relatively recent and mostly younger than 1 million years (Fig. 2, inset). The same pattern—the presence of young TE copies across the angiosperm phylogeny—holds for other rare lineages (Bianca, Ikeros, TAR, and Galadriel), suggesting that these TEs survive and replicate over long evolutionary times, even at low copy numbers.
Figure 2.
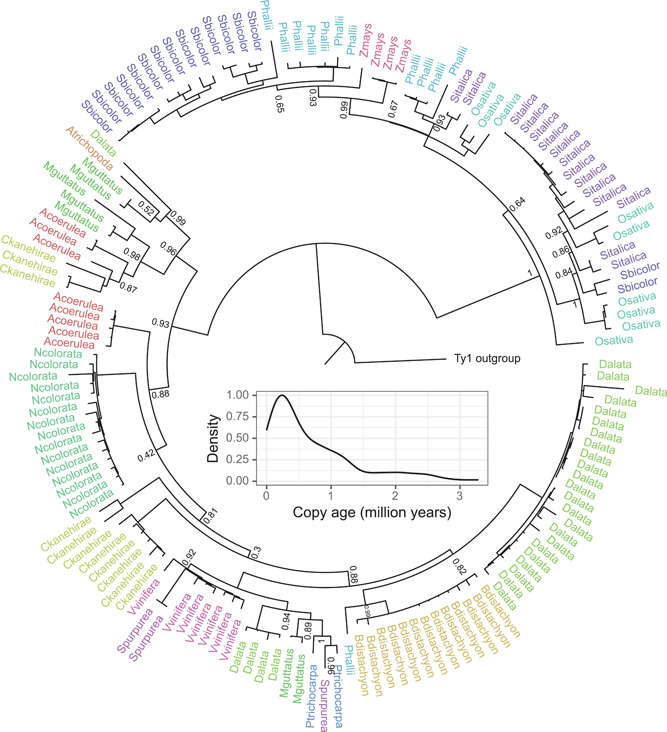
Rooted reverse transcriptase tree for the 151 Alesia elements, estimated with FastTree 2.1.11 (Price et al., 2010). The 15 plant species in which this TE lineage is present are distinguished by different colors (see Fig. 1 for the genus names). The lineage was not found in Medicago truncatula, Arabidopsis thaliana, Capsella rubella, Utricularia gibba, and Zostera marina. The inset shows the age distribution of the copies in the tree, estimated from the divergence of the LTRs: as the two LTR of a copy are identical upon its insertion, their divergence can be used to estimate the copy age, here assuming a mutation rate of 1.3 × 10−8 substitutions per base pair per generation
HOW ARE TES MAINTAINED DURING EVOLUTION?
Why are these elements not disabled by the host, crippled by mutations, or lost through genetic drift or purifying selection? A standard explanation, inspired by the P element in Drosophila, can be found in descriptions of “the TE life cycle” (e.g., Kidwell and Lisch, 2001). This cycle begins with the invasion of a genome, passes through the maturation and decay of a lineage when more and more copies become nonfunctional, and begins anew when a copy manages to invade a new, “naïve” genome. In this scenario, the long‐term maintenance of TEs is achieved through horizontal transfers of copies between populations or species.
For some time, horizontal transfers were the default explanation for discrepancies between TE and host phylogenies. Horizontal transfers, however, are difficult to distinguish from ancestral polymorphisms and stochastic loss in some species but not others (Capy et al., 1994). For the Alesia phylogeny (Fig. 2), recent horizontal transfers are unlikely: DNA sequence similarities between copies of different species are far below (maximum 77%, between copies of the closely related Salix purpurea and Populus trichocarpa) the 85% threshold previously used to detect horizontal transfers (El Baidouri et al., 2014). While horizontal transfers in the distant past are difficult to exclude, this increased level of sequence divergence between species is consistent with an evolutionary history of largely vertical descent, with ancestral polymorphisms and stochastic loss explaining phylogenetic discordance.
The life cycle analogy reveals a curious neglect in many discussions of TE dynamics: the ability of TEs to evolve and adapt. There is a widespread notion that a TE's “natural fate is inactivation, degradation and loss from the host genome as a consequence of the natural selection and/or genetic drift” (Wallau et al., 2016, p. 1094), or that TEs are “a quintessential example of neutrality” (Arkhipova, 2018, p. 1332). It is true that a large proportion of TEs in any genome is nonfunctional and slowly morphing into genomic “dark matter”. Besides these numerous dead leaves, however, many TE phylogenies contain living branches stretching into the present, as illustrated in Fig. 2. It is not clear why TEs are assumed to lose out against the host silencing machinery. On the contrary, TEs may adapt rapidly since mutations affect them directly (Orgel and Crick, 1980), and they may evolve strategies to survive at low copy numbers over long periods of time.
ADAPTIVE TRAITS OF TRANSPOSABLE ELEMENTS
From the perspective of intragenomic conflict, TEs are maintained over evolutionary time because they have been shaped by evolution to do exactly that: to produce copies of themselves and to enhance their transmission at the cost of normal genes (Burt and Trivers, 2006). Thinking about TE evolution in terms of adaptive traits and strategies of TEs provides a fascinating alternative to the widespread tendency to interpret TE activity in terms of effects on host biology. Different evolutionary strategies of TEs and the molecular traits involved were recently reviewed by Cosby et al. (2019), and we here highlight a few aspects particularly relevant for rare TEs or not covered by the authors.
Self‐regulation is likely to be an important aspect of the evolutionary strategy of rare TEs and might include mechanisms such as suboptimal enzyme efficiency and the timing and locality of expression. Expression niches are particularly intriguing for plant TEs: since plants lack a dedicated germ line (Schoen and Schultz, 2019), they provide a wide range of signals and niches that TEs might use to leave copies in pollen or ovules and thus be passed on through the generations. In addition to self‐regulation, other traits might increase the chance of survival and replication at low copy numbers, including insertion site preferences for genic regions to increase the chance of expression (Baucom et al., 2009), or a reduced length of internal repeats, which decreases the chance of crippling intra‐element recombination (Stritt et al., 2020).
Perhaps the most intriguing scenario for rare TEs is that they have turned symbiotic. Referred to as evolved dependency (Werren, 2011) or TE addictions (Cosby et al., 2019), this scenario differs from the better‐known TE domestication in that TEs maintain their ability to self‐replicate, while domestication refers to situations in which parts of the enzymatic repertoire of TEs are incorporated into host genes (Jangam et al., 2017). To our knowledge, evolved dependency has so far only been shown for the telomere‐maintaining TEs in Drosophila melanogaster (Pardue and DeBaryshe, 2011). It is tempting to speculate that the single Alesia copy identified in A. trichopoda is beneficial, although its possible function escapes our imagination.
CONCLUSIONS
For the Alesia lineage, nothing is known about adaptive traits or possible mechanisms of self‐regulation. Clearly, these elements have been successful in leaving copies in pollen or ovules, as shown by their ample representation across the angiosperm phylogeny. Like most types of TEs, the Alesia lineage so far has a name and is known to occur in various species. Beyond this basic certificate of existence, little is known about the biology of these elements. Comparative genomics is the most promising approach to change this. Currently, most TE research is conducted within the boundary of single organisms. Within these model systems, TEs are categorized into families by sequence similarity and given random names that do not allow connecting them to related families in other species. Comparing TEs across species, in a phylogenetic framework, would be particularly valuable for rare lineages, for which there might be insufficient data in single species and which are easily overlooked amid the dominant TEs in a genome.
AUTHOR CONTRIBUTIONS
C.S. performed the analysis and designed the figures. C.S., M.T., and A.C.R. wrote the manuscript. All authors approved the final version of the manuscript.
ACKNOWLEDGMENTS
We thank the Editor‐in‐Chief, Pamela Diggle, for taking an interest in transposable elements and the two reviewers for helping us to clarify our ideas. We also thank the Swiss National Science foundation and the Research Priority Program “Evolution in Action” of the University of Zürich. Open access funding provided by Universitat Zurich.
Stritt, C. , Thieme M., and Roulin A. C.. 2021. Rare transposable elements challenge the prevailing view of transposition dynamics in plants. American Journal of Botany. 108(8): 1310–1314. 10.1002/ajb2.1709
REFERENCES
- Arkhipova, I. R. 2018. Neutral theory, transposable elements, and eukaryotic genome evolution. Molecular Biology and Evolution 35: 1332–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucom, R. S. , Estill J. C., Chaparro C., Upshaw N., Jogi A., Deragon J. M., Westerman R. P., et al. 2009. Exceptional diversity, non‐random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genetics 5: e1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt, A. , and Trivers R.. 2006. Genes in conflict. Harvard University Press, Cambridge, MA, USA. [Google Scholar]
- Capy, P. , Anxolabéhère D., and Langin T.. 1994. The strange phylogenies of transposable elements: Are horizontal transfers the only explanation? Trends in Genetics 10: 7–12. [DOI] [PubMed] [Google Scholar]
- Cosby, R. L. , Chang N. C., and Feschotte C.. 2019. Host–transposon interactions: conflict, cooperation, and cooption. Genes and Development 33: 1098–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Baidouri, M. , Carpentier M. C., Cooke R., Gao D., Lasserre E., Llauro C., Mirouze M., et al. 2014. Widespread and frequent horizontal transfers of transposable elements in plants. Genome Research 24: 831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, T. A. , and Gregory T. R.. 2015. What's in a genome? The C‐value enigma and the evolution of eukaryotic genome content. Philosophical Transactions of the Royal Society, B, Biological Sciences 370: 20140331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangam, D. , Feschotte C., and Betrán E.. 2017. Transposable element domestication as an adaptation to evolutionary conflicts. Trends in Genetics 33: 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens, S. B. , Couvreur T. L. P., Mertens A., Dauby G., Dagallier L. P. M. J., Vanden Abeele S., Vandelook F., et al. 2020. A large‐scale species level dated angiosperm phylogeny for evolutionary and ecological analyses. Biodiversity Data Journal 8: e39677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell, M. G. , and Lisch D. R.. 2001. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution 55: 1. [DOI] [PubMed] [Google Scholar]
- Neumann, P. , Novák P., and Ho N.. 2019. Systematic survey of plant LTR‐ retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mobile DNA 10: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel, L. E. , and Crick F. H. C.. 1980. Selfish DNA: the ultimate parasite. Nature 288: 645–646. [DOI] [PubMed] [Google Scholar]
- Ou, S. , Su W., Liao Y., Chougule K., Agda J. R. A., Hellinga A. J., Lugo C. S. B., et al. 2019. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biology 20: 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue, M. L. , and DeBaryshe P. G.. 2011. Retrotransposons that maintain chromosome ends. Proceedings of the National Academy of Sciences, USA 108: 20317–20324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, M. N. , Dehal P. S., and Arkin A. P.. 2010. FastTree 2 – Approximately maximum‐likelihood trees for large alignments. PLoS One 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoen, D. J. , and Schultz S. T.. 2019. Somatic mutation and evolution in plants. Annual Review of Ecology, Evolution, and Systematics 50: 49–73. [Google Scholar]
- Stritt, C. , Wyler M., Gimmi E. L., Pippel M., and Roulin A. C.. 2020. Diversity, dynamics and effects of long terminal repeat retrotransposons in the model grass Brachypodium distachyon . New Phytologist 227: 1736–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte, C. , Fustier M. A., Alix K., and Tenaillon M. I.. 2014. The bright side of transposons in crop evolution. Briefings in Functional Genomics and Proteomics 13: 276–295. [DOI] [PubMed] [Google Scholar]
- Wallau, G. L. , Capy P., Loreto E., Le Rouzic A., and Hua‐Van A.. 2016. VHICA, a new method to discriminate between vertical and horizontal transposon transfer: Application to the mariner family with in Drosophila . Molecular Biology and Evolution 33: 1094–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren, J. H. 2011. Selfish genetic elements, genetic conflict, and evolutionary innovation. PNAS 108: 10863–10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker, T. , and Keller B.. 2007. Genome‐wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Research 17: 1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]


